Abstract
The roles of UvrD and Rep DNA helicases of Escherichia coli are not yet fully understood. In particular, the reason for rep uvrD double mutant lethality remains obscure. We reported earlier that mutations in recF, recO or recR genes suppress the lethality of uvrD rep, and proposed that an essential activity common to UvrD and Rep is either to participate in the removal of toxic recombination intermediates or to favour the proper progression of replication. Here, we show that UvrD, but not Rep, directly prevents homologous recombination in vivo. In addition to RecFOR, we provide evidence that RecA contributes to toxicity in the rep uvrD mutant. In vitro, UvrD dismantles the RecA nucleoprotein filament, while Rep has only a marginal activity. We conclude that UvrD and Rep do not share a common activity that is essential in vivo: while Rep appears to act at the replication stage, UvrD plays a role of RecA nucleoprotein filament remover. This activity of UvrD is similar to that of the yeast Srs2 helicase.
Keywords: helicase, RecA, RecF, Rep, UvrD
Introduction
DNA helicases are present in all kingdoms of life, and are probably important in most reactions involving DNA such as replication, repair, recombination and transcription. Their DNA unwinding activities are well characterized at the biochemical and structural levels (for a review, see Delagoutte and von Hippel, 2002a, 2002b). However, much remains to be understood about their roles in vivo. In particular, UvrD and Rep are two Escherichia coli DNA helicases of the SF1 family that share 40% amino-acid identity, and that are also remarkably similar to the PcrA helicase of Gram-positive bacteria. Some well-defined activities of UvrD, PcrA and Rep have been characterized both in vivo and in vitro. (i) They participate in the replication of various extragenic elements. The Rep helicase is needed during bacteriophages M13 and ΦX174 replication (Takahashi et al, 1979), the UvrD helicase ensures the replication of Gram-negative rolling-circle plasmids (Bruand and Ehrlich, 2000) and the PcrA helicase ensures the replication of Gram-positive rolling-circle plasmids (Petit et al, 1998; Anand et al, 2004). All these elements share the common characteristic that replication of their leading and lagging strands is uncoupled, and the helicase mediates DNA unwinding ahead of the DNA polymerase during leading strand replication. (ii) UvrD has a role in UV repair where it allows the removal of a 12-nt-long DNA segment containing a UV lesion, after its incision by the combined action of UvrA, UvrB and UvrC (Orren et al, 1992). This activity is efficiently complemented by PcrA (Petit et al, 1998). (iii) UvrD is involved in mismatch repair, where it promotes the removal of the DNA segment containing the erroneous nucleotide after its incision by the combined action of MutS, MutL and MutH (Modrich, 1994).
Other phenotypes of the rep, uvrD and pcrA mutants are less well understood: (i) Replication fork progression is twice slower in a rep mutant than in the Rep+ strain (Lane and Denhardt, 1975; Colasanti and Denhardt, 1987). (ii) Homologous recombination is increased in a uvrD mutant (Zieg et al, 1978; Arthur and Lloyd, 1980; Bierne et al, 1997a) and decreased in a UvrD overproducing strain (Maples and Kushner, 1982; Petranovic et al, 2001). Increased recombination in the uvrD mutant could be due to a replication defect, as reported for some replication mutants (Bierne et al, 1997b; Flores et al, 2001), or to a direct role of UvrD as an antirecombinase as was found in vitro (Morel et al, 1993). (iii) The pcrA single mutant of Staphylococcus aureus and of Bacillus subtilis is dead (Iordanescu, 1993; Petit et al, 1998), as is the rep uvrD double mutant of E. coli (Taucher-Scholtz et al, 1983).
Previously, we took a genetic approach to investigate the essential role of PcrA in B. subtilis, and UvrD and Rep in E. coli. We reported that the heterologous expression of pcrA in the E. coli rep uvrD double mutant strain restored its viability (Petit et al, 1998). This led to the hypothesis that pcrA, and rep or uvrD shared a common important function in vivo. We then found that lethality of the pcrA mutant is suppressed by mutations in the recF, recO, recL or recR genes of B. subtilis. Remarkably, in E. coli, lethality of the uvrD rep double mutant was also suppressed by mutations in the recF, recO or recR genes (Petit and Ehrlich, 2002).
The key protein for homologous recombination in bacteria, RecA, is loaded onto single-stranded DNA (ssDNA) through the mediation of either one of the two complexes, RecFOR or RecBCD (Kuzminov, 1999). Each complex acts and processes a specific DNA substrate so as to permit RecA binding: RecFOR proteins promote the formation of RecA nucleofilaments on ssDNA gaps covered with SSB (Umezu et al, 1993; Webb et al, 1997; Morimatsu and Kowalczykowski, 2003). RecBCD enzymatic complex is a double-stranded DNA (dsDNA) exonuclease that converts into a recombination enzyme upon encountering a specific DNA sequence, CHI (Kowalczykowski et al, 1994). Its entry point into DNA is a double-stranded end. The suppression of helicase mutations lethality by recF, recO or recR mutations suggested a toxic effect due to the RecFOR recombination proteins in the helicase mutants. Two formal possibilities were proposed to explain these data (Petit and Ehrlich, 2002). The helicases may act either during the recombination process to reverse toxic-blocked recombination intermediates, or at a step upstream of recombination, possibly during replication. In the latter case, their absence would allow recombination to occur at a high and toxic level. The question of whether or not Rep and UvrD are acting at the same step in a redundant fashion was left unanswered.
The Srs2 helicase of Saccharomyces cerevisiae is the closest found orthologue to UvrD, Rep and PcrA in eukaryotes (Aboussekhra et al, 1989). Like the uvrD mutant, the srs2 mutant exhibits a high rate of spontaneous recombination (Aguilera and Klein, 1988). A number of srs2 mutant phenotypes are suppressed by mutations that prevent formation of Rad51 nucleofilaments. These include UV sensitivity of single srs2 mutants (Aboussekhra et al, 1992) and the synthetic lethality or sickness of certain double mutants involving srs2, like the srs2 sgs1 double helicase mutant (Gangloff et al, 2000; Klein, 2001; Fabre et al, 2002). These observations led to the idea that one role of Srs2 is to prevent the formation of toxic recombination intermediates by eliminating inappropriate Rad51 nucleofilaments. In vitro data showed that Srs2 is indeed able to perform this activity (Krejci et al, 2003; Veaute et al, 2003).
The suppression of srs2 phenotypes by mutations in genes controlling the Rad51 pathway is reminiscent of the suppression of the lethality of pcrA or rep uvrD mutants by mutations affecting the RecF pathway. The aim of this work was to examine the possibility that UvrD, Rep or both helicases act in a way similar to Srs2, by disrupting RecA filaments. An in vivo assay based on conjugation, allowing the concomitant measurement of replication and recombination activity, showed that the rep and uvrD mutants behaved differently, with Rep acting principally on replication and UvrD on recombination intermediates. We also gained evidence that in addition to RecFOR the RecA protein was toxic in the rep uvrD mutant. Finally, the effects of the Rep and UvrD helicases on in vitro strand exchange assays as well as their ability to disrupt preformed RecA nucleoprotein filaments were studied. We found that UvrD, but not Rep, dismantles the RecA nucleoprotein filament. Our set of results supports the view that the lethality of the rep uvrD mutant is not due to a redundant function of these two helicases, but rather that UvrD is essential for the viability of the rep mutant. This suggests that toxic RecA filaments are formed in the rep mutant, possibly at the fork, and need to be removed by UvrD.
Results
In vivo monitoring of replication and recombination in real time
Because the UvrD and Rep helicases were suspected to affect recombination and/or replication in vivo, and because both processes are often intimately connected, an experimental system was set up for concomitant monitoring of replication and recombination as a function of time. It is based on Hfr conjugation, a process during which chromosomal DNA is transferred from a donor to a recipient strain. DNA enters the recipient strain as a single strand starting from a 5′ extremity, and is converted into dsDNA by replication. Replication depends on DNA polymerase III, the main replicative polymerase, and also on DnaB, the replicative helicase (Willetts and Wilkins, 1984; our unpublished data). The role of DnaB is likely to allow primase loading, but nothing is known about the way DnaB itself is loaded. Once replicated, DNA recombines at high rates with the recipient chromosome by homologous recombination (up to 50% of recipient cells may integrate a given allele). To follow replication and recombination in the recipient cell, two slightly different Hfr donors were constructed, and used with recipient strains containing the helicase mutation to be tested.
The replication assay is depicted in Figure 1A. The donor contained a lacIs mutation, which encodes a hyper-repressor LacIs molecule (Willson et al, 1964). As a consequence, the donor strain, although harbouring lacZ, produces a low level of β-galactosidase in the presence of the IPTG inducer (see Figure 2B, the ‘control’ curve is Hfr3000 lacIs). When the lacIs-lacZ region of the Hfr donor enters the recipient strain, which is LacZ−, it is first replicated, and then lacZ is fully transcribed, for about 1 h (see Figure 2B), the time needed for LacIs to accumulate and repress lacZ. This is designated ‘zygotic induction’. During this time period, the β-galactosidase activity of extracts mostly depends on the replication efficiency.
Figure 1.
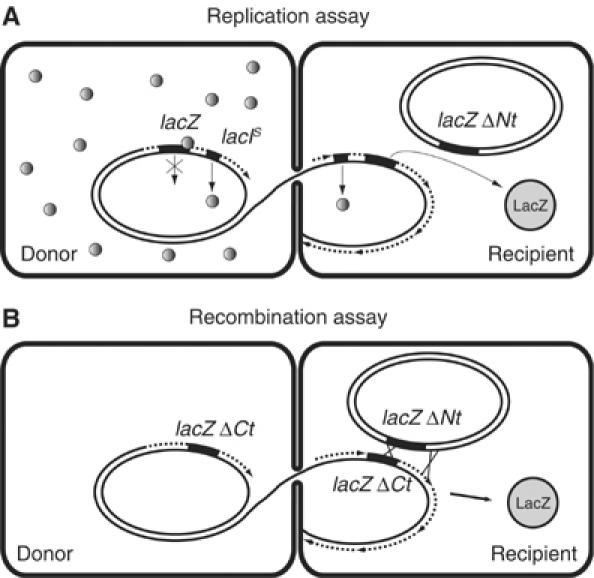
(A) Scheme of the replication assay. ssDNA is passing from the donor cell (left side) to the recipient cell (right side), in which it is converted to dsDNA (dotted arrow lines, oriented 5′–3′). In the donor strain (left side), the LacIs protein (small grey dots) represses strongly the lacZ promoter, so that almost no LacZ is produced. In the recipient strain, as soon as the incoming DNA is replicated, a burst of LacZ synthesis occurs (big grey circle), due to the initial absence of LacIs. After a 1 h delay, sufficient amounts of LacIs are produced to shut down lacZ transcription. (B) Scheme of the recombination assay. Recombination occurs between the incoming lacZΔCt allele and the lacZΔNt allele of the recipient chromosome (black boxes), and leads to a functional gene and enzyme production (big grey circle).
Figure 2.
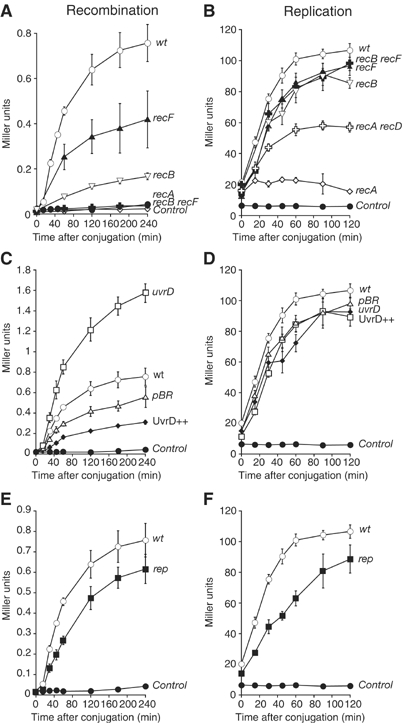
Kinetics of recombination (A, C, E) and replication (B, D, F) after conjugation in various mutants. Replication and recombination were monitored by using donor strains Nec226 for the replication assay and Nec224 for the recombination assay, as described in the text. All experiments were repeated at least three times. The recipient strains are designated by an alias drawn near each curve: ‘wt’ Nec223 (○), ‘recA’ Nec229 (◊), ‘recB’ Nec228 (∇), ‘recF’ Nec227 (▴), ‘recB recF’ Nec232 (✗), ‘recA recD’ Nec230 ( ), ‘uvrD’ MAC1058 (□), ‘pBR’ MAC1068 (Δ), ‘UvrD++’ MAC1071 (⧫) and ‘rep’ MAC1065 (▪). The ‘control’ curves in panels A, C and E (•) represent the cross between two identical lacZΔP alleles, Nec223 and Nec225 strains, which cannot result in a Lac+ recombinant. The ‘control’ curve in panels B, D and E represents the Hfr3000 lacIs donor strain without any recipient strain (•): it indicates the amount of transcription leakage in the presence of IPTG, with the lacIs allele (around 5 units). After a 40 min conjugation period on filter, β-galactosidase activity of cell extracts was measured as a function of incubation time at 28°C.
The recombination assay, which is depicted in Figure 1B, was inspired from an earlier report (Birge and Low, 1974), except that other lacZ alleles were used. The donor strain contained a lacZ C-terminal deletion, and shared 2.7 kb homology within lacZ with the recipient strain, which contained a lacZ N-terminal deletion. When the ΔlacZ region from the donor enters the recipient strain, if it replicates and recombines into the 2.7 kb lacZ region with the recipient chromosome, an intact gene is restored. As a result, the β-galactosidase activity of the extracts will depend on both replication and recombination efficiencies.
Before testing helicase mutants, strains mutated in recA, recB and recF genes, three major players in homologous recombination, were monitored for recombination during 4 h after conjugation (Figure 2A). The recA mutant exhibited no recombination activity, as expected (Birge and Low, 1974). The recB mutant exhibited a strong defect in recombination (10% of wild-type (WT) activity, as measured by comparing slopes) (Table I) and the recF mutant a moderate defect (30% of WT). In the double recB recF mutant, recombination was completely abolished. These observations are also in agreement with previously reported data (Birge and Low, 1974; Lloyd et al, 1987b), and confirm that both RecF and RecB promote recombination in this assay. We concluded that the assay was reliable to examine recombination in real time.
Table 1. Quantifications of the kinetics of recombination and replication as shown Figure 1.
| Recombinationa |
Replicationb |
Recomb./Replic. | |||
|---|---|---|---|---|---|
| Ratio/wt | Ratio/pBR | Ratio/wt | Ratio/pBR | ||
| wt | 1.00 | 1.00 | 1.00 | ||
| uvrD | 1.94 | 0.93 | 2.09 | ||
| Rep | 0.58 | 0.60 | 0.98 | ||
| recF | 0.31 | 0.92 | 0.34 | ||
| recB | 0.10 | 0.82 | 0.12 | ||
| recB recF | 0.01 | 0.78 | 0.01 | ||
| pBR | 0.63 | 1.00 | 0.87 | 1.00 | |
| UvrD++ | 0.55 | 0.79 | 0.70 | ||
| aThe slopes of the kinetics were measured between 15 and 60 min, except for strains recB, recF, and recB recF, where values between 0 and 120 min were taken. R-values were between 0.93 and 0.99. Each slope value was then divided by the slope value of the WT strain (column ratio/wt), or of the strain containing pBR322 (column ratio/pBR). | |||||
| bSlopes were measured between 0 and 60 min. R-values ranged between 0.92 and 0.99. | |||||
In a similar way, dnaEts (dnaE encodes the α-subunit of DNA polymerase III) and dnaBts (dnaB encodes the replicative helicase) mutants were used to measure replication with our assay. β-Galactosidase activity was reduced compared to the WT strain (Delmas and Matic, manuscript in preparation). We then proceeded to analyse replication (Figure 2B) in the rec strains. Compared to the WT strain, replication was slightly affected in the recB, recF and recB recF mutants (80–90% of WT) (Table I). The difference of β-galactosidase activity between the recB recF strain, in which no recombination took place, and the WT strain, in which incoming DNA did recombine, could reflect the contribution of recombination events to total β-galactosidase activity. In this case, the contribution of recombination is low and does not mask the replication efficiency. In sharp contrast with the recB recF strain, almost no β-galactosidase activity was detected in the recA strain extracts. Rather than reflecting a replication defect, we suspect that this is due to efficient degradation of replicated DNA by RecBCD in the absence of RecA, as previously observed (Skarstad and Boye, 1993). Indeed, in the recA recD double mutant, which is devoid of RecBCD exonuclease activity, β-galactosidase activity was partially restored. We concluded that the replication assay was a reliable means of detecting the fate of incoming DNA in the recipient strain, reflecting both its replication and its degradation, with little interference, if any, of recombination events.
UvrD directly prevents homologous recombination, and Rep promotes replication in vivo
Replication and recombination were then monitored in a uvrD mutant. We found that recombination was increased by a factor of two as compared to the WT strain (Figure 2C and Table I). In the replication assay, β-galactosidase activity in the uvrD strain was only slightly diminished (93% of WT), thus showing that the uvrD strain did not suffer any major replication defect. A strain overproducing UvrD was also tested (Figure 2C, UvrD++); replication was slightly affected (80% of WT) in this context, while recombination was reduced by a factor of two, as compared to the WT strain harbouring the control plasmid pBR322. Taken together, these data suggest that UvrD does not participate in DNA replication, but primarily and directly prevents recombination in vivo.
Replication and recombination were also analysed in a rep mutant. Replication was reduced (60% of WT; Figure 2F and Table I), suggesting that Rep is needed for replication of incoming DNA. Recombination was reduced as well (60% of WT; Figure 2E and Table I); this effect may simply be due to the decrease of replication.
As helicases are known to play various roles in a cell, it seemed important to test whether transcription and translation of the lacZ gene were affected in the uvrD and rep mutants and in the UvrD overproducing strain. Upon IPTG induction, lacZ expression was similar in WT, uvrD, rep and UvrD overproducing strains (not shown). We conclude that neither the helicase mutants nor the UvrD overproducing strain interfere with transcription/translation of lacZ.
The recA730 mutation severely affects viability of the rep uvrD recF mutant
Genetic data revealed that lethality of the uvrD rep double mutant is suppressed by mutations in recF, recO or recR genes (Petit and Ehrlich, 2002). The RecFOR proteins promote formation of RecA nucleofilaments on ssDNA gaps (Umezu et al, 1993; Morimatsu and Kowalczykowski, 2003). In uvrD rep cells, we suspected that the cause of toxicity may be unprocessed RecA nucleoprotein filaments themselves. In this scheme, the lethality of the rep uvrD mutant should be suppressed also by a recA mutation, but we have already shown that this was not the case (Petit and Ehrlich, 2002). The recA gene has a second role in E. coli, that is, induction of the SOS response, by stimulating cleavage of the LexA repressor. We therefore asked whether a strain in which the SOS response is constitutively expressed, due to inactivation of lexA, would allow rep uvrD recA cells to be viable. To test this, an experiment was conducted with the rep-encoding plasmid pAMrep, whose replication (based on pAM34; Gil and Bouche, 1991) relies on an IPTG-dependent promoter. The principle consists of constructing strains with the desired mutations in the presence of the plasmid providing Rep, to overcome the putative lethality of the rep uvrD combination. The plasmid is then chased from the strain by growing cells in the absence of IPTG. If cells having segregated the plasmid continue to grow, the resulting mutant is viable. The results of segregation experiments are presented in Supplementary Figure 1S. It was not possible to recover plasmidless cells from the lexA rep uvrD recA pAMrep strain grown at 37°C (MAC1107), indicating that, even in an SOS constitutive background, the rep uvrD recA combination is lethal. The rep recA double mutant was reported to plate efficiently at 42°C and not at 30°C (Bredeche et al, 2001). The segregation experiment was therefore repeated at 42°C for the rep recA lexA uvrD and its RecA+parent. Again, no plasmidless cell could be recovered at 42°C in this background. However, to our surprise, the lexA rep uvrD parent strain was viable at 42°C (see Discussion). Therefore, while removal of the RecFOR complex is beneficial in the rep uvrD strain, RecA is essential in this context. RecA filaments are formed with the help of either RecBCD or RecFOR. In uvrD rep mutants, the recF, recO and recR mutations might prevent the formation of RecA filaments at some places where they would be toxic, but not on RecBCD-generated structures that would require RecA for repair. This would explain the differential effect of recF and recA mutations on uvrD rep lethality.
To test the hypothesis that toxicity of RecFOR in uvrD rep mutant relates to RecA binding, we used the recA730 allele that is a partial suppressor of recF (Wang et al, 1993). Suppression is due to the ability of RecA730 to compete with SSB, thereby bypassing a need for RecFOR to do so (Lavery and Kowalczykowski, 1992). We asked whether the rep uvrD recF strain (MAC1136) would be affected by addition of the recA730 allele (strain MAC1146). This proved to be the case: the rep uvrD recF recA730 mutant was still viable but it grew more slowly than the RecA+ isogenic strain (Figure 3), and also accumulated suppressors. We therefore concluded that the RecA nucleoprotein filament was a factor of toxicity in the rep uvrD background.
Figure 3.
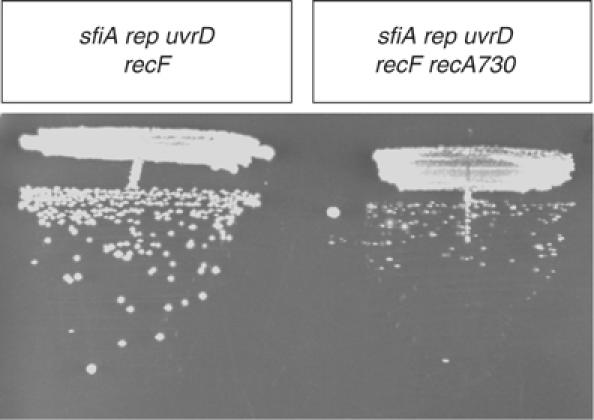
The sfiA rep uvrD recF recA730 (MAC1159) grows poorly compared to its recA+ parent (MAC1168). Both strains were streaked on an LB plate and incubated for 48 h at 37°C.
UvrD but not Rep inhibits RecA-mediated strand exchange in vitro
The above in vivo experiments led to the view that UvrD helicase, but not Rep helicase, plays a role directly connected with homologous recombination, and eventually with RecA itself. To test this hypothesis more directly, we compared the effects of UvrD and Rep helicases on an in vitro RecA-promoted DNA strand exchange reaction. In this assay, circular ΦX174 ssDNA was first incubated for 10 min with RecA and SSB proteins to form the nucleoprotein filament. The helicase and 5′ end-labelled MfeI-linearized ΦX174 dsDNA were then sequentially introduced (Figure 4A). In agreement with an earlier report (Morel et al, 1993), we found that addition of increasing amounts of UvrD to the reaction strongly inhibited the strand transfer. Almost no nicked circular duplex was detected at a UvrD concentration of 600 nM (Figure 4B and D). In contrast, Rep protein did not affect strand exchange reaction even at concentrations up to 1.4 μM (Figure 4C and D).
Figure 4.
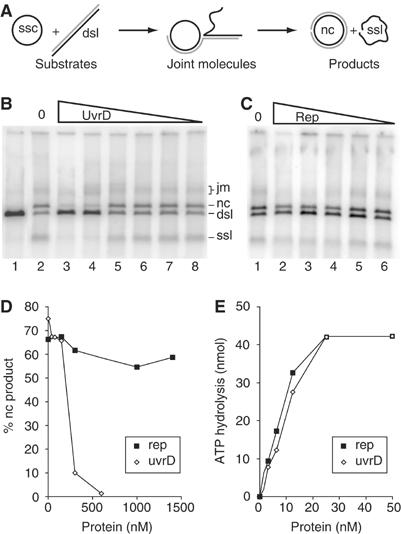
UvrD helicase but not Rep helicase inhibits strand exchange catalysed by RecA. (A) Scheme of DNA strand exchange reaction (ssc, single-stranded circular DNA; dsl, double-stranded linear DNA; jm, joint molecules; nc, nicked circular double-stranded DNA; ssl, single-stranded linear DNA). (B) After RecA nucleoprotein filament formation by preincubation of ssDNA with RecA and SSB proteins, various amounts of UvrD helicase were added simultaneously with 32P end-labelled linear dsDNA, which initiates the strand exchange (lane 1: labelled dsl; lanes 2–7 correspond to 0, 600, 300, 150, 75, 37.5 and 18.75 nM UvrD, respectively). After incubation for 40 min at 37°C, reaction mixture was deproteinized and resolved onto a 0.8% agarose gel. (C) Same as in (B) except that Rep was used in place of UvrD (lanes 1–6 correspond to 0, 1400, 1000, 600, 300 and 150 nM Rep, respectively). (D) Quantification of the reactions shown in panels B and C. (E) Comparable ATPase activities of UvrD and Rep proteins. Reactions containing 1 mM ATP, saturating amount of synthetic oligo(dT)55 (5.5 μM nucleotides) and increasing amount of proteins were incubated at 37°C for 15 min. The calculated amount of ATP hydrolysed linked to the oxidation of NADH was determined by measuring the OD at 340 nm.
We observed an abrupt change between 150 and 300 nM UvrD helicase concentrations. This effect was not due to ATP depletion because increasing three times the ATP regenerating system did not change the shape of the curve (data not shown).
Rep and UvrD were both similarly active in this assay, as revealed by their specific ATPase activities (Figure 4E). Thus, the fact that only UvrD inhibits the strand exchange reaction reflects an intrinsic property of this helicase.
RecA–ssDNA nucleoprotein filament is efficiently dismantled by UvrD but not by Rep
The yeast Srs2 helicase performs its antirecombinase action by disrupting Rad51 nucleoprotein filaments (Krejci et al, 2003; Veaute et al, 2003). We considered the possibility that similarly to Srs2, UvrD inhibits RecA recombinase function by releasing this protein from the presynaptic filament. We also hypothesized that Rep does not disrupt the RecA nucleoprotein filament because of the absence of any inhibitory effect on strand exchange. Electron microscopy was used to characterize the action of UvrD and Rep on the RecA presynaptic filament. RecA was first incubated for 5 min with ΦX174 ssDNA in the presence of SSB protein, in order to assemble nucleoprotein filaments (Figure 5A). The reaction was then diluted five times before introduction of the helicase. Subsequent to the addition of UvrD, we observed the appearance of ssDNA molecules covered by SSB (such molecules appear as small bushes, shown with a white arrow, as opposed to the well-spread RecA nucleofilaments), resulting from disruption of the RecA filament (Figure 5B–D). Dilution did not perturb the RecA filament (Figure 5D). Progressive loss of RecA filaments, coupled with concomitant formation of SSB–ssDNA complexes, was observed as a function of the amount of UvrD introduced into the reaction. At 200 nM UvrD, more than 95% of the observed DNA molecules were covered by SSB. This amount of UvrD is about 10 times lower than that of RecA, indicating that UvrD acts catalytically. In vivo, the UvrD concentration is predicted to range from 1.4 to 4 μM (1000–3000 molecules/cell) (George et al, 1994). Therefore, the range of UvrD concentrations active in vitro is relevant to the physiological conditions. The dilution step, after RecA filament assembly, was needed to see UvrD activity: when the helicase was added to the undiluted reaction, the same range of UvrD concentrations had no apparent effect. Only with a higher concentration of 1 μM UvrD could we detect partial RecA removal. We believe that, in the absence of dilution, a rapid reassociation of RecA with the ssDNA precludes SSB binding to ssDNA.
Figure 5.
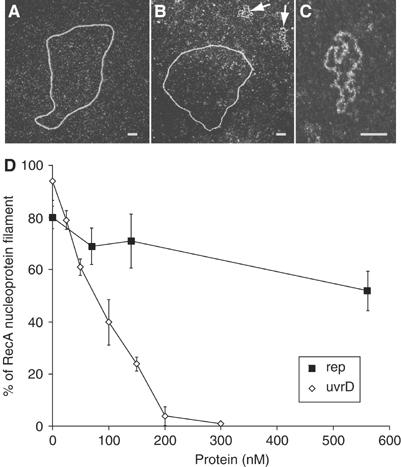
UvrD helicase, but not Rep helicase, efficiently disrupts RecA–ssDNA nucleoprotein filament. (A) RecA–ssDNA nucleoprotein filaments. (B) Preformed RecA–ssDNA complexes were incubated for 15 min with UvrD. The arrows point to the ssDNA covered with SSB. (C) Blow-up of ssDNA covered with SSB. (D) The percentage of disrupted RecA presynaptic filament was determined for different amounts of both helicases. For each concentration, mean values and standard deviations were obtained from two series of 300 molecules counted. Scale bars, 50 nm.
Unlike the effect of UvrD, introduction of the Rep helicase to the reaction did not severely perturb the RecA presynaptic filaments. Large amounts of Rep were needed to trigger a small effect; less than 30% of the nucleoprotein filaments were dislocated after incubation with 560 nM Rep (Figure 5D). In vivo, the Rep concentration is predicted to be around 70 nM (50 molecules/cell) (Scott and Kornberg, 1978). This suggests that the effect of Rep in vitro does not reflect a physiological function of this helicase.
Taken together, the data from strand exchange experiments and from electron microscopic analyses indicate that the UvrD but not the Rep helicase acts as an antirecombinase by disrupting RecA nucleoprotein filaments.
Discussion
We found that UvrD, but not Rep, removes RecA from DNA. Keeping this in mind, some of the uvrD phenotypes may be interpreted as follows: the hyper-recombination nature of the uvrD mutant would result from a shift towards a higher level of recombination products, because RecA nucleoprotein filaments are not removed by UvrD and the essential activity of UvrD in the rep mutant would be due to its role as ‘RecA remover’ at a critical place or moment in the cell.
UvrD removes RecA from DNA
We report here that UvrD helicase disrupts RecA nucleofilament in vitro. Morel et al (1993) previously showed that UvrD could block the progression of a RecA-mediated strand exchange reaction, and proposed that the role of UvrD was to unwind the dsDNA and thereby reverse the reaction. Our electron microscopic studies suggest rather that strand exchange is stopped because RecA is wiped away from DNA. This model provides a simple explanation for many earlier observations, showing that uvrD mutants are hyper-recombinogenic. Until the impact of the uvrD mutation on DNA replication was studied, one could not exclude the possibility that the absence of UvrD impedes DNA replication, leading to frequent recombinogenic structures, as observed for various replication mutants (Bierne et al, 1997b; Flores et al, 2001). Our in vivo system, based on DNA conjugation, brings evidence that replication is almost unperturbed in the uvrD mutant, while recombination is increased. We can therefore propose a new role for UvrD in vivo, which is to remove RecA nucleoprotein filaments. UvrD had already been found to remove proteins from DNA, but always in a situation coupled with DNA unwinding: in the context of UvrABC-dependent UV repair, after the incision step mediated by UvrABC, the UvrD helicase was found to remove both the 12-mer containing the lesion and the UvrC protein (Orren et al, 1992). UvrD also removes the Tus protein bound to its ter site in vitro (Hiasa and Marians, 1992), the topoisomerase IV from its cleaved DNA–substrate intermediate (Howard et al, 1994) and LacI from its DNA operator site (Yancey-Wrona and Matson, 1992). The present work provides evidence that UvrD is also active through its translocase activity on ssDNA to remove RecA nucleoprotein filaments. More generally, DNA and RNA helicases may not limit themselves to a role in DNA or RNA unwinding, but bring also an activity of protein removers, while tracking on their RNA or DNA substrate (Jankowsky et al, 2001; Fairman et al, 2004). One may also interpret in this light the surprising recent observation that UvrD has a strand-switching activity, which results in reannealing the portion of dsDNA it has unwound (Dessinges et al, 2004). During such a strand switching in vivo, all RecA molecules bound, for example to a stalled replication fork, could be displaced, so that the final balance of the operation is non-nil (see below).
UvrD behaves like Srs2
Our results with UvrD generalize to prokaryotes what has been reported recently concerning the Srs2 helicase of S. cerevisiae: These two helicases remove RecA or Rad51 nucleoprotein filament, respectively. Furthermore, Srs2 helicase was shown to disrupt RecA nucleoprotein filaments (Krejci et al, 2003; Veaute et al, 2003). We found that UvrD can also dismantle presynaptic filaments of Rad51 (data not shown). UvrD and Srs2 helicases are therefore efficient at removing nucleoprotein filaments across species. Srs2 was found to interact physically with Rad51 (Krejci et al, 2003). However, the precise way these helicases recognize the filament structure remains to be determined.
Rep acts at the replication stage
We found that Rep cannot replace UvrD in the RecA displacement activity in vitro. In vivo, the behaviour of the rep mutant is completely different from that of the uvrD mutant. Replication of transferred DNA during conjugation is reduced in the rep mutant (Figure 2F). Chromosome replication forks are reportedly twice slower in a rep mutant (Lane and Denhardt, 1975; Colasanti and Denhardt, 1987). Based on the observation that Rep efficiently unwinds LacI-bound dsDNA in vitro (Yancey-Wrona and Matson, 1992), it is generally assumed that Rep acts ahead of the replicative DNA polymerase to remove protein road-blocks. However, during conjugative replication, the DNA ahead of the DNA polymerase is single stranded rather than double stranded, and there is no replication fork. It seems therefore difficult to assume that Rep acts at the same level ‘ahead of the fork’. Alternatively, the defect of the rep mutant could be related to a default of DnaB loading. Replication during conjugation depends on DnaB (Willetts and Wilkins, 1984), whose role is probably to bring the DnaG primase to the DNA. The proteins needed to load DnaB onto the DNA in this particular case are not known, but Rep, together with PriC, has been proposed as an alternate pathway for DnaB loading, in the absence of PriA (Sandler, 2000). A role of Rep during Okazaki fragment synthesis cannot be excluded, but so far our attempts to detect it have failed (data not shown). Moreover, strains defective for lagging strand synthesis are reportedly hyper-recombinogenic (Flores et al, 2001), which is not the case of the rep mutant. Clearly, understanding the role of Rep in vivo remains a challenge for the future.
What is the essential role of UvrD in the rep mutant?
Based on the above-mentioned observations, it seems unlikely that Rep and UvrD substitute for each other in an essential process in vivo, which leads to the death of the rep uvrD mutant. On the contrary, as Rep acts in replication, we suggest that, in its absence, structures are generated that require UvrD for processing. Interestingly, it was recently reported that UvrD is needed for replication fork reversal in two different DNA polymerase III mutants (Flores et al, 2004). This points to the capacity for the UvrD helicase to be active at a fork. Fork reversal designates the capacity of a blocked fork to unwind its two nascent strands and anneal them together (Higgins et al, 1976). Such a process could permit replication restart. It was proposed to be frequent in the rep mutant (Seigneur et al, 1998). The direct study of fork reversal in the rep uvrD mutant is prevented by its inviability. However, if UvrD also acts in fork reversal in the rep mutant, and considering that in this mutant UvrD is important for the removal of RecA nucleoprotein filaments, the following scenario can be envisaged: (1) The replication fork is sometimes pausing due to the absence of Rep, and the replication apparatus collapses. (2) A particular DNA structure, unknown at present, is generated at the fork, containing ssDNA, which we will call ‘forked DNA’. (3) A RecA nucleoprotein filament is loaded by RecFOR on the single-stranded part of the ‘forked DNA’ and this complex cannot be further processed by recombination. (4) UvrD is essential at this moment to remove RecA, and permit either fork reversal or the loading of a new replication apparatus. The toxicity conferred by the recA730 mutation to the rep uvrD recF cells is explained by the ability of RecA730 protein to bind ssDNA at the ‘forked DNA’ in the absence of RecFOR. This scenario assumes that RecF, RecO and RecR can bind to ssDNA at a fork, which is different from a ‘bona fide’ single-stranded gap. In support of this view, it was observed that RecFOR protects newly replicated DNA extremities after UV irradiation (Courcelle et al, 1997; Chow and Courcelle, 2004).
A recF, but not a recA mutation suppresses rep uvrD lethality
The above-mentioned scenario does not explain why a recA mutation does not suppress rep uvrD lethality. If RecA had only a toxic effect in the rep uvrD background, its complete removal should save the strain. It could be that RecA is important for RecBCD-dependent recombination in the rep strain, in a situation unrelated to fork pausing. A simple possibility is that, due to the absence of UvrD, which plays a role in mismatch and UV repair, nicks are more frequent in the chromosome. Bradshaw and Kuzminov (2003) reported that replication ‘across’ such nicks leads to fork collapse, formation of double-stranded ends and subsequent repair by RecBCD and RecA. Although the uvrD recA double mutant is viable, it is possible that the additive rep defect provokes a cell crisis. Finally, a whole set of observations leads to the conclusion that growth at 42°C is beneficial to some helicase mutant derivatives: the rep recA strain, the rep uvrD strain (the observation made in the sfiA11 lexA rep uvrD was reproduced in the sfiA11 rep uvrD and rep uvrD strains: all strains, which were not viable at 37°C, could be constructed at 42°C, data not shown) and also the lexA uvrD recA strain (M-A Petit, unpublished observation). These observations are paradoxical given the fact that more replication forks are present at 42°C compared to 37°C, so that more replication pausing and more problems are expected. This is clearly an exciting field of investigations for the future.
In conclusion, our data show that the lethality in rep uvrD mutants is not a result of the overlapping functions of both helicases. UvrD, but not Rep, plays a role of RecA nucleoprotein filament remover in E. coli. The UvrD helicase proves therefore to play a new role, unrelated to DNA melting, in vivo. While studying the precise tracking mechanism of the PcrA helicase, Wigley and his collaborators had envisaged that an important part of the energy consumed while translocating may be used for some other purpose, like protein removal (Soultanas and Wigley, 2001). By extending the results obtained recently for the S. cerevisiae Srs2 helicase, which disrupts Rad51 nucleoprotein filaments (Krejci et al, 2003; Veaute et al, 2003), our data suggest effectively that DNA helicases play important roles as protein removers in vivo.
Materials and methods
Strains and plasmids
Strains and plasmids used are listed in Table II. Several mutations were constructed using a technique relying on the Lambda recombination genes (Datsenko and Wanner, 2000): A new uvrD null allele was needed for this study, because of the instability of the transposon insertion alleles, such as uvrD∷Tn5. The phleomycin resistance gene of pUB110, fused to the B. subtilis sacB promoter, was amplified by PCR from the pUC-phleo plasmid (a gift from E Dervyn), using two long primers containing also 50-nt-long regions homologous to the start and end regions of the uvrD gene. This fragment was then used to replace the WT uvrD gene, between positions −80 and +2600 relative to the +1 start codon of uvrD. For the same reason of transposon instability, a lexA∷KmR allele was also constructed, in which the lexA gene, between positions +1 and +608 relative to the +1 start codon of lexA, was replaced by the KmR gene of pKD4 (Datsenko and Wanner, 2000). Two precise deletion alleles of lacZ, used for the conjugation assays, lacZΔP and lacZΔT, were also constructed. In lacZΔP, the region between −138 and +10, relative to the +1 start codon of lacZ, was replaced by the CmR cassette of pKD3 (Datsenko and Wanner, 2000), and in lacZΔT, the same cassette was placed between +2798 and +3051 of lacZ. The lacZΔP deletion deleted also the C-terminal part of LacI. These alleles were transduced into the relevant strains by P1 transduction, and the CmR cassette was removed by site-specific recombination between FRT sites flanking the gene, using pCP20, as described (Datsenko and Wanner, 2000). To verify the introduction of the recA730 allele in strain MAC1146, two oligonucleotides ending at the position of the differing nucleotide (a G in WT recA, and an A in recA730, at +115 relative to the start codon) and specific for the WT recA or the recA730 allele were used in a PCR with a compatible downstream oligonucleotide.
Table 2. E. coli strains and plasmids.
| Strain | Relevant genotype or phenotype | Source/construction |
|---|---|---|
| MG1655 | Wild type | Radman laboratory stock |
| Nec220 | As MG1655 but NalR | Spontaneous NalR clone |
| JJC40 | AB1157 hsdR | B Michel laboratory stock |
| Hfr3000 | oriT near 0 min | Radman laboratory stock |
| CGSC6378 | lacI s | Willson et al (1964) |
| JJC451a | recF400∷Tn5 | B Michel |
| JJC760a | rep∷CmR (pGB2Tsrep) | B Michel |
| JJC1397a | sfiA11 | B Michel |
| JM103recA730 | recA730 srl∷Tn10 sfiA∷Tn5 | R Fuchs |
| GY5902 | ΔrecA306 srl∷Tn10 (miniF recA) | S Sommer |
| N2101 | recB268∷Tn10 | Lloyd et al (1987a) |
| DB1318 | recA938∷Tn9-200 | Wertman et al (1986) |
| CGSC7429 | recD1901∷Tn10 | Genetic stock centre |
| FR705 | proC∷Tn5 | Radman laboratory strain |
| Nec235 | proC∷Tn5 lacIs | P1 FR705*CGSC6378 |
| Nec221b | lacZΔP∷CmR | Red-Gam-mediated gene replacement |
| Nec222b | lacZΔT∷CmR | Red-Gam-mediated gene replacement |
| MAC833a | uvrD∷PhleoR | Red-Gam-mediated gene replacement |
| MAC810a | sfiA11 lexA∷KanR | Red-Gam-mediated gene replacement |
| MAC448a | rep∷CmR | P1 JJC760*JJC40 |
| Nec223b | lacZΔP | Excision of the CmR cassette from Nec221 |
| Nec227b | lacZΔP recF400∷Tn5 | P1 JJC451*Nec223 |
| MAC1058b | lacZΔP uvrD∷PhleoR | P1 MAC833*Nec223 |
| MAC1065b | lacZΔP rep∷CmR | P1 JJC760*Nec223 |
| MAC1068b | lacZΔP (pBR322) | Plasmid transformation |
| MAC1071b | lacZΔP (pGT26) | Plasmid transformation |
| Nec228b | lacZΔP recB268∷Tn10 | P1 N2101*Nec223 |
| Nec229b | lacZΔP recA938∷Tn9-200 | P1 DB1318*Nec223 |
| Nec231b | lacZΔP recD1901∷Tn10 | P1 CGSC7429*Nec223 |
| Nec230b | lacZΔP recA938∷Tn9-200 recD∷Tn10 | P1 DB1318*Nec231 |
| Nec232b | lacZΔP recB268∷Tn10 recF400∷Tn5 | P1 N2101*Nec227 |
| Nec224 | Hfr3000 lacZΔT∷CmR | P1 Nec221*Hfr3000 |
| Nec225 | Hfr3000 lacZΔP∷CmR | P1 Nec222*Hfr3000 |
| Nec226 | Hfr3000 proC∷Tn5 lacIs | P1 Nec235*Hfr3000 |
| MAC680a | sfiA11 rep∷CmR | P1 JJC760*JJC1397 |
| MAC1059a | sfiA11 rep∷CmR (pAMrep) | Plasmid transformation |
| MAC1077a | sfiA11 rep∷CmR uvrD∷PhleoR (pAMrep) | P1 MAC833*MAC1059 |
| MAC1089a | sfiA11 rep∷CmR uvrD∷PhleoR lexA∷KanR (pAMrep) | P1 MAC 810*MAC1077 |
| MAC1107a | sfiA11 lexA∷KanR rep∷CmR uvrD∷PhleoR ΔrecA306 srl∷Tn10 (pAMrep) | P1 GY5902*MAC1089 |
| MAC1136a | sfiA11 rep∷CmR uvrD∷PhleoR recF400∷Tn5 (pAMrep) | P1 JJC451*MAC1077 |
| MAC1146a | sfiA11 rep∷CmR uvrD∷PhleoR recF400∷Tn5 recA730 srl∷Tn10 (pAMrep) | P1 JM103recA730*MAC1136 |
| MAC1168a | sfiA11 rep∷CmR uvrD∷PhleoR recF400∷Tn5 | MAC1136 cured of pAMrep |
| MAC1159a | sfiA11 rep∷CmR uvrD∷PhleoR recF400∷Tn5 recA730 srl∷Tn10 | MAC1146 cured of pAMrep |
| MAC1165a | sfiA11 rep∷CmR uvrD∷PhleoR | MAC1077 cured of pAMrep at 42°C |
| Plasmids | ||
| pGT26 | pBR328 derivative encoding uvrD | Taucher-Scholz and Hoffmann-Berling (1983) |
| PAMrep | pAM34 derivative encoding rep | B Michel |
| aJJC40 background (AB1157 derivative). | ||
| bNec220 background (MG1655 derivative). | ||
Conjugational crosses and β-galactosidase assays
Overnight cultures were diluted 50-fold in LB for donor cells, or LB supplemented with IPTG (100 μM) for recipient cells, and grown to an OD530 of 0.9 (about 108 cells/ml). When the recipient strain contained a plasmid, ampicillin (100 μg/ml) was present in the medium. In all, 2 ml of donor and 2 ml of recipient cells were then mixed, and deposited on a nitrocellulose filter. Conjugation was allowed to proceed by placing the filter on a prewarmed LB plate containing 100 μM IPTG, for 40 min at 37°C. After this 40 min period, conjugation was interrupted and this corresponded to time zero of the experiment. Cells were resuspended in 2 ml of a chemically defined medium M9 lacking a carbon source, to reduce cell growth to its minimum (M9 was supplemented with 30 μg/ml thiamine, 0.002% uracil, 3 mM MgSO4 and 0.1% casaminoacids), and separated by vortexing. Nalidixic acid (40 μg/ml) was added to kill donor cells, as well as IPTG (100 μM), and the cultures were maintained at 37°C under agitation for the time course of the experiment. Samples of 0.1 ml (replication assay) or 0.2 ml (recombination assay) were withdrawn at appropriate time points, and treated for the β-galactosidase assay as described (Miller, 1972). To test whether lacZ transcription and/or translation were affected in helicase mutant and helicase overproducing strains, a time course of β-galactosidase activity after IPTG induction was performed in parallel in the JJC40 strain, the MAC448 rep derivative and MAC833 uvrD derivative. Induction of lacZ expression was equally efficient in all three strains (not shown).
Protein and DNA reagents
UvrD and Rep were gifts from Era Cassuto and Ken Marians, respectively. RecA was purchased from Roche and SSB protein was from Amersham Biosciences. ΦX174 viral (+) ssDNA and ΦX174 RF1 dsDNA were purchased from Biolabs.
DNA strand exchange reaction
ΦX174 ssDNA (4.5 μM nucleotides) was incubated in buffer A (30 mM Tris–HCl (pH 7.6), 9 mM MgCl2, 1.8 mM DTT, 1.1 mM ATP, 7.2 mM phosphocreatine, 9 U/ml phosphocreatine kinase) for 3 min at 37°C before the addition of 1.5 μM RecA protein and 0.26 μM SSB. The reaction was kept at 37°C for 10 min. Strand exchange was started by addition of 5′ 32P end-labelled MfeI-linearized ΦX174 dsDNA (4.5 μM nucleotides) and variable amounts of helicases. After 40 min incubation, reaction mixture was deproteinized and analysed by electrophoresis in 0.8% agarose gel in TAE buffer. Gels were dried and quantified by ImageQuant software. Percentage of nicked circular products was calculated as follows:
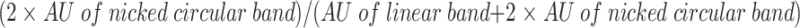 |
The signal of the nicked circular band was multiplied by 2, because only one labelled strand was transferred to the single-stranded circular DNA. AU stands for arbitrary units given by the ImageQuant software.
ATPase assay
ATPase activity was assayed by linking ATP hydrolysis to NADH oxidation, as described previously (Pullman et al, 1960). The reactions were carried out at 37°C for 15 min in 50 mM Tris–HCl (pH 7.6), 100 mM NaCl, 7 mM MgCl2, 80 μg/ml BSA in the presence of 1 mM ATP and saturating amount of synthetic oligo(dT)55 (5.5 μM nucleotides). The concentrations of proteins UvrD and Rep used in this assays are indicated in the figure legend.
Electron microscopy
RecA filaments on ssDNA were formed by incubation of 2.46 μM RecA and 0.41 μM SSB protein in DNA strand exchange buffer with 4.9 μM (nucleotides) ΦX174 viral (+) single strand for 5 min at 37°C. Reactions mixtures were then diluted five times in TNM (10 mM Tris–HCl (pH 7.6), 50 mM NaCl, 5 mM MgCl2) or buffer A before subsequent addition of variable amounts of helicase for 15 min. After an additional dilution (four times) in TNM, the products of the reaction were analysed by electron microscopy as described previously (Beloin et al, 2003).
Supplementary Material
Supplementary Figure S1
Acknowledgments
We are grateful to Miroslav Radman, in whose laboratory part of the work was conducted, for his constant support and enthusiasm. B Michel, M El Karoui, S Gangloff and A Gruss are thanked for their critical comments on the manuscript. We thank Era Cassuto and Danielle Canceill for purified UvrD, Bénédicte Michel for the pAMrep plasmid, and Peter McGlynn and Ken Marians for purified Rep protein. This work was partially supported by grants from the European Community (XV, FF, LSHG-CT-2003-503303) and the Ministère de la Recherche (XV, JJ, ELeC, FF, DRAB03/68). SD was financed by the MENRT, the ‘ligue contre le cancer’ and the ‘fondation pour la recherche médicale’.
References
- Aboussekhra A, Chanet R, Adjiri A, Fabre F (1992) Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol 12: 3224–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F (1989) RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res 17: 7211–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Klein HL (1988) Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand SP, Mitra P, Naqvi A, Khan SA (2004) Bacillus anthracis and Bacillus cereus PcrA helicases can support DNA unwinding and in vitro rolling-circle replication of plasmid pT181 of Staphylococcus aureus. J Bacteriol 186: 2195–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur HM, Lloyd RG (1980) Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol Gen Genet 180: 185–191 [DOI] [PubMed] [Google Scholar]
- Beloin C, Jeusset J, Revet B, Mirambeau G, Le Hegarat F, Le Cam E (2003) Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J Biol Chem 278: 5333–5342 [DOI] [PubMed] [Google Scholar]
- Bierne H, Seigneur M, Ehrlich SD, Michel B (1997a) uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol Microbiol 26: 557–567 [DOI] [PubMed] [Google Scholar]
- Bierne H, Vilette D, Ehrlich SD, Michel B (1997b) Isolation of a dnaE mutation which enhances RecA-independent homologous recombination in the Escherichia coli chromosome. Mol Microbiol 24: 1225–1234 [DOI] [PubMed] [Google Scholar]
- Birge EA, Low KB (1974) Detection of transcribable recombination products following conjugation in rec+, reCB- and recC-strains of Escherichia coli K12. J Mol Biol 83: 447–457 [DOI] [PubMed] [Google Scholar]
- Bradshaw JS, Kuzminov A (2003) RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol Microbiol 48: 1711–1725 [DOI] [PubMed] [Google Scholar]
- Bredeche MF, Ehrlich SD, Michel B (2001) Viability of rep recA mutants depends on their capacity to cope with spontaneous oxidative damage and on the DnaK chaperone protein. J Bacteriol 183: 2165–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Ehrlich SD (2000) UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol 35: 204–210 [DOI] [PubMed] [Google Scholar]
- Chow KH, Courcelle J (2004) RecO acts with RecF and RecR to protect and maintain replication forks blocked by UV-induced DNA damage in Escherichia coli. J Biol Chem 279: 3492–3496 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Denhardt DT (1987) The Escherichia coli rep mutation. X. Consequences of increased and decreased Rep protein levels. Mol Gen Genet 209: 382–390 [DOI] [PubMed] [Google Scholar]
- Courcelle J, Carswell-Crumpton C, Hanawalt PC (1997) recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA 94: 3714–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delagoutte E, von Hippel PH (2002a) Helicase mechanisms and the coupling of helicases within macromolar machines. Part I: structures and properties of isolated helicases. Q Rev Biophys 35: 431–478 [DOI] [PubMed] [Google Scholar]
- Delagoutte E, von Hippel PH (2002b) Helicase mechanisms and the coupling of helicases within macromolar machines. Part II: integration of helicases into cellular processes. Q Rev Biophys 36: 1–69 [DOI] [PubMed] [Google Scholar]
- Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V (2004) Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci USA 101: 6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA 99: 16887–16892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E (2004) Protein displacement by DExH/D ‘RNA helicases’ without duplex unwinding. Science 304: 730–734 [DOI] [PubMed] [Google Scholar]
- Flores MJ, Bidnenko V, Michel B (2004) The DNA repair helicase UvrD is essential for replication fork reversal in replication mutants. EMBO Rep 5: 983–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores MJ, Bierne H, Ehrlich SD, Michel B (2001) Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks. EMBO J 20: 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet 25: 192–194 [DOI] [PubMed] [Google Scholar]
- George JW, Brosh RM Jr, Matson SW (1994) A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II. A biochemical and genetic characterization. J Mol Biol 235: 424–435 [DOI] [PubMed] [Google Scholar]
- Gil D, Bouche JP (1991) ColE1-type vectors with fully repressible replication. Gene 105: 17–22 [DOI] [PubMed] [Google Scholar]
- Hiasa H, Marians KJ (1992) Differential inhibition of the DNA translocation and DNA unwinding activities of DNA helicases by the Escherichia coli Tus protein. J Biol Chem 267: 11379–11385 [PubMed] [Google Scholar]
- Higgins NP, Kato K, Strauss B (1976) A model for replication repair in mammalian cells. J Mol Biol 101: 417–425 [DOI] [PubMed] [Google Scholar]
- Howard MT, Neece SH, Matson SW, Kreuzer KN (1994) Disruption of a topoisomerase–DNA cleavage complex by a DNA helicase. Proc Natl Acad Sci USA 91: 12031–12035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanescu S (1993) Characterization of the Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid pT181 replication. Mol Gen Genet 241: 185–192 [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM (2001) Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science 291: 121–125 [DOI] [PubMed] [Google Scholar]
- Klein HL (2001) Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Δ with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157: 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev 58: 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309 [DOI] [PubMed] [Google Scholar]
- Kuzminov A (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev 63: 751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HE, Denhardt DT (1975) The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J Mol Biol 97: 99–112 [DOI] [PubMed] [Google Scholar]
- Lavery PE, Kowalczykowski SC (1992) Biochemical basis of the constitutive repressor cleavage activity of recA730 protein. A comparison to recA441 and recA803 proteins. J Biol Chem 267: 20648–20658 [PubMed] [Google Scholar]
- Lloyd RG, Buckman C, Benson FE (1987a) Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J Gen Microbiol 133: 2531–2538 [DOI] [PubMed] [Google Scholar]
- Lloyd RG, Evans NP, Buckman C (1987b) Formation of recombinant lacZ+ DNA in conjugational crosses with a recB mutant of Escherichia coli K12 depends on recF, recJ, and recO. Mol Gen Genet 209: 135–141 [DOI] [PubMed] [Google Scholar]
- Maples VF, Kushner SR (1982) DNA repair in Escherichia coli: identification of the uvrD gene product. Proc Natl Acad Sci USA 79: 5616–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J (1972) Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Modrich P (1994) Mismatch repair, genetic stability, and cancer. Science 266: 1959–1960 [DOI] [PubMed] [Google Scholar]
- Morel P, Hejna JA, Ehrlich SD, Cassuto E (1993) Antipairing and strand transferase activities of E. coli helicase II (UvrD). Nucleic Acids Res 21: 3205–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu K, Kowalczykowski SC (2003) RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell 11: 1337–1347 [DOI] [PubMed] [Google Scholar]
- Orren DK, Selby CP, Hearst JE, Sancar A (1992) Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC–DNA complex by helicase II and DNA polymerase I. J Biol Chem 267: 780–788 [PubMed] [Google Scholar]
- Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Bruand C (1998) PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol 29: 261–273 [DOI] [PubMed] [Google Scholar]
- Petit MA, Ehrlich D (2002) Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J 21: 3137–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranovic M, Zahradka K, Zahradka D, Petranovic D, Nagy B, Salaj-Smic E (2001) Genetic evidence that the elevated levels of Escherichia coli helicase II antagonize recombinational DNA repair. Biochimie 83: 1041–1047 [DOI] [PubMed] [Google Scholar]
- Pullman ME, Penefsky HS, Datta A, Racker E (1960) Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase. J Biol Chem 235: 3322–3329 [PubMed] [Google Scholar]
- Sandler SJ (2000) Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics 155: 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JF, Kornberg A (1978) Purification of the rep protein of Escherichia coli. An ATPase which separates duplex DNA strands in advance of replication. J Biol Chem 253: 3292–3297 [PubMed] [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B (1998) RuvAB acts at arrested replication forks. Cell 95: 419–430 [DOI] [PubMed] [Google Scholar]
- Skarstad K, Boye E (1993) Degradation of individual chromosomes in recA mutants of Escherichia coli. J Bacteriol 175: 5505–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P, Wigley DB (2001) Unwinding the ‘Gordian knot’ of helicase action. Trends Biochem Sci 26: 47–54 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Hours C, Chu A, Denhardt DT (1979) The rep mutation. VI. Purification and properties of the Escherichia coli rep protein, DNA helicase III. Can J Biochem 57: 855–866 [DOI] [PubMed] [Google Scholar]
- Taucher-Scholtz G, Abdel-Monem M, Hoffmann-Berling H (1983) Functions of helicases in E. coli. In Mechanisms of DNA Replication and Recombination, Cozzarelli NR (ed) pp 65–76. New York: Alan R Liss Inc. [Google Scholar]
- Taucher-Scholz G, Hoffmann-Berling H (1983) Identification of the gene for DNA helicase II of Escherichia coli. Eur J Biochem 137: 573–580 [DOI] [PubMed] [Google Scholar]
- Umezu K, Chi NW, Kolodner RD (1993) Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc Natl Acad Sci USA 90: 3875–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312 [DOI] [PubMed] [Google Scholar]
- Wang TC, Chang HY, Hung JL (1993) Cosuppression of recF, recR and recO mutations by mutant recA alleles in Escherichia coli cells. Mutat Res 294: 157–166 [DOI] [PubMed] [Google Scholar]
- Webb BL, Cox MM, Inman RB (1997) Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell 91: 347–356 [DOI] [PubMed] [Google Scholar]
- Wertman KF, Wyman AR, Botstein D (1986) Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49: 253–262 [DOI] [PubMed] [Google Scholar]
- Willetts N, Wilkins B (1984) Processing of plasmid DNA during bacterial conjugation. Microbiol Rev 48: 24–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson C, Perrin D, Cohn M, Jacob F, Monod J (1964) Non-inducible mutants of the regulator gene in the ‘lactose’ system of Escherichia coli. J Mol Biol 141: 582–592 [DOI] [PubMed] [Google Scholar]
- Yancey-Wrona JE, Matson SW (1992) Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res 20: 6713–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieg J, Maples VF, Kushner SR (1978) Recombinant levels of Escherichia coli K-12 mutants deficient in various replication, recombination, or repair genes. J Bacteriol 134: 958–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1


