In neurons, mRNAs are transported along dendrites as large RNA granules. Localization of these granules was found to be developmentally regulated, and Staufen1, a major component of RNA transport granules, was critical for activity-dependent synaptic localization.
Abstract
In neurons, RNA transport is important for local protein synthesis. mRNAs are transported along dendrites as large RNA granules. The localization and dynamics of Puralpha and Staufen1 (Stau1), major components of RNA transport granules, were investigated in cultured hippocampal neurons. Puralpha-positive granules were localized in both the shafts and spines of dendrites. In contrast, Stau1-positive granules tended to be localized mainly in dendritic shafts. More than 90% of Puralpha-positive granules were positive for Stau1 in immature dendrites, while only half were positive in mature dendrites. Stau1-negative Puralpha granules tended to be stationary with fewer anterograde and retrograde movements than Stau1-positive Puralpha granules. After metabotropic glutamate receptor 5 activation, Stau1-positive granules remained in the dendritic shafts, while Puralpha granules translocated from the shaft to the spine. The translocation of Puralpha granules was dependent on myosin Va, an actin-based molecular motor protein. Collectively our findings suggest the possibility that the loss of Stau1 in Puralpha-positive RNA granules might promote their activity-dependent translocation into dendritic spines, which could underlie the regulation of protein synthesis in synapses.
INTRODUCTION
In neurons, the intracellular transport of cargoes such as organelles, protein complexes, and mRNAs in axons and dendrites is critical for development and plasticity (Hirokawa et al., 2010). Persistent forms of synaptic plasticity require dendritic protein synthesis independent of transcription (Kang and Schuman, 1996; Huber et al., 2000). The presence of mRNA, ribosomes, and translation factors in dendrites, including dendritic spines, suggests that synapses could be modified directly, and perhaps individually, through regulation of local protein synthesis (Steward and Levy, 1982; Sutton and Schuman, 2006). Localized mRNAs are present in RNA granules, which are transported to distal dendrites by conventional kinesin (KIF5), a member of the kinesin superfamily of proteins (KIFs) (Kanai et al., 2004). These granules are large complexes composed of various proteins and could be important for the activity-dependent transport of mRNA. However, the mechanisms that regulate their transport are largely unknown.
To gain insight into the regulation and dynamics of RNA granules in neurons, we focused on two major RNA-binding proteins present in these granules—Puralpha and Staufen1 (Stau1) (Kanai et al., 2004). Puralpha is a ubiquitous RNA-binding protein. Among the various RNA-binding proteins constituting neuronal RNA transport granules, Puralpha is the most important because it is a direct binding partner of the molecular motor KIF5 (Kanai et al., 2004), and it is involved in many crucial biological processes, such as transcriptional gene control, initiation of DNA replication, and RNA translation (Gallia et al., 2000). Stau1 is implicated in RNA transport, translational regulation, and mRNA decay (Tang et al., 2001).
We investigated the localization and translocation of Puralpha and Stau1 in dendrites. Our findings suggest that RNA granules are not uniform and that their movement and localization are dependent on their constituent molecules, indicative of the diversity of RNA granules that might underlie the activity-dependent modulation of synaptic efficacy.
RESULTS AND DISCUSSION
Puralpha is localized in both the shafts and spines of dendrites, whereas Stau1 is found mainly in dendritic shafts
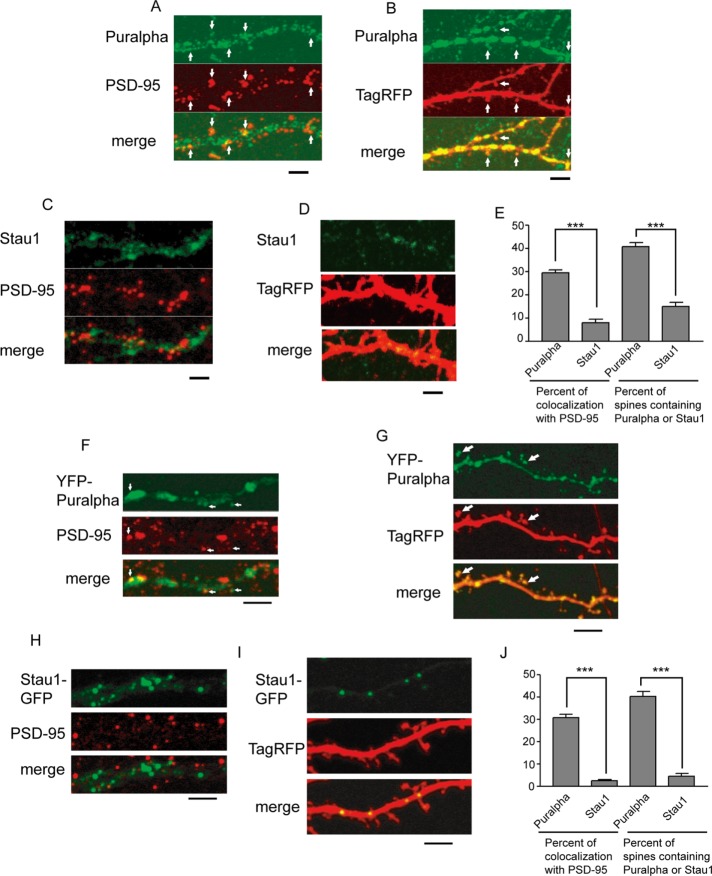
Puralpha and Stau1 are present in RNA transport granules as components of the cargo of KIF5 (Kanai et al., 2004). To investigate their localization, we immunostained primary cultures of mature hippocampal neurons (15 d in vitro [DIV]) with anti-Puralpha and anti–postsynaptic density protein-95 (PSD-95, a postsynaptic marker) antibodies (Figure 1A).
FIGURE 1:
(A) Mature hippocampal neurons were immunostained with anti-Puralpha and anti–PSD-95 antibodies. White arrows indicate the colocalization of Puralpha and PSD-95 clusters. (B) Neurons were transfected with TagRFP and immunostained with anti-Puralpha antibody. Note that some Puralpha clusters are localized in dendritic spines (arrows). (C) Neurons were immunostained with anti-Stau1 and anti–PSD-95. (D) Neurons were transfected with TagRFP and immunostained with anti-Stau1 antibody. (E) Quantification. Error bars indicate SEM; 10–15 cells from three mice were examined. (F) Mature hippocampal neurons were transfected with YFP-Puralpha and immunostained with anti–PSD-95. White arrows indicate the colocalization of Puralpha and PSD-95 clusters. (G) TagRFP was coexpressed with YFP-Puralpha. Some Puralpha clusters localize to dendritic spines (arrows). (H) Neurons were transfected with Stau1-GFP and were subsequently immunostained with anti–PSD-95. (I) TagRFP is coexpressed with Stau1-GFP. (J) Quantification. Error bars indicate SEM; 20–27 cells from three mice were examined. ***, p < 0.001; Student’s t test. Scale bars: 5 μm.
Puralpha immunoreactivity exhibited a granular staining pattern along dendrites (Figure 1A). About one-third of Puralpha granules were colocalized with PSD-95 (Figure 1, A, arrows, and E). This suggests that some of the Puralpha-positive RNA granules are localized in postsynaptic spines, given that they are specifically localized to dendrites but not axons (Kanai et al., 2004). We confirmed the presence of Puralpha in dendritic spines by visualizing the outline of neurons using overexpression of tag red fluorescent protein (TagRFP; Figure 1B, arrows). This revealed that 40.6 ± 1.8% of TagRFP-containing spines were Puralpha-positive (Figure 1E).
Fewer Stau1 signals were localized in dendritic spines compared with Puralpha signals. The percentage of Stau1 clusters positive for PSD-95 was 7.9 ± 1.4% (Figure 1, C and E), and 14.8 ± 1.8% of spines expressing TagRFP were Stau1-positive (Figure 1, D and E). These results suggest that Puralpha granules are localized in both dendritic spines and shafts, whereas Stau1 granules are localized mainly in dendritic shafts.
For assessment of whether transfected tagged Puralpha and Stau1 protein constructs were correctly localized, mature hippocampal neurons were transfected with yellow fluorescent protein (YFP)-Puralpha or Stau1–green fluorescent protein (GFP) and colabeled with PSD-95 antibody or visualized for TagRFP (Figure 1, F–I). The localization of these constructs was similar to that of the endogenous proteins (Figure 1, A–E); Puralpha granules were more likely to be localized in postsynaptic regions than Stau1 granules (Figure 1, F–J).
Presence of Stau1-positive and Stau1-negative Puralpha granules
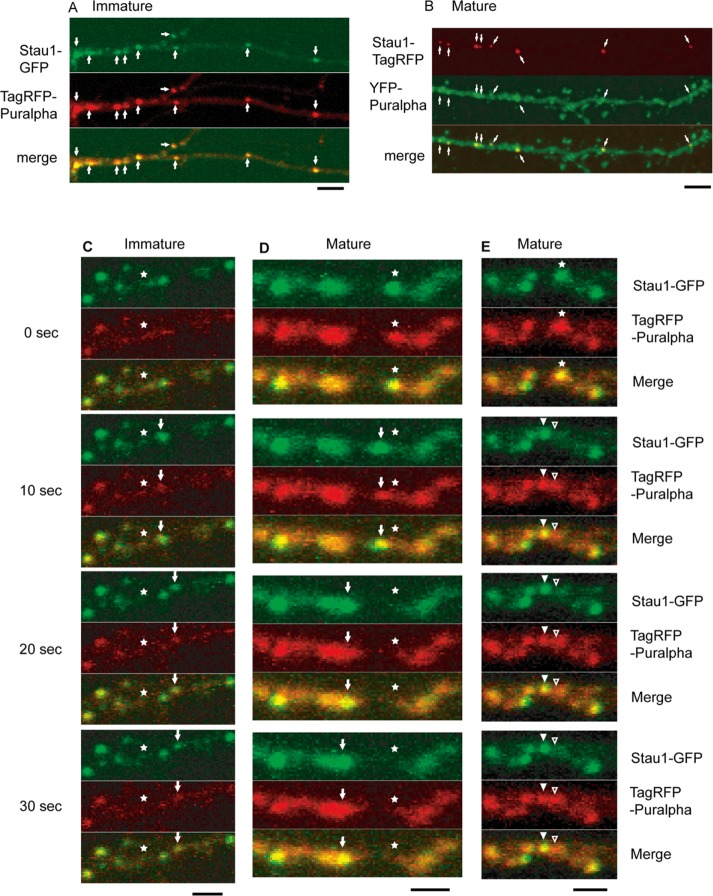
For evaluation of the dendritic localization of Puralpha and Stau1 in more detail, tagged Stau1 and tagged Puralpha were coexpressed in primary cultures of immature (8 DIV; Figure 2A) or mature (15 DIV; Figure 2B) hippocampal neurons. In immature dendrites, more than 90% of TagRFP-Puralpha granules were positive for Stau1-GFP, and more than 90% of Stau1-GFP granules contained TagRFP-Puralpha (Figure 2A and Table 1), as reported previously (Kanai et al., 2004). Conversely, in dendrites of mature neurons, about half of the YFP-Puralpha–labeled granules contained Stau1-TagRFP, while more than 90% of Stau1-TagRFP–labeled granules were positive for YFP-Puralpha (Figure 2B and Table 1). These results indicate that the proportion of Stau1-negative Puralpha clusters increases during neuronal maturation. This suggests that there are different types of RNA-containing granules that change in their composition during maturation, and that Stau1 is present only in a subset of Puralpha-containing granules localized preferentially in dendritic shafts.
FIGURE 2:
(A) In immature dendrites (8 DIV), almost all of the TagRFP-Puralpha clusters colocalized with Stau1-GFP clusters (white arrows). (B) In mature dendrites (15 DIV), YFP-Puralpha clusters only partially colocalized with Stau1-TagRFP clusters (white arrows). p < 0.001; Student’s t test. Scale bars: 5 μm. (C–E) In immature (C) or mature (D) dendrites, TagRFP-Puralpha clusters were cotransported with Stau1-GFP clusters (white arrows). Asterisks indicate initial position. (E) Puralpha-positive granules (asterisks) were classified into Stau1-positive/Puralpha-positive granules (solid arrowheads) and Stau1-negative/Puralpha-positive clusters (open arrowheads). Scale bars: 2 μm.
TABLE 1:
Percent colocalization of Puralpha and Stau1 tagged with fluorescent proteins.
| Immature neurons | Mature neurons | |
|---|---|---|
| Puralpha-positive/Stau1-positive clusters in total Puralpha-positive clusters | 91.6 ± 1.8% (mean ± SEM) | 52.0 ± 3.7% (mean ± SEM) |
| Puralpha-positive/Stau1-positive clusters in total Stau1-positive clusters | 96.1 ± 1.4% (mean ± SEM) | 96.3 ± 0.9% (mean ± SEM) |
| Chi-square test | Not significant | *p < 0.05 |
| Number of total Puralpha clusters analyzed | 462 | 906 |
| Number of total Stau1 clusters analyzed | 433 | 496 |
| Number of neurons analyzed | 20 | 22 |
A time-lapse assay was carried out 24 h after cotransfection of TagRFP-Puralpha and Stau1-GFP vectors in immature neurons. Images were recorded every 10 s over a 3-min period (Figure 2C and Table 2). In immature neurons, TagRFP-Puralpha–positive granules were largely positive for Stau1-GFP, and 63% of TagRFP-Puralpha–positive/Stau1-GFP–positive granules were stationary, while 37% were motile (Table 2). In the motile granules, TagRFP-Puralpha and Stau1-GFP signals comigrated (Figure 2C). The motile granules displayed two types of movement—oscillatory (to-and-fro movements over short distances) or unidirectional (continuous anterograde or retrograde movements) (Table 2).
TABLE 2:
Movement of Puralpha and Stau1 granules tagged with fluorescent proteins.
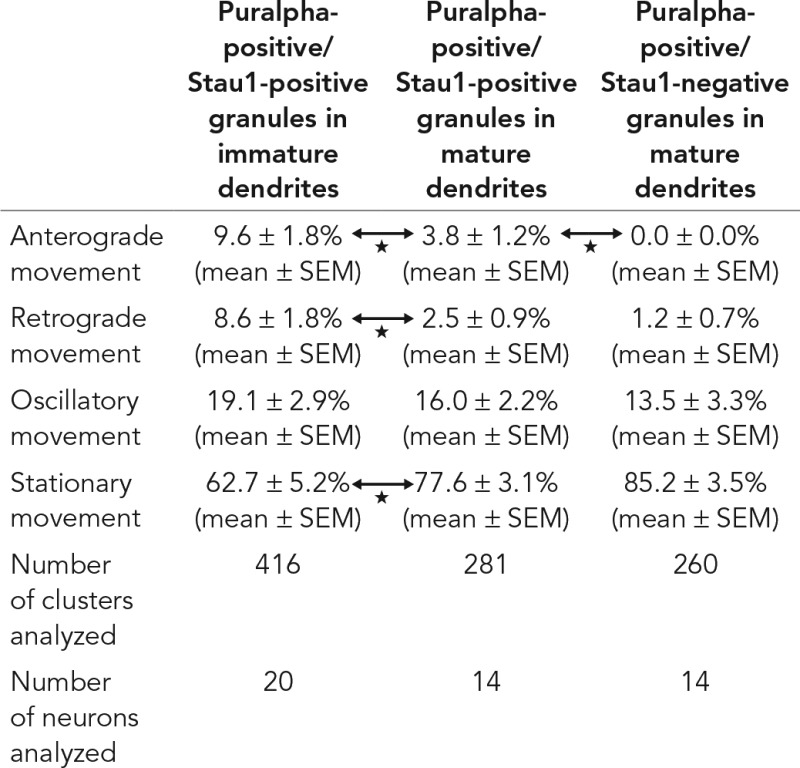 |
The movement was monitored for 3 min.
p < 0.05 (Student’s t test).
Next we analyzed the movement of granules in dendrites of mature neurons. TagRFP-Puralpha–positive granules were less dynamic compared with those in immature neurons (Table 2). We compared two types of granules in mature neurons: TagRFP-Puralpha–positive/Stau1-GFP–positive granules and TagRFP-Puralpha–positive/Stau1-GFP–negative granules. TagRFP-Puralpha–positive/Stau1-GFP–negative granules exhibited less anterograde motility compared with TagRFP-Puralpha–positive/Stau1-GFP–positive granules (Table 2).
TagRFP-Puralpha and Stau1-GFP signals comigrated in dendrites of mature neurons (Figure 2D). Separation of TagRFP-Puralpha–positive/Stau1-GFP–negative granules from TagRFP-Puralpha–positive/Stau1-GFP–positive granules was occasionally observed in dendrites of mature neurons (Figure 2E).
These data suggest that the composition and motility of Puralpha-positive granules change during neuronal development. Puralpha granules move dynamically within dendrites before neuronal maturation, whereas translocation of Puralpha granules along dendrites occurs more rarely once the neurons mature.
Activity-dependent Puralpha translocation to dendritic spines
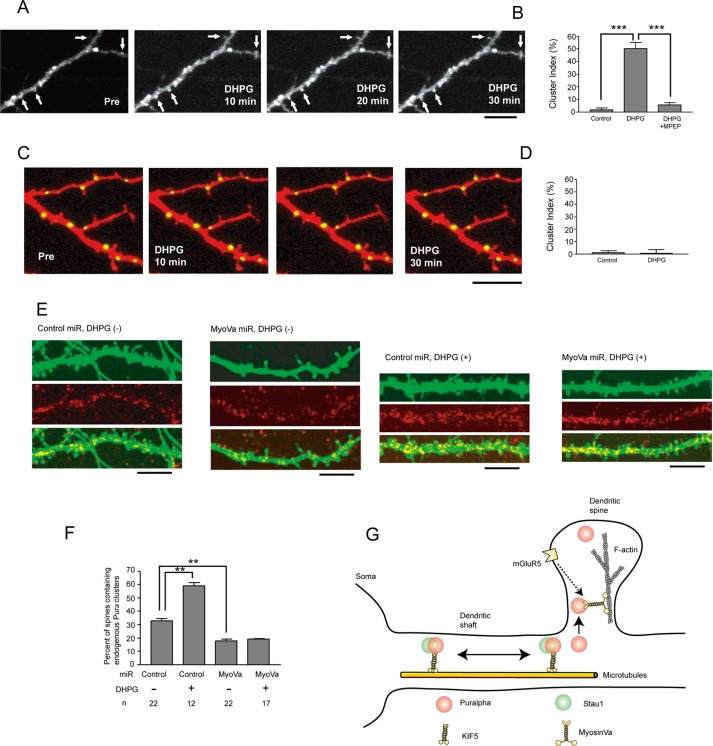
The localization of Puralpha in dendritic spines raised the possibility that Puralpha is transported to spines in an activity-dependent manner, as a previous study reported that TLS (translocated in liposarcoma), another RNA-binding protein, is translocated to dendritic spines by metabotropic glutamate receptor 5 (mGluR5) activation (Fujii et al., 2005). Because local protein synthesis subsequent to the translocation of mRNA to dendrites is known to be stimulated by DHPG ((S)-3,5-dihydroxyphenylglycine), a group 1 mGluR agonist (Job and Eberwine, 2001), we tested the effect of DHPG on the postsynaptic accumulation of YFP-Puralpha. When cultured hippocampal neurons expressing YFP-Puralpha were stimulated with DHPG (100 μM) over a 30-min period, the relative fluorescence intensity of spines (i.e., the cluster index, which is the increased percentage of relative fluorescence intensity after stimulation divided by the relative fluorescence intensity before stimulation) was significantly increased, compared with that of control neurons (Figure 3, A and B). The mGluR5 antagonist MPEP (2-methyl-6-[phenylethynyl]pyridine hydrochloride) blocked the effect of DHPG (Figure 3B), indicating that Puralpha accumulation in spines is induced upon postsynaptic activation of signaling cascades initiated by mGluR5.
FIGURE 3:
(A) Time-lapse recording of YFP-Puralpha clusters after DHPG treatment reveals that YFP-Puralpha accumulates within spines. Scale bar: 10 μm. (B) The average cluster index is increased 30 min after treatment with 100 μM DHPG (22 clusters from 12 neurons from two mice were examined). The DHPG-dependent spine accumulation is abolished by MPEP, an mGluR5 antagonist (18 clusters from 11 neurons from two mice were examined). For the control, 22 clusters from 12 neurons from two mice were examined. Error bars indicate SEM. ***, p < 0.001; Student’s t test. (C) Time-lapse recording after DHPG treatment reveals that Stau1-GFP clusters remained in dendritic shafts. Scale bar: 10 μm. (D) The average cluster index is not increased 30 min after treatment with DHPG (12 clusters from five neurons from two mice were examined). For control, 12 clusters from six neurons from two mice were examined. Error bars indicate SEM. (E) Forty-eight hours after transfection with miR vectors, neurons (15 DIV) were treated with DHPG for 1 h, fixed, and stained with anti-Puralpha antibodies. Scale bars: 10 μm. (F) Statistical analysis of E. Percentage of spines containing endogenous Puralpha clusters was increased by DHPG treatment and was decreased by myosin Va silencing. Error bars represent SEM. **, p < 0.01; Student’s t test. (G) Schematic model. Puralpha clusters are localized in both dendritic spines and shafts, whereas Stau1 clusters are present only in dendritic shafts. Some of these clusters are transported along dendrites by microtubule-based molecular motors (KIF5). Stau1-negative clusters are preferentially translocated to dendritic spines by an actin-based molecular motor (myosin Va). This oriented translocation involves activation of the mGluR5 pathway.
In contrast, localization of Stau1-GFP did not respond to DHPG treatment (100 μM, 30 min) (Figure 3, C and D). Stau1-GFP granules were exclusively localized to dendritic shafts and remained there over the 30-min period of DHPG treatment (Figure 3, C and D). These results, together with those shown in Figure 1, suggest that Stau1-negative Puralpha granules tend to be translocated to spines in response to mGluR5 activation, but Stau1-positive Puralpha granules do not respond to DHPG.
Role of myosin Va in the accumulation of Puralpha in dendritic spines
Next the molecular mechanisms of Puralpha translocation were investigated. Because dendritic spines rarely contain microtubules but do contain actin (Harris, 1999; Kaech et al., 2001), we examined whether myosin, a motor protein that migrates along actin tracts, might participate in the transport of Puralpha from dendritic shafts into spines. Indeed, a previous study indicated that the translocation of the RNA-binding protein TLS into spines is dependent on myosin Va (Yoshimura et al., 2006). Therefore we performed an RNA interference (RNAi) experiment (Supplemental Figure S1). Mature neurons were transfected with knockdown or control microRNA (miR) vectors for 48 h and immunostained with anti-Puralpha antibodies (Figure 3E). Expression of these miR vectors did not change the morphology of dendritic spines or the density of spines along dendrites (control neurons: 0.64 ± 0.05/μm [mean ± SEM], 21 neurons from two mice were examined; myosin Va-knockdown neurons: 0.59 ± 0.04/μm, 21 neurons from two mice were examined; Mann-Whitney test, not significant). In neurons expressing the control miR, the percentage of spines containing Puralpha was 32.7 ± 1.9/μm (average ± SEM; Figure 3F). Transfection of the myosin Va miR reduced this value to 17.5 ± 1.6/μm (Figure 3F).
DHPG treatment significantly increased the percentage of Puralpha-containing spines (Figure 3, E and F). Knockdown of myosin Va inhibited this DHPG-dependent synaptic accumulation of Puralpha (Figure 3, E and F). These data suggest that translocation of Puralpha into spines is mediated by myosin Va, both under basal and DHPG-stimulated conditions.
Localization and dynamics of Puralpha
In this study, we investigated the dynamics and localization of Puralpha in neurons. We demonstrate that Puralpha is localized not only in dendritic shafts, but also in spines, of mature neurons (Figure 1). This suggests that Puralpha plays a key role in the synaptic delivery of mRNA for local protein synthesis.
Another important aspect of this study is the relationship between Puralpha and Stau1 in RNA granules. In immature dendrites, more than 90% of Puralpha granules are Stau1-positive. However, in mature neurons, only about half of the Puralpha granules are positive for Stau1 (Figure 2, A and B, and Table 1). That is, the fraction of Puralpha granules lacking Stau1 increases during neuronal maturation. It is also noteworthy that Stau1-positive granules are localized mainly in dendritic shafts, and Stau1 is rarely found in dendritic spines (Figure 1, D and E). Therefore Stau1 in Puralpha clusters might prevent Puralpha-containing granules from entering postsynaptic regions.
Activity-dependent translocation of Puralpha
Metabotropic glutamate receptors (mGluRs) have diverse functions in neuronal signaling. Group 1 mGluRs, including mGluR1 and mGluR5, are localized at the periphery of the postsynaptic junction membrane of principal neurons in the hippocampus (Shigemoto et al., 1993). mGluR5 is especially abundant in dendrites and dendritic spines (Lujan et al., 1996). DHPG, a selective group 1 mGluR agonist, increased the number of YFP-Puralpha granules in the spines of mature dendrites within 30 min of stimulation (Figure 3, A and B). The mGluR5 antagonist MPEP blocked this effect of DHPG (Figure 3B). This suggests that a postsynaptic mGluR5-mediated signal stimulates the translocation of Puralpha. Group 1 mGluRs couple to phospholipase C, causing downstream activation of protein kinase C (PKC) and the release of calcium from intracellular stores (Nakanishi, 1994). Metabotropic GluR-dependent long-term depression (LTD) requires both PKC activation and an increase in postsynaptic calcium concentration (Oliet et al., 1997), and maintenance of LTD requires Arc protein synthesis following the release of translational inhibition by fragile X mental retardation protein. Increased Arc enhances the internalization of alpha-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA) receptors via endocytosis (Chowdhury et al., 2006), resulting in LTD induction. These observations, together with our present findings, suggest that Puralpha-containing RNA granules might be transported to synapses for the activity-dependent local synthesis of proteins such as Arc.
Myosin Va–dependent transport of Puralpha to synapses
Myosin Va, an isoform of class V myosins, is expressed in neurons (McCaffrey and Lindsay, 2012). Biochemical analysis has shown that the mRNA/protein complex contains myosin Va (Ohashi et al., 2002). The localization of mRNA depends on actin filaments and myosin Va (Balasanyan and Arnold, 2014). Accumulation of the mRNA/protein complex containing TLS is facilitated by myosin Va (Yoshimura et al., 2006). Therefore myosin Va transports not only extended endoplasmic reticulum (Wagner et al., 2011), but also mRNA/protein complexes. Our current results suggest that Puralpha-positive granules are transported to spines by myosin Va (Figure 3, E and F).
In mature neurons, Puralpha granules transported anterogradely and retrogradely within shafts are mainly positive for Stau1, and those lacking Stau1 enter preferentially into spines (Figure 1). This suggests that the presence or absence of Stau1 may affect the association of Puralpha granules with molecular motors such as myosin Va. Future studies should examine this intriguing possibility.
MATERIALS AND METHODS
Hippocampal dissociated cultures
Hippocampi were dissected from the brains of embryonic day 16 (E16) ICR mouse fetuses, treated with 0.25% trypsin for 15 min at 37°C, washed in Ca/Mg-free Hank’s balanced salt solution, and dissociated by repeated pipetting. Cells were added to coverglasses coated with poly-l-lysine in minimum essential medium (MEM) containing 1 mM pyruvic acid, 0.6% glucose, and 10% horse serum (Life Technologies). After 4 h at 37°C, when the cells had attached to the substrate, 70% of the media was replaced with MEM containing 1 mM pyruvic acid, 0.6% glucose, and B27 (Life Technologies).
Antibodies
The primary antibodies used in this study were Puralpha and Stau1 rabbit polyclonals (Kanai et al., 2004), postsynaptic density-95 mouse monoclonal (Affinity BioReagents), and myosin Va polyclonal (Sigma).
Immunofluorescence
For immunofluorescence analysis, cultured neurons were fixed in phosphate-buffered saline (PBS) containing 4% paraformaldehyde for 10 min at 37°C and then permeabilized with PBS/0.25% Triton X-100 for 5 min at room temperature. The cells were further washed in PBS and blocked in PBS/5% skim milk for 30 min at room temperature. Primary antibodies were diluted in PBS/5% skim milk and incubated overnight at 4°C. After three washes in PBS, cells were incubated with corresponding Alexa Fluor 488– or Alexa Fluor 568–conjugated secondary antibodies (Molecular Probes) diluted in PBS/5% skim milk for 1 h at room temperature. Finally, the cells were washed three times in PBS and were observed with an LSM510 confocal laser-scanning microscope (Carl Zeiss, Oberkochen, Germany) using a 40×/1.2 NA water-immersion objective.
Transfection and expression of tagged proteins
EYFP-Puralpha and Staufen1-EGFP plasmids were gifts from Y. Kanai, Tokyo University (Kanai et al., 2004). TagRFP plasmid was purchased from Evrogen. DNA was purified using the EndoFree plasmid maxikit (Qiagen, Hilden, Germany). Transfection of hippocampal-dissociated neurons was performed using a calcium phosphate protocol optimized for neuronal cultures (Jiang and Chen, 2006). At 24 h after transfection, neurons were used for immunocytochemistry or time-lapse imaging.
Time-lapse imaging
For time-lapse imaging, live cells were submerged in Leibovitz’s L-15 medium (Life Technologies) containing 20-mM HEPES (Life Technologies) that was heated to 37°C. Images were obtained with an LSM510 confocal laser-scanning microscope using a 40×/1.2 NA water-immersion objective. Observations were carried out for 3 min for each experiment. Unidirectional movements away from or toward the soma were defined as anterograde or retrograde, respectively. If the direction reversed more than once during 3 min of imaging time, the movement was defined as oscillatory. Classification of the movements was performed by direct observation of the sequential imaging data.
Reagents
DHPG and MPEP were purchased from Tocris (Ellisville, MO).
RNAi assay
The BLOCK-iT Pol II miR RNAi Expression vector kit (Invitrogen) was used according to the manufacturer’s protocol. Target sequences were designed using the Invitrogen website. For myosin Va knockdown, the insert sequence was 5′-TGCTGTTGACTACCTGCTTGATTAGCGTTTTGGCCACTGACTGACGCTAATCACAGGTAGTCAA-3′. For the negative control included in the kit, the insert was 5′-TGCTGAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′. Nucleotides in bold indicate antisense target sequences. Invitrogen pcDNA6.2-GW/EmGFP-miR vectors were used.
Data analysis
Maximum-intensity projection images were prepared for each image stack, and these were used for the quantitative analyses. A defined region of interest was traced within a dendritic spine, and the fluorescence intensity was measured.
Supplementary Material
Acknowledgments
This work was supported by a grant-in-aid for specially promoted research and a grant-in-aid for scientific research (S) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to N.H. This research was also partially supported by the Strategic Research Program for Brain Sciences and the Brain Mapping by Integrated Neurotechnologies for Disease Studies from the Japan Agency for Medical Research and Development and a grant from the Uehara Memorial Foundation.
Abbreviations used:
- DHPG
(S)-3,5-dihydroxyphenylglycine
- DIV
days in vitro
- GFP
green fluorescent protein
- KIF5
kinesin
- KIFs
kinesin superfamily of proteins
- LTD
long-term depression
- MEM
minimum essential medium
- mGluR5
metabotropic glutamate receptor 5
- miR
microRNA
- MPEP
2-methyl-6-[phenylethynyl]pyridine hydrochloride
- PBS
phosphate-buffered saline
- PKC
protein kinase C
- PSD-95
postsynaptic density protein-95
- RNAi
RNA interference
- Sta1
Staufen1
- TagRFP
tag red fluorescent protein
- TLS
translocated in liposarcoma
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-07-0497) on April 12, 2017.
REFERENCES
- Balasanyan V, Arnold DB. Actin and myosin-dependent localization of mRNA to dendrites. PLoS One. 2014;9:e92349. doi: 10.1371/journal.pone.0092349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishimura T, Hicks GG, Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- Gallia GL, Johnson EM, Khalili K. PurA: a multifunctional single-stranded DNA and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–3205. doi: 10.1093/nar/28.17.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM. Structure, development, and plasticity of dendritic spines. Curr Opin Neurobiol. 1999;9:343–348. doi: 10.1016/s0959-4388(99)80050-6. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent LTD. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- Job C, Eberwine J. Localization and translation of mRNA in dendrites and axons. Nat Rev Neurosci. 2001;2:889–898. doi: 10.1038/35104069. [DOI] [PubMed] [Google Scholar]
- Kaech S, Parmar H, Roelandse M, Bornmann C, Matus A. Cytoskeletal microdifferentiation: a mechanism for organizing morphological plasticity in dendrites. Proc Natl Acad Sci USA. 2001;98:7086–7092. doi: 10.1073/pnas.111146798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Lujan R, Nusser Z, Roberts JD, Shigemoto R, Somogyi P. Perisynaptic localization of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur J Neurosci. 1996;8:1488–1500. doi: 10.1111/j.1460-9568.1996.tb01611.x. [DOI] [PubMed] [Google Scholar]
- McCaffrey MW, Lindsay AJ. Roles for myosin Va in RNA transport and turnover. Biochem Soc Trans. 2012;40:1416–1420. doi: 10.1042/BST20120172. [DOI] [PubMed] [Google Scholar]
- Nakanishi S. Metabotropic glutamate receptors: synaptic transmission, modulation, and plasticity. Neuron. 1994;13:1031–1037. doi: 10.1016/0896-6273(94)90043-4. [DOI] [PubMed] [Google Scholar]
- Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato TA, Anzai K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- Oliet SHR, Malenka RC, Nicoll RA. Two distinct forms of long-term depression coexist in CA1 hippocampal pyramidal cells. Neuron. 1997;18:969–982. doi: 10.1016/s0896-6273(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Shigemoto R, Nomura S, Ohishi H, Sugihara H, Nakanishi S, Mizuno N. Immunohistochemical localization of a metabotropic glutamate receptor, mGluR5, in the rat brain. Neurosci Lett. 1993;163:53–57. doi: 10.1016/0304-3940(93)90227-c. [DOI] [PubMed] [Google Scholar]
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of Staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Wagner W, Brenowitz SD, Hammer JA., III Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat Cell Biol. 2011;13:40–48. doi: 10.1038/ncb2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Fujii R, Watanabe Y, Okabe S, Fukui K, Takumi T. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr Biol. 2006;16:2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.