Abstract
A critical step during human microRNA maturation is the processing of the primary microRNA transcript by the nuclear RNaseIII enzyme Drosha to generate the ∼60-nucleotide precursor microRNA hairpin. How Drosha recognizes primary RNA substrates and selects its cleavage sites has remained a mystery, especially given that the known targets for Drosha processing show no discernable sequence homology. Here, we show that human Drosha selectively cleaves RNA hairpins bearing a large (⩾10 nucleotides) terminal loop. From the junction of the loop and the adjacent stem, Drosha then cleaves approximately two helical RNA turns into the stem to produce the precursor microRNA. Beyond the precursor microRNA cleavage sites, approximately one helix turn of stem extension is also essential for efficient processing. While the sites of Drosha cleavage are determined largely by the distance from the terminal loop, variations in stem structure and sequence around the cleavage site can fine-tune the actual cleavage sites chosen.
Keywords: Drosha, microRNA, RNA III enzymes, RNA interference, RNA processing
Introduction
First discovered in Caenorhabditis elegans, microRNAs (miRNAs) are a large family of ∼22-nucleotide (nt)-long RNAs widely expressed in metazoan eukaryotes (Lee et al, 1993, 2004; Ruvkun et al, 2004). An estimated 1% of animal genes encode miRNAs (Bartel, 2004). While the functions of miRNAs are only beginning to be appreciated, it is generally believed that miRNAs regulate gene expression at the post-transcriptional level by inhibiting the expression of mRNAs bearing fully or partly homologous target sequences (Carrington and Ambros, 2003; Bartel, 2004; He and Hannon, 2004; Novina and Sharp, 2004).
Like most other cellular RNAs, miRNAs undergo a maturation process (Murchison and Hannon, 2004). miRNAs are initially transcribed as part of a long primary miRNA (pri-miRNA) transcript, which contains the mature miRNA as part of a predicted RNA hairpin. In animal cells, the first notable step in miRNA processing occurs when Drosha, a nuclear RNaseIII enzyme, excises the upper part of this RNA hairpin to generate the precursor miRNA (pre-miRNA), which is ∼60 nt long with a 3′ 2 nt overhang (Lee et al, 2002, 2003). The pre-miRNA is then exported to the cytoplasm by the nuclear export factor Exportin 5 and the Ran-GTP cofactor (Yi et al, 2003; Bohnsack et al, 2004; Lund et al, 2004). The second cleavage step takes place in the cytoplasm where Dicer, another RNaseIII enzyme, cuts near the hairpin loop to release an ∼22-base-pair (bp) imperfect RNA duplex intermediate with ∼2 nt 3′ overhangs at both ends (Grishok et al, 2001; Hutvágner et al, 2001; Ketting et al, 2001). Usually, only one of the two RNA strands is stable in vivo. This polarity arises from the fact that the RNA-induced silencing complex (RISC), or a related complex, identifies the strand within the duplex with weaker hydrogen bonding at its 5′ end and then selectively incorporates this strand into RISC (Khvorova et al, 2003; Schwarz et al, 2003). The opposite strand, denoted as miRNA*, is released by RISC and generally rapidly degraded. However, in the rare cases where hydrogen bonding at the two ends of the miRNA duplex intermediate is equivalent, either strand may be randomly incorporated into RISC.
The expression of several hundred miRNAs has been experimentally verified in numerous organisms and cells, and computer programs have been designed to predict more miRNAs on a genome-wide basis (Bartel, 2004). Although mature miRNAs are all ∼22 nt in size, and the secondary structures of pre-miRNAs are broadly similar, the sequences of both miRNAs and pre-miRNAs are very diverse, and the predicted pre-miRNA secondary structures can be quite different in detail. For example, the stem regions may contain different numbers of unpaired residues at different locations, and computer-predicted terminal loops range from 3 nt to well over 10 nt in size (Lagos-Quintana et al, 2001; Lau et al, 2001; Lee and Ambros, 2001). This raises the obvious question of how these unrelated RNA hairpins can be recognized by the same processing enzymes and then precisely processed to yield mature miRNAs.
Drosha has emerged as a key determinant of which part of the pri-miRNA will become the mature miRNA. As mentioned above, Drosha cleaves pri-miRNAs to yield pre-miRNAs and thereby generates one end of the mature miRNA. Dicer recognizes the 3′ 2 nt overhang of pre-miRNAs and then cuts ∼22 nt away to produce the miRNA:miRNA* duplex (Zhang et al, 2002, 2004). miRNAs are then selected over miRNAs* by RISC according to the 5′ end base-pairing rule (Khvorova et al, 2003; Schwarz et al, 2003). Therefore, Drosha plays a critical role in deciding the sequence and abundance of miRNAs, as the initial cleavage sites chosen by Drosha largely dictate where Dicer will cleave and, hence, which miRNA strand enters RISC (Bartel, 2004).
In this study, we have analyzed miRNA processing and expression in human cells transfected with plasmids encoding wild-type and mutant pri-miRNAs, and have complemented these in vivo experiments with in vitro Drosha processing assays. Our results indicate that, within the context of pri-miRNAs, RNA stem–loops with a large, unstructured terminal loop (⩾10 nt) are the preferred substrates for Drosha cleavage, and that Drosha then cleaves ∼22 nt away from the loop/stem junction. A continuation of the pri-miRNA stem, outside of the mature pre-miRNA, is also critical for miRNA processing and can slightly modify the precise cleavage sites used for pre-miRNA production.
Results
Pri-miR-30a processing requires a large terminal loop
We have previously used the CMV immediate-early promoter to overexpress pri-, pre-, and mature miR-30a in transfected human cells, as a means to study miRNA biogenesis (Zeng et al, 2002; Zeng and Cullen, 2003). The construct used, here termed pCMV-miR-30a, expresses the 73 nt miR-30a sequence shown in Figure 1A. This plasmid gives rise to readily detectable levels of two mature miRNAs, termed miR-30a-5p and miR-30a-3p, in transfected cells (miR-30a is unusual in giving rise to two mature miRNAs). By mutagenesis, we have previously identified two features of the pri-miR-30a hairpin that are important for miRNA expression (Zeng and Cullen, 2003). One is base pairing near the base of the stem, beyond the pre-miRNA sequence, as the pCMV-miR-30a(GAG) mutant (Figure 1A) makes no detectable miRNAs when transfected into 293T cells. The other is the terminal loop. Some RNA folding programs (e.g., MFOLD) predict that the apex of the pre-miR-30a hairpin folds to form a 5 nt bulge, a 3 bp stem, and finally a 4 nt terminal hairpin loop (Figure 1A), yet we had found that disruption of the predicted 3 bp stem had no effect, while deleting the 5 nt bulge, thereby extending the predicted stem by 3 bp and leaving the 4 nt loop intact, severely reduced miRNA expression (Zeng and Cullen, 2003). We therefore considered the possibility that pre-miR-30a might instead contain an unstructured 15 nt terminal loop.
Figure 1.
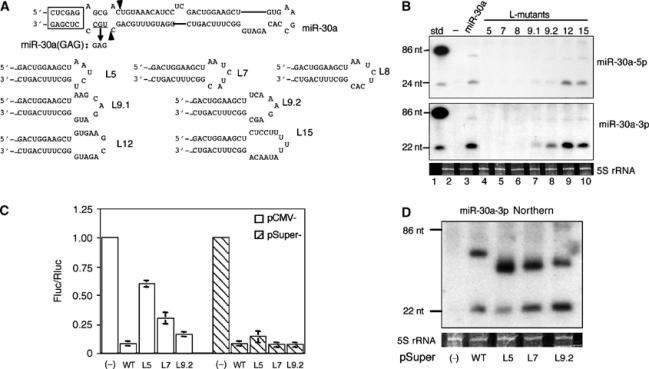
Analysis of pri-miR-30a hairpin terminal loop mutants. (A) Computer-predicted secondary structure of the pri-miR-30a hairpin (Lagos-Quintana et al, 2001) and sequences of the miR-30a terminal loop mutants. The mutation introduced into miR-30a(GAG), a miR-30a null mutant, is indicated. The boxed sequence derives from appended restriction sites and facilitates miR-30a expression. The arrowheads identify the major cleavage sites for endogenous pre-miR-30a. (B) Northern analyses of miR-30a-5p (upper panel) and miR-30a-3p (lower panel) expression in 293T cells transfected with the indicated pri-miR-30a expression plasmids (lanes 3–10). Lane 1; DNA size markers; lane 2; RNA from nontransfected 293T cells. 5S rRNA was used as a loading control. (C) A reporter assay to compare miR-30a-3p function when expressed from pCMV- or pSuper-based vectors. The firefly luciferase-based reporter, pCMV-luc-8xmiR-30a(P), was cotransfected into 293T cells along with pRL-CMV (Promega), an internal control Renilla luciferase expression plasmid, and pCMV-miR-30a or pSuper-miR-30a variants. The ratio of firefly luciferase activity relative to Renilla luciferase activity from cells transfected with an empty pCMV or pSuper vector (−) is set at 1.00. Average of three experiments with standard deviation is shown. (D) Pre-miR-30a and mature miR-30a-3p expression in 293T cells transfected with the pSuper-miR-30a plasmids was determined by Northern blotting, as shown in panel B. miRNA expression levels induced by pSuper-miR-30a and pCMV-miR-30a are comparable (data not shown).
To examine the contribution of terminal loop size and sequence in more detail, pCMV-miR-30a mutants (L5–L15) were constructed with loop sizes ranging from 5 to 15 nt. The terminal loop sequences used either represented deletions of the natural pre-miR-30a terminal loop (L12 and L9.1) or random terminal loop sequences (L5, L7, L8, L9.2, and L15) (Figure 1A). Northern analyses showed that L5, L7, and L8 made very little mature miRNA (Figure 1B, lanes 4–6). In contrast, L9 (both L9.1 and L9.2; Figure 1B, lanes 7 and 8) had improved miRNA production, while L12 and L15 were essentially wild type (Figure 1B, lanes 9 and 10). Variants of L7, L8, and L9 with different loop sequences gave similar results (Figure 1B and data not shown), thus demonstrating that the size of the loop is more important than the sequence per se, although this of course ultimately dictates the loop structure. In all the mutants that were defective in mature miRNA production, pre-miRNA levels were also diminished (Figure 1B). Primer extension experiments confirmed the Northern blotting results (data not shown). Moreover, experiments designed to measure the biological activity of miR-30a in transfected cells, using a previously described indicator construct that contains eight fully complementary target sites for miR-30a-3p linked to the firefly luciferase gene (Zeng et al, 2003), revealed that the L9.2, L7, and L5 mutants of pCMV-miR-30a were increasingly attenuated in their ability to inhibit luciferase gene expression (Figure 1C).
Production of a mature, stable miRNA from the initial pri-miRNA transcript requires nuclear processing by Drosha, nuclear export by Exportin 5, cytoplasmic processing by Dicer, and finally incorporation of the mature miRNA into RISC. While any of these steps could be affected by terminal loop size, we favored the hypothesis that the first step, that is, Drosha processing, was primarily affected in mutants of pri-miR-30a bearing small terminal loops. If this is indeed the case, then direct production, in the nucleus, of a transcript identical to the ∼63 nt pre-miR-30a intermediate, using RNA polymerase III (polIII), should rescue both mature miR-30a production and function. In fact, expression of the L5, L7, and L9.2 mutants as a pre-miR-30a transcript entirely (L7 and L9.2) or largely (L5) rescued both mature miR-30a-3p function (Figure 1C) and expression (Figure 1D) in transfected cells. These data, together with data showing that recombinant Dicer is able to process wild-type pre-miR-30a, as well as the L5 and L9.2 mutants, into mature miR-30a with equivalent efficiency in vitro (not shown) argue that terminal loop deletion mutations are inhibiting a step prior to nuclear export of the pre-miRNA, that is, most likely Drosha processing of the pri-miRNA.
Efficient processing of other pri-miRNAs also requires a large terminal loop
Our results with miR-30a indicated that a terminal loop of ⩾10 nt in size is an important determinant of efficient pri-miR-30a processing in vivo (Figure 1). If this is a general property, then other pre-miRNAs would also be predicted to contain terminal loops of ⩾10 nt. However, computer predictions of the structure of other miRNA precursors frequently predict terminal loops that are much smaller.
To determine if a large terminal loop is a general feature of pri-miRNAs, we next tested a second human miRNA termed miR-21. Computer folding programs predict that pre-miR-21 has a 5 nt terminal loop adjacent to a 4 bp stem containing two G:U base pairs (Lagos-Quintana et al, 2001; Figure 2A). Mutations were introduced to either enlarge the loop or to stabilize this stem in the context of the pri-miR-21 expression plasmid pCMV-miR-21 (Zeng and Cullen, 2003), and the resultant plasmids were transfected into cells. To detect functional miR-21, we employed a reporter assay and also performed primer extension and Northern blotting (Figure 2B). We have previously described a reporter construct that encodes the firefly luciferase gene linked to eight copies of a target sequence perfectly complementary to miR-21 and that is markedly downregulated in response to miR-21 overexpression (Zeng et al, 2003). Both the reporter assay and RNA analyses showed that while opening up the loop had no effect on mature miR-21 expression, stabilizing the predicted stem, therefore restricting the loop size, was deleterious (Figure 2B). Thus, the single nucleotide mutations M2 and M4, which are predicted to destabilize the 4 bp stem shown in Figure 2A, had no effect on either miR-21 function or expression (Figure 2B, lanes 5 and 7), while the point mutations M1 and, particularly, M5, which are predicted to stabilize the 4 bp stem, attenuated both miR-21 function and expression (Figure 2B, lanes 4 and 8). Moreover, a mutation predicted to strongly stabilize the 4 bp stem, termed miR-21(CCG), essentially abolished miR-21 production and function (Figure 2B, lane 3). These data suggest that the miR-21 terminal loop as predicted in silico (Figure 2A) is smaller than actually found in vivo, or that the loop structure is dynamic in vivo, with cellular processing factors perhaps stabilizing or inducing a larger loop (Figure 3).
Figure 2.
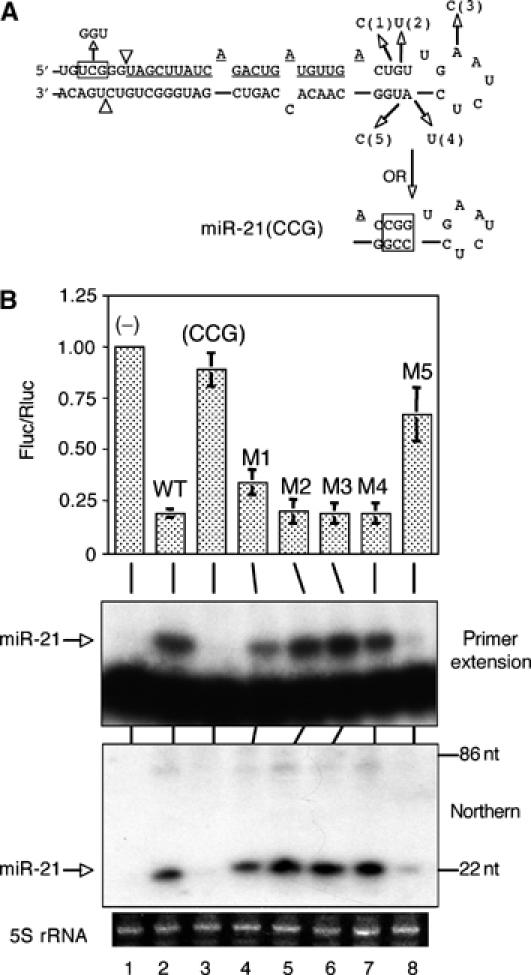
Analysis of pri-miR-21 loop mutants. (A) Schematic of the computer-predicted miR-21 secondary structure, point mutations (1, 2, 3, 4, and 5), and the miR-21(GGU) and miR-21(CCG) mutants. Mature miR-21 is underlined, and the arrowheads indicate the cleavage sites used to produce the predicted pre-miR-21. (B) Analyses of miR-21 loop mutants. Luciferase assays were performed as in Figure 1C and the similar indicator construct pCMV-luc-8xmiR-21(P) was used as the reporter. Primer extension and Northern blotting assays were performed using oligonucleotide probes described in Supplementary Table 1. 5S rRNA was used as a loading control in the Northern analysis.
Figure 3.

Alternative structures for miR-30a, miR-21, miR-27a, and miR-31. Comparison of the published, computer-predicted secondary structures (Lagos-Quintana et al, 2001) with possible new secondary structures proposing larger terminal loops for these miRNA precursors. These proposed structures are based on the mutational analyses, and transfections and in vitro assays, performed in this paper. These larger loops may form as indicated or may be opened by Drosha binding. Mature miRNA sequences are underlined, and predicted or confirmed Drosha cleavage sites are shown by arrows.
Two more human miRNAs were also analyzed for the effect of terminal loop size on Drosha processing efficiency. Genomic DNA (∼250 bp), centered on the predicted ∼80 nt pri-miR-27a and pri-miR-31 RNA hairpins, was cloned into the polIII-based expression plasmid pSuper (Brummelkamp et al, 2002). These plasmids are therefore predicted to express pri-miRNAs transcribed by polIII that are analogous to the polII-transcribed pri-miRNAs analyzed in Figures 1 and 2. As shown in Figure 4A, pre-miR-27a is predicted by computer analysis to fold into an RNA hairpin bearing an 8 nt terminal loop flanked by a single base pair, a 1 nt bulge, a 2 bp stem, and opposing 1 nt bulges. However, pre-miR-27a could instead form a 17 nt terminal loop above a 7 bp stem (Figure 3). In the case of pre-miR-31, the computer predicts an 8 nt terminal loop flanked by a 3 bp stem and three bulged nucleotides (Figure 4B), but we considered that pre-miR-31 might instead contain an unstructured 17 nt terminal loop (Figure 3). Mutations that are predicted to affect the size of the terminal loop were then introduced into the miR-27a and miR-31 expression plasmids, as shown for miR-21 in Figure 2A. Northern blotting showed that the production of both pre-miRNAs and mature miRNAs was reduced when the short stems predicted to be adjacent to either 8 nt terminal loop were stabilized or extended into the loops for both miR-27a (Figure 4A, mutants 1 and 2) and miR-31 (Figure 4B, mutants 1 and 3). In contrast, a mutation designed to destabilize the 3 bp stem predicted to be adjacent to the 8 nt terminal loop in pre-miR-31 had no effect (Figure 4B, lane 3). These data, while limited, are therefore entirely consistent with the mutational analysis of pre-miR-30a and pre-miR-21 (Figures 1 and 2) and suggest that a large (⩾10 nt) terminal loop may be a general requirement for efficient pri-miRNA processing.
Figure 4.
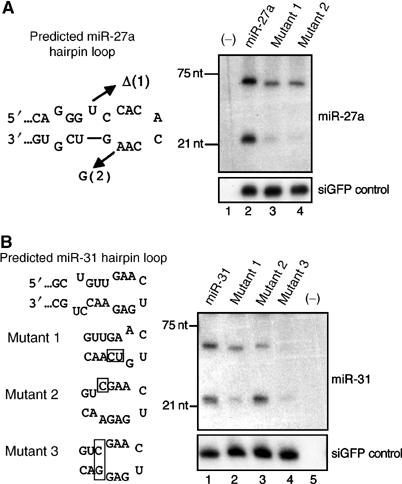
Analysis of loop mutants of human pri-miR-27a and pri-miR-31. (A) miR-27a expression. Plasmids encoding wild-type pri-miR-27a, or one of two mutants (1 and 2), were cotransfected with pH1-GFP into 293T cells. Northern blotting was performed to detect the expression of pre- and mature miR-27a (upper panel) and the control GFP siRNA (lower panel). (B) miR-31 expression. Three pri-miR-31 mutants were constructed (boxed sequences), and their miRNA expression levels compared to wild-type miR-31 in transfected 293T cells.
Drosha requires a large terminal loop for pri-miRNA processing in vitro
To test directly if Drosha discriminates against pri-miRNA substrates bearing small loops, we next performed in vitro processing assays using FLAG-tagged Drosha enzyme that had been isolated from overexpressing 293T cells by immunoaffinity purification. It is important to note that this Drosha preparation is likely to contain other cellular factors, including any proteins that remain bound to Drosha during the purification process.
Incubation of a 32P-labeled pri-miR-30a substrate RNA with FLAG immunoprecipitates obtained from FLAG-tagged Drosha overexpressing 293T cells, or from control 293T cells, yielded several RNA cleavage products that were only observed, or were much stronger, in the former case (Figure 5). The position of pre-miR-30a (‘*') was inferred by its size (63 nt) and by its complete absence in the miR-30a(GAG) lane. The miR-30a(GAG) (Figure 1A) mutant is incapable of giving rise to detectable levels of either the pre-miRNA intermediate or the mature miRNA in transfected cells (Zeng and Cullen, 2003) and a similar pri-miR-30a mutant was previously shown to be resistant to in vitro processing by whole-cell lysates (Lee et al, 2003). This mutant therefore serves as a negative control. As shown in Figure 5, there was a good correlation between the expression of pre-miR-30a in transfected cells (Figure 1) and the ability of Drosha to produce pre-miRNA in vitro. Notably, the L5 and L9.2 mutants of miR-30a, which give rise, respectively, to almost undetectable or reduced levels of mature miR-30a in transfected cells (Figure 1B) also gave rise to essentially undetectable or reduced levels of pre-miR-30a in vitro (Figure 5). We consistently observed an additional RNA band (indicated by an arrowhead at the right of Figure 5) that ran slightly higher than the pre-miR-30a marked by ‘*'. The identity of this upper band is not known, but it is not the final pre-miR-30a product, as RNA isolated from the band is not a substrate for Dicer, while RNA from the lower band is (Figure 7B). It is possible that this band is derived from one of the two flanking sequences that are predicted also to be produced by Drosha cleavage of the pri-miR-30a RNA probe used, and that are expected to be ∼78 and ∼61 nt in length.
Figure 5.
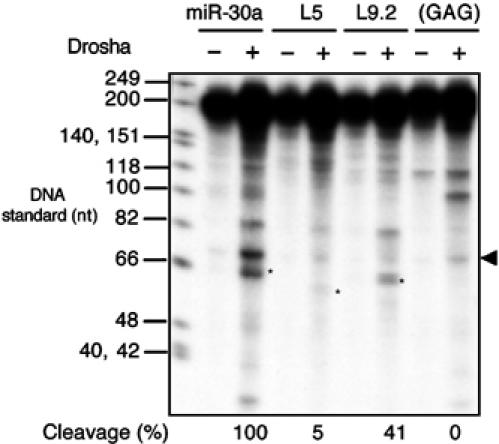
In vitro Drosha cleavage of pri-miR-30a transcripts. FLAG immunoprecipitates from mock-transfected 293T cells (−) or cells transfected with pCK-Drosha-FLAG(+) were incubated with a 32P-labeled ∼202 nt RNA probe encoding the indicated miR-30a variants. RNA cleavage products were recovered and resolved on a denaturing 10% polyacrylamide gel. The pre-miRNA band is indicated by an asterisk, and the background band running slightly above by an arrowhead. Cleavage efficiency was calculated as the intensity of the pre-miRNA band divided by that of the remaining substrate, and set as 100% for the wild-type transcript. DNA size standards are shown next to the autoradiograph.
Figure 7.
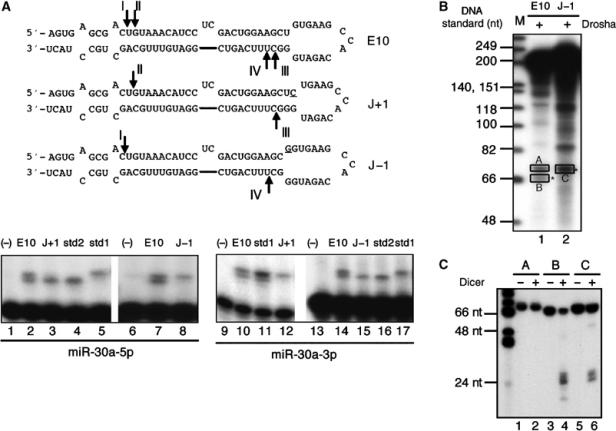
Movement of the loop/stem junction affects Drosha cleavage site selection. (A) Schematic of the predicted RNA secondary structures adopted by the J+1 and J−1 mutants of pCMV-miR-30a(E10). Mutated residues are underlined and the arrows indicate the 5′ ends of the miRNAs, as defined in the primer extension shown in the lower panel. Sequences of the size standards used are given in Supplementary Table 1. (B) Drosha processing of miR-30a transcripts in vitro. M: DNA markers. Pre-miRNA bands are indicated by asterisks. (C) To test the identities of Drosha cleavage products, RNAs from bands A, B, and C in panel B were isolated, digested with recombinant human Dicer, and resolved on a 15% denaturing gel.
To further confirm the hypothesis that Drosha preferentially cleaves pri-miRNAs bearing a large (⩾10 nt) terminal loop, we performed additional in vitro cleavage assays using RNA probes derived from wild-type and selected mutant forms of the pri-miR-21 (Supplementary Figure 1A) or pri-miR-31 (Supplementary Figure 1B) RNAs. These data demonstrated that pri-miR-21 (Figure 2) and pri-miR-31 (Figure 4B) terminal loop mutants that inhibit mature miRNA production in transfected cells also inhibit Drosha cleavage of the pri-miRNA precursor in vitro.
Role of the pri-miRNA stem extension during miRNA expression
As noted above, in addition to an optimal terminal loop, a continuation of base pairing beyond the base of the pre-miRNA stem is also critical for miRNA processing, as pCMV-miR-30a(GAG) (Figure 1A) and pCMV-miR-21(GGU) (Figure 2A) made no pre-miRNAs or mature miRNAs in transfected cells (Zeng and Cullen, 2003), and both also failed to give rise to pre-miRNAs in Drosha cleavage assays in vitro (Figure 5 and Supplementary Figure 1A). To test how long a stem extension is required, and how it might affect pri-miRNA processing, more miR-30a variants (Figure 6A) were made. These mutants were named according to the distance from the first predicted base pair outside the pre-miRNA to the 5′ end of the endogenous pre-miR-30a intermediate (position shown in E17). These extensions are predicted to be mostly double stranded but also include small bulges (Figure 6A). Thus, the E17 mutant contains an extension beyond the endogenous pre-miR-30a intermediate that is predicted to add 14 bp, and several unpaired nucleotides, to the base of the pre-miR-30a RNA hairpin (Figure 6A). Pri-miRNA sequences located outside the structures shown were predicted to be largely single stranded.
Figure 6.
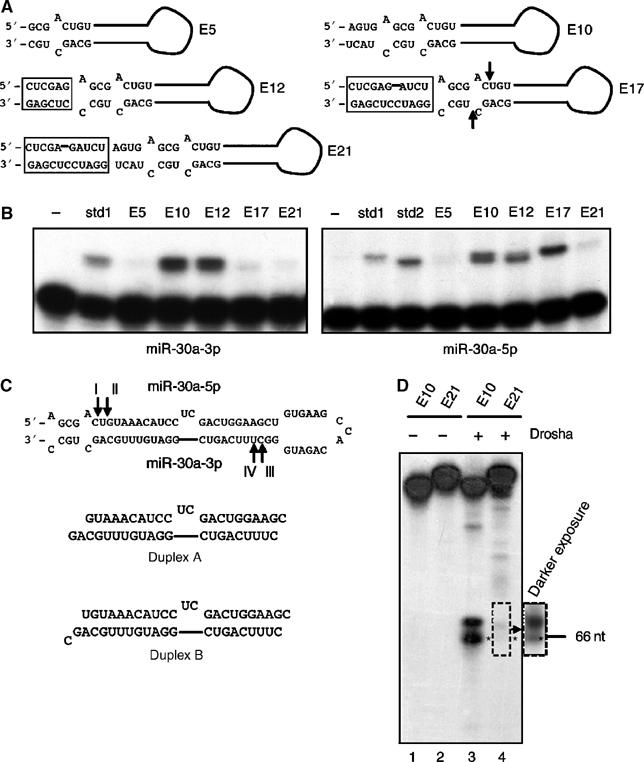
Analysis of the effects of changes in pri-miR-30a stem length. (A) Predicted hairpin RNA structures adopted by pri-miR-30a variants with different stem extensions beyond the pre-miR-30a structure. Boxed sequences represent added restriction sites. The positions of the 5′ and 3′ ends of the endogenous pre-miR-30a are indicated by arrows in E17. (B) Primer extension experiments to determine the expression level and 5′ ends of mature miR-30a-3p (left panel) or miR-30a-5p (right panel) variants. The sequences of the miR-30a-3p std1 and miR-30a-5p std1 and 2 size standards are given in Supplementary Table 1. (C) The 5′ cleavage sites mapped in panel B are indicated as I, II, III, and IV on the predicted pri-miR-30a RNA structure. Two potential duplex intermediates formed after cleavage are indicated (3′ ends are inferred). (D) In vitro Drosha processing of transcripts encoding E10 and E21. The asterisk indicates the pre-miRNA band. The position of a 66 nt DNA size marker is indicated.
Plasmids encoding these pri-miR-30a variants, driven by the CMV promoter (Zeng et al, 2002), were transfected into 293T cells, and primer extension experiments were performed to determine the expression levels and 5′ ends of overproduced miR-30a-5p and miR-30a-3p. As shown in Figure 6B, E5 produced few mature miRNAs. E10 (which is similar to wild-type pri-miR-30a) and E12 gave expression patterns that were similar, in terms of both expression level and the 5′ ends of the observed miRNAs. In contrast, E17 yielded mostly miR-30a-5p. The last variant tested, E21, was largely defective in producing any miRNAs, indicating that too long a stem extension is also undesirable. Northern analyses confirmed the expression patterns of these variants, and pre-miRNA levels correlated with mature miRNA levels (data not shown). Based on published reports (Lee et al, 2003; Zeng and Cullen, 2003) and the data presented here, we therefore conclude that a modest, ⩾8 bp stem extension, beyond the 5′ end of a pre-miRNA, is required for optimal miRNA processing from a long pri-miRNA.
The primer extension analysis (Figure 6B) showed that the three pri-miR-30a stem mutants able to produce mature miRNA effectively, that is, E10, E12, and E17, had minor but distinct differences in their processing sites. Specifically, both E10 and E12 actually produced both forms of miR-30a, while E17 produced very little miR-30a-3p (Figure 6B). The two miR-30a RNAs produced by E10 and E12 appear to be almost equal mixtures of miRNAs that differ by 1 nt, with the longer form being full-length miR-30a-3p (cleavage site III; Figure 6C) while the second form is 1 nt shorter (cleavage site IV; Figure 6C). The limited level of miR-30a-3p produced by E17 appears to be of the shorter form.
Analysis of the miR-30a-5p strand by primer extension again showed two products, differing by 1 nt, for E10. E12 appeared to produce primarily the shorter form, while E17 produced only the longer (Figure 6B). Based on the size standards used, E10 appears to be cleaved at sites I and II (Figures 6C and 7A), while E12 is predominantly cleaved at site II and E17 at site I (Figure 6C).
Based on this analysis, it is possible that E10 could give rise to four different duplex intermediates, E12 to two, and E17 to one. The duplex intermediate predicted for E17 is shown as duplex B in Figure 6C. As may be observed, this duplex contains an A:U base pair at one end and a G:C base pair at the other, thus strongly favoring the incorporation into RISC of the miR-30a-5p strand, whose 5′ end forms part of an A:U base pair, over the miR-30a-3p strand, whose 5′ end is predicted to form part of a more stable G:C base pair (Schwarz et al, 2003). In contrast, duplex A, which is predicted to be produced by processing of both the E10 and E12 pri-miRNAs, should form G:C base pairs at both ends, thus predicting similar levels of incorporation of miR-30a-3p and miR-30a-5p into RISC, and similar stability, as is indeed observed (Figure 6B).
As the E17 mutation moves the cleavage site chosen by Drosha 1 nt away from the terminal loop, does this imply that longer stem lengths invariably have this effect? The answer to this question is clearly no, as the E21 mutant, bearing the longest stem extension tested, actually produced a low but detectable level of miR-30a-3p and miR-30a-5p that appeared to have 5′ ends very similar or identical to those observed in the E10, E12, and E17 transfected cells (Figure 6B). Moreover, an in vitro Drosha processing assay showed that the E21 variant yielded greatly reduced levels of pre-miR-30a, which was however still of the same size as the pre-miR-30a produced from the ‘wild-type' E10 variant (Figure 6D, lanes 3 and 4). These data therefore suggest that the precise cleavage site used by Drosha can be slightly modified by the stem architecture around the cleavage site. As even a 1 nt change in cleavage site can have a profound effect on which strand of the duplex intermediate is then incorporated into RISC (Figure 6B and C), this minor effect can nevertheless have important consequences.
Drosha cleavage site selection is influenced by the position of the pri-miRNA loop to stem junction
The data presented so far suggest that efficient pri-miRNA processing by Drosha is facilitated by a large, unstructured terminal loop and a stem of less than 40 bp and more than 26 bp in length. However, these data do not address how Drosha determines where it should cleave. The yeast RNaseIII enzyme Rnt1p binds to RNA stem–loops bearing a terminal tetraloop and then cleaves a fixed 14–16 nt distance away from the loop (Chanfreau et al, 2000). We therefore considered the possibility that Drosha might also cleave the pri-miRNA stem at a set distance away from the junction of the terminal loop and stem. If this is the case, then moving this junction up or down the pri-miRNA stem–loop should result in movement of the Drosha cleavage site.
To test this hypothesis, we modified the E10 variant of pCMV-miR-30a, used in Figure 6, such that the loop–stem junction was predicted to move either 1 bp up the stem (mutant J+1; Figure 7A) or 1 bp down the stem (mutant J−1; Figure 7A) without changing the overall size of the pri-miRNA stem–loop. Transfection of 293T cells with these constructs, followed by primer extension analysis using size standards, revealed data entirely consistent with the above hypothesis. As previously shown in Figure 6B, the pCMV-miR-30a/E10 construct gave rise to two 5′ cleavages on each strand, that is, I and II at the 5′ end of the miR-30a-5p strand and III and IV at the 5′ end of the miR-30a-3p strand (Figure 7A). However, in the case of the J−1 mutant, both loop-proximal cleavage sites, that is, site II for miR-30a-5p and site III for miR-30a-3p, are entirely lost while sites I and IV are used efficiently (compare lanes 7 and 14 in Figure 7A with lanes 8 and 15, respectively). Similarly, for mutant J+1, both loop-distal cleavage sites are lost, that is, site I for miR-30a-5p and site IV for miR-30a-3p (compare lanes 2 and 10 in Figure 7A with lanes 3 and 12, respectively).
To further confirm the hypothesis that a change in the location of the loop/stem junction can directly lead to changes in the sites of Drosha cleavage, in vitro assays were performed. Figure 7B demonstrates directly that Drosha cleavage indeed produces a slightly larger pre-miRNAs from J−1 relative to wild-type miR-30a (compare lanes 1 and 2, bands marked by ‘*'). The identity of the inferred pre-miRNA bands (bands B and C) was confirmed by digesting the gel-isolated RNAs with recombinant Dicer, which produced the predicted small RNA species (Figure 7C). In contrast, the slower RNA (band A) was unaffected by Dicer. We note that the RNA cleavage products produced by Dicer appear to be of the same size, even though the ‘C' substrate is larger than the ‘B' substrate (Figure 7C). This implies, as expected (Lee et al, 2003), that Dicer cleaves at a set distance from the base of both of these pre-miRNA intermediates, thus also moving the Dicer cleavage site away from the terminal loop, as is indeed observed (Figure 7A). In total, these in vivo and in vitro data are fully consistent with the hypothesis that the position of the pri-miRNA loop to stem junction acts as a major determinant of the sites of pri-miRNA stem cleavage chosen by Drosha and, hence, by Dicer.
How does Drosha process pri-miRNAs?
Because Drosha processes hundreds of pri-miRNAs with vastly different sequences, it likely recognizes common structural features. In Figure 3, we list possible secondary structures for the four human miRNA precursors tested in this paper: miR-30a, miR-21, miR-27a, and miR-31. Although computer programs predict that these miRNA precursors all have terminal loops smaller than 10 nt (Lagos-Quintana et al, 2001), our data suggest terminal loops ranging from 15 to 17 nt in size (Figure 3). The confirmed or predicted positions of endogenous Drosha cleavages are marked by arrows. The 5′ cleavage sites for miR-30a, miR-21, and miR-27a are 24, 21, and 23 nt, respectively, including both single- and double-stranded residues, away from the junction of the terminal loop and the stem. This distance is ∼2 RNA helical turns, if allowance is made for distortions caused by small RNA bulges and/or interior loops. The 5′ cleavage site for miR-31, as deduced from the published sequence of miR-31 (Lagos-Quintana et al, 2001), is however only 19 nt from the loop. To test if pri-miR-31 was indeed cut at such a short distance from the loop, a primer extension assay was performed (Supplementary Figure 2). Surprisingly, we found that both endogenous miR-31 and overexpressed miR-31 were in fact 1 nt longer at the 5′ end than the reported sequence. The bona fide 5′ cleavage site in pri-miR-31 therefore is 20 nt from the terminal loop, as shown in Figure 3. Other non-human miR-31 orthologs have been reported to start at the analogous position (www.sanger.ac.uk/Software/Rfam/mirna).
Thus, our results suggest that Drosha cleaves ∼22 nt from the large terminal loop of an extended RNA hairpin. To provide additional support for this hypothesis, we designed a pri-miRNA transcript containing an artificial sequence (ARTI) whose secondary structure (Figure 8A) resembles a pre-miRNA plus an extended stem structure. ARTI is predicted to fold into an RNA hairpin that is structurally similar to the miR-30a E10 variant shown in Figure 7A, but with a different underlying sequence and a smaller, 12 nt terminal loop. As a negative control, ARTI(CUA), a predicted null mutation that is similar in structure to miR-30a(GAG) (Figure 1A), was also tested. Since ARTI follows the structural rules for Drosha substrates uncovered in this study, its primary transcript should be processed by Drosha in the same way as an authentic pri-miRNA in vitro. As shown in Figure 8B, major bands of 60–70 nt long were indeed generated from the artificial ART1 transcript (lane 1) but were absent when the ARTI(CUA) mutant was analyzed (lane 2). Two of the RNA bands in lane 1, marked as A and B, were excised from the gel, eluted, and treated with Dicer. Only band B was digested by Dicer to yield the predicted ∼22 nt RNA products, as well as a small fragment likely derived from the terminal loop (lane 6), thus confirming that band B indeed represents a functional artificial pre-miRNA.
Figure 8.
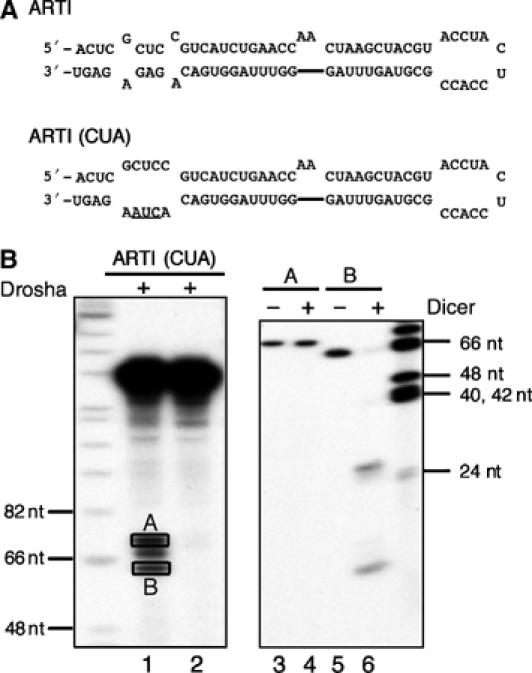
Drosha cleavage of an artificial pri-miRNA transcript. (A) Predicted secondary structures adopted by an artificial RNA hairpin, ARTI, and its mutant, ARTI(CUA). (B) In vitro processing of ARTI transcripts. Lanes 1 and 2: Drosha cleavage of the primary pri-miRNA transcripts; lanes 3–6: Dicer cleavage of the two bands, A and B, recovered from lane 1. DNA size makers are indicated.
Discussion
A fundamental question in miRNA biogenesis is how does the miRNA processing machinery identify the miRNA stem–loop present in the long pri-miRNA precursor? What are the shared characteristics that allow Drosha to bind and precisely process the RNA hairpins present in pri-miRNAs, yet ignore the many RNA hairpins found in other, irrelevant coding and noncoding RNAs? This discrimination is difficult to understand given that pri-miRNA hairpins are highly variable in sequence and, moreover, are predicted to fold into RNA hairpins that, while similar in size and overall structure, nevertheless differ greatly in terms of the sizes and locations of computer-predicted RNA bulges and loops.
In this paper, we propose that Drosha recognition and cleavage of the miRNA hairpins present in pri-miRNAs is dependent on the characteristic structure and size of these hairpins. Specifically, we propose that Drosha preferentially recognizes a large (⩾10 nt) terminal loop located on an imperfect RNA stem that is ∼30 bp in length (Figure 9). RNA hairpins having smaller terminal loops, or bearing stems that are significantly shorter or longer than ∼30 bp, are not effectively recognized by Drosha. Moreover, we propose that Drosha recognizes the terminal hairpin loop and then measures two helical turns (∼22 bp) from the loop/stem junction along the stem (Figure 9). The precise cleavage sites chosen by Drosha are however fine-tuned by the structure and, possibly, sequence of the stem around this nominally optimal ∼22 bp distance. Importantly, Drosha will not cleave effectively in the absence of an additional helical RNA turn beyond the cleavage site (Figure 9). While we have throughout referred to Drosha as the determinant of this structural recognition, our data are entirely consistent with the possibility that Drosha acts in concert with other cellular proteins or as part of a larger protein complex, as has been recently proposed (Denli et al, 2004).
Figure 9.
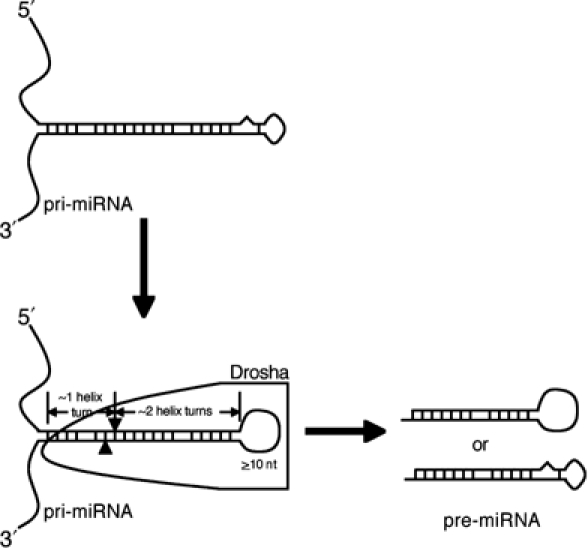
A model of how a pri-miRNA is processed to produce a pre-miRNA. In this model, Drosha, or a holoenzyme with Drosha providing the catalytic activity, selects an RNA hairpin bearing a terminal loop that is ⩾10 nt long, and cuts ∼22 nt, or ∼2 helix turns, from the terminal loop/stem junction to produce a pre-miRNA. Efficient processing, and possibly recognition, also requires an extended (∼10 bp), mostly double-stranded region located beyond the pre-miRNA stem. See text for detailed discussion.
Our studies of hairpin loop mutants of numerous human miRNAs have led to the unexpected finding that a large terminal loop facilitates miRNA maturation. Computer predictions of pre-miRNA secondary structures have some terminal loops as small as 3 nt, but the most adjacent predicted stems frequently contain relatively unstable G:U or A:U base pairs flanked by bulges or internal loops (Figure 3). Thus, although those predictions are considered thermodynamically favorable, there likely is some structural plasticity in the loop regions, intrinsically or aided by binding proteins in vivo. We show here that restricting the terminal loops to below ∼10 nt by introducing mutations that stabilize the predicted smaller loops invariably reduced pre-miRNA and mature miRNA production in transfected human cells (Figures 1, 2 and 4). We therefore believe that the proposed larger terminal loops, while certainly tentative, have more validity in vivo for pri-miRNA processing than the previously predicted small terminal loops. In vitro, Drosha also cleaved pri-miRNAs that have small terminal loops less effectively than wild-type pri-miRNAs (Figure 5 and Supplementary Figure 1). Our data therefore argue that Drosha either acts on pri-miRNA conformers with a large terminal loop or melts small adjacent stems to create such a loop. The sequence of Drosha does not contain an obvious helicase domain, so it is unclear whether Drosha alone can enlarge a small terminal loop, or can only interact with a structure already bearing a big loop, or whether it needs help from other cellular protein(s) to achieve this selection.
The terminal loop is not only involved in Drosha selection, but it also serves as a yardstick to determine the location of the pre-miRNA cleavage sites. Several lines of evidence support the notion that Drosha cuts ∼22 nt away from the junction of the terminal loop and the adjacent stem. Firstly, the E10, E12, and E17 variants of miR-30a have different stem lengths, but they all yield miRNAs with very similar (±1 nt) ends (Figure 6B). Secondly, the even longer stem present in the E21 variant of pri-miR-30a was cleaved by Drosha in vitro to produce, albeit weakly, a pre-miRNA identical in size to wild type (Figure 6D). Thus, the relative location of the base of the pri-miRNA stem is not a determinant of Drosha cleavage site selection. Lastly, and most importantly, mutations that moved the junction of the stem and terminal loop 1 bp up or down the stem simultaneously moved the cleavage sites chosen by Drosha, and hence by Dicer, by 1 nt up or down the stem (Figure 7).
Drosha would not be unique among RNaseIII enzymes in using a terminal loop as an anchor for cleavage, as the budding yeast RNaseIII, Rnt1p, uses a tetraloop as a ruler to cut 14–16 nt away into the stem (Chanfreau et al, 2000). We propose that Drosha prefers a much larger, and apparently unstructured, terminal loop and then cuts ∼22 nt away. Dicer, another RNaseIII-type enzyme, also cuts every ∼22 nt, but unlike Drosha, Dicer measures from RNA termini (Zhang et al, 2002, 2004). Recently, it has been proposed that the two RNaseIII domains of Dicer form an intramolecular dimer, with the larger, N-terminal RNaseIII domain participating in measuring the distance from the terminus of double-stranded RNAs to the cleavage sites (Zhang et al, 2004). The two RNaseIII domains in Drosha are similar in size, and Drosha does not have the PAZ protein domain that mediates Dicer recognition of the base of the pre-miRNA stem (Lingel et al, 2003; Song et al, 2003; Yan et al, 2003). It is therefore currently unclear how Drosha determines where to cleave the pri-miRNA stem. It is to be noted that the hypothesis that Drosha cuts ∼22 nt away from the loop/stem junction of pri-miRNAs, while Dicer cuts ∼22 nt from the newly generated pre-miRNA termini, implies that the location of the stem/loop junction signals both the beginning of the Drosha measurement and the end of a similar measurement by Dicer, that is, the Dicer cleavage sites should be at or near the loop/stem junction, as is indeed observed (Figure 3).
Besides structural features within the eventual pre-miRNAs, sequences outside the pre-miRNA are also important for efficient miRNA processing. We favor the hypothesis that these flanking sequences provide the optimal environment for the ‘correct' pri-miRNA structure to form and be recognized by Drosha or a Drosha-containing complex. Animal pre-miRNAs are ∼60 nt long hairpin RNAs, yet structural conservation among pri-miRNAs extends beyond the pre-miRNAs and RNA folding programs predict that many endogenous pri-miRNAs form RNA structures that extend beyond the pre-miRNA stem-loop per se see Lagos-Quintana et al, 2001). Published reports (Lee et al, 2003) and our data have confirmed that a modest extension of the stem outside a pre-miRNA is critical for miRNA maturation from a pri-miRNA. This requirement at least partly explains the observation that genomic sequences extending beyond the pre-miRNA are needed to overexpress some miRNAs from a PolIII promoter (Chen et al, 2004). More specifically, we propose that an ∼10 bp extension beyond the 5′ Drosha cleavage site, allowing and including minor mismatches, facilitates efficient pre-miRNA processing (Figure 6). Indeed, disruption of base pairing close to the ends of the eventual pre-miRNA eliminated Drosha cleavage in vitro (Figures 5 and 8B) and miRNA production in transfected human cells (Zeng and Cullen, 2003). Interestingly, minor structural variations in this stem extension can also fine-tune the positions where Drosha cuts in pri-miRNAs (Figure 6B). Thus, although the position of the loop/stem junction is likely the primary determinant of where cleavage occurs, the stem region (within the pre-miRNA and beyond), with all its distortions by bulges and internal loops, can fine-tune the processing site chosen by Drosha.
The model proposed in Figure 9 suggests that for efficient miRNA maturation in human cells, a significantly larger RNA structure than previously thought is needed. The requirements for efficient processing include a ⩾10 nt terminal loop, ∼2 helix turns that encode the miRNA:miRNA* duplex, and ∼1 helix turn of stem extension. We propose that Drosha, possibly acting together with other cellular factors, recognizes the universal structural features of such an RNA element, rather than its sequence, as an entirely artificial sequence (ARTI) that fulfills these structural requirements was readily processed by Drosha in vitro (Figure 8B). Consistent with this result, the stem region of the pri-miR-30a hairpin can be substituted by heterologous sequences as long as the structure of the hairpin is maintained, thus giving rise to artificial miRNAs with an entirely novel sequence (Zeng et al, 2002; Rangasamy et al, 2004).
In summary, we have identified structural features that are common to human pri-miRNAs and that play a critical role in pri-miRNA recognition and processing by Drosha. These observations should help guide the design of computer programs designed to predict and identify endogenous miRNA genes and allow the entirely de novo design of artificial pri-miRNA precursors that can serve as substrates for efficient Drosha, and Dicer, processing (Figure 8).
Materials and methods
Plasmid construction
pCMV-miR-30a, pCMV-miR-30a(GAG), pCMV-miR-21, pCMV-miR-21(GGU), pSuper-miR-30a, pCMV-luc-8xmiR-30a(P), pCMV-luc-8xmiR-21(P), and pH1-GFP have been described (Zeng and Cullen, 2003, 2004; Zeng et al, 2003). Other variants of miR-30a were constructed by annealing, extending, and cloning complementary oligonucleotides, or by using the Quickchange method (Stratagene) from existing plasmids. Mutants of miR-21 were constructed by Quickchange from pCMV-miR-21. To generate pSuper-gmiR-27a and pSuper-gmiR-31, ∼250 bp of DNA encoding the respective miRNAs was amplified from human genomic DNA (Clontech) and cloned into the HindIII and XhoI sites in a modified pSuper vector (Brummelkamp et al, 2002) with seven consecutive T's added after the XhoI site. Mutants of miR-27a and miR-31 were then constructed using Quickchange.
Cell transfection and RNA analysis
293T cells were maintained in DMEM supplemented with glutamine and 10% fetal bovine serum, transfected as previously described (Zeng and Cullen, 2003), and analyzed 2 days later. Dual-luciferase assays were performed according to instructions from Promega. RNAs were isolated with Trizol reagent (Invitrogen), and Northern analyses and primer extension experiments were performed as described previously (Zeng et al, 2002; Zeng and Cullen, 2004). The oligonucleotides used to detect the various miRNAs by Northern analysis or primer extension are listed in Supplementary Table 1.
Drosha and Dicer assays
To prepare RNA substrates for in vitro assays, DNA fragments were amplified by PCR from pCMV-miR-30a, pCMV-miR-21, and pSuper-gmiR-31, gel-isolated, and then transcribed by T7 RNA polymerase (Promega) in the presence of [α-32P]CTP. 293T cells were transfected with pCK-Drosha-FLAG, which expresses a FLAG-tagged human Drosha protein, and in vitro Drosha processing experiments were performed according to Lee et al (2003). After the reactions, RNAs were phenol/chloroform extracted, ethanol precipitated, and resolved on a 10% denaturing polyacrylamide gel. Quantification was achieved with a PhosphorImager. Selected bands were excised from the gel, and RNAs were eluted in 0.4 ml of 0.5 M NaAc, 0.1% SDS, and 40 μg of glycogen with shaking at 37°C overnight. RNAs were phenol/chloroform extracted, ethanol precipitated, and treated with Dicer (Invitrogen) for ∼7 min at 37°C. RNAs were then extracted and precipitated again, and run on a 15% denaturing gel.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Table 1
Acknowledgments
The authors thank Narry Kim for reagents used in this research. This work was supported by the Howard Hughes Medical Institute and by NIH grant 1R01GM071408.
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Görlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNA in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Buckle M, Jacquier A (2000) Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc Natl Acad Sci USA 97: 3142–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86 [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops B, Plasterk RHA, Ketting RF, Hannon GJ (2004) Processing of pri-microRNAs by the microprocessor complex. Nature, in press [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531 [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, vanLuenen HG, Plasterk RHA (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V (2004) A short history of a short RNA. Cell 116: S89–S92 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M (2003) Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469 [DOI] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Murchison E, Hannon GJ (2004) miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Biol 16: 223–229 [DOI] [PubMed] [Google Scholar]
- Novina CD, Sharp PA (2004) The RNAi revolution. Nature 430: 161–164 [DOI] [PubMed] [Google Scholar]
- Rangasamy D, Greaves I, Tremethick DJ (2004) RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol 11: 650–655 [DOI] [PubMed] [Google Scholar]
- Ruvkun G, Wightman B, Ha I (2004) The 20 years it took to recognize the importance of tiny RNAs. Cell 116: S93–S96 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L (2003) The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol 10: 1026–1032 [DOI] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM (2003) Structure and conserved RNA binding of the PAZ domain. Nature 426: 468–474 [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR (2003) Sequence requirements for microRNA processing and function in human cells. RNA 9: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR (2004) Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res 32: 4776–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR (2002) Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell 9: 1327–1333 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR (2003) MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA 100: 9779–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W (2004) Single processing center models for human dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Table 1


