Abstract
CUB domain‐containing protein‐1 (CDCP1) is a trans‐membrane protein predominantly expressed in various cancer cells and involved in tumor progression. CDCP1 is phosphorylated at tyrosine residues in the intracellular domain by Src family kinases and recruits PKCδ to the plasma membrane through tyrosine phosphorylation‐dependent association with the C2 domain of PKCδ, which in turn induces a survival signal in an anchorage‐independent condition. In this study, we used our cell‐free screening system to identify a small compound, glycoconjugated palladium complex (Pd‐Oqn), which significantly inhibited the interaction between the C2 domain of PKCδ and phosphorylated CDCP1. Immunoprecipitation assays demonstrated that Pd‐Oqn hindered the intercellular interaction of phosphorylated CDCP1 with PKCδ and also suppressed the phosphorylation of PKCδ but not that of ERK or AKT. In addition, Pd‐Oqn inhibited the colony formation of gastric adenocarcinoma 44As3 cells in soft agar as well as their invasion. In mouse models, Pd‐Oqn markedly reduced the peritoneal dissemination of gastric adenocarcinoma cells and the tumor growth of pancreatic cancer orthotopic xenografts. These results suggest that the novel compound Pd‐Oqn reduces tumor metastasis and growth by inhibiting the association between CDCP1 and PKCδ, thus potentially representing a promising candidate among therapeutic reagents targeting protein–protein interaction.
Keywords: CDCP1, chemical screening, metastasis, PKCδ, Src
CUB‐domain containing protein 1 (CDCP1), a type I transmembrane glycoprotein with three CUB domains in the extracellular region, was initially reported as being predominantly expressed in colorectal cancers.1 During the investigation of phosphoproteins associated with anchorage‐independent growth of lung adenocarcinoma cells, we previously discovered that CDCP1 protein plays a major role in the induction of anoikis resistance in metastatic tumors as a substrate of Src family kinases (SFKs).2 Specifically, CDCP1 phosphorylation at tyrosines in the intercellular domain upon SFK activation mediates PKCδ recruitment to the plasma membrane and its subsequent activation through specific interaction of the PKCδ C2 domain with tyrosine‐phosphorylated CDCP1.2, 3, 4 Thus, CDCP1 phosphorylation promotes anchorage‐independent cell survival and degradation of the extracellular matrix through the interaction with PKCδ.2, 5, 6 In addition, it has been revealed that CDCP1 suppresses anchorage‐independent cell death by inhibiting autophagy,7 inhibits the epithelial phenotype in pancreatic cancer cells, and is involved in the trastuzumab resistance of HER2‐positive breast cancer.8, 9, 10 Furthermore, the expression of CDCP1 is induced by activated Ras and correlates with poor prognosis in lung, pancreatic, renal, and ovarian cancer.6, 11, 12, 13 Together, such studies have indicated that the CDCP1‐PKCδ signaling pathway is involved in various features of tumor progression such as invasion and metastasis in multiple types of cancers.
Numerous experiments using mouse models have demonstrated that CDCP1 represents a potent therapeutic target for various cancers. For example, the suppression of CDCP1 reduced invasion and the peritoneal dissemination of gastric scirrhous carcinoma,14 whereas blockage of the CDCP1 pathway using specific antibodies suppressed the growth of lung and breast cancer cells15 or of ovarian cancer tissues transplanted in mice.16 Furthermore, a cytotoxin saporin‐conjugated anti‐CDCP1 antibody reduced the growth and dissemination of subcutaneous tumors in a mouse xenograft model of prostate cancer cells.17
Therefore, based on the previous work by ourselves and others, we considered that inhibiting the specific interaction between CDCP1 and PKCδ might represent an alternative therapeutic approach for targeting the CDCP1 pathway in addition to the antibody‐mediated models that directly target CDCP1 listed above. Accordingly, in the current study we performed in vitro screening of compounds that inhibited the association of phosphorylated CDCP1 with the C2 domain of PKCδ to identify a potent small molecule inhibitor. The ability of the candidate molecule to block the CDCP1‐PKCδ signaling pathway and inhibit cancer cell proliferation and invasion was assessed in vitro, as was its ability to reduce tumor growth and peritoneal dissemination in a mouse model.
Materials and Methods
Cell culture
The human gastric cancer 44As3 and 58As9 cells have been previously described.18 BxPC3, MiaPaCa‐2 (pancreatic cancer), and A549 (lung cancer) cell lines were obtained from the American Type Culture Collection. The cells were cultured in RPMI1640 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific) at 37°C in a humidified atmosphere containing 5% CO2. Human normal gastric fibroblast (NF‐37) cells were kindly gifted from Dr. Yashiro.19 NF‐37 cells were cultured in DMEM‐high glucose (Thermo Fisher Scientific) supplemented with 10% FBS. HEK293T cells were cultured in DMEM supplemented with 10% FBS.
Antibodies
An anti‐GFP antibody (#598) was purchased from MBL, Nagoya, Japan. An anti‐FLAG M2 antibody conjugated with peroxidase was purchased from Sigma (St. Louis, MO, USA). The anti‐β‐actin antibody (N21) and horse radish peroxidase (HRP)‐conjugated anti‐goat IgG antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti‐Akt (#9272), anti‐phospho‐Akt (Ser473) (#9271), anti‐CDCP1 (#4115), anti‐phospho‐CDCP1 (Tyr734) (#9050), anti‐ERK1/2 (#9102), anti‐phospho‐ERK1/2 (Thr202/Tyr204) (#9101), anti‐PKCδ (#2058), and anti‐phospho‐PKCδ (Tyr311) (#2055) antibodies were purchased from Cell Signaling Technology (Denvers, MA, USA). Anti‐CDCP1 antibody for immunoprecipitation was originally developed as described previously.2
Biding assay for CDCP1‐PKCδ interaction and chemical screening
Biotinylated peptide diluted with phosphate buffered saline calcium‐ and magnesium‐free (PBS(−)) at 1 μg/mL was incubated in streptavidin‐coated 96‐well plates (Thermo Fisher Scientific) at 37°C for 1 h. After washing twice with PBS(−) containing 0.05% Tween‐20 (PBS‐T), 50 μL C2‐FLAG diluted with blocking buffer (3% bovine serum albumin Fraction V [BSA; Roche, Mannheim, Germany] in PBS‐T) was added. For chemical screening, a 2 nM concentration of C2‐FLAG was used and each compound was added at 20 μM, then the plates were incubated at 37°C for 2 h. After the wells were washed with PBS‐T four times, anti‐FLAG M2 conjugated with peroxidase (Sigma) diluted at 1:10 000 by blocking buffer was added and incubated at room temperature (RT) for 30 min. After washing with PBS‐T four times, the bound antibody was detected by colorimetric reaction using 50 μL 3,3′,5,5′‐tetramethylbenzidine (TMB) (Wako, Osaka, Japan), then the reaction was stopped by adding the same volume of 1 M HCl. The absorbance of the wells at 450 nm was measured using an iMark microplate reader (BioRad, Hercules, CA, USA). The deposited chemical library was obtained from the Screening Committee of Anticancer Drugs, Japan. The peptide sequence used in the present study is shown in Table S1.
HTRF assay for CDCP1‐PKCδ interaction
The biotinylated peptide with phospho‐tyrosine (bio‐762pY) and a C2‐FLAG recombinant protein were used for HTRF assay. Binding was detected using an anti‐FLAG antibody conjugated with Eu (anti‐FLAG‐Eu) (Cisbio, Codolet, France) and streptavidin‐d2 (Cisbio). All of the components were diluted with HTRF assay buffer (20 mM sodium‐potassium phosphate (pH7.5), 0.5% BSA, 0.25 M potassium fluoride, 0.05% Tween‐20). The components of C2‐FLAG, bio‐762pY, anti‐FLAG‐Eu, and streptavidin‐d2 were mixed in 20 μL total volume in a 384‐well plate (784075; Greiner, Kremsmünster, Austria) and incubated at RT for 1 h. The signal was measured using SynergyH1 with the HTRF filter set (Bio Tek Instruments, Inc., Winooski, VT, USA).
Immunoprecipitation
HEK293T cells were transfected with p3xFLAG‐CMV‐14‐CDCP1, pEGFP‐N1‐PKCδ, and/or pLNCX‐vSrc using Lipofectamine2000 according to the manufacture's manual (Thermo Fisher Scientific). After a day, the cells were harvested in PLC buffer (50 mM HEPES [pH 7.0], 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 1% Triton X‐100, and 5 mM ethylene glycol‐bis(β‐aminoethyl ether)‐N,N,N′,N′‐tetraacetic acid) containing protease (Wako) and phosphatase inhibitor cocktails (Nakalai, Kyoto, Japan) and incubated on ice for 15 min. After centrifugation at 15 000 g, the supernatant was subjected to immunoprecipitation as the input cell lysate. The protein concentration of the lysate was determined using the BCA protein assay (Thermo Fisher Scientific) with BSA as a standard. The cell lysate (2 mg) was mixed and incubated with anti‐FLAG agarose (SIGMA) (15 μL bed volume) at 4°C with rotation for 2 h. The agarose was then washed five times with PLC buffer and boiled in sodium dodecyl sulfate (SDS) sample buffer. Immunoprecipitation of endogenous proteins were described previously.2 Briefly, 44As3 cells treated with DMSO or 10 μM Pd‐Oqn were lysed in PLC buffer. The cell lysate was incubated with Protein G Sepharose (GE Healthcare, Chicago, IL, USA) that was bound with anti‐CDCP1 antibody or normal rabbit IgG. The beads were washed with PLC buffer extensively and the bound proteins were finally eluted with SDS‐sample buffer.
Western blotting
The cell lysate (20 μg) or immunoprecipitated samples were subjected to SDS‐10% polyacrylamide gel electrophoresis (PAGE) and then transferred to a polyvinylidene fluoride membrane (Millipore, Darmstadt, Germany). The membrane was incubated with the Blocking One (Nakalai) and then incubated with primary antibody: anti‐GFP (1/5000), anti‐FLAG (1/5000), anti‐CDCP1 (1/1000), anti‐β‐actin (1/1000), anti‐PKCδ (1/1000), anti‐p‐PKCδ (1/1000), anti‐p‐ERK1/2 (1/1000), anti‐ERK1/2 (1/1000), anti‐p‐AKT (1/1000), or anti‐AKT (1/1000) antibody at 4°C for 16 h. The bound antibody was detected using Pierce ECL Plus Western Blotting Substrate (Thermo Fisher Scientific) after binding of anti‐rabbit or goat IgG antibody conjugated with HRP (GE Healthcare). For immunoprecipitated proteins, TrueBlot anti‐rabbit IgG (Rockland Immunochemicals, Limerick, PA, USA) was used as a secondary antibody. The chemiluminescent signals were visualized and analyzed using an Amersham Imager 600 (GE Healthcare).
Cell proliferation assay
Cells were plated onto 96‐well plates at 3 × 103 cells per well and cultured for 2 days in the presence or absence of chemical compounds. Viable cells were determined using the Cell Counting Kit (Dojindo, Kumamoto, Japan).
Soft‐agar colony formation assay
44As3 cells (1 × 104 cells) suspended with the complete medium containing 0.35% agarose were plated on the bottom layer of the complete medium with 0.5% agarose. RPMI1640‐10%FBS containing Pd‐Oqn or dimethyl sulfoxide (DMSO) was added on top of the soft‐agar and then placed in a 37°C incubator. The medium was changed to fresh medium twice a week. After 21 days, the colonies were stained with crystal violet and the numbers of visible colonies per well were counted.
Migration and invasion assay
For the invasion assay, 100 μL of 0.1 mg/mL ice‐cold Matrigel (Corning, Corning, NY, USA) was added onto a transwell chamber (8.0 μm pore size) (Corning) and dried. 44As3 (1 × 105 cells) suspended in RPMI1640 without serum was added to the top of the chamber. RPMI1640‐10% FBS was added in the bottom chamber. After 48 h, the cells on the chamber top were removed using a cotton swab and the invaded or migrated cells were fixed by 4% paraformaldehyde. The cells were stained with DAPI and visualized using a fluorescent microscope (BZ‐9000, KEYENCE) with a 20× objective lens. The stained nuclei were counted using ImageJ free software.
Preparation of Pd‐Oqn and Pt‐Oqn
Pd‐Oqn {= Chloror{N‐(hydroxo‐quinoline‐2‐ylmethylidene)‐β‐D‐glucosamine} palladium(II)} and Pt‐Oqn {= Chloror{N‐(hydroxo‐quinoline‐2‐ylmethylidene)‐b‐D‐glucosamine} platinum(II)} were synthesized as described in Yano et al.20 Briefly, 8‐hydroxy‐2‐quinolinecarbaldehyde was added to a methanol solution of D(+)‐glucosamine hydrochloride with tert‐BuOK at RT to give Schiff base and a stoichiometric amount of Na2PdCl4 or Pt(DMSO)2Cl2 was added to give Pd‐Oqn or Pt‐Oqn, individually. Each compound was purified by recrystallization in MeOH/dimethyl formamide. The structures of the final products were confirmed by nuclear magnetic resonance analysis. The compounds were dissolved in DMSO at 100 mM and stored at −20°C as small aliquots until use.
Mouse models
Pd‐Oqn was dissolved in DMSO at 100 mg/mL and then diluted in 0.5% methylcellulose containing 0.1% tween‐80. 44As3 cells (5 × 105) were inoculated intraperitoneally into 6‐week‐old BALB/c‐nu/nu male mice purchased from Charles River Laboratories Japan, Inc. Pd‐Oqn (15 mg/kg) or the same volume of DMSO was administered by intraperitoneal injection starting at 1 day after the inoculation as described in. At 14 days after inoculation, the mice were sacrificed by cervical dislocation under isoflurane anesthesia and dissected. The number of mesentery nodules visible by eye was determined. For the pancreatic cancer model, MiaPaCa‐2 cells were inoculated in the pancreas of 8‐week‐old C.B‐17/lcr‐scid/scid Jcl (SCID) female mice purchased from CLEA Japan, Inc. Pd‐Oqn (15 mg/kg) or DMSO was administrated by intraperitoneal injection starting at 1 day after the inoculation as described in. After 23 days, the mice were sacrificed and the pancreatic tumor was removed and weighed. These experiments were approved by the Committee for Ethics of Animal Experimentation and conducted in accordance with the guidelines for Animal Experiments in the National Cancer Center.
Statistical analysis
Statistical analysis was performed using the Student's t‐test with Excel software. Data are expressed as the means ± standard deviation (SD).
Results
Establishment of a screening system for CDCP1 pathway inhibitor identification
To identify small molecules that inhibit the CDCP1 signaling pathway, we established an assay system to quantitatively detect the interaction of phosphorylated CDCP1 with PKCδ. It has been reported that the C2 domain of PKCδ binds to the sequence motif containing phosphorylated tyrosine 762 of CDCP1 whereas the SH2 domain of Src binds to CDCP1 phosphorylated tyrosine at 734.4 Therefore, biotinylated peptides (23 a.a.) derived from the C2‐binding motif containing the phosphorylated tyrosine (Y762) of CDCP1 along with several controls such as non‐phosphorylated peptides were synthesized and immobilized on streptavidin‐coated plates (Table S1). The C2 domain of PKCδ (a.a. 1–160) tagged with FLAG (C2‐FLAG) was purified as described in Supplementary Doc. S1 and protein purity was confirmed by SDS‐PAGE (Fig. 1a,b).
Figure 1.
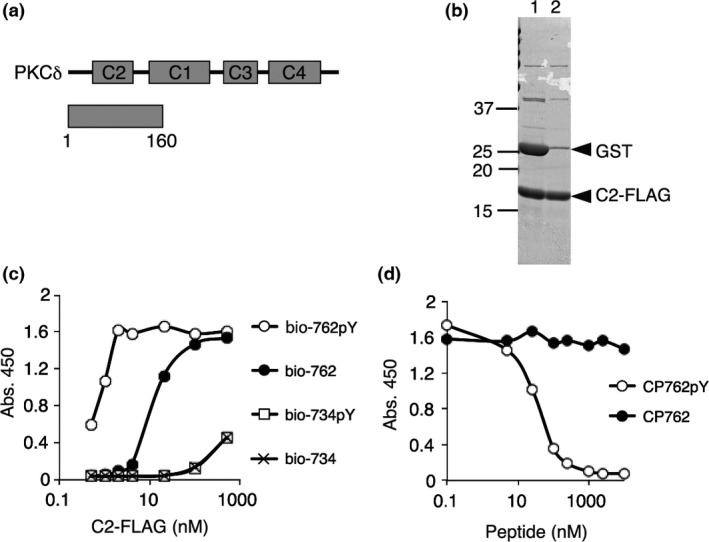
Establishment of an assay system for the interaction of CDCP1 with PKCδ. (a) Domain structure of PKCδ and the region of the C2 recombinant protein (1–160) that was used for the C2‐CDCP1 binding assay. (b) Purification of C2‐FLAG. GST‐C2‐FLAG protein purified from bacterial lysates was proteolyticaly digested (lane 1) and the GST was removed (lane 2). The proteins were subjected to SDS‐12.5% PAGE and Coomassie brilliant blue (CBB) staining. (c) Selective binding of the C2 domain to the phosphorylated site at Y762 of CDCP1. Biotinylated peptide, bio‐762pY, bio‐762, bio‐734pY, or bio‐734, immobilized on a 96‐well plate coated with streptavidin and C2‐FLAG (0.5–500 nM) was used for the binding assay. (d) Competitive inhibition in the C2‐CDCP1 binding assay by free CP762pY peptide. C2‐FLAG (2 nM) and bio‐762pY were used for the assay and the binding reaction was performed in the presence of 0.1–10 000 nM CP762pY or CP762 peptide that was not biotinylated.
The association of CDCP1 peptides with the C2‐FLAG protein was detected by colorimetric reaction with TMB following binding of an anti‐FLAG antibody conjugated with peroxidase (Fig. S1A). To confirm the specificity of the assay system, we first checked whether the C2 domain preferentially bound to a phosphotyrosine‐containing CDCP1 peptide corresponding to the PKCδ binding site. The results showed that C2‐FLAG bound to the CDCP1 peptide phosphorylated at Y762 (bio‐762pY) but not obviously to the peptide phosphorylated at Y734 (bio‐734pY). The interaction with the non‐phosphorylated peptide (bio‐762Y) was substantially weaker than bio‐762pY (Fig. 1c). In addition, a competitive peptide (11 a.a.) of 762pY (CP762pY) but not 762 (CP762) inhibited the interaction of C2 with the bio‐762pY peptide (Fig. 1d). These findings demonstrated that this assay system was capable of detecting specific association of the phosphorylated Y762 of CDCP1 with the C2 domain of PKCδ and exhibited sufficient sensitivity for use in screening molecules that inhibited the interaction of PKCδ with phosphorylated CDCP1.
Identification of an inhibitor of the CDCP1‐PKCδ interaction
The first screening library used to identify inhibitory molecules toward the interaction of CDCP1 with PKCδ consisted of 495 deposited chemical compounds. Two compounds (5‐3C and 5‐3D) were identified based on their outstanding inhibitory effect on the interaction (Fig. S1B). We then separately confirmed that these two compounds significantly inhibited the interaction in a dose‐dependent manner (Fig. 2a), compared with the effect of an adjacent unrelated compound (5‐3B) as a control. The small compound corresponding to 5‐3C was a palladium complex (Pd‐Oqn) with glucose and quinolinol and 5‐3D was a platinum complex (Pt‐Oqn) with the same structure (Figs 2b and S2). In these molecules, each metal atom is surrounded by four donor atoms, two nitrogen atoms, a phenolic oxygen atom, and a chloride ion to form an approximately square planar structure. Next, we proceeded to further screen another chemical library consisting of 10 000 compounds using the same assay system for the purpose of identifying several additional inhibitory compounds that are not as potent as the Pd‐Oqn and Pt‐Oqn compounds.
Figure 2.
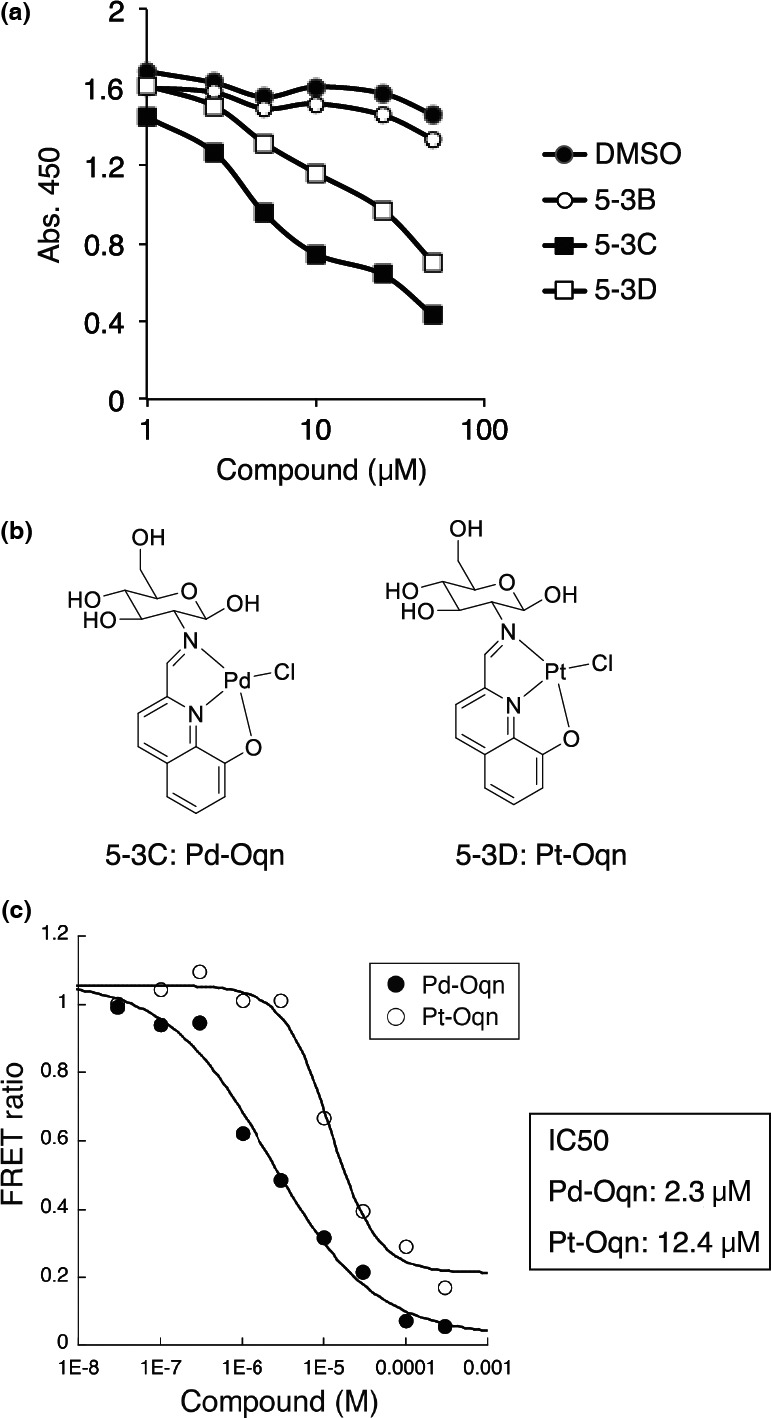
Identification of small molecules that inhibit C2‐CDCP1 binding. (a) Inhibitory effect of small compounds on the C2‐CDCP1 binding assay. The small compounds 5‐3B, 5‐3C, and 5‐3D (1.0–50 μM) or DMSO were used in the C2‐CDCP1 binding assay as inhibitory compounds. (b) Structures of 5‐3C and 5‐3D, identified as inhibitory compounds on C2‐CDCP1 binding. 5‐3C; Pd‐Oqn. 5‐3D; Pt‐Oqn. (c) Inhibition kinetics of Pd‐Oqn and Pt‐Oqn on C2‐CDCP1 binding. HTRF assays with Pd‐Oqn (Pd) or Pt‐Oqn (Pt) were performed and IC50 values were calculated.
We also developed another binding assay based on homogeneous time‐resolved fluorescence resonance energy transfer (HTRF) technology to confirm the inhibitory effect of these compounds on the CDCP1 with PKCδ interaction. For this assay, we used C2‐FLAG and bio‐762pY and binding was detected using anti‐FLAG‐Eu and Streptavidin‐d2 (Fig. S3A). The interaction of C2‐FLAG with bio‐762pY was detected in a dose‐dependent manner using the HTRF assay (Fig. S3B). In addition, the interaction was inhibited by the non‐biotinylated CP762pY peptide with an IC50 value calculated as 91 nM (Fig. S3C). The inhibition of the interaction by Pd‐Oqn and Pt‐Oqn was also detected using this assay with IC50 values calculated as 2.3 and 12.4 μM, respectively (Fig. 2c). These results suggested that Pd‐Oqn was more effective at inhibiting the interaction of phosphorylated CDCP1 with PKCδ.
Inhibitory effect of Pd‐Oqn on the CDCP1‐PKCδ pathway
We next tested whether the PKCδ‐phosphorylated CDCP1 interaction inhibition effected by Pd‐Oqn was also observed in cells. Accordingly, HEK293T cells were transfected with expression plasmids of CDCP1‐FLAG and PKCδ‐GFP with or without v‐Src. Immunoprecipitation of HEK293T cells with an anti‐FLAG antibody revealed that PKCδ co‐precipitated with CDCP1 in cells expressing both proteins but not in control cells that expressed PKCδ without CDCP1. Furthermore, the amount of PKCδ bound with CDCP1 was increased by v‐Src expression owing to phosphorylation of CDCP1 (Fig. 3a, lanes 4 and 6). Co‐precipitated PKCδ with CDCP1 was decreased by Pd‐Oqn treatment in the presence of v‐Src (lanes 6 and 7) but was not significantly affected in the absence of v‐Src (lanes 4 and 5).
Figure 3.
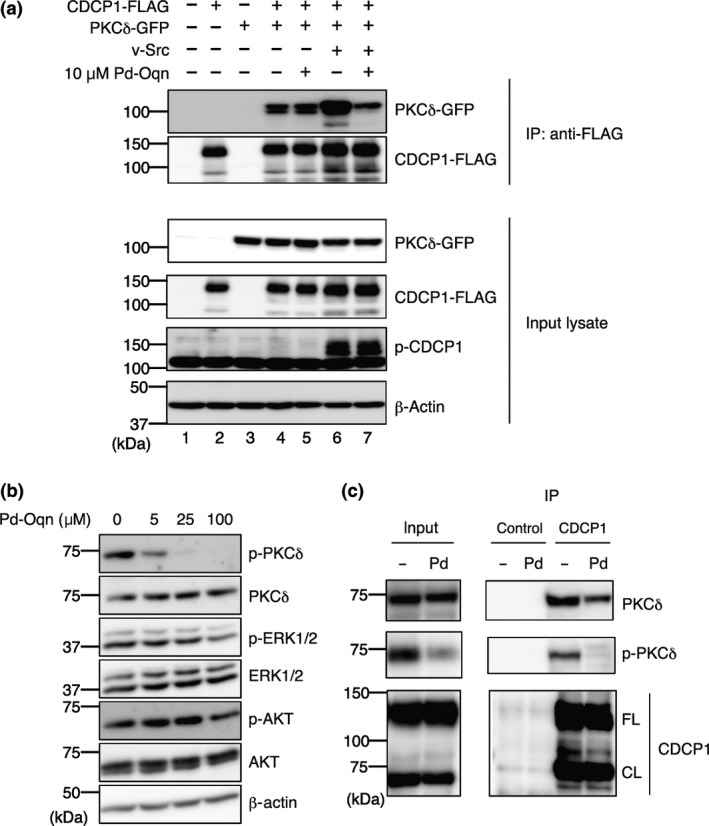
Inhibition of the CDCP1‐PKCδ pathway by Pd‐Oqn. (a) Blockage of the CDCP1‐PKCδ interaction by Pd‐Oqn in cells. Immunoprecipitation was performed as described in Materials and Methods. Immunoprecipitated proteins and input lysates were subjected to western blotting with the antibodies as indicated at the right side. The top indicates the transfection plasmids and Pd‐Oqn addition. (b) Inhibition of the phosphorylation of PKCδ by Pd‐Oqn. 44As3 cells were treated with Pd‐Oqn and the lysates were subjected to western blotting with the antibodies as indicated at the right side. (c) Inhibition of the endogenous CDCP1‐PKCδ interaction by Pd‐O qn. 44As3 cells were treated with DMSO (‐) or Pd‐Oqn (Pd) for 24 h and the cell lysates were subjected to immunoprecipitation with anti‐CDCP1 antibody or control IgG. The precipitated proteins (IP) and the cell lysate (input) were subjected to western blotting with the antibodies as indicated at right side. CDCP1 bands showed both full‐length (FL) and cleaved form (CL) at about 135 and 70 kDa respectively. Molecular weight is indicated at the left.
Following interaction with phosphorylated CDCP, PKCδ is transferred to the plasma membrane and activated by SFK phosphorylation.6 Thus, as expected, inhibition of the PKCδ‐phosphorylated CDCP1 interaction by Pd‐Oqn suppressed tyrosine phosphorylation of PKCδ (Y311) in a dose‐dependent manner whereas phosphorylation of AKT and ERK were not affected (Fig. 3b). We further examined whether Pd‐Oqn affected on the interaction of endogenous CDCP1 with PKCδ. Immunoprecipitation of 44As3 cell lysate with anti‐CDCP1 antibody revealed that endogenous PKCδ co‐precipitated with CDCP1 was decreased by Pd‐Oqn treatment (Fig. 3c). In addition, Pd‐Oqn did not affect on the cleavage of CDCP1 although it has been recently reported that the cleaved isoform rather than full‐length of CDCP1 activates the downstream signaling.21 These results demonstrated that Pd‐Oqn‐induced disruption of PKCδ interaction with CDCP1 could suppress the downstream signaling in cells.
Effect of Pd‐Oqn and Pt‐Oqn on the growth and invasion of cancer cells
Next, we tested whether the addition of Pd‐Oqn and Pt‐Oqn affects anchorage‐independent growth of 44As3 cells, comprising diffuse‐type gastric adenocarcinoma cells in which CDCP1 is highly phosphorylated, using a colony formation assay in soft agar. The number 44As3 cell colonies in soft agar significantly decreased in the presence of 5 μM Pd‐Oqn as compared to that reported for vehicle treatment and was inhibited completely at 25 and 100 μM Pd‐Oqn (Fig. 4a,b). Furthermore, both Pd‐Oqn and Pt‐Oqn could inhibit proliferation of A549, BxPC3, MiaPaCa‐2, and 44As3 cells in a dose‐dependent manner under normal culture conditions with the inhibitory effect of Pd‐Oqn being significantly higher than that of Pt‐Oqn (Fig. S4A). The inhibitory effect of Pd‐Oqn on the proliferation of 44As3 cells was most significant among the cell lines examined, while the proliferation of normal fibroblast NF‐37 cells was least affected by Pd‐Oqn (Fig. 4c). This difference in sensitivity to Pd‐Oqn among cell lines may indicate the differences in expression and phosphorylation levels of CDCP1 in individual cell lines (Fig. S4B).
Figure 4.
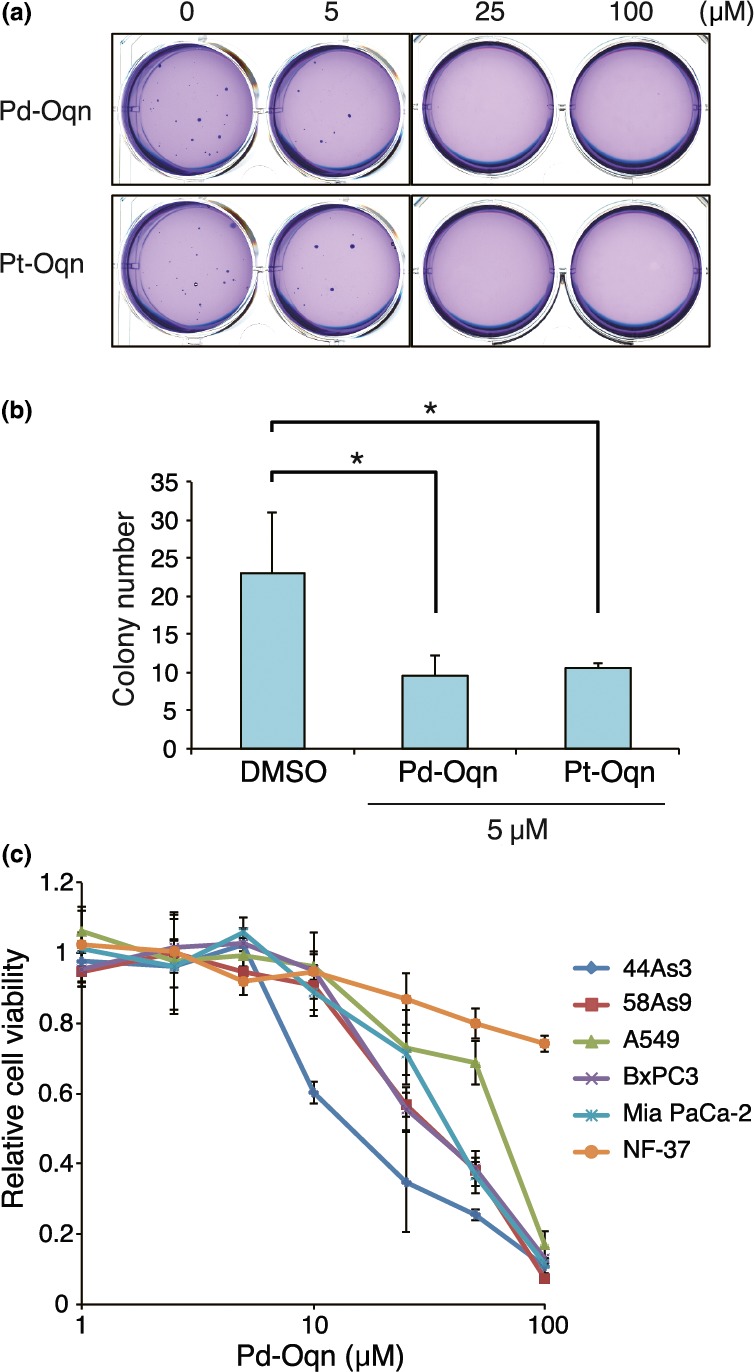
Inhibitory effect of Pd‐Oqn on the growth of cancer cells. (a, b) Inhibition of colony formation of 44As3 cells in soft agar by Pd‐Oqn and Pt‐Oqn. The soft agar colony formation assay was performed as described in Materials and Methods. Representative photos of the colonies in soft agar with DMSO, Pd‐Oqn, or Pt‐Oqn (5, 25, and 100 μM) are shown in (a). The bar graph indicates the number of colonies in soft agar with DMSO, 5 μM Pd‐Oqn, or Pt‐Oqn as the means ± SD of three biological replicates. Statistical significance was determined using the standard Student's t‐test. *P < 0.05. (d) Inhibition of cell proliferation of various cancer cells by Pd‐Oqn. The cells were cultured with Pd‐Oqn (1.0–100 μM) or DMSO (control) and cell viability was determined using the CCK assay after 48 h. The graph indicates the relative value to DMSO treated cells of each cell line as the means ± SD of three biological replicates.
We also examined whether Pd‐Oqn could affect cell motility and invasion using a Boyden chamber‐based assay. The addition of Pd‐Oqn reduced the number of cells that moved to the bottom area through the membrane coated with Matrigel extracellular matrix but not significantly without Matrigel (Fig. 5). These results indicated that Pd‐Oqn inhibited the invasion of 44As3 cells, possibly by degradation of the extracellular matrix, a process also regulated by the CDCP1 pathway as reported.5, 6
Figure 5.
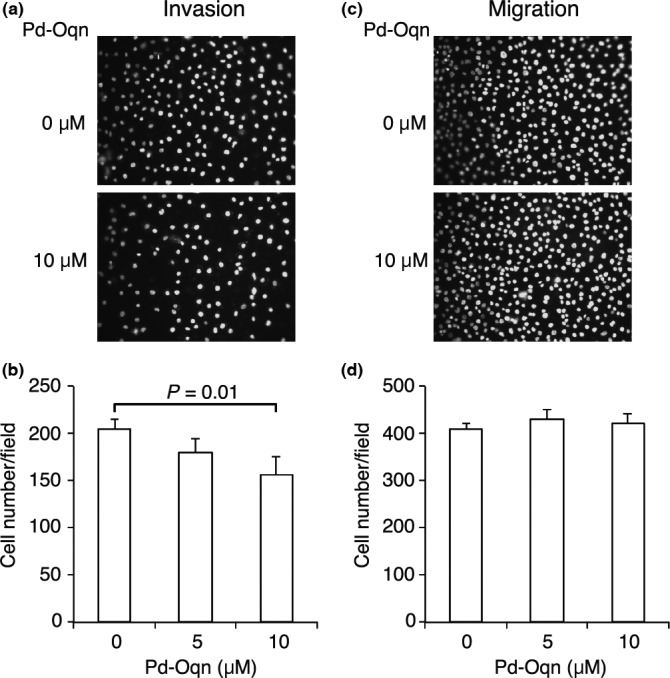
Inhibitory effect of Pd‐Oqn on the invasion of cancer cells. 44As3 cells were subjected to invasion (a, b) and migration (c, d) assays in the presence or absence of Pd‐Oqn. Representative photos of invaded (a) or migrated (c) cells are shown as nuclear staining with DAPI. The graph indicates the cell number of invaded (b) or migrated (d) cells upon DMSO or Pd‐Oqn treatment as the means ± SD of three biological replicates. Statistical significance was determined using the Student's t‐test.
In vivo effect of Pd‐Oqn on tumor metastasis and growth
Finally, we examined the effect of Pd‐Oqn on mouse models of peritoneal dissemination of gastric cancer and tumor growth of pancreatic cancer. For the metastatic model, 44As3 cells were injected into the peritoneal cavity of BALB/c‐nu/nu mice. Pd‐Oqn (15 mg/kg) was injected into the mouse peritoneum six times (Fig. 6a). The weights of the mice were not affected by Pd‐Oqn treatment (Fig. S5). After 14 days, the mice were sacrificed and the metastatic nodules on the mesenterium were counted. Notably, the metastatic nodules of the mice were markedly decreased or diminished by Pd‐Oqn treatment (Fig. 6b,c).
Figure 6.
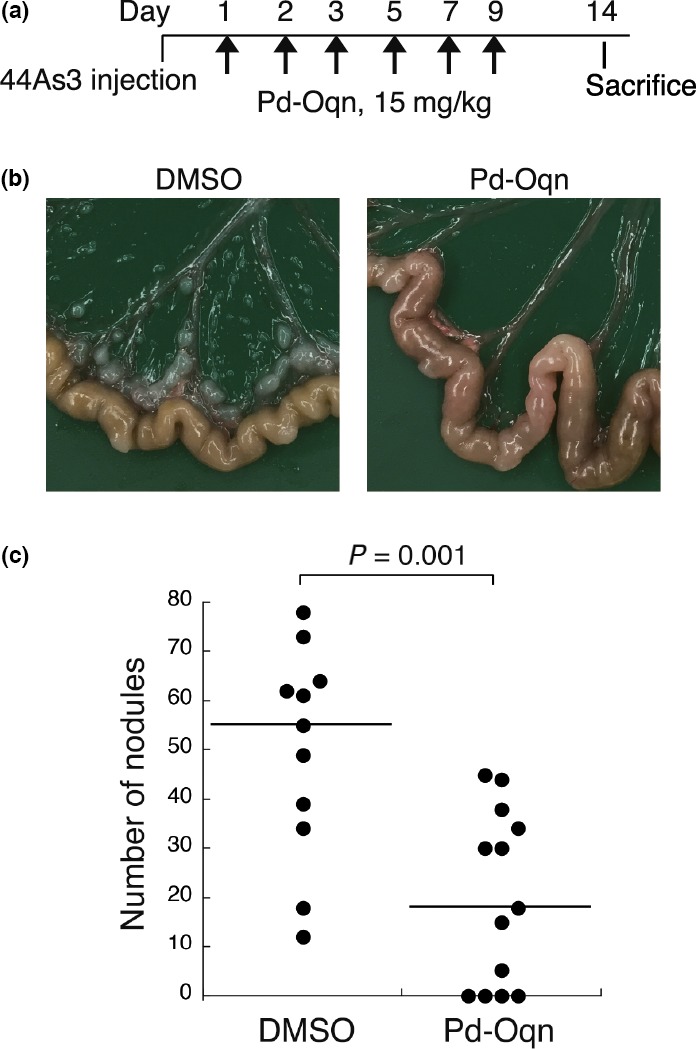
Effect of Pd‐Oqn on the peritoneal dissemination of gastric adenocarcinoma. (a) Schedule of the experiment for injection of Pd‐Oqn or DMSO into the mouse model of peritoneal dissemination using 44As3 cells. (b) Representative photos of the nodules on the mouse mesenterium. (c) The dot plot indicates the number of nodules in each mouse treated with Pd‐Oqn (n = 13) or DMSO (n = 10). The bars in the plot indicate the median value. Statistical significance was determined using the Student's t‐test.
For the pancreatic tumor model, MiaPaCa‐2 cells were transplanted into the pancreas of SCID mice. Pd‐Oqn (15 mg/kg) was injected into the mouse peritoneum seven times (Fig. 7a). Pancreatic tumors were dissected from the mice after 23 days and the weights of the pancreatic tumor tissues were determined. The pancreatic tumor weights of the mice treated with Pd‐Oqn were slightly but significantly lower than those of the control mice were (Fig. 7b,c). These results indicated that Pd‐Oqn suppressed peritoneal dissemination as well as tumor growth in vivo through inhibition of the CDCP1‐PKCδ signaling pathway.
Figure 7.
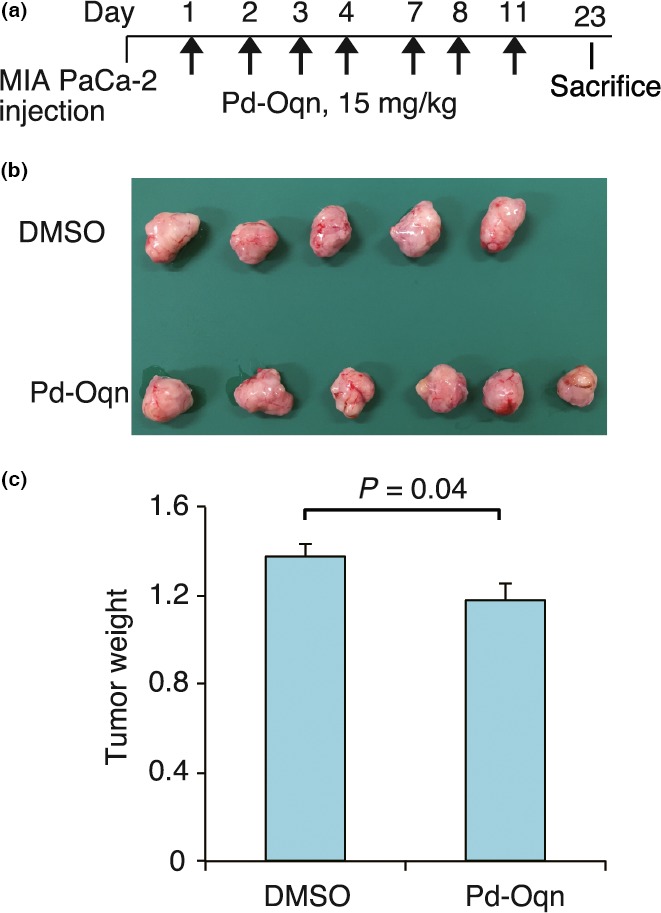
Effect of Pd‐Oqn on pancreatic cancer of orthotopic xenografts. (a) Schedule of the experiment for the injection of Pd‐Oqn or DMSO into the orthotopic xenograft model of pancreatic cancer. (b) Photo of the pancreatic tumor from the dissected mouse. (c) The bar graph indicates the weight of the tumor treated with Pd‐Oqn (n = 6) or DMSO (n = 5) as the means ± SD. Statistical significance was determined using the Student's t‐test.
Discussion
In this study, we first identified small molecules that inhibited the interaction of a phosphorylated site of CDCP1 with the C2 domain of PKCδ. The small molecule, Pd‐Oqn, inhibited the growth and invasion of cancer cells in vitro and also reduced the peritoneal dissemination of gastric adenocarcinoma and pancreatic adenocarcinoma tumor growth in mouse models. The C2 domain of PKCδ has previously been characterized as a binding domain for tyrosine‐phosphorylated sites of CDCP1.4 In addition, we have reported that the CDCP1‐PKCδ signaling pathway plays important roles in tumor progression such as the anchorage‐independent growth and metastasis of cancer cells.2, 6 Our findings in the current study further showed that Pd‐Oqn could reduce tumor metastasis and growth through inhibition of the recruitment of PKCδ to phosphorylated CDCP1 and SFK‐mediated phosphorylation of PKCδ (Fig. S6).
Src family kinases are activated in multiple cancer cells and plays a critical role in cancer progression through the regulation of various substrate proteins and pathways.22, 23, 24 Accordingly, SFK inhibitors have been developed and tested in clinical trials.25, 26 However, SFK is also ubiquitously expressed in normal tissues and involved in normal tissue development and cell differentiation through the conditional activation and phosphorylation of various physiological substrates.27, 28, 29 Therefore, compounds inhibiting the catalytic activity of SFK might naturally show significant side‐effects.30 Conversely, CDCP1, a substrate of SFK, is predominantly expressed in various cancer cells whereas it exhibits limited expression in normal tissues including hematopoietic and neural stem/progenitor cells31, 32 although its functional roles in these tissues have not yet been elucidated. Furthermore, CDCP1 deficiency in mouse shows no obvious effects on development or adult tissues as compared to wild‐type animals (Nakashima K, Sakai R et al, unpubl. data). These findings suggest that inhibition of the CDCP1 pathway by Pd‐Oqn may preferentially affect tumor progression but not normal development and cell differentiation. Consistent with this supposition, the growth inhibitory effect of Pd‐Oqn was selectively shown in cell lines such as A549 or 44As3 in which the CDCP1 pathway is activated rather than in fibroblasts and in vivo Pd‐Oqn administration did not significantly affect general condition of mice at doses shown to be effective against cancers.
The PKC family is implicated in cancer pathology including proliferation, metastasis, and angiogenesis.33, 34 PKCδ in particular plays a critical role in proapoptotic events through its phosphorylation and organelle translocation including to the nucleus and mitochondria although it also can act as an antiapoptotic protein.35, 36 The function of PKCδ depends on various factors including its localization, phosphorylation, and the presence of signaling molecules.36 However, although PKC inhibitors have been subjected to clinical trials for cancer therapy, they have mostly failed owing to lack of efficacy on tumors and severe damage on normal tissues.37 Alternatively, as previously stated, PKCδ localization and activation on cell membranes occurs through interaction with phosphorylated CDCP1 and may contribute to the induction of signaling for cancer progression. Thus, Pd‐Oqn may selectively act as a suppressor for cancer progression that is triggered by PKCδ association with CDCP1.
In the current study, we found that Pd‐Oqn inhibited the invasion but not the migration of 44As3 cells in a chamber assay. We have previously found that CDCP1 regulates the activation of matrix metallopeptidase (MMP)2 and secretion of MMP9.38 In addition, CDCP1 is an essential regulator of the trafficking and function of MT1‐MMP and the invadopodia‐mediated invasion of cancer cells.5 Here, we showed that in a mouse model, Pd‐Oqn reduced the peritoneal dissemination of gastric adenocarcinoma. These results suggested that Pd‐Oqn inhibited the invasion and metastasis of cancer cells through suppression of the extracellular matrix degradation that is promoted by the CDCP1 pathway.
Pd‐Oqn was found to clearly block the interaction of phosphorylated CDCP1 with PKCδ and inhibit PKCδ phosphorylation. Pd‐Oqn is classified under the same category as metal complex compounds such as cisplatin, which induce DNA damage. Pd‐Oqn did not, however, induce the phosphorylation of histone H2AX (γ‐H2AX), a marker of DNA damage response (Fig. S7). Although Pd‐Oqn notably inhibited the interaction of the C2 domain with the 762pY peptide, the mechanism of Pd‐Oqn action on the complex remains to be clarified.
Overall, the results of our study indicate that the novel compound Pd‐Oqn reduces tumor metastasis and growth by inhibiting the association between CDCP1 and PKCδ, thus potentially representing a promising candidate among therapeutic reagents targeting protein–protein interaction. Furthermore, whereas previous attempts to identify compounds inhibiting specific protein–protein interactions have not generally been very successful, the application of metal‐containing compounds such as Pd‐Oqn might provide a means toward overcoming this limitation.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Doc. S1. Materials and Methods.
Fig. S1. Chemical Screening.
Fig. S2. Structures of the inhibitory compounds for the C2‐CDCP1 interaction.
Fig. S3. HTRF assay.
Fig. S4. Effect of the compounds on proliferation of cancer cells.
Fig. S5. Body weight of the mice treated with Pd‐Oqn.
Fig. S6. Model for molecular action of Pd‐Oqn.
Fig. S7. No DNA damage response by Pd‐Oqn.
Table S1. Peptide sequence.
Acknowledgments
We thank Chizuru Tsuda for technical assistance. We thank the SCADS Deposited Chemical Library, Screening Committee of Anticancer Drugs supported by a Grant‐in‐Aid for Scientific Research on Innovative Areas, Scientific Support Programs for Cancer Research, from The Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was supported by the Program for Development of Innovative Research on Cancer Therapeutics (P‐Direct) from the Japan Agency for Medical Research and Development (AMED) (RS), Grand‐in‐Aid for Challenging Exploratory Research By the MEXT of Japan (RS), the Pancreas Research Foundation of Japan (KN), and JSPS KAKENHI grant number 19350031 (SY).
Cancer Sci 108 (2017) 1049–1057
Funding Information
Japan Society for the Promotion of Science (KAKENHI19350031) Japan Agency for Medical Research and Development (P‐Direct) Pancreas Research Foundation of Japan
References
- 1. Scherl‐Mostageer M, Sommergruber W, Abseher R, Hauptmann R, Ambros P, Schweifer N. Identification of a novel gene, CDCP1, overexpressed in human colorectal cancer. Oncogene 2001; 20: 4402–8. [DOI] [PubMed] [Google Scholar]
- 2. Uekita T, Jia L, Narisawa‐Saito M, Yokota J, Kiyono T, Sakai R. CUB domain‐containing protein 1 is a novel regulator of anoikis resistance in lung adenocarcinoma. Mol Cell Biol 2007; 27: 7649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Uekita T, Sakai R. Roles of CUB domain‐containing protein 1 signaling in cancer invasion and metastasis. Cancer Sci 2011; 102: 1943–8. [DOI] [PubMed] [Google Scholar]
- 4. Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell 2005; 121: 271–80. [DOI] [PubMed] [Google Scholar]
- 5. Miyazawa Y, Uekita T, Ito Y, Seiki M, Yamaguchi H, Sakai R. CDCP1 regulates the function of MT1‐MMP and invadopodia‐mediated invasion of cancer cells. Mol Cancer Res 2013; 11: 628–37. [DOI] [PubMed] [Google Scholar]
- 6. Miyazawa Y, Uekita T, Hiraoka N et al CUB domain‐containing protein 1, a prognostic factor for human pancreatic cancers, promotes cell migration and extracellular matrix degradation. Cancer Res 2010; 70: 5136–46. [DOI] [PubMed] [Google Scholar]
- 7. Uekita T, Fujii S, Miyazawa Y et al Suppression of autophagy by CUB domain‐containing protein 1 signaling is essential for anchorage‐independent survival of lung cancer cells. Cancer Sci 2013; 104: 865–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miura S, Hamada S, Masamune A, Satoh K, Shimosegawa T. CUB‐domain containing protein 1 represses the epithelial phenotype of pancreatic cancer cells. Exp Cell Res 2014; 321: 209–18. [DOI] [PubMed] [Google Scholar]
- 9. Boyer AP, Collier TS, Vidavsky I, Bose R. Quantitative proteomics with siRNA screening identifies novel mechanisms of trastuzumab resistance in HER2 amplified breast cancers. Mol Cell Proteomics 2013; 12: 180–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alajati A, Guccini I, Pinton S et al Interaction of CDCP1 with HER2 enhances HER2‐driven tumorigenesis and promotes trastuzumab resistance in breast cancer. Cell Rep 2015; 11: 564–76. [DOI] [PubMed] [Google Scholar]
- 11. Ikeda J, Oda T, Inoue M et al Expression of CUB domain containing protein (CDCP1) is correlated with prognosis and survival of patients with adenocarcinoma of lung. Cancer Sci 2009; 100: 429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emerling BM, Benes CH, Poulogiannis G et al Identification of CDCP1 as a hypoxia‐inducible factor 2alpha (HIF‐2alpha) target gene that is associated with survival in clear cell renal cell carcinoma patients. Proc Natl Acad Sci USA 2013; 110: 3483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He Y, Wu AC, Harrington BS et al Elevated CDCP1 predicts poor patient outcome and mediates ovarian clear cell carcinoma by promoting tumor spheroid formation, cell migration and chemoresistance. Oncogene 2016; 35: 468–78. [DOI] [PubMed] [Google Scholar]
- 14. Uekita T, Tanaka M, Takigahira M et al CUB‐domain‐containing protein 1 regulates peritoneal dissemination of gastric scirrhous carcinoma. Am J Pathol 2008; 172: 1729–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kollmorgen G, Niederfellner G, Lifke A et al Antibody mediated CDCP1 degradation as mode of action for cancer targeted therapy. Mol Oncol 2013; 7: 1142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrington BS, He Y, Davies CM et al Cell line and patient‐derived xenograft models reveal elevated CDCP1 as a target in high‐grade serous ovarian cancer. Br J Cancer 2016; 114: 417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Siva AC, Wild MA, Kirkland RE et al Targeting CUB domain‐containing protein 1 with a monoclonal antibody inhibits metastasis in a prostate cancer model. Cancer Res 2008; 68: 3759–66. [DOI] [PubMed] [Google Scholar]
- 18. Yanagihara K, Takigahira M, Tanaka H et al Development and biological analysis of peritoneal metastasis mouse models for human scirrhous stomach cancer. Cancer Sci 2005; 96: 323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hasegawa T, Yashiro M, Nishii T et al Cancer‐associated fibroblasts might sustain the stemness of scirrhous gastric cancer cells via transforming growth factor‐beta signaling. Int J Cancer 2014; 134: 1785–95. [DOI] [PubMed] [Google Scholar]
- 20. Yano S, Shibahara T, Ogura S. Amino sugar‐bound anti‐cancerous noble metal complex. US patent 20140378402. 2014; 25 Dec.
- 21. Wright HJ, Arulmoli J, Melson LJ et al CDCP1 cleavage is necessary for homodimerization‐induced migration of triple‐negative breast cancer. Oncogene 2016; 35: 4762–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patel A, Sabbineni H, Clarke A, Somanath PR. Novel roles of Src in cancer cell epithelial‐to‐mesenchymal transition, vascular permeability, microinvasion and metastasis. Life Sci 2016; 157: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guarino M. Src signaling in cancer invasion. J Cell Physiol 2010; 223: 14–26. [DOI] [PubMed] [Google Scholar]
- 24. Reynolds AB, Kanner SB, Bouton AH et al SRChing for the substrates of Src. Oncogene 2014; 33: 4537–47. [DOI] [PubMed] [Google Scholar]
- 25. Roskoski R Jr. Src protein‐tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol Res 2015; 94: 9–25. [DOI] [PubMed] [Google Scholar]
- 26. Liu W, Kovacevic Z, Peng Z et al The molecular effect of metastasis suppressors on Src signaling and tumorigenesis: new therapeutic targets. Oncotarget 2015; 6: 35522–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang X, Simerly C, Hartnett C, Schatten G, Smithgall TE. Src‐family tyrosine kinase activities are essential for differentiation of human embryonic stem cells. Stem Cell Res 2014; 13: 379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lowell CA, Soriano P. Knockouts of Src‐family kinases: stiff bones, wimpy T cells, and bad memories. Genes Dev 1996; 10: 1845–57. [DOI] [PubMed] [Google Scholar]
- 29. Chojnacka K, Mruk DD. The Src non‐receptor tyrosine kinase paradigm: new insights into mammalian Sertoli cell biology. Mol Cell Endocrinol 2015; 415: 133–42. [DOI] [PubMed] [Google Scholar]
- 30. Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol 2009; 6: 587–95. [DOI] [PubMed] [Google Scholar]
- 31. Conze T, Lammers R, Kuci S et al CDCP1 is a novel marker for hematopoietic stem cells. Ann N Y Acad Sci 2003; 996: 222–6. [DOI] [PubMed] [Google Scholar]
- 32. Bühring HJ, Kuci S, Conze T et al CDCP1 identifies a broad spectrum of normal and malignant stem/progenitor cell subsets of hematopoietic and nonhematopoietic origin. Stem Cells 2004; 22: 334–43. [DOI] [PubMed] [Google Scholar]
- 33. Garg R, Benedetti LG, Abera MB, Wang H, Abba M, Kazanietz MG. Protein kinase C and cancer: what we know and what we do not. Oncogene 2014; 33: 5225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jain K, Basu A. The multifunctional protein kinase C‐ε in cancer development and progression. Cancers (Basel) 2014; 6: 860–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao M, Xia L, Chen GQ. Protein kinase Cδ in apoptosis: a brief overview. Arch Immunol Ther Exp (Warsz) 2012; 60: 361–72. [DOI] [PubMed] [Google Scholar]
- 36. Basu A, Pal D. Two faces of protein kinase Cδ: the contrasting roles of PKCδ in cell survival and cell death. Sci World J 2010; 10: 2272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mochly‐Rosen D, Das K, Grimes KV. Protein kinase C, an elusive therapeutic target? Nat Rev Drug Discov 2012; 11: 937–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uekita T, Fujii S, Miyazawa Y et al Oncogenic Ras/ERK signaling activates CDCP1 to promote tumor invasion and metastasis. Mol Cancer Res 2014; 12: 1449–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Doc. S1. Materials and Methods.
Fig. S1. Chemical Screening.
Fig. S2. Structures of the inhibitory compounds for the C2‐CDCP1 interaction.
Fig. S3. HTRF assay.
Fig. S4. Effect of the compounds on proliferation of cancer cells.
Fig. S5. Body weight of the mice treated with Pd‐Oqn.
Fig. S6. Model for molecular action of Pd‐Oqn.
Fig. S7. No DNA damage response by Pd‐Oqn.
Table S1. Peptide sequence.


