ABSTRACT
APOBEC3A cytidine deaminase induces site-specific C-to-U RNA editing of hundreds of genes in monocytes exposed to hypoxia and/or interferons and in pro-inflammatory macrophages. To examine the impact of APOBEC3A overexpression, we transiently expressed APOBEC3A in HEK293T cell line and performed RNA sequencing. APOBEC3A overexpression induces C-to-U editing at more than 4,200 sites in transcripts of 3,078 genes resulting in protein recoding of 1,110 genes. We validate recoding RNA editing of genes associated with breast cancer, hematologic neoplasms, amyotrophic lateral sclerosis, Alzheimer disease and primary pulmonary hypertension. These results highlight the fundamental impact of APOBEC3A overexpression on human transcriptome by widespread RNA editing.
KEYWORDS: Cytidine deaminase, disease genes, epitranscriptomics, RNA editing, RNA seq
RNA editing enzymatically alters transcript sequences encoded by DNA to meet physiological demands and occurs in single cell organisms, plants and animals.1 Adenine and cytidine deamination are the 2 major types of RNA editing in mammals. Deamination of adenine to inosine (A > I) by adenosine deaminase enzymes occurs in hundreds of thousands of sites, but such deamination mostly targets non-coding and intronic regions, especially those containing Alu repeat sequences.2,3 Protein recoding A > I RNA editing affects dozens of genes but occurs mostly in the brain.4 Recoding RNA editing by cytidine deamination was thought to be rare in humans, physiologically occurring only in the APOB RNA in intestinal cells mediated by the APOBEC1 cytidine deaminase enzyme.5 We recently found that transcripts of hundreds of genes coordinately acquire site-specific C-to-U (C > U) RNA editing in peripheral blood monocytes that are exposed to hypoxia and during M1 (proinflammatory) macrophage differentiation.6 This editing predicts protein recoding in dozens of genes. Interferons (IFNs) and hypoxia induce C > U RNA editing in an additive manner in monocytes, increasing the RNA editing levels to above 80% for several tested genes. Gene expression, transfection, knockdown, site-directed mutagenesis studies and in vitro analysis by purified protein showed that APOBEC3A (A3A), a cytidine deaminase which is structurally related to APOBEC1 and expressed primarily in myeloid cells including monocytes and macrophages,7 catalyzes this RNA editing.6 Thus, A3A is a novel C > U RNA editing enzyme.
APOBEC3s (A3s) are comprised of 7 structurally related genes (3A, 3B, 3C, 3DE, 3F, 3G and 3H) located in tandem in the long arm of chromosome 22.8 The identification of A3G as an HIV-1 restriction factor9 and subsequent studies have led to an increasing recognition of A3 enzymes in antiviral host defenses.10 It is thought that cytidine deamination mutations introduced by A3G in HIV single stranded DNA during reverse transcription explains the mechanism of HIV-restriction.11 A3A has been implicated in the inhibition of retrotransposons and several viruses of public health concern including HIV-1, HTLV-1, HPV, parvoviruses and hepatitis B.12-19 The virus-restricting functions of A3A are typically demonstrated by reduced infectivity of the virus upon exogenous co-expression of the plasmids encoding the enzyme and viruses in a cell line such as HEK293T human embryonic kidney cells.12-17,19 Although A3A's cytidine deaminase function is essential for its antiviral activities,6,12,20,21 the mechanism by which A3A inhibits viruses is not understood well. A3A efficiently mutates single stranded DNA oligonucleotides in vitro11 and transfected plasmid DNAs in overexpression systems.22 However, DNA mutations in the infectious agents restricted by A3A could not be conclusively demonstrated.12,13,17,19,23
In this study, we examine the impact of A3A overexpression in the human transcriptome by characterizing the transcriptome wide C > U RNA editing and gene expression changes upon exogenous expression in HEK293T cells. Our results show an unprecedented extent of RNA editing by A3A. We validate editing of selected sites in primary monocytes and demonstrate that widespread RNA editing triggered by overexpression of A3A also occurs in primary cells under physiologically relevant conditions.
We have previously shown that the overexpression of A3A by transfecting plasmid A3A expression vector (pA3A) into 293T cells induces SDHB c. 136C > U RNA editing.6 Higher editing levels are observed with transfecting increased amounts of pA3A (Fig. 1A). Similarly, higher editing levels are observed upon transfection of U2OS osteosarcoma cells with increased amounts of pA3A. However, editing levels were lower in U2OS cells than in 293T cells for a given pA3A level and did not increase above 10% even with 0.75 µg of transfected pA3A despite high transfection efficiency (Fig. S1). These results suggest that, although exogenous expression of A3A induces RNA editing in 2 distinct cell types, the editing levels might be influenced by cell type specific factors. To examine transcriptome-wide RNA editing events, we transfected 1 µg of pA3A into 293T cells (293T/A3A) and confirmed high SDHB c. 136C > U RNA editing levels (mean = 50.55% compared to 0.87% with control empty vector, n = 3). To identify transcriptome-wide RNA editing sites, we initially performed a low-stringency bioinformatic analysis examining sites that have at least an average of 5% editing level and at least 1 sequence read with a variant base call. This initial analysis revealed over 11,000 sites with C > U RNA editing in 2,254 genes (detailed list is available upon request).
Figure 1.
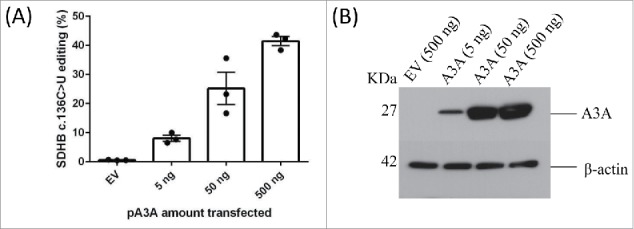
Overexpression of A3A in 293T cells induces c.136 C >U SDHB RNA editing proportional to the increased amounts of A3A plasmid. (A) SDHB c.136 C > U editing levels in 293T cells are shown. The cells are transfected with control empty vector (500 ng) or pA3A at amounts indicated in a 12-well tissue culture plate. Mean and its standard error for n = 3 are shown. (B) Immunoblot showing A3A levels in whole-cell lysates (20 µg protein) of 293T cells transiently transfected with an empty vector control (EV) or increasing amounts of plasmid DNA construct for expression of A3A (pA3A). The A3A protein expression was detected by anti-DDK tag antibody. β-Actin is used as a loading control.
To evaluate validity of the general experimental and bioinformatic approach to identify the C > U RNA editing sites, we first examined the 31 RNA editing sites previously identified in monocytes.6 Transcripts of 4 genes (C1QA, ITGB2, LGALS9 and LRP10) were not detected in 293T/A3A cells, likely reflecting low expression. Of the remaining 27 genes, RNA editing was confirmed at the same cytidine site in transcripts of 26 genes (Supplementary material Table 1). RNA editing of EVI2B at C119T (chromosome 17:29632509, hg19) was not observed in 293T/A3A cells. These data suggest that the overall approach to identify RNA editing targets in 293T/A3A cells was sound.
To validate novel RNA editing sites identified by the initial low-stringency RNA seq analysis of 293T/A3A cells, we performed Sanger sequencing of 19 new protein recoding C > U RNA editing sites in 19 genes (Table 1) associated with various diseases. We validated RNA editing both in 293T/A3A cells subjected to RNA seq analysis and/or in monocyte enriched PBMCs (MEPs) jointly exposed to interferon type 1 (IFN1) and hypoxia. RNA editing was validated by Sanger sequencing in 13 of 15 (∼87%) tested sites in 293T/A3A cells (Fig. 2 and Fig. S2). RNA editing of OPTN and RAD51C, which occurred respectively at 9% and 11% in RNA seq analysis, could not be validated by Sanger sequencing in 293T/A3A cells (Fig. S2), possibly due to low editing levels. High editing levels in RNA seq analysis of 293T/A3A cells correlated with successful validation by Sanger sequencing in MEPs jointly exposed to IFN1 and hypoxia (Table 1). Only one of 11 sites that had editing levels ≤ 20% in 293T/A3A cells by RNA seq was validated in MEPs (Table 1). In contrast, 6 of 8 sites that had editing levels >20% in 293T/A3A cells by RNA seq were validated by Sanger sequencing of MEPs cDNAs (p = 0.006, Fisher's exact test, 2-sided). Notably, several genes associated with breast cancer (BARD1, PTEN, SF3B1), hematologic neoplasms (SF3B1, KMT2A (MLL)), amyotrophic lateral sclerosis (ATXN2), Alzheimer disease (NCSTN) and primary pulmonary hypertension (BMPR2) were validated as recoding RNA editing targets at highly conserved amino acids in MEPs jointly exposed to IFN1 and hypoxia. Recoding RNA editing of classical tumor suppressor genes for breast cancer including BRCA1, BRCA2, TSC2, ATM and MSH2 could be validated in 293T/A3A cells but not in the MEP cells (Fig. S2).
Table 1.
Validation of novel C > U recoding RNA editing sites identified in 293T/A3A cells by Sanger sequencing.
| Genea | Chromosomal positionb | cDNA and amino acid changec | Editing level (%) in MEPs with Hypoxia/IFN (Sanger)d | Editing level (%) in 293T/A3A (Sanger)d | Editing level (%) in 293T/A3A (RNA Seq)e | Disease associationf |
|---|---|---|---|---|---|---|
| APH1A | 1: 150268720 | C91T, R91C | — | 25 | 27 | AD |
| ATM | 11: 108332026 | C7777T, Q2593X | — | 9 | 16 | AT |
| ATXN2 | 12: 111516348 | C1900T, H634Y | 37 | NA | 54 | ALS,SCA2 |
| BARD1 | 2: 214728731 | C2279T, S760L | 42 | 58 | 61 | Br./Ov. Ca |
| BMPR2 | 2: 202530820 | C994T, R332X | 34 | NA | 28 | PH |
| BRCA1 | 17: 43093994 | C1537T, H513Y | — | 10 | 30 | Br./Ov. Ca |
| BRCA2 | 13: 32398616 | C10103T, S3368F | — | 6 | 9 | Br./Ov. Ca |
| BRIP1 | 17: 61683858 | C3188T, S1063L | — | 9 | 12 | Br./Ov. Ca |
| FTO | 16: 53873854 | C964T, R322X | — | 6 | 13 | Obesity |
| KMT2A | 11: 118472106 | C947T, S316L | 32 | NA | 54 | Leukemia |
| MDM2 | 12: 68828924 | C677T, S226L | — | 16 | 14 | Cancers |
| MSH2 | 2: 47414318 | C644T, S215L | — | 10 | 17 | Colon ca. |
| MTHFR | 1: 11794386 | C1319T, S440L | — | 7 | 20 | Miscellaneous |
| NCSTN | 1: 160344811 | C115T, Q39X | 25 | 12 | 21 | AD |
| OPTN | 10: 13125989 | C1192T, Q398X | — | — | 9 | ALS |
| PTEN | 10: 87957858 | C640T, Q214X | 6 | 8 | 12 | Cancers |
| RAD51C | 17: 58709925 | C772T, R258C | — | — | 11 | Br./Ov. Ca |
| SF3B1 | 2: 197393060 | C3668T, S1223F | 18 | NA | 29 | Leukemia |
| TSC2 | 16: 2058820 | C922T, R308W | — | 19 | 19 | TS |
Genes selected from initial low-stringency bioinformatic analysis. The underlined genes remained after high-stringency filtering.
Based on the UCSC hg38 genome assembly used for mapping reads with the Subread subjunc aligner.
Nucleotide numbering for the shortest transcript isoform, with A of the ATG translation initiation codon at position 1.
Editing levels in monocyte-enriched PBMCs (MEPs) exposed to hypoxia and interferon type 1 or 239T/A3A cells are calculated from Sanger traces via Sequencher™ software (average of 3 donors for MEPs or 3 technical replicates for 293T/A3A). Normoxic MEPs showed no evidence of editing for any site although low levels of editing are seen for BMPR2 (6%) in MEPs. Negative sign indicates that editing is either absent or too low to be detected by Sanger sequencing. NA = Not analyzed.
Editing levels are averages estimated by Tophat and Subread alignment softwares.
AD = Alzheimer disease; AT= Ataxia Telengiectasia; ALS,SCA2 = Amyotrophic Lateral Sclerosis, Spinocerebellar Ataxia Type 2; Br./Ov. Ca = Breast or Ovarian cancer; PH=Pulmonary Hypertension; TS = Tuberous Sclerosis.
Figure 2.
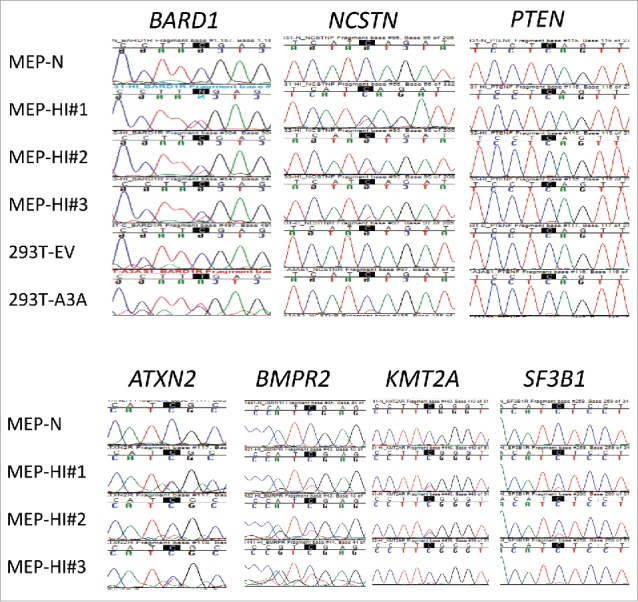
Sanger validation of C > U RNA editing sites in primary cells identified by low-stringency RNA seq analysis of 293T/A3A cells. RNA editing at the highlighted Cs was validated in monocyte/enriched PBMCs (MEPs; n = 3 donors) treated with hypoxia and IFN1 (HI) for 24 h, in 7 of 19 tested genes. C > T(U) editing is characterized by emergence of a secondary T peak (red) accompanied by a reduction in height of C peak (blue). MEP-N shows lack of editing in a representative sequence chromatogram from control MEP cells maintained in normoxia without IFN1 treatment. Chromatograms for the remaining 12 RNA editing sites that could not be validated in MEPs are shown Fig. S2.
Since sites that have low editing levels in our initial low-stringency RNA seq analysis were less likely to be validated by Sanger sequencing, especially in the primary MEP cells exposed to IFN1 and hypoxia, a high-stringency filtering was applied to increase the likelihood of experimental validation of the edited sites. The increased stringency criteria involved at least 15% editing level and at least 2 sequence read with a variant base call. Approximately 25–35 million reads were obtained for each sample in RNA sequencing (Supplementary material Table 2). Most of these reads (81%–92%) could be mapped by Subread or Tophat alignment softwares. The mapped reads mostly (55%–69%) located in coding or untranslated regions of genes (Supplementary material Table 3). The average depth of coverage by mapped reads among the samples was at least 9 for 31–35 million genomic nucleotide positions. These positions were examined for RNA sequence variation.
In analyses of RNA sequencing data to identify single-nucleotide sequence variations, significant differences in RNA sequences of the two groups of A3A and control transfectants were identified for 4,373 genomic positions (Supplementary material Table 4). At all these positions, the sequence variation was of C > U but not any other type. Average levels of such putative C > U RNA editing were 0 in all the control samples and >15% in A3A transfectant samples for all 4,373 sites. Average C > U RNA editing levels at these sites were between 15% and 73% (mean = 25%, SD = 9%). Editing levels were >25% and >50% respectively for 1,752 (40%) and 88 (2%) sites (Fig. 3A). Since we were able to validate all sites in HEK293T cells that have editing levels above 11%, we expect that sites remaining after increased stringency will be mostly validatable in 293T/A3A cells. In MEPs, increased stringency retained 8 sites, 6 (ATXN2, BARD1, BMPR2, KMT2A, NCSTN, SF3B1) of which could be validated (75%), compared to 7 of 19 sites (∼37%) validatable in the initial low stringency analysis (Table 1). However, increased stringency also eliminated PTEN which was validated in MEPs. Thus although sites remaining after increasing the stringency are more likely to be validated in MEPs, some truly edited sites are absent in the final list.
Figure 3.

Salient characteristics of C > (U)RNA editing by A3A in 293T cells (A) Mean and range of editing level at the 4,373 sites identified as targets for A3A-mediated editing are shown for the 3 A3A transfectant samples. The sites are ordered by the mean editing level. (B) Logo indicating sequence conservation and base frequency for sequences bearing the editing sites (at position 0). (C) Histogram of lengths in bases of inverted repeat sequences flanking the editing sites.
4,285 (98%) of the 4,373 sites occur in the known human (RefSeq) transcriptome. Of these 4,285 sites, 2,851 (67%) are in the known exonic RNA sequences (Table 2). C > U editing of RNA at the 4,373 sites is predicted to result in 1,499 (34%) synonymous, 989 (23%) missense and 299 (7%) nonsense changes in RNA translation (Table 2). The 4,285 editing sites that are in the known transcriptome are transcribed for a total of 3,078 genes. The highest number of editing sites (7) was seen for two genes, AFF4 and FLNA. Five, 10, 51, 199 and 594 genes respectively had 6, 5, 4, 3 and 2 editing sites (Supplementary material Table 5). These results expand the number of A3A' s C > U editing targets by an order of magnitude and suggest that thousands of genes may acquire site-specific C > U RNA editing when A3A is overexpressed in non-immune cells or in monocytes jointly exposed to hypoxia and IFN1.
Table 2.
| 5′ untranslated region | 153 | ||
|---|---|---|---|
| Exonic | Synonymous | 1499 | |
| Non-synonymous | Nonsense | 299 | |
| Stop loss | 0 | ||
| Missense | 989 | ||
| Unknown | 5 | ||
| Non-coding RNA | 59 | ||
| 3' untranslated region | 1166 | ||
| Intronic | Coding RNA | 74 | |
| Non-coding RNA | 41 | ||
| Untranscribedb | 36 | ||
| Intergenic | 52 | ||
As reported by the ANNOVAR annotation tool.
Within 1 kb up- or down-stream respectively of a known transcription start or end site.
To identify common features of sequence contexts of the editing sites, we examined 12 base-long sequences flanking the edited C residue. The edited C has a U, C, G or A at the immediate 5′ position for 4,276 (98%), 68 (1.6%), 21 (0.5%) and 8 (0.2%) sites, respectively. This observation and sequence logo analysis (Fig. 3B) suggests [AUC] [AUC] [CU]C[AG] as a sequence motif that is commonly targeted by A3A. (The residues within brackets are different possibilities for a position. Edited C is underlined). AUC, UUC and CUC are respectively seen for 2,144 (49%), 1,109 (25%) and 855 (20%) of the 4,373 editing sites. We previously noticed that edited Cs were frequently flanked by inverted repeats.6 Here, analysis of 25 nts containing the edited C in the middle for all edited sites show that the edited C was flanked by a pair of inverted repeat sequences of 3–9 b for 4,153 (95%) of the editing sites. Sequences of 4 bases were most common, seen for 1,529 (35%) sites (Fig. 3C). GCC, CCA and CCU were the 3 most common repeat sequences, seen for 83 (1.9%), 69 (1.6%) and 69 (1.6%) of editing sites. Inverted repeat sequences of 4 bases or longer flanked 62.7% of edited Cs, but only 28.6% of randomly obtained 25 nt sequences from human GRCh38 RefSeq transcriptome. These results suggest that both sequence context, especially at the immediate 5′-end, and the presence of long flanking inverted repeats play an important role in selection of edited Cs by A3A.
Among the 6 transfectant samples of the study, 18,593 genes were considered as expressed and were analyzed for differential expression. Of the 7,376 (39.7%) genes that were differentially expressed (P < 0.05), 46 and 383 were respectively down- and up-regulated with ≥2 fold-change in the A3A transfectants compared to controls (Supplementary material Table 6). Correlation between editing and gene expression levels was not observed. The differential expression of genes probably occurs as both direct and indirect effects of A3A overexpression. Activation of transcriptional/translational machinery (relative to empty vector) or downstream effects of thousands of edited transcripts may contribute to differential gene expression.
We previously performed Sanger sequencing of genomic DNAs corresponding to several edited sites in gene transcripts and found no mutations.6 To further test whether transient expression of exogenous A3A induces genomic DNA mutations, we amplified SDHB exon 2, where the edited c.136 is located, using a high fidelity Taq polymerase, in 293T cells transfected either with an empty vector (EV) or pA3A for 48 hours. We cloned the SDHB exon 2 PCR products into plasmids and sequenced 45 clones transfected with EV and 46 clones with A3A vector. The amplified segment contains 22 TC/AG motifs which are preferentially deaminated by A3A in ssDNA templates as well as 16 CC/GG motifs which are known to be targeted by the enzyme albeit with lower efficiency.24 We found one T deletion out of 8,505 nucleotides in EV-transfected clones and no mutation out of 8,694 nucleotides in A3A-transfected clones. These results suggest that whereas transient expression of A3A causes widespread RNA editing, gDNA mutation is negligible.
In summary, we show that short-term overexpression of A3A in 293T cell line which does not endogenously express the enzyme is sufficient to cause C > U RNA editing in thousands of genes' transcripts. The majority of these transcripts are likely edited in primary monocytes in response to physiological relevant stimuli (combined exposure to IFN1 and hypoxia). However, we also see evidence of RNA editing in certain transcripts (e.g. BRCA1, TSC2) in 293T cells that could not be validated in primary monocytes exposed to hypoxia/IFN1. This may be due to A3A overexpression in 293T cells which might lead to spurious editing of certain transcripts or related to differences in cell type. Additionally, this study supports our previous observation that site-specific editing mediated by A3A is dependent on the sequence and secondary structure of the transcripts.6 Furthermore, we find no evidence of genomic DNA mutations upon transient expression of A3A.
Our results demonstrate that the number of RNA editing targets of A3A is substantially higher than has been initially suggested by the RNA seq analysis of primary monocytes exposed to only hypoxia.6 Since the inhibitory functions of A3A against a range of viruses are often demonstrated by exogenous co-expression of 0.1–2 µg of pA3A along with viral plasmids,12,17,20,25-27 our findings suggest that widespread editing of host gene transcripts may play an antiviral role by altering the availability of cellular proteins required by viruses to complete their infectious cycles. Previous studies showed that increased amounts of transfected pA3A increases antiviral functions of A3A in vitro.13,23 We find that higher expression of A3A also causes higher levels of RNA editing. Thus, there is a possibility of a correlation between RNA editing of host transcripts and the antiviral activity of A3A. Cellular RNA editing model of viral restriction is particularly relevant for A3A, where mutations in the inhibited virus DNAs have yet to be conclusively demonstrated.12,13,17,19,23 Nevertheless, a direct link between host gene RNA editing and antiviral function of A3A remains to be demonstrated.
We present new evidence that certain predisposition genes for amyotrophic lateral sclerosis, Alzheimer disease and primary pulmonary hypertension are targets of A3A-mediated RNA editing. Whether RNA editing of disease predisposition genes indeed occurs in non-immune cells in vivo and predisposes to disease are the subjects for future studies. Since RNA editing function of A3A is likely transiently induced by interferons, hypoxia or ectopic expression, novel experimental and bioinformatic approaches will be required to directly examine the role of intermittent A3A-mediated RNA editing in the pathogenesis of viral and chronic diseases.
Methods
Isolation and culture of cells
The TLA-HEK293T™ 293T human embryonic kidney cell-line was obtained from Open Biosystems® (Huntsville, AL). Peripheral blood mononuclear cells (PBMC) of anonymous platelet donors were isolated from Trima Accel™ leukoreduction system chambers (Terumo BCT®, Lakewood, CO) after thrombocytapheresis, in accordance with an institutional review board-approved protocol. A density gradient centrifugation method using polysucrose-containing Lymphocyte Separation Medium (Mediatech®, Manassas, VA) was used for PBMC isolation. MEPs were prepared from PBMCs using the cold aggregation method with slight modification as described.28 Briefly, PBMCs were subjected to gentle rocking at 4°C for an hour and aggregated cells that sedimented through fetal bovine serum (FBS; VWR®, Radnor, PA) were collected after 8–16 hours for mild monocyte enrichment (∼20%−40% monocytes).
Gene expression construct and transfection of plasmid DNA
Sequence-verified plasmid constructs in pCMV6 vector for CMV promoter-driven expression of human A3A with sequences matching NCBI RefSeq sequences NM_145699.2 and containing C-terminus MycDDK tag were obtained from OriGene (Rockville, MD, product number RC220995). 293T cells were transfected with plasmid DNA using the liposomal X-tremeGENE™ 9 DNA reagent (Roche®, Indianapolis, IN) or jetPRIME™ (Polyplus-transfection®, New York, NY) reagents in either 6-well tissue culture plates using 1 µg pA3A plasmid to perform RNA seq experiments or in 12-well plates using various concentrations of pA3A plasmid as per the guidelines provided by the manufacturer. Transfection efficiency with both reagents was 60%-80% as assessed by fluorescent microscopy of cells transfected with the pLemiR™ plasmid DNA (Open Biosystems®) for expression of a red fluorescent protein. Cells were harvested 2 d after transfection.
Hypoxia and interferon treatments
For hypoxia, cells were cultured under 1% O2, 5% CO2 and 94% N2 in an Xvivo™ System (Biospherix®, Lacona, NY). Human ‘universal’ type I IFN, a hybrid of N-terminal IFNα-2 and C-terminal IFNα-1, produced in E. coli were obtained from PBL Assay Science® (Piscataway, NJ), and used at 300–600 U/ml. Hypoxia and/or IFN1 treatments were for 24 hours.
Sanger sequencing
Sequencing primers (Integrated DNA Technologies®) are listed in Supplementary Table 7. Candidate C > U RNA editing sites that were validated by Sanger sequencing in PCR-amplified cDNA fragments are noted in Table 1. PCR reactions were treated with exonuclease I and shrimp alkaline phosphatase (New England Biolabs®) and then directly used for sequencing on 3130 xL Genetic Analyzer™ (Life Technologies®). The editing–related T peaks were first examined visually for emergence of a new T peak accompanied by a reduction in height of the C peak and subsequently confirmed by recognition of these peaks by Sequencher Variance Detail Reporter software in 3 replicates (i.e., biological replicates of hypoxia/interferon exposed primary monocytes or technical replicates of 293T/A3A transfectants). Major and minor chromatogram peak heights at a nucleotide position of interest were quantified with Sequencher™ 5.0 software (Gene Codes®, Ann Arbor, MI) to calculate editing level for the position. The software identifies a minor peak only if its height is ≥5% of the major peak. Thus, editing levels less than 5% could not be quantitated. Genomic PCR amplification of SDHB exon 2 from 293T cells was performed using previously described intronic primers29 FN2A (hg38 assembly: 17044934–17044956) and RN2A (hg38 assembly: 17044724–17044744) and high fidelity Herculase II Fusion Taq Polymerase with 1%-2% DMSO (Agilent Technologies, Santa Clara, CA). PCR products are subsequently cloned into a plasmid (PCR Cloning kit, New England Biolabs, Ipswich, MA).
Immunoblotting of cell lysates
Whole-cell lysates from 293T cells were prepared using M-PER reagent (Thermo Fisher, Rockford, IL) with 1X Halt protease and phosphatase inhibitor cocktail (Thermo Fisher). 20 µg proteins were suspended in Laemmli buffer and heated at 95°C for 15 mins followed by reducing and denaturing polyacrylamide gel electrophoresis on pre-cast, 4%–15% gradient polyacrylamide gels (Mini-PROTEAN TGX, Bio-Rad, Hercules, CA). Proteins from the gel were then transferred to polyvinylidene difluoride membrane with a pore-size of 0.2 µm for 7 min at 1.3 A in a Bio-Rad Trans-Blot Turbo apparatus. Membranes were incubated in Tris-buffered 0.15 M NaCl of pH 7.5 with 0.05% v/v TWEEN 20 (Sigma Aldrich, Saint Louis, MO) and 5% w/v dried, non-fat, cow milk (Carnation, Nestle´, Glendale, CA) with mouse monoclonal anti-DDK (product number TA50011-100, 4C5, 1:25,000 dilution) purchased from OriGene (Rockville, MD)/mouse monoclonal anti-β-actin (product number AM4302, 1:15,000 dilution) and incubated overnight at 4°C. Horseradish peroxidase-conjugated, goat anti-mouse IgG antibodies were obtained from Life Technologies and used at 1:2,000 dilution and incubated for one hour at room temperature. Luminata Forte Western HRP Substrate (EMD Millipore) and CL-XPosure auto-radiography films (Thermo Fisher) were used for chemiluminescent detection.
RNA sequencing
Directional RNA sequencing libraries were prepared using the TruSeq™ Stranded Total RNA Sample Prep Kit (Illumina®, San Diego, CA). Each library, indexed for multiplex sequencing of 6 libraries per flow lane, was prepared from 1 µg of DNAse I-treated total RNA (Agilent® RIN values of 7.6–9.5) after rRNA depletion with Ribo-Zero™ Gold reagents. Ten PCR cycles were used during library generation and the modal library fragment size was 300 bp. Paired-end, 101 b sequencing of libraries was done on HiSeq™ 2000 instrument with TruSeq™ SBS and PE Cluster v3 Kit reagents (Illumina®). CASAVA 1.8.2 (Illumina®) was used for base-calling and de-multiplexing to obtain raw sequencing reads. Raw read data was deposited in NCBI BioProject with accession number 261741. Reads were filtered and trimmed to remove adapter sequences and poor-quality bases using Trimmomatic 0.33 with options: HEADCROP:12 ILLUMINACLIP: TruSeq3-PE-2.fa:2:30:10:6:TRUE LEADING:5 TRAILING:5 SLIDINGWINDOW:4:15 MINLEN:30. Raw and processed read counts are provided in Supplementary material Table 2.
Mapping of RNA sequencing reads
Reads were uniquely mapped to the GRCh38 human reference genome assembly (Ensembl release 78 of December 2014) using the Subread 1.4.5-p1 aligner, with these options specified for the subjunc command: -d 30 -D 400 -uH. Reads were also uniquely mapped with the Tophat 2.0.12 aligner, with these options specified for the tophat2 command: –library-type=fr-firststrand –mate-std-dev=35 -g 1 -N 3 –no-novel-juncs -r 10 –read-edit-dist 3. Genome indices for these aligners were respectively built with Subread index (options: -BF) and Bowtie2 2.2.3 bowtie2-build commands. Transcriptome index for Tophat was built with tophat2 command using Ensembl's gene model for the genome assembly. RSeQC 2.4 was used to identify UCSC gene features of genomic regions that the reads mapped to. Mapping statistics are provided in Supplementary material Table 3. To count mapped reads at the gene level for the Ensembl gene model, Subread-aligned read data was analyzed with Subread featureCounts with options: -g gene_id -O -p -s 2 -t exon.
Identification of putative A3A-affected RNA editing sites
The clipOverlap command (option: –poolSize 10000000) of bamUtil master version of 21 September 2014 was used to clip sequence overlaps between read pair-mates in mapped read data. The mpileup command (options: -AB -d 2000 -q 1 -Q 20) of Samtools 0.1.19 was then used to obtain genome-wide read base information from the mapped sequencing data. Custom scripts were used to parse the mpileup output into base calls. Genome positions at which the total A/C/T/G base call was <6 in any of the 6 samples or <9 in any of the 2 sample-groups of this study were ignored. Positions were then identified as possibly variant if all samples of at least one sample-group had ≥2 non-reference base calls of the same base-type at a variation level ≥0.03, with group-wide averages of ≥3 and ≥0.045 respectively. Variation level was calculated as the ratio of variant base call count to the sum of variant and reference base call counts. Variant positions for which calls for base-types other than the reference and variant constituted ≥1% of all A/C/T/G calls or were >1 in any sample were ignored. Positions with range/variation value of <2 for variation levels across all 6 samples were also ignored. A two-tailed β binomial test (IBB 13.06 package for R 3.0.2) comparing variation levels of the 2 sample-groups was then performed for each of the remaining variant positions. P values obtained with the test were corrected for multiple testing by the Benjamini-Hochberg method. Positions for which the average variation level was >0 for both sample-groups or was <0.05 for ≥1 sample-group, or for which the corrected P value was ≥0.01, or which were not seen with both Subread- and Tophat-aligned data or failed a strand-bias test were ignored. The strand-bias test used base call counts across the group of A3A trasfectant samples, and was applied to only those positions for which there were ≥8 calls for each of reference or variant base-types, with <25% of base calls of either base-type from reads of either forward or reverse direction. Failure was deemed if P value in a Fisher exact test that compared reference and variant base calls from reads of the 2 directions was <0.05. Finally, putative A3A-affected RNA editing sites were identified from the remaining genome positions for which the average editing level for the 3 A3A trasfectant samples was >0.15. The editing level was the mean of variation levels calculated from Subread- and Tophat-aligned data. The directional nature of the RNA sequencing libraries was used to identify the transcribed chromosomal strand at a genomic position to assign the type of RNA editing. RNA sequences flanking an editing site were deduced from the reference human genome and corrected for homozygous single-nucleotide polymorphisms in the HEK-293T cell-line. The 22 March 2015 release of ANNOVAR tool with its RefSeq-based hg38_refGene database was used to annotate sites with information on gene, gene feature and effect of editing of protein sequence. The list of editing sites with annotations is provided in Supplementary material Table 5. The step-wise filtering of genome positions in the process to identify editing sites is detailed in Supplementary material Table 4.
Other
Sequence logos were created with the WebLogo 3 online tool. Gene set enrichment analyses were performed with PANTHER 9.0 online tool (URL: http://www.pantherdb.org). The edgeR 3.10.2 Bioconductor package was used for differential gene expression analyses. Only those genes with read counts >20 in at least 3 of the study's 6 samples were analyzed; the rowsum.filter and prior.df values for estimateCommonDisp and estimateTagwiseDisp functions were respectively set at 12 and 0.2; and, P values calculated by the Fisher exact test were corrected with the Benjamini-Hocheberg method. Differentially expressed genes with a ≥2-fold change are noted in Supplementary material Table 6. Allele specific qPCR using LightCycler 480 System (Roche) analysis is employed to measure SDHB transcript editing levels at c.C136U as previously described.6,36 Ratio of edited and total SDHB transcript levels is measured in a given cDNA using technical triplicates.
Supplementary Material
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by startup funds from the Departments of Pathology and Thoracic Surgery of Roswell Park Cancer Institute. RNA and Sanger sequencing services were provided by the institute's core facilities, which are partly supported by Cancer Center Support Grant 5P30 CA016056 of National Cancer Institute. We thank T. Ouchi for providing U2OS cell line that was obtained by his lab from ATCC.
References
- 1.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 2010; 79:321-49; PMID:20192758; http://dx.doi.org/ 10.1146/annurev-biochem-060208-105251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al.. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 2014; 24:365-76; PMID:24347612; http://dx.doi.org/ 10.1101/gr.164749.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human alu and non-alu RNA editing sites. Nature methods 2012; 9:579-81; PMID:22484847; http://dx.doi.org/ 10.1038/nmeth.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li JB, Church GM. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat Neurosci 2013; 16:1518-1522; PMID:24165678; http://dx.doi.org/ 10.1038/nn.3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol 2012; 23:258-68; PMID:22001110; http://dx.doi.org/ 10.1016/j.semcdb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma S, Patnaik SK, Taggart RT, Kannisto ED, Enriquez SM, Gollnick P, Baysal BE. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun 2015; 6:6881; PMID:25898173; http://dx.doi.org/ 10.1038/ncomms7881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: Implications for HIV-1 restriction. Nucleic Acids Res 2010; 38:4274-84; PMID:20308164; http://dx.doi.org/ 10.1093/nar/gkq174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 2002; 79:285-96; PMID:11863358; http://dx.doi.org/ 10.1006/geno.2002.6718 [DOI] [PubMed] [Google Scholar]
- 9.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral vif protein. Nature 2002; 418:646-50; PMID:12167863; http://dx.doi.org/ 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- 10.Willems L, Gillet NA. APOBEC3 interference during replication of viral genomes. Viruses 2015; 7:2999-3018; PMID:26110583; http://dx.doi.org/ 10.3390/v7062757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol 2004; 4:868-77; PMID:15516966; http://dx.doi.org/ 10.1038/nri1489 [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol 2006; 16:480-5; PMID:16527742; http://dx.doi.org/ 10.1016/j.cub.2006.01.031 [DOI] [PubMed] [Google Scholar]
- 13.Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog 2009; 5:e1000439; PMID:19461882; http://dx.doi.org/ 10.1371/journal.ppat.1000439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiegand HL, Cullen BR. Inhibition of alpharetrovirus replication by a range of human APOBEC3 proteins. J Virol 2007; 81:13694-9; PMID:17913830; http://dx.doi.org/ 10.1128/JVI.01646-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger G, Durand S, Fargier G, Nguyen XN, Cordeil S, Bouaziz S, Muriaux D, Darlix JL, Cimarelli A. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog 2011; 7:e1002221; PMID:21966267; http://dx.doi.org/ 10.1371/journal.ppat.1002221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, et al.. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014; 343:1221-8; PMID:24557838; http://dx.doi.org/ 10.1126/science.1243462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and alu retrotransposition. Proc Natl Acad Sci USA 2006; 103:8780-5; PMID:16728505; http://dx.doi.org/ 10.1073/pnas.0603313103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 2008; 320:230-3; PMID:18403710; http://dx.doi.org/ 10.1126/science.1153201 [DOI] [PubMed] [Google Scholar]
- 19.Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res 2007; 35:2955-64; PMID:17439959; http://dx.doi.org/ 10.1093/nar/gkm181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulliard Y, Narvaiza I, Bertero A, Peddi S, Rohrig UF, Ortiz M, Zoete V, Castro-Diaz N, Turelli P, Telenti A, et al.. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J Virol 2011; 85:1765-76; PMID:21123384; http://dx.doi.org/ 10.1128/JVI.01651-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitra M, Hercik K, Byeon IJ, Ahn J, Hill S, Hinchee-Rodriguez K, Singer D, Byeon CH, Charlton LM, Nam G, et al.. Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties. Nucleic Acids Res 2014; 42:1095-110; PMID:24163103; http://dx.doi.org/ 10.1093/nar/gkt945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol 2010; 17:222-9; PMID:20062055; http://dx.doi.org/ 10.1038/nsmb.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem 2006; 281:22161-72; PMID:16735504; http://dx.doi.org/ 10.1074/jbc.M601716200 [DOI] [PubMed] [Google Scholar]
- 24.Love RP, Xu H, Chelico L. Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A. J Biol Chem 2012; 287:30812-22; PMID:22822074; http://dx.doi.org/ 10.1074/jbc.M112.393181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooms M, Krikoni A, Kress AK, Simon V, Munk C. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J Virol 2012; 86:6097-108; PMID:22457529; http://dx.doi.org/ 10.1128/JVI.06570-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, Lambert PF, Santiago ML, Pyeon D. APOBEC3A functions as a restriction factor of human papillomavirus. J Virol 2015; 89:688-702; PMID:25355878; http://dx.doi.org/ 10.1128/JVI.02383-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt K, Guo K, Katuwal M, Wilson D, Prochnow C, Bransteitter R, Chen XS, Santiago ML, Stephens EB. Lentivirus restriction by diverse primate APOBEC3A proteins. Virology 2013; 442:82-96; PMID:23648232; http://dx.doi.org/ 10.1016/j.virol.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baysal BE, De Jong K, Liu B, Wang J, Patnaik SK, Wallace PK, Taggart RT. Hypoxia-inducible C-to-U coding RNA editing downregulates SDHB in monocytes. PeerJ 2013; 1:e152; PMID:24058882; http://dx.doi.org/ 10.7717/peerj.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baysal BE. A recurrent stop-codon mutation in succinate dehydrogenase subunit B gene in normal peripheral blood and childhood T-cell acute leukemia. PLoS One 2007; 2:e436; PMID:17487275; http://dx.doi.org/ 10.1371/journal.pone.0000436 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


