ABSTRACT
In recent years, the realization that most of the genome is transcribed has transformed the study of mammalian gene expression. Much effort has gone into investigating how this pervasive transcription is regulated and what the functions of the resulting transcripts are, if any. We recently discovered that stress-induced transcriptional readthrough generates very long downstream of gene containing transcripts (DoGs), which may explain up to 20% of intergenic transcription. DoGs are induced by osmotic stress at the level of transcription by a mechanism that depends on calcium release from the endoplasmic reticulum mediated by IP3 receptors. Here, we discuss DoG induction and function in the context of the literature, with special focus on 2 outstanding questions. First, we discuss possible molecular mechanisms underlying DoG induction through reduced transcription termination. Second, we explore how DoGs may function in maintaining euchromatin after nuclear scaffold stress. In short, we review important aspects of DoG biogenesis and function, and provide an outlook for continued DoG study.
KEYWORDS: DoG, redthrough transcription, transcription termination
Pervasive transcription
In recent years, it has become evident that most of the human genome is transcribed, mainly into different types of non-coding RNAs (ncRNAs).1,2 Classical ncRNAs include small nuclear RNAs, tRNAs and rRNAs (reviewed in1). More recently, a wealth of new ncRNAs has been discovered. Long intervening ncRNAs (lincRNAs) are similar to mRNAs in that they are transcribed by RNA polymerase II (Pol II) from independent genes, carry 5′ caps, and are spliced and polyadenylated.3 Enhancer RNAs and promoter-associated RNAs are shorter Pol II transcripts derived from active enhancers4,5 and promoters,6 respectively. Still, these above-mentioned types of transcripts together do not account for the ∼87% of the genome reported to be transcribed,7 suggesting the existence of other transcript classes. We have recently shown that readthrough transcription can generate very long transcripts, which we refer to as DoGs for downstream of gene containing transcripts, that can account for up to 20% of intergenic transcription.8
Identification and characterization of DoGs
We came across DoG transcripts by chance when studying a putative long ncRNA (lncRNA) associated with bad prognosis in neuroblastoma, a childhood tumor of the sympathetic nervous system.9,10 We had found one such lncRNA to be potently upregulated ∼30-fold by osmotic stress. To identify the gene encoding this transcript, we undertook a thorough annotation of intergenic transcripts in our model neuroblastoma cell line SK-N-BE(2)C before and after osmotic stress. This annotation was performed by combining RNA-Seq of total RNA and an analysis of capped sequence tags using a procedure (Cap-Seq) that we recently developed.8,11 These analyses revealed that our candidate lncRNA is in fact part of an RNA generated by readthrough from an upstream protein-coding gene – thus, we had discovered the first DoG. This discovery prompted us to search our transcript annotation for other, similar transcripts. Indeed, we found DoGs to be a prevalent transcript type – using bioinformatics methods, we identified DoGs downstream of more than 10% of protein-coding genes. In addition to this large number of different DoGs – more than 2000 DoGs genome-wide – each DoG is long (often >45 kb). The diversity and length of DoGs can explain as much as 20% of intergenic transcription.
The Cap-Seq analysis included in our transcript annotation process was important for concluding that DoGs are not initiated at downstream, stress-inducible transcription start sites (TSS), but instead result from transcriptional readthrough. We further confirmed DoGs as readthrough transcripts by using catalytically inactive CRISPR/Cas9 to inhibit transcription of genes found to generate DoGs.8,12 Inhibiting transcription of the upstream gene reduced the levels of the mRNA derived from the associated gene as expected, and importantly, also prevented DoG generation. This observation demonstrated that DoG transcription depends on the transcription of the upstream gene.8 Additionally, we pulled down a transcript that has a downstream DoG by using biotinylated antisense probes and streptavidin beads, followed by qRT-PCR detection of both the upstream transcript and the DoG after pulldown. Thus, we demonstrated that DoGs and upstream coding regions are part of the same transcript.8
By performing cellular fractionation and RNA fluorescence in situ hybridization (FISH), we showed that DoG transcripts remain chromatin bound. Further, using RNA FISH simultaneously for a DoG and for introns of the upstream transcript, we demonstrated that DoGs remain at their site of transcription. RNA FISH also confirmed the robust induction of DoGs by osmotic stress observed by qRT-PCR and RNA-Seq. DoG abundance increases some 10- to 100-fold when osmotic stress is induced by treatment of cells with moderate concentrations of KCl, NaCl, or sucrose. This induction is dependent on IP3 receptor (IP3R) mediated calcium release from the endoplasmic reticulum, which we interpret to cause decreased transcription termination of the upstream transcript.8
The identification of stress-inducible, chromatin-bound DoGs generated by decreased transcription termination of upstream transcripts raises 2 main questions: 1) How is the general mechanism of transcription termination regulated to give rise to DoGs? and 2) What is the function of the simultaneous induction of thousands of long, chromatin-bound ncRNAs? We have partly addressed these questions in our recent publication8 and here discuss them further, providing a more complete overview of the relevant literature.
Stress-Regulated transcription termination generates DoGs
In our recent study,8 we presented 2 main experiments arguing that DoGs are induced at the level of transcription after stress. First, treating cells with actinomycin D, which inhibits transcription and thus allows an assessment of RNA decay, showed that DoG half-lives of ∼1h did not change after osmotic stress. Second, we used the uridine analog 5-ethynyl uridine to label newly synthesized RNA. After incorporation and cell lysis, 5-ethynyl uridine can be attached to biotin (Click-IT, Life Technologies) and labeled RNA is isolated on streptavidin beads. Newly transcribed DoGs isolated using this method showed full induction by osmotic stress after 25 min of labeling, whereas the levels of newly transcribed RNA from the upstream genes were not affected. This lack of effect on upstream transcripts means that DoG induction cannot be due to increased transcription initiation at upstream genes. Instead, the observed induction must result from reduced transcription termination of the upstream transcripts.8 This mechanism of inducing transcription – allowing Pol II engaged in productive elongation to continue past normal termination sites rather than requiring new transcription initiation – is a strikingly simple way for cells to quickly generate new transcripts. However, the question of exactly how this effect on termination is achieved remains to be addressed mechanistically.
Transcription termination requires elongation through a cleavage and polyadenylation (poly(A)) signal, which is most commonly AAUAAA, but can also be AUUAAA or a number of other sequence variants.13 The nascent RNA is cleaved ∼10-40 nucleotides downstream of the poly(A) signal during the process of cleavage and polyadenylation.13 The poly(A) signal is recognized by the cleavage and polyadenylation specific factor (CPSF), which contains several subunits including the catalytic subunit CPSF73 that cleaves the nascent RNA. The cleavage complex also includes cleavage stimulation factor (CstF), cleavage factors I and II, and symplekin. Next, poly(A) polymerase adds non-templated A residues to the 3′-end of the mRNA, and the emerging poly(A) tail is bound and protected by poly(A) binding protein (PABP). Meanwhile, Pol II continues transcribing downstream of the poly(A) signal, generating an RNA with an unprotected 5′-end that results from the cleavage and polyadenylation process. Transcription termination, whereby Pol II releases both the nascent RNA and the template DNA, occurs within nucleotides to kilobases from the poly(A) signal. Two molecular models have been forwarded to explain the termination mechanism, referred to as the allosteric model and the torpedo model. According to the allosteric model, transcription through the poly(A) signal induces conformation changes in Pol II, causing it to dissociate from the RNA and template DNA. The torpedo model instead proposes that the exonuclease Xrn2 gains access to the unprotected 5′-end of the downstream transcript and processively degrades it, likewise causing Pol II dissociation from the template DNA.14,15 Recent evidence has been presented in favor of both models, which are not mutually exclusive and may act together.16,17 In addition to transcription of the poly(A) signal, Pol II pausing near the end of the gene is thought to promote transcription termination.14,15
In further support of our conclusion that DoG induction is caused by decreased transcription termination after stress, knockdown of CPSF73 yielded a modest increase in DoG transcription (3- to 4-fold).8 However, the question of how osmotic stress, through IP3R signaling, results in 10-fold or greater inhibition of cleavage and polyadenylation/termination downstream of many protein-coding genes remains to be addressed. Some potential hypotheses are discussed below, and an overview of factors that affect transcription termination efficiency is provided in Fig. 1.
Figure 1.
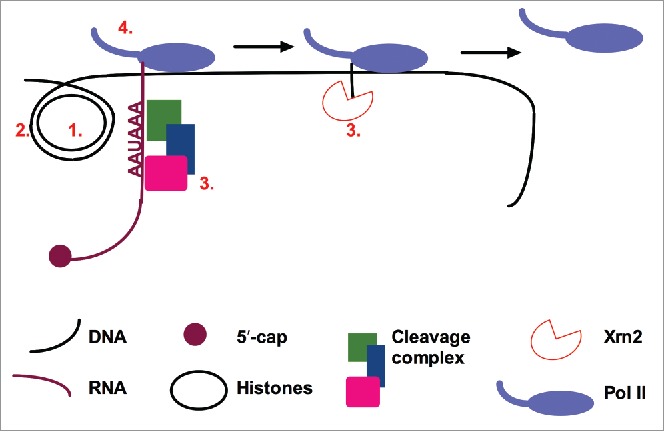
Factors that may affect transcription termination efficiency. 1) Histone acetylation and 2) other chromatin modifications can affect Pol II elongation rate. As Pol II pausing is associated with transcription termination, Pol II elongation rate may affect the efficiency of termination. Further, modification of 3) factors involved in cleavage and polyadenylation or in downstream events, and 4) Pol II itself could affect the efficiency of transcription termination.
One possible antitermination mechanism is suggested by the effect of calcium signaling on Pol II elongation reported by Sharma and colleagues.18 They found that calcium signaling leads to nuclear exclusion of class II histone deacetylases (HDACs), which in turn results in increased histone acetylation. Histone acetylation causes relaxed chromatin structure, making it easier for Pol II to transcribe through nucleosomes. In this way, histone acetylation could increase the Pol II elongation rate. Indeed, the authors found increased Pol II elongation through a number of regions that undergo alternative splicing, which was the focus of their study.18 As transcription termination is enhanced by Pol II pausing near the ends of genes, it is plausible that an enhanced Pol II elongation rate would result in decreased termination. However, we do not observe an osmotic stress-inducible shift in the nuclear/cytoplasmic distribution of a number of class II HDACs investigated in our neuroblastoma cell line SK-N-BE(2)C (our unpublished results), making this hypothesis less likely to explain DoG production. Additionally, it is unclear that an effect on reduced histone deacetylation would be fast enough to explain the rapid kinetics of full DoG induction within 25 minutes.8
On the other hand, calcium signaling is known to induce histone acetylation through other pathways.19-21 Other effects on chromatin structure could also affect termination, as other chromatin modifications are important for Pol II transcription activity (reviewed in22). Therefore, investigating the effects of histone acetylation on DoG induction, as well as correlating DoGs with various chromatin marks, may lead to insights into the mechanism of DoG transcription.
It is possible that one or more of the factors involved in transcription termination, or Pol II itself, is regulated by calcium signaling to allow DoG transcription. For example, calcium signaling causes the activation of several protein kinases (reviewed in23), and many of the factors involved in cleavage and polyadenylation are targeted by phosphorylation.24 Yet, the potential relevance of these modifications and the kinases responsible for them remain unknown. So far, the importance of individual termination factors in DoG generation remains to be tested, with 2 exceptions: Knockdown of CPSF73 mildly promotes DoG transcription,8 while our preliminary results on Xrn2 knockdown found no effect on DoG generation. However, a recent study reports that Xrn2 knockdown generates robust termination defects only when combined with the expression of a dominant-negative Xrn2 variant,17 suggesting that this latter negative result may be inconclusive. In addition to these termination factors, calcium signaling could regulate phosphorylation of the Pol II C-terminal domain (CTD), whose serine 2 phosphorylation peaks close to transcription end sites (TES).25
Another outstanding question concerns why DoGs are transcribed downstream of certain, but not all, genes. We sought to identify characteristics distinguishing DoG-generating genes from other genes. Motif searches on the 200 nt surrounding annotated TESs using MEME/DREME software26 did not reveal any particular motif that could serve as a binding site for gene-specific regulatory factors. However, our bioinformatic analyses suggested that genes exhibiting low or no readthrough after stress are likely to have exceptionally strong poly(A) sites, defined as sites more likely to contain the canonical poly(A) signal AAUAAA, as well as having an increased frequency of additional sequences reported to promote cleavage and polyadenylation (see13 for details). Conversely, we found that the 5′-most 5 kb of DoG regions are relatively depleted in poly(A) signals.8 Further support for a link between weaker/fewer poly(A) sites and DoG generation comes from a recent study of transcriptional readthrough in response to Herpes Simplex virus 1 (HSV-1) infection: cellular genes exhibiting higher levels of readthrough were less likely to possess the canonical poly(A) site AAUAAA than genes exhibiting less readthrough.27 These observations further implicate the process of cleavage and polyadenylation/transcription termination as central for DoG induction. However, the exact details of how stress acts to inhibit termination, and why this inhibition affects some genes more than others, remain to be elucidated.
Putative DoG functions
DoG retention at their sites of transcription, combined with the fact that DoGs are induced all over euchromatin, suggests that after stress, euchromatin is largely covered by long transcripts of diverse sequence. This observation supports the notion that DoG function is sequence independent and acts on euchromatin in response to stress. The literature offers a few examples of such sequence independent functions of RNAs in chromatin. It was recently shown that chromosome territories – the part of the nuclear scaffold that surrounds individual chromosomes – depend on diverse repetitive-element containing RNA for their integrity.28 Such RNAs are necessary for maintaining euchromatin as shown by chromatin collapse in response to transcription inhibition in cells that had divided and therefore had lost these exceptionally stable RNAs, but not in undivided cells with these RNAs intact. Another report identified a function for a group of nuclear RNAs in maintaining euchromatin29 by observing chromatin collapse in response to nuclear injection of RNases. These authors further identified the RNA species responsible for maintaining normal chromatin structure as a diverse set of RNAs including nuclear-retained mRNAs with long 3′UTRs.29 Such features – nuclear transcripts derived from coding genes that have long 3′-extensions – raise the possibility that some of those RNAs may actually have been DoGs. Taken together, these reports of ncRNAs functioning in the nuclear scaffold suggests the possibility of a similar role for DoGs.
The nuclear scaffold supports chromatin and maintains nuclear structure. The scaffold is thought to be comprised largely of lamin proteins, which are capable of generating a meshwork that forms inside of the nuclear envelope and contacts chromatin.30 Maintaining chromatin structure is essential for DNA repair, replication and transcription, underscoring the importance of an intact nuclear scaffold.31 Notably, mice lacking all lamin proteins survive embryonic development, revealing the importance of other nuclear scaffold components.30 Interestingly, the 2 stress conditions – osmotic stress and heat shock – which we found to induce a number of DoGs,8 are both associated with nuclear scaffold stress.31,32 Hyperosmotic stress, as used in our study,8 induces nuclear stress by causing water to leave the cell, leading to nuclear shrinkage and chromatin condensation.31 Additionally, the nuclear scaffold is sensitive to heat shock because many of its protein constituents are heat labile and denature at temperatures above 43 degrees.32 See Fig. 2 for illustration.
Figure 2.
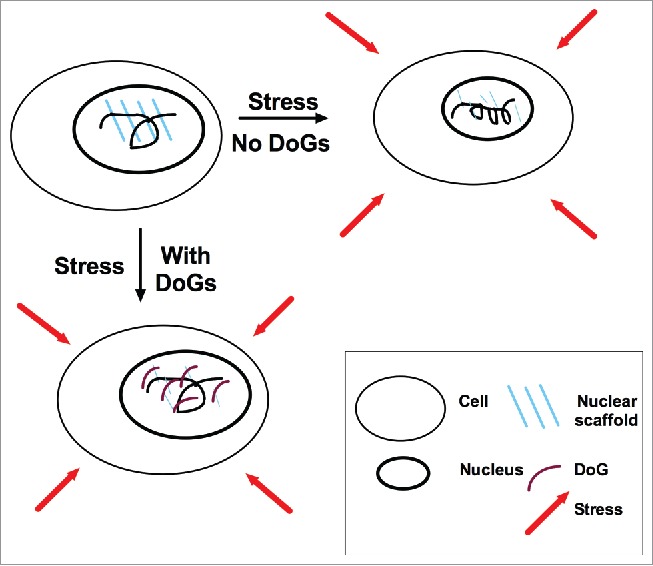
Model for putative DoG function. After exposure to stress – including osmotic stress – the nuclear scaffold is compromised due to stress sensitivity of its protein constituents, and DoGs are induced as reinforcement. The upper-left panel depicts an unstressed nucleus. The upper-right panel depicts a hypothetical nucleus that lacks DoG induction in response to stress: the nuclear scaffold is weakened, the nucleus shrinks and the chromatin condenses. The lower-left panel depicts a stressed nucleus with DoG induction: the DoGs reinforce the nuclear scaffold, resulting in the maintenance of euchromatin and nuclear size.
The fact that DoG-inducing stress compromises the nuclear scaffold, in combination with roles for diverse RNAs in supporting chromatin, as mentioned above, suggests that DoGs are induced to function in nuclear scaffold reinforcement. This hypothesis prompted us to investigate the effect of osmotic stress on the nuclear scaffold in the absence of DoGs. We subjected SK-N-BE(2)C neuroblastoma cells to osmotic stress in the presence of the IP3R inhibitor 2-APB, which prevents DoG induction, and found that nuclear phenotypes associated with osmotic stress were aggravated.8 Specifically, we observed chromatin condensation and decreased nuclear size in response to osmotic stress after 2-APB treatment,8 supporting a role for DoGs in maintaining chromatin integrity after nuclear stress. While this experiment, which relies on general IP3R inhibition, needs to be complemented with more specific methods, it nonetheless suggests an important role for DoGs in the nuclear scaffold stress response with implications for DoG function in cell and tissue homeostasis. A putative model for DoG function is presented in Fig. 2.
Mounting evidence suggests that DoG induction may not be limited to osmotic conditions, but may be a more general response to stress. As mentioned above, our results indicate increased expression of a number of DoGs also in response to heat shock.8 Further, a recent study found widespread induction of readthrough in response to HSV-1 infection,27 suggesting that viral stress may also induce DoG transcription. Such findings are in line with previous observations on the prevention of 3′ cleavage and polyadenylation by the influenza virus protein NS1.33 Additionally, a recent report found a correlation between readthrough and bad prognosis in renal cancer.34 It remains to be shown whether this observation is indeed a correlation between readthrough and malignancy, or if osmotic stress experienced specifically by kidney cells accentuates this correlation in renal cancer. However, in the light of this report and the fact that we identified the first DoG when studying a transcript associated with bad prognosis in neuroblastoma,8,10 the study of DoGs in oncogenic stress is certainly worth pursuing. Together, all these observations suggest a more widespread occurrence of readthrough transcription in response to stress than previously appreciated.
Outlook
Pervasive transcription generates a plethora of ncRNA transcripts. Even if only a small fraction of these are functional RNA molecules, we can expect many new roles for ncRNAs to be revealed in the future. So far, ncRNAs exhibit surprising diversity in their modes of action. Strikingly, some ncRNAs – most likely including DoGs – have functions that are largely independent of sequence. We envision that deepened understanding of the transcription and function of DoGs, along with other classes of ncRNAs, will continue to generate surprises in the years to come. Ultimately, these unexpected ncRNA functions may provide entirely new angles for approaching questions of cellular stress and disease.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Johanna Withers and Paulina Pawlica for critical discussion and Angela Miccinello for editorial assistance.
Funding
This work was supported by grant GM026154 from the National Institutes of Health. A.V. was supported by the Wenner-Gren Foundations, the Swedish Society for Medical Research and the Sweden-America Foundation. J.A.S is an Investigator at the Howard Hughes Medical Institute.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014; 157:77-94; PMID:24679528; http://dx.doi.org/ 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 2.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013; 154:26-46; PMID:23827673; http://dx.doi.org/ 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al.. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458:223-7; PMID:19182780; http://dx.doi.org/ 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 2010; 8:e1000384; PMID:20485488; http://dx.doi.org/ 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010; 465:182-7; PMID:20393465; http://dx.doi.org/ 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science 2008; 322:1849-51; PMID:19056940; http://dx.doi.org/ 10.1126/science.1162253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 2013; 9:e1003569; PMID:23818866; http://dx.doi.org/ 10.1371/journal.pgen.1003569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA. Widespread inducible transcription downstream of human genes. Mol Cell 2015; 59:449-61; PMID:26190259; http://dx.doi.org/ 10.1016/j.molcel.2015.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maris JM. Recent advances in neuroblastoma. N Engl J Med 2010; 362:2202-11; PMID:20558371; http://dx.doi.org/ 10.1056/NEJMra0804577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mestdagh P, Fredlund E, Pattyn F, Rihani A, Van Maerken T, Vermeulen J, Kumps C, Menten B, De Preter K, Schramm A, et al.. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene 2010; 29:3583-92; PMID:20383195; http://dx.doi.org/ 10.1038/onc.2010.106 [DOI] [PubMed] [Google Scholar]
- 11.Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Šestan N, Steitz JA. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell 2013; 155:1568-80; PMID:24360278; http://dx.doi.org/ 10.1016/j.cell.2013.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al.. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013; 154:442-51; PMID:23849981; http://dx.doi.org/ 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian B, Graber JH. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA 2012; 3:385-96; PMID:22012871; http://dx.doi.org/ 10.1002/wrna.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Gen Dev 2009; 23:1247-69; PMID:19487567; http://dx.doi.org/ 10.1101/gad.1792809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porrua O, Libri D. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol 2015; 16:190-202; PMID:25650800; http://dx.doi.org/ 10.1038/nrm3943 [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Rigo F, Martinson HG. Poly(A) Signal-dependent transcription termination occurs through a conformational change mechanism that does not require cleavage at the poly(A) site. Mol Cell 2015; 59:437-48; PMID:26166703; http://dx.doi.org/ 10.1016/j.molcel.2015.06.008 [DOI] [PubMed] [Google Scholar]
- 17.Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, Nguyen T, Karp S, Bentley DL. Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol Cell 2015; 60:256-67; PMID:26474067; http://dx.doi.org/ 10.1016/j.molcel.2015.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, Lou H. Calcium-mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc Natl Acad Sci U S A 2014; 111:E4920-8; PMID:25368158; http://dx.doi.org/ 10.1073/pnas.1408964111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maharana C, Sharma KP, Sharma SK. Depolarization induces acetylation of histone H2B in the hippocampus. Neuroscience 2010; 167:354-60; PMID:20167251; http://dx.doi.org/ 10.1016/j.neuroscience.2010.02.023 [DOI] [PubMed] [Google Scholar]
- 20.Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem 2004; 279:40545-59; PMID:15273246; http://dx.doi.org/ 10.1074/jbc.M402229200 [DOI] [PubMed] [Google Scholar]
- 21.Chandramohan Y, Droste SK, Arthur JS, Reul JM. The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-D-aspartate/mitogen- and stress-activated kinase signalling pathway. Eur J Neurosci 2008; 27:2701-13; PMID:18513320; http://dx.doi.org/23085255 10.1111/j.1460-9568.2008.06230.x [DOI] [PubMed] [Google Scholar]
- 22.Mischo HE, Proudfoot NJ. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochimica et Biophys Acta 2013; 1829:174-85; PMID:23085255; http://dx.doi.org/ 10.1016/j.bbagrm.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clapham DE. Calcium signaling. Cell 2007; 131:1047-58; PMID:18083096; http://dx.doi.org/ 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- 24.Ryan K, Bauer DL. Finishing touches: post-translational modification of protein factors involved in mammalian pre-mRNA 3′ end formation. Int J Biochem Cell Biol 2008; 40:2384-96; PMID:18468939; http://dx.doi.org/ 10.1016/j.biocel.2008.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 2010; 17:1272-8; PMID:20818391; http://dx.doi.org/ 10.1038/nsmb.1903 [DOI] [PubMed] [Google Scholar]
- 26.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucl Acids Res 2009; 37:W202-8; PMID:19458158; http://dx.doi.org/ 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutkowski AJ, Erhard F, L'Hernault A, Bonfert T, Schilhabel M, Crump C, Rosenstiel P, Efstathiou S, Zimmer R, Friedel CC, et al.. Widespread disruption of host transcription termination in HSV-1 infection. Nat Commun 2015; 6:7126; PMID:25989971; http://dx.doi.org/ 10.1038/ncomms8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, Fackelmayer FO, Lawrence JB. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell 2014; 156:907-19; PMID:24581492; http://dx.doi.org/ 10.1016/j.cell.2014.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caudron-Herger M, Muller-Ott K, Mallm JP, Marth C, Schmidt U, Fejes-Toth K, Rippe K. Coding RNAs with a non-coding function: maintenance of open chromatin structure. Nucleus 2011; 2:410-24; PMID:21983088; http://dx.doi.org/ 10.4161/nucl.2.5.17736 [DOI] [PubMed] [Google Scholar]
- 30.Hampoelz B, Lecuit T. Nuclear mechanics in differentiation and development. Curr Opin Cell Biol 2011; 23:668-75; PMID:22079175; http://dx.doi.org/ 10.1016/j.ceb.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 31.Finan JD, Guilak F. The effects of osmotic stress on the structure and function of the cell nucleus. J Cell Biochem 2010; 109:460-7; PMID:20024954; http://dx.doi.org/ 10.1002/jbc.22437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roti Roti JL, Kampinga HH, Malyapa RS, Wright WD, vanderWaal RP, Xu M. Nuclear matrix as a target for hyperthermic killing of cancer cells. Cell Stress Chaperones 1998; 3:245-55; PMID:9880237; http://dx.doi.org/ 10.1379/1466-1268(1998)003%3c0245:NMAATF%3e2.3.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell 1998; 1:991-1000; PMID:9651582; http://dx.doi.org/ 10.1016/S1097-2765(00)80099-4 [DOI] [PubMed] [Google Scholar]
- 34.Grosso AR, Leite AP, Carvalho S, Matos MR, Martins FB, Vitor AC, Desterro JM, Carmo-Fonseca M, de Almeida SF. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. eLife 2015; 4:e09214; PMID:26575290 [DOI] [PMC free article] [PubMed] [Google Scholar]


