Abstract
Diffuse large B‐cell lymphoma (DLBCL) is the most common B‐cell lymphoma subtype, and the Epstein–Barr virus (EBV)‐positive subtype of DLBCL is known to show a more aggressive clinical behavior than the EBV‐negative one. BTB and CNC homology 2 (BACH2) has been highlighted as a tumor suppressor in hematopoietic malignancies; however, the role of BACH2 in EBV‐positive DLBCL is unclear. In the present study, BACH2 expression and its significance were studied in 23 EBV‐positive and 43 EBV‐negative patient samples. Immunohistochemistry revealed BACH2 downregulation in EBV‐positive cases (P < 0.0001), although biallelic deletion of BACH2 was not detected by FISH. Next, we analyzed the contribution of BACH2 negativity to aggressiveness in EBV‐positive B‐cell lymphomas using FL‐18 (EBV‐negative) and FL‐18‐EB cells (FL‐18 sister cell line, EBV‐positive). In BACH2‐transfected FL‐18‐EB cells, downregulation of phosphorylated transforming growth factor‐β‐activated kinase 1 (pTAK1) and suppression in p65 nuclear fractions were observed by Western blot analysis contrary to non‐transfected FL‐18‐EB cells. In patient samples, pTAK1 expression and significant nuclear p65, p50, and p52 localization were detected immunohistochemically in BACH2‐negative DLBCL (P < 0.0001, P = 0.006, and P = 0.001, respectively), suggesting that BACH2 downregulation contributes to constitutive activation of the nuclear factor‐κB pathway through TAK1 phosphorylation in BACH2‐negative DLBCL (most EBV‐positive cases). Although further molecular and pathological studies are warranted to clarify the detailed mechanisms, downregulation of BACH2 may contribute to constitutive activation of the nuclear factor‐κB pathway through TAK1 activation.
Keywords: BACH2, diffuse large B‐cell lymphoma, Epstein–Barr virus, NFκB, TAK1
The Epstein–Barr virus (EBV), a linear double‐stranded DNA virus and member of the Herpesviridae family, was initially discovered from a Burkitt lymphoma‐derived cell line in 1964.1 Epstein–Barr virus was subsequently found to induce the proliferation of human B cells2 and is currently considered to contribute to the oncogenesis of several types of B‐cell lymphoma, including Burkitt lymphoma and diffuse large B‐cell lymphoma (DLBCL).3 Of these, DLBCL is the most common subtype accounting for 40% of all non‐Hodgkin lymphoma cases. EBV‐positive DLBCL accounts for 8–10% of DLBCL cases in Asian countries and is frequently detected in immunocompetent people aged 50 years or older.4 Compared with EBV‐negative DLBCLs, EBV‐positive DLBCLs show an aggressive clinical behavior and were accordingly designated as a separate category in the 2008 WHO classification. Immunophenotypically, most EBV‐positive DLBCLs are of the non‐germinal center B cell type;5 in such cases, nuclear factor‐κB (NFκB) activation through TNFAIP3, CARD11, and other gene mutations has been reported to play a pathogenic role.6, 7 In addition, several groups also described NFκB activation through latent infection membrane protein 1 (LMP1) in EBV‐positive lymphoma cells.8, 9
BTB and CNC homology 2 (BACH2) has been highlighted as a transcriptional factor in the regulation of plasma cell differentiation. Specifically, BACH2 inhibits plasma cell differentiation by repressing PRDM1 expression, thus allowing B cells to undergo class switch recombination and somatic hypermutation.10, 11 Sakane‐Ishikawa et al.12 reported that patients with higher levels of BACH2 expression had a better prognosis in DLBCL, although there seemed to be little consensus regarding the significance of BACH2 as a prognostic factor.13 As BACH2 was reported to mediate the negative selection of pre‐B cells by p53 upregulation, the tumor‐suppressive function of BACH2 has been highlighted.14 However, the role of BACH2 expression and its tumor‐suppressive function in EBV‐positive DLBCL is still unclear.
In the present study, we examined BACH2 expression in EBV‐positive DLBCLs and found it to be downregulated in 78.3% of cases. In addition, we sought to investigate how BACH2 negativity contributes to aggressiveness in EBV‐positive DLBCL using B‐cell lymphoma‐derived EBV‐positive and ‐negative cell lines.
Materials and Methods
Patient selection
Formalin‐fixed, paraffin‐embedded tissues (FFPET) were obtained from 23 patients with EBV‐positive DLBCL who were diagnosed from 2008 to 2011 and 43 patients with EBV‐negative DLBCL who were diagnosed from 2004 to 2012 at Okayama University Graduate School of Medicine (Okayama, Japan). Three hematopathologists diagnosed the cases according to the criteria described in the 2008 WHO classification. The study protocol was approved by the Institutional Review Board of Okayama University (IRB No. 493, November 29, 2011). All study procedures were undertaken in accordance with the guidelines of the Declaration of Helsinki.
Immunohistochemical analysis and in situ hybridization
All immunohistochemical analyses of FFPET were carried out using an automated immunostainer (Bond‐max; Leica Microsystems, Wetzlar, Germany). The following primary antibodies and dilutions were used: CD20 (L26, 1:200), CD3 (PS‐1, 1:50), CD10 (56C6, 1:50), CD5 (4C7, 1:100), Ki‐67 (MIB‐1, 1:5000), LMP1 (1:10) (all Leica Microsystems); multiple myeloma oncogene 1 (MUM1) (MUM1p, 1:50), NFκB p65 (1:1000) (both from Dako, Glostrup, Denmark); BCL‐6 (D‐8, 1:100) (Santa Cruz Biotechnology, Santa Cruz, CA, USA); germinal center B‐cell expressed transcript 1 (GCET1) (RAM341, 1:100), NFκB p105/p50 (E381, 1:250) (Abcam, Cambridge, UK); NFκB2 p100/p52 (18D10, 1:100) (Cell Signaling Technology, Danvers, MA, USA); forkhead box protein P1 (FOXP1) (JC12, 1:500), and BACH2 (1:400) (both from Life Span Biosciences, Seattle, WA, USA). In situ hybridization with EBV‐encoded small RNA probes (Leica Microsystems) was used to detect EBV. A sample was scored as positive if >30% of the lymphoma cells were positively stained.
Fluorescence in situ hybridization for BACH2
The FFPET were subjected to FISH using a spectrum red‐labeled BACH2 probe and spectrum green‐labeled centromeric probe for chromosome 6 (CEP6) (Vysis/Abbott Molecular Laboratories, Des Plaines, IL, USA) according to the manufacturer's instructions. To identify the BACH2 gene, we prepared specific probes using the BAC clones RP11‐16C2, RP11‐402C18, and RP11‐147G14, which cover approximately 500 kb of the BACH2 gene, and incubated samples with these probes at 37°C for approximately 48 h in a Hybridizer (Dako). Cells with two CEP6 signals were scored, and the signal ratio of BACH2 to CEP6 was calculated. BACH2 biallelic and monoallelic deletions were defined as having signal ratios of 20–60% and 60–80%, respectively, as previously described.15
Cell lines, RNA extraction, and RT‐PCR
We prepared human non‐Hodgkin's lymphoma cell lines (FL‐18, FL‐218, and FL‐318) and an EBV‐positive sister cell line (FL‐18‐EB),16, 17 all provided by Dr. Ohno at Kyoto University (Kyoto, Japan). RNA was extracted from cultured cells using the miRNeasy Mini kit (Qiagen, Hilden, Germany), and cDNA was produced using SuperScript VILO MasterMix (Life Technologies, Palo Alto, CA, USA). BACH2 and β‐actin were amplified as previously described;18 the primers are listed in Table S1.
Transfection assay
FL‐18‐EB cells were transfected with a pIRES2‐EGFP plasmid containing a BACH2 sequence using the Neon transfection system (Life Technologies). The optimum transfection conditions (pulse voltage, 1100 V; pulse width, 30 ms; once) were set using the control plasmid pmaxGFP (Amaxa Bioscience, Basel, Switzerland). Transfected cells were grown in RPMI 1640 medium containing 10% (v/v) FBS at 37°C in an atmosphere with 5% CO2.
Western blot analysis
Whole cell lysates and nuclear and cytoplasmic fractions were resolved by SDS‐PAGE and transferred onto nitrocellulose membranes using a Trans‐Blot Turbo Blotting System (Bio‐Rad, Hercules, CA, USA). The nuclear and cytoplasmic fractions were separated using a nuclear/cytosol fractionation kit (BioVision, Milpitas, CA, USA) according to the manufacturer's instructions. Antibody reactions were carried out as previously described;19 the primary antibodies and conditions are listed in Table S2 (phosphorylated transforming growth factor‐β‐activated kinase 1 [pTAK1] was the resource from the previous report.20). ImageJ (NIH, Bethesda, MD, USA) was used to quantify the protein expression and the ratio to control was calculated.
Statistical analysis
statcel3 software (OMS, Saitama, Japan) was used to undertake χ2‐test and t‐test analyses. Kaplan–Meier plots were made using spss version 14.0 (IBM, Chicago, IL, USA). P‐values <0.05 were considered statistically significant.
Results
Significant downregulation of BACH2 expression in EBV‐positive DLBCL
The clinicopathological features of 23 cases of EBV‐positive DLBCL and 43 cases of EBV‐negative DLBCL are summarized in Table 1. The histological features and immunohistochemical findings of representative cases are shown in Figure 1. The EBV‐positive cases had a significantly poorer prognosis than EBV‐negative cases (P = 0.04; Table 1, Fig. S1). In addition, 15 of the 18 cases (83%) of EBV‐positive DLBCL showed an activated B‐cell‐like (ABC) phenotype according to Choi's criteria (P = 0.04; Table 1, Fig. 2). There were no other significant differences in clinicopathological characteristics between EBV‐positive and ‐negative cases of DLBCL.
Table 1.
Clinicopathological features of patients with diffuse large B‐cell lymphoma (DLBCL), grouped according to Epstein–Barr virus (EBV) positivity
| EBV‐positive DLBCL (n = 23) | EBV‐negative DLBCL (n = 43) | P‐value | |
|---|---|---|---|
| Clinical features | |||
| Median age, years (range) | 77 (2–88) | 71 (30–91) | 0.66 |
| Male : female | 1.6:1.0 | 1.0:1.0 | 0.50 |
| Clinical stage | |||
| I–II (%) | 14/23 (61) | 24/40 (60) | 0.84 |
| III–IV (%) | 9/23 (39) | 16/40 (40) | |
| Median LDH, IU/L (range) | 290 (141–1255) | 243.5 (158–970) | 0.31 |
| Median sIL2R, U/mL (range) | 1932 (340–11000) | 1483.5 (165–25400) | 0.63 |
| Anemia | 8/11 (73) | 13/31 (42) | 0.16 |
| Three‐year survival rate, % | 38.9 | 72.2 | 0.04 |
| Immunohistochemical features | |||
| CD5‐positive (%) | 0/15 (0) | 5/43 (12) | 0.40 |
| Median Ki‐67 labelling index (range) | 49.95 (20.5–76.1) | 60 (27.9–83.7) | 0.08 |
| ABC type (%) | 15/18 (83) | 22/43 (51) | 0.04 |
| GCB type (%) | 3/18 (17) | 21/43 (49) | |
| Treatment | |||
| R‐CHOP or R‐CHOP‐like regimen (%) | 8/13 (62) | 23/32 (72) | |
| CHOP or CHOP‐like regimen (%) | 2/13 (15) | 6/32 (19) | |
| Radiotherapy (%) | 0/13 (0) | 1/32 (3) | |
| Other (%) | 3/13 (23) | 2/32 (6) | |
Bold indicates statistical significance (P < 0.05). ABC, activated B‐cell‐like; CHOP, cyclophosphamide, doxorubicin, vincristine, and prednisolone; GCB, germinal centre B‐cell‐like; LDH, lactate dehydrogenase; R, rituximab; sIL2R, soluble interleukin‐2 receptor.
Figure 1.
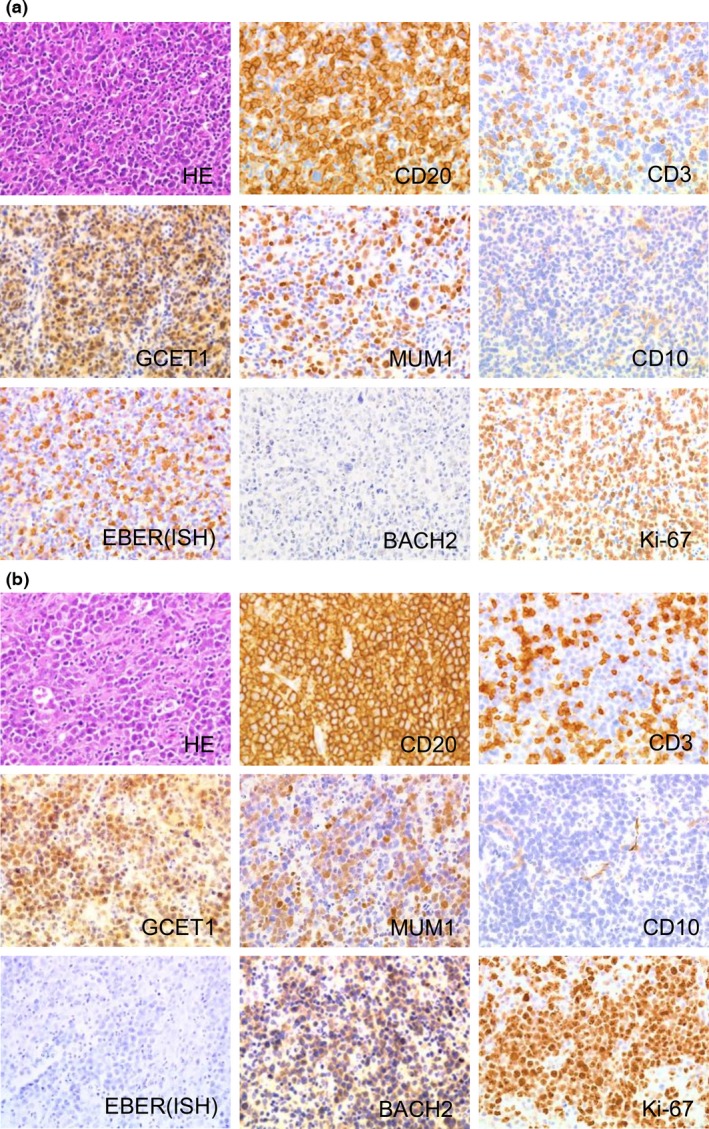
Histological features and immunohistochemical findings of representative cases of diffuse large B‐cell lymphoma (DLBCL). (a) Immunohistochemical analysis of Epstein–Barr virus (EBV)‐positive DLBCL, activated B‐cell‐like (ABC) type reveals that the sample is CD20‐positive, CD3‐negative, germinal center B‐cell expressed transcript 1 (GCET1)‐positive, multiple myeloma oncogene 1 (MUM1)‐positive, CD10‐negative, EBV‐encoded small RNA in situ hybridization (EBER (ISH))‐positive, and BACH2‐negative with a Ki‐67 labeling index of 62.8%. (b) Immunohistochemical analysis of EBV‐negative DLBCL, ABC type indicates that the sample is CD20‐positive, CD3‐negative, GCET1‐positive, MUM1‐positive, CD10‐negative, EBER(ISH)‐negative, and BACH2‐positive with a Ki‐67 labeling index of 81.2%.
Figure 2.
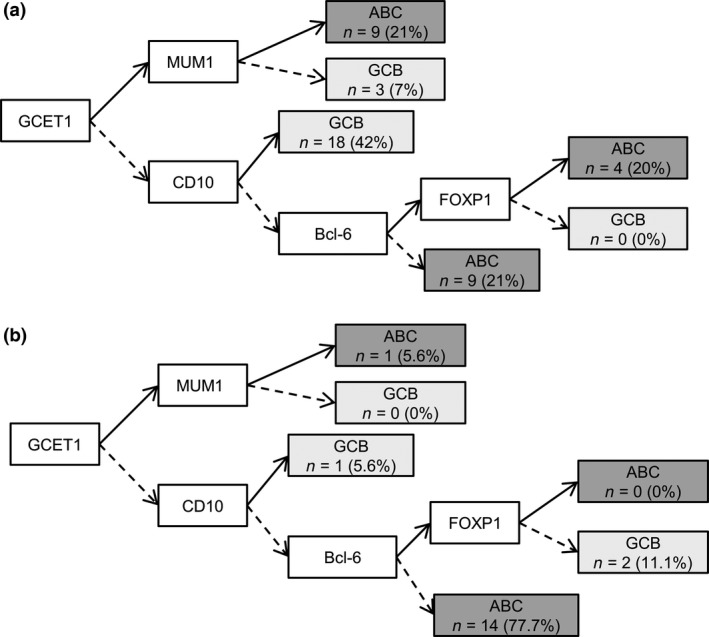
Distribution of Epstein–Barr virus (EBV)‐negative (a) and EBV‐positive (b) diffuse large B‐cell lymphoma (DLBCL) molecular phenotypes. →, Positive; ⇢, negative. ABC, activated B‐cell‐like; FOXP1, forkhead box protein P1; GCB, germinal center B‐cell‐like; GCET1, germinal center B‐cell expressed transcript 1; MUM1, multiple myeloma oncogene 1.
Immunohistochemically, 38 of 43 (88.4%) EBV‐negative DLBCL cases were BACH2‐positive, compared to only 5 of 23 (21.7%) EBV‐positive cases (P < 0.0001; Table 2a). When the analysis was restricted to ABC cases, EBV‐positive DLBCL cases also showed a significantly lower BACH2 expression rate relative to EBV‐negative cases (P < 0.0001; Table 2b), suggesting that the downregulation of BACH2 expression is associated with the presence of EBV. When compared with ABC‐type (22 cases) or germinal center B‐cell‐type (21 cases) EBV‐negative DLBCL cases, EBV‐positive cases showed a significantly lower rate of BACH2 positivity (P < 0.0001 and P < 0.0001, respectively; Table S3).
Table 2.
Differential BTB and CNC homology 2 (BACH2) expression in Epstein–Barr virus (EBV)‐positive and EBV‐negative diffuse large B‐cell lymphoma (DLBCL) and EBV‐positive and EBV‐negative DLBCL, activated B‐cell‐like (ABC) type (b)
| Diagnosis | BACH2 expression | P‐value | |
|---|---|---|---|
| Positive | Negative | ||
| (a) | |||
| EBV‐positive DLBCL (n = 23) | 5 | 18 | <0.0001 |
| EBV‐negative DLBCL (n = 43) | 38 | 5 | |
| (b) | |||
| EBV‐positive DLBCL, ABC type (n = 15) | 3 | 12 | <0.0001 |
| EBV‐negative DLBCL, ABC type (n = 22) | 20 | 2 | |
According to the criteria reported by Kikuchi et al.,21 all five BACH2‐positive cases of EBV‐positive DLBCL were strongly positive for BACH2. Among 38 BACH2‐positive cases of EBV‐negative DLBCL, 10 cases were weakly positive and 28 cases were strongly positive for BACH2.
In accordance with a previous report,21 we categorized our data into negative, weakly positive, or strongly positive BACH2 expression (Table S4). In total, BACH2 expression was negative, weakly positive, and strongly positive in 34.8%, 15.2%, and 50% of cases, respectively. In EBV‐negative cases, BACH2 negative, weakly positive, and strongly positive expression constituted 11.6%, 23.3%, and 65.1%. In EBV‐positive cases, all five BACH2‐positive cases were strongly positive, and in EBV‐negative cases, 10 cases of 38 BACH2‐positive cases were weak and 28 cases of BACH2‐positive cases were strongly positive. There was no significant difference between EBV status and the staining intensity of BACH2 (P = 0.247).
Biallelic BACH2 deletion was not detected in EBV‐positive DLBCL cases and human B‐cell lymphoma cell lines
We used FISH for the 18 EBV‐positive DLBCL cases that were immunohistochemically negative for BACH2. Of these, 15 cases (83.3%) had no evidence of BACH2 gene deletion (Fig. 3a). The other three cases (16.7%) showed monoallelic deletion; no cases had biallelic deletion. Notably, BACH2 gene loss was not detected in any EBV‐negative case (Fig. S2). Further studies using human B‐cell lymphoma‐derived cell lines (FL‐18, FL‐218, FL‐318, and the FL‐18 sister cell line FL‐18‐EB) were carried out. Through RT‐PCR analysis, BACH2 mRNA was detected in FL‐18, FL‐218, and FL‐318 but not in the EBV‐infected line, FL‐18‐EB (Fig. 3b). An immunohistochemical study yielded consistent results (Fig. 3c). Notably, biallelic BACH2 deletion was not detected in either FL‐18‐EB or FL‐18 using FISH (Fig. 3d), indicating that BACH2 expression was downregulated in the former despite the lack of gene deletion. This result was concordant with the findings from patient sample analyses.
Figure 3.
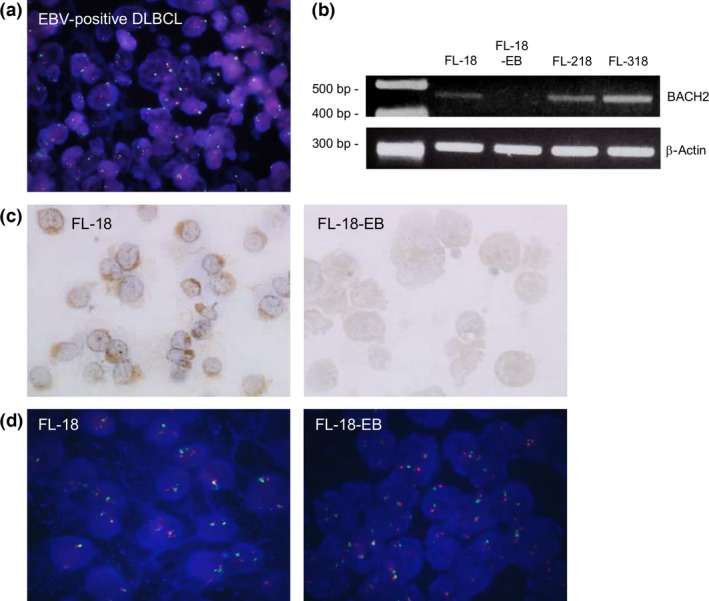
(a) Fluorescence in situ hybridization (FISH) analysis using a spectrum red labeled BACH2 probe and spectrum green labeled centromeric probe for chromosome 6 (CEP6). Epstein–Barr virus (EBV)‐positive diffuse large B‐cell lymphoma (DLBCL) yielded a signal ratio of 90.5%. (b–d) BACH2 expression in human non‐Hodgkin's lymphoma cell lines. (b) BACH2 expression was not detected by PCR in the FL‐18‐EB cell line. (c) BACH2 was expressed in FL‐18 cells but not in FL‐18‐EB cells. (d) FL‐18 and FL‐18‐EB showed signal ratios of 94% and 93%, respectively, by FISH analysis with the BACH2 probe.
Transfection of the BACH2 gene into FL‐18‐EB inactivated the TAK1–NFκB pathway
We transfected the BACH2 gene into FL‐18‐EB cells to analyze the effect of BACH2 on the expression of other genes. First, we confirmed the overexpression of BACH2 in FL‐18 cells using RT‐PCR (Fig. 4a), and we examined the NFκB pathway protein expression by Western blot analysis. From real‐time PCR analysis, NFKB2 (encodes p52) and RELA (encodes p65), the products of which comprise the NFκB pathway, were significantly downregulated in the BACH2‐transfected FL‐18‐EB cells (Fig. S3). Then we particularly focused on the expression of TAK1, a representative NFκB pathway molecule in EBV‐positive B cells.22 FL‐18‐EB cells contained pTAK1, an active form of TAK1, whereas this form was not detected in FL‐18 cells. Notably, the transfection of BACH2 into FL‐18‐EB cells repressed the activation of TAK1, as shown by the reduced levels of pTAK1 (Fig. 4b). No significant difference was observed in the expression of total TAK1. Moreover, p65 expression of the nuclear fractions was suppressed in BACH2‐transfected FL‐18‐EB cells, suggesting that BACH2 represses the TAK1–NFκB pathway (Fig. 4c,d).
Figure 4.

(a) BACH2 was detected in BACH2‐transfected FL‐18‐EB cells (FL‐18‐EB [BTB and CNC homology 2 (BACH2)+]) by PCR. (b) Phosphorylated transforming growth factor‐β‐activated kinase 1 (pTAK1) expression was downregulated in FL‐18‐EB (BACH2+) cells as shown by Western blot analysis. (c) p65 expression was downregulated in nuclear fractions of FL‐18‐EB (BACH2+) cells compared to FL‐18‐EB and FL‐18‐EB (control vector) cells. (d) The ratio of BACH2 expression level to LaminB1 is shown. (e) Immunohistochemistry in BACH2‐negative diffuse large B‐cell lymphoma (DLBCL) shows p65 positivity in the nucleus, pTAK1 positivity, and EBV‐encoded small RNA in situ hybridization (EBER (ISH)) positivity. (f) Immunohistochemistry in BACH2‐positive DLBCL shows p65 positivity in the cytoplasm, pTAK1 negativity, and EBER (ISH) negativity. (g) Immunohistochemistry in BACH2‐negative DLBCL shows nuclear staining of p50 and p52. (h) Immunohistochemistry in BACH2‐positive DLBCL shows cytoplasmic staining of p50 and p52.
Phosphorylated TAK1 expression and p65, p50, and p52 nuclear localization in BACH2‐negative DLBCL
Finally, we immunohistochemically evaluated the localization of p65, which was significantly downregulated in BACH2‐transfected FL‐18‐EB cells, by real‐time PCR, in 36 cases of BACH2‐positive DLBCL and 14 cases of BACH2‐negative DLBCL that were available for further examination and successfully stained. Among the BACH2‐positive DLBCLs, only 2 of 36 cases (5.6%) showed nuclear p65 localization, whereas 10 of 14 cases (71.4%) of BACH2‐negative DLBCL showed nuclear p65 localization (P < 0.0001; Table 3, Fig. 4e,f). The BACH2‐negative cases were found to express pTAK1, whereas BACH2‐positive cases did not (Fig. 4e,f). Furthermore, BACH2‐positve DLBCL showed significant cytoplasmic staining of p50 and p52 (P = 0.006 and P = 0.001, respectively; Fig. 4g,h, Table 3), although the total number of examined cases was limited due to shortage of samples. In addition, significant nuclear localization of p65, p50, and p52 was observed in EBV‐positive cases (P < 0.0001, P < 0.01, and P < 0.01, respectively; Table S5).
Table 3.
Differential p65, p50, and p52 expression in BTB and CNC homology 2 (BACH2)‐positive and BACH2‐negative diffuse large B‐cell lymphoma (DLBCL)
| Diagnosis | Cytoplasmic | Nuclear | P‐value |
|---|---|---|---|
| p65 | |||
| BACH2‐positive DLBCL (n = 36) | 34 | 2 | <0.0001 |
| BACH2‐negative DLBCL (n = 14) | 4 | 10 | |
| p50 | |||
| BACH2‐positive DLBCL (n = 17) | 16 | 1 | 0.0060 |
| BACH2‐negative DLBCL (n = 16) | 8 | 8 | |
| p52 | |||
| BACH2‐positive DLBCL (n = 14) | 14 | 0 | 0.0010 |
| BACH2‐negative DLBCL (n = 16) | 7 | 9 | |
Discussion
Despite previous reports of the aggressive behavior of EBV‐positive DLBCL, no reports have addressed the tumor‐suppressive factor BACH2. The findings from the present study reveal significant downregulation of BACH2 expression in EBV‐positive DLBCL and suggest its importance in EBV‐positive B‐cell lymphomas. In our study, we used the FL‐18 and EBV‐positive FL‐18‐EB cell lines, established from a single patient diagnosed with follicular lymphoma, which harbor t(14;18) and the MYC‐related translocation t(8;22)(q24;q13).16, 17 Therefore, these paired cell lines were considered a good model to analyze the impact of EBV in aggressive B‐cell lymphoma. Additionally, FL‐18‐EB showed partial LMP1 positivity in our examination (Fig. S4).
A member of the CNC (Cap'n'collar) group of transcription factors, BACH2 forms heterodimers with small Maf family proteins. These heterodimers repress the expression of target genes by binding to a DNA element termed the Maf recognition element.10, 23 Expression of BACH2 has been described in hematopoietic cells and nerve cells,24, 25 and the function of BACH2 has been well investigated in B lymphocytes. BACH2 represses plasma cell differentiation by repressing BLIMP1 transcription, and studies of BACH2‐deficient mice have led to the description of this protein as indispensable for the induction of class switch recombination.11, 26
In high‐grade lymphomas, a loss of heterozygosity on the long arm of chromosome 6 is frequently detected.27 BACH2 is accordingly among the candidate genes associated with lymphomagenesis,28, 29 and several research groups have recently described the tumor‐suppressive function of its protein product. For example, Swaminathan et al.14 reported that BACH2‐positive B cells resist MYC‐induced leukemic transformation through p53 upregulation. In chronic myeloid leukemia, Yoshida et al.30 reported that BACH2 phosphorylation, which occurred downstream of BCR/ABL signaling, repressed nuclear translocation and led to the expression of the anti‐apoptotic factor heme oxygenase‐1. Furthermore, Chen et al.31 reported that, in mantle cell lymphoma, BACH2 localization determined cell viability in response to oxidative stress. In the present study, significant downregulation of BACH2 expression was observed in EBV‐positive DLBCL, suggesting that BACH2 negativity contributes to the presence of EBV.
In previous reports, immunohistochemical BACH2 expression was described as negative, weakly positive, and strongly positive. The negative‐case ratio of our series (34.8%) is very similar to that of the series (35%) by Kikuchi et al.21 Sakane‐Ishikawa et al.12 and Ichikawa et al.13 reported no BACH2‐negative case in their series, which seems to be different from our data. However, in our series, only 11.6% of EBV‐negative cases were completely negative for BACH2. Therefore, we suppose this difference is not crucial. We focused on EBV status, and our proportion of EBV‐positive cases is different from other reports. We suppose that the higher negative ratio of BACH2 was due to the larger number of EBV‐positive DLBCLs in our series. Moreover, the anti‐BACH2 antibody used was different among the studies, which might affect the results. In our cases, there was no significant difference between EBV status and the staining level of BACH2, indicating BACH2 positivity or negativity is more important than BACH2 immunostaining intensity as to EBV status.
Previous reports have not examined the mechanism underlying the downregulation of BACH2 protein expression in EBV‐positive DLBCL. Notably, Takakuwa et al.32 reported that the integration of EBV into BACH2 induced a loss of BACH2 expression in the Raji Burkitt lymphoma cell line. As FISH did not detect biallelic deletion in any patient samples or cell lines used in our study, viral integration may have occurred in BACH2. Epigenetic silencing is another possible mechanism underlying the downregulation of BACH2 expression, as was previously reported in gastric cancer.33 Although we treated FL‐18‐EB cells with the DNA methylation inhibitor 5‐azacytidine, we were unable to recover BACH2 expression (data not shown), thus failing to support a methylation‐based mechanism of reduced expression. In contrast, proteolytic underlying mechanisms remain worth considering. Another member of the CNC transcription factor group, Nrf2, has been widely investigated, and the role of Keap1 in Nrf2 ubiquitination and subsequent proteasomal degradation has attracted considerable attention. Many reports have revealed an association between abnormal Keap1–Nrf2 signaling and tumorigenesis in many types of carcinomas.34, 35 Therefore, further evaluation of BACH2 proteolysis may be merited.
In this report, we also attempted to elucidate the association between BACH2 downregulation and NFκB activation. In EBV‐infected cells, LMP1, which is produced by EBV and expressed in the cell membrane, has been reported to affect neoplastic transformation through the ligand‐independent activation of several signaling pathways that induce cell growth.36, 37 As the NFκB pathway was among these oncogenic pathways, we focused on the association between BACH2 and the NFκB pathway.38 In canonical NFκB activation, tumor necrosis factor receptor‐associated factor 6 ubiquitination by the C‐terminal activation region 2 of LMP1 induces TAK1 binding protein 2/3 activation. Transforming growth factor‐β‐activated kinase 1 forms a complex with TAK1 binding protein 2/3 and TAK1 is subsequently activated by phosphorylation and polyubiquitination, leading to phosphorylation of the IκB kinase complex and degradation of IκBα. Subsequently, p50 and p65 transfers to the nucleus and induces the expression of genes related to cell proliferation and survival.39
Although the target protein of BACH2 in the NFκB pathway has not yet been clarified, BACH2 expression led to a decrease in TAK1 phosphorylation, which occurs upstream of the NFκB pathway. Therefore, BACH2 was suggested as a repressive factor upstream of TAK1 phosphorylation. Immunohistochemistry of pTAK1 was not detected in BACH2‐positive DLBCL cases and it was determined that these cases showed cytoplasmic p65 expression, whereas pTAK1 positivity and nuclear p65 expression were observed in BACH2‐negative DLBCL. Furthermore, nuclear p65 expression was repressed in the BACH2‐transfected cells by Western blot analysis.
Compagno et al.40 and Montes‐Moreno et al.5 used anti‐p50 and ‐p52 antibody to evaluate NFκB activation, and Ok et al.41 reported that expression of p65 showed no difference between EBV‐positive and ‐negative DLBCLs, contrary to the results of p50 and p52 in Caucasian. In our present study, p50, p52, and p65 had very similar status, and p65 frequently localized to the nucleus in EBV‐positive cases (P < 0.0001; Table S5). There are some possible factors for the discrepancy, such as differences in race and antibodies (although the antibody information was not available for the previous report). EBV‐associated lymphoproliferative disorder is more frequent in Asian than in Caucasian, that is, vast majority of patients with chronic active EBV infection, or EBV‐associated natural killer cell lymphoma of nasal type is almost exclusively found in Asia. Therefore, this discrepancy could be potentially explained by the difference of the population. All 66 DLBCL cases in our series are from Japanese patients.
Figure 5 shows a schema of the relationships between BACH2 and the NFκB pathway, showing that BACH2 repressed TAK1 phosphorylation and induced the inactivation of the NFκB signaling pathway. In EBV‐positive B cells, BACH2 downregulation contributes to the activation of the NFκB pathway through TAK1 activation. From a recent report, C‐Rel (an NFκB subunit) regulates BACH2 expression and the NFκB subunit binds to the region of the BACH2 gene in an EBV‐positive B cell line.42 Further study needs to clarify the relationships of TAK1, BACH2 and C‐Rel.
Figure 5.
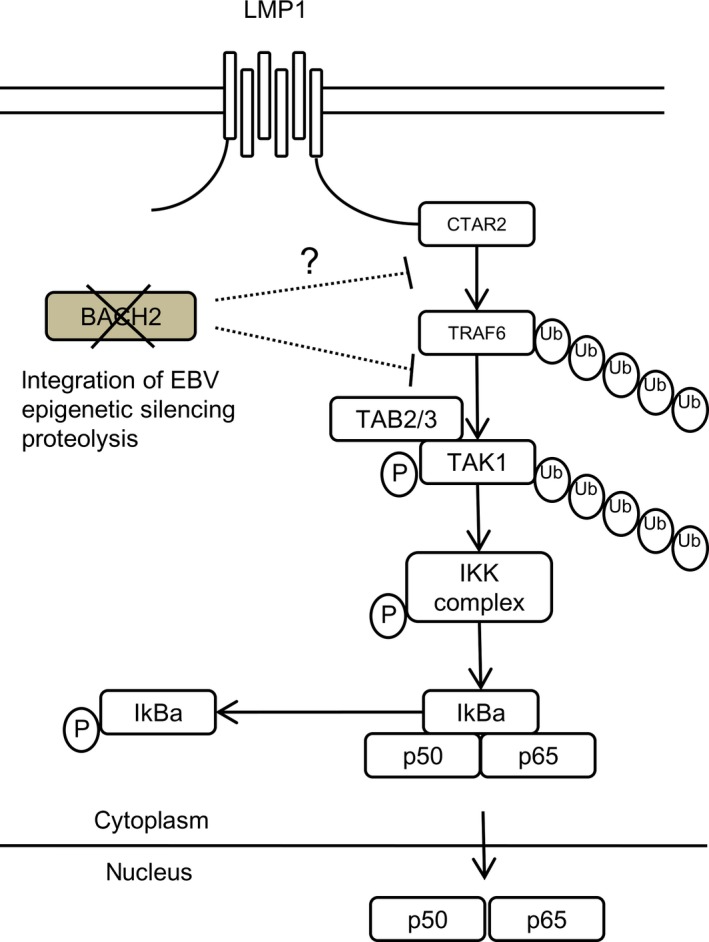
Schema of BTB and CNC homology 2 (BACH2) interaction in the nuclear factor‐κB (NFκB) signaling pathway. BACH2 repressed the transforming growth factor‐β‐activated kinase 1 (TAK1) phosphorylation and induced the inactivation of the NFκB signaling pathway. In Epstein–Barr virus (EBV)‐positive B‐cells, BACH2 downregulation contributes to the activation of the NFκB pathway through TAK1 activation. CTAR2, C‐terminal activation region 2; IKK, IκB kinase; LMP1, latent infection membrane protein 1; P, phosphorylation; TAB2/3, TAK1 binding protein 2/3; TRAF6, tumor necrosis factor receptor associated factor 6; Ub, ubiquitination.
In conclusion, our study reported the downregulation of BACH2 expression in the majority of EBV‐positive DLBCL cases, and indicated that it contributes to constitutive activation of the NFκB pathway in an EBV‐positive B‐cell lymphoma cell line. Points such as the target genes of BACH2 and the mechanism by which BACH2 expression is downregulated in EBV‐positive B‐cell lymphomas remain unresolved. Accordingly, further molecular and pathological studies are warranted to clarify the significance of BACH2 in EBV‐positive B‐cell lymphomas.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Table S1. Primer sequences used to amplify BACH2 and β‐actin.
Table S2. Antibodies and conditions for Western blot analysis.
Table S3. Differential BTB and CNC homology 2 (BACH2) expression in Epstein–Barr virus (EBV)‐positive and EBV‐negative diffuse large B‐cell lymphoma (activated B‐cell‐like [ABC] and germinal center B‐cell‐like [GCB] types).
Table S4. BTB and CNC homology 2 (BACH2) expression level based on staining intensity.
Table S5. Differential p65 expression in Epstein–Barr virus (EBV)‐positive and EBV‐negative diffuse large B‐cell lymphoma.
Fig. S1. Kaplan–Meier curves for overall survival in Epstein–Barr virus (EBV)‐positive and EBV‐negative diffuse large B‐cell lymphoma.
Fig. S2. Fluorescence in situ hybridization analysis using a spectrum red labeled BACH2 probe and spectrum green labeled centromeric probe for chromosome 6 (CEP6) in Epstein–Barr virus‐negative diffuse large B‐cell lymphoma.
Fig. S3. NFKB1, NFKB2, and RELA expression in FL‐18‐EB (BTB and CNC homology 2 [BACH2]+) cells analyzed by real‐time PCR.
Fig. S4. Immunohistochemical analysis of latent infection membrane protein 1 (LMP1) in FL‐18‐EB cells.
Acknowledgments
We thank the following doctors for providing patient samples: Kunihiro Omonishi, Kazuo Hamaya, Soichiro Nose, Wakako Oda, Ichiro Yamadori, Yoko Shinno, Koichi Mizobuchi, Toshiaki Morito, Midori Ando, Shin Ishizawa, Rie Yamasaki, Eiko Hayashi, Nobuya Ohara, Yuka Takahashi, Koji Taguchi, Keita Kobayashi, Katsuya Miyatani, Toshiharu Maeda, Tadayoshi Kunitomo, Yoshimi Bando, Yasushi Terasaki, Yoshinobu Maeda, Yumi Oshiro, Akira Hida, Eiji Ikeda, and Aya Ishii.
Cancer Sci 108 (2017) 1071–1079
Funding Information
This work was supported by a grant from the Japan Society for the Promotion Science (JSPS no. 24790350 and 15K19053).
References
- 1. Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet 1964; 1: 702–3. [DOI] [PubMed] [Google Scholar]
- 2. Nilsson K. Human B‐lymphoid cell lines. Hum Cell 1992; 5: 25–41. [PubMed] [Google Scholar]
- 3. Zhang T, Fu Q, Gao D, Ge L, Sun L, Zhai Q. EBV associated lymphomas in 2008 WHO classification. Pathol Res Pract 2014; 210: 69–73. [DOI] [PubMed] [Google Scholar]
- 4. Oyama T, Yamamoto K, Asano N et al Age‐related EBV‐associated B‐cell lymphoproliferative disorders constitute a distinct clinicopathologic group: a study of 96 patients. Clin Cancer Res 2007; 13: 5124–32. [DOI] [PubMed] [Google Scholar]
- 5. Montes‐Moreno S, Odqvist L, Diaz‐Perez JA et al EBV‐positive diffuse large B‐cell lymphoma of the elderly is an aggressive post‐germinal center B‐cell neoplasm characterized by prominent nuclear factor‐kB activation. Mod Pathol 2012; 25: 968–82. [DOI] [PubMed] [Google Scholar]
- 6. Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell‐like diffuse large B cell lymphoma cells. J Exp Med 2001; 194: 1861–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan WJ. Pathogenesis of diffuse large B cell lymphoma. Int J Hematol 2010; 92: 219–30. [DOI] [PubMed] [Google Scholar]
- 8. Kilger E, Kieser A, Baumann M, Hammerschmidt W. Epstein–Barr virus‐mediated B‐cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J 1998; 17: 1700–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hammarskjold ML, Simurda MC. Epstein–Barr virus latent membrane protein transactivates the human immunodeficiency virus type 1 long terminal repeat through induction of NF‐kappa B activity. J Virol 1992; 66: 6496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Igarashi K, Ochiai K, Muto A. Architecture and dynamics of the transcription factor network that regulates B‐to‐plasma cell differentiation. J Biochem 2007; 141: 783–9. [DOI] [PubMed] [Google Scholar]
- 11. Muto A, Ochiai K, Kimura Y et al Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J 2010; 29: 4048–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sakane‐Ishikawa E, Nakatsuka S, Tomita Y et al Prognostic significance of BACH2 expression in diffuse large B‐cell lymphoma: a study of the Osaka Lymphoma Study Group. J Clin Oncol 2005; 23: 8012–7. [DOI] [PubMed] [Google Scholar]
- 13. Ichikawa S, Fukuhara N, Katsushima H et al Association between BACH2 expression and clinical prognosis in diffuse large B‐cell lymphoma. Cancer Sci 2014; 105: 437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swaminathan S, Huang C, Geng H et al BACH2 mediates negative selection and p53‐dependent tumor suppression at the pre‐B cell receptor checkpoint. Nat Med 2013; 19: 1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ando M, Sato Y, Takata K et al A20 (TNFAIP3) deletion in Epstein–Barr virus‐associated lymphoproliferative disorders/lymphomas. PLoS ONE 2013; 8: e56741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohno H, Doi S, Fukuhara S, Nishikori M, Uchino H, Fujii H. A newly established human lymphoma cell line, FL‐18, carrying a 14;18 translocation. Jpn J Cancer Res 1985; 76: 563–6. [PubMed] [Google Scholar]
- 17. Doi S, Ohno H, Tatsumi E et al Lymphoma cell line (FL‐18) and Epstein–Barr virus‐carrying cell line (FL‐18‐EB) obtained from a patient with follicular lymphoma: monoclonal derivation and different properties. Blood 1987; 70: 1619–23. [PubMed] [Google Scholar]
- 18. Nakamura N, Kuze T, Hashimoto Y et al Analysis of the immunoglobulin heavy chain gene variable region of CD5‐positive and ‐negative diffuse large B cell lymphoma. Leukemia 2001; 15: 452–7. [DOI] [PubMed] [Google Scholar]
- 19. Oka T, Ouchida M, Koyama M et al Gene silencing of the tyrosine phosphatase SHP1 gene by aberrant methylation in leukemias/lymphomas. Cancer Res 2002; 62: 6390–4. [PubMed] [Google Scholar]
- 20. Singhirunnusorn P, Suzuki S, Kawasaki N, Saiki I, Sakurai H. Critical roles of threonine 187 phosphorylation in cellular stress‐induced rapid and transient activation of transforming growth factor‐beta‐activated kinase 1 (TAK1) in a signaling complex containing TAK1‐binding protein TAB 1 and TAB 2. J Biol Chem 2005; 280: 7359–68. [DOI] [PubMed] [Google Scholar]
- 21. Kikuchi T, Tokunaka M, Kikuti YY et al Over‐expression of BACH2 is related to ongoing somatic hypermutation of the immunoglobulin heavy chain gene variable region of de novo diffuse large B‐cell lymphoma. Pathol Int 2013; 63: 339–44. [DOI] [PubMed] [Google Scholar]
- 22. Wu L, Nakano H, Wu Z. The C‐terminal activating region 2 of the Epstein–Barr virus‐encoded latent membrane protein 1 activates NF‐kappaB through TRAF6 and TAK1. J Biol Chem 2006; 281: 2162–9. [DOI] [PubMed] [Google Scholar]
- 23. Igarashi K, Ochiai K, Itoh‐Nakadai A, Muto A. Orchestration of plasma cell differentiation by Bach2 and its gene regulatory network. Immunol Rev 2014; 261: 116–25. [DOI] [PubMed] [Google Scholar]
- 24. Nakamura A, Ebina‐Shibuya R, Itoh‐Nakadai A et al Transcription repressor Bach2 is required for pulmonary surfactant homeostasis and alveolar macrophage function. J Exp Med 2013; 210: 2191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoshino H, Igarashi K. Expression of the oxidative stress‐regulated transcription factor bach2 in differentiating neuronal cells. J Biochem 2002; 132: 427–31. [DOI] [PubMed] [Google Scholar]
- 26. Muto A, Tashiro S, Nakajima O et al The transcriptional programme of antibody class switching involves the repressor Bach2. Nature 2004; 429: 566–71. [DOI] [PubMed] [Google Scholar]
- 27. Guan XY, Horsman D, Zhang HE, Parsa NZ, Meltzer PS, Trent JM. Localization by chromosome microdissection of a recurrent breakpoint region on chromosome 6 in human B‐cell lymphoma. Blood 1996; 88: 1418–22. [PubMed] [Google Scholar]
- 28. Sasaki S, Ito E, Toki T et al Cloning and expression of human B cell‐specific transcription factor BACH2 mapped to chromosome 6q15. Oncogene 2000; 19: 3739–49. [DOI] [PubMed] [Google Scholar]
- 29. Scholtysik R, Kreuz M, Hummel M et al Characterization of genomic imbalances in diffuse large B‐cell lymphoma by detailed SNP‐chip analysis. Int J Cancer 2015; 136: 1033–42. [DOI] [PubMed] [Google Scholar]
- 30. Yoshida C, Yoshida F, Sears DE et al Bcr‐Abl signaling through the PI‐3/S6 kinase pathway inhibits nuclear translocation of the transcription factor Bach2, which represses the antiapoptotic factor heme oxygenase‐1. Blood 2007; 109: 1211–9. [DOI] [PubMed] [Google Scholar]
- 31. Chen Z, Pittman EF, Romaguera J et al Nuclear translocation of B‐cell‐specific transcription factor, BACH2, modulates ROS mediated cytotoxic responses in mantle cell lymphoma. PLoS ONE 2013; 8: e69126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takakuwa T, Luo WJ, Ham MF, Sakane‐Ishikawa F, Wada N, Aozasa K. Integration of Epstein–Barr virus into chromosome 6q15 of Burkitt lymphoma cell line (Raji) induces loss of BACH2 expression. Am J Pathol 2004; 164: 967–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haam K, Kim HJ, Lee KT et al Epigenetic silencing of BTB and CNC homology 2 and concerted promoter CpG methylation in gastric cancer. Cancer Lett 2014; 351: 206–14. [DOI] [PubMed] [Google Scholar]
- 34. Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1‐Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011; 16: 123–40. [DOI] [PubMed] [Google Scholar]
- 35. Jaramillo MC, Zhang DD. The emerging role of the Nrf2‐Keap1 signaling pathway in cancer. Genes Dev 2013; 27: 2179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dawson CW, Tramountanis G, Eliopoulos AG, Young LS. Epstein–Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3‐kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J Biol Chem 2003; 278: 3694–704. [DOI] [PubMed] [Google Scholar]
- 37. Roberts ML, Cooper NR. Activation of a ras‐MAPK‐dependent pathway by Epstein–Barr virus latent membrane protein 1 is essential for cellular transformation. Virology 1998; 240: 93–9. [DOI] [PubMed] [Google Scholar]
- 38. Liebowitz D. Epstein–Barr virus and a cellular signaling pathway in lymphomas from immunosuppressed patients. N Engl J Med 1998; 338: 1413–21. [DOI] [PubMed] [Google Scholar]
- 39. Ersing I, Bernhardt K, Gewurz BE. NF‐kappaB and IRF7 pathway activation by Epstein–Barr virus Latent Membrane Protein 1. Viruses 2013; 5: 1587–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Compagno M, Lim WK, Grunn A et al Mutations of multiple genes cause deregulation of NF‐kappaB in diffuse large B‐cell lymphoma. Nature 2009; 459: 717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ok CY, Li L, Xu‐Monette ZY et al Prevalence and clinical implications of Epstein–Barr virus infection in de novo diffuse large B‐cell lymphoma in Western countries. Clin Cancer Res 2014; 20: 2338–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hunter JE, Butterworth JA, Zhao B et al The NF‐kappaB subunit c‐Rel regulates Bach2 tumour suppressor expression in B‐cell lymphoma. Oncogene 2016; 35: 3476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences used to amplify BACH2 and β‐actin.
Table S2. Antibodies and conditions for Western blot analysis.
Table S3. Differential BTB and CNC homology 2 (BACH2) expression in Epstein–Barr virus (EBV)‐positive and EBV‐negative diffuse large B‐cell lymphoma (activated B‐cell‐like [ABC] and germinal center B‐cell‐like [GCB] types).
Table S4. BTB and CNC homology 2 (BACH2) expression level based on staining intensity.
Table S5. Differential p65 expression in Epstein–Barr virus (EBV)‐positive and EBV‐negative diffuse large B‐cell lymphoma.
Fig. S1. Kaplan–Meier curves for overall survival in Epstein–Barr virus (EBV)‐positive and EBV‐negative diffuse large B‐cell lymphoma.
Fig. S2. Fluorescence in situ hybridization analysis using a spectrum red labeled BACH2 probe and spectrum green labeled centromeric probe for chromosome 6 (CEP6) in Epstein–Barr virus‐negative diffuse large B‐cell lymphoma.
Fig. S3. NFKB1, NFKB2, and RELA expression in FL‐18‐EB (BTB and CNC homology 2 [BACH2]+) cells analyzed by real‐time PCR.
Fig. S4. Immunohistochemical analysis of latent infection membrane protein 1 (LMP1) in FL‐18‐EB cells.


