A network of interactions for human adenine nucleotide translocases, required for oxidative phosphorylation, is reported. Of particular interest is an evolutionarily conserved and functionally important association with respiratory supercomplexes, which is surprising because the respirasomes of yeast and mammals are different.
Abstract
Members of the adenine nucleotide translocase (ANT) family exchange ADP for ATP across the mitochondrial inner membrane, an activity that is essential for oxidative phosphorylation (OXPHOS). Mutations in or dysregulation of ANTs is associated with progressive external ophthalmoplegia, cardiomyopathy, nonsyndromic intellectual disability, apoptosis, and the Warburg effect. Binding partners of human ANTs have not been systematically identified. The absence of such information has prevented a detailed molecular understanding of the assorted ANT-associated diseases, including insight into their disparate phenotypic manifestations. To fill this void, in this study, we define the interactomes of two human ANT isoforms. Analogous to its yeast counterpart, human ANTs associate with heterologous partner proteins, including the respiratory supercomplex (RSC) and other solute carriers. The evolutionarily conserved ANT–RSC association is particularly noteworthy because the composition, and thereby organization, of RSCs in yeast and human is different. Surprisingly, absence of the major ANT isoform only modestly impairs OXPHOS in HEK293 cells, indicating that the low levels of other isoforms provide functional redundancy. In contrast, pharmacological inhibition of OXPHOS expression and function inhibits ANT-dependent ADP/ATP exchange. Thus ANTs and the OXPHOS machinery physically interact and functionally cooperate to enhance ANT transport capacity and mitochondrial respiration.
INTRODUCTION
The inner mitochondrial membrane (IMM) is a densely packed environment that houses nuclear- and mitochondrial-encoded proteins alike, many of which are built into protein complexes. For instance, the oxidative phosphorylation (OXPHOS) machinery, which lies at the core of energy metabolism, is composed of the multisubunit respiratory complexes and the dimeric/oligomeric ATP synthase in addition to mobile electron carriers and solute carriers (Hatefi, 1985). Moreover, higher-order structural organizations are observed in the IMM, as best exemplified by the presence of respiratory supercomplexes (RSCs), which are believed to optimize substrate channeling and facilitate electron flow between the individual participating respiratory complexes for enhanced OXPHOS efficiency (Cruciat et al., 2000; Schagger and Pfeiffer, 2000; Acin-Perez et al., 2008; Gu et al., 2016; Letts et al., 2016).
The ADP/ATP carrier (AAC) is one of the most abundant proteins in the IMM, a reflection of its vital role as the gatekeeper of energy flux—by facilitating ADP import into the mitochondrial matrix and ATP export to the cytosol, AAC enables succeeding rounds of ATP production and delivery to the rest of the cell (Klingenberg, 2008). It also mediates metabolic cycling and proton leak (Brand et al., 2005). Similar to other members of the mitochondrial carrier family (MCF), AAC is a membrane protein made up of ∼300 amino acids forming six transmembrane helices and exhibiting threefold pseudosymmetry (Palmieri, 2004). It has been extensively studied, with sequence annotations and crystal structures available, and is thus often regarded as the archetype of mitochondrial carriers (Pebay-Peyroula et al., 2003; Nury et al., 2005; Klingenberg, 2008).
For decades, AACs had been modeled to carry out their tasks in physical isolation (Klingenberg, 2009). However, a study that focused on Aac2p, the major isoform in Saccharomyces cerevisiae, determined the existence of Aac2p complexes, which were initially believed to correspond to Aac2p homodimers and homotetramers (Jiang et al., 2000). Further analysis revealed that Aac2p interacts with other IMM proteins, including RSCs, and additional MCF members, such as the phosphate carrier (Pic1p and Pic2p), the GDP/GTP carrier (Ggc1p), and the dicarboxylate carrier (Dic1p), and that this assembly is dependent on the presence of the mitochondrial-resident lipid, cardiolipin (CL; Claypool et al., 2008). In addition, Aac2p has also been shown to bind to the TIM23 translocon (Dienhart and Stuart, 2008; Mehnert et al., 2014). The critical role of CL in maximizing respiratory complex activity and maintaining the assembly and/or stability of high–molecular weight assemblies that include Aac2p (Zhang et al., 2002; Pfeiffer et al., 2003; Claypool et al., 2008), together with the observation that lack of CL in yeast results in reduced Aac2p function and OXPHOS efficiency under optimal conditions and abolished OXPHOS under stress (Jiang et al., 2000; Claypool et al., 2008), supports the physiological importance of the Aac2p interactome. Nonetheless, whether these interactions are necessary for normal mitochondrial function or important only under specialized circumstances, and to what extent their destabilization serves as grounds for mitochondrial impairment, are all unexplored territories.
AAC’s mammalian equivalent, called the adenine nucleotide translocase (ANT), exists as four isoforms, each with distinct transport activity and tissue-specific and yet overlapping expression. ANT1 is the main isoform in the heart and skeletal muscle, ANT2 is predominant in the kidney and liver, ANT3 is ubiquitously expressed at low levels (Stepien et al., 1992; Doerner et al., 1997), and ANT4 is found only in lung and germ tissues (Dolce et al., 2005; Rodic et al., 2005; Lim et al., 2015). Whether all ANT isoforms are functionally equivalent or instead have unique functions remains a fundamental and unanswered question.
ANT1 deficiency is implicated in various pathological states, such as hypertrophic cardiomyopathy, mitochondrial myopathy, lactic acidosis, progressive external opthalmoplegia, fascioscapulohumeral muscular dystrophy, and Sengers syndrome (Kaukonen et al., 2000; Jordens et al., 2002; Komaki et al., 2002; Fontanesi et al., 2004; Sharer, 2005; Palmieri, 2008; Echaniz-Laguna et al., 2012). Mutations in ANT2 have also been associated with nonsyndromic intellectual disability (Vandewalle et al., 2013) and cardiac noncompaction (Kokoszka et al., 2016) and its dysregulation associated with a Warburg metabolic phenotype (Maldonado et al., 2016). These diseases have disparate phenotypic manifestations bound by an underlying mitochondrial dysfunction, albeit without a clear molecular explanation. In terms of human ANT interactors, essentially nothing is known. Identification of ANT binding partners and subsequent characterization of the physiological significance of these associations could provide unexpected insights into ANT function at the molecular and cellular levels. Defining the ANT interactome may also help to reveal functional benefits of quaternary interactions and ultimately augment our understanding of the mitochondrion as a whole system.
We therefore set out to define the human ANT1 and ANT2 interactomes through an unbiased proteomics approach. We find that, similar to yeast Aac2p, human ANTs form supramolecular associations with heterologous proteins. Of particular note is the evolutionarily conserved ANT–RSC interaction, which was surprising because the composition of RSCs in yeast (complexes III and IV) and humans (complexes I, III, and IV) is different. The maintenance of ANT–RSC interaction over time and across species despite a divergence in RSC’s structural organizations (Schagger and Pfeiffer, 2000) suggests that this partnership is functionally important. To dissect the interaction, we used 293 Flp-In cells as a model to reciprocally query the effects of ANT deletion on OXPHOS and the consequences of OXPHOS inhibition on ANT transport. Our results demonstrate that the minor ANT isoforms are not functionally minor and indicate that the cooperation of ANTs and RSCs can enhance transport capacity of the former, leading to improved mitochondrial respiration and thereby affirming the functional importance of this evolutionarily conserved interaction.
RESULTS
Defining the interactome of human ANT1 and ANT2
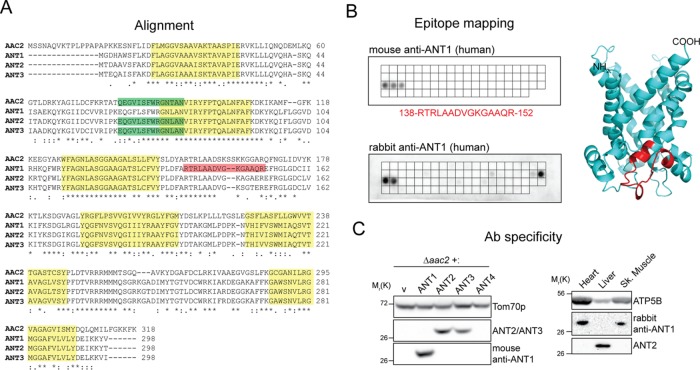
Owing to the absence of quality ANT-specific antibodies, we generated antibodies against recombinant human ANT1 that are highly specific for this isoform (Figure 1, A–C). To detect ANT2 and ANT3, we used a monoclonal antibody (mAb) raised against yeast Aac2p, 5H7, that conveniently cross-reacts with both ANT2 and ANT3 but not ANT1 (Figure 1, A and C).
FIGURE 1:
Isoform-specific antibodies are capable of distinguishing the highly homologous ANT proteins. (A) Human adenine nucleotide translocases are highly conserved and share significant similarity with Aac2p, the major isoform in S. cerevisiae. Highlighted in red and green are epitopes recognized by ANT1 (mouse mAb) and ANT2/ANT3 mAbs, respectively; transmembrane helices are in yellow. (B) The epitope recognized by the newly generated ANT1-specific mAb was mapped using a peptide array (top) and found to be RTRLAADVGKGAAQR. The epitope is also highlighted in red in the structure of human ANT1 modeled using bovine ANT1 (Protein Data Bank 1OKC) as template. A peptide array (bottom) was also used to determine the epitopes recognized by the corresponding rabbit polyclonal ANT1. (C) Yeast extracts (left) derived from Δaac2 heterologously expressing yhANTs or empty vector were resolved by SDS–PAGE and immunoblotted with the ANT1-specific mAb or the ANT2/ANT3 mAb to test for isoform specificity. Tom70p, an outer mitochondrial membrane protein, serves as a loading control. Mitochondria (50 μg) from mouse liver, heart, and skeletal muscle (right) were similarly resolved to test the specificity of the rabbit ANT1 antiserum.
With these two reagents, we first determined whether human ANTs assemble in high–molecular weight complexes like yeast Aac2p (Claypool et al., 2008). Mitochondrial extracts from 293 Flp-In (wild type [wt]) and 293 Flp-In stably overexpressing ANTs with or without an N-terminal consecutive nondenaturing affinity purification (CNAP) tag were analyzed by two-dimensional (2D) blue native (BN)/SDS–PAGE to monitor the assembly of ANT2/3 and ANT1 (Figure 2, A–C). Of note, both endogenous (Figure 2, A–C) and overexpressed ANTs (Figure 2, B and C) assembled in a range of complexes from 67 to >669 kDa, some of which comigrated with complex I and complex IV assemblies (Figure 2A). Murine ANTs in heart and liver mitochondria similarly engaged in a range of high–molecular weight assemblies in vivo (Figure 2, B and C), consistent with observations in rat muscle and kidney mitochondria (Faustin et al., 2004).These results suggest that mammalian ANT isoforms participate in multiple distinct macromolecular complexes and thus may associate with heterologous proteins and/or protein complexes.
FIGURE 2:
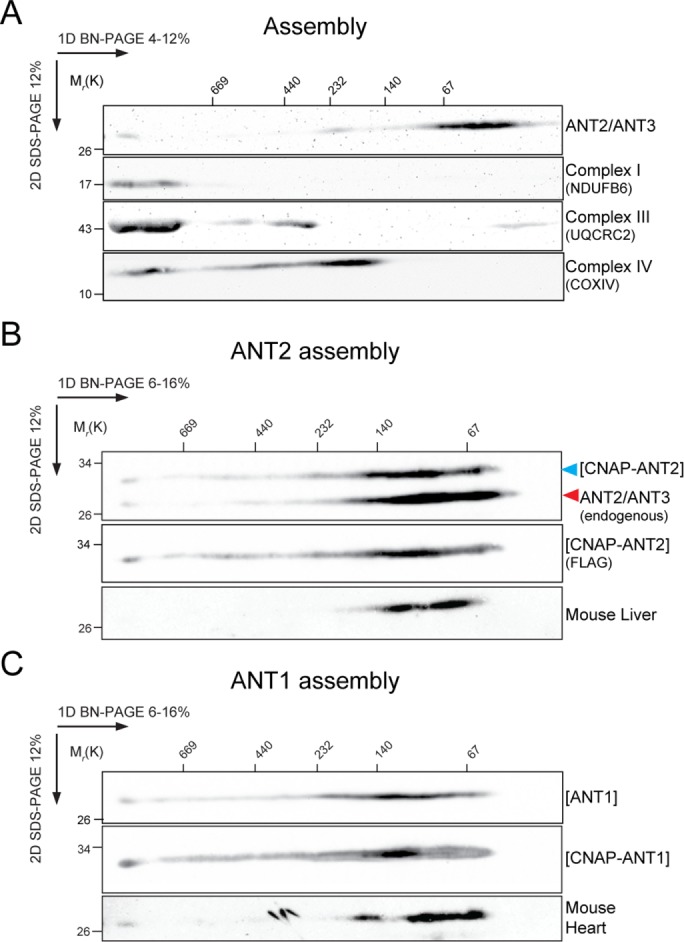
ANTs assemble in high–molecular weight complexes. Mitochondria (200 μg) isolated from HEK293 Flp-In cells (wt) expressing endogenous ANT2/3 (A) or overexpressing (B, C) the indicated ANT constructs or from the indicated mouse tissues were solubilized with digitonin, resolved by 2D BN/SDS–PAGE, and immunoblotted for ANTs using (A, B) ANT2/ANT3, (C) ANT1, or (B) FLAG antibodies. UQCRC2, NDUFB6, and COX4 are presented to show ANT comigration with RSCs. In B, overexpressed CNAP-ANT2 and endogenous ANT2/3 are indicated.
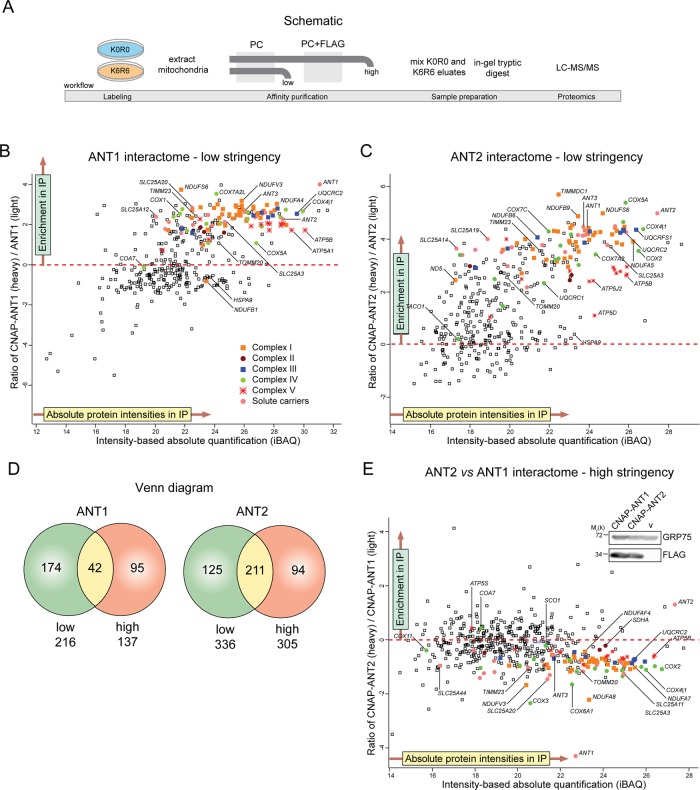
To define the interactome of human ANTs, we used stable isotope labeling by amino acids in cell culture (SILAC)–based quantitative proteomics analysis (Ong et al., 2002). CNAP-ANTs were affinity purified from mitochondria isolated from 293 Flp-In cells overexpressing CNAP-tagged or untagged ANTs and differentially labeled by growing them either in heavy medium containing [13C6]lysine and [13C6]arginine or in light medium with standard 12C-containing amino acids. The derived eluates were mixed in a 1:1 ratio and resolved by gradient SDS–PAGE before in-gel trypsin digest, liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses (Figure 3A). Similar to yeast Aac2p (Claypool et al., 2008; Dienhart and Stuart, 2008; Mehnert et al., 2014), the proteomics analyses revealed that human ANT1 and ANT2 each associated with many respiratory components, translocase machineries, and mitochondrial carriers (Figure 3, B and C, and Supplemental Tables S1–S3). When the stringency of the experiment was increased by including a second, nondenaturing FLAG affinity purification, the number of interacting proteins identified with each of the ANT isoforms was reduced (Supplemental Figure S1, A and B, and Supplemental Tables S1–S3). However, many of the same interactors were identified between the two immunoprecipitation (IP) stringencies, illustrating the specificity of our approach (Figure 3D). Of interest, among the strongest hits for ANT1 and ANT2 were other ANT isoforms (ANT2 and ANT3 for CNAP-ANT1 and ANT1 and ANT3 for CNAP-ANT2). Overall the interactomes of ANT1 and ANT2 are highly similar and include subunits of respiratory complexes I–IV (Supplemental Table S1). To compare directly the ANT1 and ANT2 interactomes, we differentially labeled and copurified together CNAP-ANT1 (light medium)– and CNAP-ANT2 (heavy medium)–overexpressing cells (Figure 3E). Given that CNAP-ANT1 and CNAP-ANT2 are overexpressed to similar levels (Figure 3E, inset), this analysis indicated that in the 293 Flp-In setting, neither isoform engaged in any overtly unique interactions, although ANT1 displayed a modestly increased affinity for OXPHOS proteins.
FIGURE 3:
Overexpressed ANT1 and ANT2 in HEK293 Flp-In cells interact with numerous heterologous proteins, including respiratory subunits and other solute carriers. (A) Experimental workflow to define the ANT interactome. Scatter plot of proteins identified to interact with overexpressed CNAP-ANT1 (B) or CNAP-ANT2 (C) under a low-stringency IP. Identified hits that lie above the red dashed line are enriched proteins with a heavy/light isotopic ratio >0. (D) Venn diagram showing the number of proteins identified (heavy/light isotopic ratio >0) that are shared or unique to the low- and high-stringency IPs. (E) Scatter plot of proteins identified when CNAP-ANT2 (labeled) was queried against CNAP-ANT1 (unlabeled). Inset, relative expression of CNAP-ANT1 and CNAP-ANT2, using a FLAG antibody.
Generation of an ant2TALEN cell model
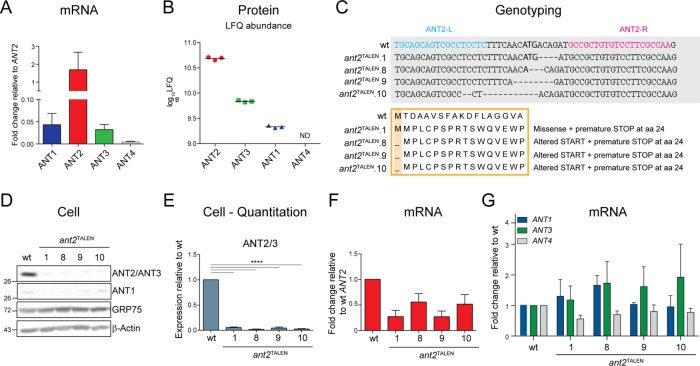
To dissect the functional relevance of ANT’s interaction with respiratory components, we generated an ant-null cell culture model. Because 293 Flp-Ins primarily express ANT2 at both the mRNA level (ANT2 represents >90% of all ANT transcripts, ANT1 and ANT3 are expressed only at 5% of ANT2 levels, and ANT4 is at an even lower relative expression; Figure 4A) and protein level (Figure 4B), we used transcription activator–like effector nuclease (TALEN)–mediated genome editing to introduce genetic lesions within exon 1 of ANT2. Four ant2TALEN clones were established with disruptions in the targeted locus (Figure 4C). Indeed, the levels of ANT2 protein was reduced by ∼95% in all four clones (Figure 4, D and E). We speculate that the remaining signal detected by immunoblot can be attributed to ANT3, which has lower expression at the mRNA level (Figure 4A), is significantly less abundant in 293 Flp-Ins based on proteomics, and shares the epitope detected by the utilized mAb (Figure 1C). Further, there is a concomitant reduction in ANT2 transcript levels (Figure 4F) without a compensatory increase in the mRNA expression of ANT1, ANT3, or ANT4 (Figure 4G).
FIGURE 4:
Generation of an ant2-null cell model using TALEN-mediated gene disruption. (A) Relative mRNA level of ANT isoforms in 293 Flp-In (wt). Data were analyzed by the comparative CT (ΔΔCT) method, represented as mean fold change (2-ΔΔCT) ± SEM (n = 4) relative to ANT2 expression, which was set to 1. (B) Mass spectroscopy–based quantitation of relative protein abundance of ANT isoforms in wt mitochondria. The log10 label-free quantification (LFQ) signal intensities. ANT4 was not detected. n = 3. (C) TALEN-mediated disruption of ANT2 in wt cells introduces nucleotide deletions that alter the start site and result in premature ANT2 translation termination in all four ant2TALEN clones. (D) Whole-cell extracts (50 μg) from wt and ant2TALEN clones immunoblotted with ANT2/ANT3- and ANT1-specific antibodies. (E) Densitometry analyses of ANT2/ANT3 expression in cell extracts from D. Expression relative to wt ± SEM (n ≥ 4). (F) Relative mRNA levels of ANT2 in ant2TALEN clones analyzed by the ΔΔCT method, represented as mean fold change compared with wt ± SEM (n = 4). (G) Relative mRNA levels of ANT1, ANT3, or ANT4 in ant2TALEN clones analyzed by the ΔΔCT method, represented as mean fold change compared with wt ± SEM (n ≥ 4).
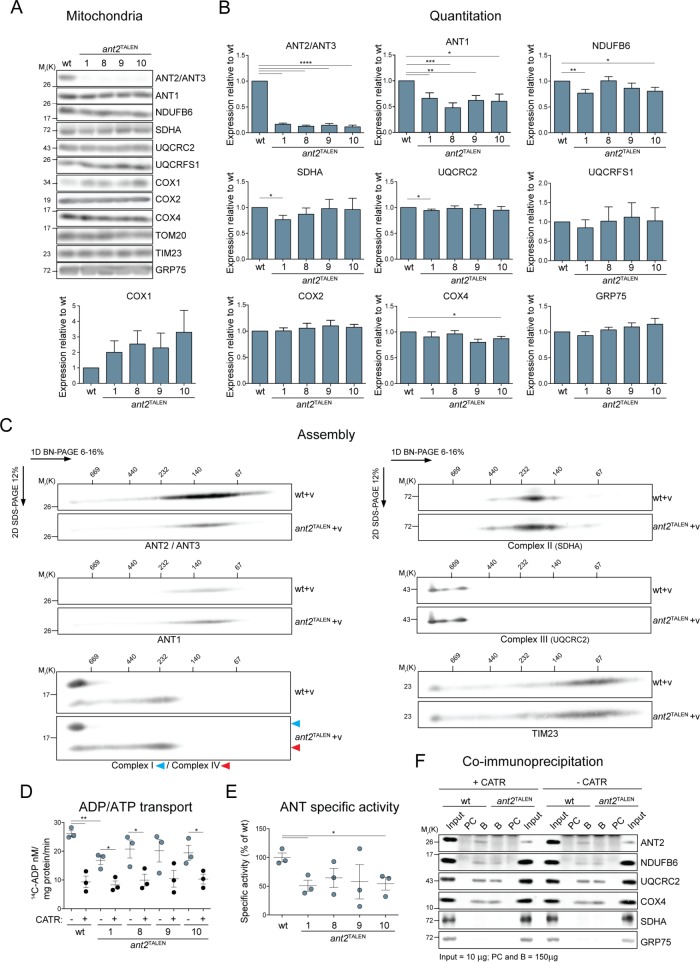
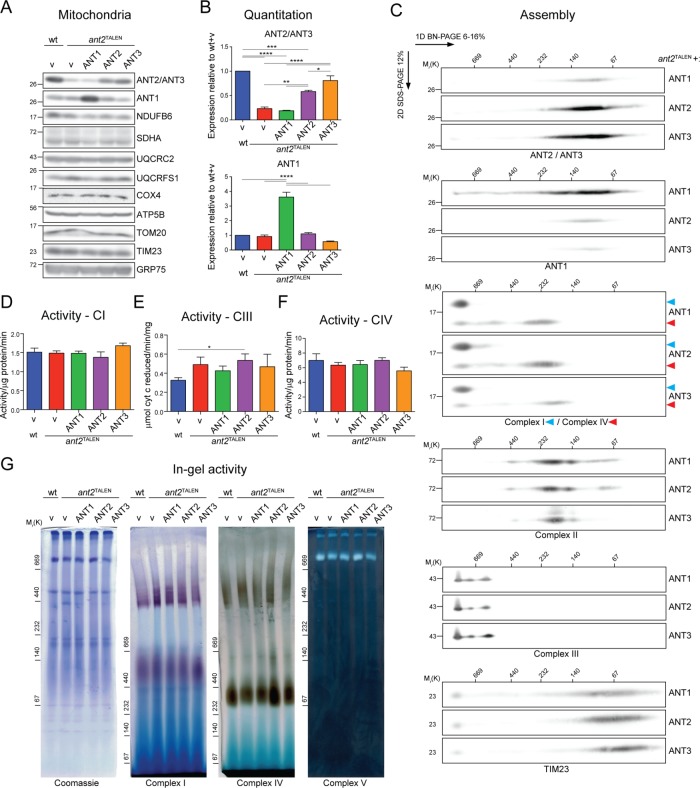
It was previously noted in mice lacking ANT1 that transcripts of nuclear and mitochondrially encoded OXPHOS components were up-regulated (Murdock et al., 1999). To test whether deletion of the major ANT isoform in 293 Flp-In alters mitochondrial protein levels, we analyzed the steady-state amounts of mitochondrial proteins (respiratory complexes I–IV; TOM20, TIMM23, and GRP75—markers for outer mitochondrial membrane [OMM], IMM, and matrix compartments, respectively; and COX1 and COX2—mitochondrially encoded subunits of complex IV) and found them to be largely unaffected in the null clones (Figure 5, A and B). Further, no assembly defect was noted for respiratory complexes I–IV in the absence of ANT2 (Figure 5C), and in-gel activity assays indicated that complexes I, IV, and V were assembled properly and functional in the absence of ANT2 (Figure 8G).
FIGURE 5:
ant2TALEN cells have reduced ADP/ATP transport but preserved expression and assembly of RSCs subunits. (A) Mitochondria (50 μg) from the indicated strains were immunoblotted as listed. (B) Densitometry analyses of protein steady state levels from mitochondrial extracts from A. Expression relative to wt ± SEM (n ≥ 4). (C) Mitochondria (250 μg) from empty vector–transfected wt or ant2TALEN cells were solubilized with 1.25% (wt/vol) digitonin, resolved by 2D BN/SDS–PAGE, and immunoblotted for ANT2/ANT3, NDUFB6 (complex I) and COX4 (complex IV), SDHA (complex II), UQCRC2 (complex III), or TIM23 (loading control). n = 3. (D) Mitochondria (100 μg) from wt and ant2TALEN cells were fused with liposomes internally preloaded with 5 mM unlabeled ADP. The uptake of [14C]ADP (nM/mg protein/min ± SEM; n = 3) was followed in the presence and absence of 40 μM CATR. (E) ANT-specific activity was calculated from D by subtracting CATR-sensitive exchange from ADP/ATP transport in the absence of inhibitor and expressed as a percentage of wt (100%). (F) CoIP of endogenous ANT2 with COX4 in the presence or absence of 10 μM CATR. B, bound; PC, preclear.
FIGURE 8:
ANT overexpression in ant2TALEN cells does not alter the expression, assembly, or activity of RSCs subunits. (A) SDS–PAGE-resolved mitochondria (50 μg) from the indicated strains were immunoblotted as listed. (B) Densitometry analyses of protein steady-state levels from mitochondrial extracts from A. Expression relative to wt ± SEM (n = 3). (C) ant2TALEN mitochondria (250 μg) expressing the various ANT isoforms were solubilized with 1.25% (wt/vol) digitonin, resolved by 2D BN/SDS–PAGE, and immunoblotted for ANT2/ANT3, ANT1, NDUFB6 (complex I) and COX4 (complex IV), SDHA (complex II), UQCRC2 (complex III), and TIM23. n = 3. (D) NADH dehydrogenase (activity/μg/min ± SEM; n = 6, (E) cytochrome c reductase (μmol cytochrome c reduced/min/mg ± SEM; n = 8), and (F) cytochrome c oxidase (activity/μg/min ± SEM; n ≥ 3) activity of DDM-solubilized mitochondrial extracts. (G) In-gel activity assays for complexes I, IV, and V. Mitochondria (100 μg) from empty vector –transfected wt cells or ant2TALEN cells transfected as indicated were solubilized with 1.25% (wt/vol) digitonin, resolved by 3–10% (complexes I and IV), or 6–16% (Coomassie and complex V) one-dimensional BN-PAGE and incubated with complex I, IV, or V substrates. Coomassie staining of the gel served as a loading control.
Next we assessed the transport capacity of the ant2TALEN clones by measuring [14C]ADP uptake into liposome-fused mitochondria in the presence or absence of carboxyatractyloside (CATR), a potent pan-ANT inhibitor (Figure 5D; Brandolin et al., 1974). Although mitochondria from all four ant2TALEN clones exhibited the same trend, only clones 1 and 10 had a significantly reduced ANT specific transport activity relative to wt (Figure 5E). On the basis of these results, we decided to proceed with ant2TALEN.1 for the remainder of the experiments.
Endogenous ANT2 associates with respiratory subunits
Armed with an ant2-null cell model, we next sought to confirm the ANT2 interaction with respiratory complexes by coimmunoprecipitation and immunoblot analyses of endogenous proteins. Because the ANT2/3 mAb does not immunoprecipitate, we instead determined whether endogenous ANT2 coimmunoprecipitated with COX4, a core component of complex IV. Endogenous ANT2 was copurified with COX4, albeit at low amounts, in mitochondrial extracts from wt but not ant2TALEN (Figure 5F). Of importance, this interaction was preserved upon inclusion of CATR, which locks ANTs in an extreme transition state, thus preventing it from unfolding. COX4 was able to pull down NDUFB6 (complex I) and UQCRC2 (complex III) but not SDHA (complex II) and GRP75, which are excluded from RSCs (Figure 5F). These results indicate that endogenous ANT2 interacts with RSC-participating and/or free complex IV.
The minor ANT isoforms are not functionally minor
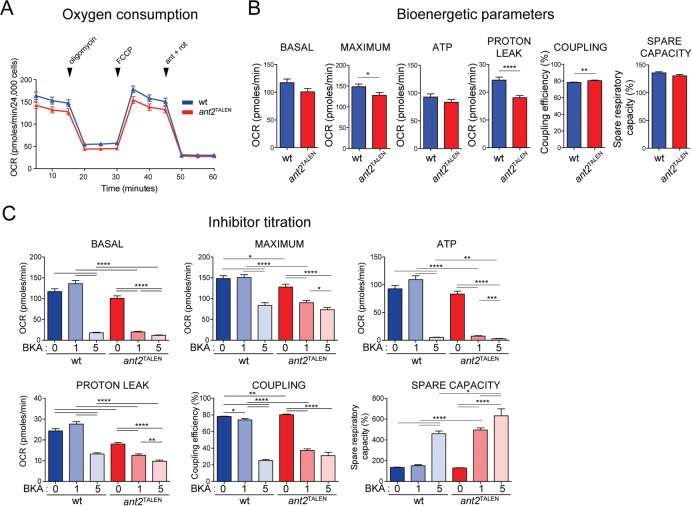
Given the central bioenergetic role of ANTs (Klingenberg, 2008), we hypothesized that the absence of ANT2, the major ANT isoform expressed in 293 Flp-In cells (Figure 4, A and B), would severely compromise cellular respiration by limiting the supply of ADP, the substrate used by complex V for ATP synthesis. Indeed, in comparison to wt cells, ant2TALEN had significantly depressed maximal respiration after addition of the uncoupler FCCP (Figure 6B). However, basal respiration in the absence of inhibitors and spare capacity—the ability to respond to increased energy need—were not affected by the absence of ANT2 (Figure 6B). In addition, respiration linked with ATP production (oxygen consumption rate [OCR] after injection of the complex V inhibitor oligomycin) was also not significantly different between wt and ant2TALEN (Figure 6B). Non–ATP-generating respiration attributed to proton leak was reduced in the absence of ANT2 (Figure 6B), in agreement with the documented role of ANTs in mediating at least half of the basal proton conductance across the mitochondrial inner membrane (Brand et al., 2005). Because of this drop in proton leak in the context of normal ATP production, the coupling efficiency, or the ratio of ATP turnover to basal respiration, was higher in ant2TALEN (Figure 6B).
FIGURE 6:
ant2TALEN cells have altered bioenergetics parameters, reduced respiration, and increased sensitivity to ANT inhibitor. (A) OCR (pmol O2/min ± SEM; n ≥ 40) were measured using a Seahorse XF96e FluxAnalyzer with the Mito Stress Test kit under indicated conditions. (B) Basal and maximal OCR were obtained under glucose stimulation or after FCCP treatment to uncouple mitochondria. ATP production is expressed as basal OCR subtracted by postoligomycin oxygen consumption. Proton-leak OCR details oxygen consumed by the mitochondrion after oligomycin treatment. Degree of coupling (percentage) and spare respiratory capacity (as percentage of wt) obtained under glucose stimulation or after FCCP treatment to uncouple mitochondria. Data represent mean ± SEM. n ≥ 79. (C) Basal and maximal OCRs, ATP production, proton-leak OCR, degree of coupling, and spare respiratory capacity of wt and ant2TALEN cells after overnight incubation with the indicated concentrations of BKA. Data represent mean ± SEM. n ≥ 79.
Clearly, the degree of mitochondrial impairment was smaller than expected. We decided to determine whether the other ANT isoforms, whose mRNA (Figure 4G) and protein (Figure 4B) levels are very low compared with ANT2, were supporting residual ADP/ATP exchange in the ant2TALEN cells. Consistent with this notion, ant2TALEN cells were more sensitive to bongkrekic acid (BKA), a well-characterized pan-ANT inhibitor (Henderson and Lardy, 1970), than wt cells (Figure 6C). We conclude that the remaining BKA-sensitive ADP/ATP exchange detected in ant2TALEN mitochondria reflects the normal contribution supplied by the minor isoforms (ANT1 and ANT3) in the 293 cell model.
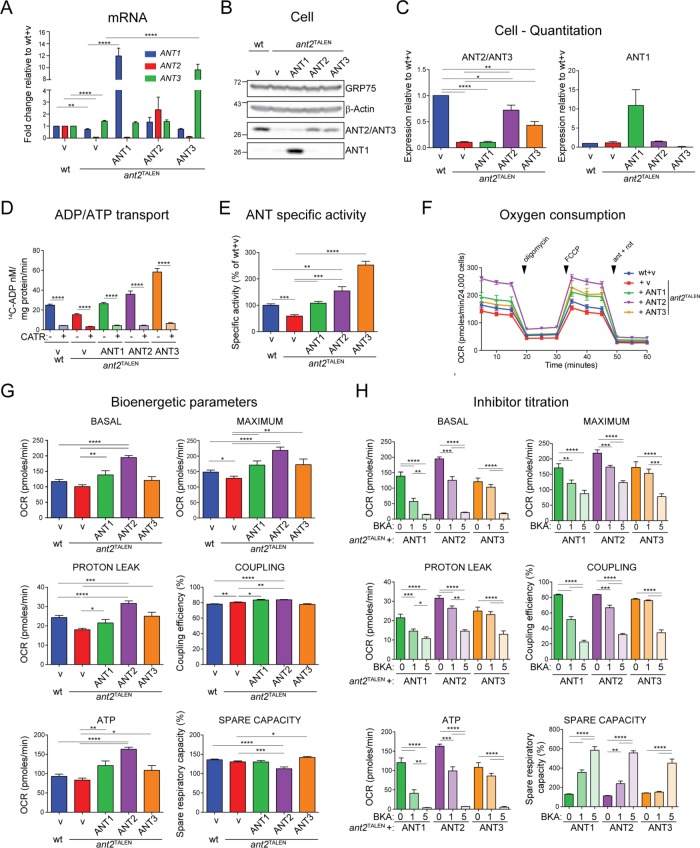
Overexpressed ANT alleles are functional and can restore bioenergetic defects
We determined the ability of ANT2, as well as of ANT1 or ANT3, to rescue the mild bioenergetic defects of the ant2TALEN cells. Overexpression of each ANT isoform in ant2TALEN cells resulted in increased mRNA transcripts (Figure 7A) and protein amounts (Figures 7, B and C, and 8, A and B) for the relevant isoform compared with empty vector (v)–transfected cells. ANT overexpression increased ADP/ATP exchange (Figure 7, D and E) and rescued the impaired maximal respiration of the ant2TALEN cells (Figure 7G). Moreover, overexpression of any of the ANT isoforms increased basal respiration, ATP production, and proton leak relative to ant2TALEN. ANT overexpression slightly increased or maintained coupling efficiency compared with ant2TALEN. As expected, ANT overexpression decreased the sensitivity of the ant2TALEN cells to BKA treatment (Figure 7H). It is interesting to note that although the ANT2 rescue only restored protein abundance to slightly less than wt levels (Figure 7B), this cell line showed the highest values for the three parameters just mentioned (Figure 7G). This suggests that ANT2 overexpression causes the 293 cells to work at or near maximal respiration, as reflected by the significantly lower spare capacity (Figure 7G). Of interest, the ANT2 reintroduced into ant2TALEN cells, which was cloned from HeLa cells, has a naturally occurring Leu111Arg polymorphism (Dorner et al., 2006). Thus our results suggest that the Leu111Arg ANT2 allele may be better able to support OXPHOS than the Leu111 variant that is normally expressed by 293 Flp-In cells (unpublished data).
FIGURE 7:
Overexpression of ANTs in ant2TALEN cells rescues ADP/ATP exchange and improves bioenergetic efficiency. (A) Relative mRNA level of ANTs in ANT overexpressors and the relevant wt and ant2TALEN vector controls. Data were analyzed by the ΔΔCT method, represented as mean fold change (2-ΔΔCT) ± SEM (n ≥ 3) relative to ANT expression in wt transfected with empty vector, which was set to 1. (B) Whole-cell extracts (50 μg) from wt, ant2TALEN, and ant2TALEN clones overexpressing various ANT isoforms immunoblotted with ANT1- and ANT2/ANT3-specific antibody. GRP75 and β-actin served as loading controls. (C) Densitometric analyses of ANT2/ANT3 (left) or ANT1 (right) expression in cell extracts from B. Expression relative to wt ± SEM (n ≥ 7). (D) Mitochondria (100 μg) from wt, ant2TALEN, and ant2TALEN clones overexpressing various ANT isoforms were fused with liposomes internally preloaded with 5 mM unlabeled ADP. Uptake of [14C]ADP (nM/mg protein/min ± SEM; n = 6) in the presence and absence of 60 μM CATR. (E) ANT-specific activity was calculated from D by subtracting CATR-sensitive exchange from ADP/ATP transport in the absence of inhibitor and expressed as a mean percentage of wt (100%) ± SEM (n ≥ 3). (F) OCR and (G) calculated basal, maximal, proton-leak OCR (pmol O2/min ± SEM; n ≥ 21), degree of coupling, spare respiratory capacity (as percentage of wt + v), and ATP production of ant2TALEN cells overexpressing ANT1, ANT2, or ANT3 (± SEM; n ≥ 21). Note that the values obtained from vector-transfected wt and ant2TALEN cells are the same as under 0 μM BKA treatment in Figure 6C. (H) Basal, maximal, and proton-leak OCR, as well as ATP production, degree of coupling, and spare respiratory capacity, obtained after overnight incubation with the indicated concentrations of BKA (μM).
Next the molecular basis for the modestly reduced bioenergetics capacity of the ant2TALEN cells was investigated using the ANT rescue cell lines as additional controls. Overexpression of any ANT isoform in ant2TALEN cells did not significantly affect the levels (Figure 8, A and B) or assembly (Figure 8C) of respiratory components or of select marker proteins of the OMM, the IMM, and the mitochondrial matrix (Figure 8, A and C). The individual activities of complexes I, III, and IV (Figure 8, D–F), measured after solubilization with dodecylmaltoside (DDM), which dissociates RSCs, or complex I, IV, and V, determined after BN-PAGE with digitonin solubilization (Figure 8G), were constant regardless of the absence or presence of overexpressed ANTs. Thus, while the absence of ANT2 does not compromise the steady-state amount, assembly, or function of the respiratory complexes, it does impair integrated OXPHOS function in the context of living cells with intact membranes.
Reversible destabilization of the ANT–RSC interaction
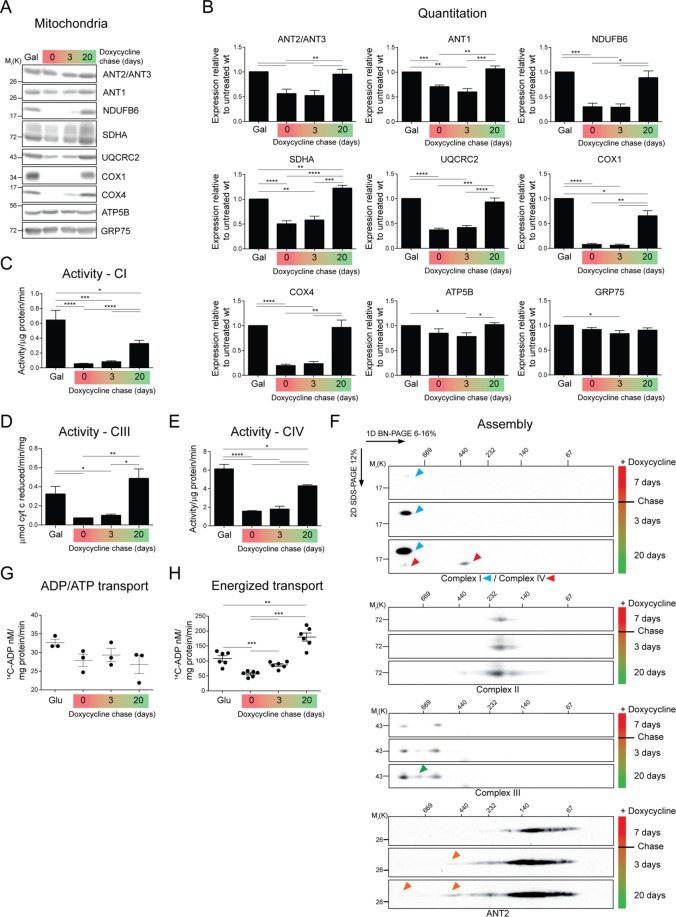
The antibiotic doxycycline (Dox) reversibly inhibits translation of mitochondrial DNA (mtDNA)–encoded proteins, most of which make up the highly hydrophobic core of respiratory complexes I, III, IV, and V (Moreno-Lastres et al., 2012). Therefore wt cells were treated with Dox for 7 d and then chased with medium lacking Dox for 3 and 20 d. After Dox treatment, COX1, a mtDNA-encoded subunit of complex IV, was undetectable by immunoblot (Figure 9, A and B). In addition, nuclear-encoded respiratory subunits such as NDUFB6, COX4, and UQCRC2, whose stability is compromised in the absence of mtDNA-encoded core respiratory complex subunits (Nijtmans et al., 1995b), were also lacking or reduced after translation inhibition (Figure 9, A and B). Consistently, complex I, III, and IV activity mirrored the steady-state protein levels of individual subunits and was reversibly inhibited by Dox (Figure 9, C–E). ATP5B, which is present in unassembled pools or intermediate subassemblies in the absence of mtDNA-encoded ATP6 and ATP8 (Nijtmans et al., 1995a), and GRP75, a matrix-localized heat shock protein, were unresponsive to Dox. Surprisingly, the amount of SDHA, a subunit of complex II that is entirely nuclear encoded, was reversibly affected by Dox. The basis for this observation is not clear. Of interest, the expression of endogenous ANTs (ANT2/3 and ANT1) was reduced after Dox treatment and fully restored after 20 d of chase.
FIGURE 9:
Reduced energized ANT transport in the absence of OXPHOS assembly and function. (A) Mitochondria (50 μg) from wt cells treated with 15 μg/ml Dox for 7 d or from cells chased with fresh medium for 3 or 20 d were immunoblotted as listed. (B) Densitometry analyses of protein steady-state levels from A. Expression relative to untreated wt (galactose) ± SEM (n ≥ 6). (C) NADH dehydrogenase (activity/μg/min ± SEM; n ≥ 4), (D) cytochrome c reductase (μmol cytochrome c reduced/min/mg ± SEM; n = 4), and (E) cytochrome c oxidase (activity/μg/min ± SEM; n ≥ 4) activity of DDM-solubilized mitochondrial extracts. (F) Mitochondria (200 μg) from wt cells treated with 15 μg/ml Dox for 7 d or from cells chased with fresh medium for 3 or 20 d were solubilized with 1.25% (wt/vol) digitonin, resolved by 2D BN/SDS–PAGE, and immunoblotted for ANTs and the indicated respiratory complexes. (G) Mitochondria (100 μg) from wt cells untreated and grown in glucose (Glu), treated with 15 μg/ml Dox for 7 d or from cells chased with fresh medium for 3 or 20 d were fused with liposomes internally preloaded with 5 mM of unlabeled ADP. Uptake of [14C]ADP (nM/mg protein/min ± SEM; n = 3). (H) The uptake of [14C]ADP (nM/mg protein/min ± SEM; n = 6) into energized mitochondria (500 μg) was determined in the presence of 5 mM glutamate and malate.
Respiratory subunits encoded by the mtDNA are required for the assembly of respiratory complexes I, III, and IV and the RSCs in which they participate. Accordingly, RSCs containing complexes I and IV were not detected in Dox-treated cells (Figure 9F). Upon chase in Dox-free medium, the assembly of complexes I and IV into larger complexes was initiated, albeit at different rates. As RSCs began to re-form during the chase, ANT2, which migrated below 140 kDa after Dox treatment, was progressively integrated into larger assemblies that comigrated with RSCs. Consistent with a prior study (Moreno-Lastres et al., 2012), complex III, which contains one subunit encoded by the mtDNA, was still detected after Dox treatment. Still, upon chase, the abundance of complex III–containing RSCs increased, and a third, intermediate-sized supercomplex was increasingly detected. As expected, assembly of complex II (SDHA) was not affected by Dox. These results indicate that association of ANT2 into high–molecular weight complexes is sensitive to the assembly of RSCs that contain subunits encoded by the mtDNA.
ANT transport is enhanced upon interaction with respiratory supercomplexes
To ascertain whether ANT activity is affected by RSC assembly, we monitored ANT transport in mitochondria isolated after treatment with Dox or after 3- and 20-d chase minus Dox. The intrinsic capacity of ANT2 to mediate ADP/ATP exchange was not affected by the absence or presence of RSCs (Figure 9G). However, when glutamate and malate were supplied to drive substrate-dependent ADP/ATP exchange, [14C]ADP uptake was suppressed when mtDNA translation was inhibited (Figure 9H). These results suggest that whereas the intrinsic ability of ANT to transport adenine nucleotides is unaffected upon Dox treatment, its transport capacity is enhanced in the presence of assembled and functional RSCs.
DISCUSSION
The possibility of ANTs interacting with heterologous protein complexes is not a new proposition. ANTs have been described to complex with ATP synthasomes, consisting of the ATP synthase, the phosphate carrier, and ANT1 (Ko et al., 2003; Chen et al., 2004). Others have provided evidence in support of ANT’s cooperation with creatine kinase to form microcompartments for phosphocreatine exchange (Dolder et al., 2001). However, the existence of these interactions and whether all ANT isoforms partake in the same types of associations were far from resolved until our analyses. By demonstrating that human ANT1 and ANT2 interact with other MCF members, including additional ANT isoforms, and multiple subunits of complexes I–IV, as well as with the ATP synthasome, our results significantly bridge this knowledge gap. The architecture of RSCs in mammals and yeast is notably different, in that the latter lacks complex I. In spite of this, mammalian ANT1 and ANT2 copurified many mitochondrial- and nuclear-encoded complex I subunits (Figure 3, B, C, and E, Supplemental Figure S1, A and B, and Supplemental Tables S1 and S3). These results strongly suggest that ANT’s interaction with RSCs and/or individual respiratory complexes is of such functional significance that the association was maintained even after the loss of complex I, as is the case in yeast, or alternatively regained during the evolution of higher eukaryotes. Given the sheer abundance of ANTs in mitochondria, it is tempting to speculate that there exist multiple ANT-containing mitochondrial complexes that may or may not form in a dynamic manner (Faustin et al., 2004), be regulated by metabolic states, and perhaps provide a local source of ADP/ATP exchange capable of driving important metabolic processes, including, for instance, biogenesis of mitochondrial proteins and OXPHOS.
To determine the functional benefits of ANT’s evolutionarily convergent association with RSCs, we ablated the major adenine translocase isoform expressed in 293 cells, ANT2. Of interest, the decrease in ANT-specific transport in the ant2TALEN cells is less than expected based on the levels of ANT2 relative to the other isoforms. Further, the ant2TALEN cells were more sensitive to ANT inhibitors, indicating that with respect to OXPHOS, ANT1 and/or ANT3 may be minor in abundance but are not functionally insignificant. These combined results strongly suggest that ANT isoforms differ in their inherent transport properties, which affect their ability to support OXPHOS. Differences in the transport properties of ANT isoforms have been reported (De Marcos Lousa et al., 2002) but inconsistently (Hamazaki et al., 2011); a limitation with these studies is that they were performed in yeast reconstituted either with wt ANT isoforms that were poorly expressed (De Marcos Lousa et al., 2002) or chimeric molecules that contained the N-terminus of the major yeast ANT isoform (Hamazaki et al., 2011). Thus our results provide compelling evidence that different human ANT isoforms have distinct transport activities as endogenously expressed in a mammalian model.
Because ANTs are essential in supplying ADP to the ATP synthase, we were surprised and initially disappointed that elimination of ANT2 resulted in only a modest bioenergetics phenotype. Whereas uncoupler-stimulated maximal oxygen consumption rate was significantly decreased in ant2TALEN cells compared with wt, the basal oxygen consumption rate, ATP production, and spare capacity were unchanged, suggesting that under normal conditions, 293 cells respire just fine without ANT2. The significant drop in proton leak in ant2TALEN cells is consistent with reports implicating ANTs in basal proton leak (Brand et al., 2005) and inducible proton leak (Samartsev et al., 1997). Recently ANT2 and ANT1 were included in a set of mitochondrial carriers that could participate in N-acyl amino acid–associated uncoupled respiration (Long et al., 2016). In yeast that lack cardiolipin, CATR inhibits proton leak that occurs across the mitochondrial inner membrane, implicating Aac2p in this process (Baile et al., 2014). In 293 cells, the elimination of ANT-derived ADP/ATP transport using BKA reduces proton leak by only 60%; the remaining ∼40% can be attributed to ANT-independent futile proton cycling (Figure 6C), like that mediated by uncoupler proteins (Brand et al., 2005).
The functionality of ANT’s interaction with respiratory components was also reciprocally tested by measuring ANT transport when the expression of mtDNA-encoded respiratory subunits was reversibly inhibited with Dox. When mtDNA translation is inhibited by Dox treatment, the intrinsic activity of the translocases was unchanged. In contrast, under energized conditions, ADP/ATP exchange was decreased. It is likely that lack of respiratory complexes results in a weaker proton gradient that is energetically unfavorable for electrogenic ADP/ATP exchange. Whether the association of ANTs with RSCs contributes directly to the activity of ANTs, whose transport is stimulated by a robust membrane potential (Kramer and Klingenberg, 1980), is an intriguing possibility that we are actively investigating.
There are numerous ramifications of the surprisingly mild OXPHOS phenotype of our ant2TALEN cells. First, it implies that ANTs are present in huge excess over what is actually needed to support OXPHOS, at least in 293 cells. This simple fact raises the possibility that ANTs participate in non–OXPHOS-related processes that may or may not be isoform specific. Second, although accounting for >90% of total ANT levels, ANT2 is responsible for only ∼50% of ANT-specific transport in 293 cells. Thus, as already discussed, ANT1 and/or ANT3 would appear to have very different transport activities than ANT2. Third, an intriguing potential explanation for the apparent huge excess in available ANT is that a sizeable fraction is maintained in an inactive state, perhaps due to a specific posttranslational modification. There is precedent for this. Human ANTs are phosphorylated at various sites (Feng et al., 2008; Cesaro and Salvi, 2010; Zhao et al., 2011). For example, in ANT1, two of the three highly conserved tyrosine residues (Y190, Y194) modeled to guide ADP from the intermembrane space into the cavity of the translocator are phosphorylated; expressing ANT1 with phospho-null mutations in these residues in a yeast model resulted in defective OXPHOS and ATP transport (Feng et al., 2008). Fourth, it raises the possibility that other ATP-transporting solute carriers are an important source of ATP flux in 293 cells. Potential candidates include calcium-binding mitochondrial carriers (SLC25A12 and 13), which are present in the 293 proteome (unpublished data). Indeed, Sal1p, a calcium-dependent ATP-Mg/Pi carrier in yeast, has been shown to compensate for adenine nucleotide transport in the absence of Aac2p (Laco et al., 2010). Three isoforms of the ATP-Mg/Pi carrier exist in humans and thus could provide an additional means of regulating the adenine nucleotide pools in and out of the mitochondrial matrix (Fiermonte et al., 2004). Undoubtedly, understanding the role and relative contribution of the four ANT isoforms in maintaining energy homeostasis, in isolation and in tandem, will be important in unraveling the basis for the phenotypic mosaicism in ANT1-associated diseases (Kaukonen et al., 2000; Palmieri et al., 2005; Echaniz-Laguna et al., 2012).
Outside of their assumed, crucial role in OXPHOS, very little is known about the biological functions of the different ANT isoforms. ANT isoforms are categorized by their tissue-specific transcriptional expression; however, in fact, multiple ANT isoforms are often coexpressed, albeit at different levels. Whether the reported pattern of mRNA tissue expression is reflected at the protein level has not been systematically documented due to the absence of quality isoform-specific antibodies. Ablation of ANT2 in numerous models has led to varying phenotypes without causing a complete loss of respiratory function (Graham et al., 1997; Kokoszka et al., 2004; Le Bras et al., 2006; Cho et al., 2015; Prabhu et al., 2015; Gavalda-Navarro et al., 2016; Kokoszka et al., 2016); however, compensation or redundancy in transport function by other isoforms was not rigorously investigated. Our ant2TALEN cells grew in galactose, did not require uridine and/or pyruvate supplementation, and retained measurable mitochondrial OXPHOS function. Thus, even though ANT3 and ANT1 are expressed at very low levels relative to ANT2 (Figure 4, A, C, E, and F), they are sufficiently active to support fairly robust OXPHOS. Perhaps the presence of other isoforms, such as the ubiquitous ANT3, may supply a baseline level of functionality that is by and large sufficient for most ATP-dependent processes upon loss of the main isoform, as in the case of our ant2TALEN system. In fact, it is well documented that deletion of certain ANT isoforms, even in clinically relevant cells and/or tissues (Graham et al., 1997; Esposito et al., 1999; Yin et al., 2005), has disparate but nonlethal outcomes.
ANT1 has been overexpressed in myotubes (Kawamata et al., 2011), rat cardiomyocytes (Klumpe et al., 2016), and mouse muscles (Flierl et al., 2005), and ANT2 in a hepatic cell line (Kim et al., 2012). It is important to consider that the overexpressed isoform is already present in huge excess relative to the other ANTs in these models. Transient overexpression of ANT1 or ANT3, but not ANT2 and ANT4, has been described to induce apoptosis in cultured cells (Bauer et al., 1999; Zamora et al., 2004a, b). In contrast, we generated multiple stable cell lines that overexpress ANT1, ANT2, or ANT3 in either the wt or ant2TALEN background. We speculate that the different results induced upon ANT overexpression reflect the mode of transfection, for example, transient versus stable.
Overexpression of ANTs in ant2TALEN can increase both OCR and adenine nucleotide exchange, consistent with what was observed in other cell models (Kim et al., 2012). The degree by which each ANT augmented OXPHOS did not strictly correlate with either its abundance or enhanced transport activity. For example, although ANT2 and ANT3 were similarly overexpressed, ANT3 allowed more ANT-specific transport, whereas ANT2 provided significantly increased basal and maximal respiration. Whether these observations reflect intrinsic differences between these isoforms, which are 91% identical, is unclear. Of interest, the reintroduced ANT2, which was derived from HeLa cells, contains a naturally occurring polymorphism that encodes arginine at position 111 instead of leucine (Dorner et al., 2006). Occurring in the general population with a 24% frequency, a prior study concluded that Leu-111 allele was functionally silent; for example, it worked as well as the Arg-111 version of ANT2 (Dorner et al., 2006). In contrast, our results provide evidence that with respect to their ability to support OXPHOS, the Arg-111 ANT2 variant is better than the Leu-111 version. Although a direct comparison of these alleles is needed to substantiate this conclusion, it is notable that every other ANT isoform contains arginine instead of leucine at this position.
Solute carriers (SLCs), which encompass all mitochondrial carriers including ANTs and a whole slew of other facilitative transporters and secondary active transporters, play a significant role in controlling and maintaining physiological functions such as nutrient and vitamin uptake, ion exchange, cofactor transport, and waste disposal, among others. Of note, SLCs exhibit robust coexpression and clustering patterns across tissue types that are suggestive of high interdependence between the carriers to maintain the integrative metabolic networks necessary for homeostasis (Cesar-Razquin et al., 2015). The fact that ANT1 and ANT2 copurified with numerous mitochondrial carriers, such as the phosphate carrier (SLC25A3), numerous Ca2+-dependent carriers (aspartate/glutamate SLC25A13; Mg-ADP/Mg-ATP SLC25A24; Mg-ATP/Pi SLC25A25), the tricarboxylate carrier (SLC25A1), and the carnitine/acylcarnitine carrier SLC25A20, among others (Supplemental Tables S2 and S3), points not only to coopted protein expression but also interactions and likely function. Thus it is plausible that the dysregulation of ANT expression and transport in various cell and animal models or in patients disrupt not only energetic flux between mitochondria and the cytosol due to engagement of ANTs with RSCs but also influence the activities of other SLCs, thereby altering the global metabolic landscape. Systematic dissection of SLC localization, expression, function, and interactomes, as we have done for human ANTs in 293 cells, will be critical for understanding how SLCs work in concert to enable, regulate, and compartmentalize the multitude of fundamental biological processes that are separated by membranes.
MATERIALS AND METHODS
Molecular biology
Human ANT1 and ANT2 were amplified by PCR using cDNA from HeLa cells and subcloned first into pBSK (Stratagene) and then into pcDNA5/FRT (Invitrogen). Tagged-ANT1 and ANT2 amplicons were generated via primer extension containing an N-terminal CNAP tag (amino acid sequence Met-DYKDDDDK-GGAGG-EDQVDPRLIDGK; FLAG epitope tag underlined and Protein C tag in bold) using untagged ANT plasmids as template and then subcloned into pcDNA5/FRT. Human ANT3 was first amplified by PCR using genomic DNA from yeast expressing yNhANT3 (Hamazaki et al., 2011) and then replacing the first 10 amino acids with the correct human ANT3 N-terminal nucleotides by PCR, followed by subcloning into pcDNA5/FRT. The fidelity of every construct was verified by DNA sequencing. TALEN binding pairs were designed to recognize sequences in ANT2 (SLC25A5) exon 1 that are separated by 16 nucleotides and encompass the start site (Life Technologies). The two constructs, which are fused to the truncated Fok1 nuclease, were subcloned into pcDNA3.1 and pEF6/V5-HisA (Life Technologies), generating pcDNA3ANT2-R and pEF6AANT2-L, respectively.
Cell culture
HEK293 Flp-In cells (wt; Invitrogen) were grown in DMEM containing 10% fetal bovine serum (Life Technologies), 2 mM l-glutamine (Life Technologies), and 100 μg/ml Zeocin (Invitrogen) at 37°C and 5% CO2. To establish an ant2-null cell model, wt cells grown in medium lacking antibiotic but supplemented with 50 μg/ml uridine were transfected with a 1:1 ratio of pCDNA3ANT2-R and pEF6AANT2-L using FuGENE 6 (Promega) and selected with 0.5 mg/ml G418 and 5 μg/ml blasticidinS (Cellgro) for 1 wk, and single colonies were isolated by ring cloning. Individual clones were maintained in the same medium as used for the parental 293 Flp-In cells, and the inclusion of uridine was found to not be essential. Whole-cell extracts derived from candidate clones were harvested and analyzed by SDS–PAGE and immunoblot. Stable wt or ant2TALEN rescue cell lines overexpressing ANTs were generated by cotransfecting wt or ant2TALEN clone 1 cell lines with pOG44 (expressing the Flp-recombinase) and the relevant pcDNA5/FRT plasmid (vector, ANT1, ANT2, CNAP-tagged ANT1/ANT2, or ANT3) at a ratio of 9:1 using FuGENE 6. Transfected cells were selected using 200 μg/ml hygromycin B (Invitrogen), and individual clones were recovered by ring cloning, expanded, and screened by immunoblot. An equal number of four to six clones per construct per ant2TALEN host were combined to establish a pooled clonal population that was used in all subsequent analyses. Mycoplasma contamination was routinely monitored and not detected.
To inhibit mitochondrial DNA translation, 293 Flp-In cells were allowed to expand to confluency in the presence of 15 μg/ml Dox (MP Biomedicals) for 7 d. Mitochondria were isolated from cells at the end of the Dox treatment or at 3 and 20 d after chasing the cells with fresh medium without Dox.
Purification of recombinant human ANT1
The entire open reading frame of human ANT1 was cloned into the pET28a vector (Novagen) downstream of the hexahistidine tag and induced at 37°C for 4 h with 0.5 mM isopropyl-β-d-thiogalactoside in C41 (DE3) Escherichia coli. The cells were pelleted at 3030 × g and washed once with 0.9% (wt/vol) NaCl. To extract proteins, the pellet was resuspended with 40 ml of extraction buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole) with 1 mg/ml lysozyme, incubated with rocking for 30 min at 4°C, and then passed through an Avestin Homogenizer four times at 40 psi. The lysate was centrifuged at 10,000 × g at 4°C for 20 min to remove lipids and debris and enrich inclusion bodies. Inclusion bodies were solubilized with inclusion body solubilization buffer (0.1 mM EDTA, 10 mM dithiothreitol [DTT], 10 mM Tris-Cl, pH7.4, 0.05% [wt/vol] PEG 3350) with 1.67% (wt/vol) Sarkosyl by vigorous vortexing and clarified by centrifugation at 12,000 × g for 10 min at 4°C. To precipitate proteins from the supernatant (containing solubilized inclusion bodies), (NH4)2SO4 was added to a final concentration 20% (wt/vol) and the solution allowed to rotate at room temperature for 1 h. After centrifugation at 10,000 × g for 10 min, the pellet was resuspended in 2 ml of inclusion body solubilization buffer with 0.5% (wt/vol) Sarkosyl. Before affinity purification of hANT1 using 1.5 ml of packed Ni2+–nitriloacetic acid resin (Qiagen), the resuspension was further diluted to 10 ml with extraction buffer such that the remaining Sarkosyl was present at 0.1% (wt/vol). After binding of proteins to resin, the column was washed once with wash buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 20 mM imidazole, 10% [vol/vol] glycerol, 0.1% [wt/vol] Sarkosyl), followed by a high-salt wash (50 mM NaH2PO4, pH 7.0, 600 mM NaCl, 20 mM imidazole, 10% [vol/vol] glycerol, 0.1% [wt/vol] Sarkosyl) before elution of bound hANT1 with elution buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 250 mM imidazole, 10% [vol/vol] glycerol, 0.1% [wt/vol] Sarkosyl).
Antibodies
The ANT1 mAb (GenScript) and rAb (Pacific Immunology) were generated using recombinant human ANT1 as antigen. Antibodies were against the following proteins: GRP75 (75-127; Antibodies Inc.); β-actin (A5441; Sigma-Aldrich); TOM20 (sc-11415; Santa Cruz Biotechnology); TIM23 (611332; BD); NDUFB6 (ab110244), UQCRFS1 (ab14746), UQCRC2 (ab14745), COX1 (ab14705), and COX4 (ab16056; Abcam); SDHA (4592000; Invitrogen); and COX2 (A6404; Thermo Fisher). ATP5B was a kind gift from Peter Pedersen (Johns Hopkins School of Medicine), and the ANT2/3 (Panneels et al., 2003) antibody was raised against the yeast ADP/ATP carrier, Aac2p, but conveniently cross-reacts with human and murine ANT2 and ANT3. Goat anti-rabbit or mouse secondary antibodies conjugated to horseradish peroxidase were also used (31460 and 62-6520; Pierce).
Two-dimensional BN SDS–PAGE
Mitochondria were solubilized for 30 min on ice in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–KOH, 10% (vol/vol) glycerol, 50 mM NaCl, 1 mM EDTA, and 2.5 mM MgCl2, pH 7.4, supplemented with 1% (wt/vol) digitonin (293 mitochondria in Figure 2A and mouse tissue mitochondria in Figure 2, B and C) or 1.25% (wt/vol) digitonin (293 mitochondria in Figure 2, B and C; Biosynth International) and protease inhibitors. Extracts were clarified by centrifugation for 30 min at 21,000 × g at 4°C and analyzed by 2D BN/SDS–PAGE exactly as described (Claypool et al., 2008).
Epitope mapping
A peptide scan consisting of 15-mer peptides with an 11–amino acid overlap between adjacent peptides that spans the entire 298–amino acid human ANT1 protein was generated by JPT Peptide Technologies. The membrane was hydrated with methanol before immunoblotting, and the epitopes recognized by both the hANT1 mAb and rabbit antisera were determined by densitometry analysis of the immunoreactive PepSpots.
Immunoblotting
SDS–PAGE and immunoblotting were performed as previously described (Lu et al., 2016). Images were captured with a Fluorchem Q (Cell Biosciences) quantitative digital imaging system and quantitation performed using the indicated software. Images were processed in Adobe Photoshop, with only linear adjustments in contrast and brightness, and assembled in Adobe Illustrator.
SILAC
Isotopic 13C6-lysine-HCl (0.8 mM) and 13C6-arginine-HCl (0.4 mM; Cambridge Isotopes) or unlabeled lysine (0.8 mM) and arginine (0.4 mM) were exogenously added to DMEM formulated without lysine, arginine, and sodium pyruvate (Invitrogen). CNAP-tagged ANTs were cultured in “heavy,” labeled medium, and untagged ANTs were correspondingly cultured in “light,” unlabeled medium for several passages to ensure isotopic lysine and arginine incorporation. Two days before mitochondrial isolation (Lu et al., 2016), cells were switched to medium lacking glucose but containing 10 mM galactose along with the appropriate amino acid supplementation. Note that in the experiment in which CNAP-ANT1 was compared with CNAP-ANT2 (Figure 3E), the latter was grown in heavy medium. Equivalent amounts of freshly prepared mitochondrial extracts (∼10 mg) isolated from cells grown in heavy or light medium were solubilized with lysis buffer containing 1.25% (wt/vol) digitonin (20 mM HEPES-KOH, pH 7.4, 100 mM NaCl, 20 mM imidazole, 1 mM CaCl2, and 10% [vol/vol] glycerol spiked with protease inhibitors), and clarified supernatants were diluted to 10 ml with protease inhibitor–spiked equilibration buffer (20 mM HEPES-KOH, pH 7.4, 100 mM NaCl, 1 mM CaCl2), loaded onto columns containing 1 ml of packed anti–Protein C (Roche) affinity resin, and rotated overnight at 4°C. The flowthrough was collected for analysis, and the bound material was washed twice with 10 bed volumes (10 ml) of ice-cold equilibration buffer with 0.1% (wt/vol) digitonin. To elute bound material, the resin was incubated with 1 ml of warmed EDTA elution buffer (20 mM HEPES-KOH, pH 7.4, 100 mM NaCl, 5 mM EDTA, 0.1% [wt/vol] digitonin) and allowed to rotate for 30 min at room temperature. After three additional elutions, the eluates (4 ml/each) from both the light and heavy mitochondrial IPs were mixed and concentrated using an Amicon Ultra-4 10,000 MWCO filter device (Millipore) and trichloroacetic acid (TCA) precipitated (20% [vol/vol] TCA with 0.07% [wt/vol] deoxycholate) for 1 h on ice. TCA pellets were then resuspended in a 1:1 mix of 2× reducing sample buffer and 0.1 M NaOH, boiled for 5 min, and resolved by 8–16% SDS–PAGE. In the case of two consecutive nondenaturing IPs (high stringency), bound material on anti–Protein C resin was incubated with 10 ml of warmed EDTA elution buffer for 30 min at room temperature and the eluate allowed to drain directly into a separate column containing 0.5 ml of packed anti-FLAG (GenScript) affinity resin. After the addition of protease inhibitors, the FLAG IP was allowed to proceed for 1 h at 4°C with rotation. The flowthrough was collected for analysis, and the resin was washed as described with anti–Protein C resin, except that the bound material was eluted thrice with 0.5 ml of FLAG elution buffer (100 µg/ml FLAG peptide, 0.1% [wt/vol] digitonin) and then again with two bed volumes of the FLAG elution buffer with rotation for 30 min at room temperature. Equal volumes of the eluate from the light and heavy immunoprecipitates were mixed and concentrated as described.
In-gel trypsin digestion and C18 desalting
Silver staining was carried out to visualize eluted proteins in the gel. Briefly, the gel was sensitized with 0.02% (wt/vol) sodium thiosulfate for 2 min, incubated in 0.1% (wt/vol) silver nitrate at 4°C for 40 min, developed in 0.04% (vol/vol) formaldehyde/Formalin and 2% (wt/vol) sodium carbonate, and finally quenched in 1% (vol/vol) acetic acid. The stained gel was divided into 11–18 fractions, depending on silver staining banding patterns. The subsequent destaining was carried out in 15 mM potassium ferricyanide and 50 mM sodium thiosulfate and then conditioned in 40 mM ammonium bicarbonate. For reduction, divided gel cubes were incubated in 5 mM DTT at 60°C for 20 min, followed by alkylation using 20 mM iodoacetamide at room temperature in the dark for 10 min. Gel cubes were dehydrated in 100% acetonitrile at room temperature for 10 min, cooled on ice for 10 min, and rehydrated in 0.001% trypsin on ice for 1 h and 37°C for 16 h. To extract tryptic peptides, gel cubes were incubated in 3% (vol/vol) acetonitrile, 0.1% (wt/vol) trifluoroacetic acid at room temperature for 10 min twice, 50% (vol/vol) acetonitrile, 0.1% (wt/vol) trifluoroacetic acid at room temperature for 10 min, and 100% acetonitrile at room temperature for 10 min. Extracted peptides underwent C18 stage tip desalting, and were vacuum dried and then analyzed in LC-MS/MS.
LC-MS/MS analysis
Purified peptides from gel digests were analyzed on an Orbitrap-Elite mass spectrometer interfaced with Easy nanoLC II liquid chromatography system. The peptides were loaded on a desalting column packed in-house (2 cm, 5-µm particle size, and 200-Å pore size; Magic C18 material; Michrom Biosciences) at a flow rate of 5 µl/min and resolved on an analytical column packed in-house (25 cm, 3-µm particle size, and 200-Å pore size; Magic C18 material) at a flow rate on 300 nl/min. The eluted peptides were ionized by applying 2.1-kV voltage using a Flexion source operated at 200°C. The duration of LC gradient was 45 min, and the total run time to acquire the data was 60 min. The full MS scans were acquired at 120,000 resolution at 400 m/z and measured using an Orbitrap mass analyzer. The MS/MS spectra were acquired at 30,000 resolution at 400 m/z and measured using an Orbitrap mass analyzer. The top 10 peptide precursors were selected in a data-dependent manner and subjected to 32% normalized higher-energy collisional dissociation. The AGC targets were 1 × 106 and 5 × 104 and maximum ion filling times were 120 and 300 ms for MS and MS/MS, respectively. Polysilaxolane ion m/z 445.1200024 was used for internal calibration.
Mass spectrometry data analysis
The raw data corresponding to each SILAC experiment were searched together using MaxQuant (version 1.5.3.8) and deposited into the PRIDE database through ProteomeXchange (accession number 1-20160616-130067). The raw data were processed and searched against the Human Refseq 73 protein database by including known contaminants using the built-in Andromeda search engine of MaxQuant. The following search parameters were used for the database searches. Strict trypsin specificity to K or R of C-terminal of a peptide by allowing two missed cleavages. The minimum peptide length was set as seven amino acids. Carbamidomethylation of cysteine (57.021464 Da) was set as fixed modification. Oxidation of methionine (42.010565 Da) and protein N-terminal acetylation (15.994915 Da) were selected as variable modifications. Quantification of SILAC pairs was performed by enabling multiplicity of 2, and to ensure reliable quantification, the ratio count was set as a minimum of 2. For the absolute quantification purpose, the iBAQ option was enabled. Peptide precursor mass tolerance was set as 20 ppm for first-pass search and 4.5 ppm for main search. Fragment mass tolerance was set as 20 ppm. The Andromeda search algorithm uses a target-decoy approach for the identification of peptides and proteins, and identifications were filtered by applying 1% false discovery rate. The protein-group output files from MaxQuant are given in Supplemental Table S3.
Preparation of mitochondria
Cells were seeded onto four or more 150 mm × 25 mm tissue-culture dishes and allowed to expand to confluency. Two days before mitochondrial isolation, the cells were switched from dextrose- to galactose-based medium (DMEM without glucose, Life Technologies; supplemented with 10 mM galactose, Sigma-Aldrich). Mitochondria were isolated as described in Lu et al. (2016) and, if not used immediately, aliquoted, snap frozen with liquid N2, and stored at −80°C for downstream analyses.
Relative abundance of ANT isoforms by proteomics
Three independent 293 Flp-In mitochondrial preparations were used. Proteins were reduced by adding 10 mM DTT, incubated at 56°C for 20 min, alkylated with 20 mM iodoacetamide, and incubated in the dark at room temperature for 30 min, followed by acetone precipitation. Proteins were resolubilized in 6 M urea buffer (6 M urea, 50 mM triethylammonium bicarbonate) and processed for trypsin digestion. Trypsin was added at 1:50 enzyme-to-substrate ratio and incubated at 37°C for 16 h. The tryptic peptides were cleaned using Sep-Pak cartridges, and the eluted peptides were vacuum dried and stored at −80°C until the LC-MS/MS analysis. A 1-μg amount from each of the replicates was injected twice on an Orbitrap Fusion Lumos tribrid mass spectrometer coupled to Easy nanoLC II Liquid Chromatography System 1200. A linear gradient of 5–30% acetonitrile in 0.1% (vol/vol) formic acid was used for 85 min and total run time of 120 min. Both MS and MS/MS were measured using an Orbitrap mass analyzer in top speed mode. The raw data were analyzed using the MaxQuant software suite as described. The LFQ and iBAQ algorithms were enabled for precursor ion quantification. Perseus was used for determining the relative abundance of ANTs in 293 cells.
mRNA
Total RNA was isolated from wt and ant2TALEN- and ant2TALEN-overexpressing ANT alleles using the PureLink RNA Mini Kit with DNase treatment (Invitrogen). A 25-ng amount of RNA was analyzed in 20-μl reactions using the EXPRESS One-Step SYBR GreenER quantitative real-time PCR (qRT-PCR) Kit (Life Technologies) with QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher) according to the manufacturer’s instructions. Each reaction, including nontemplate controls, was performed at least in duplicate with two different biological replicates and contained 50 nM ROX and 200 nM each of forward and reverse gene-specific primers designed with Primer3 (Untergasser et al., 2012). The reaction conditions were as follows: 5 min at 50°C and 2 min at 95°C, followed by 40 two-temperature cycles (15 s at 95°C and 1 min at 60°C) and melt curve profiling. Expression of the ANT isoforms was analyzed by the comparative CT (ΔΔCT) method (XTest/XGAPDH = 2-ΔΔCT, where GAPDH signifies glyceraldehyde 3-phosphate dehydrogenase) with GAPDH as an endogenous reference gene. Values were represented as fold change (2ΔΔCT) ± SEM relative to ANT2 in wt cells (Figure 4, A and F) or fold change (2ΔΔCT) ± SEM relative to ANT expression in the appropriate wt setting for Figures 4G and 7A (Prism 6; GraphPad).
Genotyping ant2TALEN cells
Genomic DNA was extracted using the Gentra Puregene Cell Kit (Qiagen), and the genomic region surrounding the target site (exon1 encompassing the ATG) was PCR amplified, digested with XbaI/HindIII (NEB), and ligated into pBSK(–). Transformants were analyzed by Sanger sequencing. Because 293 cells are hypotriploid, sequences of at least 10 individual clones for each of the four ant2TALEN cell lines were obtained, revealing disruptions in and around ANT2’s start site.
Whole-cell extraction
Proteins were extracted from confluent six-well tissue culture dishes using RIPA lysis buffer (1% [vol/vol] Triton X-100, 20 mM HEPES-KOH, pH 7.4, 50 mM NaCl, 1 mM EDTA, 2.5 mM MgCl2, 0.1% [wt/vol] SDS) spiked with 1 mM phenylmethylsulfonyl fluoride and quantified using the bicinchoninic acid assay (Pierce) as described in Lu et al. (2016).
Endogenous IP
To coimmunoprecipitate endogenous ANT2, 5 µl of normal rabbit serum (NRS) or 8 µl of antiserum against COX4 (ab16056; Abcam) was bound to 20 µl of Protein A–conjugated magnetic beads (Bio-Rad) by incubating the sera and magnetic beads at room temperature for 1 h with rotation in a final volume of 300 µl of IP buffer (20 mM HEPES-KOH, pH 7.4, 100 mM NaCl, 1 mM EDTA, 2.5 mM CaCl2). We solubilized 150 µg of mitochondrial extracts with 1.25% (wt/vol) digitonin in the IP buffer base spiked with protease inhibitors and precleared the clarified supernatants (diluted to 500 µl with 0.01% [wt/vol] digitonin in IP buffer) with NRS-bound magnetic beads for 1 h with rotation at 4°C. After preclear, the supernatant was transferred into tubes containing COX4-bound magnetic beads. The volume was again adjusted to 500 µl with 0.01% (wt/vol) digitonin in IP buffer and spiked with protease inhibitors, and the IP was allowed to proceed for 4 h with rotation at 4°C. The beads were washed thrice with 500 µl of 0.01% (wt/vol) digitonin in IP buffer, and bound material was eluted by the addition of 40 µl of a 1:1 mix of 2× reducing sample buffer and 0.1 M NaOH, boiled for 5 min, and resolved by 12% SDS–PAGE.
ADP/ATP transport
Adenine nucleotide influx was monitored at 20°C as described previously (Ruprecht et al., 2014), with slight modifications. Briefly, mitochondrial matrix was uniformly labeled with adenine nucleotides by fusion with 5 mM cold ADP-loaded liposomes. Liposome-fused mitochondria were extruded seven times through a 1-µm membrane filter (Avanti), centrifuged at 100,000 × g for 30 min at 4°C, and resuspended in 50 mM KPi buffer, pH 8.0. To measure the [14C]ADP uptake, 100 µl of fused membranes (0.1 mg of proteins) was added to 300 µl of KPi buffer containing 1.34 µM [14C]ADP (0.02 µCi/µl; PerkinElmer) at 20°C. The reaction was stopped after 20 min by the addition of 2 ml of cold KPi buffer, and the entire reaction volume was applied onto a 0.2-µm cellulose nitrate filter under vacuum. Filtered membranes were washed with 4 ml of KPi buffer, and the radioactivity retained on the membrane filters was quantified using a liquid scintillation counter. OXPHOS-coupled ADP uptake was performed as previously described (Aprille and Austin, 1981). Briefly, 0.5 mg of isolated mitochondria resuspended in assay buffer (0.6 M mannitol, 5 mM malate, 5 mM glutamate, 5 mM MgCl2, 2 mM KPi, 10 mM KCl) was mixed with [14C]ADP (1.34 µM in 300 µl of assay buffer), and the uptake was measured as described.
Oxygen consumption measurements
The OCR was measured on adherent cells using a Seahorse XF96e analyzer and the Seahorse XF Cell Mito Stress Kit (Seahorse Bioscience, North Billerica, MA). Forty-eight hours before assays, 293 Flp-In cells were seeded at 24,000 cells/well in XF96 V3 PS cell culture microplates coated with 0.001% (wt/vol) poly-d-lysine (Sigma) to improve cell adherence. Twenty-four hours before assays, cells were treated overnight with 0, 1, or 5 µM BKA. Before XF measurements, cells were washed twice using the XF base medium supplemented with glucose, sodium pyruvate, and l-glutamine and preincubated with the appropriate BKA concentration for 1 h in a 37°C, humidified, CO2-free incubator.
The Mito Stress Test was performed in accordance with the manufacturer’s instructions. Briefly, OCR measurements were made approximately every 9 min under basal conditions, after the addition of oligomycin (2 µM), after the addition of FCCP (250 nM), and after the addition of rotenone and antimycin A (1 mM). OCR values were collapsed from at least six independent experiments and represent the average measurement of a minimum of 20 (and up to 79) wells for each strain and BKA condition.
Complex activity
The activity of complex I (NADH dehydrogenase) or complex IV (cytochrome c oxidase) was measured by using Abcam’s Complex I (ab109721) or Complex IV (ab109909) Human Enzyme Activity Microplate Assay Kit, respectively, according to manufacturer’s instructions (Figures 8, D and F, and 9, C and E). Briefly, frozen mitochondrial extracts were solubilized using the provided detergent at a final concentration of 5 mg/ml spiked with protease inhibitors. The clarified extracts (50 µg for complex I, 10 µg for complex IV) were added to the supplied buffer and then loaded onto microplate wells precoated with complex I– or complex IV–specific capture antibody. Complex activity was then monitored by following the oxidation of NADH to NAD+ by complex I at 450 nm or the oxidation of cytochrome c by complex IV at 550 nm by the immobilized target.
Isolated complex III activities were measured as previously described (Tzagoloff et al., 1975; Figures 8E and 9D). Briefly, 10–30 µg of mitochondria solubilized in 0.5% (wt/vol) n-dodecyl β-d-maltoside (Anatrace) and spiked with protease inhibitors was added to reaction buffer (50 mM KPi, 2 mM EDTA, pH 7.4) with 0.008% (wt/vol) equine heart cytochrome c (Sigma-Aldrich) and 1 mM KCN. The reaction was initiated with the addition of 100 µM decylubiquinol, and the reduction of cytochrome c was followed at 550 nm.
Data analysis
Band densitometry analyses were performed using Image J (Schneider et al., 2012). All comparisons (ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001) were analyzed by unpaired two-tailed t test (or paired for Figure 4B) assuming Gaussian distributions and the same SDs in Prism 6. All graphs report the mean ± SEM.
Supplementary Material
Acknowledgments
We are grateful to Ya-Lin Lu (Washington University) for help in designing ANT isoform-specific primers used in qRT-PCR, Kim-Vy Nguyen (formerly of Andrew Ewald’s lab) for providing all of the mouse tissues used for experiments in this article, Naohiro Terada (University of Florida) for the yNhANT3 yeast strain, and Pete Pederson (Johns Hopkins University School of Medicine) for ATP5B antisera. This work was supported by National Institutes of Health Grant R01HL108882 to S.M.C. and predoctoral fellowships from the American Heart Association (12PRE11910004 to Y.-W.L. and 16PRE31140006 to M.G.A.).
Abbreviations used:
- AAC
ADP/ATP carrier
- ANT
adenine nucleotide translocase
- BKA
bongkrekic acid
- CATR
carboxyatractyloside
- CL
cardiolipin
- CNAP
consecutive nondenaturing affinity purification
- Dox
doxycycline
- IMM
inner mitochondrial membrane
- IP
immunoprecipitation
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- mAb
monoclonal antibody
- MCF
mitochondrial carrier family
- OCR
oxygen consumption rate
- OMM
outer mitochondrial membrane
- OXPHOS
oxidative phosphorylation
- RSC
respiratory supercomplex
- SILAC
stable isotope labeling by amino acids in cell culture
- wt
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-03-0195) on April 12, 2017.
REFERENCES
- Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Aprille JR, Austin J. Regulation of the mitochondrial adenine nucleotide pool size. Arch Biochem Biophys. 1981;212:689–699. doi: 10.1016/0003-9861(81)90413-6. [DOI] [PubMed] [Google Scholar]
- Baile MG, Sathappa M, Lu YW, Pryce E, Whited K, McCaffery JM, Han X, Alder NN, Claypool SM. Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J Biol Chem. 2014;289:1768–1778. doi: 10.1074/jbc.M113.525733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MK, Schubert A, Rocks O, Grimm S. Adenine nucleotide translocase-1, a component of the permeability transition pore, can dominantly induce apoptosis. J Cell Biol. 1999;147:1493–1502. doi: 10.1083/jcb.147.7.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD, Pakay JL, Ocloo A, Kokoszka J, Wallace DC, Brookes PS, Cornwall EJ. The basal proton conductance of mitochondria depends on adenine nucleotide translocase content. Biochem J. 2005;392:353–362. doi: 10.1042/BJ20050890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandolin G, Meyer C, Defaye G, Vignais PM, Vignais PV. Partial purification of an atractyloside-binding protein from mitochondria. FEBS Lett. 1974;46:149–153. doi: 10.1016/0014-5793(74)80356-x. [DOI] [PubMed] [Google Scholar]
- Cesaro L, Salvi M. Mitochondrial tyrosine phosphoproteome: new insights from an up-to-date analysis. BioFactors. 2010;36:437–450. doi: 10.1002/biof.123. [DOI] [PubMed] [Google Scholar]
- Cesar-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, Bai X, Reithmeier RA, Hepworth D, Hediger MA, Edwards AM, et al. A call for systematic research on solute carriers. Cell. 2015;162:478–487. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Chen C, Ko Y, Delannoy M, Ludtke SJ, Chiu W, Pedersen PL. Mitochondrial ATP synthasome: three-dimensional structure by electron microscopy of the ATP synthase in complex formation with carriers for Pi and ADP/ATP. J Biol Chem. 2004;279:31761–31768. doi: 10.1074/jbc.M401353200. [DOI] [PubMed] [Google Scholar]
- Cho J, Seo J, Lim CH, Yang L, Shiratsuchi T, Lee MH, Chowdhury RR, Kasahara H, Kim JS, Oh SP, et al. Mitochondrial ATP transporter Ant2 depletion impairs erythropoiesis and B lymphopoiesis. Cell Death Differ. 2015;22:1437–1450. doi: 10.1038/cdd.2014.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- De Marcos Lousa C, Trezeguet V, Dianoux AC, Brandolin G, Lauquin GJ. The human mitochondrial ADP/ATP carriers: kinetic properties and biogenesis of wild-type and mutant proteins in the yeast S. cerevisiae. Biochemistry. 2002;41:14412–14420. doi: 10.1021/bi0261490. [DOI] [PubMed] [Google Scholar]
- Dienhart MK, Stuart RA. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol Biol Cell. 2008;19:3934–3943. doi: 10.1091/mbc.E08-04-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner A, Pauschinger M, Badorff A, Noutsias M, Giessen S, Schulze K, Bilger J, Rauch U, Schultheiss HP. Tissue-specific transcription pattern of the adenine nucleotide translocase isoforms in humans. FEBS Lett. 1997;414:258–262. doi: 10.1016/s0014-5793(97)01000-4. [DOI] [PubMed] [Google Scholar]
- Dolce V, Scarcia P, Iacopetta D, Palmieri F. A fourth ADP/ATP carrier isoform in man: identification, bacterial expression, functional characterization and tissue distribution. FEBS Lett. 2005;579:633–637. doi: 10.1016/j.febslet.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Dolder M, Wendt S, Wallimann T. Mitochondrial creatine kinase in contact sites: interaction with porin and adenine nucleotide translocase, role in permeability transition and sensitivity to oxidative damage. Biol Signals Recept. 2001;10:93–111. doi: 10.1159/000046878. [DOI] [PubMed] [Google Scholar]
- Dorner A, Giessen S, Gaub R, Grosse Siestrup H, Schwimmbeck PL, Hetzer R, Poller W, Schultheiss HP. An isoform shift in the cardiac adenine nucleotide translocase expression alters the kinetic properties of the carrier in dilated cardiomyopathy. Eur J Heart Fail. 2006;8:81–89. doi: 10.1016/j.ejheart.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Echaniz-Laguna A, Chassagne M, Ceresuela J, Rouvet I, Padet S, Acquaviva C, Nataf S, Vinzio S, Bozon D, Mousson de Camaret B. Complete loss of expression of the ANT1 gene causing cardiomyopathy and myopathy. J Med Genet. 2012;49:146–150. doi: 10.1136/jmedgenet-2011-100504. [DOI] [PubMed] [Google Scholar]
- Esposito LA, Melov S, Panov A, Cottrell BA, Wallace DC. Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci USA. 1999;96:4820–4825. doi: 10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustin B, Rossignol R, Rocher C, Benard G, Malgat M, Letellier T. Mobilization of adenine nucleotide translocators as molecular bases of the biochemical threshold effect observed in mitochondrial diseases. J Biol Chem. 2004;279:20411–20421. doi: 10.1074/jbc.M314259200. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhu M, Schaub MC, Gehrig P, Roschitzki B, Lucchinetti E, Zaugg M. Phosphoproteome analysis of isoflurane-protected heart mitochondria: phosphorylation of adenine nucleotide translocator-1 on Tyr194 regulates mitochondrial function. Cardiovasc Res. 2008;80:20–29. doi: 10.1093/cvr/cvn161. [DOI] [PubMed] [Google Scholar]
- Fiermonte G, De Leonardis F, Todisco S, Palmieri L, Lasorsa FM, Palmieri F. Identification of the mitochondrial ATP-Mg/Pi transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem. 2004;279:30722–30730. doi: 10.1074/jbc.M400445200. [DOI] [PubMed] [Google Scholar]
- Flierl A, Chen Y, Coskun PE, Samulski RJ, Wallace DC. Adeno-associated virus-mediated gene transfer of the heart/muscle adenine nucleotide translocator (ANT) in mouse. Gene Ther. 2005;12:570–578. doi: 10.1038/sj.gt.3302443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Palmieri L, Scarcia P, Lodi T, Donnini C, Limongelli A, Tiranti V, Zeviani M, Ferrero I, Viola AM. Mutations in AAC2, equivalent to human adPEO-associated ANT1 mutations, lead to defective oxidative phosphorylation in Saccharomyces cerevisiae and affect mitochondrial DNA stability. Hum Mol Genet. 2004;13:923–934. doi: 10.1093/hmg/ddh108. [DOI] [PubMed] [Google Scholar]
- Gavalda-Navarro A, Mampel T, Vinas O. Changes in the expression of the human adenine nucleotide translocase isoforms condition cellular metabolic/proliferative status. Open Biol. 2016;6:150108. doi: 10.1098/rsob.150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BH, Waymire KG, Cottrell B, Trounce IA, MacGregor GR, Wallace DC. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- Gu J, Wu M, Guo R, Yan K, Lei J, Gao N, Yang M. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- Hamazaki T, Leung WY, Cain BD, Ostrov DA, Thorsness PE, Terada N. Functional expression of human adenine nucleotide translocase 4 in Saccharomyces cerevisiae. PLoS One. 2011;6:e19250. doi: 10.1371/journal.pone.0019250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- Henderson PJ, Lardy HA. Bongkrekic acid. An inhibitor of the adenine nucleotide translocase of mitochondria. J Biol Chem. 1970;245:1319–1326. [PubMed] [Google Scholar]
- Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem. 2000;275:22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- Jordens EZ, Palmieri L, Huizing M, van den Heuvel LP, Sengers RC, Dorner A, Ruitenbeek W, Trijbels FJ, Valsson J, Sigfusson G, et al. Adenine nucleotide translocator 1 deficiency associated with Sengers syndrome. Ann Neurol. 2002;52:95–99. doi: 10.1002/ana.10214. [DOI] [PubMed] [Google Scholar]
- Kaukonen J, Juselius JK, Tiranti V, Kyttala A, Zeviani M, Comi GP, Keranen S, Peltonen L, Suomalainen A. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–785. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- Kawamata H, Tiranti V, Magrane J, Chinopoulos C, Manfredi G. adPEO mutations in ANT1 impair ADP-ATP translocation in muscle mitochondria. Hum Mol Genet. 2011;20:2964–2974. doi: 10.1093/hmg/ddr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Je JH, Son TG, Park HR, Ji ST, Pokharel YR, Jeon HM, Kang KW, Kang HS, Chang SC, et al. The hepatoprotective effects of adenine nucleotide translocator-2 against aging and oxidative stress. Free Radic Res. 2012;46:21–29. doi: 10.3109/10715762.2011.636042. [DOI] [PubMed] [Google Scholar]
- Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta. 2008;1778:1978–2021. doi: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Klingenberg M. Cardiolipin and mitochondrial carriers. Biochim Biophys Acta. 2009;1788:2048–2058. doi: 10.1016/j.bbamem.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Klumpe I, Savvatis K, Westermann D, Tschope C, Rauch U, Landmesser U, Schultheiss HP, Dorner A. Transgenic overexpression of adenine nucleotide translocase 1 protects ischemic hearts against oxidative stress. J Mol Med (Berl) 2016;94:645–653. doi: 10.1007/s00109-016-1413-4. [DOI] [PubMed] [Google Scholar]
- Ko YH, Delannoy M, Hullihen J, Chiu W, Pedersen PL. Mitochondrial ATP synthasome. Cristae-enriched membranes and a multiwell detergent screening assay yield dispersed single complexes containing the ATP synthase and carriers for Pi and ADP/ATP. J Biol Chem. 2003;278:12305–12309. doi: 10.1074/jbc.C200703200. [DOI] [PubMed] [Google Scholar]
- Kokoszka JE, Waymire KG, Flierl A, Sweeney KM, Angelin A, MacGregor GR, Wallace DC. Deficiency in the mouse mitochondrial adenine nucleotide translocator isoform 2 gene is associated with cardiac noncompaction. Biochim Biophys Acta. 2016;1857:1203–1212. doi: 10.1016/j.bbabio.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoszka JE, Waymire KG, Levy SE, Sligh JE, Cai J, Jones DP, MacGregor GR, Wallace DC. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–465. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki H, Fukazawa T, Houzen H, Yoshida K, Nonaka I, Goto Y. A novel D104G mutation in the adenine nucleotide translocator 1 gene in autosomal dominant progressive external ophthalmoplegia patients with mitochondrial DNA with multiple deletions. Ann Neurol. 2002;51:645–648. doi: 10.1002/ana.10172. [DOI] [PubMed] [Google Scholar]
- Kramer R, Klingenberg M. Modulation of the reconstituted adenine nucleotide exchange by membrane potential. Biochemistry. 1980;19:556–560. doi: 10.1021/bi00544a025. [DOI] [PubMed] [Google Scholar]
- Laco J, Zeman I, Pevala V, Polcic P, Kolarov J. Adenine nucleotide transport via Sal1 carrier compensates for the essential function of the mitochondrial ADP/ATP carrier. FEMS Yeast Res. 2010;10:290–296. doi: 10.1111/j.1567-1364.2010.00606.x. [DOI] [PubMed] [Google Scholar]
- Le Bras M, Borgne-Sanchez A, Touat Z, El Dein OS, Deniaud A, Maillier E, Lecellier G, Rebouillat D, Lemaire C, Kroemer G, et al. Chemosensitization by knockdown of adenine nucleotide translocase-2. Cancer Res. 2006;66:9143–9152. doi: 10.1158/0008-5472.CAN-05-4407. [DOI] [PubMed] [Google Scholar]
- Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- Lim CH, Brower JV, Resnick JL, Oh SP, Terada N. Adenine nucleotide translocase 4 is expressed within embryonic ovaries and dispensable during oogenesis. Reprod Sci. 2015;22:250–257. doi: 10.1177/1933719114542026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Svensson KJ, Bateman LA, Lin H, Kamenecka T, Lokurkar IA, Lou J, Rao RR, Chang MR, Jedrychowski MP, et al. The secreted enzyme PM20D1 regulates lipidated amino acid uncouplers of mitochondria. Cell. 2016;166:424–435. doi: 10.1016/j.cell.2016.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YW, Galbraith L, Herndon JD, Lu YL, Pras-Raves M, Vervaart M, Van Kampen A, Luyf A, Koehler CM, McCaffery JM, et al. Defining functional classes of Barth syndrome mutation in humans. Hum Mol Genet. 2016;25:1754–1770. doi: 10.1093/hmg/ddw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado EN, DeHart DN, Patnaik J, Klatt SC, Gooz MB, Lemasters JJ. ATP/ADP turnover and import of glycolytic ATP into mitochondria in cancer cells is independent of the adenine nucleotide translocator. J Biol Chem. 2016;291:19642–19650. doi: 10.1074/jbc.M116.734814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert CS, Rampelt H, Gebert M, Oeljeklaus S, Schrempp SG, Kochbeck L, Guiard B, Warscheid B, van der Laan M. The mitochondrial ADP/ATP carrier associates with the inner membrane presequence translocase in a stoichiometric manner. J Biol Chem. 2014;289:27352–27362. doi: 10.1074/jbc.M114.556498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Lastres D, Fontanesi F, Garcia-Consuegra I, Martin MA, Arenas J, Barrientos A, Ugalde C. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 2012;15:324–335. doi: 10.1016/j.cmet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdock DG, Boone BE, Esposito LA, Wallace DC. Up-regulation of nuclear and mitochondrial genes in the skeletal muscle of mice lacking the heart/muscle isoform of the adenine nucleotide translocator. J Biol Chem. 1999;274:14429–14433. doi: 10.1074/jbc.274.20.14429. [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Klement P, Houstek J, van den Bogert C. Assembly of mitochondrial ATP synthase in cultured human cells: implications for mitochondrial diseases. Biochim Biophys Acta. 1995a;1272:190–198. doi: 10.1016/0925-4439(95)00087-9. [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Spelbrink JN, Van Galen MJ, Zwaan M, Klement P, Van den Bogert C. Expression and fate of the nuclearly encoded subunits of cytochrome-c oxidase in cultured human cells depleted of mitochondrial gene products. Biochim Biophys Acta. 1995b;1265:117–126. doi: 10.1016/0167-4889(94)00203-q. [DOI] [PubMed] [Google Scholar]
- Nury H, Dahout-Gonzalez C, Trezeguet V, Lauquin G, Brandolin G, Pebay-Peyroula E. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 2005;579:6031–6036. doi: 10.1016/j.febslet.2005.09.061. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- Palmieri F. Diseases caused by defects of mitochondrial carriers: a review. Biochim Biophys Acta. 2008;1777:564–578. doi: 10.1016/j.bbabio.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Palmieri L, Alberio S, Pisano I, Lodi T, Meznaric-Petrusa M, Zidar J, Santoro A, Scarcia P, Fontanesi F, Lamantea E, et al. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum Mol Genet. 2005;14:3079–3088. doi: 10.1093/hmg/ddi341. [DOI] [PubMed] [Google Scholar]
- Panneels V, Schussler U, Costagliola S, Sinning I. Choline head groups stabilize the matrix loop regions of the ATP/ADP carrier ScAAC2. Biochem Biophys Res Commun. 2003;300:65–74. doi: 10.1016/s0006-291x(02)02795-x. [DOI] [PubMed] [Google Scholar]
- Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJ, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- Prabhu D, Goldstein AC, El-Khoury R, Rak M, Edmunds L, Rustin P, Vockley J, Schiff M. ANT2-defective fibroblasts exhibit normal mitochondrial bioenergetics. Mol Genet Metab Rep. 2015;3:43–46. doi: 10.1016/j.ymgmr.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodic N, Oka M, Hamazaki T, Murawski MR, Jorgensen M, Maatouk DM, Resnick JL, Li E, Terada N. DNA methylation is required for silencing of ant4, an adenine nucleotide translocase selectively expressed in mouse embryonic stem cells and germ cells. Stem Cells. 2005;23:1314–1323. doi: 10.1634/stemcells.2005-0119. [DOI] [PubMed] [Google Scholar]
- Ruprecht JJ, Hellawell AM, Harding M, Crichton PG, McCoy AJ, Kunji ER. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc Natl Acad Sci USA. 2014;111:E426–E434. doi: 10.1073/pnas.1320692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samartsev VN, Mokhova EN, Skulachev VP. The pH-dependent reciprocal changes in contributions of ADP/ATP antiporter and aspartate/glutamate antiporter to the fatty acid-induced uncoupling. FEBS Lett. 1997;412:179–182. doi: 10.1016/s0014-5793(97)00667-4. [DOI] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharer JD. The adenine nucleotide translocase type 1 (ANT1): a new factor in mitochondrial disease. IUBMB Life. 2005;57:607–614. doi: 10.1080/15216540500217735. [DOI] [PubMed] [Google Scholar]
- Stepien G, Torroni A, Chung AB, Hodge JA, Wallace DC. Differential expression of adenine nucleotide translocator isoforms in mammalian tissues and during muscle cell differentiation. J Biol Chem. 1992;267:14592–14597. [PubMed] [Google Scholar]
- Tzagoloff A, Akai A, Needleman RB. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem. 1975;250:8228–8235. [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewalle J, Bauters M, Van Esch H, Belet S, Verbeeck J, Fieremans N, Holvoet M, Vento J, Spreiz A, Kotzot D, et al. The mitochondrial solute carrier SLC25A5 at Xq24 is a novel candidate gene for non-syndromic intellectual disability. Hum Genet. 2013;132:1177–1185. doi: 10.1007/s00439-013-1322-3. [DOI] [PubMed] [Google Scholar]
- Yin H, Stahl JS, Andrade FH, McMullen CA, Webb-Wood S, Newman NJ, Biousse V, Wallace DC, Pardue MT. Eliminating the Ant1 isoform produces a mouse with CPEO pathology but normal ocular motility. Invest Ophthalmol Vis Sci. 2005;46:4555–4562. doi: 10.1167/iovs.05-0695. [DOI] [PubMed] [Google Scholar]
- Zamora M, Granell M, Mampel T, Vinas O. Adenine nucleotide translocase 3 (ANT3) overexpression induces apoptosis in cultured cells. FEBS Lett. 2004a;563:155–160. doi: 10.1016/S0014-5793(04)00293-5. [DOI] [PubMed] [Google Scholar]
- Zamora M, Merono C, Vinas O, Mampel T. Recruitment of NF-kappaB into mitochondria is involved in adenine nucleotide translocase 1 (ANT1)-induced apoptosis. J Biol Chem. 2004b;279:38415–38423. doi: 10.1074/jbc.M404928200. [DOI] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- Zhao X, Leon IR, Bak S, Mogensen M, Wrzesinski K, Hojlund K, Jensen ON. Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol Cell Proteomics. 2011;10:M110.000299. doi: 10.1074/mcp.M110.000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.