Abstract
Pc2 is a polycomb protein, which has SUMO E3 activity for the corepressors CtBP and CtBP2. Here we demonstrate that, in vivo, Pc2 adapter function contributes to enhancement of CtBP sumoylation. Mutation of the CtBP binding site on Pc2 abolishes E3 activity toward CtBP. However, a carboxyl-terminal fragment of Pc2 that recruits both Ubc9 and CtBP lacks E3 activity. We identify a second domain, which, when coexpressed with the carboxyl-terminal adapter region, restores E3 function. In vitro, this domain has E3 activity in isolation, suggesting that it is a functional domain, and that adapter function is required to selectively corecruit E2 and substrate in vivo. These results demonstrate the presence of two domains in Pc2 that contribute to full in vivo E3 activity, and suggest that SUMO E3s are more than simple platforms to which E2 and substrate bind.
Keywords: CtBP, E3, Polycomb, Pc2, SUMO
Introduction
Modification of proteins by covalent attachment of ubiquitin and related molecules is implicated in numerous cellular processes (Hershko and Ciechanover, 1998; Melchior, 2000; Yeh et al, 2000; Pickart, 2001; Hay, 2001; Schwartz and Hochstrasser, 2003). Attachment of ubiquitin to a substrate occurs via a two- or three–enzyme cascade (Hershko and Ciechanover, 1998; Pickart, 2001): The activating enzyme (E1) forms a thiol ester with the carboxyl-terminal glycine of ubiquitin. Ubiquitin is then transferred to the conjugating enzyme (E2), again via a thiol ester linkage. The E3 ligating enzyme, of which there are two classes, transfers the ubiquitin to a target lysine residue. RING domain E3s contain a cysteine/histidine zinc coordinating domain, and act as adapters, which bring together E2 and substrate (Lorick et al, 1999; Jackson et al, 2000). Many RING domain E3s function as part of larger E3 complexes, where it is the RING finger that contacts the E2. A direct catalytic role for the RING finger is unlikely, since the E2 active site is relatively far from the RING domain (Zheng et al, 2000). In contrast, for HECT domain E3s, the ubiquitin is transferred to a conserved cysteine, and then on to the substrate lysine (Scheffner et al, 1993; Huibregtse et al, 1995). Thus the HECT domain E3s contain both adapter function and enzymatic activity, whereas the RING domain proteins are simple adapters.
Substrate modification by SUMO is analogous to ubiquitination (Melchior, 2000; Hay, 2001): The E1 activating enzyme, consisting of a heterodimer of Aos1 and Uba2, passes SUMO to the E2 (Ubc9) and in both cases, the SUMO is linked via a thiol ester to conserved cysteine residues (Dohmen et al, 1995; Desterro et al, 1997; Johnson and Blobel, 1997; Johnson et al, 1997). Ubc9 then interacts directly with substrate to modify the target lysine. In contrast to the multiple ubiquitin E2s, which help determine substrate specificity, there is only a single SUMO E2. The sumoylation consensus motif (ΨKxE, where Ψ is hydrophobic) is a major determinant of substrate specificity (Sampson et al, 2001; Bernier-Villamor et al, 2002). Structural and mutational analyses have demonstrated the importance of this motif for Ubc9 interaction and sumoylation (Bernier-Villamor et al, 2002). A further level of specificity within the SUMO pathway was revealed with the discovery of SUMO E3s. In Saccharomyces cerevisiae Siz1p and Siz2p are essential for normal levels of sumoylation in vivo, and have E3 activity in vitro (Johnson and Gupta, 2001; Takahashi et al, 2001). Siz proteins bind E2 and substrate, and stimulate the transfer of SUMO to substrate in vitro. Siz proteins contain a divergent RING-like motif, which is present in mammalian PIAS (Protein Inhibitor of Activated STAT) proteins. PIAS family members are SUMO E3s for numerous proteins, including LEF1, p53, c-Jun, Smad4 and CtBP (Kahyo et al, 2001; Sachdev et al, 2001; Schmidt and Muller, 2002; Lee et al, 2003; Lin et al, 2003; Ohshima and Shimotohno, 2003; Long et al, 2004). The nuclear pore complex (NPC)-associated protein RanBP2 (or Nup358) is a SUMO E3, which is involved in the modification of Sp100 and HDAC4 (Kirsh et al, 2002; Pichler et al, 2002). RanBP2 contains a zinc finger domain, different from the RING fingers present in both ubiquitin E3s and Siz/PIAS SUMO E3s, which is dispensable for E3 activity.
We identified the polycomb group protein, Pc2, as a SUMO E3 for the transcriptional corepressors CtBP and CtBP2 (Kagey et al, 2003). Pc2 is a member of the polycomb group of proteins, which were first identified in Drosophila, as regulators of segment identity (Simon et al, 1992; Kennison, 1995; Simon, 1995; Simon and Tamkun, 2002). Human Pc2 was identified by its homology to Drosophila Pc, but shares only limited sequence similarity, primarily in the amino-terminal chromodomain and a small region at the extreme carboxyl-terminus (Satijn et al, 1997). Pc2 is a transcriptional repressor, and this is likely due in part to interactions with other polycomb proteins, such as RING1 (Muller, 1995; Satijn et al, 1997; Satijn and Otte, 1999; Poux et al, 2001). In mammalian cells, many polycomb proteins, including Pc2, have a distinct subnuclear localization, forming discrete foci, termed polycomb bodies (Gerasimova and Corces, 1998; Saurin et al, 1998). Pc2 interacts with the transcriptional corepressor, CtBP, via a PLDLS-like motif and recruits it to polycomb bodies (Sewalt et al, 1999; Kagey et al, 2003). Pc2 has no obvious sequence similarity to other known E3s, suggesting that there are at least three families of SUMO E3, with no apparent unifying structural features.
Here we report the functional analysis of Pc2 E3 activity. A domain of Pc2, which acts as a docking site for both Ubc9 and CtBP, does not have E3 activity in vivo, and we identify a separate domain of Pc2, which in vitro has E3 activity on its own. In vivo, both domains of Pc2 contribute to E3 activity, suggesting that SUMO E3s are more complex than simple adapter proteins.
Results
CtBP interaction with Pc2 is necessary for enhanced sumoylation
Pc2 recruits CtBP to polycomb bodies via a PIDLR motif (Sewalt et al, 1999). To determine if this motif is important for CtBP SUMO modification, COS-1 cells were cotransfected with T7-tagged CtBP, six-histidine (H6)-tagged SUMO1 and Flag-tagged Pc2. Cell lysates were separated by SDS–PAGE and Western blotted with a T7 antibody to detect CtBP (Figure 1A). In contrast to full-length Pc2, no enhancement of CtBP sumoylation was seen with the PIDLR deletion, indicating that CtBP interaction with Pc2 is required. Interestingly, no increase in sumoylation was observed with a Pc2 deletion that lacks the first 400 amino acids even though the PIDLR motif is still present (Figure 1A; Kagey et al, 2003). This suggests that the amino-terminus of Pc2 is crucial for enhancement of CtBP sumoylation. To ensure that the Pc2(401–558) construct interacts with CtBP, COS-1 cells were cotransfected with various Fl-Pc2 constructs and T7-CtBP. Proteins were collected on anti-Flag-agarose, and co-precipitating T7-CtBP was detected by Western blotting (Figure 1B). T7-CtBP clearly interacted with both full-length Pc2 and Pc2(401–558), indicating that deletion of the amino-terminus of Pc2 does not affect Pc2 with respect to CtBP binding. Deletion of the PIDLR motif significantly weakened the association with CtBP. These results suggest that CtBP interaction with Pc2 is required but not sufficient for Pc2 E3 activity.
Figure 1.
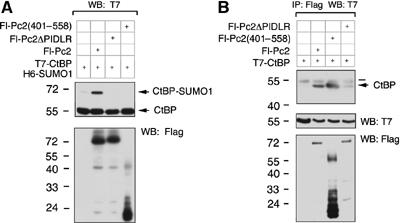
The amino-terminus of Pc2 is required for enhanced sumoylation. (A) COS-1 cells were cotransfected with T7-CtBP, H6-SUMO1 and the indicated Flag-tagged Pc2 expression constructs. Cells were lysed directly by boiling in SDS–PAGE loading buffer. Proteins were separated by SDS–PAGE and analyzed by Western blot with a T7 or Flag antibody. Sumoylated and unsumoylated CtBP are shown. (B) COS-1 cells were transfected with T7-CtBP alone or with the indicated Flag-tagged Pc2 expression constructs. Following lysis, proteins were precipitated with anti-Flag-agarose, separated by SDS–PAGE and analyzed by Western blot with a T7 antibody. Expression of T7 and Flag constructs is shown below. The bar indicates the immunoglobulin heavy chain. The positions of molecular weight markers (kDa) are indicated.
To begin to address the role of the amino-terminal 400 amino acids of Pc2, we tested localization to polycomb bodies. YFP fusions of full-length Pc2 or Pc2(401–558) were transfected into COS-1 cells and visualized by fluorescence microscopy. Both formed distinct foci indicative of polycomb bodies, whereas YFP alone displayed a diffuse nuclear and cytosolic localization (Figure 2A). Next we tested whether Pc2(401–558) could recruit CtBP and Ubc9 to polycomb bodies. Both Fl-Pc2(401–558) and Fl-Pc2 recruited CFP-CtBP to polycomb bodies, consistent with the presence of the PIDLR motif (Figure 2B). As expected, coexpression of Fl-Pc2ΔPIDLR did not result in a significant portion of YFP-CtBP being localized to polycomb bodies. Interestingly, when YFP-Ubc9 was cotransfected with Fl-Pc2, Fl-Pc2ΔPIDLR or Fl-Pc2(401–558), a significant amount of YFP-Ubc9 was brought to polycomb bodies by all three Pc2 constructs (Figure 2C). Thus, recruitment of Ubc9 is independent of the first 400 amino acids of Pc2 and the PIDLR motif. Furthermore Fl-Pc2(401–558), which does not have E3 activity for CtBP, colocalized both CFP-CtBP and YFP-Ubc9 to polycomb bodies, suggesting that E3 activity may be more complex than simple corecruitment of substrate and enzyme by Pc2 (Figure 2D).
Figure 2.
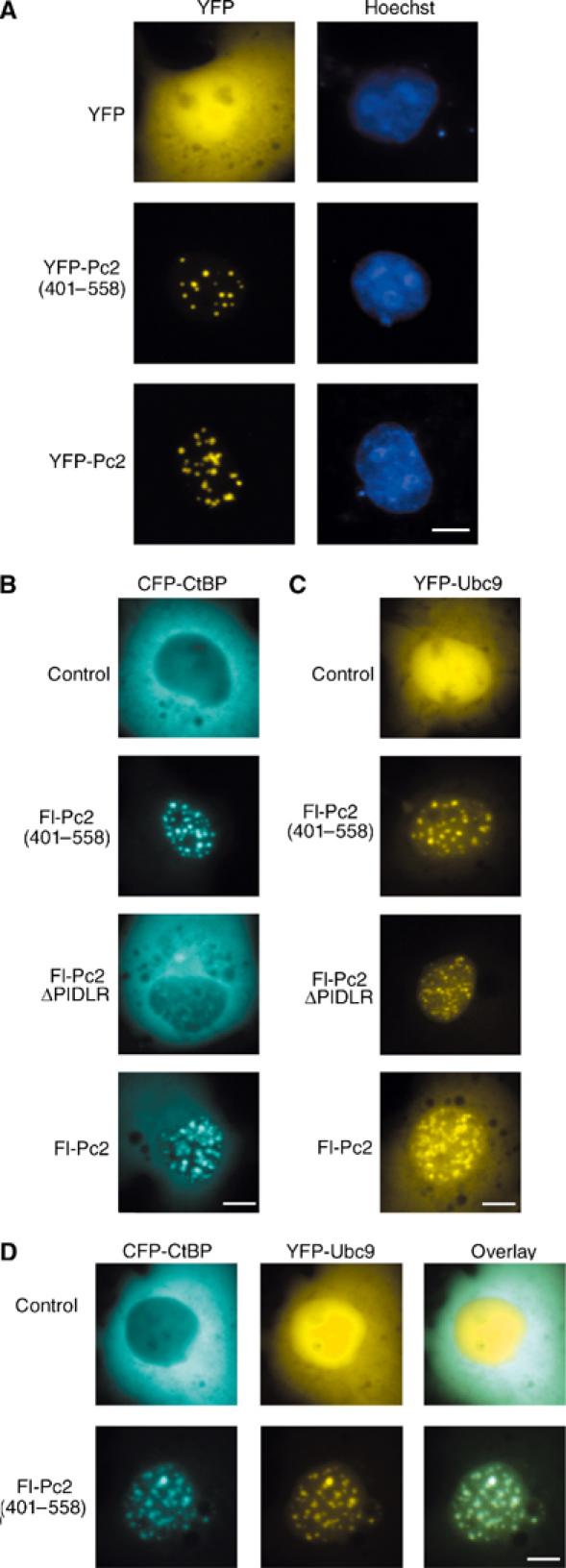
Pc2(401–558) colocalizes Ubc9 and CtBP. COS-1 cells were transfected with the indicated expression constructs and visualized by live cell fluorescence microscopy 24 h later. (A) YFP, YFP-Pc2(401–558) or YFP-Pc2 was expressed and cells were stained with Hoechst before visualization. YFP and Hoechst images are shown. (B, C) CFP-CtBP or YFP-Ubc9 was expressed alone (upper panel) or with the indicated Fl-Pc2 constructs (lower panels). CFP and YFP images are shown. (D) CFP-CtBP and YFP-Ubc9 were coexpressed in the presence or absence of Fl-Pc2(401–558). Individual CFP and YFP images along with the merged (overlay) are shown. Scale bar=10 μm.
The amino-terminus of Pc2 is required for enhanced CtBP sumoylation
We next tested the effect of increasing amino-terminal deletions on Pc2 E3 activity. COS-1 cells were cotransfected with T7-CtBP and H6-SUMO1 with various Fl-Pc2 deletions, and cell lysates were analyzed with a T7 antibody to determine the level of SUMO modified CtBP (Figure 3A and C). Deleting the chromodomain or the first 168 amino acids of Pc2 did not significantly reduce E3 activity. However, further truncation to amino acid 288 dramatically impaired enhancement of CtBP sumoylation. We next tested two amino-terminal fragments of Pc2 for E3 activity. Neither the amino-terminal 288 nor 375 amino acids of Pc2 alone had E3 activity, consistent with the requirement to recruit CtBP (Figure 3B). Interestingly, when Pc2(401–558), which interacts with both CtBP and Ubc9, was cotransfected with an amino-terminal fragment of Pc2 (residues 2–288 or 2–375), along with T7-CtBP and H6-SUMO1, E3 activity was significantly restored (Figure 3B). These results demonstrate that in addition to a recruitment function in the carboxyl-terminus, the amino-terminus of Pc2 is required for E3 activity, and suggest that these functions can be separated.
Figure 3.
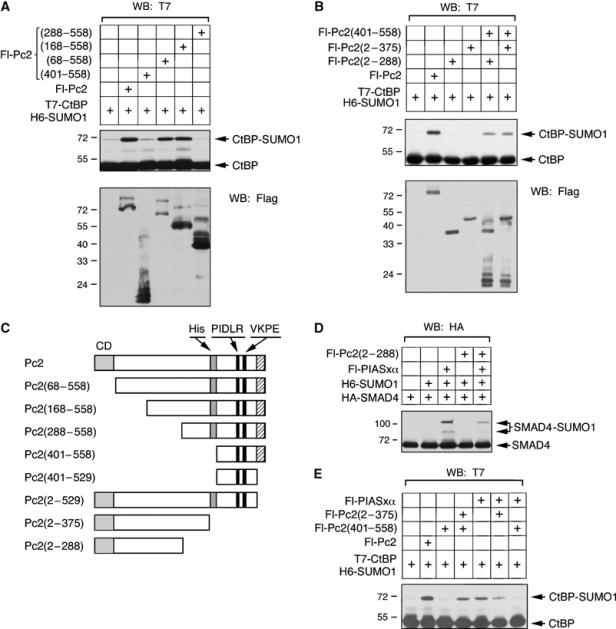
The amino-terminus of Pc2 is required for E3 activity. COS-1 cells were transfected with the indicated expression constructs and analyzed as in Figure 1. (A, B) T7-CtBP and H6-SUMO1 were coexpressed alone or with the indicated amino- or carboxyl-terminal Fl-Pc2 fragments. Lysates were blotted for CtBP and CtBP-SUMO with a T7 antibody, and expression of Fl-Pc2 constructs is shown below. (C) The Flag-tagged Pc2 expression constructs used here, and in other figures, are shown schematically: CD: chromodomain; His: polyhistidine stretch; PIDLR: CtBP interaction motif; VKPE: sumoylation consensus. The hatched region at the extreme carboxyl-terminus is the region required for correct localization (see Figure 4) and interaction with RING1. (D) HA-Smad4 was expressed with H6-SUMO1, Fl-PIASxα and Fl-Pc2(2–288) as indicated. Sumoylated and unmodified Smad4 were detected by HA Western blot. (E) T7-CtBP and H6-SUMO1 were coexpressed in the presence of the indicated combinations of Fl-Pc2, Fl-Pc2(401–558), Fl-Pc2(2–375) and Fl-PIASxα. CtBP and sumoylated CtBP detected by T7 Western blot are shown. The positions of molecular weight markers (kDa) are indicated.
Smad4 is sumoylated and PIASxα has been shown to be an E3 for Smad4 (Lee et al, 2003; Ohshima and Shimotohno, 2003; Long et al, 2004). To test the specificity of the amino-terminus of Pc2, the effect of Pc2(2–288) on Smad4 sumoylation was determined. Analysis of COS-1 cells that were cotransfected with HA-tagged Smad4 and H6-SUMO1 revealed that SUMO modification of Smad4 was only observed when PIASxα was also overexpressed (Figure 3D). Cotransfection of Fl-Pc2(2–288) did not significantly increase Smad4 sumoylation levels even when Fl-PIASxα was also transfected, indicating that the amino-terminus of Pc2 is only functional together with the carboxyl-terminus of Pc2, or that it is specifically influencing CtBP sumoylation.
PIAS proteins are E3s for many substrates, including CtBP (Lin et al, 2003), suggesting that they may have a more general role in the SUMO conjugation pathway. Potentially, a PIAS protein could interact with Ubc9 and stimulate sumoylation, provided that the substrate and Ubc9 had been brought together by an adapter. We, therefore, tested the effect of PIASxα on CtBP sumoylation in the presence of amino- or carboxyl-terminal fragments of Pc2 (Figure 3E). Cotransfection of PIASxα with T7-CtBP and H6-SUMO1 results in an increase in SUMO modified CtBP, suggesting that in vivo PIASxα has E3 activity for CtBP. Cotransfection of either Fl-Pc2(401–558) or Fl-Pc2(2–375) with T7-CtBP, H6-SUMO1 and Fl-PIASxα did not increase CtBP sumoylation over that seen with PIASxα alone (Figure 3E). Together, these results suggest that the amino- and carboxyl-terminal regions of Pc2 cooperate to function as a SUMO E3, and that this cooperativity is specific.
Localization to polycomb bodies is not required for Pc2 E3 activity
Together, Pc2(2–375) and Pc2(401–558) enhance sumoylation of CtBP, suggesting either an interaction or perhaps corecruitment to polycomb bodies. To determine the importance of localization to polycomb bodies, we deleted the carboxyl-terminal 29 amino acids from Pc2(401–558) to generate Pc2(401–529). When Fl-Pc2(401–529) was cotransfected with Fl-Pc2(2–375), T7-CtBP and H6-SUMO1, E3 activity was not restored, suggesting an important role of the extreme carboxyl-terminus (Figure 4A). As shown in Figure 4B, YFP Pc2(401–529) is delocalized from polycomb bodies, suggesting that correct localization may be important for E3 activity. However, deletion of this region from full-length Pc2 did not affect E3 activity, despite the fact that YFP-Pc2(2–529) did not localize to polycomb bodies (Figure 4B and C). As with full-length Pc2, this enhancement was abolished when the PIDLR motif was deleted from Pc2(2–529) (Figure 4C). These results indicate that Pc2 localization to polycomb bodies is not required for E3 activity.
Figure 4.
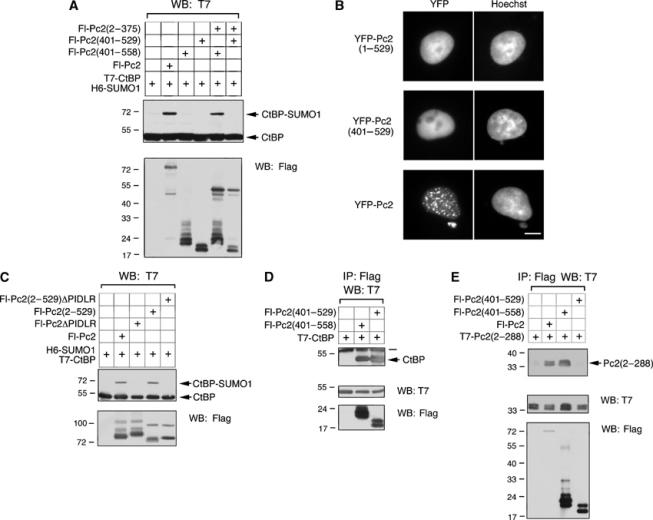
Pc2 localization to polycomb bodies is not required for E3 activity. (A) T7-CtBP and H6-SUMO1 were coexpressed alone or with the indicated Fl-Pc2 constructs. Cell lysates were analyzed by Western blot with a T7 antibody to detect SUMO modified and unmodified CtBP. Expression of Flag constructs is shown below. (B) YFP-Pc2(1–529), YFP-Pc2(401–529) or YFP-Pc2 was expressed in COS-1 cells and visualized by live cell fluorescence microscopy 24 h later. YFP and Hoechst images are shown. (C) T7-CtBP and H6-SUMO1 were expressed without or with Fl-Pc2, Fl-Pc2ΔPIDLR, Fl-Pc2(2–529) or Fl-Pc2(2–529)ΔPIDLR. Cell lysates were analyzed by Western blot with a T7 antibody to detect sumoylated and unsumoylated CtBP, and with a Flag antibody for expression controls. (D) COS-1 cells were transfected with the indicated Fl-Pc2 expression constructs and T7-CtBP. Protein complexes were precipitated with anti-Flag-agarose and analyzed by Western blot for T7-CtBP. The bar denotes the immunoglobulin heavy chain. (E) COS-1 cells were transfected with T7-Pc2(2–288) and the indicated Flag-tagged Pc2 constructs. Proteins were collected on anti-Flag-agarose and analyzed for co-precipitating T7-Pc2(2–288). Expression of Flag and T7 constructs in the lysates is shown below. The positions of molecular weight markers (kDa) are indicated.
Pc2(401–529) did not enhance CtBP sumoylation together with Pc2(2–375), despite the fact that localization to polycomb bodies is not required. To confirm that Pc2(401–529) binds CtBP, COS-1 cells were transfected with T7-CtBP and either Fl-Pc2(401–558) or Fl-Pc2(401–529) and proteins were precipitated with anti-Flag-agarose. T7-CtBP co-precipitated with both Fl-Pc2(401–558) and Fl-Pc2(401–529), indicating that truncation of the last 29 amino acids of Pc2 does not disrupt CtBP binding (Figure 4D). An alternative possibility is that the carboxyl-terminal region of Pc2 binds to the amino-terminus of the protein, bringing together the two polypeptides for SUMO modification of CtBP. To address this possibility, COS-1 cells were transfected with T7-Pc2(2–288) and Flag-tagged Pc2 constructs. Proteins were precipitated with anti-Flag-agarose and analyzed with a T7 antibody to detect interacting T7-Pc2(2–288). T7-Pc2(2–288) co-precipitated with full-length Fl-Pc2 and Fl-Pc2(401–558) (Figure 4E). Interestingly, T7-Pc2(2–288) did not interact with Pc2(401–529), suggesting that coexpression of Pc2(2–375) and Pc2(401–529) do not enhance CtBP sumoylation because of failure to interact with each other.
Contribution of the amino-terminus of Pc2 to E3 activity in vivo
The first 288 amino acids of Pc2 are crucial for E3 activity, as indicated by Pc2 amino-terminal deletions. In order to further elucidate the function of the amino-terminus of Pc2 in stimulating SUMO transfer to CtBP, a fusion protein with the first 288 amino acids of Pc2 and the CtBP-interacting motif (PIDLR) was generated (see Figure 5E). This construct was transfected into COS-1 cells with T7-CtBP, H6-SUMO1 and Fl-Ubc9. When cell lysates were analyzed by Western blotting with a T7 antibody, it was evident that Pc2(2–288)-PIDLR was able to enhance sumoylation of CtBP (Figure 5A). The increase in sumoylation of CtBP by Pc2(2–288)-PIDLR was not as strong as with full-length Pc2, but it was significantly higher than with Pc2(2–288). This increased SUMO modification of CtBP by Pc2(2–288)-PIDLR was dependent on overexpression of Ubc9 (Figure 5B). A YFP fusion with the PLDLS and NLS from the adenovirus E1A protein failed to stimulate CtBP sumoylation (Figure 5A and B). The inability of the YFP-PLDLS construct to enhance sumoylation of CtBP is not due to a lack of interaction, since binding of the two proteins can be detected by immunoprecipitation (Figure 5C). The E3 activity of Pc2(2–288) requires an interaction with CtBP since CtBP co-precipitated with Fl-Pc2(2–288)-PIDLR but not Fl-Pc2(2–288) (Figure 5D). Importantly, CtBP interacted much less efficiently with Pc2(2–288)-PIDLR than with the YFP-PLDLS fusion.
Figure 5.
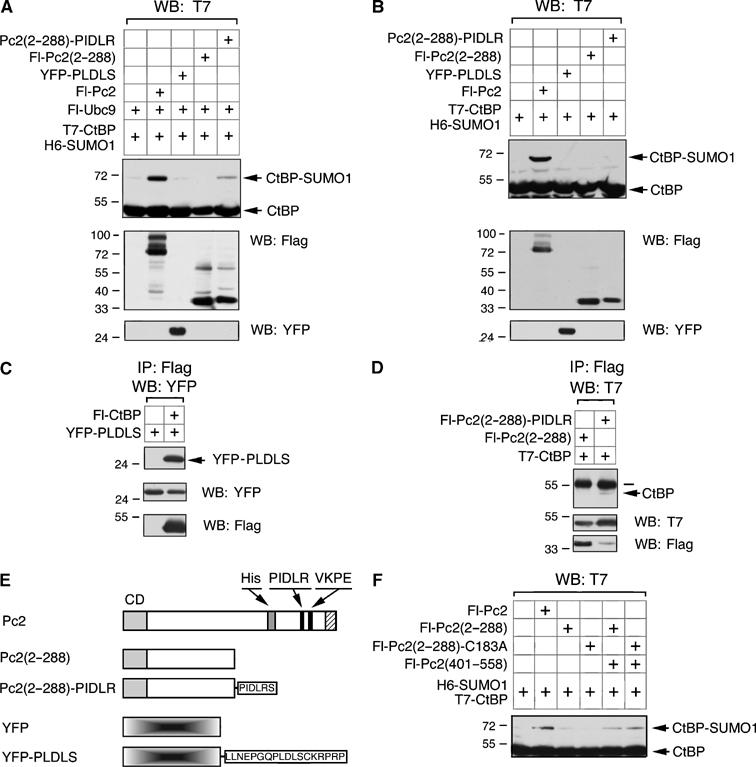
Pc2(2–288) enhances sumoylation of recruited CtBP. (A) COS-1 cells were cotransfected with T7-CtBP, H6-SUMO1 and Fl-Ubc9 either alone or with the indicated Fl-Pc2 or YFP constructs. Following lysis, proteins were separated by SDS–PAGE and analyzed with a T7 antibody to determine relative levels of CtBP sumoylation. Expression of the Flag and YFP constructs is shown below. (B) COS-1 cells were transfected and analyzed as in panel A, except that Fl-Ubc9 was not present. (C, D) COS-1 cells were transfected with the indicated CtBP, Pc2 and YFP expression constructs. Cell lysates were precipitated with anti-Flag-agarose and analyzed for co-precipitating YFP-PLDLS (C) or T7-CtBP (D). Expression of YFP, Flag and T7 constructs is shown below. The bar denotes the immunoglobulin heavy chain. (E) The expression constructs used in (A–D) are shown schematically (labeling as in Figure 3). (F) T7-CtBP and H6-SUMO1 were coexpressed alone or with Fl-Pc2, Fl-Pc2(401–558), Fl-Pc2(2–288) or a mutant in which cysteine 183 has been altered to alanine. Cell lysates were analyzed by Western blot with a T7 antibody to detect SUMO modified and unmodified CtBP. The positions of molecular weight markers (kDa) are indicated.
It appears that the amino-terminal 288 amino acids of Pc2 play an important role in CtBP sumoylation, but are unable to recruit CtBP by themselves. For HECT domain E3s, the ubiquitin is transferred to a cysteine in the E3 and then onto the substrate. Within the amino-terminal region of Pc2, there is only a single cysteine, at amino acid 183. To test whether Pc2 may function similarly to HECT E3s, we altered C183 to alanine within the context of the Pc2(2–288) construct (creating Pc2(2–288)-C183A). As shown in Figure 5F, when coexpressed with Pc2(401–588), this mutant stimulated CtBP sumoylation as efficiently as the wild-type Pc2(2–288), suggesting that this region of Pc2 does not function in a manner analogous to HECT domain ubiquitin E3s.
In vitro E3 activity of the Pc2 amino-terminal region
To test whether Pc2(2–288)-PIDLR has SUMO E3 activity, we performed in vitro sumoylation reactions using GST-Pc2(2–288)-PIDLR or GST-Pc2(2–288) that had been expressed and purified from bacteria. Recombinant E1 (Aos1/Uba2), T7-CtBP and SUMO1 were incubated with increasing concentrations of GST-Pc2(2–288)-PIDLR in the presence or absence of E2 (Ubc9). Increasing amounts of GST-Pc2(2–288)-PIDLR resulted in enhanced sumoylation of CtBP, indicating that in vitro this Pc2 amino-terminal fusion has E3 activity (Figure 6A). Interestingly, GST-Pc2(2–288) also functioned in a similar manner in vitro, suggesting that this region of Pc2 is sufficient to promote sumoylation once it has access to substrate (Figure 6B). No modified CtBP was observed in the absence of either SUMO1 or Ubc9, even with maximal amounts of either Pc2 construct (Figure 6C). In vivo, Pc2(2–288) requires the artificial recruitment of CtBP through the PIDLR motif, but in vitro, recruitment does not appear to be necessary. Most likely this is due to the high concentration of proteins in the in vitro reactions bypassing the need for active recruitment of CtBP by Pc2.
Figure 6.
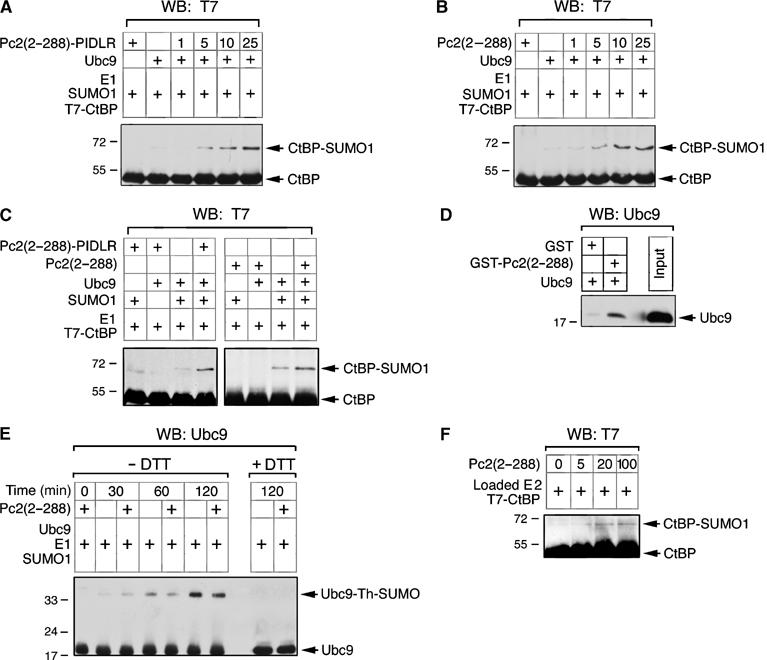
Pc2(2–288) stimulates CtBP sumoylation in vitro. Recombinant T7-CtBP, E1, SUMO1 and Ubc9 (as indicated) were incubated with increasing amounts (1, 5, 10 or 25 ng) of GST-Pc2(2–288)-PIDLRS (A) or GST-Pc2(2–288) (B). In (C), ‘+' indicates 25 ng GST-Pc2(2–288)-PIDLRS or GST-Pc2(2–288). Reactions were terminated by the addition of SDS loading buffer, and SUMO modified CtBP and unmodified CtBP were detected by T7 Western blot. (D) Either GST or GST-Pc2(2–288) bound to glutathione Sepharose was incubated with recombinant Ubc9. Following extensive washing, bound Ubc9 was analyzed by Western blot with a Ubc9 antibody. The input lane is 100 ng Ubc9. (E) Ubc9, E1 and SUMO1 were incubated in vitro for the indicated times with or without GST-Pc2(2–288), and analyzed by nonreducing SDS–PAGE and Western blot with a Ubc9 antibody. The positions of Ubc9 and Ubc9 with SUMO1 attached via a thioester linkage are indicated. The right-hand lanes are reducing SDS–PAGE to disrupt the thioester. (F) SUMO1-loaded E1 and T7-CtBP were incubated with apyrase and the indicated amounts of Pc2(2–288). CtBP and sumoylated CtBP were detected by Western blot with a T7 antibody. The positions of molecular weight markers (kDa) are indicated.
The enhancement of CtBP sumoylation by GST-Pc2(2–288) in vitro suggests that this region of Pc2 is contacting Ubc9 directly. To test this possibility, we incubated purified Ubc9 with glutathione Sepharose bound either with GST alone or a GST-Pc2(2–288) fusion. The Sepharose was washed extensively and bound Ubc9 was analyzed by Western blotting with a Ubc9 antibody. As shown in Figure 6D, Ubc9 binding to GST-Pc2(2–288) was clearly detectable, whereas little Ubc9 bound to GST alone, suggesting a direct interaction between this domain of Pc2 and Ubc9. In contrast, we were unable to detect a direct interaction between GST-Pc2(2–288) and CtBP (data not shown).
The stimulation of in vitro CtBP sumoylation by GST-Pc2(2–288) suggests that this region of Pc2 is either promoting loading of Ubc9 with SUMO or the transfer of SUMO from a loaded Ubc9. To distinguish between these possibilities, we first analyzed loading of Ubc9 with SUMO1 in vitro. We set up in vitro sumoylation reactions, lacking substrate, with and without GST-Pc2(2–288). Over time, an increasing amount of the SUMO-loaded Ubc9 was detected, and when exposed to reducing conditions, which disrupt the thiol ester linkage, this species was lost (Figure 6E). The rate of Ubc9 loading was not affected by the presence of GST-Pc2(2–288), suggesting that it is functioning at a stage after Ubc9 has become loaded with SUMO. To confirm this, we performed Ubc9 loading reactions, and then added apyrase to deplete ATP and inactivate the E1 preventing further Ubc9 loading. The preloaded Ubc9 was then incubated with CtBP in the absence of ATP. As shown in Figure 6F, increasing amounts of GST-Pc2(2–288) resulted in a small but reproducible stimulation of transfer of SUMO from the preloaded Ubc9 to CtBP. Together, these results suggest that the amino-terminal region of Pc2 has E3 activity in vitro.
The carboxyl-terminal region of Pc2 recruits Ubc9
The recruitment of Ubc9 and CtBP by Pc2(401–558) in vivo suggests that this region of Pc2 may act as a platform that can bring together E2 and substrate. Alternatively, the interaction with Ubc9 may simply reflect a substrate–E2 interaction. To distinguish between these possibilities, we analyzed the function of this domain in more detail. Pc2 contains a single consensus sumoylation motif (VKPE, amino acids 491–494). Mutation of lysine 492 to arginine, in the context of full-length Pc2, resulted in a dramatic reduction in the amount of sumoylated Pc2, when coexpressed in COS-1 cells with SUMO1 (Figure 7A). Thus K492 appears to be the major sumoylation site in Pc2. Neither the K492R mutation nor a 10-amino-acid deletion surrounding the consensus site affected the ability of full-length Pc2 to promote CtBP sumoylation in transfected COS-1 cells (Figure 7B). To test whether the carboxyl-terminus of Pc2 contributes anything other than interaction with CtBP via the PIDLR motif to in vivo E3 function, we created a series of carboxyl-terminal deletions (see Figure 7E). Deletion to amino acid 486, which removes the sumoylation site but not the PIDLR motif, did not affect E3 activity in transfected COS-1 cells, whereas further truncation to amino acid 469, removing the PIDLR, abolished E3 activity toward CtBP (Figure 7C). These results suggest that if a Ubc9 docking site exists, it must lie between amino acids 401 and 469.
Figure 7.
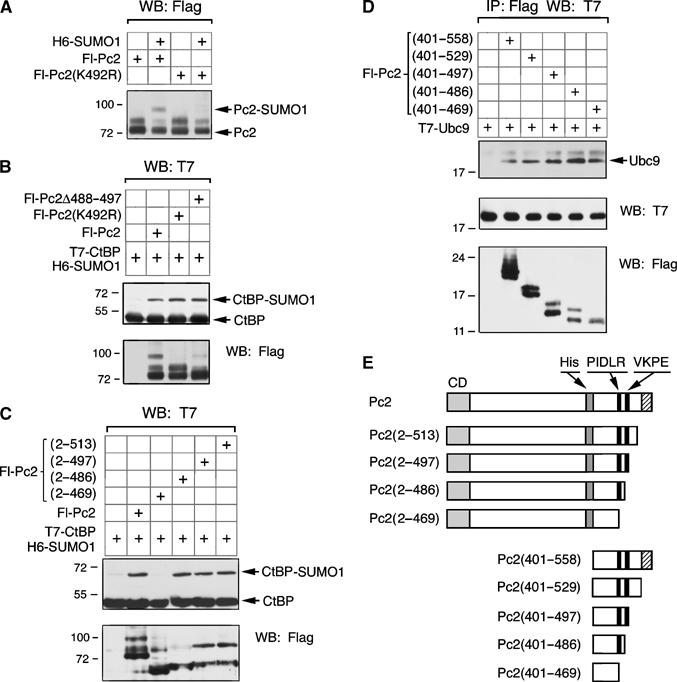
Analysis of the carboxyl-terminal region of Pc2. (A) COS-1 cells were transfected with Fl-Pc2 or a mutant form in which lysine 492 is altered to arginine, either with or without H6-SUMO1. Lysates were analyzed by Flag Western blot and the unmodified and sumoylated Pc2 are indicated. (B) COS-1 cells were transfected with T7-CtBP, H6-SUMO1 and the indicated Flag-Pc2 constructs. Cell lysates were analyzed by Western blot with a T7 antibody to detect SUMO modified and unmodified CtBP. Expression of Flag constructs is shown below. (C) A series of carboxyl-terminal deletions of Pc2 was tested for their ability to enhance SUMO modification of CtBP in COS-1 cells. (D) COS-1 cells were transfected with the indicated Fl-Pc2 expression constructs and T7-Ubc9. Protein complexes were precipitated with anti-Flag-agarose and analyzed by Western blot for T7-Ubc9. Expression of Ubc9 and the Fl-Pc2 constructs in the lysate was analyzed by direct Western blot. The positions of molecular weight markers (kDa) are indicated. (E) The Pc2 deletion constructs tested in panels C and D are shown schematically (labeling as in Figure 3).
To determine whether Ubc9 interacts with the carboxyl-terminal region of Pc2 outside the VKPE motif, we transfected COS-1 cells with T7-Ubc9 and a series of Flag-tagged carboxyl -terminal Pc2 deletions, in the context of Pc2(401–558) (see Figure 7E) and isolated proteins on anti-Flag-agarose. All of the deletion constructs tested interacted with T7-Ubc9, including the smallest, which contains only amino acids 401–469, and lacks both the sumoylation consensus site and the CtBP recruitment motif (Figure 7D). This suggests that Ubc9 interaction with this construct is independent of sumoylation of Pc2 itself and is not dependent on CtBP recruitment.
Ubc9 recruitment by the carboxyl-terminal region of Pc2 contributes to E3 activity
Since it appears that the carboxyl-terminal region of Pc2 interacts with Ubc9 independent of the sumoylation motif, we next wanted to test the importance of this interaction for Pc2 E3 activity. In an in vitro sumoylation reaction, GST-Pc2(401–558) caused a small but significant increase in CtBP sumoylation when relatively high levels of GST-Pc2(401–558) were added (Figure 8A). To confirm that the GST-Pc2(401–558) fragment was functioning after loading of Ubc9 with SUMO1, we performed Ubc9 loading reactions, then depleted ATP with apyrase, and incubated the preloaded Ubc9 with CtBP and increasing amounts of GST-Pc2(401–558) or GST. Addition of GST-Pc2(401–558) resulted in increased transfer of SUMO1 to CtBP, suggesting that it was acting after the loading of Ubc9. In contrast, the addition of GST alone did not increase CtBP sumoylation. This suggests that even in vitro, adapter function can contribute to sumoylation of CtBP.
Figure 8.
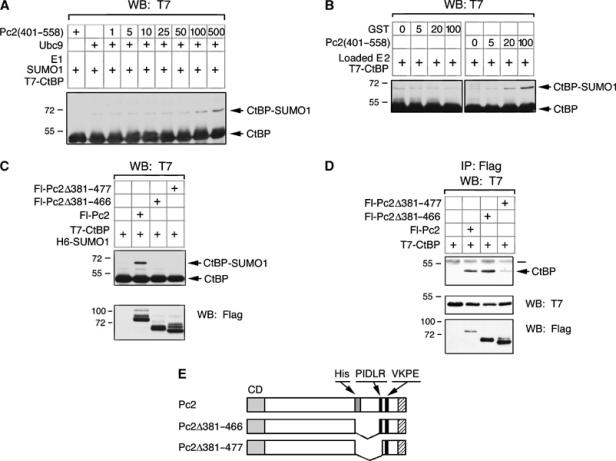
A Ubc9 binding domain required for enhancement of CtBP sumoylation. (A) Recombinant T7-CtBP, E1, SUMO1 and Ubc9 (as indicated) were incubated with increasing amounts (1–500 ng) of GST-Pc2(401–558). (B) SUMO1-loaded E1 and T7-CtBP were incubated with apyrase and the indicated amounts of GST-Pc2(401–558) or GST. Reactions were terminated by the addition of SDS loading buffer and SUMO modified and unmodified CtBP were detected by T7 Western blot. (C) COS-1 cells were transfected with T7-CtBP, H6-SUMO1 and the indicated Flag-Pc2 constructs. Cell lysates were analyzed by Western blot with a T7 antibody to detect SUMO modified and unmodified CtBP. (D) The Fl-Pc2 constructs used in (B) were coexpressed with T7-CtBP in COS-1 cells. Lysates were precipitated with anti-Flag-agarose and analyzed for co-precipitating T7-CtBP. The positions of molecular weight markers (kDa) are indicated. (E) The constructs used in panels C and D are shown schematically.
To determine whether the Ubc9 binding region of Pc2 is important for CtBP modification in transfected COS-1 cells, we expressed CtBP and SUMO1 with either full-length Pc2 or a mutant version lacking amino acids 381–466, which removes the putative Ubc9 recruitment domain, but retains the PIDLR motif. We also tested a second mutant, which additionally removes the PIDLR (Pc2Δ381–477; see Figure 8E). Neither of these deletion mutants stimulated CtBP sumoylation to any detectable level (Figure 8C). To ensure that the Pc2Δ381–466 construct was functional for CtBP interaction, we cotransfected COS-1 cells with the same Flag-Pc2 constructs and T7-CtBP and precipitated proteins on anti-Flag-agarose. As shown in Figure 8D, Pc2Δ381–466, which failed to stimulate CtBP sumoylation, clearly interacted with CtBP. Together, these results suggest the presence of a Ubc9 docking site within the carboxyl-terminal domain of Pc2, which is separable from the Pc2 sumoylation site, but is required for E3 activity in vivo toward CtBP.
Discussion
We previously demonstrated that Pc2 has SUMO E3 activity, both in vivo and in vitro (Kagey et al, 2003). Here we have begun to elucidate the mechanisms by which Pc2 regulates sumoylation. We show that, in vivo, adapter function is necessary for Pc2 to increase modification of CtBP, but is not on its own enough: A second domain in Pc2, which does not interact with CtBP, is required for E3 activity. In vitro, this domain interacts with Ubc9 and has E3 activity, suggesting that the adapter function of Pc2, which stably recruits CtBP and Ubc9, is not required when there is a high concentration of both proteins.
Pc2 adapter function
No enzymatic activity has been demonstrated for the known SUMO E3s, and it is possible that they act primarily as adapters, which bring together substrate and E2 (Johnson and Gupta, 2001; Seeler and Dejean, 2003; Muller et al, 2004). This is mechanistically similar to the function of RING domain ubiquitin E3s, which often form part of a larger E3 complex in which they recruit specific E2s. A carboxyl-terminal fragment of Pc2 (amino acids 401–558) interacts with both Ubc9 and CtBP, and recruits both proteins to polycomb bodies. This polypeptide appears to fulfill the requirements of an adapter, but does not have E3 activity in vivo. Additionally, this region of Pc2 is dispensable for E3 activity in vitro, suggesting that with purified components at relatively high concentration, adapter function is not absolutely necessary, although we cannot rule out the possibility that within the context of full-length Pc2, adapter function contributes to E3 activity in vitro. Structural data on the Ubc9–RanGAP complex and in vitro sumoylation assays in the absence of E3s clearly show that Ubc9 can bind to and modify a sumoylation motif in a target protein, without the need for an adapter (Bernier-Villamor et al, 2002). Thus, adapter function may be an important part of SUMO E3 function in vivo, where substrate and Ubc9 may not be present together at high enough local concentration that efficient sumoylation can occur. In this context, it is interesting that simple corecruitment of CtBP and Ubc9 is not enough to stimulate CtBP sumoylation. One possibility is that the recruitment of Ubc9 to the carboxyl-terminal region of Pc2 is not for CtBP sumoylation but for sumoylation of Pc2 itself. Pc2 is sumoylated and contains a single consensus site within the carboxyl-terminal adapter region. However, our results suggest that recruitment of Ubc9 can be independent of the sumoylation consensus site in Pc2, and that there is a second Ubc9-interacting domain in the carboxyl-terminal region of Pc2. Additionally, we show that deletion of this Ubc9-interacting domain from Pc2 abolishes in vivo E3 function, suggesting that this represents a Ubc9 docking site, which contributes to full E3 activity.
PIASy is a SUMO E3 for the transcription factor LEF1, and it has been shown that deletion of the LEF1-interacting region of PIASy abolishes in vivo SUMO E3 activity (Sachdev et al, 2001). Based on the minimal E3 domain of the related PIAS1, this deletion is unlikely to affect E3 activity other than as a consequence of substrate recruitment (Liang et al, 2004). Additionally, in vitro interaction of purified Ubc9 and PIAS proteins is detectable (Kotaja et al, 2002), suggesting that, as for Pc2, the PIAS proteins perform an adapter function. RanBP2 interacts with Ubc9, and a small domain (the I1I2 domain) can sequester Ubc9, suggesting a stable interaction (Pichler et al, 2002; Saitoh et al, 2002). In contrast to Pc2 and PIAS proteins, there is no evidence for specific substrate binding sites on RanBP2, although it is possible that the docking of transport factors onto RanBP2 as they pass through the NPC serves a substrate recruitment function (Pichler et al, 2002). Based on our results, it appears that adapter function is not the only component of SUMO E3 activity, but with all three classes of SUMO E3, it appears to play a role, although perhaps to differing degrees.
A second domain in Pc2 contributes to E3 activity
In addition to adapter activity, we show that a region in the amino-terminus of Pc2 has E3 activity in vitro. This domain stimulates transfer of SUMO from Ubc9 to CtBP in vitro and is required for E3 activity in vivo. This domain interacts directly with Ubc9 and stimulates CtBP sumoylation in vitro, but we have been unable to detect an interaction with CtBP. The I1I2 domain of RanBP2 appears to function similarly, in that it stimulates sumoylation in vitro, but does not interact with substrate (Pichler et al, 2002). This domain of RanBP2 stimulates the transfer of SUMO1 from a preformed Ubc9-SUMO complex, and we show that the amino-terminal domain of Pc2 also performs this function. It has been suggested that RanBP2 allosterically alters Ubc9, such that it has increased affinity for specific substrates or lysine residues in a specific context. As yet we have no evidence that the amino-terminal domain of Pc2 alters the affinity of Ubc9 for substrate, but this is clearly a possibility. An alternative is that the amino- and carboxyl-terminal domains cooperate to recruit Ubc9. Comparison in vitro of the amino-terminal domain with larger fragments, which also contained the Ubc9 docking site, did not reveal a significant difference in activity (data not shown). However, both domains are clearly important in vivo where Ubc9 is more limiting, so we cannot rule this out.
E3 activity of PIAS proteins appears to require a relatively large minimal region, including the RING-like domain, and mutations that disrupt the structure of the RING domain prevent E3 activity (Liang et al, 2004). However, it is not known how these proteins stimulate sumoylation. Sequence comparison of Pc2, the I1I2 region from RanBP2 and PIAS proteins has not revealed any convincing similarity. Neither Pc2 nor the RanBP2 E3 domain contains an obvious zinc coordinating domain. It is possible that all three proteins have a similar effect on Ubc9 activity, but interact via different motifs, or that they function differently from each other. However, it is likely that at least Pc2 and RanBP2 contain functional domains that are capable of stimulating sumoylation reactions. Mutation of the cysteine residues within the I1I2 domain of RanBP2 demonstrated that this region is unlikely to act in a manner analogous to HECT domain ubiquitin E3s, where the ubiquitin is transferred to the catalytic cysteine (Pichler et al, 2002). Mutation of the single cysteine residue within the amino-terminal domain of Pc2 did not affect its activity in vivo, suggesting that like RanBP2, there is no requirement for transfer of SUMO from the E2 to the E3 prior to substrate sumoylation.
Sumoylation and polycomb complexes
Coexpression of the amino- and carboxyl-terminal domains of Pc2 as separate polypeptides results in a functional E3 complex in vivo, and we show that these regions of Pc2 interact in cells. This interaction is dependent on the extreme carboxyl-terminal region of Pc2, which has also been shown to be important for interaction with the polycomb protein, RING1 (Satijn and Otte, 1999). We do not know whether Pc2–Pc2 interaction is direct or requires RING, or another protein, raising the possibility that, in vivo, Pc2 functions as part of a larger E3 complex. Clearly, other proteins are not absolutely required, since Pc2, or its isolated amino-terminal domain, has E3 function in vitro. However, other polycomb proteins may play a structural or regulatory role in vivo, or may recruit other substrates to Pc2 for sumoylation. It is also possible that other polycomb proteins are themselves targets for SUMO modification, dependent on interaction with Pc2.
Deletion of carboxyl-terminal 29 amino acids of Pc2 does not prevent E3 activity toward CtBP in the context of an otherwise wild-type Pc2 protein. This deletion does, however, delocalize Pc2 from polycomb complexes, suggesting that Pc2 might regulate sumoylation at other locations in the nucleus. It is possible that Pc2 is recruited to a specific DNA response element via interaction with a DNA binding protein and enhances sumoylation of DNA-bound transcriptional regulatory complexes. Additionally, SUMO modification of histone H4 has been demonstrated (Shiio and Eisenman, 2003), and based on the likely interaction of the Pc2 chromodomain with methylated lysine 27 of histone H3 (Fischle et al, 2003; Min et al, 2003), it is tempting to speculate that Pc2 could be recruited to methylated lysine 27 of H3 and then stimulate sumoylation of H4.
Two functional domains in Pc2
Taken together, our results demonstrate that the carboxyl-terminal region of Pc2 recruits E2 (Ubc9) and substrate (CtBP), and also interacts with the amino-terminal domain. Thus this region of Pc2 may form a scaffold, which positions the E2, substrate and the Pc2 amino-terminal domain, such that transfer of SUMO from E2 to substrate is facilitated by the action of the amino-terminal domain. The amino-terminus of Pc2 is functionally separable from the carboxyl-terminal scaffold region, and in isolation has E3 activity, at least in vitro. However, in vivo, this domain minimally also needs a substrate recruitment motif. It will now be of interest to determine whether other Pc2 substrates exist and if so, how they are recruited to the E3 complex.
In summary, we show that in vivo Pc2 requires two separate activities to act as a SUMO E3: an adapter function, which recruits substrate and E2, and a second activity, which is functionally distinct and in isolation has E3 activity in vitro. These results suggest that SUMO E3s are more than simple adapters.
Materials and methods
Cell culture and transfection
COS-1 cells were grown in DMEM supplemented with 10% FBS. Transfections were carried out in six-well plates or 60 mm dishes with LipofectAMINE (Invitrogen) according to the manufacturer's protocol.
Plasmids
T7-CtBP, H6-SUMO1, Fl-Ubc9, T7-Ubc9, Fl-PIASxα, HA-Smad4 and all Fl-Pc2 constructs were expressed from pCMV5 plasmids with amino-terminal T7, Flag, six histidine or HA epitope tags (Kagey et al, 2003). Fl-PIASxα was generated by PCR from an EST clone. Amino- and carboxyl-terminal Pc2 deletions were generated by PCR. Internal Pc2 deletions were created by PCR and replaced with an Asp718 site. Mutations and deletions were verified by sequence analysis as necessary. PIDLR fusions to Pc2(2–288) or YFP were constructed by inserting a double-stranded oligonucleotide into pCS2 with YFP. For the Pc2(2–288) fusions, the YFP was removed by digestion and replaced with Pc2(2–288). Fluorescent proteins (eYFP, eCFP; Clontech) or fusions with the YFP or CFP at the amino-terminus were expressed from pCS2. Bacterial expression constructs are as previously described (Kagey et al, 2003), except for GST fusions to Pc2(2–288), Pc2(2–288)-PIDLR and Pc2(401–558), which were expressed from pGEX-4T1 (Amersham Pharmacia).
Co-immunoprecipitation and Western blot
COS-1 cells were lysed by sonication in phosphate-buffered saline with 1% NP-40 (PBS-N) and protease inhibitors (Complete MINI; Roche) or MSLD buffer (50 mM Hepes, pH 7.4, 10% glycerol, 100 mM NaCl, 0.1% Tween 20 and protease inhibitors). Cell debris was removed by centrifugation and lysates were subjected to immunoprecipitation with anti-Flag-agarose (Sigma). The anti-Flag-agarose was washed and precipitating and co-precipitating proteins were removed by boiling in SDS–PAGE loading buffer. A portion of the lysate was removed prior to the addition of anti-Flag-agarose for expression controls. Proteins were separated by SDS–PAGE and transferred to Immobilon-P (Millipore) membrane. Blots were incubated with anti-T7 (Novagen) or anti-GFP antisera (Covance) and HRP-conjugated goat anti-mouse antibody (Pierce). Proteins were visualized by ECL (Amersham Pharmacia). For direct Western, cells were boiled in SDS–PAGE loading buffer. Proteins were separated by SDS–PAGE, transferred to Immobilon-P and visualized with the appropriate antisera (anti-Flag M2, Sigma; anti-T7, Novagen; anti-HA, Covance; anti-GFP, Covance) and HRP-conjugated goat anti-mouse antibody (Pierce).
Recombinant proteins and in vitro sumoylation
GST fusion proteins were expressed in BL21 and proteins were purified on a 1 ml Hi-Trap GST column (Amersham Biosciences) using a linear glutathione gradient (0–40 mM). E1 (Aos1/Uba2), T7-CtBP-H6, SUMO1 and Ubc9 were expressed in BL21 or BL21-CodonPlus-RP (Stratagene) and purified as described (Kagey et al, 2003). In vitro sumoylation reactions were performed basically as described (Schmidt and Muller, 2002). Briefly, reactions were carried out at 30°C for 2 h with 100 ng of E1, 200 ng of Ubc9, 2 μg of SUMO1 and 250 ng of T7-CtBP-H6 in the presence or absence of increasing amounts of GST-Pc2(2–288), GST-Pc2(2–288)-PIDLR or GST-Pc2(401–558). Reactions were terminated by addition of SDS–PAGE loading buffer. Ubc9 loading and transfer reactions were performed essentially as described by Pichler et al (2002). In vitro binding between Ubc9 and GST-Pc2(2–288): GST or GST-Pc2(2–288) was bound to glutathione Sepharose and incubated with 1 μg of purified Ubc9 at 4°C in transport buffer with 0.2% NP-40. Sepharose was washed extensively and bound proteins were analyzed by Western blot with a Ubc9 antibody (Boston Biochem).
Live cell imaging
COS-1 cells were transfected, stained and imaged as previously described (Kagey et al, 2003). Images were captured with a Hammamatsu Orca II cooled CCD camera controlled by Openlab 3.1.4 (Improvision) and manipulated in Photoshop 6.0.
Supplementary Material
Supplemental Figure 1
Supplemental Figure 1 Legend
Acknowledgments
This work was supported by an NIH grant to DW (HD 39926).
References
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002) Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 [DOI] [PubMed] [Google Scholar]
- Desterro JM, Thomson J, Hay RT (1997) Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett 417: 297–300 [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A (1995) An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem 270: 18099–18109 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (2003) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521 [DOI] [PubMed] [Google Scholar]
- Hay RT (2001) Protein modification by SUMO. Trends Biochem Sci 26: 332–333 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA 92: 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD (2000) The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol 10: 429–439 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272: 26799–26802 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J 16: 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Wotton D (2003) The polycomb protein Pc2 is a SUMO E3. Cell 113: 127–137 [DOI] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, Yasuda H (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell 8: 713–718 [DOI] [PubMed] [Google Scholar]
- Kennison JA (1995) The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet 29: 289–303 [DOI] [PubMed] [Google Scholar]
- Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, Dejean A (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J 21: 2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Janne OA, Palvimo JJ (2002) PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol 22: 5222–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Chang C, Liu D, Derynck R (2003) Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J Biol Chem 278: 27853–27863 [DOI] [PubMed] [Google Scholar]
- Liang M, Melchior F, Feng XH, Lin X (2004) Regulation of Smad4 sumoylation and transforming growth factor-{beta} signaling by protein inhibitor of activated STAT1. J Biol Chem 279: 22857–22865 [DOI] [PubMed] [Google Scholar]
- Lin X, Sun B, Liang M, Liang YY, Gast A, Hildebrand J, Brunicardi FC, Melchior F, Feng XH (2003) Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol Cell 11: 1389–1396 [DOI] [PubMed] [Google Scholar]
- Long J, Wang G, He D, Liu F (2004) Repression of Smad4 transcriptional activity by SUMO modification. Biochem J 379: 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 96: 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F (2000) SUMO—nonclassical ubiquitin. Annu Rev Cell Dev Biol 16: 591–626 [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM (2003) Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J (1995) Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J 14: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Ledl A, Schmidt D (2004) SUMO: a regulator of gene expression and genome integrity. Oncogene 23: 1998–2008 [DOI] [PubMed] [Google Scholar]
- Ohshima T, Shimotohno K (2003) Transforming growth factor-beta-mediated signaling via the p38 MAP kinase pathway activates Smad-dependent transcription through SUMO-1 modification of Smad4. J Biol Chem 278: 50833–50842 [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Poux S, McCabe D, Pirrotta V (2001) Recruitment of components of Polycomb group chromatin complexes in Drosophila. Development 128: 75–85 [DOI] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev 15: 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Pizzi MD, Wang J (2002) Perturbation of SUMOlation enzyme Ubc9 by distinct domain within nucleoporin RanBP2/Nup358. J Biol Chem 277: 4755–4763 [DOI] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ (2001) The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem 276: 21664–21669 [DOI] [PubMed] [Google Scholar]
- Satijn DP, Otte AP (1999) RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol 19: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn DP, Olson DJ, van der Vlag J, Hamer KM, Lambrechts C, Masselink H, Gunster MJ, Sewalt RG, van Driel R, Otte AP (1997) Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol Cell Biol 17: 6076–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Shiels C, Williamson J, Satijn DP, Otte AP, Sheer D, Freemont PS (1998) The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol 142: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75: 495–505 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Muller S (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA 99: 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Hochstrasser M (2003) A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci 28: 321–328 [DOI] [PubMed] [Google Scholar]
- Seeler JS, Dejean A (2003) Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4: 690–699 [DOI] [PubMed] [Google Scholar]
- Sewalt RG, Gunster MJ, van der Vlag J, Satijn DP, Otte AP (1999) C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol Cell Biol 19: 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN (2003) Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA 100: 13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J (1995) Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol 7: 376–385 [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W (1992) Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114: 493–505 [DOI] [PubMed] [Google Scholar]
- Simon JA, Tamkun JW (2002) Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev 12: 210–218 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Kahyo T, Toh EA, Yasuda H, Kikuchi Y (2001) Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J Biol Chem 276: 48973–48977 [DOI] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Kamitani T (2000) Ubiquitin-like proteins: new wines in new bottles. Gene 248: 1–14 [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102: 533–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1
Supplemental Figure 1 Legend


