A force-calibrated microneedle setup and controlled biochemical perturbation reveal that chromatin acts as a spring-like mechanical module that controls the rigidity of cell nuclei. The underlying molecular mechanism involves linker DNA and internucleosomal interaction via histone tails.
Abstract
Cells, as well as the nuclei inside them, experience significant mechanical stress in diverse biological processes, including contraction, migration, and adhesion. The structural stability of nuclei must therefore be maintained in order to protect genome integrity. Despite extensive knowledge on nuclear architecture and components, however, the underlying physical and molecular mechanisms remain largely unknown. We address this by subjecting isolated human cell nuclei to microneedle-based quantitative micromanipulation with a series of biochemical perturbations of the chromatin. We find that the mechanical rigidity of nuclei depends on the continuity of the nucleosomal fiber and interactions between nucleosomes. Disrupting these chromatin features by varying cation concentration, acetylating histone tails, or digesting linker DNA results in loss of nuclear rigidity. In contrast, the levels of key chromatin assembly factors, including cohesin, condensin II, and CTCF, and a major nuclear envelope protein, lamin, are unaffected. Together with in situ evidence using living cells and a simple mechanical model, our findings reveal a chromatin-based regulation of the nuclear mechanical response and provide insight into the significance of local and global chromatin structures, such as those associated with interdigitated or melted nucleosomal fibers.
INTRODUCTION
The cell nucleus is a highly organized intracellular organelle that packages DNA and is the site for genomic functions such as transcription, replication, and repair/recombination in eukaryotes. Decades of studies have established that this micrometer-sized organelle is subjected to a range of mechanical forces while maintaining its structural and functional stability (Wang et al., 2009; Hampoelz and Lecuit, 2011; Zwerger et al., 2011; Davidson and Lammerding, 2014). This is particularly evident in migrating processes, in which a cell penetrates tissues by squeezing its body through spaces smaller than the size of the nucleus (Friedl et al., 2011). In skeletal and cardiac muscles, the nuclei are exposed to repetitive contractile forces generated by the cells (Davidson and Lammerding, 2014). Failure to resist such mechanical distortion results in substantial damage to nuclear structure and subsequently perturbs genomic integrity, which has been linked to tumorigenesis and apoptosis (Zink et al., 2004; Isermann and Lammerding, 2013; Davidson and Lammerding, 2014). Nuclear deformability has also been implicated in directing stem cells to commit to specific lineages (Engler et al., 2006; Buxboim et al., 2010). The nucleus may thus possess mechanisms that precisely define and tune its deformability to resist and respond to mechanical force.
A key to understand the mechanical properties of the nucleus is the nongenetic function of chromatin as a load-bearing molecule (Bustin and Misteli, 2016). Chromatin is a thread-like macromolecular complex, the primary structure of which comprises a linear polymer of DNA wrapped around octamers of core histones (Luger et al., 1997). This linear nucleosomal array—the so-called 10-nm fibers—folds into a compacted 30-nm chromatin fiber in vitro (Schalch et al., 2005; Robinson et al., 2006; Song et al., 2014). Force spectroscopy analyses demonstrate that this compacted fiber generates elastic restoring forces against mechanical stretch (Cui and Bustamante, 2000; Kruithof et al., 2009). Although the 30-nm fiber had long been assumed to be a relevant structure in vivo, recent studies suggest that within the nucleus, the 10-nm fibers are rather irregularly folded into higher-order structures such as those with interdigitated nucleosomal fibers that resemble a “polymer melt” (Maeshima et al., 2010, 2016; Fussner et al., 2012; Joti et al., 2012; Gan et al., 2013; Hsieh et al., 2015; Ricci et al., 2015; Sanborn et al., 2015; Chen et al., 2016) and undergo compacted domain–like organization (Markaki et al., 2010; Bian and Belmont, 2012; Smallwood and Ren, 2013; Dekker and Heard, 2015). Furthermore, individual chromosomes occupy discrete “territories” in the nucleus (Cremer and Cremer, 2001). It is largely unknown how chromatin bears load as such an entity.
Early biophysical measurements demonstrated that nuclei with a condensed form of chromatin generate substantial mechanical resistance when aspirated by a micropipette (Dahl et al., 2005; Pajerowski et al., 2007) or indented by an atomic force microscopy cantilever (Krause et al., 2013). The change in nuclear deformability was also detected by an optical tweezers–based fluctuation analysis (Mazumder et al., 2008). Genetic manipulation studies further revealed that changes in the chromatin’s compaction level and spatial localization impair the nucleus’ ability to withstand mechanical force (Furusawa et al., 2015; Schreiner et al., 2015). Therefore it is widely acknowledged that the rigidity of the nucleus depends on the organization properties of chromatin. Quantitative determination of chromatin’s mechanical contribution, together with comprehensive characterization of its biochemical properties, will provide detailed information on how local and global chromatin architecture contributes to regulating the nuclear mechanical response.
Here we combined a microneedle-based force measurement setup (Shimamoto et al., 2011) with controlled biochemical assays (Takata et al., 2013) to examine the mechanical response of human cell nuclei and their dependence on biochemical manipulations of chromatin. The use of force-calibrated thin microneedles (∼1-µm diameter) allowed for quantitatively determining the mechanical response of isolated nuclei to localized forces as well as for micromanipulating those nuclei in living cells. Our data suggest that linker DNA and internucleosomal interaction via histone tails play a predominant role in providing nuclear rigidity, thus highlighting the mechanical function of condensed chromatin in the nucleus.
RESULTS
Nuclei are highly rigid and elastic at millimolar levels of Mg2+
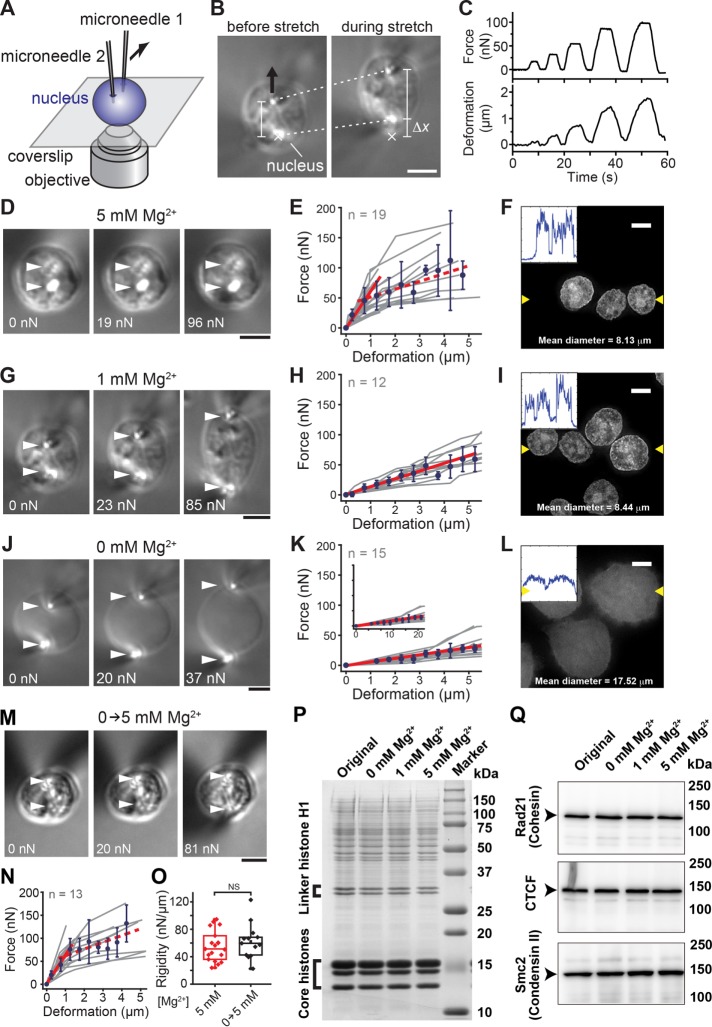
We examined the mechanical properties of nuclei using a microneedle-based force-measurement setup that we previously developed for characterizing the viscoelastic properties of vertebrate meiotic spindles (Shimamoto et al., 2011; Shimamoto and Kapoor, 2012; Figure 1A and Supplemental Movie S1). Briefly, single interphase nuclei isolated from HeLa cells (Lewis and Laemmli, 1982; Takata et al., 2013) were captured using a pair of glass microneedles and then stretched by moving one microneedle away from the other. This resulted in an extensional deformation in the nuclei (dotted lines, Figure 1B) and the development of a sustained resisting force, which was monitored based on the deflection of the stiffness-calibrated flexible microneedle tip (Δx; Figure 1B). By repeatedly applying mechanical stretches with different force magnitudes in controlled directions (Figure 1C), we measured nuclear mechanical response under various biochemical conditions.
FIGURE 1:
Mg2+-induced stiffening of nuclei associated with chromatin compaction. (A) Schematic of the setup for measuring the mechanical response of nuclei. (B) Bright-field images of a micromanipulated nucleus. The nucleus isolated from a HeLa cell line was deformed by moving the upper microneedle (black arrow) while monitoring the magnitude of applied force, which was estimated based on deflection of the force-calibrated microneedle tip (Δx) from its equilibrium (marked ×). Dotted lines show positions of the microneedle tips, which were used to estimate the extent of deformation. (C) Typical time recordings of the measurement showing cycles of stretch-hold-release with varying force magnitudes. (D, G, J) Images of isolated nuclei exposed to buffers with indicated Mg2+ levels and stretched at indicated force magnitudes. Arrowheads, microneedle tip positions. (E, H, K) Force–deformation plots obtained at Mg2+ levels of 5 mM (n = 19) (E), 1 mM (n = 12) (H), and 0 mM (n = 15) (K). Data from individual nuclei (gray lines) were pooled for 0.5-µm bins and averaged (bars represent SD). Slopes are 63.1, 13.5, and 5.5 for E, H, and K, respectively (red solid lines; R2 > 0.97). The red broken line shows the region of predominant viscous deformation (slope, 14.5). (F, I, L) Images of DAPI-stained nuclei. Insets, intensity line profiles generated across two yellow arrowheads in each image. (M–O) Reversibility of nuclear mechanical response. (M) Images showing an isolated nucleus, which was preincubated in 0 mM Mg2+ buffer and then exposed to 5 mM Mg2+ buffer. The nucleus was stretched using the microneedle-based setup at indicated force magnitudes. (N) Force–deformation plot. Data from those nuclei (gray lines, n = 15) were pooled for 0.5-µm bins and averaged (blue circles; bars represent SD). Slope, 60.2 (R2 = 0.99; red solid line). (O) Nuclear rigidity of untreated control nuclei (red) and nuclei once exposed to 0 mM Mg2+ buffer (black). NS, not significant by Student’s t test (p = 0.42). (P) CBB staining of overall nuclear components at different Mg2+ levels. (Q) Western blot of major interphase chromatin assembly factors. Scale bars, 5 µm.
We first examined the mechanical response of isolated nuclei in the presence of 5 mM Mg2+ (Figure 1, D and E), at which chromatin is assembled into highly compacted domains (Figure 1F and Supplemental Figure S1A; Maeshima et al., 2016). Mg2+ ions are abundant in nuclei (>1 mM; Strick et al., 2001). We observed that nuclei under a small stretching force (∼20 nN) underwent submicrometer-scale deformation while largely retaining their round shape (Figure 1D, middle). The deformation was restored as soon as the applied force was released, showing predominantly elastic response (Supplemental Figure S2A). On the other hand, the nuclei exhibited more noticeable deformation as a larger force was applied (∼100 nN; Figure 1D, right), and the deformation persisted after release of applied force, showing more viscous response (Supplemental Figure S2A). The force–deformation plot, which was obtained by applying several stretch–release cycles with different force magnitudes, revealed that nuclei developed a resisting force that increased in proportion to the magnitude of deformation before yielding at ∼50 nN (red solid line in Figure 1E). The rigidity of the nuclei, as determined by linear regression analysis within the elastic deformation range, was 55.4 ± 21.9 nN/µm (mean ± SD, n = 19). Therefore nuclei with condensed chromatin have significant elastic rigidity and resist a load on the order of tens of nanonewtons.
Nuclear rigidity decreases with Mg2+-dependent chromatin decompaction
Because chromatin is a complex made up of a negatively charged polymer and various associated proteins, its compaction state varies with the concentration of cations such as Mg2+ (Earnshaw and Laemmli, 1983; Widom, 1986; Hansen, 2002; Eltsov et al., 2008; Visvanathan et al., 2013; Maeshima et al., 2016). At 5 mM Mg2+, chromatin becomes highly condensed and forms compacted domains in vitro (Supplemental Figure S1A), whereas, in the absence of Mg2+, it is significantly stretched (Supplemental Figure S1C). At 0.8–1 mM Mg2+, chromatin is slightly decondensed and forms an intermediate structure between the stretched fibers and compacted domains (Supplemental Figure S1B).
To test the effect of changes in chromatin structure on the mechanical response of nuclei, we examined the dependence of the nuclear mechanical response on buffer Mg2+ concentrations. Fluorescence microscopy imaging revealed that the size of nuclei exposed to 1 mM Mg2+ increased by ∼4% relative to their size at 5 mM Mg2+ (Figure 1I; compare with Figure 1F), indicative of chromatin decondensation. We found that the mechanical response of nuclei also differed at 1 and at 5 mM Mg2+. Specifically, at the lower Mg2+ concentration, the magnitude of nuclear deformation was severalfold larger at a comparable level of loading (Figure 1G), and the nuclei were deformed in a predominantly elastic manner over an extended range of deformation (>5 µm; Figure 1H). The rigidity of the nuclei was greater-than-threefold lower at 1 than at 5 mM Mg2+ (14.3 ± 5.3 nN/µm, n = 12).
The observed change in nuclear rigidity was even more prominent when nuclei were exposed to 1 mM EDTA (i.e., at ∼0 mM Mg2+). Under this condition, the nuclei were more swollen (>200%) and optically less dense, and a much smaller force was required for their deformation (Figure 1J). Their structure was predominantly elastic and extremely soft over a broader range of deformations (Figure 1K and inset). Under this condition, the fluorescence signal of chromatin appeared diffuse and evenly spread over the entire nucleus (Figure 1L), indicating an extended fiber structure (Supplemental Figure S1C; Maeshima et al., 2016). The rigidity of nuclei with the decondensed chromatin was 5.5 ± 2.1 nN/µm (n = 15), which was >10-fold lower than the value at 5 mM Mg2+.
To verify whether the Mg2+-dependent change in nuclear rigidity was due to mechanical damage or dissociation of components, we examined the reversibility of nuclear rigidity upon change in the buffer composition. To this end, we exposed isolated nuclei preincubated in 1 mM EDTA and swollen by severalfold to 5 mM Mg2+ buffer and measured their mechanical response. These nuclei regained their morphology and force dependences to a level comparable to nuclei that were immersed in the 5 mM Mg2+ buffer throughout (Figure 1, M–O). Further analysis using SDS–PAGE revealed no obvious differences in the abundance of major nuclear components, including core and linker histones, over the tested range of Mg2+ concentrations (Figure 1P). A Western blot analysis also confirmed that the putative interphase DNA-cross-linking proteins cohesin (Nasmyth and Haering, 2005; Uhlmann, 2016), CTCF (Ghirlando and Felsenfeld, 2016), and condensin II (Hirano, 2012) remained intact (Figure 1Q). Together the results show that nuclei alter their mechanical rigidity upon Mg2+-dependent chromatin compaction with minimal modulations of the composition of nuclear components and chromatin assembly factors.
Digestion of linker DNA results in loss of nuclear rigidity
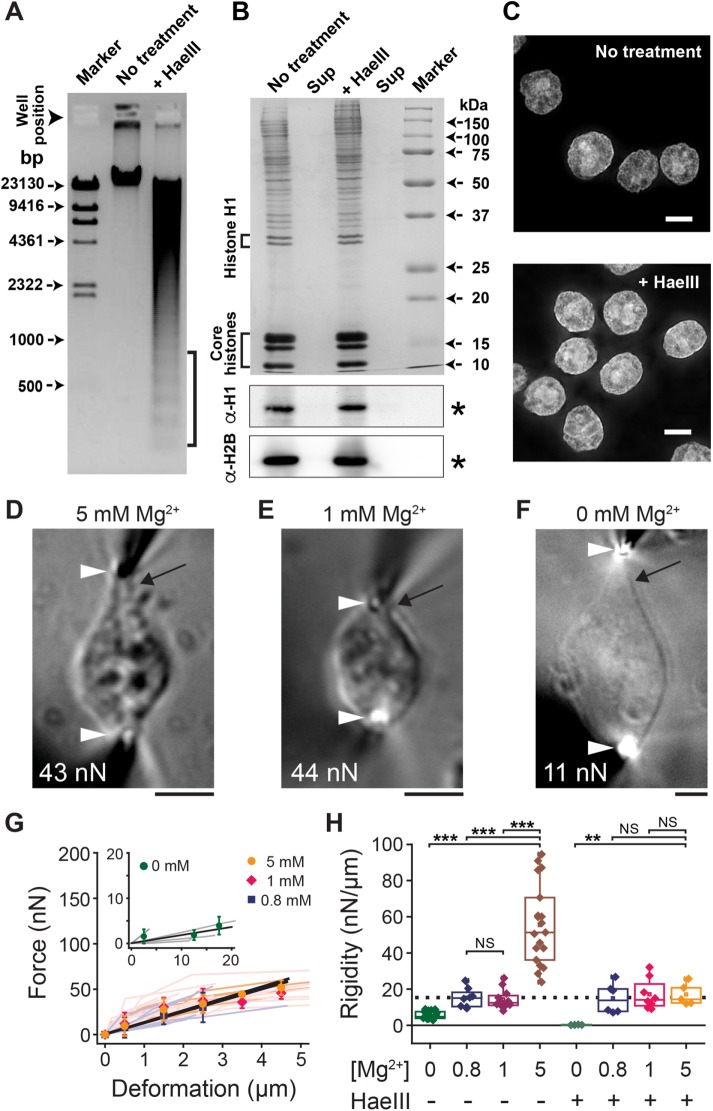
To determine quantitatively the contribution of chromatin to the nuclear mechanical response, we treated the isolated nuclei with a restriction endonuclease, HaeIII, and then immersed them in buffer solutions of various Mg2+ concentrations. Agarose gel electrophoresis of purified DNA from HaeIII-treated nuclei revealed a ladder pattern characteristic of linker DNA digestion (Figure 2A, brackets). In contrast, the protein composition of HaeIII-treated nuclei, including linker and core histones (Figure 2B), and overall chromatin distribution patterns (Figure 2C and Supplemental Figure S3, A and B) were similar to those of undigested samples.
FIGURE 2:
Linker DNA digestion results in loss of nuclear rigidity. (A) Agarose gel electrophoresis analysis showing significant DNA fragmentation after HaeIII treatment of nuclei (brackets). (B) CBB staining (top) and Western blotting with anti–histone H1 and H2B antibodies (bottom) of nuclear proteins in pellet and supernatant fractions of the nuclei without (left) or with HaeIII nuclease (right). (C) DAPI-stained nuclei for untreated control (reproduced from Figure 1F) and HaeIII-treated groups. (D–F) Bright-field images of HaeIII-treated nuclei exposed to buffers with indicated Mg2+ concentrations and stretched at indicated force magnitudes. Black arrows highlight a prominent extension from the nuclear main body. Other marks are as in Figure 1. (G) Force–deformation plots obtained at 5, 1, and 0.8 mM Mg2+ (n = 5, 8, and 6, respectively) and 0 mM Mg2+ (inset, n = 4). Data from individual nuclei (lines with light colors) were pooled for 0.5-µm bins and averaged (bars represent SD). Slopes are 12.6, 11.5, 12.5, and 0.41, respectively (black solid lines; R2 > 0.87). (H) Summary of nuclear rigidity measurements performed at different Mg2+ levels with or without HaeIII treatment. **p < 0.001, ***p < 0.0001 (Student’s t test); NS, not significant (p > 0.1). Broken line shows the level of the Mg2+-independent “baseline” fraction (see the text). Data for 5 mM Mg2+ control correspond to those in Figure 1O. Scale bars, 5 µm.
We found that at 5 mM Mg2+, HaeIII-treated nuclei were more deformable than untreated control samples, and their structure was predominantly elastic over an extended range of deformations (>4 µm; Figure 2, D and G). Further, in contrast to untreated samples, a structure near the nuclear periphery was stretched out of the optically dense main body, revealing an elastic membranous material tapered at both ends (black arrow in Figure 2D). The rigidity of these nuclei was greater-than-threefold lower than that of the untreated control (17.1 ± 6.0 nN/µm, n = 5; vs. >55 nN/µm). Therefore linker DNA significantly contributes to the mechanical rigidity of nuclei.
Of note, the rigidity of HaeIII-treated nuclei did not change further when the buffer Mg2+ concentration was reduced to 1 mM (17.1 ± 8.4 nN/µm, n = 8; Figure 2, E and G). The nuclei exhibited deformation morphology that was similar to that observed at 5 mM Mg2+ and generated a predominantly elastic restoring force (Figure 2E). Similar results were obtained at an even lower Mg2+ concentration of 0.8 mM (14.9 ± 8.3 nN/µm, n = 6; Figure 2G and Supplemental Figure S2, B–E). In other words, upon linker DNA digestion, nuclear rigidity became largely insensitive to changes in buffer Mg2+ concentration. Figure 2H summarizes the force measurements, indicating that there are two mechanical components within nuclei: 1) a Mg2+-sensitive fraction that is coupled with chromatin compaction (brown column in Figure 2H), and 2) a Mg2+-insensitive “baseline” that is independent of chromatin (dotted line in Figure 2H). When HaeIII-treated nuclei were exposed to 1 mM EDTA, their structure became highly deformable compared with their deformability under other conditions (0.3 ± 0.2 nN/µm, n = 4; Figure 2F–H), suggesting a substantial weakening of the overall nuclear structure.
Histone acetylation leads to loss of nuclear rigidity
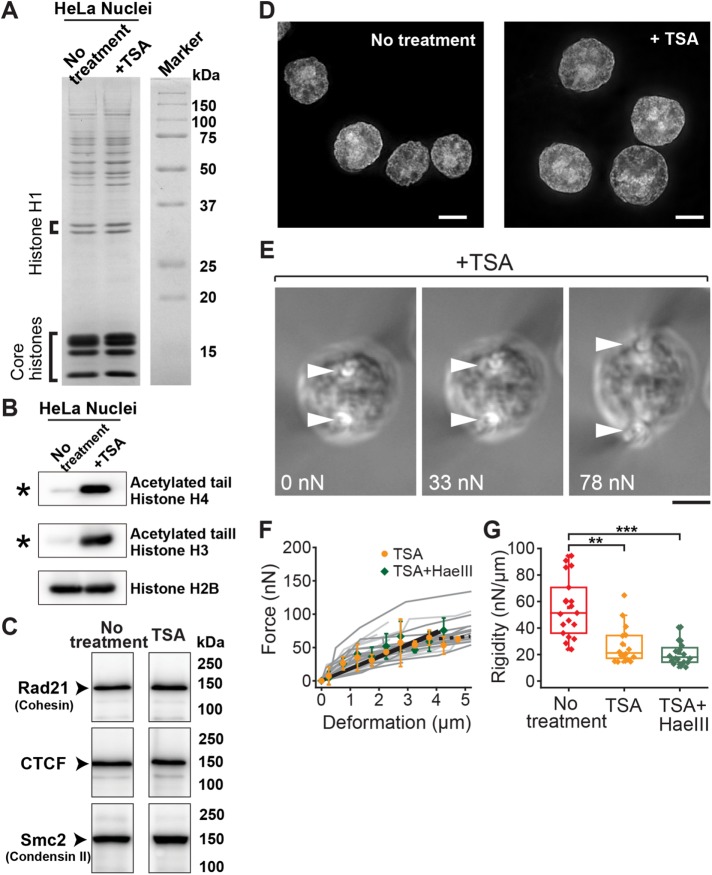
We next examined the role of another major factor regulating chromatin structure: nucleosome–nucleosome interactions mediated by histone tails (Bannister and Kouzarides, 2011; Kalashnikova et al., 2013; Funke et al., 2016). It was previously shown that treatment of cells with the histone deacetylase inhibitor trichostatin A (TSA; Yoshida et al., 1990) leads to decondensation of chromatin (Gorisch et al., 2005), presumably because the nucleosome–nucleosome interactions are weakened by blocking the binding of the N-terminal tails of histones, including H4, to the neighboring nucleosomes (Kalashnikova et al., 2013). To obtain histone-acetylated nuclei, we treated cells with TSA for 3 h (Gorisch et al., 2005) and isolated their nuclei using the same purification protocol as employed earlier. A biochemical analysis confirmed that the protein compositions, including linker and core histones, of nuclei isolated from TSA-treated cells were indistinguishable from those of untreated controls (Figure 3A and Supplemental Figure S4A). However, the TSA-treated nuclei had a marked increase of tail acetylation in histones, including H3 and H4 (Figure 3B, asterisks). Mass spectrometry data further confirmed the sites of tail acetylation (Supplemental Table S1). Western blot analysis revealed that the amounts of cohesin, CTCF, and condensin II in all samples were largely identical (Figure 3C and Supplemental Figure S4B). The chromatin of TSA-treated nuclei was slightly decondensed compared with that of untreated samples, as determined by nucleus size (Figure 3D).
FIGURE 3:
Histone tail acetylation leads to loss of nuclear rigidity. (A) CBB staining showing abundance of major nuclear components, including linker and core histones, for untreated control and TSA-treated cells. (B, C) Western blot analyses showing hyperacetylation of histone H3 and H4 tails in TSA-treated nuclei (B; asterisk) and abundance of key chromatin assembly factors (C; no treatment data correspond to the right-most lane in Figure 1Q; see Supplementary Figure S4B for full-size scan of blots). (D) Representative images of DAPI-staining in untreated control (left; reproduced from Figure 1F) and TSA-treated (right) nuclei. (E) Bright-field images of a TSA-treated nucleus exposed to 5 mM Mg2+ buffer and stretched at indicated force magnitudes. Marks are as in Figure 1. (F) Force–deformation plots obtained for nuclei treated with TSA (orange circles, n = 16) and those treated with TSA and HaeIII (green diamonds, n = 20). Individual measurements (dark and light gray lines, respectively) were pooled for 0.5-µm bins and averaged (bars represent SD). Slopes are 18.7 and 19.0, respectively (black solid lines; R2 > 0.92). The broken line shows the region of predominant viscous deformation. (G) Summary of nuclear rigidity measurements of untreated control (red; corresponding to data in Figure 1O), TSA-treated (orange), and TSA/HaeIII-treated (green) nuclei. **p < 0.001, ***p < 0.0001 (Student’s t test). Scale bars, 5 µm.
We measured the rigidity of histone-acetylated nuclei using the microneedle-based setup and buffer composition for which chromatin was highly compacted and provided significant rigidity to nuclei (i.e., 5 mM Mg2+; Figure 3E). We found that histone-acetylated nuclei exhibited substantial deformation even against a relatively small stretching force (Figure 3E vs. Figure 1D). The force–deformation plot revealed that TSA treatment reduced nuclear rigidity by approximately twofold (26.0 ± 13.5 nN/µm, n = 19) compared with that of the untreated control (Figure 3, F and G). To determine whether TSA treatment affects the “baseline” rigidity (Figure 2H, broken line), we treated histone-acetylated nuclei with HaeIII and measured their mechanical response. These double-treated nuclei exhibited a rigidity that was comparable to the “baseline” value (21.0 ± 8.9 nN/µm, n = 20; Figure 3, F and G), whereas the level of DNA digestion and composition of major nuclear components were unchanged (Supplemental Figure S4, C and D). Although TSA might enhance the cell’s transcriptional activity and increase the RNA content in nuclei, treatment of hyperacetylated nuclei with RNase did not significantly alter the nuclear mechanical response (26.7 ± 8.6 nN/µm, n = 10; Supplemental Figure S5). Taken together, our data indicate that histone tail acetylation decreases the rigidity of nuclei while having little effect on nuclear composition and suggest that the nucleosome–nucleosome interactions via histone tails play a major role in regulating the mechanical properties of the nucleus.
To verify whether the measured nuclear mechanical response is also observed in living cells, we examined the rigidity of nuclei in situ (Figure 4). To this end, we maintained HeLa cells from a suspension culture, which is the cell line we used for the mechanical measurements of isolated nuclei, in a culture medium and micromanipulated their nuclei by inserting the tips of microneedle probes into the cells’ interior (Figure 4A). Differential interference contrast (DIC) images showed the successful capture and stretch of a nucleus in response to motion of the microneedle tip (Figure 4B). We observed that the applied stretching force induced an extensional deformation in the nucleus. The deformation increased with the magnitude of applied force, whereas the overall cell shape remained relatively intact (Figure 4B). Approximately 60% of cells were susceptible to such micromanipulation, particularly when microneedles of very fine tips (<1 µm in diameter) were used. The remaining ∼40% exhibited extreme blebbing or flattening (Figure 4C), likely due to mechanical damage to the cell cortex. Subsequent analyses were performed for cells that survived this micromanipulation. The force–deformation plot obtained from n = 15 cells revealed that the in situ nuclear mechanical response was approximately linear (Figure 4G), and the rigidity was 23.7 ± 8.6 nN/µm (Figure 4H). Of note, treatment of cells with TSA resulted in a marked decrease of in situ nuclear rigidity (9.7 ± 4.2 nN/µm, n = 14), whereas cellular morphology and susceptibility against mechanical manipulation were similar to those of untreated control cells (Figure 4, D, G, and H). To examine the contribution of the cell cortex and cytoplasm, including its cytoskeleton, to the mechanical measurement, we inserted the microneedle tips into the cells but outside of their nuclei (Figure 4E). This micromanipulation yielded a significant extension of cell peripheral structure, revealing a fusiform-like body (Figure 4F). The rigidity measured by this experiment was greater-than-fourfold lower (5.6 ± 2.0 nN/µm, n = 3) than the value measured based on nucleus capture (Figure 4, G and H). Together these results support the relevance of our nuclear mechanical measurements between in situ and in vitro conditions.
FIGURE 4:
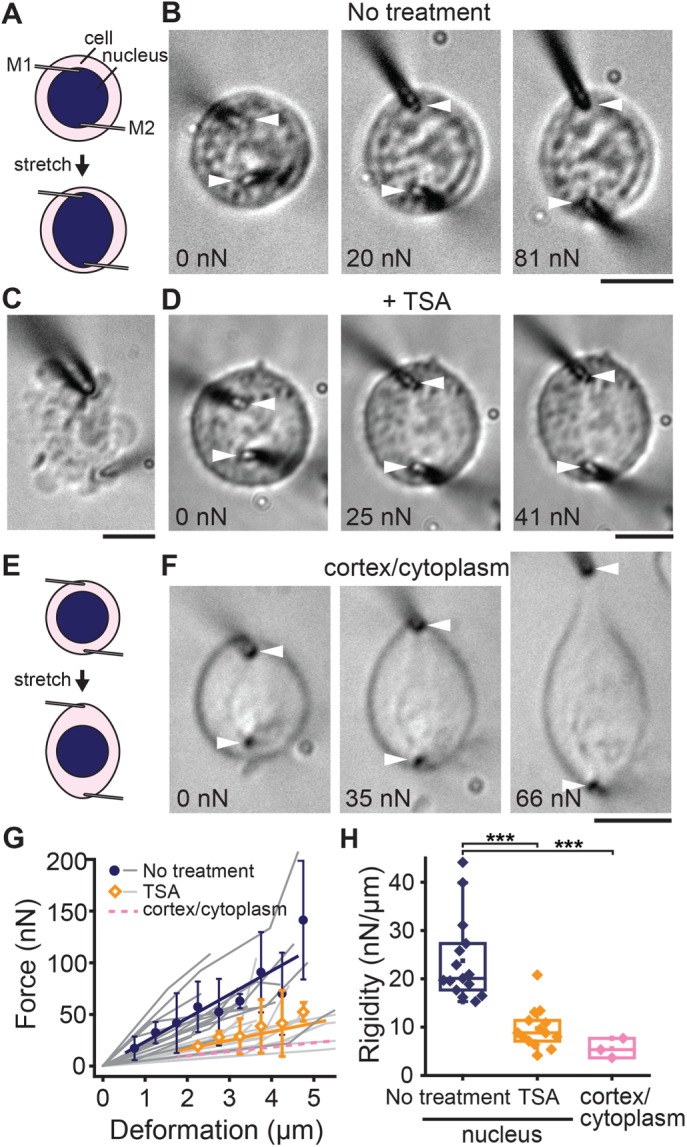
In situ measurement of the nuclear mechanical response. (A) Schematic showing the experimental setting for measuring the nuclear mechanical response in living cells. (B) DIC images of a HeLa cell from suspension culture, the nucleus of which was captured by inserting the tips of microneedles (white arrowheads) into the cells and then stretched at indicated force magnitudes. (C) An unsuccessful measurement. A cell exhibited significant blebbing upon micromanipulation. (D) A TSA-treated HeLa cell, the nucleus of which was stretched as in B and at indicated force magnitudes. Schematic (E) and representative images (F) of a HeLa cell stretched at near the cell periphery but not capturing the nucleus. (G) Force–deformation plot obtained from measurements in untreated control cells (blue circles, n = 20) and cells treated with TSA (orange diamonds, n = 15). Plots are mean ± SD. Linear regression yields slopes of 22.9 and 8.3, respectively. Data from individual samples are shown in dark and light gray, respectively. Also shown is the level of cytoplasm stiffness estimated as in F (pink dashed line). (H) Summary of in situ nuclear rigidity measurement for untreated control cells (n = 20), cells treated with TSA (n = 15), and cytoplasm (n = 3). ***p < 0.0001. Scale bars, 5 µm.
DISCUSSION
The mechanical properties of nuclei lie at the heart of maintaining cellular integrity in the face of mechanical force. In this study, we demonstrated that chromatin supports a significant fraction of nuclear rigidity and that the underlying molecular mechanism involves linker DNA and nucleosome–nucleosome interaction via histone tails. Of note, these changes require minimal compositional alterations in linker histone and loop formation proteins, highlighting the importance of other chromatin features, presumably an interdigitated or “melted” state of nucleosome fibers, as discussed later.
On the basis of our findings, we propose a simple model for the chromatin-based control of nuclear mechanical response (Figure 5). In this model, the mechanical resistance of nuclei is governed by two linear mechanical springs: one attributable to chromatin (κCh) and the other attributable to the nuclear envelope structure (κNE), which is connected in parallel to κCh (Figure 5A, right). The stiffness of κCh decreases severalfold upon histone tail acetylation (Figure 5B, right) and is reduced to nearly zero after linker DNA digestion (Figure 5C, right). κCh is also connected in series to a viscous dashpot element (γCh), whose contribution to nuclear mechanics becomes evident when chromatin forms compacted domains and is deformed by an extremely large load. γCh is likely linked to a relaxation of topological constraint between DNA polymers, given that inhibition of topoisomerase II activity (Wang, 1996) using a well-characterized drug compound (VM-26) significantly suppressed the viscous softening of the nucleus (Supplemental Figure S6). In contrast, the stiffness of κNE is resistant to perturbations to chromatin and thus provides the “baseline” stiffness. We predict that κNE is related to lamin-based structures underlying the nuclear envelope (Butin-Israeli et al., 2012; Gruenbaum and Medalia, 2015; Osmanagic-Myers et al., 2015; Stephens et al., 2017). Consistent with this prediction, lamin A/C levels in nuclei were maintained upon biochemical perturbations of chromatin (Supplemental Figure S4B). Previous studies reported that nuclei in cells have a significant elasticity (Guilak et al., 2000; Caille et al., 2002; Krause et al., 2013) and can be irreversibly deformed by certain types of force perturbations (Dahl et al., 2005; Guilluy et al., 2014). Our model suggests that the mechanical responses of nuclei largely depend on the strength of the chromatin spring κCh, which varies in response to the surrounding ionic conditions and posttranslational histone modifications.
FIGURE 5:
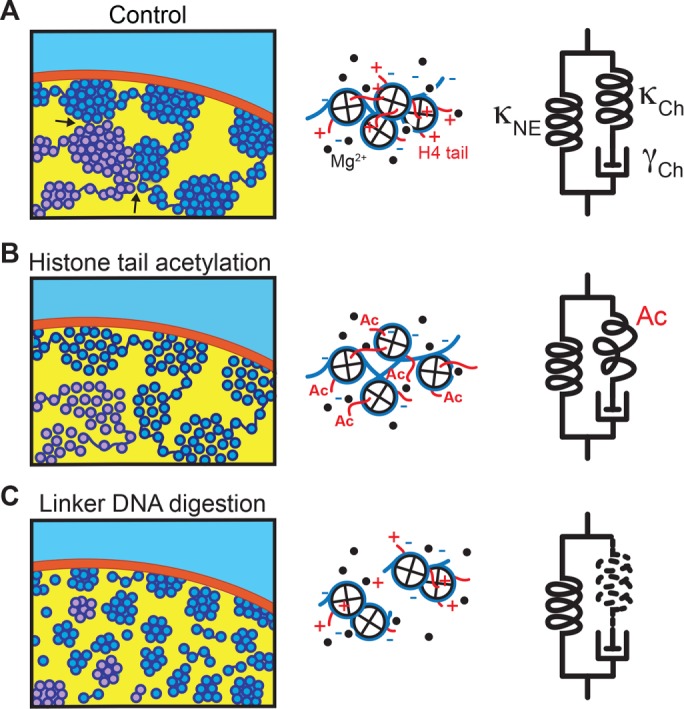
Model for the nuclear mechanical response. Left, possible folding architecture of chromatin within a nucleus. DNA (dark blue) is wrapped around core histones (light blue), forming compacted domains at high Mg2+ levels (A). Also shown is a chromatin strand of another chromosome (light purple). Histone tail acetylation weakens the nucleosome–nucleosome interaction and induces chromatin decompaction (B). Linker DNA digestion leads to disassembly of chromatin domains (C). Middle, molecular views of the chromatin strand. Nucleosomes are packed at high Mg2+ levels and provide spring-like elasticity (A). Histone tail acetylation (Ac) weakens the internucleosomal interaction, resulting in smaller restoring force (B). Linker DNA digestion disrupts the connection between nucleosomes, and no restoring force is generated (C). Right, model of nuclear mechanics, which is composed of two parallel units: 1) an elastic spring attributable to the nuclear envelope (κNE), and 2) a series connection of a spring and a viscous dashpot attributable to chromatin (κCh and γCh, respectively) (A). Histone tail acetylation reduces the stiffness of κCh (B), and linker DNA digestion leads to its complete loss (C). Neither of these perturbations alters κNE, thereby preserving the “baseline” stiffness (Figure 2H).
Our model agrees with a recent study by Stephens et al. (2017) but with two critical differences: in their model, 1) the two elastic springs are nonlinear, and each acts at a different deformation length scale, and 2) the values of spring stiffness were >10 times smaller than those in our study. These inconsistencies likely stem from exposing the nuclei to different buffer conditions (they used cell culture medium; we used buffers of defined chemical composition) and using different micromanipulation systems (they used a glass micropipette to suck the nuclear periphery; we used thin microneedles to “hook” the nuclei). Examining the consequence of these differences should help in developing a more comprehensive model of the nuclear mechanical response.
How does chromatin generate an elastic restoring force in the nucleus? Our analysis reveals that the linker DNA and nucleosome–nucleosome interactions via histone tails act together to generate a restoring force that opposes deformation and maintains the condensed chromatin structure. In the presence of divalent cations, a nonacetylated form of histone tails mediates nucleosome–nucleosome interactions (Funke et al., 2016) and contributes to local compaction of chromatin fibers (Figure 5B; Bannister and Kouzarides, 2011; Kalashnikova et al., 2013). Linker DNA contributes to the maintenance of large-scale globular chromatin assembly (Figure 5C; Maeshima et al., 2016). A lack of either of these two factors results in nearly complete loss of chromatin force generation in the nucleus, suggesting the importance of local and global chromatin organization properties. Of note, chromatin force generation in the nucleus is not evident in its fibrous form, such as the regular 30-nm fibers that are present at low salt concentrations (Figure 5 and Supplemental Figure S1; Maeshima et al., 2016). The elastic restoring force in the nucleus is likely generated by higher-order forms of chromatin, such as interdigitated or “melted” 10-nm nucleosome fibers, which provide the biological relevance for the irregular folding of chromatin observed in various cells (Maeshima et al., 2010; Fussner et al., 2012; Joti et al., 2012; Gan et al., 2013; Hsieh et al., 2015; Ricci et al., 2015; Sanborn et al., 2015; Chen et al., 2016). Irregular folding of chromatin (McDowall et al., 1986; Eltsov et al., 2008; Nishino et al., 2012) can also contribute to mitotic chromosomal integrity because the mechanical rigidity of isolated mitotic chromosomes is highly sensitive to nuclease treatment (Poirier and Marko, 2002) and salt concentration (Poirier et al., 2002). This is advantageous for chromosome segregation and transmission processes in mitosis, during which chromosomes are subjected to significant pulling and shearing stresses.
Within the nucleus, chromosomes are packaged into discrete regions that occupy individual “territories” (Cremer and Cremer, 2001). A localized pulling force acting on the nucleus should be transmitted to one or a few chromosomes that are directly associated with the area at which the force is applied. If these chromosomes are easily separated from other chromosomes that do not have a direct interaction with the force-acting surface, no strain will be developed in each chromosome, and thus the chromatin will generate no resisting force. At the interface of neighboring chromosomes, chromatin fibers engage in topological interaction or intermingling (Branco and Pombo, 2006; Zhao et al., 2006). Our data indicate that this interchromosomal interaction is strong enough to allow for the transmission of the force across multiple chromosomes (Figure 5A, left, arrows). We propose that the DNA polymer has an effective continuity across the entire genome in a mechanical sense.
It is particularly interesting that highly condensed chromatin can function as a strong mechanical “spring” that provides elasticity to the nucleus. We previously reported that condensed chromatin is more resistant to radiation damage than its extended form, presumably because condensed chromatin has a lower potential for reactive radical generation on exposure to ionizing irradiation (Takata et al., 2013). The condensed state also protects genomic DNA from chemical attack (Takata et al., 2013). Taken together, our findings suggest that condensed chromatin domains play essential roles in maintaining genomic integrity in the face of various mechanical and biochemical perturbations.
Consistent with this notion, recent chromosome conformation capture (3C) and Hi-C (an extension of 3C) studies show that genomic DNA forms numerous packed domains called “topologically associating domains” (Smallwood and Ren, 2013; Dekker and Heard, 2015), which may function as the universal building blocks of chromosomes. Similar condensed chromatin features were also observed by pulse labeling of a megabase-sized genomic DNA and are suggested to act as DNA replication foci in nuclei (Rouquette et al., 2009; Markaki et al., 2010). Of note, chromatin condensation also plays a role in controlling the optical properties of rod photoreceptor cells (Solovei et al., 2009). Our study suggests that higher-order condensed chromatin domains might have an evolutionary advantage in maintaining genomic DNA integrity, as well as in performing nongenetic functions, which has not been well appreciated.
MATERIALS AND METHODS
Sample preparation
Nuclei were isolated as previously described (Lewis and Laemmli, 1982; Takata et al., 2013) with minor modifications. Briefly, HeLa cells were maintained in suspension culture at 37°C and 5% CO2 in RPMI 1640 medium (Sigma-Aldrich) supplemented with 5% fetal bovine serum (Nichirei Biosciences) using spinner flasks (Bellco). Collected cells were washed with nucleus isolation buffer composed of 3.75 mM Tris-HCl (pH 7.5), 20 mM KCl, 0.5 mM EDTA, 0.05 mM spermine, 0.125 mM spermidine, 0.1% Trasylol, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich) by two cycles of centrifugal spins at 193 × g for 7 min at 23°C. The pellets were then resuspended in nucleus isolation buffer containing 0.05% Empigen (Sigma-Aldrich) (nucleus isolation buffer+) and immediately homogenized with 10 downward strokes using a tight Dounce pestle. After 5-min centrifugation of the cell lysates at 433 × g, the pellets were washed once with nucleus isolation buffer+ and then stored at −20°C in the same buffer but with 50% glycerol added. For enzyme inhibition experiments, exponentially growing HeLa cells were treated with either 500 nM TSA (Sigma-Aldrich) for 3 h or 20 µM VM-26 (teniposide; Tokyo Chemical Industry) for 2 h. The cells were then either transferred to the nucleus isolation procedure described here or subjected to in situ mechanical measurement. Before the mechanical measurement of isolated nuclear samples, the buffer was replaced with assay buffer comprising 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–KOH (pH 7.4), 0.1 mM PMSF, 0.005% digitonin, and either 0.8–5 mM MgCl2 or 1 mM EDTA by two cycles of centrifuge spins (800 × g for 5 min).
Mechanical measurement and analysis
The experimental setup used to measure the mechanical properties of nuclei was built in an inverted microscope (Ti-U; Nikon) equipped with a 60× objective (Plan Fluor, 0.60 numerical aperture; Nikon), an AxioCam MRc5 (Zeiss) or Neo (Andor) camera, and a pair of glass microneedles each held by a hydraulic three-axis micromanipulator (MHW-3; Narishige). The tips of the microneedles were microfabricated using a capillary puller (PD-10; Narishige) and microforge (World Precision Instruments) such that they had a nearly even and cylindrical shape with a diameter and length of ∼1 and ∼100 µm, respectively. The stiffness of the microneedle tip was precalibrated (elastic constant: 9.0–16.0 nN/μm) according to a previously described method (Shimamoto and Kapoor, 2012). The stiffness calibration was performed by two independent experimenters, and the error was confirmed to be within 10%.
To measure the mechanical response of isolated nuclei, 5 µl of nucleus suspension was spread onto an open experimental chamber, which was assembled by adhering a glass coverslip to the bottom of a 1-mm-thick rubber plate with a central aperture. The coverslip surface was precoated with a thin layer of silicone (SL2; Sigma-Aldrich) to minimize the nonspecific adsorption of nuclear samples. After covering of the nucleus suspension with mineral oil (M5310; Sigma-Aldrich), a single isolated nucleus of typical size and shape was captured using a pair of microneedles. The attachment was achieved by applying the microneedle tips near the edges of the nucleus at an approach angle of ∼80°. The captured nucleus was then lifted off the coverslip surface and stretched by cyclic application of calibrated force, which was exerted by moving the microneedle at a velocity of 1–2 µm/s. A typical force application cycle consisted of sequential phases of stretch, hold, and release. In some cases, stretching of the nucleus caused the microneedle tip to slip over the nuclear surface and eventually become detached. These data were excluded from subsequent analyses because the initial capture geometry was lost. Measurements were performed at room temperature (23 ± 2°C) and within 30–120 min of completing the final buffer exchange, over which no detectable change in nuclear mechanical response was observed.
To measure the mechanical response of nuclei in living cells, a suspension culture of HeLa cells maintained in RPMI 1640 medium at 37°C and 5% CO2 was transferred to a glass-bottom cell imaging dish (740.017; Eppendorf) and placed on the micromanipulation microscope stage. After confirmation of the location of the nucleus within a cell using DIC imaging, the tips of microneedles were inserted into the cell near the edges of the nucleus. Cells exhibiting significant blebbing or membrane rupturing during micromanipulation were excluded from subsequent analyses. Measurements were performed in a temperature-controlled room at 26 ± 1°C and completed within 30 min after transferring cells into dishes, over which no detectable changes in cell morphology and mechanical properties were observed.
The nuclear rigidity was determined by dividing the applied force (F) by the magnitude of deformation (D) that developed within the nucleus. F was estimated based on the microneedle tip’s displacement from the equilibrium point (Δx) and its precalibrated stiffness (kf) according to the equation F = kf Δx. D was measured as the change in distance between the tips of the two microneedles, the positions of which were determined based on a line-scan analysis of time-lapse images acquired during the measurement. These values were obtained at the end of each hold phase after stretch, at which the response reached a nearly steady state. The analysis was performed using NIS-Elements (version 4.20; Nikon) or ImageJ (version 1.48) software. Statistical analysis was performed in Origin Pro 9.0 (Origin Lab). Box plots were drawn where the median, first and third quartiles, and minimum and maximum values are presented for each variable.
Fluorescence imaging
To visualize the morphology of nuclei and nuclear chromatin, isolated nuclei were resuspended in H10Mg5 buffer (10 mM HEPES-KOH, pH 7.4, 5 mM MgCl2, and 0.1 mM PMSF) and attached to poly-l-lysine–coated coverslips by centrifugation at 2380 × g for 15 min. The nuclei on the coverslips were transferred to one of the following buffers: H10Mg5, H10Mg1 (10 mM HEPES-KOH, pH 7.4, 1 mM MgCl2, and 0.1 mM PMSF), or H10E (0 mM Mg2+ buffer; 10 mM HEPES-KOH, pH 7.4, 1 mM EDTA, pH 8.0, and 0.1 mM PMSF). The nuclei were then fixed with 2% formaldehyde prepared in the corresponding buffers. After staining of DNA with 4′,6-diamidino-2-phenylindole (DAPI), the coverslips were sealed with nail polish, and optical sections at a thickness of 200 nm were imaged using a DeltaVision microscope (Applied Precision). Acquired images were deconvolved to remove out-of-focus information. The Softworx package of DeltaVision was used to measure average nuclear sizes (n = ∼20 for each condition) and generate intensity line profiles of DAPI-stained nuclei for chromatin distribution.
Biochemical analyses of nuclei
The nuclei (weight, 50 µg) were incubated on ice for 15 min in a series of buffers: HE (0 mM Mg2+ buffer), H10Mg1, and H10Mg5. After incubation, centrifugation was performed to recover nuclei. The nuclear pellets were suspended in a final sample buffer (Laemmli, 1970) and subjected to 12.5% SDS–PAGE. Subsequently, the gels were subjected to Coomassie brilliant blue (CBB) staining and Western blotting using the following antibodies: mouse anti-Rad21 (05-908; Millipore), rabbit anti-CTCF (07-729; Millipore), rabbit anti-Smc2 (ab10412; Abcam), rabbit anti-H2B (ab1790; Abcam), rabbit anti–acetyl histone H4 (06-866; Millipore), rabbit anti–acetyl histone H3 (06-599; Millipore), and goat anti–lamin A/C (sc-6215; Santa Cruz Biotechnology).
Enzymatic digestion experiments
Digestion of DNA in nuclei was performed by incubating 50 µg (DNA) of nuclei with 50 U of HaeIII (Takara) in H10Mg5 buffer containing 1 mM dithiothreitol (DTT) and 0.05% digitonin at 16°C for 1 h. To analyze the extent of digestion, DNA was purified by treating the nuclei with a mixture of RNase A (Wako, Japan), SDS, and proteinase K (Wako), followed by phenol/chloroform extraction and ethanol precipitation according to the molecular cloning method. Purified DNA (500 ng) with or without digestion was analyzed by 0.8% agarose gel electrophoresis.
For protein composition analysis, the same amounts of the nuclei with or without digestion were separated into nuclear pellets and supernatant fractions by centrifugation. The proteins in the supernatant fractions were precipitated using 17% trichloroacetic acid (Wako) and cold acetone. Both pellets were suspended in the final sample buffer and subjected to 14% SDS–PAGE and subsequent CBB staining and Western blotting using rabbit anti-H1.0 antibody (GTX114462; Sigma-Aldrich) and rabbit anti-H2B antibody.
RNase-treated nuclei were prepared by adding 100 µg/ml DNase-free RNase A to nuclear suspension prepared from TSA-treated HeLa cells in H10Mg5 buffer containing 0.05% digitonin (Sigma-Aldrich), 1 µM PMSF, and 1 µM TSA and incubated at 15°C for 1 h. Prior to mechanical measurements, the reagents were washed by two cycles of centrifugal spins with the identical buffer but without RNase.
Protein identification by mass spectrometry analysis
For analyzing the histone acetylation profile, TSA-treated nuclear proteins were first fractionated in 15% SDS polyacrylamide gel (ATTO, Japan), and protein bands were detected using CBB staining (Wako). The protein bands containing histone H3 and H4 were cut out from the gel. The protein in the gel was treated by trypsin and eluted from the gel following the manufacturer’s protocol (Bruker Daltonics). Briefly, the sliced gel was destained (Silver Stain MS Kit; Wako), completely dehydrated with acetonitrile, and then treated with the reduction buffer (10 mM DTT, 25 mM ammonium bicarbonate [ABC]) at 56°C for 45 min. After removal of the reduction buffer and a wash with 25 mM ABC, the gel was treated with alkylating solution (55 mM iodoacetamide, 25 mM ABC) at room temperature for 30 min. After removal of the alkylating buffer and a wash with 25 mM ABC, the gel was completely dehydrated with acetonitrile again. The gel was then soaked with 20 ng/µl Trypsin Gold (Promega) and incubated at 37°C overnight. The gel was mixed with the elution buffer (50% acetonitrile, 5% trifluoroacetate), sonicated for 3 min, and vortexed for 30 min. The elution containing trypsin-digested peptides was recovered, and then its volume was reduced to less than10 μl by an evaporator. The peptide sample was desalinated with a Zip-Tip C18 column (Millipore). The desalinated sample was mixed with equal volume of 0.7 mg/ml α-cyano-4-hydroxycinnamic acid (HCCA; Bruker Daltonics) as a matrix. The HCCA-mixed sample was analyzed to obtain m/z peak data by UltraflexIII matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry with the equipped software (flexcontrol, version 3.3, and flexanalysis, version 3.3; Bruker Daltonics). The m/z signal data were analyzed to identify acetylated peptides in histones using biotools, version 3.2 (Bruker Daltonics), and the matrix server (Matrix Science), as well as a public database (Swiss-Prot).
Supplementary Material
Acknowledgments
We thank members of the Shimamoto and Maeshima labs for helpful discussions and assistance and are grateful to anonymous referees for their invaluable input. This research was supported by PRIME, AMED, Japan Society for the Promotion of Science KAKENHI 15K14515, and the Japan Science and Technology Agency Disseminate Tenure-Track System (Y.S.) and Japan Society for the Promotion of Science KAKENHI 16H04746 and the Japan Science and Technology Agency, CREST JPMJCR15G2 (K.M.).
Abbreviations used:
- 3C
chromosome conformation capture
- DIC
differential interference contrast
- TSA
trichostatin A.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-11-0783) on April 20, 2017.
REFERENCES
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Belmont AS. Revisiting higher-order and large-scale chromatin organization. Curr Opin Cell Biol. 2012;24:359–366. doi: 10.1016/j.ceb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M, Misteli T. Nongenetic functions of the genome. Science. 2016;352:aad6933. doi: 10.1126/science.aad6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet. 2012;28:464–471. doi: 10.1016/j.tig.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A, Ivanovska IL, Discher DE. Matrix elasticity, cytoskeletal forces and physics of the nucleus: how deeply do cells “feel” outside and in. J Cell Sci. 2010;123:297–308. doi: 10.1242/jcs.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caille N, Thoumine O, Tardy Y, Meister JJ. Contribution of the nucleus to the mechanical properties of endothelial cells. J Biomech. 2002;35:177–187. doi: 10.1016/s0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- Chen C, Lim HH, Shi J, Tamura S, Maeshima K, Surana U, Gan L. Budding yeast chromatin is dispersed in a crowded nucleoplasm in vivo. Mol Biol Cell. 2016;27:3357–3368. doi: 10.1091/mbc.E16-07-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc Natl Acad Sci USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Engler AJ, Pajerowski JD, Discher DE. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys J. 2005;89:2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson PM, Lammerding J. Broken nuclei—lamins, nuclear mechanics, and disease. Trends Cell Biol. 2014;24:247–256. doi: 10.1016/j.tcb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Heard E. Structural and functional diversity of topologically associating domains. FEBS Lett. 2015;589:2877–2884. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Laemmli UK. Architecture of metaphase chromosomes and chromosome scaffolds. J Cell Biol. 1983;96:84–93. doi: 10.1083/jcb.96.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke JJ, Ketterer P, Lieleg C, Schunter S, Korber P, Dietz H. Uncovering the forces between nucleosomes using DNA origami. Sci Adv. 2016;2:e1600974. doi: 10.1126/sciadv.1600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, Bustin M. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun. 2015;6:6138. doi: 10.1038/ncomms7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussner E, Strauss M, Djuric U, Li R, Ahmed K, Hart M, Ellis J, Bazett-Jones DP. Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep. 2012;13:992–996. doi: 10.1038/embor.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Ladinsky MS, Jensen GJ. Chromatin in a marine picoeukaryote is a disordered assemblage of nucleosomes. Chromosoma. 2013;122:377–386. doi: 10.1007/s00412-013-0423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorisch SM, Wachsmuth M, Toth KF, Lichter P, Rippe K. Histone acetylation increases chromatin accessibility. J Cell Sci. 2005;118:5825–5834. doi: 10.1242/jcs.02689. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Medalia O. Lamins: the structure and protein complexes. Curr Opin Cell Biol. 2015;32:7–12. doi: 10.1016/j.ceb.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Guilak F, Tedrow JR, Burgkart R. Viscoelastic properties of the cell nucleus. Biochem Biophys Res Commun. 2000;269:781–786. doi: 10.1006/bbrc.2000.2360. [DOI] [PubMed] [Google Scholar]
- Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol. 2014;16:376–381. doi: 10.1038/ncb2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampoelz B, Lecuit T. Nuclear mechanics in differentiation and development. Curr Opin Cell Biol. 2011;23:668–675. doi: 10.1016/j.ceb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isermann P, Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr Biol. 2013;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joti Y, Hikima T, Nishino Y, Kamada F, Hihara S, Takata H, Ishikawa T, Maeshima K. Chromosomes without a 30-nm chromatin fiber. Nucleus. 2012;3:404–410. doi: 10.4161/nucl.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J R Soc Interface. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Te Riet J, Wolf K. Probing the compressibility of tumor cell nuclei by combined atomic force-confocal microscopy. Phys Biol. 2013;10:065002. doi: 10.1088/1478-3975/10/6/065002. [DOI] [PubMed] [Google Scholar]
- Kruithof M, Chien FT, Routh A, Logie C, Rhodes D, van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat Struct Mol Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lewis CD, Laemmli UK. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982;29:171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maeshima K, Hihara S, Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo. Curr Opin Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Maeshima K, Rogge R, Tamura S, Joti Y, Hikima T, Szerlong H, Krause C, Herman J, Seidel E, DeLuca J, et al. Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers. EMBO J. 2016;35:1115–1132. doi: 10.15252/embj.201592660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markaki Y, Gunkel M, Schermelleh L, Beichmanis S, Neumann J, Heidemann M, Leonhardt H, Eick D, Cremer C, Cremer T. Functional nuclear organization of transcription and DNA replication: a topographical marriage between chromatin domains and the interchromatin compartment. Cold Spring Harb Symp Quant Biol. 2010;75:475–492. doi: 10.1101/sqb.2010.75.042. [DOI] [PubMed] [Google Scholar]
- Mazumder A, Roopa T, Basu A, Mahadevan L, Shivashankar GV. Dynamics of chromatin decondensation reveals the structural integrity of a mechanically prestressed nucleus. Biophys J. 2008;95:3028–3035. doi: 10.1529/biophysj.108.132274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall AW, Smith JM, Dubochet J. Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J. 1986;5:1395–1402. doi: 10.1002/j.1460-2075.1986.tb04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanagic-Myers S, Dechat T, Foisner R. Lamins at the crossroads of mechanosignaling. Genes Dev. 2015;29:225–237. doi: 10.1101/gad.255968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MG, Marko JF. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc Natl Acad Sci USA. 2002;99:15393–15397. doi: 10.1073/pnas.232442599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MG, Monhait T, Marko JF. Reversible hypercondensation and decondensation of mitotic chromosomes studied using combined chemical-micromechanical techniques. J Cell Biochem. 2002;85:422–434. doi: 10.1002/jcb.10132. [DOI] [PubMed] [Google Scholar]
- Ricci MA, Manzo C, Garcia-Parajo MF, Lakadamyali M, Cosma MP. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the "30-nm" chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, Fakan S. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res. 2009;17:801–810. doi: 10.1007/s10577-009-9070-x. [DOI] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SG, King MC. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun. 2015;6:7159. doi: 10.1038/ncomms8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y, Kapoor TM. Microneedle-based analysis of the micromechanics of the metaphase spindle assembled in Xenopus laevis egg extracts. Nat Protoc. 2012;7:959–969. doi: 10.1038/nprot.2012.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y, Maeda YT, Ishiwata S, Libchaber AJ, Kapoor TM. Insights into the micromechanical properties of the metaphase spindle. Cell. 2011;145:1062–1074. doi: 10.1016/j.cell.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol. 2013;25:387–394. doi: 10.1016/j.ceb.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell. 2017;28 doi: 10.1091/mbc.E16-09-0653. mbc.E16-09-0653 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick R, Strissel PL, Gavrilov K, Levi-Setti R. Cation-chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J Cell Biol. 2001;155:899–910. doi: 10.1083/jcb.200105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H, Hanafusa T, Mori T, Shimura M, Iida Y, Ishikawa K, Yoshikawa K, Yoshikawa Y, Maeshima K. Chromatin compaction protects genomic DNA from radiation damage. PLoS One. 2013;8:e75622. doi: 10.1371/journal.pone.0075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol. 2016;17:399–412. doi: 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- Visvanathan A, Ahmed K, Even-Faitelson L, Lleres D, Bazett-Jones DP, Lamond AI. Modulation of higher order chromatin conformation in mammalian cell nuclei can be mediated by polyamines and divalent cations. PLoS One. 2013;8:e67689. doi: 10.1371/journal.pone.0067689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Widom J. Physicochemical studies of the folding of the 100 A nucleosome filament into the 300 A filament. Cation dependence. J Mol Biol. 1986;190:411–424. doi: 10.1016/0022-2836(86)90012-4. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- Zwerger M, Ho CY, Lammerding J. Nuclear mechanics in disease. Annu Rev Biomed Eng. 2011;13:397–428. doi: 10.1146/annurev-bioeng-071910-124736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.