Abstract
Ovarian cancer is the most lethal gynecologic malignancy. Recently, several molecularly targeted anticancer agents have been developed for ovarian cancer; however, its prognosis remains extremely poor. The development of molecularly targeted therapy, as well as companion diagnostics, is required to improve outcomes for patients with ovarian cancer. In this study, to identify microRNAs (miRNAs) involved in the progression of ovarian cancer we analyzed serum miRNAs in patients with ovarian cancer using miRNA array and quantitative RT‐PCR and examined the anticancer properties of miRNA expression in ovarian cancer cells. In patients with ovarian cancer, high amount of miR‐135a‐3p in serum samples was significantly associated with favorable clinical prognosis. The amount of miR‐135a‐3p was significantly decreased in patients with ovarian cancer compared with patients with ovarian cysts or normal ovaries. In SKOV‐3 and ES‐2 human ovarian cancer cells, enhanced expression of miR‐135a‐3p induced drug sensitivity to cisplatin and paclitaxel and suppressed cell proliferation and xenograft tumor growth. These findings suggest that miR‐135a‐3p may be considered as a biomarker and a therapeutic agent in ovarian cancer.
Keywords: Biomarker, miR‐135a‐3p, molecular target, nucleic acid therapy, ovarian cancer
MicroRNAs (miRNAs) are small non‐coding RNAs (18–22 nt) that modulate gene expression. They contribute to many biological processes including cell proliferation, differentiation, and death, and are also involved in pathologies such as cancer, diabetes, cardiovascular, and gynecologic diseases.1, 2, 3 MicroRNAs are present in exosomes in various biofluids such as plasma, serum, urine, semen, menstrual blood, and amniotic fluid, and consequently have been proposed as ideal candidates for disease biomarkers.4 Recently, they have also been proposed as drug targets for many human diseases.5, 6, 7
Ovarian cancer, the most lethal malignancy in gynecologic oncology, is characterized by an extremely poor prognosis from the extensive spread of ovarian cancer cells into the peritoneal cavity.8 Cytoreductive surgery and the introduction of platinum plus taxane‐based chemotherapies in the past decade have improved survival in patients with epithelial ovarian cancer.9, 10 In the ICON7 trial, standard chemotherapy with bevacizumab did not increase overall survival in patients with newly diagnosed ovarian cancer, although it did improve progression‐free survival (PFS) and peritoneal fluid retention.11 Progression‐free survival, which is involved in chemosensitivity, has been recognized as cancer‐specific survival data.12, 13, 14, 15, 16 Previous studies have shown that patients have poor prognosis when PFS ranges within 6 months.17, 18 In clinical trials, poly(ADP‐ribose) polymerase inhibitors did not improve overall survival, whereas PFS was significantly prolonged.19 Accordingly, the development of molecularly targeted therapeutics and their diagnostic companion tests is required to improve these poor outcomes.
Many reports have investigated alterations in miRNA levels as biomarkers in ovarian cancer.20, 21, 22, 23 Currently, inconsistencies in results do not support the use of miRNA as a diagnostic tool in tissue or serum specimens from patients with ovarian cancer. In many studies, miRNA expression in ovarian cancer tissue was compared with that in normal ovarian tissue. However, typically, the ovarian epithelium was not microdissected or scraped, meaning that the difference in miRNA expression between ovarian cancer tissue and ovarian stroma was evaluated, even though ovarian stroma compromises more than 95% of the ovarian tissue in normal specimens.21 Moreover, because of major differences in serum miRNA levels between malignant and benign tumor status, comparisons of normal versus tumor tissue are considered inadequate to identify miRNAs regulating carcinogenesis. To explore key miRNAs as biomarkers for cancer progression, proper selection and high quality tissue or serum specimens are essential.
In this study, to identify candidate miRNAs as biomarkers, and as the potential therapeutic targets among circulating miRNAs, we compared miRNA array data of sera between patients with poor and favorable prognosis, and then evaluated the relationship between the selected miRNAs and PFS in 98 patients with ovarian cancer. Additionally, we examined the further clinical significance of expression of the selected miRNAs in peritoneal fluid and cancer tissues. Using ovarian cancer cell lines, we also examined the relationship between selected miRNAs and chemosensitivity or tumor formation. Our data suggest that miR‐135a‐3p represents a biomarker and a potential therapeutic target among the circulating miRNAs in ovarian cancer.
Materials and Methods
Patients
To explore candidate miRNAs involved in clinical prognosis in serum samples, 12 patients with ovarian cancer (Table S1) were enrolled for miRNA array analysis. They were divided into two groups: the poor prognosis group (group A) consisted of six patients who had recurrence of ovarian cancer within 6 months after completion of primary treatment including both platinum and taxane, and the favorable prognosis group (group B) included six patients with ovarian cancer who had no recurrence or recurrence more than 6 months after completion of primary treatment containing both platinum and taxane. Ninety‐eight patients with ovarian cancer (Table S2, group C) were also enrolled to assess the relationship between the levels of these candidate miRNAs in serum samples and PFS. In addition, to assess the amount of miR‐135a‐3p in serum and the expression of cancer tissue samples in ovarian cancer, 30 patients with ovarian cancer were examined (Table S2, group D). Seventeen patients ovarian cancer (Table S2, group E) were examined for the levels of miR‐135a‐3p in serum, peritoneal fluid, and cancer tissue samples. As a control, seven patients with benign gynecologic diseases (mean age ± SD, 65.2 ± 4.8 years) were examined for miR‐135a‐3p levels in serum samples, and 24 patients with benign ovarian tumor (age, 38.0 ± 17.6 years) were also examined for miR‐135a‐3p levels in serum and peritoneal fluid samples. Additionally, the expression of miR‐135a‐3p was examined in five RNA samples of normal ovary tissue obtained from OriGene Technologies (catalog nos. CR561675, CR561070, CR561721, CR560512, and CR562355; Kanagawa, Japan). The present study was approved by the Ethics Committee of Fukuoka University Hospital (Fukuoka, Japan) and Kyusyu University Hospital (Fukuoka, Japan). All individuals provided written informed consent. Sera, peritoneal fluid, or tissue samples from ovarian cancer patients were collected from 2009 to 2015 at Fukuoka University and Kyusyu University. Clinical and pathologic information for ovarian cancer patients was collected from clinical records. Tumors were staged using the International Federation of Gynecology and Obstetrics staging system. All ovarian cancer patients had been diagnosed histopathologically without radiotherapy or chemotherapy prior to study participation. These patients included newly diagnosed cases and those undergoing follow‐up.
Serum, peritoneal fluid, and tissue samples
All clinical samples were obtained from Fukuoka University Hospital (n = 88) or Kyusyu University Hospital (n = 98). Serum samples were placed at 4°C no more than 24 h after collection and separated from clots by centrifugation at 3000 g for 20 min at 4°C. Peritoneal fluids and tissue samples were collected at primary surgery and immediately placed on ice. Peritoneal fluids were centrifuged at 3000 rpm for 20 min at 24°C to separate debris within 1 h of collection. Tissue samples were soaked in RNAlater solution (Life Technologies, Kanagawa, Japan) and incubated at 4°C overnight (for ≤24 h). All sera, peritoneal fluids, and tissue samples were stored at −80°C for further RNA extraction.
Total RNA extraction
Total RNA was isolated from sera and peritoneal fluids using QIAzol reagent (Qiagen, Tokyo, Japan) and the miRNeasy Serum/Plasma Kit (Qiagen). Briefly, 300 μL serum or peritoneal fluid sample was mixed with 1.5 mL QIAzol Lysis Reagent and incubated at room temperature for 5 min. As a spike‐in control, 3.5 μL of 1.6 × 108 copies/μL miR‐39‐3p from Caenorhabditis elegans (Cel‐miR‐39‐3p) was added. RNA was purified according to the manufacturer's protocol and eluted in 38 μL RNase‐free water. Total RNA was isolated from tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. RNA concentrations and 260–280 nm absorbance ratios were measured using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Reverse transcription and quantitative PCR
For cDNA generation for miRNAs, 5 μL total RNA was used for each RT reaction with specific primers for miR‐135a‐3p, miR‐630, miR‐1207, Cel‐miR‐39‐3p, and RNU6B, according to the manufacturer's protocol (Life Technologies). For cDNA generation from mRNA, total RNA from tissues was reverse transcribed using PrimeScript II reverse transcriptase (TaKaRa Bio, Otsu, Japan) according to the manufacturer's protocol. The RT product was stored at −80°C. Quantitative PCR was subsequently carried out with an ABI Prism 7500 Fast Real‐Time PCR system (Applied Biosystems, Foster City, CA, USA) in a 15‐ or 20‐μL reaction with TaqMan Universal Master Mix (Applied Biosystems). Amplification was carried out by denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Exponential processes (Ct‐values) of miRNAs from serum and peritoneal fluids were converted to linear comparisons relative to the control group and normalized to the spike‐in Cel‐miR‐39‐3p using 2−∆∆Ct methods. Gene expression was analyzed using the difference in cycle threshold (2−∆∆Ct) method. The Ct values of tissue miRNA were normalized to RNU6B, and those of mRNA were normalized to GAPDH as internal controls. The ∆∆Ct values were plotted and statistical analysis was carried out using GraphPad Prism (GraphPad Software, San Diego, CA, USA). The TaqMan primer and probes (Applied Biosystems, Kanagawa, Japan) used were as follows: miR‐135a‐3p, 002232; miR‐630, 001563; miR‐1207, 002826; cel‐miR‐39; 000200; RNU6B, 001093; baculoviral IAP repeat‐containing protein 3 (BIRC3), Hs00985031_g1; gamma‐aminobutyric acid receptor subunit α3 (GABRA3), Hs00968132_m1; sperm protein associated with the nucleus on the X chromosome B1/2 (SPANXB1/2), Hs02387419_gH; enteropeptidase (TMPRss15), Hs00160491_m1; neural cell adhesion molecule 2 (NCAM2), Hs00189850_m1; synaptotagmin‐4 (SYT4), Hs01086433_m1; C9orf152, Hs00418117_m1; interleukin‐1 receptor accessory protein (IL1RAP), Hs00895050_m1; Ras‐related and estrogen‐regulated growth inhibitor (RERG), Hs01556349_m1; polyadenylate‐binding protein‐interacting protein 1 (PAIP1), Hs01925976_s1; protein phosphatase 2 regulatory subunit β, PPP2R2B, Hs00270227_m1; and GAPDH, Hs02758991.
MicroRNA microarray
MicroRNA microarray profiling was carried out using Agilent Human miRNA Microarray (Agilent Technologies, Tokyo, Japan), containing probes for 1523 human and 364 viral miRNAs from the Sanger database. Total RNA was extracted as above and purified using the SV Total RNA Isolation System (Promega, Tokyo, Japan). Complementary RNA was amplified and labeled with Cy3 using a Low Input Quick Amp Labeling Kit (Agilent Technologies) then hybridized to a 60 K 60‐mer oligomicroarray (SurePrint Human miRNA Microarray Release 16.0, 8 × 60 K; Agilent Technologies) according to the manufacturer's instructions. Hybridized microarray slides were scanned using an Agilent scanner. Relative hybridization intensities and background hybridization values were calculated using Feature Extraction Software version 9.5.1.1 (Agilent Technologies). Scanned images were analyzed with Feature Extraction Software 9.5.1.1 (Agilent Technologies) using default parameters to obtain background subtracted and spatially detrended processed signal intensities. Raw signal intensities and flags for each probe were calculated from hybridization intensities and spot information according to procedures recommended by Agilent Technologies using Flag criteria in GeneSpring Software (Agilent Technologies). The mean values for each miRNA in group A or B were estimated, and then the fold change was computed by dividing the mean value in group A by the mean value in group B. The complete dataset was deposited into the NCBI Gene Expression Omnibus repository under the accession number GSE79943.
Cell culture
Human ovarian cancer cell lines SKOV‐3, ES‐2, OVCAR‐3, and RMG‐1 were obtained from ATCC (Manassas, VA, USA). Cells were cultured in RPMI‐1640 medium supplemented with 10% FBS (ICN Biochemicals, Irvine, CA, USA), 100 U/mL penicillin G, and 100 μg/mL streptomycin (Invitrogen) in a humidified atmosphere of 5% CO2 at 37°C.
Generation of stably transfected cell lines
The hsa‐miR‐135a coding region was PCR‐amplified from 293T genomic DNA using the following primers: miR‐135a_Fw_735, 5′‐gcgaagcttAGGCCTCGCTGTTCTCTATGG‐3′; and miR‐135a_Rv, 5′‐catggatccTGTCCCCGCCGTGCGCCACGG‐3′. The fragment was inserted into HindIII/BamHI sites of pmR‐ZsGreen‐1 (TaKaRa, Clontech, Shiga, Japan) vector. Expression vectors for pmR‐ZsGreen1/miR‐135a or pmR‐ZsGreen1 (as control) were transfected into SKOV‐3, ES‐2, OVCAR‐3, and RMG‐1 cells using Lipofectamine 2000 (Invitrogen). At 48–72 h post‐transfection, the culture medium was changed to RPMI‐1640 supplemented with 10% FBS and puromycin to select transfected cells.
Cell viability assay
The effect of miR‐135a‐3p on SKOV‐3, ES‐2, OVCAR‐3, or RMG‐1 cell viability was determined using the WST‐8 assay kit (Cell Counting Kit‐8; Dojindo, Kumamoto, Japan). To calculate the IC50 value of cisplatin (CDDP) and paclitaxel (PTX) in SKOV‐3 and ES‐2 cells transfected with the miR‐blank or miR‐135a‐3p expression vector, cells were seeded in Poly‐l‐Lysine 96‐well plates (Corning, Tokyo, Japan) and allowed to grow for 48 h. After 48 h, WST‐8 was added to each well for 1 h before the first measurement. Absorbance at 450 nm was measured using a microplate reader according to the manufacturer's instructions. To exclude the effects of transfection on cells, untransfected cells were used as controls. Transfected cells were exposed to the following treatments: 1, 10, 25, 50, 75, 1 × 102, 2.5 × 102, 5 × 102, 1 × 103, or 2 × 103 μM of CDDP, or 7 × 10−3, 3.5 × 10−2, 7 × 10−2, 1.75 × 10−1, 3.5 × 10−1, 7 × 10−1, 1.75, 3.5, 7, or 70 μM of PTX. Each IC50 value was calculated by determining the concentration that caused 50% inhibition of cell growth. All experiments were carried out in triplicate. Absorbance at 450 nm was measured using a microplate reader according to the manufacturer's instructions.
Antitumor effects of miR‐135a‐3p in a mouse xenograft model
For the s.c. implanted ovarian cancer cell lines, out of the four ovarian cancer cell lines, we used two cell lines (SKOV‐3 and ES‐2) that showed lower growth inhibition despite the high transfection efficiency of the miR‐135a‐3p vector in in vitro assays. Subconfluent cell cultures were detached from plates with trypsin–EDTA. A total volume of 250 μL containing 1 × 106 cells suspended in serum‐free RPMI‐1640 was injected s.c. into 4‐week‐old nude mice (Charles River Laboratories Japan, Yokohama, Japan). To assess the inhibitory effects of miR‐135a‐3p on tumor growth, cells transfected with miR‐blank or miR‐135a‐3p were injected s.c. and the tumor volume was estimated from 2‐D tumor measurements as follows: tumor volume (mm3) = (length × width2)/2. The animal protocol was approved according to the guidelines of the Animal Care of Fukuoka University (Approval No. 1306662) and the ethics committee. Animals were observed daily. Humane endpoints were defined as loss of more than 10% of body mass, tumor diameter >20 mm, or inability to ambulate or rise for food and water. If animals reached these endpoints, they were killed by exsanguination. Animal surgery and euthanasia using decapitation were carried out under inhalation (isoflurane) anesthesia, and all efforts were made to minimize suffering.
Gene expression array analysis
Expression array profiling was carried out using the Affymetrix Human Gene 2.0 ST Array as described in the manufacturer's protocol. The analysis was undertaken using SKOV‐3 and ES‐2 cells with forced expression of miR‐135a using an expression vector. Total RNA extraction was carried out using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's instructions. Complementary DNA synthesis, amplification, and labeling with biotin were carried out with the WT PLUS Reagent Kit (Affymetrix). RNA integrity number values of extracted RNA were determined with an Agilent 2100 Bioanalyzer (Agilent Technologies). To ensure optimal data quality, only samples with RNA integrity number ≥9.0 were included in the analysis. Following fragmentation, 4.7 μg cDNA was hybridized on the GeneChip Human Gene 2.0 ST Array using the GeneChip Hybridization Oven 645. GeneChips were washed and stained in the GeneChip Fluidics Station 450. GeneChips were scanned using the GeneChip Scanner 3000 7G. Data were analyzed with Expression Console version 1.4 (Affymetrix) and Transcriptome Analysis Console version 3.0 (Affymetrix). Robust multichip average (RMA) was used as a summarization algorithm. Value definition was log2 GC‐RMA signal. Microarray data were analyzed using GeneSpring software. The mean fold changes of mRNA expression in cells with miR‐135a‐3p overexpression or miR‐blank overexpression were estimated. The fold changes were computed by dividing the mean value in cells with miR‐135a‐3p overexpression by the mean value in cells with miR‐blank. A fold change <−3.0 was considered statistically significant. The complete dataset was deposited into the NCBI Gene Expression Omnibus repository under the accession number GSE 84723.
Target gene profiling
Microarray data were analyzed using GeneSpring software. A fold change <−3.0 or >3.0 was considered statistically significant. Hierarchical clustering analysis was used to organize the genes based on similarities in expression profiles.
Target prediction for miR‐135a‐3p was also undertaken using the algorithms TargetScan (http://www.targetscan.org/vert_71/), miRDB (http://mirdb.org/miRDB/), and miRanda (http://www.microrna.org/microrna/home.do) in terms of base sequence. All prediction processes were carried out using custom‐written executable files that computed the parameters between miRNAs and mRNAs based on inherent algorithms and set thresholds. The thresholds for the algorithms were context score ≤−0.4 for TargetScan, Target score ≥60 for miRDB, and seed mirSVR score >−1.25 for miRanda.
Statistical analysis
In miRNA array analyses, statistically significant differences in values between group A and B were determined using the Mann–Whitney U‐test. In gene expression analyses, statistically significant differences in values between cells with miR‐135a‐3p overexpression and miR‐blank overexpression were determined using the Mann–Whitney U‐test. All the other experiments were carried out in triplicate and quantitative values were expressed as means ± SEM. Cut‐off values of miR‐135a‐3p, miR‐630, and miR‐1207 expression were taken as the medians of expression levels in sera from 98 ovarian cancer patients. The statistical significance of differences among the groups was assessed using the Mann–Whitney U‐test. Correlation was tested and presented as Pearson's correlation coefficient (r). The survival fraction was calculated by the Kaplan–Meier test with 95% confidence intervals computed by the asymmetrical method using the log–rank test. A value of P < 0.05 was considered statistically significant. Analysis was carried out using GraphPad Prism (GraphPad Software, La Jolla, CA, USA).
Results
Serum miRNA amount profiles associated with prognosis in ovarian cancer patients
To identify serum biomarkers associated with clinical prognosis of ovarian cancer, we compared changes in serum miRNA levels between group A (poor prognosis) and group B (favorable prognosis). Three miRNAs, miR‐135a‐3p, miR‐630, and miR‐1207, that showed significant differences in the miRNA expression index in serum samples between groups A and B were identified using miRNA array analysis. The fold change of miR‐135a‐3p, miR‐630, and miR‐1207 between the groups was 0.673 (P = 0.0104), 0.405 (P = 0.0097), and 1.266 (P = 0.0236), respectively. To assess clinical significance, we calculated the median value for each miRNA in 98 ovarian cancer patients and examined PFS in patients with low or high miRNA levels based on each median value. The median expression index of miR‐135a‐3p, miR‐630, and miR‐1207 was 0.406, 0.000403, and 0.211, respectively, with miR‐135a‐3p markedly enhanced compared with the other two (Fig. S1a). Patients with high miR‐135a‐3p levels showed a favorable prognosis compared with those with low levels; in contrast, the level of miR‐630 or miR‐1207 was not associated with clinical prognosis (Fig. 1). We also found that levels of miR‐135a‐3p, miR‐630, or miR‐1207 were not associated with different histological subtypes of ovarian cancer (Fig. S1b). These results suggest that miR‐135a‐3p might be considered a biomarker in ovarian cancer.
Figure 1.
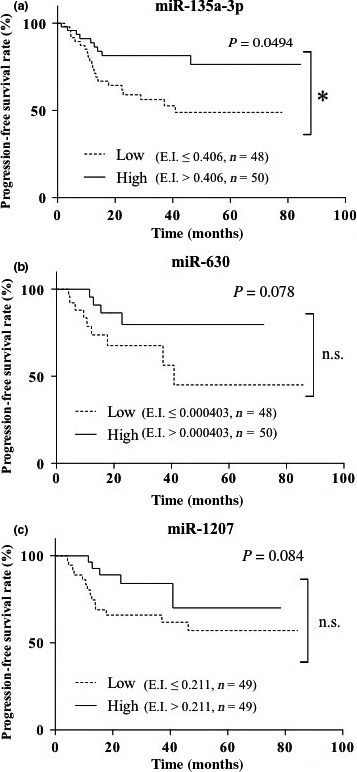
Progression‐free survival in ovarian cancer patients with high or low expression index of microRNA (miR)‐135a‐3p (a), miR‐630 (b), or miR‐1207 (c). Each solid line or dotted line indicates progression‐free survival in ovarian cancer with high or low expression index, respectively, of each miRNA. *P < 0.05. n, number of ovarian cancer patients.
Clinical significance of miR‐135‐3p in ovarian cancer patients
To provide clinical insights into the levels of miR‐135a‐3p in ovarian cancer, we examined the miR‐135a‐3p expression index in sera, peritoneal fluid, and tissue samples of patients with ovarian cancer, ovarian cysts, normal ovaries, or endometrial cancer. This serum index was significantly decreased in ovarian cancer patients compared with those with ovarian cysts or normal ovaries (Fig. 2a), while the tissue sample index was significantly reduced in ovarian cancer patients compared with those with normal ovaries (Fig. 2b). The peritoneal fluid sample expression index was significantly reduced in ovarian cancer patients compared with those with ovarian cysts (Fig. 2c). In 47 ovarian cancer patients, the serum miR‐135a‐3p expression index was significantly correlated with that in tissue samples (P < 0.05, r = 0.3671) (Fig. S2a). In 17 of the 47 ovarian cancer patients, the serum miR‐135a‐3p expression index was also significantly associated with that in peritoneal fluid samples (P < 0.05, r = 0.5703) (Fig. S2b). In 17 ovarian cancer patients, we found that the expression index values of miR‐135a‐3p were markedly higher in peritoneal fluid samples than those in serum samples, and approximately 100 times higher than those in cancer tissue samples (Fig. S3). However, in all 47 ovarian cancer patients, the serum miR‐135a‐3p expression index was not correlated with the value of CA125 in serum samples (Fig. S4). These findings suggest that miR‐135a‐p is produced in the cells and excreted into the peritoneal fluid, and may contribute to cancer progression.
Figure 2.
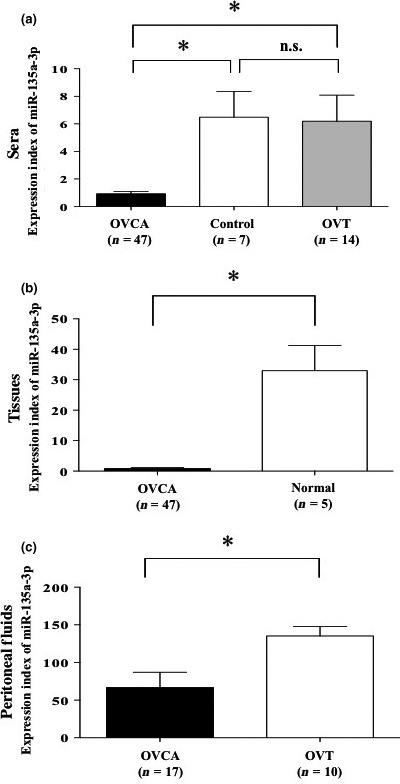
Clinical significance of microRNA (miR)‐135a‐3p in patients with ovarian cancer, other gynecologic diseases, and normal ovary. (a) Level of miR‐135a‐3p in serum samples. Closed, open, and shaded columns indicate the expression index of miR‐135a‐3p in patients with ovarian cancer (OVCA), benign ovarian tumor (OVT), or normal ovary (Control), respectively. (b) Expression of miR‐135a‐3p in tissue samples. Closed and open columns indicate the expression index of miR‐135a‐3p in patients with ovarian cancer (OVCA) or normal ovary (Normal), respectively. (c) Level of miR‐135a‐3p in peritoneal fluid samples. Closed and open columns indicate the expression index of miR‐135a‐3p in patients with ovarian cancer (OVCA) or benign ovarian tumor (OVT), respectively. All data indicate mean ± standard error. Bar indicates standard error. *P < 0.05. n, number of patients.
Contribution of miR‐135a‐p to cancer progression
To gain insights into miR‐135a‐p as a therapeutic target for ovarian cancer, we examined tumor activity including cell proliferation, chemosensitivity, and tumor formation after transfection with a miR‐135a‐3p expression vector or miR‐blank expression vector into ovarian cancer cells. Cells transfected with the miR‐135a‐3p expression vector showed marked expression of mR‐135a‐3p compared with cells transfected with miR‐blank (all P < 0.05) (Fig. 3a). Transfection of the miR‐135a‐3p expression vector into SKOV‐3, ES‐2, OVCAR‐3, or RMG‐1 cells significantly suppressed cell viability compared with miR‐blank transfection (all P < 0.05) (Fig. 3a). Transfection of the miR‐135a‐3p expression vector into SKOV‐3 or ES‐2 cells significantly reduced cell viability for each concentration of CDDP or PTX compared with transfection of miR‐blank (all P < 0.05). The IC50 value of CDDP in cells transfected with the miR‐135a‐3p expression vector showed a lower concentration than that in cells transfected with miR‐blank expression vector in both SKOV‐3 and ES‐2 cell lines: miR‐blank/SKOV‐3, 10.1 μM; miR‐135a‐3p/SKOV‐3, 6.1 μM; miR‐blank/ES‐2, 4.9 μM; and miR‐135a‐3p/ES‐2, 2.3 μM. The IC50 value of PTX in cells transfected with miR‐135a‐3p expression vector was also decreased compared with that in cells transfected with miR‐blank expression vector in both cell lines: IC50 miR‐blank/SKOV‐3, 14.6 μM; miR‐135a‐3p/SKOV‐3, 9.7 μM; miR‐blank/ES‐2, 5.4 μM; and miR‐135a‐3p/ES‐2, 3.9 μM (Fig. 3b). In SKOV‐3 cells, transfection with the miR‐135a‐3p expression vector completely blocked tumor formation compared with miR‐blank transfection (Fig. 3c). In ES‐2 cells, transfection of the miR‐135a‐3p expression vector markedly suppressed tumor growth compared with miR‐blank transfection (Fig. 3c). There were no adverse events, such as a weight loss, in either examination (Fig. S5). These findings indicate that miR‐135a‐3p attenuates malignant properties of ovarian cancer cells, including cell proliferation, drug sensitivity, and tumor formation, suggesting that miR‐135a‐3p itself may be considered a therapeutic agent in nucleic acid medicine for ovarian cancer.
Figure 3.
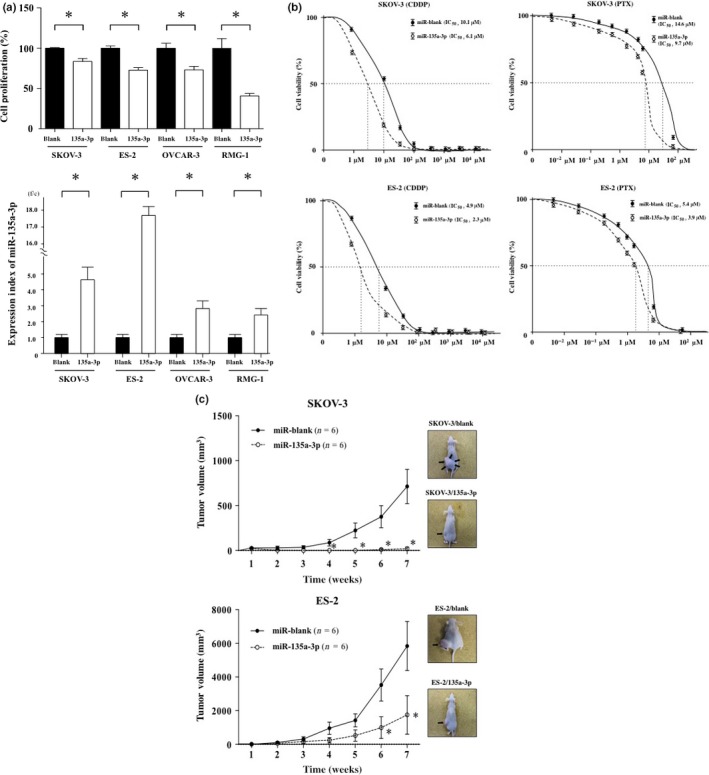
Assessment of antitumor properties induced by the introduction of miR‐135a‐3p gene into ovarian cancer cells. Alterations in cell proliferation and the expression index of microRNA (miR)‐135a‐3p induced by the transfection of miR‐135a‐3p expression vector or miR‐blank expression vector into SKOV‐3, ES‐2, OVCAR‐3, or RMG‐1 cells. (a) Closed and open columns indicate the cell proliferation of miR‐blank or miR‐135a‐3p in the upper figure, and the expression index of miR‐blank or miR‐135a‐3p of each cell line in lower figure, respectively. *P < 0.05. All data indicate mean ± standard error. (b) Closed (solid line) and open (dotted line) circles indicate the cell viability of SKOV‐3 and ES‐2 cells transfected with miR‐blank or miR‐135a‐3p expression vectors. Alteration in cell viability of SKOV‐3 or ES‐2 cells transfected with miR‐blank or miR‐135a‐3p expression vector in the presence of various concentrations of cisplatin (CDDP) or paclitaxel (PTX). In addition, the IC 50 values of CDDP and PTX were calculated. (c) Closed (solid line) and open (dotted line) circles indicate the tumor volume of SKOV‐3 and ES‐2 cells transfected with miR‐blank or miR‐135a‐3p. Each tumor was inoculated 7 weeks after the s.c. injection of the transfected cells. Arrows indicate tumor. *P < 0.05. n, number of mice. All data indicate mean ± standard error.
Identification of genes regulated by miR‐135a‐3p expression in ovarian cancer
To identify the genes and specific pathways altered by miR‐135a‐3p expression, we examined cell proliferation and gene expression changes between cells transfected with expression vectors for miR‐135a‐3p and miR‐blank and then analyzed the specific pathways that were affected. We observed a lower inhibition of cell proliferation by the transfection of the miR‐135a‐3p expression vector in SKOV‐3 and ES‐2 cells compared with OVCAR‐3 and RMG‐2 cells, even with higher miR‐135a‐3p levels in the transfected ES‐2 or SKOV‐3 cells compared with transfected RMG‐2 or OVCAR‐3 cells. To examine chemosensitivity and tumor formation mediated by miR‐135a‐3p, we thus selected SKOV‐3 and ES‐2 cells for subsequent experiments. Comparison of common gene expression changes in SKOV‐3 and ES‐2 cells after transfection with miR‐135a‐3p or miR‐blank expression vectors identified five genes that showed >3‐fold expression changes in cells transfected with miR‐135a‐3p compared with miR‐blank as candidate targeted genes (group 1) (Table 1). Additionally, genes showing >2‐fold change between cells transfected with miR‐135a‐3p and miR‐blank were also analyzed according to specific pathways. A total of 20 genes showed >2‐fold upregulation and 58 genes showed >2‐fold downregulation in cells transfected with miR‐135a‐3p compared with miR‐blank (Fig. 4a). However, analysis of specific pathways revealed that no more than three genes were involved in the same pathway.
Table 1.
Target candidate genes by microarray analysis
| Gene symbol | Gene name | Fold change |
|---|---|---|
| BIRC3 | Baculoviral IAP repeat containing 3 | −3.3256896 |
| GABRA3 | Gamma‐aminobutyric acid type A receptor alpha 3 | −3.3673131 |
| SPANXB1/2 | SPANX family member B1/2 | −5.5325503 |
| TMPRSS15 | Transmembrane protease, serine 15 | −10.862935 |
| NCAM2 | Neural cell adhension molecule 2 | −11.769233 |
Figure 4.
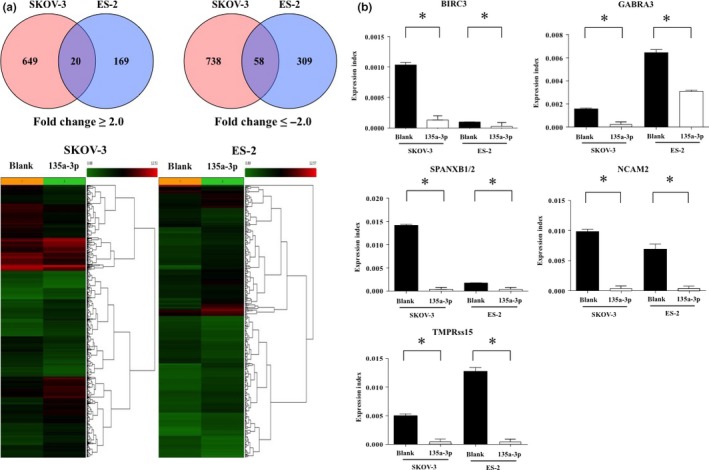
Expression changes in genes targeted by the transfection of miR‐135a‐3p gene in ovarian cancer. Gene profiling between SKOV‐3 and ES‐2 cells transfected with microRNA (miR)‐135a‐3p or miR‐blank. (a) Alterations in the expression of commonly targeted genes between SKOV‐3 and ES‐2 cells transfected by miR‐135a‐3p. Red and blue circles indicate SKOV‐3 and ES‐2 in Venn diagram, respectively. Numbers indicate the number of targeted genes. The number of intersections in the Venn diagram is the number of targeted genes of fold change ≥2.0 or fold change <2.0. In the heatmap in the lower figure, the red signal indicates high expression of targeted genes, and green signal indicates low expression of genes. (b) Closed and open columns indicate the expression of targeted genes in SKOV‐3 and ES‐2 cells transfected by miR‐blank or miR‐135a‐3p. All data indicate mean ± standard error. Bar indicates standard error. *P < 0.05.
To confirm the expression of the five genes showing >3‐fold change in expression in miR‐135a‐3p‐transfected cells, we examined the expression index of each gene in SKOV‐3 or ES‐2 cells transfected with miR‐135a‐3p or miR‐blank expression vectors using RT‐PCR. Expression of all five genes was reduced by the introduction of miR‐135a‐3p compared with miR‐blank in both ovarian cancer cell lines (all P < 0.05) (Fig. 4b). To verify the clinical importance of these genes in ovarian cancer, we examined their expression in tissue samples of 20 out of 47 patients with ovarian cancer and five patients with normal ovaries using RT‐PCR. Among the five genes in group 1, the expression index of BIRC3, GAGRA3, and SPANXB1/SPANXB2 was significantly increased in ovarian cancer patients compared with patients with normal ovaries (all P < 0.05), whereas no expression of NCAM2 or TMPRSS15 was found in ovarian cancer patients or those with normal ovaries (Fig. 5a).
Figure 5.

Association of targeted genes involved in microRNA (miR)‐135a‐3p with ovarian cancer. (a) Alterations in the expression of genes targeted by the transfection of miR‐135a‐3p gene in patients with ovarian cancer (OVCA) or normal ovary (Normal) in tissue samples. (b) Alterations in the expression of targeted genes predicted by TargetScan, miRDB, and miRanda in patients with ovarian cancer (OVCA; closed columns) or normal ovary (Normal; open columns) in tissue samples. All data indicate mean ± standard error. Bar indicates standard error. *P < 0.05. n, number of patients.
Through combined analyses using TargetScan, miRDB, and miRanda for the sequence of miR‐135a‐3p, six genes were identified as theoretical targets for miR‐135a‐3p (group 2) (Fig. S6, Table S3). To assess the clinical importance of these genes in ovarian cancer we examined their expression as described above for the six genes. PPP2R2B expression was significantly increased in ovarian cancer patients compared with patients with normal ovaries (P < 0.05) (Fig. 5b). No significant difference was observed in the expression of SYT4, IL1RAP, RERG, C9orf152, or PAIP1 between ovarian cancer patients and those with normal ovaries (Fig. 5b). According to these findings, miR‐135a‐3p appears to directly regulate PPP2R2B expression and indirectly regulate that of BIRC3, GAGRA3, or SPNAXB1/SPANXB2, resulting in the progression of ovarian cancer.
Discussion
In this study, we found that miR‐135a‐3p is abundantly present in cells and biofluids, including serum and peritoneal fluid, and that miR‐135a‐3p suppresses malignant properties in ovarian cancer. It is therefore plausible that miR‐135a‐3p, which is already recognized as a serum biomarker and a nucleic acid agent for cancer therapy, is a promising miRNA for the treatment of ovarian cancer.
The level of miR‐135a‐3p in peritoneal fluid was enriched up to 100‐fold compared with that in cells or sera of any patient. Additionally, the level was attenuated with the increase in peritoneal fluid during ovarian cancer progression. Accordingly, it is possible that miR‐135a‐3p affects the circulation of peritoneal fluid. Many studies have investigated the relationship between miR‐135a‐5p and cancer,24, 25, 26, 27, 28, 29, 30 but have not examined miR‐135a‐3p.31, 32 MicroRNA‐135a‐5p has often been reported to function as a potential tumor suppressor.24, 25, 26, 27, 28, 29, 30 In prostate cancer cells, miR‐135a‐3p was shown to mimic the enhanced apoptosis induced by death receptor 5 with respect to increased poly(ADP‐ribose) polymerase cleavage, Bax expression, and the number of TUNEL‐positive cells following cotreatment with tanshinone I and TRAIL.32 Our present study indicates that miR‐135a‐3p may also have antitumor activities in ovarian cancer. Human miR‐135a is encoded by two genes located on different chromosomes that produce identical active sequences (chromosomes 3 and 12 for miR‐135a1 and miR‐135a2, respectively). Additionally, miR‐135a‐3p and miR‐135a‐5p are produced from the same transcript. Although contradictory results have been reported concerning the effects of miR‐135a‐3p and miR‐135a‐5p on cancer progression, the genes targeted by miR‐135a may themselves be involved in cell growth.
Combined analysis of the miR‐135a‐3p sequence by TargetScan, miRDB, and miRanda predicted that miR135a‐3p might directly regulate the expression of six genes (Fig. S6). We showed that PPP2R2B expression was significantly increased in ovarian cancer compared with normal ovaries. PPP2R2B encodes a regulatory subunit of protein phosphatase 2A, which has been implicated in breast tumorigenesis and progression,33, 34, 35 and PPP2R2B was reported to contribute to human epidermal growth factor receptor‐2 signaling and enhanced expression of estrogen receptor in breast cancer.33, 34 Our screen of transfected ovarian cancer cells identified five genes as putative targets for miR‐135a‐3p. We further showed that the expressions of BIRC3, GABRA3, and SPANXB1/SPANXB2 were significantly upregulated in ovarian cancer compared with normal ovaries. Belonging to the inhibitor of apoptosis family of proteins, BIRC3 plays pivotal roles in the regulation of nuclear factor‐κB signaling and apoptosis.36 Various somatic alterations of BIRC3 have been identified in lymphoid malignancies and many epithelial tumors, including ovarian cancer.37, 38
Gamma‐aminobutyric acid type B receptor subunit 1 (GABA) is an inhibitory neurotransmitter that functions through two types of GABA receptors: ionotropic receptors including GABAA and GABAC receptors, and the metabotropic GABAB receptor.39, 40 Of the GABAA group of receptors, GABRA3 is normally expressed in adult brain, and is inversely correlated with breast cancer survival; it activates the protein kinase B pathway to promote breast cancer cell migration, invasion, and metastasis.41 Lymphatic metastasis in lung adenocarcinoma is enhanced by GABRA3 by activating the JNK/AP‐1 signaling pathway.42 The testis‐specific gene SPANXB is normally expressed only in spermatozoa. Although sporadic SPANXB expression has previously been reported in cancer cell lines and hematologic malignancies, the function of SPANXB or related genes is poorly understood.43, 44 We also showed that 58 genes were suppressed and 30 upregulated in our gene profiling, suggesting that miR‐135a‐3p can directly or indirectly regulate the expression of many oncogenic or suppressive genes in ovarian cancer progression.
DNA and RNA are simple linear polymers that have moved into the realm of stand‐alone therapeutic agents for a variety of human diseases.45 Several types of RNA molecules with complex biological functions, including mRNA, transfer RNA, miRNA, and siRNA, have been discovered,45 while major chemical modifications in siRNA and miRNA have been identified, including the ribose 2′‐OH group modification, locked and unlocked nucleic acids, and phosphorothioate modification, leading to a diverse range of therapeutic applications for human diseases.46 Based on these findings, clinical trials of siRNA‐ and miRNA‐based drugs have already been initiated.46, 47 In this study, miR‐135a‐3p can be regarded as an integrated therapeutic agent with multiple targets for ovarian cancer therapy, and is probably associated with the retention of peritoneal fluid, chemoresistance, and tumorigenesis in ovarian cancer. In ovarian cancer patients, the presence of ascites is essentially treated through management of underlying disease. Once chemoresistance has developed, however, intractable ascites can be a major problem involving several molecules.48 Accordingly, i.v. or s.c. administration of miR‐135a‐3p mimics may improve the clinical prognosis in peritonitis carcinomatosa induced by ovarian cancer through the enhancement of chemosensitivity or the reduction of ascites.
In conclusion, the miR‐135a‐3p expression index is suitable for application as a biomarker in treatment of ovarian cancer, and miR‐135a‐3p mimics are promising agents for ovarian cancer therapy.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. (A) Distribution of expression index of microRNA (miR)‐135a‐3p, miR‐630, or miR‐1207 in serum samples from patients with ovarian cancer or normal ovary. Alterations in the expression index of miR‐135a‐3p, miR‐630, and miR‐1207 in high and low level groups in 98 patients with ovarian cancer or controls. In patients with ovarian cancer, the mean ± standard error expression index of miR‐135a‐3p, miR‐630, or miR‐1207 in serum samples is shown. Closed circle indicates the expression index of patients. Long or short bar indicates the value of mean or standard error value, respectively. n, number of patients. (B) Comparison of the expression levels of miR‐135a‐3p in each histological subtype of ovarian cancer. The expression index of miR‐135a‐3p, miR‐630, and miR‐1207 in ovarian cancer patients with various histological subtypes. Closed circle indicates the expression index of patients. Long or short bar indicates the value of mean or standard error value, respectively. n, number of patients.
Fig. S2. Relationship between expression index of microRNA (miR)‐135a‐3p in patients with ovarian cancer among serum, peritoneal fluid, and tissue samples. The correlation in expression index between tissue and serum samples (A), and between peritoneal fluid and serum samples (B). Closed circle indicates the expression index of patients. Bar indicates the correlation index. R, value of correlation index. n, number of patients.
Fig. S3. Association of expression index of microRNA (miR)‐135a‐3p in tissue, peritoneal fluid, and serum samples in the same patient with ovarian cancer. Closed circle indicates the expression index of miR‐135a‐3p in 17 patients with ovarian cancer. The mean ± standard error expression index of miR‐135a‐3p in tissue, peritoneal fluid, and serum samples is shown.
Fig. S4. Relationship between the expression index of microRNA (miR)‐135a‐3p in serum samples and serum CA125 in 47 patients with ovarian cancer. R, value of correlation index. n, number of patients.
Fig. S5. Alterations in body weight of NOD/SCID mice with xenograft tumors of SKOV‐3 or ES‐2 ovarian cancer cells transfected with microRNA (miR)‐135a‐3p or miR‐blank gene. The body weight of NOD/SCID mice (n = 6/group) with SKOV‐3 or ES‐2 cells transfected with miR‐135a‐3p or miR‐blank. Closed (solid line) or open (dotted line) circle indicates the body weight of mice with xenograft tumors of SKOV‐3 and ES‐2 cells transfected with miR‐blank or miR‐135a‐3p, respectively. n, number of mice. All data indicate mean ± standard error.
Fig. S6. Gene profiling predicted by miR‐135a‐3p gene as a target using TargetScan (black circle), miRDB (red circle), and miRanda (blue circle). The number indicates the number of commonly targeted genes. The context + score by TargetScan evaluates the binding of miRNAs to the context of entire 3′‐UTR of a gene by summing over contributions made by individual sites that have perfect sequence complementarities to the miRNA seed. Target Score by miRDB is used to scan all available miRNA sequences for a given genome against 3′‐UTR sequences of that genome. mirSVR score by miRanda trained on sequence and contextual features of the predicted miRNA. Expression profiles are derived from a comprehensive sequencing project of a large set of mammalian tissues and cell lines of normal and disease origin.
Table S1. Clinical characteristics of patients with primary ovarian cancer for microarray analysis.
Table S2. Clinical characteristics of patients with ovarian cancer for RT‐PCR analysis.
Table S3. Target prediction by TargetScan, miRDB, and miRanda.
Acknowledgments
This work was supported in part by a Grant‐in‐Aid for Young Scientists (B) (no. 227790536) Challenging Exploratory Research (no. 26670731), Scientific Research (B) (no. 26293362), Scientific Research (C) (no. 23592470), and funds from the Central Research Institute of Fukuoka University (141011), in addition to The Center for Advanced Molecular Medicine, Fukuoka University from the Ministry of Education, Culture, Sports, Science and Technology (Tokyo, Japan), a Grant‐in‐Aid from the Kakihara Science and Technology Foundation (Fukuoka, Japan), and the Princess Takamatsu Cancer Research Fund (to S.M.).
Cancer Sci 108 (2017) 886–896
Funding Information
Grant‐in‐Aid for Young Scientists (B) (227790536) Challenging Exploratory Research (26670731), Scientific Research (B) (26293362), Scientific Research (C) (23592470); Central Research Institute of Fukuoka University (141011); Ministry of Education, Culture, Sports, Science and Technology of Japan; Kakihara Science and Technology Foundation; Princess Takamatsu Cancer Research Fund.
References
- 1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 2. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120(1): 15–20. [DOI] [PubMed] [Google Scholar]
- 3. Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012; 148: 1172–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014; 15: 509–24. [DOI] [PubMed] [Google Scholar]
- 5. Lindow M, Kauppinen S. Discovering the first microRNA‐targeted drug. J Cell Biol 2012; 199: 407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014; 20: 460–9. [DOI] [PubMed] [Google Scholar]
- 7. Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer 2015; 113: 569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan DS, Agarwal R, Kaye SB. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol 2006; 7: 925–34. [DOI] [PubMed] [Google Scholar]
- 9. Crawford SC, Vasey PA, Paul J, Hay A, Davis JA, Kaye SB. Does aggressive surgery only benefit patients with less advanced ovarian cancer? Results from an international comparison within the SCOTROC‐1 Trial. J Clin Oncol 2005; 23: 8802–11. [DOI] [PubMed] [Google Scholar]
- 10. Trimble EL, Wright J, Christian MC. Treatment of platinum‐resistant ovarian cancer. Expert Opin Pharmacother 2001; 2: 1299–306. [DOI] [PubMed] [Google Scholar]
- 11. Oza AM, Cook AD, Pfisterer J et al Standard chemotherapy with or without bevacizumab for women with newly diagnosed ovarian cancer (ICON7): overall survival results of a phase 3 randomised trial. Lancet Oncol 2015; 16: 928–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mirza MR, Monk BJ, Herrstedt J et al Niraparib maintenance therapy in platinum‐sensitive, recurrent ovarian cancer. N Engl J Med 2016; 375: 2154–64. [DOI] [PubMed] [Google Scholar]
- 13. Chan JK, Brady MF, Penson RT et al Weekly vs. Evere‐3‐week paclitaxel and carboplatin for ovarian cancer. N Engl J Med 2016; 374: 738–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ledemann J, Harter P, Gourley C et al Olaparib maintenance therapy in platinum‐sensitive relapsed ovarian cancer. N Engl J Med 2012; 366: 1382–92. [DOI] [PubMed] [Google Scholar]
- 15. Tang PA, Bentzen SM, Chen EX, Siu LL. Surrogate end points for median overall survival in metastatic colorectal cancer: literature‐based analysis from 39 randomized controlled trials of first‐line chemotherapy. J Clin Oncol 2007; 25: 4562–8. [DOI] [PubMed] [Google Scholar]
- 16. Buyse M, Burzykowski T, Carroll K et al Progression‐free survival is a surrogate for survival in advanced colorectal cancer. J Clin Oncol 2007; 25: 5218–24. [DOI] [PubMed] [Google Scholar]
- 17. Parmar MK, Ledermann JA, Colombo N et al Paclitaxel plus platinum‐based chemotherapy versus conventional platinum‐based chemotherapy in women with relapsed ovaryan cancer: the ICON4/AGO‐OVAR‐2.2 trial. Lancet 2003; 361(9375): 2099–106. [DOI] [PubMed] [Google Scholar]
- 18. Kaye SB. Chemotherapy for recurrent ovarian cancer. Lancet 2003; 361(9375): 2099–106. [DOI] [PubMed] [Google Scholar]
- 19. Wiggans AJ, Cass GK, Bryant A, Lawrie TA, Morrison J. Poly(ADP‐ribose) polymerase (PARP) inhibitors for the treatment of ovarian cancer. Cochrane Database Syst Rev 2015; (5): Cd007929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katz B, Trope CG, Reich R, Davidson B. MicroRNAs in ovarian cancer. Hum Pathol 2015; 46: 1245–56. [DOI] [PubMed] [Google Scholar]
- 21. Pal MK, Jaiswar SP, Dwivedi VN, Tripathi AK, Dwivedi A, Sankhwar P. MicroRNA: a new and promising potential biomarker for diagnosis and prognosis of ovarian cancer. Cancer Biol Med 2015; 12: 328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y, Fang Y, Liu Y, Yang X. MicroRNAs in ovarian function and disorders. J Ovarian Res 2015; 8: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, Kimura T. Clinical relevance of circulating cell‐free microRNAs in ovarian cancer. Mol Cancer 2016; 15(1): 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamada Y, Hidaka H, Seki N et al Tumor‐suppressive microRNA‐135a inhibits cancer cell proliferation by targeting the c‐MYC oncogene in renal cell carcinoma. Cancer Sci 2013; 104: 304–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi H, Ji Y, Zhang D, Liu Y, Fang P. MiR‐135a inhibits migration and invasion and regulates EMT‐related marker genes by targeting KLF8 in lung cancer cells. Biochem Biophys Res Commun 2015; 465(1): 125–30. [DOI] [PubMed] [Google Scholar]
- 26. Kroiss A, Vincent S, Decaussin‐Petrucci M et al Androgen‐regulated microRNA‐135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene 2015; 34: 2846–55. [DOI] [PubMed] [Google Scholar]
- 27. Deng YQ, Yang YQ, Wang SB et al Intranasal administration of lentiviral miR‐135a regulates mast cell and allergen‐induced inflammation by targeting GATA‐3. PLoS ONE 2015; 10(9): e0139322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tribollet V, Barenton B, Kroiss A et al miR‐135a inhibits the invasion of cancer cells via suppression of ERRalpha. PLoS ONE 2016; 11(5): e0156445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yao S, Tian C, Ding Y et al Down‐reguation of Kruppel‐like factor‐4 by microRNA‐135a‐5p promotes proliferation and metastasis in hepatocellular carcinoma by transforming growth factor‐beta1. Oncotarget 2016; 7(27): 42566–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coarfa C, Fiskus W, Eedunuri VK et al Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene 2016; 35: 2345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang B, Jing C, Wang J et al Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. Clin Transl Oncol 2014; 16: 374–9. [DOI] [PubMed] [Google Scholar]
- 32. Shin EA, Sohn EJ, Won G et al Upregulation of microRNA135a‐3p and death receptor 5 plays a critical role in Tanshinone I sensitized prostate cancer cells to TRAIL induced apoptosis. Oncotarget 2014; 5: 5624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mumby M. PP2A: unveiling a reluctant tumor suppressor. Cell 2007; 130(1): 21–4. [DOI] [PubMed] [Google Scholar]
- 34. Wong LL, Chang CF, Koay ES, Zhang D. Tyrosine phosphorylation of PP2A is regulated by HER‐2 signalling and correlates with breast cancer progression. Int J Oncol 2009; 34: 1291–301. [PubMed] [Google Scholar]
- 35. Keen JC, Zhou Q, Park BH et al Protein phosphatase 2A regulates estrogen receptor alpha (ER) expression through modulation of ER mRNA stability. J Biol Chem 2005; 280: 29519–24. [DOI] [PubMed] [Google Scholar]
- 36. Gyrd‐Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF‐kappaB, inflammation and cancer. Nat Rev Cancer 2010; 10: 561–74. [DOI] [PubMed] [Google Scholar]
- 37. Yamato A, Soda M, Ueno T et al Oncogenic activity of BIRC2 and BIRC3 mutants independent of nuclear factor‐kappaB‐activating potential. Cancer Sci 2015; 106: 1137–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jonsson JM, Bartuma K, Dominguez‐Valentin M et al Distinct gene expression profiles in ovarian cancer linked to Lynch syndrome. Fam Cancer 2014; 13: 537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Watanabe M, Maemura K, Oki K, Shiraishi N, Shibayama Y, Katsu K. Gamma‐aminobutyric acid (GABA) and cell proliferation: focus on cancer cells. Histol Histopathol 2006; 21: 1135–41. [DOI] [PubMed] [Google Scholar]
- 40. Vithlani M, Terunuma M, Moss SJ. The dynamic modulation of GABA(A) receptor trafficking and its role in regulating the plasticity of inhibitory synapses. Physiol Rev 2011; 91: 1009–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gumireddy K, Li A, Kossenkov AV et al The mRNA‐edited form of GABRA3 suppresses GABRA3‐mediated Akt activation and breast cancer metastasis. Nat Commun 2016; 7: 10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu L, Yang C, Shen J et al GABRA3 promotes lymphatic metastasis in lung adenocarcinoma by mediating upregulation of matrix metalloproteinases. Oncotarget 2016; 7(22): 32341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Z, Zhang J, Zhang Y, Lim SH. SPAN‐Xb expression in myeloma cells is dependent on promoter hypomethylation and can be upregulated pharmacologically. Int J Cancer 2006; 118: 1436–44. [DOI] [PubMed] [Google Scholar]
- 44. Wang Z, Zhang Y, Liu H, Salati E, Chiriva‐Internati M, Lim SH. Gene expression and immunologic consequence of SPAN‐Xb in myeloma and other hematologic malignancies. Blood 2003; 101: 955–60. [DOI] [PubMed] [Google Scholar]
- 45. Breaker RR, Joyce GF. The expanding view of RNA and DNA function. Chem Biol 2014; 21: 1059–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids 2015; 4: e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sridharan K, Gogtay NJ. Therapeutic nucleic acids: current clinical status. Br J Clin Pharmacol 2016; 82: 659–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kipps E, Tan DS, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat Rev Cancer 2013; 13: 273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (A) Distribution of expression index of microRNA (miR)‐135a‐3p, miR‐630, or miR‐1207 in serum samples from patients with ovarian cancer or normal ovary. Alterations in the expression index of miR‐135a‐3p, miR‐630, and miR‐1207 in high and low level groups in 98 patients with ovarian cancer or controls. In patients with ovarian cancer, the mean ± standard error expression index of miR‐135a‐3p, miR‐630, or miR‐1207 in serum samples is shown. Closed circle indicates the expression index of patients. Long or short bar indicates the value of mean or standard error value, respectively. n, number of patients. (B) Comparison of the expression levels of miR‐135a‐3p in each histological subtype of ovarian cancer. The expression index of miR‐135a‐3p, miR‐630, and miR‐1207 in ovarian cancer patients with various histological subtypes. Closed circle indicates the expression index of patients. Long or short bar indicates the value of mean or standard error value, respectively. n, number of patients.
Fig. S2. Relationship between expression index of microRNA (miR)‐135a‐3p in patients with ovarian cancer among serum, peritoneal fluid, and tissue samples. The correlation in expression index between tissue and serum samples (A), and between peritoneal fluid and serum samples (B). Closed circle indicates the expression index of patients. Bar indicates the correlation index. R, value of correlation index. n, number of patients.
Fig. S3. Association of expression index of microRNA (miR)‐135a‐3p in tissue, peritoneal fluid, and serum samples in the same patient with ovarian cancer. Closed circle indicates the expression index of miR‐135a‐3p in 17 patients with ovarian cancer. The mean ± standard error expression index of miR‐135a‐3p in tissue, peritoneal fluid, and serum samples is shown.
Fig. S4. Relationship between the expression index of microRNA (miR)‐135a‐3p in serum samples and serum CA125 in 47 patients with ovarian cancer. R, value of correlation index. n, number of patients.
Fig. S5. Alterations in body weight of NOD/SCID mice with xenograft tumors of SKOV‐3 or ES‐2 ovarian cancer cells transfected with microRNA (miR)‐135a‐3p or miR‐blank gene. The body weight of NOD/SCID mice (n = 6/group) with SKOV‐3 or ES‐2 cells transfected with miR‐135a‐3p or miR‐blank. Closed (solid line) or open (dotted line) circle indicates the body weight of mice with xenograft tumors of SKOV‐3 and ES‐2 cells transfected with miR‐blank or miR‐135a‐3p, respectively. n, number of mice. All data indicate mean ± standard error.
Fig. S6. Gene profiling predicted by miR‐135a‐3p gene as a target using TargetScan (black circle), miRDB (red circle), and miRanda (blue circle). The number indicates the number of commonly targeted genes. The context + score by TargetScan evaluates the binding of miRNAs to the context of entire 3′‐UTR of a gene by summing over contributions made by individual sites that have perfect sequence complementarities to the miRNA seed. Target Score by miRDB is used to scan all available miRNA sequences for a given genome against 3′‐UTR sequences of that genome. mirSVR score by miRanda trained on sequence and contextual features of the predicted miRNA. Expression profiles are derived from a comprehensive sequencing project of a large set of mammalian tissues and cell lines of normal and disease origin.
Table S1. Clinical characteristics of patients with primary ovarian cancer for microarray analysis.
Table S2. Clinical characteristics of patients with ovarian cancer for RT‐PCR analysis.
Table S3. Target prediction by TargetScan, miRDB, and miRanda.


