Abstract
Magnesium-protoporphyrin IX monomethyl ester cyclase (MPEC) catalyzes the conversion of MPME to divinyl protochlorophyllide (DVpchlide). This is an essential enzyme during chlorophyll (Chl) biosynthesis but details of its function in rice are still lacking. Here, we identified a novel rice mutant yellow-leaf 1 (yl-1), which showed decreased Chl accumulation, abnormal chloroplast ultrastructure and attenuated photosynthetic activity. Map-based cloning and over-expression analysis suggested that YL-1 encodes a subunit of MPEC. The YL-1 protein localizes in chloroplasts, and it is mainly expressed in green tissues, with greatest abundance in leaves and young panicles. Results of qRT-PCR showed that Chl biosynthesis upstream genes were highly expressed in the yl-1 mutant, while downstream genes were compromised, indicating that YL-1 plays a pivotal role in the Chl biosynthesis. Furthermore, the expression levels of photosynthesis and chloroplast development genes were also affected. RNA-seq results futher proved that numerous membrane-associated genes, including many plastid membrane-associated genes, have altered expression pattern in the yl-1 mutant, implying that YL-1 is required for plastid membrane stability. Thus, our study confirms a putative MPME cyclase as a novel key enzyme essential for Chl biosynthesis and chloroplast membrane stability in rice.
Introduction
Chloroplasts are specific organelles in higher plants and the site of Chl biosynthesis. Chl is extremely important in photosynthesis because it plays an essential role in harvesting light energy and converting it to chemical energy [1]. Improving the efficiency of Chl biosynthesis is an excellent way to drive the accumulation of more photoassimilates and ultimately increase crop yield [2, 3]. Hence, breeders strive to develop plants with a long stay-green period in order to increase the yield potential.
The Chl biosynthesis pathway is complex, requiring many essential enzymes and other protein regulators. 5-Aminolaevulinic acid (ALA) is the precursor of chlorophyll and other tetrapyrrole end products [4]. Currently, at least 15 enzymes have been identified as involved in the ultimate conversion of ALA to Chl a and b [5]. Glutamyl-tRNA synthetase (GluRS) is the initial enzyme that regulates the formation of ALA [6, 7] and mutation of OsGluRS caused yellow leaf and dwarf height phenotypes in rice [7]. Magnesium ion chelating enzyme (Mg-chelatase) is the second rate-limiting enzyme in chlorophyll synthesis, and catalyzes formation of the magnesium-protoporphyrin IX complex from magnesium ions and protoporphyrin IX [8, 9]. ChlD, ChlH and ChlI are the main subunits of Mg-chelatase. T-DNA insertion into OsChlH caused impediment of Chl biosynthesis and subsequent death of the plants [10]. Divinyl reductase (DVR) converts 8-vinyl groups on various chlorophyll intermediates to ethyl groups, and is indispensable for Chl biosynthesis [11–13]. The OsDVR gene mutant exhibited a yellow-green leaf phenotype, reduced Chl level, arrested chloroplast development, and retarded growth rate [13]. In the last step of Chl biosynthesis, Chl b is synthesized from Chl a through the oxidation of a methyl group on the D ring, which is catalyzed by chlorophyllide a oxygenase (CAO) [14–16]. Two homologous genes of CAO, OsCAO1 and OsCAO2, have been identified from the rice genome. OsCAO1 plays a major role in Chl b synthesis and chloroplast development under the light, whereas OsCAO2 may function in the dark. A study using T-DNA mutants found that OsCAO2 knockout mutant leaves do not differ significantly from wild-type leaves, whereas OsCAO1 knockout mutants had pale green leaves [17]. Chlorophyll (Chl) synthase catalyzes esterification of chlorophyllide to complete the last step of Chl biosynthesis [18, 19]. YGL1 is the main Chl synthase gene which has been cloned. The ygl1 mutant shows yellow-green leaves in young plants, with decreased Chl synthesis, increased level of tetrapyrrole intermediates, and delayed chloroplast development [20].
Most of enzymes inovlved in Chl biosynthesis in rice have been identified and characterized from a series of Chl-deficient mutants. One of the least understood enzymatic steps is formation of the homocyclic ring, which is a characteristic feature of all chlorophyll molecules, including those found in bacteria. In chloroplasts, formation of the isocyclic ring is an aerobic reaction catalyzed by MPEC[4], which controls the conversion of MPME to divinyl protochlorophyllide (DVpchlide). This pathway has still not been elucidated in rice. MPEC is a highly conserved protein in bacteria, green algae and plants, and was originally identified as the acsF gene in Rubrivivax gelatinosus, a photosynthetic bacterium [21]. In algae copper response defect 1 (Crd1) mutants, homologs of AcsF, encoding the catalytic component were identified, which were required for the accumulation of Photosystem I (PSI) and light-harvesting complex I (LHCI) during hypoxia or copper deficiency [22, 23]. In higher plants, a protein encoded by PNZIP was the first isolated catalytic subunit of the MPEC, and contains a leucine zipper domain [24]. PNZIP in Pharbitis nil has been found to be regulated by phytochrome and circadian rhythms [24, 25]. Xantha-l encodes a membrane-bound cyclase subunit in barley. The xantha-l mutants are leaky and able to synthesize a limited amount of chlorophyll, which gives them a pale green color. They supports the view that the aerobic cyclase is most likely composed of the products of three genes, a souble protein, a membrane-bound component encoded by Xantha-l, and another membrane-bound component encoded by Viridis-k [26]. Ycf54 has been identified as a potential componet of MPEC, stimulating its activity. Ycf54 is likely to be a membrane-bound component different from the Viridis-k and the Xantha/AcsF gene products [27]. In addition, the homology of YL-1 in Arabidopsis, CHL27 has been identified as required for the synthesis of protochlorophyllide a. The CHL27 mutant has a chlorotic phenotype with the accumulation of MPEC as a cyclase reaction substrate [28, 29]. Recently, a negative regulator of tetrapyrrole biosynthesis (FLU) NADPH: protochlorophyllide oxidoreductase (POR), and CHL27 were found to form a FLU-CHL27-POR cyclase complex, which is requred for substrate channeling from MgPME to Chlide and also ALA synthesis in chloroplasts [30]. Despite extensive investigations in many species, the function of MPEC in rice, one of main crops in the world, is still not fully elucidated. The photosynthesis and chloroplast morphology phenotypes conferred by mutations in MPEC have mostly not been reported, although recently, ygl8, a novel MPEC defect mutant in rice, has been identified. This also shows a chlorisis phenotype with faults in Chl synthesis [31].
In this study, we identified a novel Chl-defects mutant, named yellow leaf 1 (yl-1), of which the seedling leaves and panicles are deficient in Chl content, corresponding to its chlorisis phenotype through the whole developmental stages. It was shown that YL-1 encodes a subunit of MPEC, which play important roles in conversion of MPME to divinyl Dvpchlide.Transcription levels of many genes associated with the Chl biosynthesis, chloroplast development and photosynthesis were significantly changed in the yl-1 mutant, compared to wild-type. These results indicated that the function of YL-1 in MPEC complex is essential for normal Chl biosynthesis and which would used for roguing in hybrid rice production.
Materials and methods
Plant materials and planted conditions
The yl-1 mutant was identified from EMS-induced mutant library of indica super early-season rice cv. Zhongjiazao 17. An F2 population derived from the cross between yl-1 and tropic japonica cv. D50 was used for genetic analysis and gene mapping. The yl-1 and wild-type plants were grown in paddy fields during normal growing seasons at the China National Rice Research Institute, in Hangzhou (30°N latitude), as well as in a growth chamber (12/12h light/dark; light intensity 300 μmol m−2 s−1) at a constant temperature of either 30, 26 or 22°C. The F2 and transgenic plants were grown in the Hangzhou paddy fields.
Chlorophyll content and transmission electron microscopy analysis
Leaf chlorophyll contents were determined according to the method described in [20]. A 0.2g sample of fresh leaf was immersed in 10 mL ethanol for 48 h in the dark, following which plant debris was removed by centrifugation and the supernatant analyzed by spectrophotometric scanning at 665, 649 and 470 nm. Chloroplast structures were examined in the third leaf of both yl-1 and wild-type cv. Zhongjiazao 17 plants using transmission electron microscopy (TEM) as described [32] with minor modifications. Briefly, Samples of wild-type and yl-1 mutant leaves were prepared for transmission electron microscopy by cutting into small pieces, fixed in 2.5% glutaraldehyde in a phosphate buffer at 4°C for 4 h, rinsed and incubated overnight in 1% OsO4 at 4°C, dehydrated through an ethanol series, and infiltrated with a graded series of epoxy resin, and then embedded in Epon 812 resin. Thin sections were obtained using a diamond knife and a Reichert OM2 ultramicrotome, stained in 2% uranyl acetate, and 10 mM lead citrate, pH 12, and then viewed with a Hitachi H-7650 transmission electron microscope.
Photosynthetic rate and chlorophyll fluorescence analysis
An Li-6400 portable photosynthetic apparatus was used on a sunny day between 9~11 a.m, to determine the net photosynthetic rate (Pn) of wild-type and mutant plants at the beginning of the tillering stage. The red and blue light source were used for determination of photosynthetic rates of three representative leaves of both mutant and wild-type. The light quantum density was 1200 μmol m-2 s-1, velocity 500 μmol s-1. Chlorophyll fluorescence analysis of the third leaf at the 3-leaf stage were performed with a PAM-2000 portable chlorophyll fluorometer (MINIPAM; Walz; http://walz.com).
Mapping and cloning of YL-1
Initially, 36 yellow plants of yl-1/D50 F2 progeny were used for coarse linkage analysis, and subsequently 1220 homozygous mutant F2 segregants were used to fine mapping of yl-1. The microsatellite and In-Del markers used to genotype these progeny were given in Electronic Supplementary Material (ESM) S3 Table. The positional cloning strategy were according to [33]. The total volume of the PCR reaction was 10μL, containing 1μL of 10×PCR buffer (25 mM/L MgCl2), 0.1 mM of dNTPs, 0.1 mM of each primer, 0.25μL of Taq DNA polymerase (2 U/μL), 1μL of genomic DNA and 4.95μL of dd H2O. The amplification protocol included an initial denaturation at 95°C for 3 min, 35 cycles of 45 s at 95°C, 30 s at 55°C and 45 s at 72°C, and a final extension step at 72°C for 8 min. PCR products less than 300 bp were separated by 6% native polyacrylamide gel and silver-stained for visualization. Finally, the genetype of each plant was identified.
Vector construction and rice transformation
The full-length coding region of YL-1 was isolated from Zhongjiazao 17 by PCR (primers sequences were listed in S3 Table), and then subcloned into the binary vector pCAMBIA1305::Ubiqutin:GFP under the control of the rice Ubiqutin 1 promoter. The resultant plasmid was introduced into Agrobacterium tumefaciens strain EHA105, and then was introduced into yl-1 mutant by means of Agrobacterium mediated transformation. The genotypes of transgenic plants were determined by the Neomycin phosphotransferase II (nptII) resistance detection and the phenotype analysis of transgenic lines.
RNA extraction and qRT-PCR analysis
RNA of various organs harvested from yl-1 and wild-type plants were isolated using a RNA extraction kit (Beijing Dingguo Biotechnology Co. Ltd.; http://www.dingguo.com), following the manufacturer’s instructions. The cDNA first strand was reverse-transcribed by using oligo (dT) as primer. The transcriptions of YL-1 and a series of chlorophyll synthesis- and photosynthesis-associated genes were assessed using quantitative real-time PCR (qRT-PCR), with the rice ubiquitin gene (GenBank accession AF184280) as the control. The relevant primer sequences are listed in S3 Table.
Subcellular localization of YL-1
The coding sequence of YL-1 was amplified by PCR using the specific primer pairs shown in S3 Table. The PCR products were cloned into the pAN580-GFP vector between the cauliflower mosaic virus 35S promoter and the NOS terminator to form translational fusions with the N terminus of GFP. All transient expression constructs were separately transformed into rice protoplasts and incubated in the dark at 28°C for 16 h before examination according to the protocols described previously[34, 35]. The fluorescence of GFP was observed using a confocal laser scanning microscope (Leica TCS SP5).
RNA-sequencing analysis
4-week-old leaves of both yl-1 and wild type were sampled, and total RNA was extracted using Trizol reagent (Invitrogen, CA, USA) following the manufacturer's procedure. The total RNA quantity and purity were analyzed by Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with RIN number >7.0. RNA-sequencing analysis was then performed on an Illumina Hiseq2000/2500 (LC Sciences, USA) following the vendor's recommended protocol.
Results
The yl-1 mutant shows a yellow leaf and panicle phenotype with reduced Chl accumulation through the whole growth phases
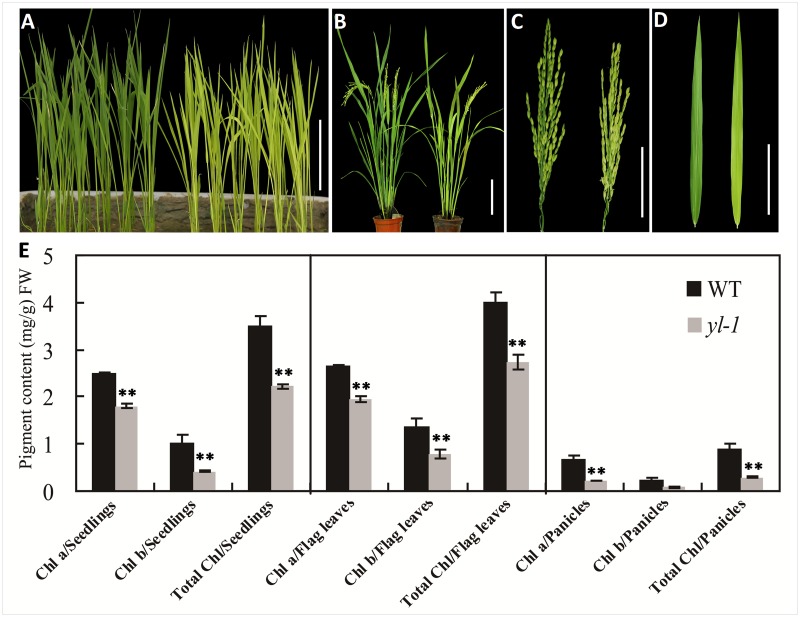
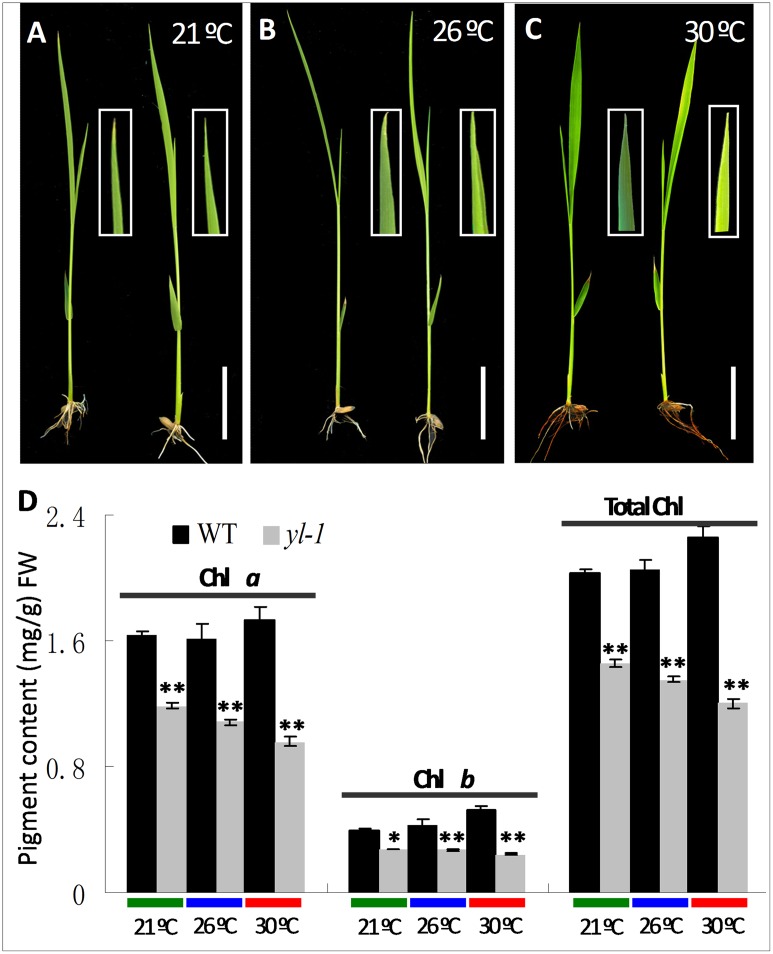
We measured Chl a, Chl b and total Chl accumulation in the leaves and young panicles of both yl-1 mutant and wild-type plants at different developmental stages. The former exhibited a chlorosis leaf and yellow panicle phenotype throughout all developmental stages (Fig 1A–1D). Significantly reduced levels of Chl were detected in yl-1, with 30%, 35% and 60% reductions in Chl a and 65%, 50% and 70% reductions in Chl b of 4-week-old leaves, flag leaves and young panicles, respectively (Fig 1E). Furthermore, compared to the wild-type, the yl-1 mutant exhibited significantly reduced plant height, grains per panicles and grain yield per plant, but with increased tiller number and seed setting rate (S1 Fig). Taken together, this suggests that the yl-1 mutant phenotype results from reduced rates of Chl accumulation. To further identify whether the yellow leaf phenotype of yl-1 is affected by external temperature changes, we analysed the Chl a, Chl b and total Chl content at 21°C, 26°C and 30°C. Our results showed that yl-1 exhibited the yellow leaf phenotype under each of the three different temperature conditions and Chl contents were always much lower than those of the wild-type (Fig 2). However, Chl accumulation was much lower at 30°C than at 21°C, and also a more obviously chlorotic phenotype was observed at 30°C (Fig 2), which indicates that the yl-1 mutant is intolerant of higher temperature environments.
Fig 1. Phenotypic characterization of wild-type (WT) and yl-1 mutants.
A, Phenotypes of 3-week-old wild-type and yl-1 seedlings (from left to right), bar = 10 cm. B, Phenotypes of wild-type and yl-1 at the heading stage grown in paddy field, bar = 20 cm. C-D, Enlarged views of spikelets (C) and flag leaves (D) of wild-type and yl-1 in B, bars = 5 cm. E, Chlorophyll content of leaves in wild-type and yl-1 mutants at three different developmental stages: the seedlings stage; flag leaves and young panicles of the heading stage. Data are means ± SD (n = 5). Chl.a, Chlorophyll a; Chl.b, chlorophyll b; FW, fresh weight. The asterisks indicate statistical significance between the wild-type and the mutants, as determined by the Student’s t–test (* P < 0.05; ** P < 0.01).
Fig 2. Chlorophyll content of leaves in wild-type and yl-1 mutant at three different temperatures.
A-C, 10-day-old seedlings of wild-type and yl-1 mutant at 21°C (A), 26°C (B) and 30°C (C); bars = 2 cm. D, Chlorophyll content of leaves in wild-type and yl-1 mutant at 21°C (A), 26°C (B) and 30°C (C). Data are means ± SD (n = 5). The asterisks indicate statistical significance between the wild-type and the mutants, as determined by the Student’s t–test (* P < 0.05; ** P < 0.01).
The yl-1 mutant presents abnormal chloroplast development and attenuated photosynthetic efficiency
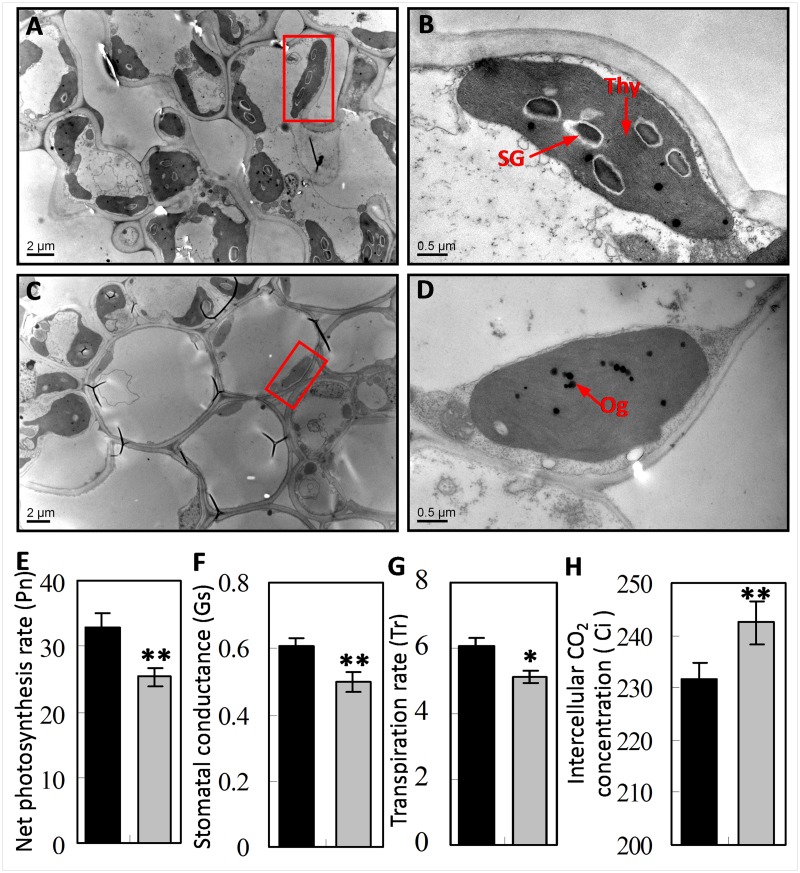
To investigate whether the lack of chlorophyll synthesis in the y1-1 mutant was accompanied by defective chloroplast development, we compared the ultrastructure of plastids between yl-1 mutant and wild-type plants using transmission electron microscopy (TEM). Chloroplast ultrastructures were observed in the 3rd leaves of L4 seedlings. The results revealed that chloroplasts in the leaves of wild-type plants displayed well-developed membrane structure, with dense thylakoids arranged in the grana and in the membranes interconnecting of grana (Fig 3A and 3B). However, granas stacked in the yl-1 mutant appear less dense and with many osmiophilic plastoglobuli. Thus, unlike the wild-type, leaves of young yl-1 mutant plants display characteristics of ongoing cell senescence (Fig 3C and 3D).
Fig 3. Chloroplast ultrastructures and photosynthetic parameters in wild-type and yl1-1 mutant.
A-D, Transmission electron microscopy (TEM) of chloroplasts from Leaf 4 of the wild-type (A-B) and yl-1 mutant (C-D) seedlings at 3-week-old. E-H, photosynthetic parameters in wild-type (left) and yl1-1 mutant (right). Data are presented as means ± SD (n = 5). Thy thylakoid lamellar; SG starch granule; OG osmiophilic plastoglobuli. Asterisk indicates significant difference between the wild-type and yl-1 mutant. The asterisks indicate statistical significance between the wild-type and the mutants, as determined by the Student’s t–test (* P < 0.05; ** P < 0.01).
The photosynthetic parameters of yl-1 and wild-type plants grown in the field at the heading stage were measured. As expected, significantly reduced net photosynthetic rate (Pn), stomatal conductance (Gs) and transpiration rate (Tr) and higher intercellular CO2 concentration (Ci), were detected in yl-1 compared to wild-type plants (Fig 3E–3H). To test whether the photosynthetic apparatus was affected in yl-1, some key light-induced chlorophyll fluorescence parameters of PSI and PSII were compared in yl-1 and wild-type plants. Distinct decreases in the electron transport rate (ETR), photochemicalquenching (Qp) and maximal efficiency of PSII photochemistry (Fv/Fm) were oberserved in the yl-1 mutant (Table 1). These observations indicated that the yl-1 mutant suffer from defects affecting photosynthesis.
Table 1. Main photosynthetic parameters of wild-type and yl-1 mutant, The asterisks indicated statistical significance between the wild-type and yl-1, as determined by the Student’s t–test (* P < 0.05; ** P < 0.01).
| Fm | Fo | Fv/Fm | NPQ | Qp | ETR | |
|---|---|---|---|---|---|---|
| WT | 0.33±0.02 | 0.10±0.006 | 0.70±0.04 | 0.26±0.03 | 0.74±0.02 | 63.93±1.97 |
| yl-1 | 0.21±0.03** | 0.11±0.01 | 0.48±0.02** | 0.27±0.04 | 0.55±0.01** | 55.38±1.29** |
Map-based cloning of YL-1
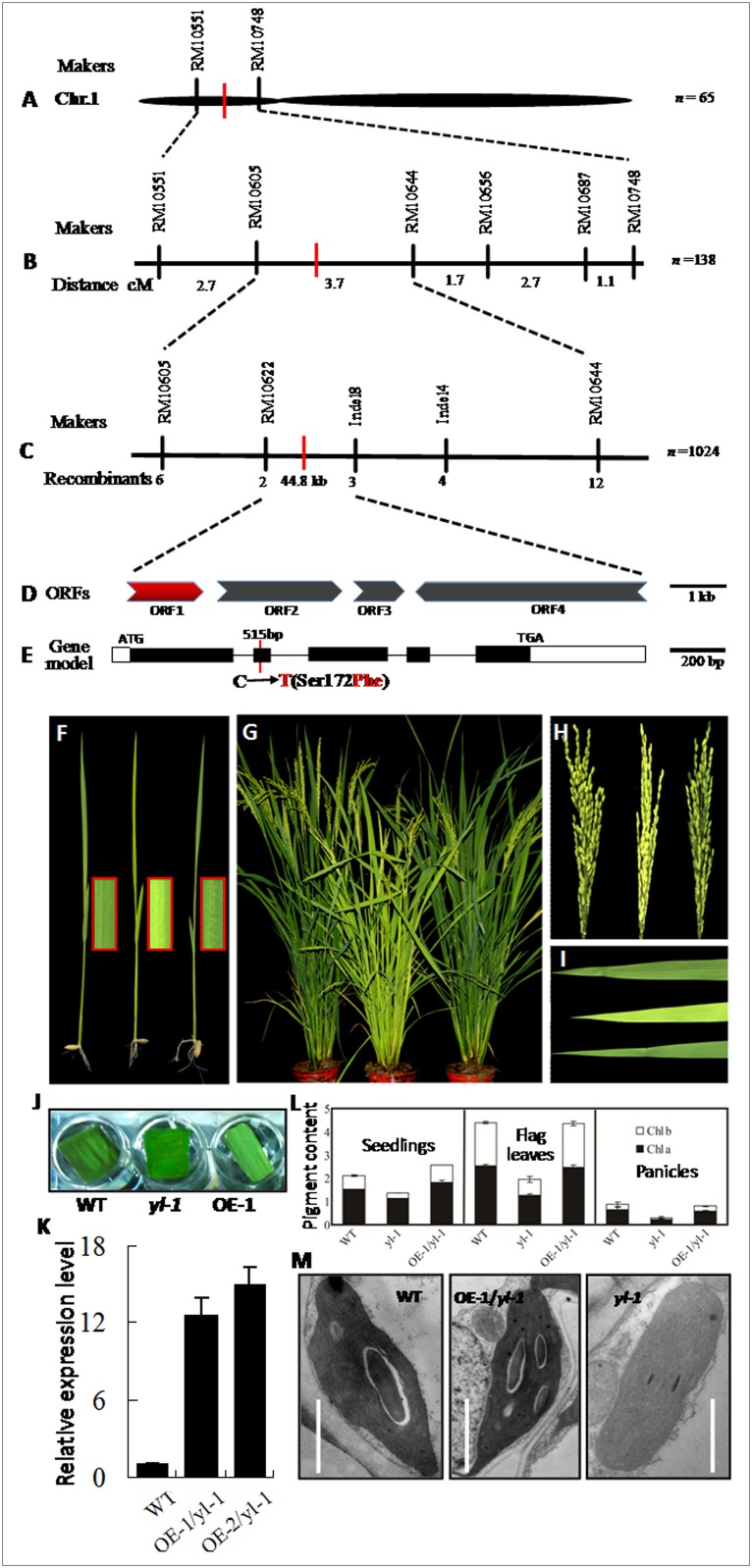
The F1 young leaves of yl-1×cv. D50 were similar to those of wild-type cv.Zhongjiazao17, while the segregation behavior in the derived F2 population was consistent with the Mendelian monogenic ratio of three wild-type phenotype to one chlorisis phenotype (S1 Table). Linkage analysis fixed the location of the yl-1 into a 3.7-cM interval on chromosome 1, between the SSR markers RM10605 and RM10644 (Fig 4A and 4B). Using 1220 mutation phenotype plants from the F2 population derived from yl-1 × cv. D50, the location of yl-1 was then narrowed down to a 44.8 kb physical region between SSR marker RM10622 and Indel marker Indel8 (Fig 4C). Within this region, 4 open reading frames (ORFs) were predicted (http://www.gramene.org/; Fig 4D). Genomic sequencing analysis of the 4 ORFs revealed that the first ORF (LOC_Os01g17170) carries a single-nucleotide transition (C→T) at the position 515 bp from the start codon ATG in yl-1 (Fig 4E), leading to the transition of an amino acid residue from serine (Ser) in wild-type to phenylalanine (Phe) in yl-1 (Fig 4E). Thus, ORF1 was the candidate gene of yl-1.
Fig 4. Map-based cloning of YL-1 gene.
A-B, Position of the YL-1 locus in an 3.7 cM interval on Chr. 1 between SSR markers RM10605 and RM10644. C, YL-1 was narrowed to a 44.8 kb region between the SSR marker RM10622 and Indel marker Indel 18. D, 4 putative ORFs in the 44.8 kb genomic region. D, Gene structure of the YL-1 candidate gene (ORF1). The black boxes represent exons, the lines between them represent introns, and the white boxes means 5,UTR and 3,UTR of YL-1. F-I, Phenotype of wild-type, yl-1 and T1 plant of transgenic yl-1expressing YL-1 at the seedling (F) and heading (G-I) stages. Hygromycin resistance detection (J) and qRT-PCR of YL-1 (K). L-M, Chlorophyll content (L) and chloroplast ultrastructures (M) in cells of third leaf of wild-type, yl-1 and T1 transgenic plants expressing YL-1. Data in (K, L) are shown as means ± SD from 5 individual replicates.
To verify the identity of yl-1, the plasmid Ubi:YL-1 containing wild-type YL-1 cDNA region driven by the maize Ubiquitin promoter was transformed into yl-1 via the Agrobacterium-mediated method. As expected, the yellow-leaf seedlings, flag leaves and panicles of yl-1 containing YL-1 cDNA recovered to a normal green phenotype (Fig 4F–4K). Moreover, chlorophyll content, chloroplast ultrastructural structure and the relative expression of YL-1 in the transgenic positive lines returned to normal levels (Fig 4L and 4M). Thus, ORF1 (LOC_Os01g17170) indisputably corresponds to YL-1.
YL-1 encodes an MgPME cyclase
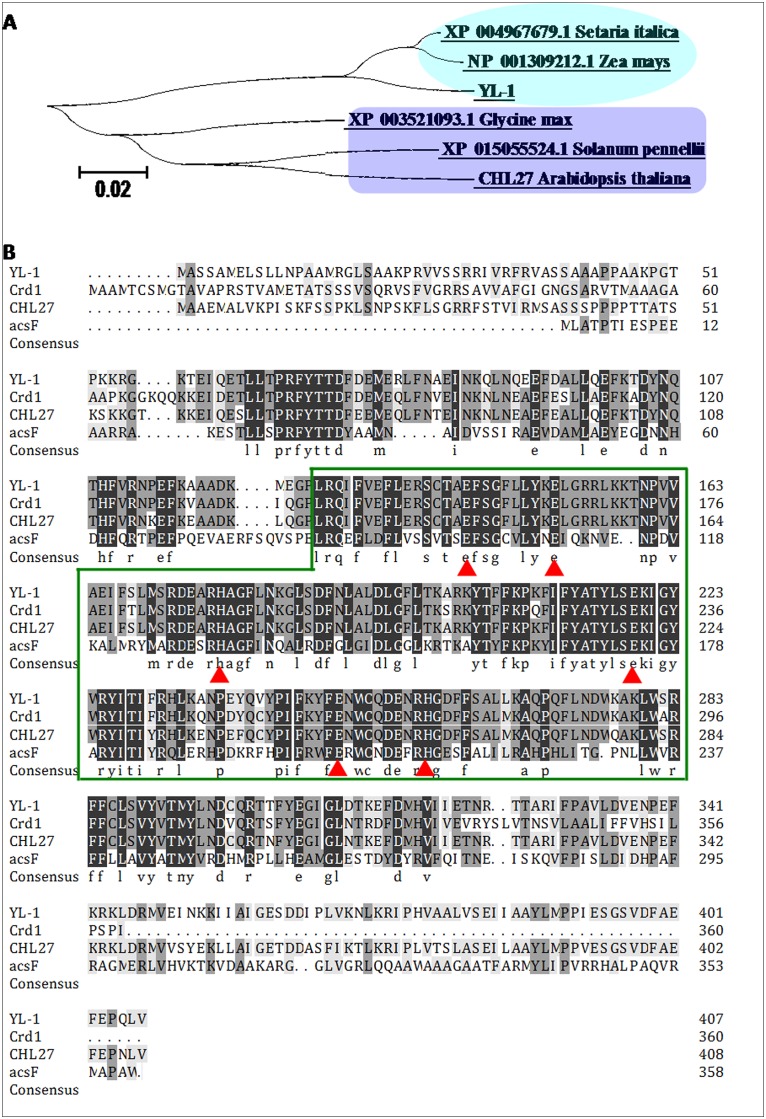
The coding sequence of YL-1 is 1227 bp and contains a 409-residue polypeptide with a molecular mass of approximately 47 kDa (http://rice.plantbiology.msu.edu). The YL-1 sequence as a whole has homologs among other higher plants, particularly with sequences encoded by monocotyledonous species such as maize and Setaria italica (Fig 5A). Furthermore, YL-1 encodes an MPMC with a AcsF domain (Fig 5B) which is highly conserved in bacteria, green algae and plants and often found in proteins which bind metal ion (Fig 5B).
Fig 5. Phylogeny of the YL-1 protein family.
A, phylogenetic tree representing alignment of YL-1 proteins. The oval-cyan box presents monocotyledons, and the rectangle-blue box means dicotyledons. The phylogenetic tree was constructed using the neighbor-joining algorithm. Bootstrap values were shown at each node. Bar Indicator of genetic distance based on branch length. B, Comparison of YL-1 protein sequences from both bacteria, green algae and plants. Black shading identical residues, gray shading similar residues. The green box presents the AcsF domain in different species, and the red-triangle box stands for the conserved residues of a diiron center (ion binding site) which is present in the AcsF domain.
Transcription profiling of YL-1 and subcellular localization of its product
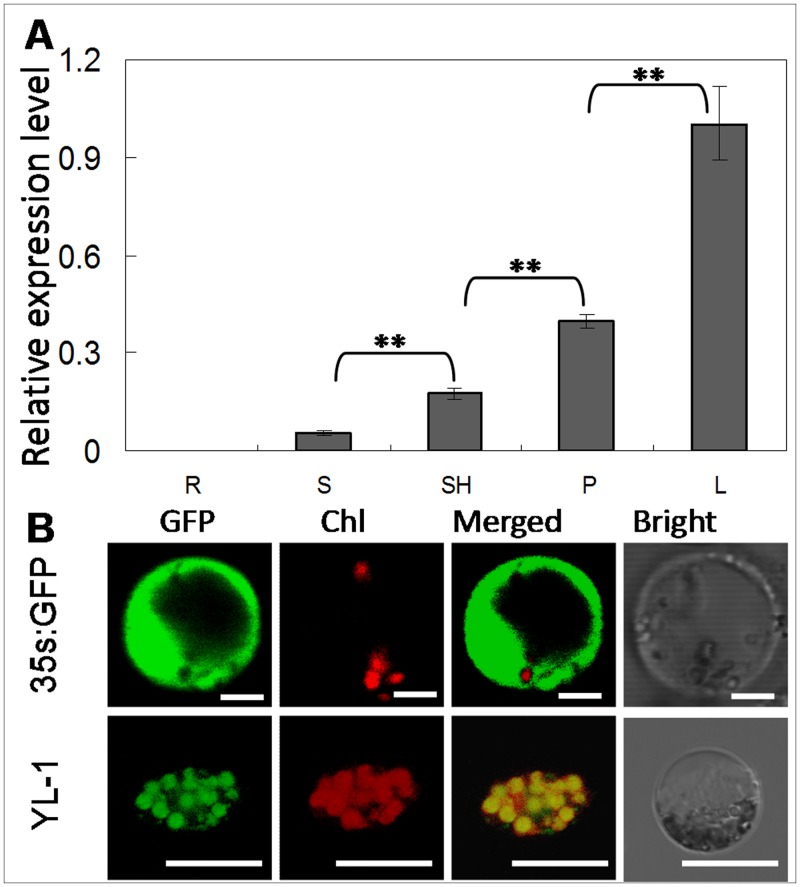
Given that yl-1 encodes a key enzyme subunit for chlorophyll biosynthesis, we determined its transcription levels in leaf photosynthetic tissues. When grown in the Hangzhou fields, YL-1 were transcribed in the leaf, leaf sheath, culm, young panicle, and root. The expression profile of YL-1 in these various tissues was investigated by qRT-PCR and the results showed that YL-1 is constitutively expressed in the examined tissues, with higher levels in leaves, young panicles and leaf sheaths (Fig 6A), which corresponds to its protein function in chlorenchyma.
Fig 6. Expression pattern analysis and chloroplast localization of YL-1.
A, Tissue-specific expression of YL-1 at heading stage in wild-type. RNA was isolated from flag leaves (L), leaf sheaths (SH), culms (C), young panicles (P) and roots (R). B, GFP signal in tissue transformed with the empty GFP vector or YL-1-GFP fusion protein. Data in (A) are shown as means ± SD from 3 individual replicates. Bars = 2 μm in (B). The asterisks indicate statistical significance between the wild-type and the mutants, as determined by the Student’s t–test (* P < 0.05; ** P < 0.01).
To identify the subcellular localization of the YL-1 protein, a transient expression system was conducted by delivering a 35S::YL-1:GFP construct into rice protoplasts cells. The result showed that the GFP signal was localized in chloroplasts, while the signal pattern of 35S::GFP control plastid was distributed over the whole cell (Fig 6B).
The yl-1 affects mRNA accumulation levels of Chl biosynthesis process
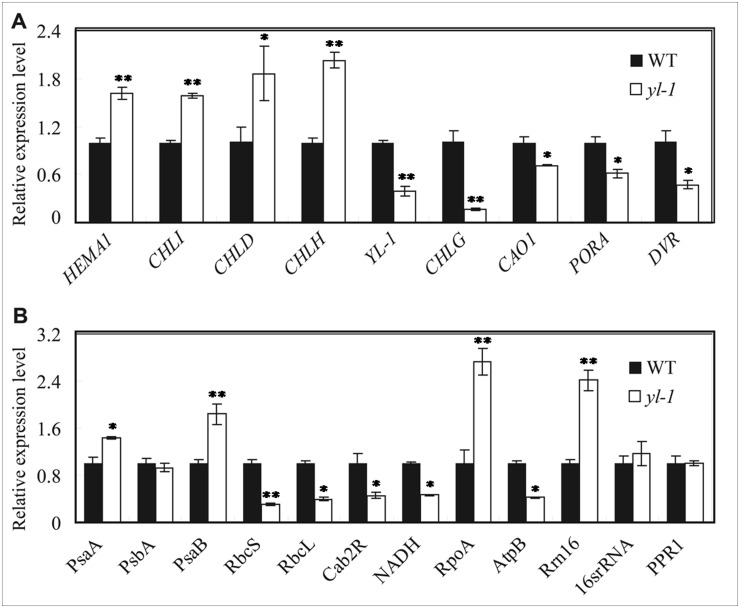
MPEC catalyzes the conversion of MPME to DVpchlide, which plays an important role in the central process of Chl biosynthesis. Therefore, the expression of genes involved in Chl biosynthesis were examined by qRT-PCR in both yl-1 and wild-type plants. The genes, upstream of Chl biosynthesis, such as glutaml-tRNA reductase (HEMA1), Mg chelatase D subunit (CHLD), Mg chelatase H subunit (CHLH) and Mg chelatase I subunit (CHLI) were all up-regulated in the yl-1 mutant, compared to wild-type (Fig 7A). On the other hand, the genes down-stream of Chl biosynthesis, such as chlorophyllide a oxygenase (CHLG, YGL1), chlorophyllide a oxygenase 1 (CAO1), protochlorophyllide A (PORA) and divinylreductase (DVR) were significantly decreased in the yl-1 mutant (Fig 7A). In addition, we investigated the expression levels of genes involved in chloroplast development and photosynthesis. The chloroplast development depends on the regulation of two major RNA polymerase: nuclear-encoded RNA polymerase (NEP) and plastid-encoded RNA polymerase (PEP). In yl-1 plants we found significant changes in the expression of PEP-dependent genes, such as the subunit of photosystem I gene psaA, a core component of Photosystem II gene psbA, the photosynthesis apparatus Rubisco large subunit gene rbcL and Rubisco small subunit gene rbcS. The NEP-dependent genes, such as rpoA and rrn16 which highly expressed at early stages of chloroplast development were significantly increased in yl-1 (Fig 7B). Moreover, transcription of photosynthesis-related genes Oscab2R and NADH were remarkably down-regulated in the yl-1 mutant (Fig 7B). These data suggest that abnormal expressions of these chloroplast associated genes may be responsible for the chlorisis phenotype of yl-1.
Fig 7. Transcriptional changes of chlorophyll biosynthesis-related genes and chloroplast development -associated genes between wild-type and yl-1 mutant.
A, Expression levels of genes related to Chlorophyll biosynthesis. B, Expression levels of Photosynthesis- and chloroplast development-related genes. Data are shown as means ± SD from 3 individual replicates. The asterisks indicate statistical significance between the wild-type and yl-1, as determined by the Student’s t–test (* P < 0.05; ** P < 0.01).
Discussion
Chl is extremely important in high-photosynthesis and plays an essential role in harvesting light energy and converting it to chemical energy. Generally, Chl deficiency affects the chloroplast development and photosynthesis, leading to abnormal leaf morphology including virescent (v), stripe (st), albino, chlorina, zebra, and yellow-variegated [36, 37]. Some of these Chl-deficient mutants are seedling-specific and gradually become normal during later growth stages, and some can affect the plants for the entire life cycle. The yss1 rice mutant shows striated leaves at the seedling stage, particularly in leaf 3, but produces normal wild-type leaves later [38]. While, the vyl mutant develops decreased Chl levels throughout the whole developmental stages [39]. Here, we identified a novel Chl-deficient mutant yl-1 with yellow leaf and yellow panicle during its entire growth duration, which is compatible with its reduced chlorophyll content, abnormal chloroplast morphology, and reduced photochemical efficiency.
Certain Chl-deficient mutants show abnormal phenotypes in response to temperature changes. Some are sensitive to lower temperature, including v1, v2 and wlp1, these all show albino phenotypes at a restrictive low temperature [40–42]. Others often show yellowing, stripe or completely albinism when subjected to high temperature. Although the yl-1 mutant presents chlorisis under a range of temperature conditions, at higher temperature, there is a more serious yellow leaf phenotype with much more defects in Chl synthesis, which indicated high temperature may aggravate the effect of mutant phenotype of yl-1, due to YL-1 could likely involved in the temperature-regulated chlorophyll biosynthesis.
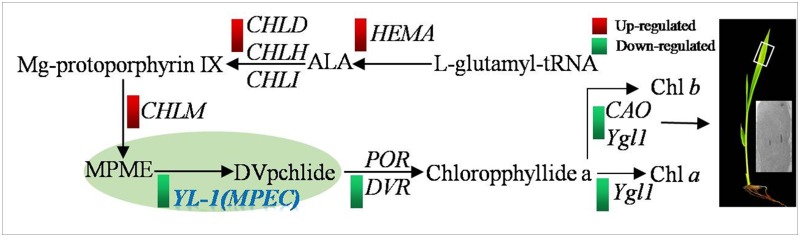
Map-based cloning suggests that YL-1 encodes an enzyme, MPMC, essential for during Chl synthesis, which catalyzes the conversion of MgPME to Dvpchlide. Chl synthesis is a complex process starting with the formation of ALA and proceeding to the synthesis of Chl a and Chl b. In this study, we found extensive up-regulation of HEMA, which encodes the first enzyme that controls the formation of ALA, and of three genes encoding subunits of magnesium-ion chelating enzyme (CHLD, CHLH, CHLI), which play roles upstream of Chl synthesis. In addition, expression levels of down-stream genes including YGL1, CAO1, PORA and DVR were all significantly reduced. The results indicates that normal function of YL-1 is essential for Chl biosynthesis, where it plays central role. In addition, we found that transcription level of YL-1was dramatically reduced in the yl-1 mutant, which may interfere with its enzyme activity (Fig 8). Recently, ygl8, a novel gene was reported in rice, which also shows defects in Chl synthesis and chloroplast development with chlorisis phenotype [31]. Taken these evidences, we concluded that YL-1 may play a pivotal function during Chl synthesis (Fig 8).
Fig 8. Model showing mutation of YL-1 caused the abnormal Chl biosynthesis and leaf development in the mutant plant.
The subcellular localization of YL-1 shows it to be a chloroplast-located protein. Meanwhile, Arabidopsis CHL27 also encodes a chloroplastic protein which is distributed in the inner-envelope and thylokoid membranes[28, 29]. Moerover, MPMC has been reported to exist in chloroplasts as a membrane-interacting multi-subunit enzyme containing at least one soluble and two membrane bound components [26]. In this study, RNA-seq analysis showed that expression of many membrane-associated genes, including plastid membrane-associated genes, was altered in the yl-1 mutant, compared to wild-type, including the chloroplast outer envelope 24 kDa protein, and the membrane-associated 30 kDa protein (S2 Fig and S2 Table). It has been reported that the synthesis of ALA and Mg-protoporphyrin IX take place in chloroplast stroma, while, the subsequent processes of Chl synthesis including the conversion of MPME to Dvpchlide, and Chl a/b synthesis, are mostly catalyzed by membrane bound enzymes such as chlorophyllide a oxygenase, divinyl reductase and Chl synthase, which are located in the thylakoid membrane. In the yl-1 mutant, loose thylakoid membranes and less dense grana stacks were observed, which indicated that the mutation of YL-1 results in structure damage to thylakoid membranes. Furthermore, in Arabidopsis, CHL27, PORB, PORC and Geranylgeranyl reductase (GGR/CHLP) were identified as components of a FLU-CHL27-POR complex in chloroplast membranes, which plays important roles in Chl biosynthesis [30]. Thus, YL-1 has been demonstrated to be essential for chloroplast membrane stability in rice, even though there are no putative transmembrane domains in the YL-1 or CHL27 proteins.
There are many reports of rice leaf color mutants, including v1, v2, wlp1 [40–42] and yss1 [38]. Most of them were used as morphological marker applied in roguing of hybrid rice seed production and parents seed multiplication. However, many of the leaves color mutant showed their color change phenotype only under particular temperature regimes or specific development periods. Therefore they are not useful for identifying variation. In addition if roguing in hybrid rice seed production or parents seed multiplication was not carried out in a timely manner, these morphological markers would be useless and cause huge losses. For yl-1, the yellow leaf and panicle phenotype was stable and lasted the whole growth duration, which makes it suitable for roguing during the plant breeding processes. Furthermore, compared to wild-type, yl-1 showed excellent agronomic traits of increased tiller number and reduced plant height, which is desirable in hybrid rice, especially in male sterile line rice.
In conclusion, we identified YL-1, function as MPEC subunit in rice, which might be essential for Chl biosynthesis and chloroplast development in rice. And the yellow leaf and panicle phenotype, together with improved agronomic traits of yl-1 would be useful in male sterile line rice.
Supporting information
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
This research was financially supported by the Zhejiang Science and technology projects (Grant No: 2015C32045), Zhejiang Natural Science Foundation (Grant No: LY14C130009), China Natural Science Foundation (No. 31501285), the Central level, non-profit, scientific research institute basic R and D operations Special Fund (2014RG002-1), and National Key Research and Development Program of China (2016YFD0101801).
Data Availability
The RNA-seq data have been submitted to the database Gene Expression Omnibus (GEO), and the accession number is GSE96073.
Funding Statement
This research was financially supported by the Zhejiang Natural Science Foundation (Grant No: LY14C130012, LY14C130009), Zhejiang Science and technology projects (Grant No: 2015C32045), China Natural Science Foundation (No. 31501285), and the Central level, non-profit, scientific research institute basic R and D operations Special Fund (2014RG002-1).
References
- 1.Fromme P, Melkozernov A, Jordan P, Krauss N. (2003) Structure and function of photosystem I: interaction with its soluble electron carriers and external antenna systems. FEBS Letters 555: 40–44. 10.1016/S0014-5793(03)01124-4 . [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Qin F, Zang G, Kang Z, Zou H, Hu F, Yue C, Li X, Wang G. (2013) Mutation of OsDET1 increases chlorophyll content in rice. Plant Sci 210: 241–249. 10.1016/j.plantsci.2013.06.003 . [DOI] [PubMed] [Google Scholar]
- 3.Mitchell PL, Sheehy JE. (2006) Supercharging rice photosynthesis to increase yield. New phytol 171: 688–693. 10.1111/j.1469-8137.2006.01855.x . [DOI] [PubMed] [Google Scholar]
- 4.Lange BM, Ghassemian M. (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51: 925–948. 10.1023/A:1023005504702 . [DOI] [PubMed] [Google Scholar]
- 5.Masuda T, Fujita Y. (2008) Regulation and evolution of chlorophyll metabolism. Photochemical &Photobiological Sci 7: 1131–1149. 10.1039/b807210h . [DOI] [PubMed] [Google Scholar]
- 6.Eckhardt U, Grimm B, Hortensteiner S. (2004) Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol Biol 56: 1–14. 10.1007/s11103-004-2331-3 . [DOI] [PubMed] [Google Scholar]
- 7.Liu W, Fu Y, Hu G, Si H, Zhu L, Wu C, Sun Z. (2007) Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L). Planta 226: 785–795. 10.1007/s00425-007-0525-z . [DOI] [PubMed] [Google Scholar]
- 8.Larkin RM, Alonso JM, Ecker JR, Chory J. (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906. 10.1126/science.1079978 . [DOI] [PubMed] [Google Scholar]
- 9.Zhou S, Sawicki A, Willows RD, Luo M. (2012) C-terminal residues of oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Letters 586: 205–210. 10.1016/j.febslet.2011.12.026 . [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Li J, Yoo JH, Yoo SC, Cho SH, Koh HJ, Seo HS, Paek NC. (2006) Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol 62: 325–337. 10.1007/s11103-006-9024-z . [DOI] [PubMed] [Google Scholar]
- 11.Islam MR, Watanabe K, Kashino Y, Satoh K, Koike H. (2013) Spectral properties of a divinyl chlorophyll a harboring mutant of Synechocystis sp. PCC6803. Photosynthesis Res 117: 245–255. 10.1007/s11120-013-9877-3 . [DOI] [PubMed] [Google Scholar]
- 12.Ito H, Tanaka A. (2011) Evolution of a divinyl chlorophyll-based photosystem in Prochlorococcus. Proc Natl Acad Sci USA 108: 18014–18019. 10.1073/pnas.1107590108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez F, Garrido JL, Sobrino C, Johnsen G, Riobo P, Franco J, Aamot I, Ramilo I, Sanz N, Kremp A. (2016) Divinyl chlorophyll a in the marine eukaryotic protist Alexandrium ostenfeldii (Dinophyceae). Environmental Microbiology 18: 627–643. 10.1111/1462-2920.13042 . [DOI] [PubMed] [Google Scholar]
- 14.Espineda CE, Linford AS, Devine D, Brusslan JA. (1999) The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 10507–10511. 10.1073/pnas.96.18.10507 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper AL, von Gesjen SE, Linford AS, Peterson MP, Faircloth RS, Thissen MM, Brusslan JA. (2004) Chlorophyllide a Oxygenase mRNA and Protein Levels Correlate with the Chlorophyll a/b Ratio in Arabidopsis thaliana. Photosynthesis Res 79: 149–159. 10.1023/B:PRES.0000015375.40167.76 . [DOI] [PubMed] [Google Scholar]
- 16.Lee S, Kim JH, Yoo ES, Lee CH, Hirochika H, An G. (2005) Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol Biol 57: 805–818. 10.1007/s11103-005-2066-9 . [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Xu J, Huang L, Leng Y, Dai L, Rao Y, Chen L, Wang Y, Tu Z, Hu J, Ren D, Zhang G, Zhu L, Guo L, Qian Q, Zeng D. (2016) PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. Journal of Experimental Botany 67: 1297–1310. 10.1093/jxb/erv529 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Sun X, Deng X. (2010) Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice. Plant Physiol 153: 994–1003. 10.1104/pp.110.158477 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Wan C, Xu Z, Wang P, Wang W, Sun C, Ma X, Xiao Y, Zhu J, Gao X, Deng X. (2013) One divinyl reductase reduces the 8-vinyl groups in various intermediates of chlorophyll biosynthesis in a given higher plant species, but the isozyme differs between species. Plant Physiol 161: 521–534. 10.1104/pp.112.208421 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z, Zhang X, He B, Diao L, Sheng S, Wang J, Guo X, Su N, Wang L, Jiang L, Wang C, Zhai H, Wan J. (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145: 29–40. 10.1104/pp.107.100321 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinta V, Picaud M, Reiss-Husson F, Astier C. (2002) Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. Journal of Bacteriology 184: 746–753. 10.1128/JB.184.3.746-753.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moseley J, Quinn J, Eriksson M, Merchant S. (2000) The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J 19: 2139–2151. 10.1093/emboj/19.10.2139 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moseley JL, Page MD, Alder NP, Eriksson M, Quinn J, Soto F, Theg SM, Hippler M, Merchant S. (2002) Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688. 10.1105/tpc.010420 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng CC, Porat R, Lu P, O'Neill SD. (1998) PNZIP is a novel mesophyll-specific cDNA that is regulated by phytochrome and the circadian rhythm and encodes a protein with a leucine zipper motif. Plant physiol 116: 27–35. 10.1104/pp.116.1.27 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang YT, Yu YL, Yang GD, Zhang JD, Zheng CC. (2009) Tissue-specific expression of the PNZIP promoter is mediated by combinatorial interaction of different cis-elements and a novel transcriptional factor. Nucleic Acids Res 37, 2630–2644. 10.1093/nar/gkp126 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rzeznicka K, Walker CJ, Westergren T, Kannangara CG, von Wettstein D, Merchant S, Gough SP, Hansson M. (2005) Xantha-l encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc Natl Acad Sci USA 102: 5886–5891. 10.1073/pnas.0501784102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bollivar D, Braumann I, Berendt K, Gough SP, Hansson M. (2014) The Ycf54 protein is part of the membrane component of Mg-protoporphyrin IX monomethyl ester cyclase from barley (Hordeum vulgare L.). FEBS J 281: 2377–2386. 10.1111/febs.12790 . [DOI] [PubMed] [Google Scholar]
- 28.Bang WY, Jeong IS, Kim DW, Im CH, Ji C, Hwang SM, Kim SW, Son YS, Jeong J, Shiina T, Bahk JD. (2008) Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant and Cell Physiol 49: 1350–1363. 10.1093/pcp/pcn111 . [DOI] [PubMed] [Google Scholar]
- 29.Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Scheller HV, Merchant S, Jensen PE. (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA 100: 16119–16124. 10.1073/pnas.2136793100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kauss D, Bischof S, Steiner S, Apel K, Meskauskiene R. (2012) FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg(++)-branch of this pathway. FEBS Letters 586: 211–216. [DOI] [PubMed] [Google Scholar]
- 31.Kong WY, Yu XW, Chen HY, Liu LL, Xiao YJ, Wang YL, Wang CL, Lin Y, Yu Y, Wang CM, Jiang L, Zhai HQ, Zhao ZG and Wan JM. (2016) The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice. Plant Mol Biol 92: 177–191. 10.1007/s11103-016-0513-4 . [DOI] [PubMed] [Google Scholar]
- 32.Inada N, Sakai A, Kuroiwa H, Kuroiwa T. (1998) Three-dimensional analysis of the senescence program in rice (Oryza sativa L.) coleoptiles. Investigations of tissues and cells by fluorescence microscopy. Planta 205: 153–164. 10.1007/s004250050307 . [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Shen BZ, Dai XK, Mei MH, Saghai MA, Li ZB. (1994) Using bulked extremes and recessive classes to map genes for photoperiod-sensitive genic male sterility in rice. Proc. Natl Acad. Sci. 91: 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen SB, Tao LZ, Zeng LR, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427. 10.1111/j.1364-3703.2006.00346.x . [DOI] [PubMed] [Google Scholar]
- 35.Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. (1996) Engineered GFP as a vital reporter in plants. Current Biol 6: 325–330. 10.1016/S0960-9822(02)00483-9 . [DOI] [PubMed] [Google Scholar]
- 36.Jung KH, Hur J, Ryu CH, Choi Y, Chung YY, Miyao A, Hirochika H, An GH. (2003) Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant and Cell Physiol 44: 463–472. 10.1093/pcp/pcg064 . [DOI] [PubMed] [Google Scholar]
- 37.Zhu XY, Guo S, Wang ZW, Du Q, Xing YD, Zhang TQ, Shen WQ, Sang XC, Ling YH, He GH. (2016) Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa). BMC Plant Biol 16: 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou K, Ren Y, Zhou F, Wang Y, Zhang L, Lyu J, Wang Y, Zhao S, Ma W, Zhang H, Wang L, Wang C, Wu F, Zhang X, Guo X, Cheng Z, Wang J, Lei C, Jiang L, Li Z, Wan J. (2016) Young Seedling Stripe 1 encodes a chloroplast nucleoid-associated protein required for chloroplast development in rice seedlings. Planta 244: 1–16. [DOI] [PubMed] [Google Scholar]
- 39.Dong H, Fei GL, Wu CY, Wu FQ, Sun YY, Chen MJ, Ren YL, Zhou KN, Cheng ZJ, Wang JL, Jiang L, Zhang X, Guo XP, Lei CL, Su N, Wang HY, Wan JM. (2013) A Rice Virescent-Yellow Leaf Mutant Reveals New Insights into the Role and Assembly of Plastid Caseinolytic Protease in Higher Plants. Plant Physiol 162: 1867–1880. 10.1104/pp.113.217604 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kusumi K, Hirotsuka S, Shimada H, Chono Y, Matsuda O, Iba K. (2010) Contribution of chloroplast biogenesis to carbon-nitrogen balance during early leaf development in rice. Plant Res 123: 617–622. 10.1007/s10265-009-0277-x . [DOI] [PubMed] [Google Scholar]
- 41.Song J, Wei XJ, Shao GN, Sheng ZH, Chen DB, Liu CL, Jiao GA, Xie LH, Tang SQ, Hu PS. (2014) The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions. Plant Mol Biol 84: 301–314. 10.1007/s11103-013-0134-0 . [DOI] [PubMed] [Google Scholar]
- 42.Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K. (2007) The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J 52: 512–527. 10.1111/j.1365-313X.2007.03251.x . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The RNA-seq data have been submitted to the database Gene Expression Omnibus (GEO), and the accession number is GSE96073.