Abstract
Adenoviruses are widely used to deliver genes to a variety of cell types and have been used in a number of clinical trials for gene therapy and oncolytic virotherapy. However, several concerns must be addressed for the clinical use of adenovirus vectors. Selective delivery of a therapeutic gene by adenovirus vectors to target cancer is precluded by the widespread distribution of the primary cellular receptors. The systemic administration of adenoviruses results in hepatic tropism independent of the primary receptors. Adenoviruses induce strong innate and acquired immunity in vivo. Furthermore, several modifications to these vectors are necessary to enhance their oncolytic activity and ensure patient safety. As such, the adenovirus genome has been engineered to overcome these problems. The first part of the present review outlines recent progress in the genetic modification of adenovirus vectors for cancer treatment. In addition, several groups have recently developed cancer‐targeting adenovirus vectors by using libraries that display random peptides on a fiber knob. Pancreatic cancer‐targeting sequences have been isolated, and these oncolytic vectors have been shown by our group to be associated with a higher gene transduction efficiency and more potent oncolytic activity in cell lines, murine models, and surgical specimens of pancreatic cancer. In the second part of this review, we explain that combining cancer‐targeting strategies can be a promising approach to increase the clinical usefulness of oncolytic adenovirus vectors.
Keywords: Adenovirus, fiber knob, library, pancreatic cancer, targeting
The adenovirus family consists of non‐enveloped DNA viruses with a linear genome of 30–38 kb.1 There are 57 adenovirus serotypes, which are classified into categories A–G, based on viral properties of agglutination.2 Adenovirus vectors based on human serotype 5 of species C are beneficial as gene‐delivery vehicles that enable high‐titer production and highly efficient gene transfer into a wide spectrum of dividing and non‐dividing cells both in vitro and in vivo.2 These vectors have been used in a number of clinical trials utilizing gene therapy and oncolytic virotherapy.3 However, several concerns need to be addressed before widespread clinical use of adenovirus vectors is possible. First, selective adenovirus‐mediated delivery of a therapeutic gene to target cancer is precluded by the widespread distribution of the primary cellular receptors.4, 5 Second, i.v. injection of adenovirus vectors results in hepatic tropism, making it difficult to systemically deliver adenovirus vectors for metastasis treatment.6, 7 Third, the strong immunogenic nature of adenoviruses seriously hampers their efficacy and safety in vivo.6, 8 Fourth, clinical studies on oncolytic adenoviruses have suggested that several modifications are necessary to enhance the oncolytic activity and improve safety in the clinical setting.2, 6, 9 The adenovirus genome has been engineered in an attempt to solve these issues; as such, studies have been carried out to address cancer‐specific delivery, strong liver sequestration, immune reaction to adenovirus‐infected tissues, and oncolytic virus potency. The present review outlines recent progress in the genetic modification of adenovirus vectors. In addition, several groups have recently constructed an adenovirus library by displaying random peptides on a fiber knob to generate adenovirus vectors that target specific cell types.10, 11, 12 This review also summarizes the development of a cancer‐targeting oncolytic virus using an adenovirus library approach as an example of a technological advance made in adenovirus vector research.
Cancer‐targeting adenovirus vectors
The entry of serotype 5 adenovirus into cells requires two distinct and sequential steps. The initial step is the interaction of the fiber protein with its cellular receptor, coxsackievirus and adenovirus receptor (CAR).4, 5 The next step, internalization of the virus, is promoted by an interaction between the arginine‐glycine‐aspartic acid (RGD) sequence of the penton base and αv integrins on the cell surface.13, 14 Transductional targeting aims to affect the cellular entry of vectors. This type of targeting can create tissue tropism to new cellular targets by conferring novel ligand‐mediated binding properties to receptors and by simultaneously inhibiting binding to natural receptors (detargeting). To detarget an adenovirus vector, the AB‐loop of the fiber knob, which is the binding site for CAR, is generally modified (Fig. 1).10 The ablation of binding sites in the adenoviral capsid with integrin and heparan sulfate proteoglycans (HSPG) might be useful to further reduce native tropism (Fig. 1).15
Figure 1.
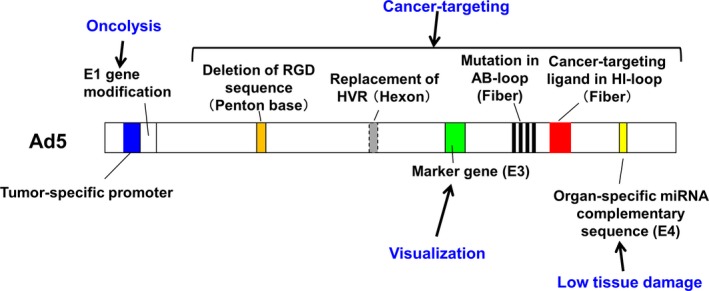
Genetic modification of recombinant adenovirus vectors. Mutations in the E1 region are introduced or the E1 gene is transcribed by a tumor‐ or tissue‐specific promoter to restrict viral replication to targeted cancer cells, without affecting normal cells. The arginine‐glycine‐aspartic acid (RGD) sequence in the penton base is depleted and mutation in the AB‐loop of the fiber knob is induced to suppress naïve tropism. Ligands are inserted into the HI‐loop and AB‐loop of the fiber to target cancer cells. The hypervariable region of the hexon is replaced to inhibit interaction with coagulation factor. Organ‐specific microRNA (miRNA) targeting sequences are inserted into the E4 region to suppress tissue damage that results from immune reactions. Marker genes such as enhanced green fluorescent protein are inserted in the deleted E3 region for visualization.
To date, several promising approaches have been used for target‐specific gene delivery. For example, CAR expression can decrease with tumor progression. Replacing part of the serotype 5 adenovirus capsid with non‐serotype 5 adenovirus capsid (fiber switching) can be used to circumvent CAR deficiency. The pseudotyping of serotype 5 adenovirus with serotype 3 adenovirus fibers was shown to result in CAR‐independent infectivity and enhanced gene transfer to a broad range of cancer cell types.2, 16 Chimeric serotype 5 vectors with serotype 35 fibers exploit the CD46 receptor for cell infection, as many cancers express high levels of CD46.1 As such, a chimeric serotype 3/serotype 11p virus was found have increased selectivity for colon cancer cells.6
The tropisms of chimeric viruses are defined by those of the replacing serotypes. Thus, retargeting adenovirus vectors by incorporating ligands into the viral capsid is preferable. Such targeting has been achieved by direct genetic modifications of the capsid proteins; specifically, targeting ligands can be incorporated into the C‐terminus and HI‐loop of fiber proteins, the L1 loop of the hexon, the RGD loop of the penton base, and into minor capsid protein IX.17, 18, 19, 20 One of the most effective fiber modifications is the incorporation of an RGD sequence into the fiber.7 However, cancer‐specific ligands that can be used for targeting adenovirus vectors are generally unknown. To overcome this problem, several groups have developed random peptide‐displaying adenovirus library technology and used this approach to produce cancer‐targeting adenovirus vectors.10, 11, 12 The details of vector development using an adenovirus library are described in a subsequent part of this review.
Inhibition of liver sequestration
Systemic administration of adenoviruses results in the majority of transduction occurring in the liver. Liver tropism was shown to be linked to a pathway different from CAR binding. Binding between the fiber knob domain and plasma proteins such as coagulation factor and complement component C4‐binding protein results in virus uptake in hepatocytes and Kupffer cells via HSPG and low‐density lipoprotein receptors in a CAR‐independent manner.6, 7
It was recently reported that coagulation factor (F) X binds the hypervariable regions (HVRs) of the adenovirus hexon, leading to enhanced liver infection.21, 22 Therefore, insertion of mutations in the FX‐binding domain of HVR and its replacement with HVR of other serotype adenoviruses markedly decreased the liver sequestration of the adenoviruses (Fig. 1). However, it should be noted that the binding of FV to HVR also plays an important role in protecting adenoviruses from the classical complementary pathway.23 In the future, HVR‐mutant adenovirus vectors that do not activate the complementary pathway need to be developed. The vectors could specifically infect certain tumors even after systemic administration.24
Suppression of adenovirus‐induced immune response
As is widely known, adenovirus vectors evoke both innate and adaptive immune responses in infected tissues; this represents an important safety consideration for the clinical development of adenovirus vectors. Systemically administered adenovirus vectors are delivered into Kupffer cells, dendritic cells, and macrophages in the liver and spleen, and elicit an innate immune reaction within a few hours.25 The innate immune response is initiated by the recognition of viral components by different mechanisms including several pattern‐recognition receptor in the cells. Adenovirus genomic DNA is recognized by Toll‐like receptor 9 and cyclic GMP‐AMP synthase (cGAS).26 Adenovirus‐derived virus‐associated RNA (VA‐RNA) are also recognized by RNA sensors and transduce intracellular signaling through interferon (IFN)‐β promoter stimulator (IPS)‐1.27 The adenovirus capsid also activates the NLRP3 inflammasome, leading to the activation of caspase‐1.28 Through these mechanisms, the expression of type I IFN and other proinflammatory cytokines such as IL‐1β, IL‐6, and TNF‐α is induced,25 resulting in the activation of natural killer cells. Subsequently, the innate immune reaction and the expression of cytokines such as type I IFN lead to the induction of an acquired immune response against adenovirus and the expression of its transgenes. Capsid antigens are largely responsible for the induction of the adenovirus‐specific cytotoxic T lymphocyte (CTL) response.8 In addition, the leaky expression of adenovirus genes, which has been observed for E2A, E4, pIX, hexon, and fiber, are thought to play a crucial role in the induction of cellular immunity.29 Such adenovirus protein‐induced cellular immunity frequently results in the elimination of adenovirus vector‐transduced cells and tissue damage.
Several approaches have been used to suppress the immune response to adenovirus vectors. Adenovirus vectors lacking the expression of VA‐RNA resulted in suppression of the innate immune response.8 To suppress leaky adenovirus genes, E2A−, E4−, and/or pIX‐deleted adenovirus vectors have been used, and this approach was shown to result in a decreased CTL response and diminished hepatotoxicity.30, 31 A helper‐dependent adenovirus vector that lacks all viral coding regions resulted in a reduced inflammatory response in the organs.32, 33 However, the development of a production system for such mutant adenovirus vectors has been hampered by technical complexity and low titers. Recently, a method based on tissue‐specific microRNA (miRNA) expression was established. For example, the incorporation of complementary sequences for miRNA‐122a, which exhibits liver‐specific expression, into the 3′‐UTR of E2A, E4, or pIX genes suppressed the leaky expression of adenovirus genes, and significantly reduced hepatotoxicity (Fig. 1).29 Mechanisms of immune response by adenovirus vectors need to be fully clarified to develop new adenovirus vectors that can suppress the induction of immune reaction in vivo.
Oncolytic adenovirus vectors
Numerous strategies have been used to augment the selectivity and efficacy of oncolytic adenovirus for cancer treatment. First, restricting viral entry and infection through transductional targeting has been accomplished using several methods as described earlier in Cancer‐targeting adenovirus vectors. Transcriptional targeting is the second key method that is used to induce tissue‐specific oncolytic adenovirus replication. To date, two methods have been used to restrict viral replication to cancer cells, and not normal cells.2, 6, 9
The first method (type I) is to introduce a mutation in the E1 region; these missing genes are functionally complemented by genetic mutations in tumor cells, such as abnormalities in the retinoblastoma (RB) pathway or p53 mutations. The ONYX‐015 and H101 viruses have deletions in the E1B55K gene, which normally inactivates p53; these defective E1B55K proteins fail to block the normal apoptotic defense pathway, thereby restricting adenovirus replication in normal cells. A recent study suggested that cancer cell‐specific replication is based on late viral mRNA transport.34 Another commonly used type I oncolytic adenovirus contains a 24‐bp mutation in the E1A gene (E1AΔ24) that disrupts the Rb‐binding domain. This results in the release of free E2F from the Rb/E2F complex, which results in activation of the remaining transcriptional units. As such, E1AΔ24 can promote viral replication in cancer cells with Rb pathway mutations but not in normal cells. The oncolytic adenovirus ICOVIR was developed to take advantage of excessive free E2F by regulating E1A transcription by the insertion into E2F binding sites.9
The second method (type II) involves the construction of viruses in which the transcription of E1 genes is restricted to tumor cells by a tumor or tissue‐specific promoter. The CV706 adenovirus was designed with the E1A gene under control of the prostate specific antigen promoter for prostate cancer treatement.9 The OBP‐301 adenovirus was engineered to use the telomerase reverse transcriptase (TERT) promoter to regulate the E1A gene for expression in cancer cells with high levels of TERT.9 For recently completed and ongoing clinical trials involving oncolytic adenoviruses, patients with head and neck cancer were treated with 5‐FU/cisplatin and H101, and showed a 78.8% response rate, compared to 39.6% in the 5‐FU/cisplatin‐only regimen. The Chinese regulatory agency approved H101 for the treatment of nasopharyngeal carcinoma in combination with chemotherapy. ONYX‐015 was evaluated in several clinical trials, but its efficacy was limited. This reagent is considered to improve clinical results as a combination therapy with other anticancer agents. Phase I and II clinical trials of ICOVIR‐5, CV706 and OBP‐301 are now in progress.6, 9, 35
The third method is to overcome the physical barrier to the dissemination of oncolytic adenoviruses in solid tumors. In the tumor microenvironment, accumulation of cancer‐associated fibroblasts and dense extracellular matrix and formation of neovasculature impede the spread of oncolytic adenoviruses throughout the entire tumor mass. To enhance viral dissemination, viruses have been generated to target the extracellular matrix (ECM).36 Oncolytic adenoviruses expressing relaxin have been shown to disrupt the ECM and successfully increase viral spreading in tumors.37 VCN‐01, which expresses hyaluronidase, disrupts the ECM and shows a strong antitumor effect in murine cancer models.38
The fourth method is to increase the potency of the oncolytic adenovirus by arming the virus with a therapeutic transgene. This combined approach, referred to as suicide gene therapy, which uses genes encoding proteins such as cytosine deaminase (CD) and HSV‐1 thymidine kinase (TK) showed enhanced antitumor efficacy in vivo when compared to that with the virus alone.39 It was recently demonstrated that tumor cell killing by oncolytic viruses produces a potent costimulatory signal and results in the release of tumor antigens that can be recognized by antigen‐presenting cells. It was previously shown that genes encoding immunostimulatory cytokines such as granulocyte‐macrophage colony stimulating factor (GM‐CSF) markedly enhance antitumor immunity of oncolytic adenoviruses by recruiting natural killer cells and priming tumor‐specific CTL.40 Arming the virus with various immune costimulatory genes will be attempted in further basic researches and clinical trials.
Screening cancer‐targeting vectors by using adenovirus libraries that display random peptides on the fiber
To overcome the paucity of cancer‐specific ligands, insertions into the adenovirus fiber have been used, as described earlier in Cancer‐targeting adenovirus vectors. Three groups, including our own, have developed a system for producing adenovirus libraries that display a variety of peptides on the fiber knob (Table 1). In our system, the adenovirus genome library is first constructed in vitro by Cre‐lox‐mediated recombination between an adenoviral fiber‐modified plasmid library and adenoviral genome DNA, and recombined DNA is transfected into cells.10, 41, 42 In terms of live viruses, the complexity of the library reached a level of 104 in a 6‐mm dish using our method.10, 41 As the complexity of an adenovirus library depends on the number of producing cells, the complexity was increased by scaling up of the number and size of culture plates. Lupold et al.11 reported a method to construct adenovirus libraries by infecting producer cells with genetically fiberless pseudotyped adenoviruses and transfecting the same cells with a plasmid containing a modified fiber gene. Miura et al.12 improved adenovirus library construction by infecting fiber‐complemented producer cells with genetically fiberless pseudotyped adenovirus and transfecting the cells with a fiber‐modified shuttle vector. Using this approach, they reported that the viral genomic diversity of the library was 5 × 108 in a 6‐cm dish.
Table 1.
Reports of peptide‐displaying adenovirus libraries
| Institution | Year | Library construction method | Peptide‐displaying site | Diversity | Library screening | Targeting vector |
|---|---|---|---|---|---|---|
| National Cancer Center | 2007 | In vitro construction of adenovirus genome‐coding fiber library10 | HI‐loop in fiber knob | >1 × 104 live viral complexity per a 6‐cm dish | Cell line, Murine tumor model | Glioma10 Pancreatic cancer43, 44 |
| 2014 | Infection of genetically fiberless pseudotyped adenovirus and transfection of shuttle plasmid‐coding fiber library in producer cells42 | HI‐loop in fiber knob | >1 × 104 live viral complexity per one well in a 6‐well dish | |||
| Johns Hopkins University | 2007 | Infection of genetically fiberless pseudotyped adenovirus and transfection of fiber‐gene shuttle in producer cells11 | HI‐loop in fiber knob | Cell line | Prostate cancer45 (PSMA‐targeting) | |
| University of Minnesota | 2013 | Infection of genetically fiberless pseuydotyped adenovirus and transfection of shuttle plasmid‐coding fiber library in fiber‐transcomplement producer cells12 | AB‐loop in fiber knob | >5 × 108 genomic complexity per a 6‐cm dish | Cell line | Pancreatic cancer12 (mesothelin‐targeting) |
PSMA, prostate‐specific membrane antigen.
The position of the targeting motif in the fiber might be important for successful targeting. Insertion into the HI‐loop does not prevent trimer formation of the fiber. In libraries constructed by our and Lupold's groups, six to seven random amino acids were inserted into the HI‐loop of the fiber, and mutations were included in the AB‐loop of the fiber knob to abolish CAR binding. Miura et al. inserted seven random amino acids into the AB‐loop of the fiber, which was shown to abolish CAR binding and result in the simultaneous display of new targeting motifs. Comparing HI‐loops, AB‐loops, and C‐termini of the fiber knobs is necessary to determine the best motif‐insertion site for the development of targeting vectors. To date, the screening of these adenovirus libraries using cultured cells and a murine peritoneal tumor model has resulted in the successful isolation of adenovirus vectors that target glioma, pancreatic cancer, and prostate cancer.10, 12, 43, 44, 45
Development of pancreatic cancer‐targeting oncolytic adenovirus
Pancreatic cancer is the leading cause of cancer‐related death in Japan as well as in Western countries.46, 47, 48 Its 5‐year survival rate is still less than 5%, and overcoming this poor prognosis remains one of the most formidable challenges in oncology.47, 49 In locally advanced cases, the development of strategies to control local lesions and to prevent distant metastases is important. Novel approaches to improve patient survival are expected.49, 50
To explore pancreatic cancer‐targeting vectors, an adenovirus library was screened using a human pancreatic cancer cell line, and an adenovirus vector displaying a SYENFSA (SYE) motif was isolated. The gene transduction efficiency of the SYE‐displaying adenovirus vector was significantly enhanced in pancreatic cancer cell lines (2.2–6.9‐fold), whereas this vector showed almost identical efficiency, compared to that of the control adenovirus vector, in other cancer cells and in normal cells such as fibroblasts. This suggests that the SYENFSA sequence enhances adenovirus infectivity for a certain population of pancreatic cancer cells (Fig. 2).44
Figure 2.
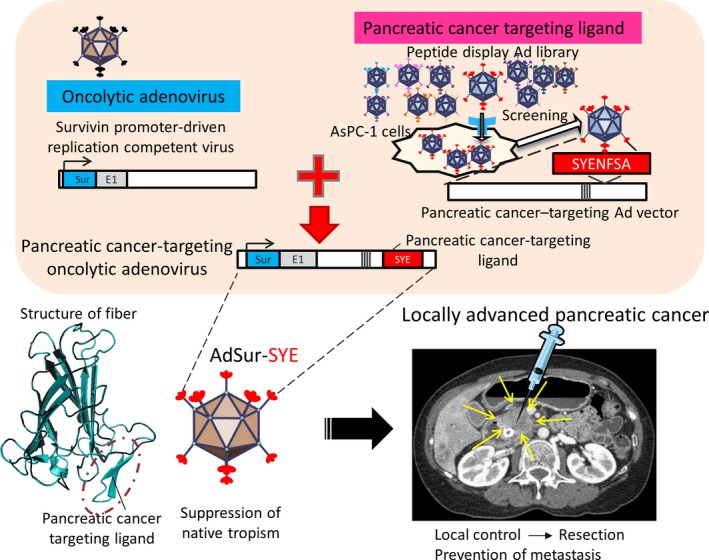
Development of pancreatic cancer‐targeting oncolytic virus by a library approach. The pancreatic cancer‐targeting sequence is combined with a survivin promoter‐driven oncolytic adenovirus (AdSur‐SYE). Locally advanced pancreatic cancer is surgically unresectable but can be accessed by ultrasound‐ or computed tomography‐guided percutaneous injection or endoscopic ultrasound‐guided injection of viruses.
The addition of tumor‐targeting potential to an oncolytic adenovirus could broaden the therapeutic window between antitumor effectiveness and systemic toxicity.51, 52 A survivin promoter was shown to have significantly enhanced activity in pancreatic cancer cell lines. We constructed a virus that displays the targeting ligand SYE on the fiber knob of a survivin‐mediated, replication‐competent adenovirus (AdSur), for which native tropism is ablated. This AdSur‐SYE effectively suppressed tumor growth, and to a greater extent than that by AdSur, in pancreatic cancer cell lines and murine tumor models.44, 53 AdSur‐SYE showed increased proliferation and dissemination in the tumors, whereas no transgene‐positive cells were detected in the adjacent skin and skeletal muscle. Analysis of adenovirus genome copy number indicated that the addition of the SYE ligand decreased ectopic and undesirable infection of organs.53
As the expression levels and types of cell surface receptors can be substantially different in the microenvironment, infectivity towards human pancreatic cancer specimens might differ from that towards cell lines. AdSur‐SYE showed significantly higher infectivity and oncolytic activity compared to those of AdSur in single cells prepared from surgical pancreatic cancer specimens, whereas the infectivity of AdSur‐SYE and AdSur was almost identical in cells from other cancers, specifically pancreas and liver.53 Immunostaining indicated that the cells infected with the virus were pancreatic ductal adenocarcinoma cells and not stromal cells.53 The results demonstrated that direct injection of pancreatic cancer‐targeting oncolytic adenovirus is a promising therapeutic strategy for locally advanced cases of pancreatic cancer (Fig. 2).
Future perspectives
Adenovirus‐based vectors and oncolytic viruses are widely used as therapeutic agents. They are especially prominent in the field of cancer gene therapy. However, there are several problems that need to be addressed before fully implementing this strategy in a clinical therapeutic platform. For future development, oncolytic adenoviruses require five characteristics: (i) high and specific infectivity in cancer tissues; (ii) cancer‐specific lysis; (iii) suppression of organ toxicity; (iv) suppression of inflammatory reactions; and (v) detectability by imaging modalities (Fig. 3). The genetic modifications reported by a variety of laboratories could be combined to fulfil these requirements.
Figure 3.
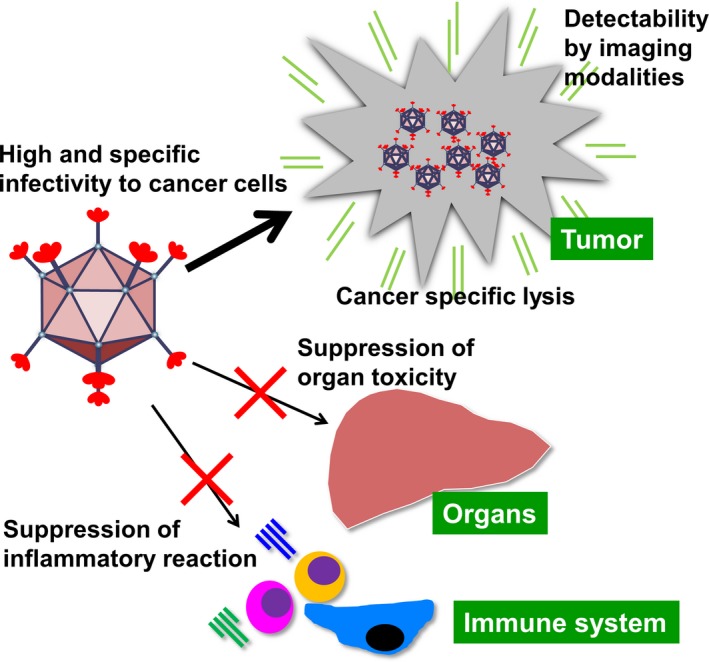
Future directions for oncolytic virus research. In future, the five characteristics would be required for progress in the development of oncolytic viruses.
A library approach has been experimentally applied to develop cancer‐targeting vectors, and several vectors have been isolated using in vitro cell culture and murine models. Extensive exploration of target peptides through in vitro and in vivo screening of adenovirus libraries might result in an extensive list of targeting peptides for each type of cancer. Future development will involve screening biopsies and surgically resected tumors, thereby leading to the generation of more clinically useful targeting adenoviruses. This library‐based technology, for specific adenovirus vector selection, could have broad implications for a variety of applications in medicine.
Oncolytic viruses, in combination with other targeting strategies, represent a promising avenue for the next generation of oncolytic virotherapy. One of the most important factors for achieving success in clinical trials is selection of patients with cancer tissues that are suitable for this targeting strategy. Selection of patients for whom high viral infectivity of biopsy samples is observed might be feasible in clinical settings. If a list of several cancer‐targeting viruses is generated and made available for clinical use, the most suitable virus for each patient could be selected based on viral infectivity of biopsy samples and patient‐derived xenografts, ultimately leading to personalized oncolytic virotherapy.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This work was supported in part by grants‐in‐aid for research from the Ministry of Health, Labour and Welfare of Japan, and by the National Cancer Center Research and Development Fund (23‐A‐9 and 26‐A‐11).
Cancer Sci 108 (2017) 831–837
Funding Information
This work was supported in part by grants‐in‐aid for research from the Ministry of Health, Labour and Welfare of Japan, and by the National Cancer Center Research and Development Fund. (23‐A‐9 26‐A‐11).
Reference
- 1. Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov 2015; 14: 642–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yamamoto M, Curiel DT. Current issues and future directions of oncolytic adenoviruses. Mol Ther 2010; 18: 243–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2012 – an update. J Gene Med 2013; 15: 65–77. [DOI] [PubMed] [Google Scholar]
- 4. Bergelson JM, Cunningham JA, Droguett G et al Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 1997; 275: 1320–3. [DOI] [PubMed] [Google Scholar]
- 5. Tomko RP, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA 1997; 94: 3352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pesonen S, Kangasniemi L, Hemminki A. Oncolytic adenoviruses for the treatment of human cancer: focus on translational and clinical data. Mol Pharm 2011; 8: 12–28. [DOI] [PubMed] [Google Scholar]
- 7. Beatty MS, Curiel DT. Chapter two–Adenovirus strategies for tissue‐specific targeting. Adv Cancer Res 2012; 115: 39–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Machitani M, Yamaguchi T, Shimizu K et al Adenovirus vector‐derived VA‐RNA‐mediated innate immune responses. Pharmaceutics 2011; 3: 338–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nat Biotechnol 2012; 30: 658–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miura Y, Yoshida K, Nishimoto T et al Direct selection of targeted adenovirus vectors by random peptide display on the fiber knob. Gene Ther 2007; 14: 1448–60. [DOI] [PubMed] [Google Scholar]
- 11. Lupold SE, Kudrolli TA, Chowdhury WH, Wu P, Rodriguez R. A novel method for generating and screening peptides and libraries displayed on adenovirus fiber. Nucleic Acids Res 2007; 35: e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miura Y, Yamasaki S, Davydova J et al Infectivity‐selective oncolytic adenovirus developed by high‐throughput screening of adenovirus‐formatted library. Mol Ther 2013; 21: 139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai M, Harfe B, Freimuth P. Mutations that alter an Arg‐Gly‐Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell‐rounding activity and delay virus reproduction in flat cells. J Virol 1993; 67: 5198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wickham TJ, Segal DM, Roelvink PW et al Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J Virol 1996; 70: 6831–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mizuguchi H, Koizumi N, Hosono T et al CAR‐ or alphav integrin‐binding ablated adenovirus vectors, but not fiber‐modified vectors containing RGD peptide, do not change the systemic gene transfer properties in mice. Gene Ther 2002; 9: 769–76. [DOI] [PubMed] [Google Scholar]
- 16. Kaufmann JK, Nettelbeck DM. Virus chimeras for gene therapy, vaccination, and oncolysis: adenoviruses and beyond. Trends Mol Med 2012; 18: 365–76. [DOI] [PubMed] [Google Scholar]
- 17. Krasnykh V, Dmitriev I, Mikheeva G, Miller CR, Belousova N, Curiel DT. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J Virol 1998; 72: 1844–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dmitriev I, Krasnykh V, Miller CR et al An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor‐independent cell entry mechanism. J Virol 1998; 72: 9706–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yoshida Y, Sadata A, Zhang W, Saito K, Shinoura N, Hamada H. Generation of fiber‐mutant recombinant adenoviruses for gene therapy of malignant glioma. Hum Gene Ther 1998; 9: 2503–15. [DOI] [PubMed] [Google Scholar]
- 20. Douglas JT, Miller CR, Kim M et al A system for the propagation of adenoviral vectors with genetically modified receptor specificities. Nat Biotechnol 1999; 17: 470–5. [DOI] [PubMed] [Google Scholar]
- 21. Kalyuzhniy O, Di Paolo NC, Silvestry M et al Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc Natl Acad Sci USA 2008; 105: 5483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Irons EE, Flatt JW, Doronin K et al Coagulation factor binding orientation and dimerization may influence infectivity of adenovirus‐coagulation factor complexes. J Virol 2013; 87: 9610–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu Z, Qiu Q, Tian J et al Coagulation factor X shields adenovirus type 5 from attack by natural antibodies and complement. Nat Med 2013; 19: 452–7. [DOI] [PubMed] [Google Scholar]
- 24. Waddington SN, McVey JH, Bhella D et al Adenovirus serotype 5 hexon mediates liver gene transfer. Cell 2008; 132: 397–409. [DOI] [PubMed] [Google Scholar]
- 25. Muruve DA. The innate immune response to adenovirus vectors. Hum Gene Ther 2004; 15: 1157–66. [DOI] [PubMed] [Google Scholar]
- 26. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP‐AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013; 339: 786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamaguchi T, Kawabata K, Kouyama E et al Induction of type I interferon by adenovirus‐encoded small RNAs. Proc Natl Acad Sci USA 2010; 107: 17286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barlan AU, Griffin TM, McGuire KA, Wiethoff CM. Adenovirus membrane penetration activates the NLRP3 inflammasome. J Virol 2011; 85: 146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shimizu K, Sakurai F, Tomita K et al Suppression of leaky expression of adenovirus genes by insertion of microRNA‐targeted sequences in the replication‐incompetent adenovirus vector genome. Mol Ther Methods Clin Dev 2014; 1: 14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lusky M, Christ M, Rittner K et al In vitro and in vivo biology of recombinant adenovirus vectors with E1, E1/E2A, or E1/E4 deleted. J Virol 1998; 72: 2022–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andrews JL, Kadan MJ, Gorziglia MI, Kaleko M, Connelly S. Generation and characterization of E1/E2a/E3/E4‐deficient adenoviral vectors encoding human factor VIII. Mol Ther 2001; 3: 329–36. [DOI] [PubMed] [Google Scholar]
- 32. Mian A, Guenther M, Finegold M, Ng P, Rodgers J, Lee B. Toxicity and adaptive immune response to intracellular transgenes delivered by helper‐dependent vs. first generation adenoviral vectors. Mol Genet Metab 2005; 84: 278–88. [DOI] [PubMed] [Google Scholar]
- 33. Reddy PS, Sakhuja K, Ganesh S et al Sustained human factor VIII expression in hemophilia A mice following systemic delivery of a gutless adenoviral vector. Mol Ther 2002; 5: 63–73. [DOI] [PubMed] [Google Scholar]
- 34. Cheng PH, Wechman SL, McMasters KM, Zhou HS. Oncolytic replication of E1b‐deleted adenoviruses. Viruses 2015; 7: 5767–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goldufsky J, Sivendran S, Harcharik S et al Oncolytic virus therapy for cancer. Oncolytic Virother 2013; 2: 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rosewell Shaw A, Suzuki M. Recent advances in oncolytic adenovirus therapies for cancer. Curr Opin Virol 2016; 21: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee SY, Park HR, Rhee J, Park YM, Kim SH. Therapeutic effect of oncolytic adenovirus expressing relaxin in radioresistant oral squamous cell carcinoma. Oncol Res 2013; 20: 419–25. [DOI] [PubMed] [Google Scholar]
- 38. Martinez‐Velez N, Xipell E, Vera B et al The oncolytic adenovirus VCN‐01 as therapeutic approach against pediatric osteosarcoma. Clin Cancer Res 2016; 22: 2217–25. [DOI] [PubMed] [Google Scholar]
- 39. Rogulski KR, Wing MS, Paielli DL, Gilbert JD, Kim JH, Freytag SO. Double suicide gene therapy augments the antitumor activity of a replication‐competent lytic adenovirus through enhanced cytotoxicity and radiosensitization. Hum Gene Ther 2000; 11: 67–76. [DOI] [PubMed] [Google Scholar]
- 40. Cerullo V, Pesonen S, Diaconu I et al Oncolytic adenovirus coding for granulocyte macrophage colony‐stimulating factor induces antitumoral immunity in cancer patients. Cancer Res 2010; 70: 4297–309. [DOI] [PubMed] [Google Scholar]
- 41. Hatanaka K, Ohnami S, Yoshida K et al A simple and efficient method for constructing an adenoviral cDNA expression library. Mol Ther 2003; 8: 158–66. [DOI] [PubMed] [Google Scholar]
- 42. Yamamoto Y, Goto N, Miura K et al Development of a novel efficient method to construct an adenovirus library displaying random peptides on the fiber knob. Mol Pharm 2014; 11: 1069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nishimoto T, Yamamoto Y, Yoshida K, Goto N, Ohnami S, Aoki K. Development of peritoneal tumor‐targeting vector by in vivo screening with a random peptide‐displaying adenovirus library. PLoS One 2012; 7: e45550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nishimoto T, Yoshida K, Miura Y et al Oncolytic virus therapy for pancreatic cancer using the adenovirus library displaying random peptides on the fiber knob. Gene Ther 2009; 16: 669–80. [DOI] [PubMed] [Google Scholar]
- 45. Wu P, Kudrolli TA, Chowdhury WH, Liu MM, Rodriguez R, Lupold SE. Adenovirus targeting to prostate‐specific membrane antigen through virus‐displayed, semirandom peptide library screening. Cancer Res 2010; 70: 9549–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Werner J, Combs SE, Springfeld C, Hartwig W, Hackert T, Buchler MW. Advanced‐stage pancreatic cancer: therapy options. Nat Rev Clin Oncol 2013; 10: 323–33. [DOI] [PubMed] [Google Scholar]
- 47. Chadha AS, Khoo A, Aliru ML, Arora HK, Gunther JR, Krishnan S. Recent advances and prospects for multimodality therapy in pancreatic cancer. Semin Radiat Oncol 2016; 26: 320–37. [DOI] [PubMed] [Google Scholar]
- 48. Mosquera C, Maglic D, Zervos EE. Molecular targeted therapy for pancreatic adenocarcinoma: a review of completed and ongoing late phase clinical trials. Cancer Genet 2016; 209: 567–81. [DOI] [PubMed] [Google Scholar]
- 49. Valsecchi ME, Diaz‐Canton E, de la Vega M, Littman SJ. Recent treatment advances and novel therapies in pancreas cancer: a review. J Gastrointest Cancer 2014; 45: 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hackert T, Buchler MW. Pancreatic cancer: advances in treatment, results and limitations. Dig Dis 2013; 31: 51–6. [DOI] [PubMed] [Google Scholar]
- 51. Chira S, Jackson CS, Oprea I et al Progresses towards safe and efficient gene therapy vectors. Oncotarget 2015; 6: 30675–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rein DT, Breidenbach M, Curiel DT. Current developments in adenovirus‐based cancer gene therapy. Future Oncol 2006; 2: 137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamamoto Y, Hiraoka N, Goto N et al A targeting ligand enhances infectivity and cytotoxicity of an oncolytic adenovirus in human pancreatic cancer tissues. J Control Release 2014; 192: 284–93. [DOI] [PubMed] [Google Scholar]


