ABSTRACT
Chromosomal fusions are common in normal and cancer cells and can produce aberrant gene products that promote transformation. The mechanisms driving these fusions are poorly understood, but recurrent fusions are widespread. This suggests an underlying mechanism, and some authors have proposed a possible role for RNA in this process. The unicellular eukaryote Oxytricha trifallax displays an exorbitant capacity for natural genome editing, when it rewrites its germline genome to form a somatic epigenome. This developmental process provides a powerful model system to directly test the influence of small noncoding RNAs on chromosome fusion events during somatic differentiation. Here we show that small RNAs are capable of inducing chromosome fusions in 4 distinct cases (out of 4 tested), including one fusion of 3 chromosomes. We further show that these RNA-mediated chromosome fusions are heritable over multiple sexual generations and that transmission of the acquired fusion is associated with endogenous production of novel piRNA molecules that target the fused junction. We also demonstrate the capacity of a long noncoding RNA (lncRNA) to induce chromosome fusion of 2 distal germline loci. These results underscore the ability of short-lived, aberrant RNAs to act as drivers of chromosome fusion events that can be stably transmitted to future generations.
KEYWORDS: Chromosome fusion, long noncoding RNA, Oxytricha, piRNAs
Abbreviations
- MAC
macronucleus
- MIC
micronucleus
- MDS
macronuclear destined sequence
- IES
internal eliminated sequence
Introduction
Chromosome fusions are common in cancers1,2 and can drive transformation3-7; however, the molecular basis of these chromosomal fusions is largely unknown. Many fusions are recurrent and associated with an underlying tumor type.2,8 In at least one instance, the presence of a chimeric trans-spliced RNA precedes the corresponding chromosomal fusion, raising the possibility that the incorrectly-spliced RNA triggers DNA rearrangement.9,10 Guide RNAs in the context of CRISPR-Cas9 can also reproduce common chromosomal translocations11,12 and small RNA molecules have been implicated in DNA damage response and repair in plants and animals.13,14 Extrachromosomal circular DNA molecules are also among the aberrant DNA structures observed in both normal and cancer cells15-17 but their mechanism of formation is unknown. Because natural RNA-guided genome editing events are abundant in ciliates, where they are tightly regulated by both small and long noncoding RNAs,18-25 these organisms provide unique model systems to test the hypothesis that transient, aberrant RNA molecules could mediate abnormal chromosome fusion events, which we demonstrate here.
The ciliate Oxytricha possesses 2 distinct types of nuclei: a germline micronucleus and a somatic macronucleus26 that develops by programmed rearrangement of a copy of the germline, beginning with loss of ∼90% of sequence complexity.27 An elaborate cascade of RNA-regulated genome rearrangement events reorganizes and joins the ∼225,000 remaining DNA segments (Macronuclear-Destined Sequences, MDSs) to assemble functional coding sequences, while a nearly equivalent number of intragenic spacer DNA segments (Internal Eliminated Sequences, IESs) undergo precise deletion. The resulting somatic genome is also fragmented at chromosome breakage sites into ∼16,000 “nanochromosomes” that average just 3.2 kb26 and typically encode a single gene. During development, the maternal somatic genome produces millions of 27 nt piRNAs that associate with a PIWI protein and specify regions of the germline genome for retention,19,20 as well as a set of of long, noncoding template RNAs that program the order18 and orientation28 of DNA segments during somatic rearrangement. We previously showed that synthetic RNA templates with permuted or inverted gene segments can reprogram the rearrangement pathway across multiple sexual generations18 and that synthetic 27 nt piRNAs can reprogram the retention of DNA regions that are normally deleted.19 Given this documented ability of RNA to restructure Oxytricha's genome, here we tested whether aberrant noncoding RNAs can also program somatic chromosomal fusions in this system, demonstrating the power of such RNA molecules to alter chromosome architecture.
Results
Induction of fused chromosomes by sRNA microinjection
We previously reported that the highly abundant endogenous class of 27 nt piRNAs that specify somatic DNA sequences for retention in Oxytricha19,20 are curiously depleted at nanochromosome ends.19,20 This led us to conjecture that somatic chromosome ends (and associated telomere addition sites) might be defined, in part, by this absence of piRNAs. In addition, terminal MDSs often cluster in the MIC, with precursor MDS sequences immediately adjacent or overlapping27 even if they reside on separate chromosomes in the MAC (i.e., the linkage between them is broken). Therefore, we tested whether microinjection of small, 27 nt RNAs that overlap these telomere addition sites at the ends of 2 gene loci in close proximity to each other in the MIC could reprogram the natural chromosomal breakage between them in favor of somatic chromosomal fusion.
These experiments produced somatic chromosomal fusions in 4 (out of 4) separate small RNA injection experiments: 1) fusing TEBPβ (Telomere End-Binding Protein β) to its adjacent locus, contig11396 (which encodes a RAS family member; Fig. 1a,b; as well as 2) fusing 2 highly scrambled chromosomes, contig9.1 (encoding a predicted calcium-activated potassium channel) to contig310.1 (which encodes a hypothetical protein; Fig. 2a, b); 3) fusing a set of 3 chromosomes (Fig. 3b, S1b,c); and 4) fusing a circularly permuted chromosome to itself (Fig. 4a,b). In each case, injection of a bridging 27 nt sRNA into the cytoplasm of mating cells at 10–15 hours post conjugation promoted chromosomal fusion, allowing the end-to-end linkage of chromosomes in the somatic nucleus that would otherwise be processed as separate nanochromosomes (Fig. 1b, Fig. 2b). Some of the fused chromosomes produced chimeric protein-coding regions (Fig. 3b, Supplemental Alignment 4). In three experiments, microinjection of a control 27 nt single-stranded DNA molecule of identical sequence failed to induce chromosomal fusion (Fig. 1b, 2c, 4b), demonstrating the requirement for RNA and that the observed effect is not simply due to hybridization as a block against chromosome breakage. Microinjection of a 22 nt RNA was also unable to induce fusion of TEBPβ to contig11396, confirming a 27 nt length preference in Oxytricha (negative data not shown). These data suggest that sRNA-mediated chromosomal fusion operates via Oxytricha's piRNA pathway, which we confirmed by sequencing PIWI-associated small RNAs that are endogenously produced during mating. The endogenous production of novel PIWI-associated RNAs by the cell provides the opportunity to propagate the fusion to the next generation (see below and Fig. 1e,f).
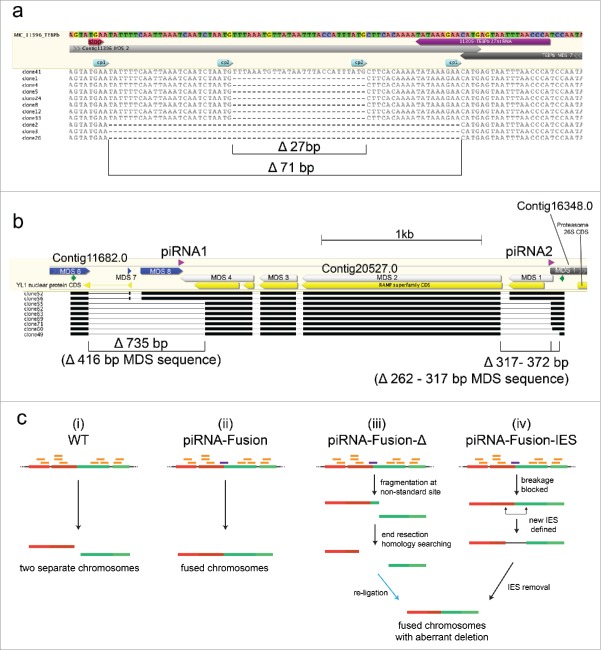
Figure 1.
Small RNA injection leads to heritable fusion of 2 somatic chromosomes encoding the non-scrambled genes, contig11396.0 and TEBPβ. (a) Schematic germline micronuclear (MIC, top) and somatic macronuclear (MAC, middle) maps are shown for each gene. The injected sRNA (purple bar), MDSs (numbered white boxes; TEBPβ MDS 1–7 are in the inverse orientation, indicated by a bar), IESs (gray boxes), somatic telomeres (black vertical rectangles), and a 4 bp overlap (CATG) between the 2 loci are not to scale. Locations of PCR primers are shown as small colored arrows; hybridization probes as thick black lines. (b) Southern analysis provides direct evidence for the presence of full-length somatic chromosome fusions in sRNA-injected but not DNA oligonucleotide-injected cells. “Strip” indicates an image of the stripped membrane before hybridizing to the TEBPβ probe. The full length WT TEBPβ chromosome is 1,858 bp, contig11396 is 1,635 bp and the fusion is predicted 3,493bp; each panel exposed to X-ray film for an equal time (24 hrs). (c) Transgenerational inheritance of the DNA fusion revealed by PCR analysis (with green primers) of a backcross (BC1) to WT strain JRB510. (d) RT-PCR with the same primers using oligo-dT (dT) or random hexamer (hex) primed cDNA from WT and fusion cells reveals conjugation-specific transcription (at 8–10 hrs) across the chromosomal fusion site, relative to asexually growing offspring of injected cells (Veg). (e) Mapping of piRNAs indicates an absence of piRNAs near the chromosomal fusion site in WT cells, but (f) the presence in BC1 cells of newly-produced piRNAs that bridge the normal chromosome ends but are distinct from the injected 27 nt sRNA (shown in purple). To normalize sequencing depth across libraries, the same number of raw, uncompressed reads (35 million) from each library were mapped onto the MIC contig containing TEBPβ and contig11396. Asterisks mark the 5′ ends of novel piRNAs and the injected sRNA (purple); contig11396, green; TEBPβ, blue.
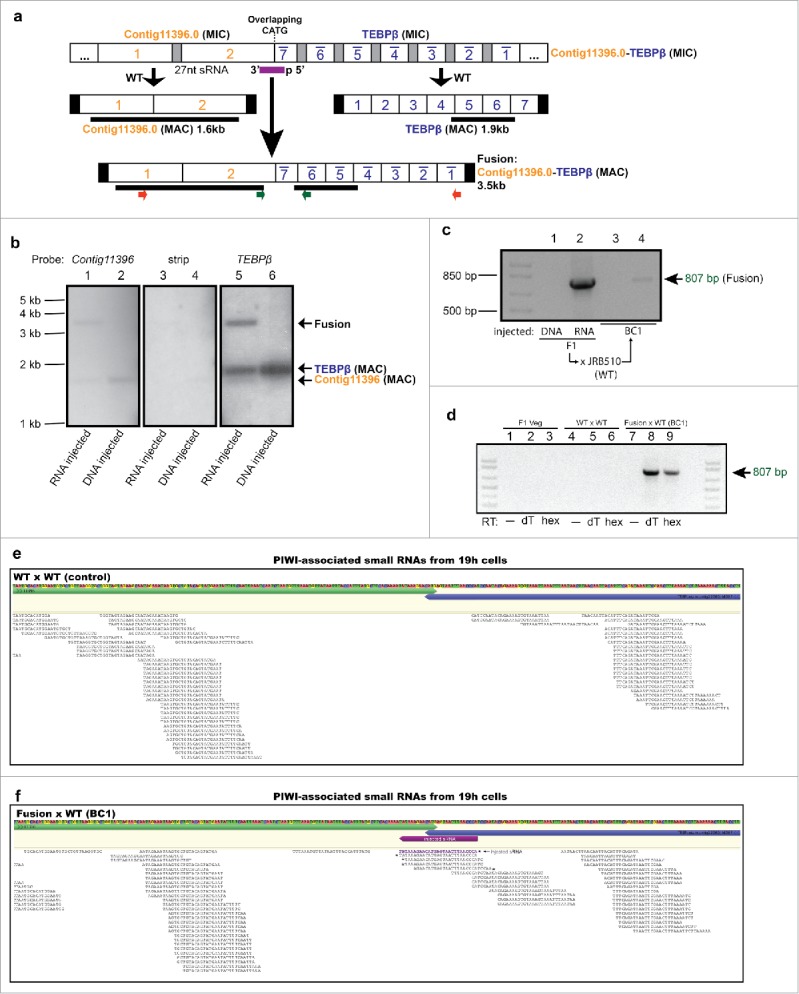
Figure 2.
Small RNA injection leads to heritable somatic fusion of 2 complex loci. (a) Fusion of 2 highly scrambled genes, contig9.1 and contig310.1, whose precursor MDS segments are intertwined in the germline on a 54 kb MIC contig (ctg7180000089708). Partial germline and somatic reference maps are shown, with segment numbers for contig9.1 in blue and contig310.1 in orange; other nomenclature as in Fig. 2 (full germline and somatic maps available: accession numbers given in Data Deposition section); PCR primers used to detect chromosomal fusion are indicated by small arrows above blue MDS 3 (inverted) and orange MDS 2; thick black bars denote Southern hybridization probes for contig9.1, spanning DNA segments 4–15, and for contig310.1 (MDS 2). (b) Southern analysis provides direct evidence for the presence of the full-length chromosome fusion induced by small RNA injection. Quantitative assessment of the phosphorimager signal in lane 4 (injected line 2 probed with contig310.1) suggests that the fusion chromosome is present at roughly half the levels of wild-type contig310.1. “Strip” is the signal remaining before hybridization to the Contig9.1 probe (exposure length and settings the same for each panel); asterisk indicates an aberrant band containing contig9.1 but not contig310.1. JRB310 and JRB510 are compatible WT mating strains of O. trifallax. (c) Transgenerational inheritance of the fusion chromosome revealed by PCR of a backcross to JRB510 cells (BC1, backcross generation 1). (d) Detailed map (not to scale) of the micronuclear locus containing intertwined precursor segments for these genes and 4 others. The upward pointing triangle represents 18 MDSs for 2 other genes (Contig15950 and Contig211.1), and the downward pointing triangle represents 20 MDSs for 3 other genes: Contig13252, Contig15950, and Contig7005.
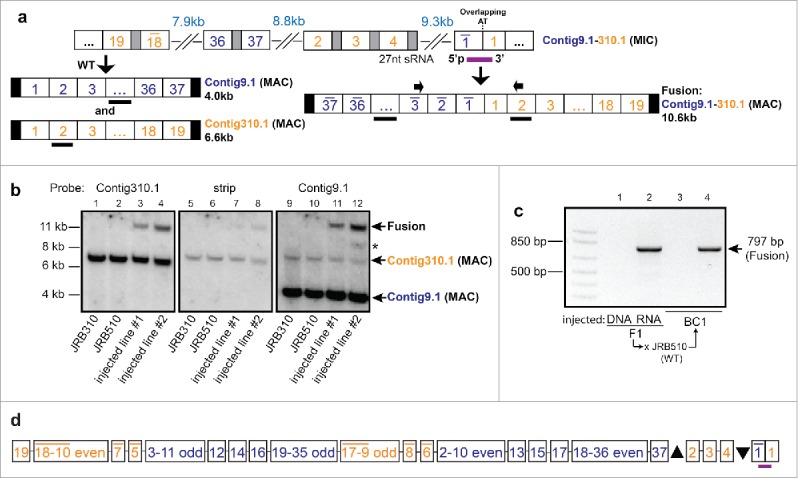
Figure 3.
Detection of aberrant deletions in some fusion products. (a) Deletions detected in the contig11396.0 - TEBPβ fusion. The injected piRNA is indicated in purple, and the clone sequence alignments are shown with dashes for deleted regions. “cp1” and “cp2” represent recombination between cryptic 3 bp pointers that resulted in 71 bp and 27 bp deletions, respectively. Only clone 41 is deletion-free. The stop codon (TGA) for the RAS homolog gene encoded on contig11396.0 is indicated in red. (b) Partial schematic representation of 3 neighboring germline loci and the locations of 2 co-injected small RNAs (purple). Cloning and sequencing of the PCR product between the 2 primers (small green arrows; gel shown in Fig. S1b) revealed fusion of all 3 somatic chromosomes in the progeny of injected cells. Partial germline structures of the 3 chromosomes (Contig11682.0, Contig20527.0, and Contig16348.0) are indicated in blue, light gray, and dark gray, respectively, with the complete gene and exon structures of these loci shown in Fig. S1c. Open reading frames are indicated in yellow (partial for the genes encoding YL1 Nuclear Protein and Proteasome 26S). Complex, combined deletions were observed in several sequenced clones. Black bars indicate sequence aligned regions, with thin black lines showing deleted regions. (c) A model for the formation of aberrant deletions during chromosome fusion. Two neighboring chromosomal loci are shown (indicated as red and green colored lines), each composed of 2 Macronuclear-Destined Segments (MDSs, indicated by slightly different shades of red or green). The injected piRNA is shown in purple and endogenous piRNAs in orange; thin black lines between MDSs indicate Internal Eliminated Sequences (IESs); thin dotted black lines mark flanking sequence. (i) Normal chromosome breakage in WT cells. (ii) piRNA-mediated chromosome fusion without deletions. (iii) piRNA mediated chromosome fusion with deletion. The re-ligation step is marked by a blue arrow. (iv) piRNA-mediated fusion containing a new IES-like deletion.
Figure 4.
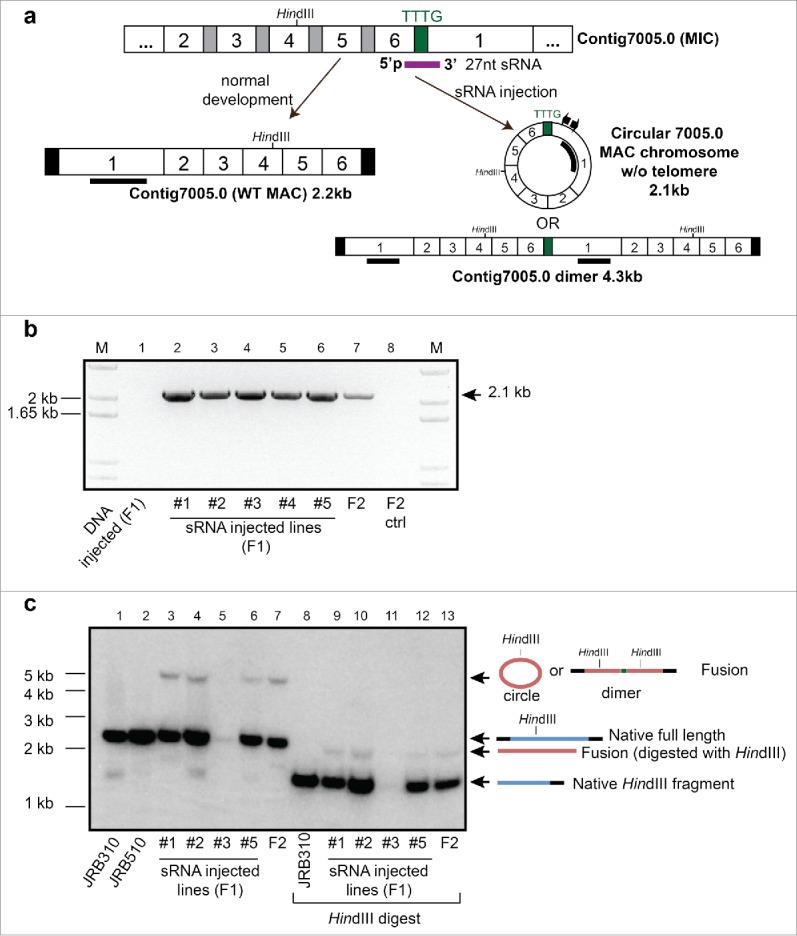
Small RNA injection leads to formation of a putative chromosome circle or dimer. (a) Schematic germline and somatic map of scrambled contig7005.0 and the injected sRNA (purple) that spans the end of the last MDS 6 and beginning of MDS 1, as well as a 4bp sequence that separates them; other nomenclature as in Fig. 2. The pair of inverse PCR primers used to detect the chromosome fusion are shown as small arrows; the thick black bar in MDS 1 denotes the Southern hybridization probe. (b) Inverse PCR confirms the formation and epigenetic inheritance of a fused chromosome in clonal lines derived from F1 and F2 cells. (c) Southern analysis of both undigested and HindIII digested total DNA provides direct evidence for the formation of the fusion in F1 progeny of sRNA-injected cells, as well as the offspring of a mixed mating between F1 lines #1-#5 (F2).
To confirm that IES removal still occurred normally in sRNA-injected cells, we amplified a long region containing most of the fusion between TEBPβ and contig11396 (red primers in Fig. 1a). Cloning and sequencing of these products, and also products from the contig9.1-contig310.1 fusion (Fig. 2c), confirmed end-to-end fusions of mature, IES-lacking somatic chromosomes (Supplemental Alignments 1 and 2). Molecules containing the TEBPβ-contig11396 fusion also contained frequent small deletions near the fusion site (Fig. 3a) that we discuss below. From these results we conclude that proper processing of internal spacer (IES) DNA and reordering of scrambled gene segments occurred independently of the fusion event itself. Southern analysis confirmed that both fusions were hybrids of the full-length wild-type somatic chromosomes (Fig. 1b, 2b) and that fusion efficiency sometimes produced relatively high levels of chimeric chromosomes—on the same order of magnitude as wild-type (Fig. 1b, 2b) (and similar to the levels of naturally occurring variation in Oxytricha chromosome structure26).
To challenge this system to test whether small RNAs can also induce fusion of multiple chromosomes, we co-injected 2 synthetic piRNAs that span the germline boundaries between 3 somatic chromosomes (Fig. 3b, Fig. S1c). While Southern analysis could only demonstrate the presence of a 2-chromosome fusion in the progeny of microinjected cells (Fig. S1a), more sensitive PCR detected the presence of the 3-chromosome fusion in 2 independent replicate experiments (Fig. S1b), confirmed by cloning and sequencing (Fig. 3b, S1c and Supplemental Alignment 4). Notably, because one of the chromosomes in this experiment (Contig11682.0) encodes 3 genes, the resulting double or triple fusions could produce somatic chromosomes bearing 4 or 5 genes (Fig. S1c), creating a level of somatic linkage which is very rare in Oxytricha: The genome sequence reported a maximum of 8 genes on a single somatic chromosome.26 Surprisingly, no consistent functional correlations have been identified among genes that are linked versus unlinked in the macronucleus.26
Aberrant deletions in some fusions support sRNA-programmed fusion
The triple chromosome fusion experiment also produced a complex set of aberrant fusion products that appear as several smaller bands in PCR (Fig. S1b), confirmed by cloning and sequencing (Fig. 3b, and Supplemental Alignment 4). A similar set of aberrant, short deletions was present among the sequences for the TEBPβ-contig11396 fusion products (Fig. 3a). The deletions in these fused chromosomes (e.g. Fig. 3a, lower 10 clones; Fig. 3b lower 7 clones) create new chromosomal architectures not present in either germline (MIC) or somatic (MAC) WT Oxytricha genomes. The deletions all occur near the piRNA binding sites, sometimes removing the piRNA binding sites entirely (Fig. 3b, clone 49 deletes both piRNA sites, while clones 55,62,63,69,71 and 60 are missing the piRNA1 region). Fig. 3a (lower 3 clones) shows that deletions can also partially eliminate a piRNA binding region, and of the 11 clones obtained, only one (clone 41) is identical to MIC sequence at the fusion junction (Fig. 3a).
These small deletions are inconsistent with a simple model of piRNA-mediated DNA retention via blocked DNA cleavage.19 Here we provide an updated model to account for the formation of these unusual sequences (described below and graphically in Fig. 3c). In our model, the novel piRNAs mark or obscure the natural chromosomal breakpoint, thereby preventing normal DNA cleavage at chromosomal termini, so cleavage occurs at nearby, unprotected locations instead (Fig. 3 c(iii). (We and others previously noted a natural depletion of piRNAs at chromosome boundaries, which might make these regions vulnerable to the non-standard cleavage events reported here.19,20) These aberrantly broken DNA molecules may be resected before repair by ligation, leading to deletion of some DNA sequences (which may remove portions of coding sequence or even the piRNA-binding sites; Fig. 3a,b and model Fig. 3c(iii). We hypothesize that the re-ligation events might occur by microhomology-mediated end joining, a non-homologous end-joining (NHEJ) pathway that involves end resection and homology searching.29 Consistent with this, we note that short regions of microhomology are present in all deletion-fusion cases (microhomologies marked in light blue in Fig. 3a, denoted cp1 and cp2, for cryptic pointers, and Supplemental Alignment 4), similar to the intrachromosomal aberrant deletions previously reported in Oxytricha during genome rearrangement events.18,30
Because re-ligation of broken DNA containing deletions requires an extra DNA processing step compared to WT cells (blue arrow in model Fig. 3c(iii)), the presence of deletions in some of the fusions (Fig. 3a,b) suggests that the microinjected piRNA favors re-ligation. DNA molecules that remain broken (even if healed by telomeres) would appear very similar to wild-type chromosomes and not be detected in our Southern or PCR analyses (compare Fig. 3c(i) and Fig. 3 c(iii),(iv)). Long template RNAs can program chromosome structure,18 however, there are no detectable junction-spanning template RNAs (which might guide chromosome ligation events) until after the fusion events occur (Fig. 1d and section on Transgenerational inheritance of chromosome fusions). This suggests that the artificial piRNA injected into the cell serves to reset the normal programming of 2 chromosomal units (red and green in Fig. 3c) to produce a linked chromosomal unit (Fig. 3c(ii, iii, iv) (that may produce its own template RNA, Fig. 1d). In a variation on this model, the aberrant deletions we observe may be excised as novel IESs (Fig. 3c(iv)). In this view, the synthetic piRNA protects the normal fragmentation site against breakage. Unprotected by endogenous piRNAs,19,20 the subtelomeric sequences can be excised as IESs, generating the observed fusion chromosomes containing deletions (Fig. 3c(iv)). While our data do not distinguish these explicit possibilities, we note that even the new IES model (Fig. 3c(iv)) requires an additional DNA re-joining step (after IES removal) that is not present in wild-type cells (Fig. 3c(i)).
The presence of small deletions that overlap the predicted piRNA binding sites after exposure to the new piRNA may seem paradoxical. However, such events are likely rooted in the temporal order of genome programming events. Endogenous piRNAs accumulate (and synthetic piRNAs were microinjected) relatively early in Oxytricha development (18–24 hrs after cell mating).19,20 The abundance of endogenous piRNAs decreases sharply thereafter,19,20 whereas chromosome breakage events have been reported to occur much later.30-34 We propose that Oxytricha piRNAs mediate an epigenetic change to the DNA, which programs the joining of the genic regions to define a new chromosomal unit containing the somatic fusion, even after the piRNA is no longer physically present. Reports of piRNA-mediated DNA methylation35,36 and histone modification37,38 in mice and flies are consistent with this model. So far, DNA methylation has been detected in Oxytricha,39 but as a link to DNA elimination, not DNA retention or chromosome end formation. Because piRNAs are much smaller than single nucleosome units, we propose that any epigenetic change is most likely directly on the DNA.
We note that all the deletions observed after double piRNA injection curiously preserve a reading frame, fusing the coding regions of the RAMP-superfamily gene of contig20527.0 to the YL1 nuclear protein gene of contig11682.0 (Fig. 3b, lower 7 clones). This could theoretically allow for translation of a fusion protein, although the deletion around piRNA2 would cause the RAMP-YL1 fusion protein to initiate translation at an internal ATG (for which several candidates do exist). The piRNA-induced deletions in contig11396-TEBPβ(Fig. 3a) do not fuse coding sequences, since the reading frames for contig11396 and TEBPβ are on opposite strands. Furthermore, the deletion termini occur only in non-coding regions, precluding the formation of chimeric coding sequences in the sequences, as depicted in Fig. 3a.
Transgenerational inheritance of chromosome fusions and detection of new piRNAs
In support of a mechanism whereby endogenously-produced Oxytricha piRNAs mediate transgenerational, epigenetic inheritance of chromosome architecture, we detected the presence of 2 chromosome fusions (TEBPβ-contig11396 and contig9.1-310.1) not only in the F1 progeny of injected cells, but also in a backcross between wild-type and F1 fusion-containing cells (Fig. 1c, Fig. 2c). This confirms that sRNA exposure in the parental generation can program transmission of chromosomal fusions across more than one sexual generation, without further direct experimental manipulation. RT-PCR detected conjugation-specific expression of RNA derived from the TEBPβ-contig11396 fusion in the F1-WT backcross (Fig. 1d). This demonstrates transcription during development across a fusion boundary. The RNA is only detected in the backcross to fusion-containing cells and is absent from wild-type or vegetative cells. Because this RNA is co-expressed in cells that also produce novel piRNAs (Fig. 1f), the transcripts detected in Fig. 1d could be either run-through mRNAs spanning the fusion site, the piRNA precursors, or the template RNA transcripts that span the entire fused chromosome and guide chromosome rearrangement in the next generation.18 Importantly, we next used PIWI-ChIP-sequencing19 to survey the set of pi-RNAs produced in the TEBPβ-contig11396 backcross. This unequivocally revealed the endogenous production of new, boundary-spanning PIWI-associated 27 nt RNAs (piRNAs) only in the backcross between fusion-containing cells and wild-type, but not in a control mating between wild-type cells (Fig. 1e,f). We infer that the production of these novel piRNAs in the next generation offers a mechanism for the transgenerational inheritance and propagation of the fused chromosome architecture.
Fusion of a circularly permuted locus
We also tested whether microinjection of a small RNA bridging the last and first segments of a circularly permuted scrambled gene (contig7005.0, encoding a hypothetical protein with germline DNA segment order 2-3-4-5-6-1; Fig. 4a), could produce either a circular chromosome or a chromosome dimer, fusing a copy of the chromosome to itself. Microinjection of a 27 nt RNA, but not DNA of identical sequence, led to formation of the predicted fused junction, as demonstrated by both inverse PCR (Fig. 4b) and Southern analysis (Fig. 4c).The inverse PCR product (Fig. 4b) and large band observed by Southern analysis (Fig. 4c) are consistent with either a circular chromosome (which would, if not supercoiled, migrate more slowly than a linear isoform) or chromosome dimer; our data do not distinguish these possibilities. This chromosome fusion was also epigenetically inherited in the F2 sexual progeny (Fig. 4b lane 7, and 4c lanes 7 and 13) and stably maintained for approximately 40 asexual cell divisions in the laboratory (Fig. S2).
Long, chimeric RNAs can program chromosome fusion over a distance
We have previously shown that long noncoding RNA molecules (template RNAs) can re-program chromosome structure in Oxytricha,18,28 so here we tested whether they can also cause chromosomal fusions. The genes encoding Telomere End-Binding Protein α (TEBPα) and TEBPβ reside on separate somatic nanochromosomes in wild-type cells and are located at least 49 kb apart in the germline27 (Fig. S3a). To test whether a long, noncoding RNA that bridged them would lead to the production of chimeric, fusion chromosome, we generated a long (1,375 nt) artificial chimeric template RNA fusing the 2 genes (Fig. 5a, Fig. S3b). The fusion RNA template contains the first 3 segments (MDS) of TEBPβ at the 5′ end, fused to segments 14–18 of TEBPα, a scrambled gene, at a 6 bp repeat (GTCGAC). To distinguish injected templates from rearranged endogenous molecules, a C-to-A substitution creating an AflII restriction site was present on the template 90 bp away from the fusion site. Sense (s) or antisense (as) chimeric RNA templates were injected into the cytoplasm of mating cells 10–15 hours post mixing of mating types, which initiates conjugation. Genomic DNA was harvested from the F1 progeny 10 days later, ensuring that genome rearrangement was complete and that any genomic alterations were stably transmitted during asexual growth. PCR primers specific to the chimeric chromosome amplified the fusion chromosome only in the progeny of injected cells but not uninjected controls (Fig. 5b). Cloned PCR products were sequenced (Supplemental Alignment 5) and lacked the AflII site, confirming de novo creation of the fusion chromosome. Southern hybridization using probes targeting either TEBPβ or TEBPα (Fig. 5a,c) both demonstrated chimera formation (Fig. 5c and Fig. S3c) and RT-PCR even detected the presence of RNA transcripts from the fusion chromosome (Fig. 5d). The cloned DNA and cDNA sequences also differed from the injected template at many segregating sites (Supplemental Alignment 5), which confirmed endogenous rearrangement and expression of these loci. These data raise many questions about the relationship between the long and small noncoding RNA pathways that will lead to future investigations. Taken together, the current experiments suggest a surprisingly strong ability of noncoding RNA to regulate genome structure.
Figure 5.
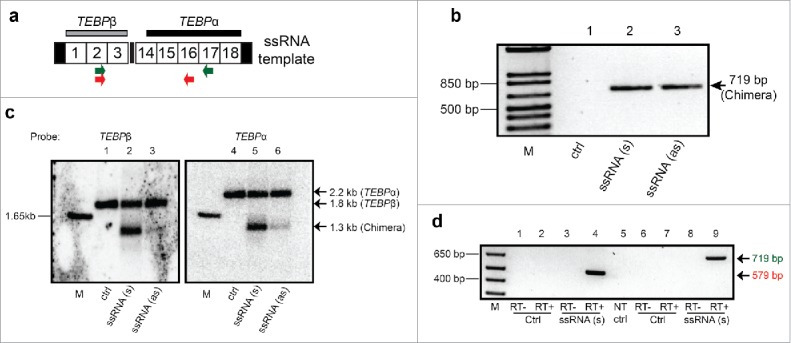
Microinjection of a long chimeric RNA leads to somatic formation of a hybrid TEBPβ/α chromosome. (a) Schematic map of injected RNA (1.375 kb): Gray and black horizontal bars denote Southern hybridization probes for TEBPβ and TEBPα, respectively; Numbered boxes are MDSs (not to scale); terminal black rectangles indicate telomeres; Locations of PCR primers to detect chimeric products are shown as colored arrows. (b) PCR confirms the formation of hybrid TEBPβ/α molecules in the progeny of sense (s) or antisense (as) RNA-injected cells but not uninjected cells (ctrl) (all primer and PCR sequences provided in Supplementary Information). (c) Southern analysis provides direct evidence for the presence of TEBPβ/α chimeric DNA molecules in the same cells used in (b). (d) Oligo-dT primed RT-PCR using either pair of primers detects chimeric RNA transcripts in the progeny of injected cells. Sequencing of the larger band confirmed that these RNA molecules do not contain the point substitution in the injected RNA. NT, no template control, RT+/− indicates the presence of reverse transcriptase enzyme; M, marker.
Discussion
Here, we have shown that aberrant noncoding RNAs can mediate chromosomal fusion in Oxytricha trifallax. Some of these fusions are stably transmitted to the next sexual generation, without further experimental manipulation (i.e., in the absence of the initial triggering molecule) (Figs. 1c, 2c,4b,4c). Notably, the production of de novo piRNAs is associated with epigenetic transmission of these fused chromosomes across generations (Fig. 1e,f). Chromosome fusions were also transmitted across multiple asexual (vegetative) divisions (Fig. S2).
In binucleate ciliates, with both germline and somatic genomes, chromosome breakage is part of a complex process of nuclear development.40 In Oxytricha, approximately 75 precursor (germline) chromosomes27 rearrange and fragment to produce over 16,000 (somatic) ‘nanochromosomes’.26 Our experiments suggest that small RNA-mediated chromosome fusion occurs as a result of complex interaction with the chromosome breakage machinery. While two of the fusions we observed (Fig. 2 and Fig. 4) derive from simple non-breakage of a precursor region (Fig. 3c(ii)), 3 out of 5 sRNAs induced a complex set of nested deletions in the fused chromosomes (Fig. 3a, 3b). The observation of piRNA-induced fusions containing deletions suggests a possible model involving DNA cleavage, end resection, and re-joining (Fig. 3c(iii)). Alternatively, the deletions may result from excision of novel IESs when a piRNA blocks ordinary chromosome fragmentation (Fig. 3c(iv)). In all cases, the microinjected piRNA clearly alters normal DNA processing with profound genomic consequences. Because 27 nt single-stranded DNA control oligonucleotide sequences that target the same regions had no effect (Fig. 1, Fig. 2, and Fig. 4) we conclude that the artificial piRNAs do not simply block access of the chromosome-fragmentation machinery to the breakage site. Oxytricha piRNAs interact with a specific PIWI protein, Otiwi119 (verified here with Otiwi1-IP-seq in Fig. 1f) and we hypothesize that piRNA-mediated recruitment of Otiwi1 is key to reprogramming chromosome breakage to fusion, and subsequent epigenetic transmission of this trait (Fig. 1f).
Unlike some ciliates with defined chromosome breakage signals,41,42 Oxytricha somatic chromosome termini (breakpoints) lack a clear sequence motif for chromosome breakage. They do, however, display a 10 bp periodic purine (A+G) skew43,27, the strength of which correlates with the strength of chromosome fragmentation at that site.27 We previously proposed that a sharp switch from pyrimidine- to purine-richness may regulate chromosome breakage.27 Oxytricha telomere addition is notably imprecise, often occurring within a range of 100 bp around the chromosome terminus in the precursor DNA.26,44 As we noted earlier, the depletion of piRNA binding sites at chromosome termini19 may be an important marker for chromosome fragmentation. Consistent with this hypothesis, the piRNA depleted regions are also purine-rich (usually ∼50 bp on either side of a breakpoint19,27). We favor a model in which the absence of piRNAs specifies regions where chromosome breakage is allowed, and a change in purine content may help target the cleavage machinery toward a more specific site. Future studies are needed to test this or alternative models and to determine whether purine-richness might act by influencing piRNA production (rather than directly guiding the breakage machinery) or whether piRNAs and purine-richness independently affect chromosome breakage site selection.
The enzyme that cuts DNA at the cleavage sites is unknown. Domesticated PiggyBac-family transposases participate in programmed DNA deletion in the distantly-related ciliates, Tetrahymena and Paramecium.45,46 While this class of transposase has not been detected in Oxytricha,26 a high copy-number germline transposase of the Tc1/mariner class has been implicated in its genome rearrangements, instead.47 Since a canonical role of the piRNA pathway is to silence germline transposons,48 a demonstration that piRNAs can potentially block transposase-mediated DNA cleavage would fundamentally extend the accepted paradigm of antagonism between pi-RNAs and transposons.
Our experiments in RNA-mediated genome editing reveal a new and fundamental role for transiently available, aberrant RNAs in disrupting chromosome maintenance and genome integrity, with the ability to trigger somatic fusions. The RNA-mediated transgenerational effects illustrate the power of such transient, triggering RNAs to interact with the entire suite of noncoding RNA molecules that sculpt and maintain genome architecture, constructing a heritable, epigenetic memory of somatic genome structure for future generations.
Our findings are also timely, given the discovery that Piwi proteins, the germline-specific binding partners of piRNAs, are re-expressed in diverse cancers.49-52 More work is needed to elucidate both the potential involvement of piRNAs in cancer52,53 and any mechanistic similarities between piRNA involvement in Oxytricha and humans. Recent reports suggest that a piRNA (piRNA-823) plays key roles in multiple myeloma and gastric cancer.54,55 Correspondingly, studies of transiently expressed, aberrant piRNAs or other noncoding chimeric RNAs56 in human somatic tissues may shed light on the poorly understood origin of a subset of chromosomal fusions and extrachromosomal circles in humans. We note that a recent study found that fusion chromosomes produce circular noncoding RNA transcripts that can drive oncogenesis.57 Our data, presented here, suggest that some fusion RNA molecules—‘trigger RNAs'—may actually precede, and drive, DNA fusion events9,10 with the potential for driving oncogenesis.9,10 By bringing disparate regions of the genome into proximity, chromosome fusion could directly increase the production of oncogenic fusion RNA molecules, either circular57 or linear.2,8,56
While ciliates and humans may be more than 2 billion years diverged58 and their underlying genomics are quite different, ciliates offer many variations on familiar themes in eukaryotic biology. For example, cytosine DNA methylation, used in animals and plants to silence gene expression,59 in Oxytricha marks DNA for elimination39—which may be viewed as an extreme form of silencing. Ciliate piRNAs, first defined in Tetrahymena,25 distinguished deleted60,61 or retained DNA segments,19,20 rather than transposons, as in mammals and flies.35,36 Like metazoa, ciliates have a distinct germ line and soma, but relegate these functions to separate nuclei.40 We therefore anticipate that ciliate-driven insights into somatic genome structure and chromosome stability may provide clues to somatic diseases of the genome, such as cancer.
Materials and methods
Cell culture and mating
Two Oxytricha trifallax mating strains JRB310 and JRB510 were cultured in Pringsheim medium (0.11 mM Na2HPO4, 0.08 mM MgSO4, 0.85 mM Ca(NO3)2, 0.35 mM KCl at pH 7.0) with the algae Chlamydomonas reinhardtii as the main food source, supplemented with Klebsiella pneumoniae. To induce mating, cells from 2 compatible mating types were starved and mixed in equal numbers. Cells began mating approximately 2 hours post-mixing with 80–90% conjugation efficiency.
Microinjection of synthetic oligonucleotides
27 nt DNA and RNA oligonucleotides were purchased from IDT DNA with standard desalting and used without further purification. Prior to each injection, 3μL of an oligonucleotide in nuclease free water (Ambion) (15μg/μL) was heated at 65 °C for 2 min, and chilled on ice for at least 2 min, then injected (Narishige IM 300) into the cytoplasm of mating cells 10–15 hours post mixing of mating types. For each trial, ∼50 pairs of cells were injected, and either pooled together or cultured individually post injection. The injected cells were allowed to separate and grow asexually for either 7 days (pooled cells) or 20–30 days (individual cell lines) before genomic DNA was harvested for subsequent PCR and Southern analyses. For single-cell analysis, individual cells were hand picked in 5μL cell culture medium and used directly as the PCR template.
To obtain F2 or F1 backcross progeny, F1 cells were cultured asexually for 20–30 days (more than 40 asexual generations) and then starved to induce either selfing or backcrossing (to a WT JRB510 parental stain). Mating pairs were hand picked and cultured together in a separate pool. We allowed conjugation to proceed and the isolated cells to divide asexually for another 7 days before genomic DNA was harvested for PCR and Southern analysis.
RT-PCR
RNA was extracted from vegetative cells using Mirvana RNA isolation kit (Ambion). Residual DNA was digested from RNA samples using Turbo-DNA free kit (Invitrogen). First strand cDNA was prepared using Superscript III (Invitrogen) and oligo-dT primer, following manufacturer's instructions. The cDNA was used as the template in subsequent PCR experiments using FastStart High Fidelity DNA polymerase (Roche Applied Science) with gene-specific primers indicated below.
PCR analysis post microinjection
PCR products were amplified with FastStart High Fidelity DNA polymerase (Roche Applied Science) with oligonucleotide primers indicated below. Typical reactions were 35 cycles with an annealing temperature of 55–60 °C and an extension time consistent with amplicon length (1 min per kb).
Southern hybridization
Genomic DNA (∼2 μg) was electrophoresed in an EtBr stained 1% agarose gel, depurinated (0.25% HCl 15 min, washed in 0.4 M NaOH for 15 min) and transferred to Hybond XL membrane in 0.4 M NaOH solution using Nytran TurboBlotter (GE Healthcare). Labeled probes were generated via random priming of corresponding wild-type Oxytricha PCR products. After overnight hybridization at 60 °C (0.5 M NaPO4, pH 7.2, 1% BSA, 1 mM EDTA, 7% SDS), the membrane was washed in 0.2 × SSC with 0.1% SDS (30 min, 60 °C).
piRNA isolation, deep sequencing, and mapping
The antibody used to immunoprecipitate (IP) piRNAs is anti-PIWIL1 (Abcam, ab12337), originally raised against the human PIWIL1 protein C-terminal peptide. Immunoprecipitation, Illumina library preparation, deep sequencing, and bioinformatic analysis of Piwi-associated small RNAs from the sexual progeny of both WT and backcrossed cells followed our published protocol.19 Small RNA sequences were mapped onto the micronuclear region containing contig11396-TEBPβ with the gmapper command in the SHRiMP software package,62 setting the threshold score to 80% of the maximum possible score using the ‘-h 80%' flag. To ensure comparable depth between control and fusion libraries, we subsampled to equivalent sequencing depth of 35 million reads per library.
Microinjection of chimeric TEBPβ/α fusion RNA template
A DNA version of the template was synthesized by ligating the SalI digested PCR products of TEBPβ and TEBPα (Fig. S3b). To distinguish injected template from endogenous chimeric molecules in the progeny of injected cells, an AflII restriction site was introduced on the template via point mutation in the PCR primer. RNA versions of each strand were produced by in vitro transcription of PCR products from the corresponding DNA template between the vector T7 promoter and the telomere on the opposite side. After DNase treatment (Invitrogen), RNA templates were injected (Narishige IM 300) into the cytoplasm of each mating cell in a conjugating pair at 10–15 hours post cell mixing, as described.18 For each RNA template, ∼40 pairs of cells were injected and pooled together at the end of injection. The pooled cells were allowed to separate and to grow asexually for 10 days before genomic DNA and RNA were harvested for subsequent PCR, Southern, and RT-PCR analyses.
Availability of supporting data
Small RNA sequences have been deposited in Gene Expression Omnibus (GEO: www.ncbi.nlm.nih.gov/geo/) under accession number (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE78993), and germline DNA sequences have been deposited to GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under accession number (ARYC00000000); data also available on the Oxytricha trifallax genome database (oxy.ciliate.org). The contig sequences used in this manuscript are given at http://trifallax.princeton.edu/cms/raw-data/genome/mic/chromosome_fusion_mic_loci.fa; the annotated MIC-MAC maps for these sequences are available at: http://trifallax.princeton.edu/cms/raw-data/genome/mic/chromosome_fusion_mic_loci.gff. MAC chromosome data and annotations available at http://oxy.ciliate.org
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Jessica Wiggins, Wei Wang, Donna Storton, and Lance Parsons for Illumina sequencing; Jaspreet Khurana and Kathryn Bracht for comments on the manuscript, Wenwen Fang for the PIWI antibody, and Charles Warden for earlier work on designing the long chimeric chromosome. We also thank Thomas Coate (Georgetown University) for sharing equipment. This work was supported by NIH grants GM59708 and GM111933 and NSF grants 0923810 and 0900544 to L.F.L. and NIH grant 1F32GM099462 to J.R.B.
Author contributions
X.W., L.F.L. and J.R.B designed the sRNA study; X.W. performed microinjections of 27 nt small RNAs and PCR analysis with help from D.M.C. and S.S.M; J.R.B. did the Southern analysis of piRNA-mediated fusions, with help from G.J.U. and E.C.C., and performed small RNA sequencing and analysis. M.N. and L.F.L. designed the lncRNA-mediated fusion experiments, which were carried out by K.S. and M.N.. X.C. provided and suggested micronuclear sequences, loci, and maps for piRNA-mediated fusion experiments. J.W. cultured cells for the lnc-RNA experiment. X.W., J.R.B, K.S., M.N. and L.F.L. analyzed the data. J.R.B., X.W., and L.F.L. wrote the manuscript.
References
- 1.Sankar S, Lessnick SL. Promiscuous partnerships in Ewing's sarcoma. Cancer Genet 2011; 204:351-65; PMID:21872822; http://dx.doi.org/ 10.1016/j.cancergen.2011.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 2007; 7:233-45; PMID:17361217; http://dx.doi.org/ 10.1038/nrc2091 [DOI] [PubMed] [Google Scholar]
- 3.Heisterkamp N, Jenster G, Tenhoeve J, Zovich D, Pattengale PK, Groffen J. Acute-Leukemia in Bcr/Abl Transgenic Mice. Nature 1990; 344:251-3; PMID:2179728; http://dx.doi.org/ 10.1038/344251a0 [DOI] [PubMed] [Google Scholar]
- 4.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The C-Myc Oncogene Driven by Immunoglobulin Enhancers Induces Lymphoid Malignancy in Transgenic Mice. Nature 1985; 318:533-8; PMID:3906410; http://dx.doi.org/ 10.1038/318533a0 [DOI] [PubMed] [Google Scholar]
- 5.Daley GQ, Vanetten RA, Baltimore D. Induction of Chronic Myelogenous Leukemia in Mice by the P210bcr/Abl Gene of the Philadelphia-Chromosome. Science 1990; 247:824-30; PMID:2406902; http://dx.doi.org/ 10.1126/science.2406902 [DOI] [PubMed] [Google Scholar]
- 6.Elefanty AG, Hariharan IK, Cory S. bcr-abl, the hallmark of chronic myeloid leukaemia in man, induces multiple haemopoietic neoplasms in mice. Embo J 1990; 9:1069-78; PMID:1691092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Garcia A, Sanchez-Martin M, Perez-Losada J, Perez-Mancera PA, Sagrera-Aparisi A, Gutierrez-Cianca N, Cobaleda C, Sánchez-García I. Selective destruction of tumor cells through specific inhibition of products resulting from chromosomal translocations. Curr Cancer Drug Targets 2001; 1:109-19; PMID:12188884; http://dx.doi.org/ 10.2174/1568009013334214 [DOI] [PubMed] [Google Scholar]
- 8.Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2013; http://cgap.nci.nih.gov/Chromosomes/Mitelman [Google Scholar]
- 9.Rowley JD, Blumenthal T. Medicine - The cart before the horse. Science 2008; 321:1302-4; PMID:18772424; http://dx.doi.org/ 10.1126/science.1163791 [DOI] [PubMed] [Google Scholar]
- 10.Li H, Wang JL, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science 2008; 321:1357-61; PMID:18772439; http://dx.doi.org/ 10.1126/science.1156725 [DOI] [PubMed] [Google Scholar]
- 11.Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nature Communications 2014; 5:3964; PMID:24888982; http://dx.doi.org/ 10.1038/ncomms4964 [DOI] [PubMed] [Google Scholar]
- 12.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, . et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 2014; 516:423-7; PMID:25337876; http://dx.doi.org/ 10.1038/nature13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell 2012; 149:101-12; PMID:22445173; http://dx.doi.org/ 10.1016/j.cell.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 14.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012; 488:231-5; PMID:22722852; http://dx.doi.org/ 10.1038/nature11179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn PJ. Molecular biology of double-minute chromosomes. Bio Essays 1993; 15:477-84 [DOI] [PubMed] [Google Scholar]
- 16.Barker PE. Double minutes in human tumor cells. Cancer Genetics Cytogenetics 1982; 5:81-94; PMID:6175392; http://dx.doi.org/ 10.1016/0165-4608(82)90043-7 [DOI] [PubMed] [Google Scholar]
- 17.Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, Dutta A. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 2012; 336:82-6; PMID:22403181; http://dx.doi.org/ 10.1126/science.1213307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 2008; 451:153-8; PMID:18046331; http://dx.doi.org/ 10.1038/nature06452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-Interacting RNAs Protect DNA against Loss during Oxytricha Genome Rearrangement. Cell 2012; 151:1243-55; PMID:23217708; http://dx.doi.org/ 10.1016/j.cell.2012.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zahler AM, Neeb ZT, Lin A, Katzman S. Mating of the stichotrichous ciliate Oxytricha trifallax induces production of a class of 27 nt small RNAs derived from the parental macronucleus. PLoS ONE 2012; 7:e42371; PMID:22900016; http://dx.doi.org/ 10.1371/journal.pone.0042371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lepere G, Betermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev 2008; 22:1501-12; PMID:18519642; http://dx.doi.org/ 10.1101/gad.473008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lepere G, Nowacki M, Serrano V, Gout JF, Guglielmi G, Duharcourt S, Meyer E. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res 2009; 37:903-15; PMID:19103667; http://dx.doi.org/ 10.1093/nar/gkn1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nowacki M, Haye JE, Fang W, Vijayan V, Landweber LF. RNA-mediated epigenetic regulation of DNA copy number. Proc Natl Acad Sci U S A 2010; 107:22140-4; PMID:21078984; http://dx.doi.org/ 10.1073/pnas.1012236107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garnier O, Serrano V, Duharcourt S, Meyer E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol Cell Biol 2004; 24:7370-9; PMID:15314149; http://dx.doi.org/ 10.1128/MCB.24.17.7370-7379.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 2002; 110:689-99; PMID:12297043; http://dx.doi.org/ 10.1016/S0092-8674(02)00909-1 [DOI] [PubMed] [Google Scholar]
- 26.Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, . et al The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol 2013; 11:e1001473; PMID:23382650; http://dx.doi.org/25171416 10.1371/journal.pbio.1001473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Bracht JR, Goldman AD, Dolzhenko E, Clay DM, Swart EC, Perlman DH, Doak TG, Stuart A, Amemiya CT, . et al. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell 2014; 158:1187-98; PMID:25171416; http://dx.doi.org/ 10.1016/j.cell.2014.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowacki M, Shetty K, Landweber LF. RNA-Mediated Epigenetic Programming of Genome Rearrangements. Annual Rev Genomics Hum Genetics 2011; 12:367-89; PMID:21801022; http://dx.doi.org/23610439 10.1146/annurev-genom-082410-101420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truong LN, Li Y, Shi LZ, Hwang PY, He J, Wang H, Razavian N, Berns MW, Wu X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A 2013; 110:7720-5; PMID:23610439; http://dx.doi.org/ 10.1073/pnas.1213431110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mollenbeck M, Zhou Y, Cavalcanti AR, Jonsson F, Higgins BP, Chang WJ, Juranek S, Doak TG, Rozenberg G, Lipps HJ, et al.. The pathway to detangle a scrambled gene. PLoS ONE 2008; 3:e2330; PMID:18523559; http://dx.doi.org/ 10.1371/journal.pone.0002330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ammermann D, Steinbruck G, von Berger L, Hennig W. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma 1974; 45:401-29; PMID:4209692; http://dx.doi.org/ 10.1007/BF00283386 [DOI] [PubMed] [Google Scholar]
- 32.Ammermann D, Muenz A. DNA and protein content of different hypotrich ciliates. European J Cell Biol 1982; 27:22-4; PMID:680610118922463 [PubMed] [Google Scholar]
- 33.Prescott DM, Murti KG, Bostock CJ. Genetic apparatus of Stylonychia sp. Nature 1973; 242:576, 597–600; PMID:4621095; http://dx.doi.org/18922463 10.1038/242576a0 [DOI] [PubMed] [Google Scholar]
- 34.Prescott DM. The DNA of ciliated protozoa. Microbiological Rev 1994; 58:233-67; PMID:807843518922463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 2008; 31:785-99; PMID:18922463; http://dx.doi.org/ 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, . et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 2011; 332:848-52; PMID:21566194; http://dx.doi.org/ 10.1126/science.1203919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Chikka MR, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 2004; 303:669-72; PMID:14752161; http://dx.doi.org/ 10.1126/science.1092653 [DOI] [PubMed] [Google Scholar]
- 38.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 2007; 450:304-8; PMID:17952056; http://dx.doi.org/ 10.1038/nature06263 [DOI] [PubMed] [Google Scholar]
- 39.Bracht JR, Perlman DH, Landweber LF. Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biol 2012; 13:R99; PMID:23075511; http://dx.doi.org/ 10.1186/gb-2012-13-10-r99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bracht JR, Fang W, Goldman AD, Dolzhenko E, Stein EM, Landweber LF. Genomes on the edge: programmed genome instability in ciliates. Cell 2013; 152:406-16; PMID:23374338; http://dx.doi.org/ 10.1016/j.cell.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao MC, Zheng K, Yao CH. A conserved nucleotide sequence at the sites of developmentally regulated chromosomal breakage in Tetrahymena. Cell 1987; 48:779-88; PMID:3815524; http://dx.doi.org/ 10.1016/0092-8674(87)90075-4 [DOI] [PubMed] [Google Scholar]
- 42.Klobutcher LA, Gygax SE, Podoloff JD, Vermeesch JR, Price CM, Tebeau CM, Jahn CL. Conserved DNA sequences adjacent to chromosome fragmentation and telomere addition sites in Euplotes crassus. Nucleic Acids Res 1998; 26:4230-40; PMID:9722644; http://dx.doi.org/ 10.1093/nar/26.18.4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavalcanti AR, Dunn DM, Weiss R, Herrick G, Landweber LF, Doak TG. Sequence features of Oxytricha trifallax (class Spirotrichea) macronuclear telomeric and subtelomeric sequences. Protist 2004; 155:311-22; PMID:15552058; http://dx.doi.org/ 10.1078/1434461041844196 [DOI] [PubMed] [Google Scholar]
- 44.Williams KR, Doak TG, Herrick G. Telomere formation on macronuclear chromosomes of Oxytricha trifallax and O. fallax: alternatively processed regions have multiple telomere addition sites. BMC Genetics 2002; 3:16; PMID:12199911; http://dx.doi.org/ 10.1186/1471-2156-3-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baudry C, Malinsky S, Restituito M, Kapusta A, Rosa S, Meyer E, Bétermier M. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev 2009; 23:2478-83; PMID:19884254; http://dx.doi.org/ 10.1101/gad.547309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng CY, Vogt A, Mochizuki K, Yao MC. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell 2010; 21:1753-62; PMID:20357003; http://dx.doi.org/ 10.1091/mbc.E09-12-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. A functional role for transposases in a large eukaryotic genome. Science 2009; 324:935-8; PMID:19372392; http://dx.doi.org/ 10.1126/science.1170023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annual Rev Biochem 2015; 84:405-33;PMID:25747396; http://dx.doi.org/16377660 10.1146/annurev-biochem-060614-034258 [DOI] [PubMed] [Google Scholar]
- 49.Lee JH, Schutte D, Wulf G, Fuzesi L, Radzun HJ, Schweyer S, Engel W, Nayernia K. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet 2006; 15:201-11; PMID:16377660; http://dx.doi.org/ 10.1093/hmg/ddi430 [DOI] [PubMed] [Google Scholar]
- 50.Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene 2002; 21:3988-99; PMID:12037681; http://dx.doi.org/ 10.1038/sj.onc.1205505 [DOI] [PubMed] [Google Scholar]
- 51.Liu JJ, Shen R, Chen L, Ye Y, He G, Hua K, Jarjoura D, Nakano T, Ramesh GK, Shapiro CL, . et al. Piwil2 is expressed in various stages of breast cancers and has the potential to be used as a novel biomarker. Int J Clin Exp Pathol 2010; 3:328-37; PMID:20490325 [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki R, Honda S, Kirino Y. PIWI Expression and Function in Cancer. Front Genet 2012; 3:204; PMID:23087701; http://dx.doi.org/ 10.3389/fgene.2012.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol 2013; 15:563-8; PMID: 23229900; http://dx.doi.org/22047710 10.1007/s12094-012-0966-0 [DOI] [PubMed] [Google Scholar]
- 54.Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett 2012; 315:12-7; PMID:22047710; http://dx.doi.org/ 10.1016/j.canlet.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 55.Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, Chen L, Chu ZB, Tang B, Wang K, . et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 2015; 29:196-206; PMID:24732595; http://dx.doi.org/ 10.1038/leu.2014.135 [DOI] [PubMed] [Google Scholar]
- 56.Fang W, Landweber LF. RNA-mediated genome rearrangement: Hypotheses and evidence. Bio Essays 2013; 35:84-7; PMID:23281134; http://dx.doi.org/27040497 10.1002/bies.201200140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016; 165:289-302; PMID:27040497; http://dx.doi.org/ 10.1016/j.cell.2016.03.020 [DOI] [PubMed] [Google Scholar]
- 58.Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A 2011; 108:13624-9; PMID:21810989; http://dx.doi.org/ 10.1073/pnas.1110633108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genetics 2010; 11:204-20; PMID:20142834; http://dx.doi.org/ 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noto T, Kataoka K, Suhren JH, Hayashi A, Woolcock KJ, Gorovsky MA, Mochizuki K. Small-RNA-Mediated Genome-wide trans-Recognition Network in Tetrahymena DNA Elimination. Mol Cell 2015; 59:229-42; PMID:26095658; http://dx.doi.org/ 10.1016/j.molcel.2015.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schoeberl UE, Kurth HM, Noto T, Mochizuki K. Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev 2012; 26:1729-42; PMID:22855833; http://dx.doi.org/ 10.1101/gad.196493.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol 2009; 5:e1000386; http://dx.doi.org/ 10.1371/journal.pcbi.1000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.