Abstract
Metastatic castration recurrent prostate cancer (mCRPC) remains incurable and is one of the leading causes of cancer-related death among American men. Therefore, detection of prostate cancer (PCa) at early stages may reduce PCa-related mortality in men. We show that lipid quantification by vibrational Raman Microspectroscopy and Biomolecular Component Analysis may serve as a potential biomarker in PCa. Transcript levels of lipogenic genes including sterol regulatory element-binding protein-1 (SREBP-1) and its downstream effector fatty acid synthase (FASN), and rate-limiting enzyme acetyl CoA carboxylase (ACACA) were upregulated corresponding to both Gleason score and pathologic T stage in the PRAD TCGA cohort. Increased lipid accumulation in late-stage transgenic adenocarcinoma of mouse prostate (TRAMP) tumors compared to early-stage TRAMP and normal prostate tissues were observed. FASN along with other lipogenesis enzymes, and SREBP-1 proteins were upregulated in TRAMP tumors compared to wild-type prostatic tissues. Genetic alterations of key lipogenic genes predicted the overall patient survival using TCGA PRAD cohort. Correlation between lipid accumulation and tumor stage provides quantitative marker for PCa diagnosis. Thus, Raman spectroscopy-based lipid quantification could be sensitive and reliable tool for PCa diagnosis and staging.
Keywords: Prostate cancer, Raman microspectroscopy, lipogenesis, TRAMP, biomarker
1. Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related death in American men [1]. Despite recent advances in targeted therapies, specifically against the androgen receptor, metastatic castration-recurrent prostate cancer (mCRPC) remains incurable [2]. By definition, mCRPC is resistant to androgen deprivation therapy (ADT), but most aggressive mCRPCs also develop resistance to various chemotherapeutic agents [3]. Thus, alternative approaches to detect this advanced disease state must be identified to prevent progression and recurrence of PCa.
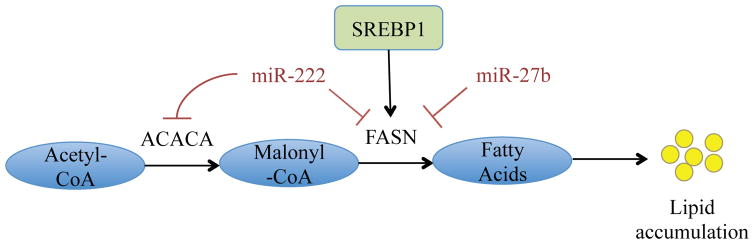
It is established that tumor cells require more energy than normal cells and metabolic alterations are required to sustain the proliferative potential of growing tumors [4, 5]. One such metabolic alteration is endogenous or de novo lipogenesis by cancer cells, which usually takes place in liver and adipose tissues but not in other normal tissues [6]. Several recent studies determined that cancer cells of numerous origins fulfill their requirement of fatty acids through de novo lipogenesis [7–9]. Due to their high proliferation rate, cancer cells require a substantial amount of metabolic energy in order to synthesize cellular and subcellular membranes, which are mainly composed of fatty acids. Lipids in cancer cells are the main fuel source for membrane synthesis and act as one of the major drivers in tumor cells, especially PCa, for growth and membrane synthesis [10]. Moreover, the rate of fatty acid synthesis and accumulation has been reported to accelerate in hypoxic PCa cells and tumor core as a survival response to hypoxic stress [11]. Following re-oxygenation, the accumulated lipids support growth and aggressiveness of PCa [8, 11]. Recently, Rysman et al. reported that de novo lipogenesis protects cancer cells from endogenous and exogenous stresses like ROS and chemotherapeutic drugs. These evidences further emphasize the importance of de novo lipogenesis in averting cancer cell death causing cancer progression [12]. For example, de novo lipogenesis protects cancer cells from lipid peroxidation and subsequent cell death due to free radicals by enhancing saturation of membrane lipids [12]. A brief overview of the key lipogenic steps is highlighted in Figure 1. Briefly, pyruvate is synthesized from glucose by glycolysis, and this pyruvate is converted to Acetyl-CoA either inside mitochondria (via pyruvate dehydrogenase complex) or in the cytosol through citrate derived from TCA cycle. In the cytosol, Acetyl-CoA is carboxylated by acetyl-CoA carboxylase (gene: ACACA, protein: ACC) to form malonyl-CoA. In the next rate-limiting step, malonyl-CoA is synthesized into palmitic acid and various other fatty acids via fatty acid synthase (FASN). These fatty acids are accumulated and stored as lipid droplets, which can be used as fuel source in proliferating tumor cells. FASN and other enzymes involved in de novo lipogenesis are under control of the lipogenic transcription factor sterol regulatory element binding protein 1 (SREBP-1). In addition, miRNAs miR-27b and miR-222 have also been predicted to negatively regulate lipogenesis, as miR-27b targets FASN and miR-222 targets both the ACACA and FASN mRNA for degradation [2].
Figure 1. Schematic of key steps in lipogenesis.
Acetyl-CoA is carboxylated by acetyl-CoA carboxylase (ACC) to form malonyl-CoA. In the rate-limiting step, malonyl-CoA is synthesized into palmitic acid and various other fatty acids via fatty acid synthase (FASN). These fatty acids are accumulated and stored as lipid droplets, which can be used as fuel source in proliferating tumor cells.
A link between lipogenesis and PCa progression has been investigated in the past few decades [8, 12–15]. Various studies have determined that inhibition of the key lipogenic enzyme FASN in PCa cells slows growth and proliferation, while increasing apoptosis of PCa cells [16–18]. In addition, ACC inhibition in PCa cells resulted in sensitization to chemotherapeutic agents [12]. De novo lipogenesis is also crucial for development of mCRPC because of reactivation of AR signaling by de novo steroidogenesis [19, 20]. In this manuscript, we implemented a systemic approach to verify the role of lipogenesis in PCa progression and validate the impact of lipogenic transcript levels for the evaluation of PCa biomarkers. Specifically, we employed vibrational Raman Microspectroscopy and Biomolecular Component Analysis (BCA) approach to quantify expression of major types of biomolecules: DNA, proteins and lipids in prostate tumor tissues [21]. This label-free optical approach has unraveled a significant correlation elevation in the lipid content during the PCa progression. We suggest that macromolecular lipid quantification by microRaman-BCA approach can serve as quantitative marker for general categorization of the lesions as well as for clinical prognosis.
2. Materials and Methods
2.1. Analysis from publicly available datasets
The TCGA prostate adenocarcinoma (PRAD) dataset (version 2016-08-16) was retrieved from the UCSC Xena Browser. The polyA+ IlluminaHiSeq gene expression dataset was downloaded in its normalized format. RNA-seq values were grouped between tumor and matched-normal samples and the tumor samples were further divided according to Gleason score or pathologic T-score. The average transcript reads were calculated for each group. The RPKM method was used to quantify gene expression from RNA sequencing data by normalizing for total read length and the number of sequencing reads. A student’s T-test was performed between matched normal and each Gleason score or pathologic T-score. cBioportal [22, 23] was used to download the Trento/Cornell/Broad Neuroendocrine (2016) dataset and the PRAD TCGA (provisional) dataset for heat map generation, OncoPrint analyses, co-expression analyses, and survivorship analysis.
2.2. Whole tissue lysates preparation and Western blotting
Murine prostate tumors (TRAMP) were generated as previously described [24]. Briefly, mice hemizygous for the Pb-Tag transgene were crossed with non-transgenic FVB mice to obtain non-transgenic (WT) and transgenic (TRAMP) males. Animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee at the department of Laboratory Animal Resources, Roswell Park Cancer Institute, Buffalo, NY. Whole tissue lysates from WT prostate and TRAMP tumor tissues were prepared by homogenization and lysed in RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 1 mM EDTA, 1 mM dithiothreitol (DTT) supplemented with protease inhibitors (1 mM PMSF, 1% aprotinin, 1 mM leupeptin, 1 mg/ml pepstatin A and 1 mg/ml chymostatin). Thirty micrograms protein samples were loaded on 4–20% SDS polyacrylamide gels for Western blotting. The membrane was probed with indicated primary antibodies and corresponding HRP-conjugated secondary antibodies followed by immunodetection using ECL reagent (BioRad, Hercules, CA). Antibodies against SREBP1 and β-Actin-HRP were procured from Santa Cruz Biotech, Dallas, TX. FASN, ACC, AceCS1, ACL, and ACSL1 antibodies were procured from Cell signaling, Danvers, MA.
2.3. Raman Micro-spectroscopy
A confocal Raman microscope used to acquire spectra from individual tissue cells is equipped with an inverted Nikon TE200 microscope, fiber-coupled MS3501i imaging monochromator/spectrograph (Solar TII), a Hamamatsu S9974 CCD camera cooled down to −60°C, and a 532 nm DPSSL excitation laser. This configuration enables the measurements within the range of Raman shifts of 600–3000 cm−1. The spectral resolution for the fixed diffraction grating position (wave number interval of 1210 cm−1) was ~1.5 cm−1. An excitation laser beam of ~20 mW power was focused onto the sample in a spot of ~0.8 μm, using a 100× NA=1.3 Nikon oil-immersion objective lens. A 100 μm pinhole ensures for confocal acquisition of Raman signal. The integration time for spectra acquisition was 20 sec for all our experiments. The measurements performed in bovine serum albumin solution demonstrated that experimental error of Raman spectra measurements does not exceed 5% within the Raman shift range of 700–1700 cm−1.
2.4. Biomolecular component analysis
For assessment of intranuclear macromolecular content the biomolecular component analysis (BCA) [27–29] was applied to pre-processed Raman spectra. This method is based on an accurate spectral fit of a model spectrum - the linear summation of the weighted spectra of the basic components (Linear Combination Modeling (LCM) [30–33]), into a pre-processed Raman spectrum of nuclei. The spectral weights (coefficients), which are varied during the fitting procedure, are considered as the specific contributions of the basic spectra into the resulting spectrum and relate directly to the concentrations of basic macromolecules. In our case LCM of the acquired Raman spectra was utilized for experimental evaluation of the local biomolecular composition tissue cells nuclei by generating a model spectrum through a linear summation of weighted Raman spectra of the basic classes of bio-molecules, DNA, RNA, proteins, lipids, which make the largest contribution to nuclear Raman spectrum [28]. In other words it means to obtain a numerical value of the fractional fit contribution (weight, Ci) of each component, i, to Raman spectrum of measured nuclear domain (rtotal): rtotal = C1r1+C2r2+C3r3+C4r4, where ri is Raman spectrum of i-component. More detailed description of BCA used in this study can be found in our previous publications [28, 29]. For initial step of BCA we used protein component (r1), obtained from our previous studies of HeLa cell culture corresponding to 100 ml/mg of bovine serum albumin (C1=1) equivalent of concentration. Preprocessed Raman spectra of extracted DNA, RNA, and lipid droplets from HeLa cells, calibrated to 20 mg/ml of calf thymus DNA, S. cerevisiae RNA and bovine heart lipid extract equivalents of concentration accordingly, were used as the reference spectra of the DNA, RNA and lipids components (ri, i=2,3,4 with Ci=1, i=2,3,4).
The LCM algorithm was coded in Matlab environment and includes preprocessing procedures subtracting the background, Savitzky-Golay smoothing (2nd polynom order, 13 points smoothing frame) and baseline correction. Background spectrum was modeled by fitting the five background components for each measurement [29].
2.5 Free fatty acid (FFA) and total cholesterol content estimation
FFA and total cholesterol content in WT prostate and TRAMP tissues were quantitated by free fatty acid fluorometric assay kit and cholesterol fluorometric assay kit, respectively (Cayman Chemical, Ann Arbor) as per manufacturer’s instructions. Briefly, 10 mg of WT prostate and TRAMP tissues were homogenized in 200 μl of Chloroform/Triton X-100 (1% v/v) and incubated on ice for 30 min for FFA extraction. Cholesterol was extracted by homogenization of 10 mg tissue in 200 μl of Chloroform:Isopropanol:NP-40 (7:11:0.1 v/v). Tissue homogenates were centrifuged at 12000 rpm for 10 min at 4°C. Lower organic phase was collected for FFA, whereas all liquid was collected for cholesterol. The collected samples were transferred to new tubes and dried on a heat block at 50°C for 2 hours followed by vacuum dry for 1 hour to remove traces of chloroform. Dried lipids were dissolved in either 200 μl of FFA assay buffer for assaying FFA content or 200 μl of cholesterol assay buffer for assaying total cholesterol content using the instructions provided in the respective assay kits.
3. Results
3.1. Upregulation of key lipogenic genes correlates with the PCa progression
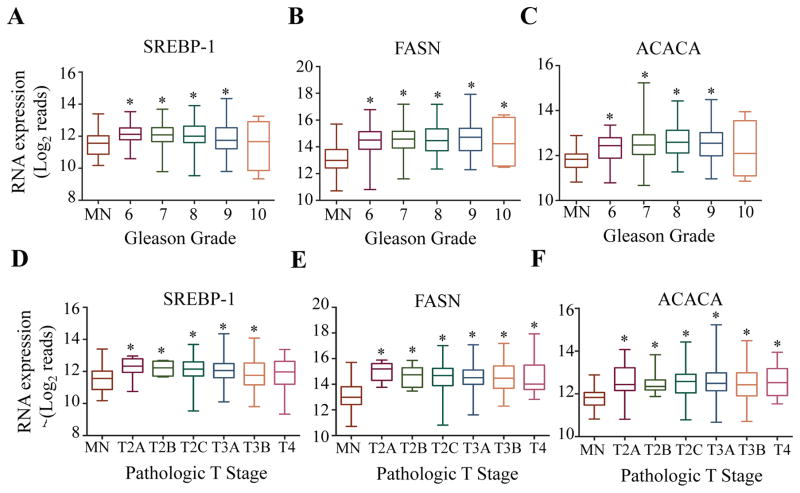
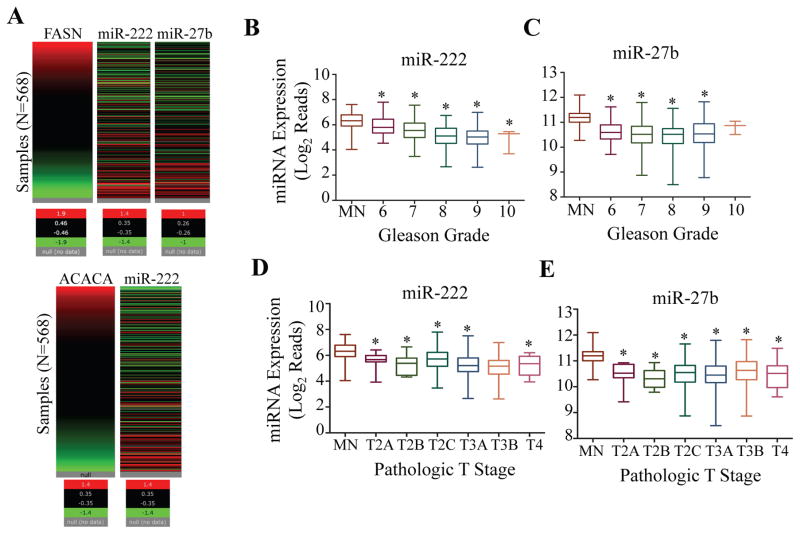
To better understand a link between increased lipogenesis and PCa progression, a large cohort of sample specimens (N = 551) were stratified based on different criteria for disease progression and was correlated to transcript levels of key lipogenic genes (Figure 2A–F). SREBP-1, FASN, and ACACA were all upregulated with respect to patient Gleason score (Figure 2A–C). Similarly, upregulation of SREBP-1, FASN, and ACACA is directly linked to patient T-score (Figure 2D–F). Thus, transcript levels of these key lipogenic genes are increased as the tumor becomes more undifferentiated and metastatic, which is characteristic of aggressive disease. Since transcript levels of these key lipogenic genes are significantly upregulated in prostate tumors, known negative regulators of these genes were assayed to understand if their suppression is correlated to increased lipogenic gene transcription. Indeed, the down regulators of lipogenesis, miR-222 and miR-27b, seem to be functional in this cohort of patients, as an increase in their level of transcript reads negatively correlated to transcript reads of FASN and ACACA and vice versa (Figure 3A). Interestingly, transcript reads of both miRNAs significantly decreased with respect to both Gleason score (Figure 3B and C) and T-score (Figure 3D and E). Thus, miRNA downregulation may contribute to an increased lipogenesis in advanced PCa.
Figure 2. Key lipogenic genes are upregulated with respect to disease progression.
RNA-seq datasets derived from human PCa patient samples were retrieved from TCGA (version 2016-08-16) in their normalized format. RNA-seq values for SREBP-1, FASN and ACACA were grouped by matched normal tissue and Gleason score (A–C) or MN and pathologic T score (D–F). A Student’s T-Test was performed to determine statistical difference between MN and other groups (*=p<0.05)
Figure 3. Negative regulators of key lipogenic genes are repressed with respect to disease progression.
A) Heatmaps representing the correlation between levels of miR-222 and miR-27b with transcript levels of FASN and ACACA, respectively, in the TCGA PRAD dataset (generated via cBioportal). Patient samples were retrieved from TCGA (version 2016-08-16) in their normalized format. miRNA-seq values for miR-222 and miR27b were grouped by matched normal tissue and Gleason score (B&C) or MN and pathologic T score (D&E). A Student’s T-Test was performed to determine statistical difference between MN and other groups (*=p<0.05)
3.2 The lipogenic pathway correlates with aggressive signatures in prostate adenocarcinoma
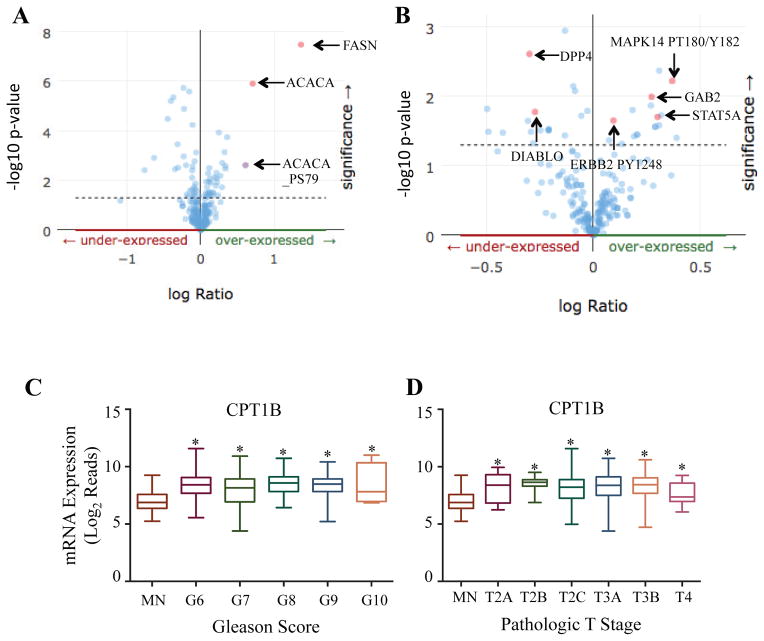
We found that when lipogenic transcription factor SREBP-1 is overexpressed, protein levels of FASN, as well as ACACA and its active state S79 phosphorylation are significantly elevated, suggesting an activation of the lipogenic pathway (Figure 4A). Additionally, when FASN protein is expressed, several notable oncogenes such as MAPK and its active state phospho-T180/Y182, as well as the active state of ERBB2 (phospho-Y1248) were overexpressed (Figure 4B). Interestingly, several tumor suppressor proteins are downregulated when FASN is upregulated, including direct IAP binding protein with low pI (DIABLO) and dipeptidyl peptidase 4 (DPP4) (Figure 4B). Similar to lipogenic genes, CPT1B (carnitine palmitoyltransferase 1B) mRNA expression was also upregulated in accordance with Gleason score and pathologic T stages. CPT1 is a mitochondrial key enzyme for fatty acid oxidation or β-oxidation and therefore important for utilizing the stored lipid for energy demands. Thus, not only does the lipogenic pathway seem to be active in this cohort of PCa patients, but also may correlate with an induction of hallmark proto-oncogenic pathways.
Figure 4. The lipogenic pathway is active in patients with prostate adenocarcinoma and correlates with induction of aggressive pathways.
Patient protein data from TCGA PRAD (Provisional) was acquired via cBioportal. A) Protein co-expression analysis was performed in patients with enriched SREBP-1 expression. Arrow identifies notable proteins that were co-expressed with SREBP-1. B) Protein co-expression analysis was performed in patients with enriched FASN expression. Arrow identifies notable proteins that were co-expressed with FASN. RNA-seq datasets derived from human PCa patient samples were retrieved from TCGA (version 2016-08-16) in their normalized format. RNA-seq values for CPT1B were grouped by matched normal tissue and Gleason score (C) or MN and pathologic T score (D). A Student’s T-Test was performed to determine statistical difference between MN and other groups (*=p<0.05)
3.3. Lipogenesis is upregulated in transgenic adenocarcinoma of mouse prostate (TRAMP) tumors
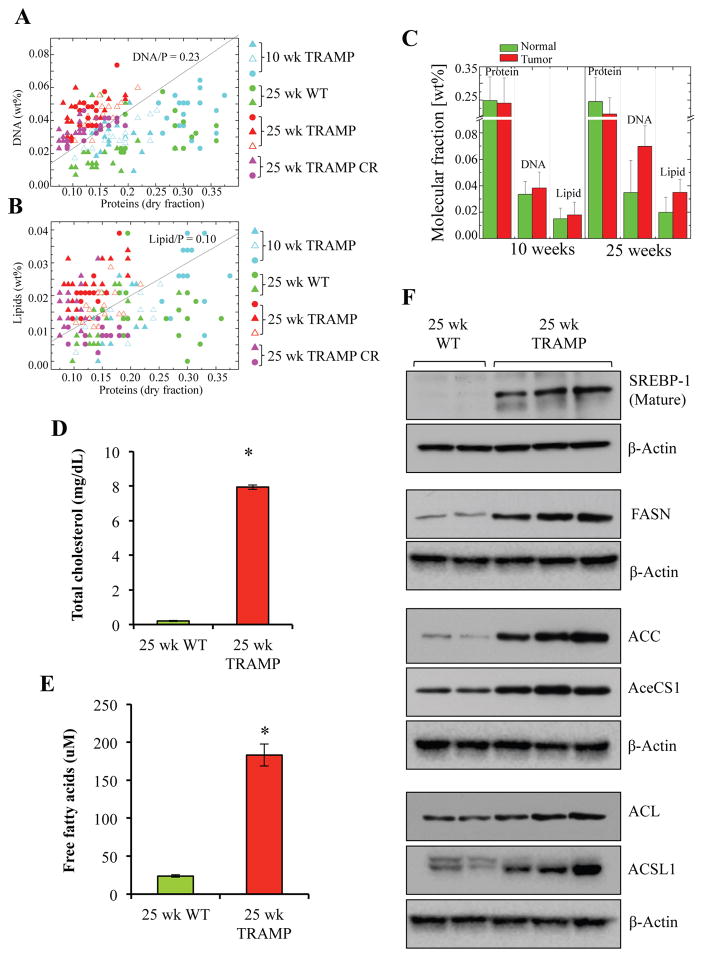
To characterize the resulting impact of lipogenesis on molecular makeup of PCa lesions, levels of lipids, as well DNA and proteins were assayed via Raman microspectroscopy (Figure 5A–C). Prostate tumors produced from TRAMP model are poorly differentiated, mimic the neuroendocrine (NEPC) phenotype of PCa, the predominant phenotype observed in the clinic in response to ADT-resistance and is extremely aggressive [24, 34–36]. Therefore, TRAMP tumors were used for this analysis. In comparison to WT normal prostatic tissues and TRAMP lesions with prostatic intraepithelial neoplasia (PIN), TRAMP tumors had highly elevated levels of both lipid and DNA content. To further correlate lipid profile in WT prostate and tumor tissues, we also assayed FFA and total cholesterol content in these tissues as secondary method to quantitate lipid content in WT and TRAMP prostatic tissues. As shown in Figure 5D and E, the levels of both forms of lipids were significantly elevated in TRAMP tissues compared to WT normal prostatic tissues, thus supporting lipid quantification by Raman microspectroscopy. Increased lipid content indicates elevation of lipogenesis and activation of the lipogenic pathways. To further confirm these results, we assayed the levels of SREBP1 and other enzymes involved in the lipogenesis pathway including FASN, ACC, AceCS1, ACL and ACSL1 in WT prostate and TRAMP tissues by Western blotting. We observed that FASN, ACC, AceCS1, ACL and ACSL were upregulated in TRAMP lysates compared to normal prostate tissues. The levels of processed or mature SREBP-1 (~ 68 kDa) were upregulated in TRAMP tissues compared to WT counterpart.
Figure 5. Lipogenesis is upregulated in poorly differentiated PCa.
A) DNA versus Protein distribution in cell nuclei of different tissue slices B) Lipid versus Protein distribution in cell nuclei of different tissue slices. Ratios DNA/P = 0.23 (A) and Lipids/P = 0.10 (B) represent interface lines between 3normal2 (lower part) and 3tumor2 (upper part) cells. C) Mean value of weight fraction of proteins, DNA and Lipids in normal and tumor cell nucleus. D) Total cholesterol content was measured and quantitates in 25 wk WT and 25 wk TRAMP prostatic tissue as described in material and methods and represented as mg/dL. E) Free fatty acid content was measured and quantitates in 25 wk WT and 25 wk TRAMP prostatic tissue as described in material and methods and represented as μM. A Student’s T-Test was performed to determine statistical difference between WT and TRAMP groups (n=3, *=p<0.05). F) Protein expression levels of SREBP-1 and key lipogenic enzymes like FASN, ACC, AceCS1, ACL and ACSL1 in 25 wk WT and 25 wk TRAMP prostatic tissue lysates were analyzed by western blotting as described in Material and Methods. Membranes were probed with β-Actin as a loading control.
3.4. Lipogenic enzymes are upregulated in human prostate cancer and predict poor prognosis
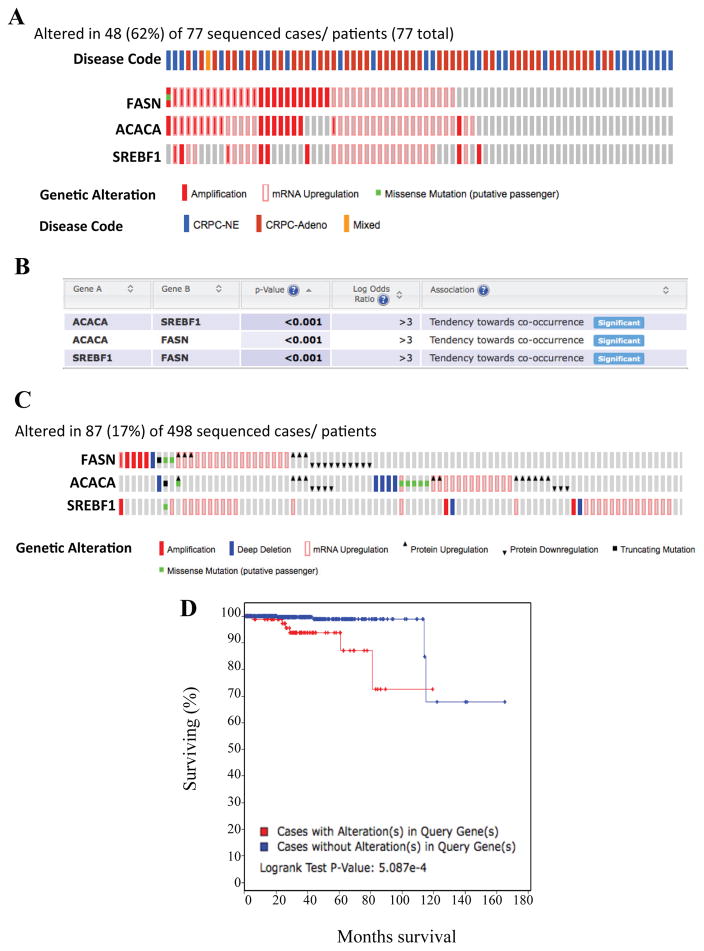
The TRAMP model mainly represents the NEPC phenotype and not adenocarcinoma, however, the cohort of specimens analyzed in previous TCGA figures displayed adenocarcinoma. Therefore, to understand the translational impact of this study related to neuroendocrine disease in humans, we validated these findings using the human NEPC cohort. Notably, gene alterations in ACACA, SREBP1 and FASN occurred in 62% of patients present with CRPC, including both adenocarcinoma and neuroendocrine phenotypes (Figure 6A). Transcript levels of multiple lipogenic genes tend to significantly co-occur together, indicating that lipogenic pathway is indeed active in these patients (Figure 6B).
Figure 6. Alterations in key lipogenic genes predict overall survival in patients with prostate adenocarcinoma.
A) Genetic and mRNA alteration of key lipogenic genes in patients with neuroendocrine PCa, made publicly available by cBioportal. B) Chart depicting the probability that alterations in the key lipogenic genes co-occur together in patients with CRPC (including both PRAD and NEPC phenotypes) made available by cBioportal. C) Genetic and mRNA alterations of key lipogenic genes in patients with prostate adenocarcinoma (TCGA PRAD, provisional), made publicly available by cBioportal. D) Kaplan-Meier curve depicting overall survival between patients with no genetic alterations (blue line) and patients with genetic alterations (red line) in the key lipogenic proteins. Kaplan-Meier curve was generated via cBioportal. Abbreviations: CRPC = castrate recurrent prostate cancer, NE = neuroendocrine, Adeno = adenocarcinoma.
As our data demonstrated that lipogenic genes positively correlate to disease progression in prostate adenocarcinoma (Figure 2), we determined if these key lipogenic genes could predict prognosis. Although genetic alterations in the genes occur in only 17% of patients in this cohort, these alterations are sufficient to predict overall survival (Figure 6C–D). Interestingly, patients without alterations in key lipogenic genes survived longer than patients with alterations. Thus this observation validates our earlier findings and our notion that lipid content measurement using this non-invasive technique can be utilized as a biomarker in PCa diagnosis.
4. Discussion
Lipids are essential macromolecules that are required for cellular viability and prostate cancer progression. Normal cells predominantly utilize circulating lipids derived from diet, but cancer cells synthesize their own lipids by activating de novo lipogenesis pathways [37, 38]. Not surprisingly, aberrant lipogenesis has been extensively studied in various cancers, including prostate to identify new therapeutic targets [8, 39, 40]. These studies also suggest that lipids are accumulated in significant quantities in tumor tissues compared to normal tissues. In this study, we provide evidence using the TRAMP model of PCa that lipid estimation by non-invasive Raman microspectroscopy can be used as a reliable biomarker tool to predict incidence of PCa. In addition, we also broadened the scope of knowledge on the role of lipogenesis in PCa by studying large cohorts of samples from publicly available datasets from TCGA and cBioportal. Through these databases, we verified the previous noteworthy observations that may play a large role in the context of PCa progression. In general, many types of tumor cells require lipids to meet their energy and growth demands, but this requirement of lipids in the case of PCa is unique because the use of lipids is required A) as an energy source, B) as building blocks for cellular and subcellular membranes and C) as precursor reagents for de novo steroidogenesis. PCa is primarily a hormone-driven cancer and need androgens to grow, which is why androgen deprivation therapy (castration and administration of anti-androgens) or ADT is mostly employed by clinicians to manage this cancer [41]. Unfortunately, PCa re-emerges with time (termed as CRPC), which is more aggressive and resistant against current therapeutics, and reactivation of AR signaling promotes the return of PCa [42–45]. Sustained activation of AR signaling following ADT has been attributed to de novo steroidogenesis by PCa cells [46–49]. De novo synthesized lipids (mainly cholesterol) are utilized as the precursor for androgen synthesis by PCa cells. In this regard, we determined that ACACA, FASN and the lipogenic transcription factor SREBP-1 are transcriptionally upregulated with respect to disease state and this upregulation has been very well correlated with the Gleason score of PCa patients. Therefore, it can be suggested that the lipogenic pathway seems to be active in patients with prostate adenocarcinoma, specifically progression to androgen independence [19, 20, 49]. Strikingly, in our dataset analysis we also noted that FASN protein expression coincided with enhanced expression of hallmark proto-oncogenes such as MAPK, ERBB2, GAB2, PI3K and STAT along with down regulation of pro-apoptotic proteins like DIABLO and DDP4 (dipeptidyl peptidase IV). Specifically, the active forms of MAPK and ERBB2 are reported to be involved with cell cycle regulation and involved in metastasis and resistance to androgen deprivation therapy, respectively [50–52]. Whereas, GAB2 is a known regulator of PI3K and STAT5A is critical for PCa cell survival and growth, and overexpression of both proteins coincides with FASN expression [53, 54]. Interestingly, DIABLO and DDP4 were under-expressed with FASN expression. These findings can explain in part the importance of FASN in PCa progression as previous reports suggest that pharmacological inhibition or siRNA-mediated downregulation of FASN in cancer cells including PCa is sufficient to induce growth arrest and apoptosis in vitro as well as in vivo tumor models [55–58]. Furthermore, the expression of SREBP1 also correlates with progression of PCa, as this transcription factor regulates the expression of all lipogenic genes that contain sterol response elements in their promoter region [49, 59, 60]. Correspondingly, it has been observed that overexpression of SREBP1 alone is sufficient to drive PCa growth and progression [13]. We have also indicated that miRNA-222 and miRNA-27b are under-expressed with respect to disease state and may contribute to the observed overexpression of the three key lipogenic genes, which coincides with the observations made by Li, et al [2].
In this study, we adjusted and employed a microRaman-BCA approach to probe PCa lesions. The use of Raman microscopy as a tool for detecting PCa progression has been previously reviewed [61]. Our contribution to this field is application of BCA approach, which uniquely enabled a quantification of proteins, lipids and DNA in the samples. This approach enabled us to validate lipid accumulation in the TRAMP model, and we highlighted the notion that lipogenesis may play an even a larger role in NEPC, as key lipogenic genes are altered in 62% of CRPC patients (which includes both adenocarcinoma and NEPC phenotypes), compared to 17% of prostate adenocarcinoma patients. Thus, Raman microscopy may be used down the line as a tool to non-invasively assess PCa progression via detection of lipid accumulation. The results of Raman microscopy were further validated by direct quantitation of lipid content in TRAMP tumor tissues by estimating FFA and cholesterol content. The non-invasive capabilities of Raman technology could be further applied for diagnostics and prognostics of PCa with minimal stress to patients. Finally, we have determined that the overall genetic alterations within SREBP-1, FASN, and ACACA add up to only 17% of patients within the TCGA prostate adenocarcinoma dataset. However, these genetic alterations, though small in number, are sufficient to predict overall survival of patients (Figure 6A and B). These results are startling because as we have previously mentioned, 62% of CRPC patients contain alteration in the three lipogenic genes. Thus, lipogenic transcripts/proteins may play an implemental role in promoting and diagnosing PCa, but may have an even larger role in poorly differentiated, neuroendocrine-like disease. Overall, these findings significantly highlight the importance of lipids in PCa growth, progression and emergence of mCRPC and suggest that detection of lipid accumulation in PCa tumor can be utilized as a predictive biomarker tool.
Highlights.
Lipid accumulation may provide quantitative marker for prostate cancer diagnosis.
Micro-Raman-BCA lipid quantification serves as a novel approach in prostate tumor staging.
Upregulation of lipogenic molecules correlates with aggressive disease in prostate cancer.
Acknowledgments
This work was supported in part by the National Cancer Institute of the National Institutes of Health under Award Number RO1CA160685, and the American Cancer Society Research Scholar Grant RSG-12-214-01-CCG; and the National Cancer Institute Center Support Grant P30 CA016056 to the Roswell Park Cancer Institute.
Abbreviations
- FASN
fatty acid synthase
- ACC
acetyl Co-A carboxylase
- SREBP1
sterol regulatory element binding protein 1
- TRAMP
transgenic adenocarcinoma of the mouse prostate
- Pca
prostate cancer
- mCRPC
metastatic castration recurrent prostate cancer
- CRPC
castration recurrent prostate cancer
- ADT
androgen deprivation therapy
- AR
androgen receptor
- BCA
biomolecular component analysis
- WT
wild-type
- FFA
free fatty acids
- CPT1
carnitine palmitoyltransferase
- PIN
prostatic intraepithelial neoplasia
- NEPC
neuroendocrine prostate cancer
- PRAD
prostate adenocarcinoma
- TCA
tricarboxylic acid
- AceCS1
acetyl-coA synthetase 1
- ACSL1
acetyl-coA synthetase long chain family member 1
- ACL
ATP-citrate lyase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Gupta S, Li J, Kemeny G, Bitting RL, Beaver J, Somarelli J, Ware KE, Gregory S, Armstrong AJ. Whole genomic copy number alterations in circulating tumor cells from men with abiraterone or enzalutamide resistant metastatic castration-resistant prostate cancer. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-1211. [DOI] [PubMed] [Google Scholar]
- 3.Wong YN, Ferraldeschi R, Attard G, de Bono J. Evolution of androgen receptor targeted therapy for advanced prostate cancer. Nat Rev Clin Oncol. 2014;11:365–376. doi: 10.1038/nrclinonc.2014.72. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis. 2006;9:230–234. doi: 10.1038/sj.pcan.4500879. [DOI] [PubMed] [Google Scholar]
- 5.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Luo L, Liu M. Adipose tissue in control of metabolism. J Endocrinol. 2016;231:R77–R99. doi: 10.1530/JOE-16-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutierrez-Pajares JL, Ben Hassen C, Chevalier S, Frank PG. SR-BI: Linking Cholesterol and Lipoprotein Metabolism with Breast and Prostate Cancer. Front Pharmacol. 2016;7:338. doi: 10.3389/fphar.2016.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deep G, Schlaepfer IR. Aberrant Lipid Metabolism Promotes Prostate Cancer: Role in Cell Survival under Hypoxia and Extracellular Vesicles Biogenesis. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17071061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, Lombardo PS, Van Nostrand JL, Hutchins A, Vera L, Gerken L, Greenwood J, Bhat S, Harriman G, Westlin WF, Harwood HJ, Jr, Saghatelian A, Kapeller R, Metallo CM, Shaw RJ. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med. 2016;22:1108–1119. doi: 10.1038/nm.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000;16:202–208. doi: 10.1016/s0899-9007(99)00266-x. [DOI] [PubMed] [Google Scholar]
- 11.Schlaepfer IR, Nambiar DK, Ramteke A, Kumar R, Dhar D, Agarwal C, Bergman B, Graner M, Maroni P, Singh RP, Agarwal R, Deep G. Hypoxia induces triglycerides accumulation in prostate cancer cells and extracellular vesicles supporting growth and invasiveness following reoxygenation. Oncotarget. 2015;6:22836–22856. doi: 10.18632/oncotarget.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, Van Veldhoven PP, Waltregny D, Daniels VW, Machiels J, Vanderhoydonc F, Smans K, Waelkens E, Verhoeven G, Swinnen JV. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70:8117–8126. doi: 10.1158/0008-5472.CAN-09-3871. [DOI] [PubMed] [Google Scholar]
- 13.Huang WC, Li X, Liu J, Lin J, Chung LW. Activation of androgen receptor, lipogenesis, and oxidative stress converged by SREBP-1 is responsible for regulating growth and progression of prostate cancer cells. Mol Cancer Res. 2012;10:133–142. doi: 10.1158/1541-7786.MCR-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang M, Mulholland DJ. Lipogenic metabolism: a viable target for prostate cancer treatment? Asian J Androl. 2014;16:661–663. doi: 10.4103/1008-682X.132947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burch TC, Isaac G, Booher CL, Rhim JS, Rainville P, Langridge J, Baker A, Nyalwidhe JO. Comparative Metabolomic and Lipidomic Analysis of Phenotype Stratified Prostate Cells. PLoS One. 2015;10:e0134206. doi: 10.1371/journal.pone.0134206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Schrijver E, Brusselmans K, Heyns W, Verhoeven G, Swinnen JV. RNA interference-mediated silencing of the fatty acid synthase gene attenuates growth and induces morphological changes and apoptosis of LNCaP prostate cancer cells. Cancer Res. 2003;63:3799–3804. [PubMed] [Google Scholar]
- 17.Kridel SJ, Axelrod F, Rozenkrantz N, Smith JW. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res. 2004;64:2070–2075. doi: 10.1158/0008-5472.can-03-3645. [DOI] [PubMed] [Google Scholar]
- 18.Sadowski MC, Pouwer RH, Gunter JH, Lubik AA, Quinn RJ, Nelson CC. The fatty acid synthase inhibitor triclosan: repurposing an anti-microbial agent for targeting prostate cancer. Oncotarget. 2014;5:9362–9381. doi: 10.18632/oncotarget.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Twiddy AL, Leon CG, Wasan KM. Cholesterol as a potential target for castration-resistant prostate cancer. Pharm Res. 2011;28:423–437. doi: 10.1007/s11095-010-0210-y. [DOI] [PubMed] [Google Scholar]
- 20.Leon CG, Locke JA, Adomat HH, Etinger SL, Twiddy AL, Neumann RD, Nelson CC, Guns ES, Wasan KM. Alterations in cholesterol regulation contribute to the production of intratumoral androgens during progression to castration-resistant prostate cancer in a mouse xenograft model. Prostate. 2010;70:390–400. doi: 10.1002/pros.21072. [DOI] [PubMed] [Google Scholar]
- 21.Yadav N, Pliss A, Kuzmin A, Rapali P, Sun L, Prasad P, Chandra D. Transformations of the macromolecular landscape at mitochondria during DNA-damage-induced apoptotic cell death. Cell Death Dis. 2014;5:e1453. doi: 10.1038/cddis.2014.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan-Lefko PJ, Chen TM, Ittmann MM, Barrios RJ, Ayala GE, Huss WJ, Maddison LA, Foster BA, Greenberg NM. Pathobiology of autochthonous prostate cancer in a pre-clinical transgenic mouse model. Prostate. 2003;55:219–237. doi: 10.1002/pros.10215. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Gogada R, Yadav N, Lella RK, Badeaux M, Ayres M, Gandhi V, Tang DG, Chandra D. Defective molecular timer in the absence of nucleotides leads to inefficient caspase activation. PLoS One. 2011;6:e16379. doi: 10.1371/journal.pone.0016379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandra D, Liu JW, Tang DG. Early mitochondrial activation and cytochrome c up-regulation during apoptosis. J Biol Chem. 2002;277:50842–50854. doi: 10.1074/jbc.M207622200. [DOI] [PubMed] [Google Scholar]
- 27.Pliss A, Kuzmin AN, Kachynski AV, Prasad PN. Biophotonic probing of macromolecular transformations during apoptosis. P Natl Acad Sci USA. 2010;107:12771–12776. doi: 10.1073/pnas.1006374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuzmin AN, Pliss A, Kachynski AV. Biomolecular component analysis of cultured cell nucleoli by Raman microspectrometry. J Raman Spectrosc. 2013;44:198–204. [Google Scholar]
- 29.Kuzmin AN, Pliss A, Prasad PN. Changes in Biomolecular Profile in a Single Nucleolus during Cell Fixation. Analytical chemistry. 2014 doi: 10.1021/ac503172b. [DOI] [PubMed] [Google Scholar]
- 30.Buschman HP, Deinum G, Motz JT, Fitzmaurice M, Kramer JR, van der Laarse A, Bruschke AV, Feld MS. Raman microspectroscopy of human coronary atherosclerosis: Biochemical assessment of cellular and extracellular morphologic structures in situ. Cardiovasc Pathol. 2001;10:69–82. doi: 10.1016/s1054-8807(01)00064-3. [DOI] [PubMed] [Google Scholar]
- 31.Shafer-Peltier KE, Haka AS, Fitzmaurice M, Crowe J, Myles J, Dasari RR, Feld MS. Raman microspectroscopic model of human breast tissue: implications for breast cancer diagnosis in vivo. J Raman Spectrosc. 2002;33:552–563. [Google Scholar]
- 32.Short KW, Carpenter S, Freyer JP, Mourant JR. Raman spectroscopy detects biochemical changes due to proliferation in mammalian cell cultures. Biophys J. 2005;88:4274–4288. doi: 10.1529/biophysj.103.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swain RJ, Kemp SJ, Goldstraw P, Tetley TD, Steyens MM. Assessment of Cell Line Models of Primary Human Cells by Raman Spectral Phenotyping. Biophys J. 2010;98:1703–1711. doi: 10.1016/j.bpj.2009.12.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipianskaya J, Cohen A, Chen CJ, Hsia E, Squires J, Li Z, Zhang Y, Li W, Chen X, Xu H, Huang J. Androgen-deprivation therapy-induced aggressive prostate cancer with neuroendocrine differentiation. Asian J Androl. 2014;16:541–544. doi: 10.4103/1008-682X.123669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jongsma J, Oomen MH, Noordzij MA, Van Weerden WM, Martens GJ, van der Kwast TH, Schroder FH, van Steenbrugge GJ. Androgen deprivation of the PC-310 [correction of prohormone convertase-310] human prostate cancer model system induces neuroendocrine differentiation. Cancer Res. 2000;60:741–748. [PubMed] [Google Scholar]
- 36.Sciarra A, Di Silverio F. Effect of nonsteroidal antiandrogen monotherapy versus castration therapy on neuroendocrine differentiation in prostate carcinoma. Urology. 2004;63:523–527. doi: 10.1016/j.urology.2003.10.043. [DOI] [PubMed] [Google Scholar]
- 37.Zadra G, Photopoulos C, Loda M. The fat side of prostate cancer. Biochim Biophys Acta. 2013;1831:1518–1532. doi: 10.1016/j.bbalip.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suburu J, Chen YQ. Lipids and prostate cancer. Prostaglandins Other Lipid Mediat. 2012;98:1–10. doi: 10.1016/j.prostaglandins.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nambiar DK, Deep G, Singh RP, Agarwal C, Agarwal R. Silibinin inhibits aberrant lipid metabolism, proliferation and emergence of androgen-independence in prostate cancer cells via primarily targeting the sterol response element binding protein 1. Oncotarget. 2014;5:10017–10033. doi: 10.18632/oncotarget.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yue S, Li J, Lee SY, Lee HJ, Shao T, Song B, Cheng L, Masterson TA, Liu X, Ratliff TL, Cheng JX. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merseburger AS, Alcaraz A, von Klot CA. Androgen deprivation therapy as backbone therapy in the management of prostate cancer. Onco Targets Ther. 2016;9:7263–7274. doi: 10.2147/OTT.S117176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spratt DE, Evans MJ, Davis BJ, Doran MG, Lee MX, Shah N, Wongvipat J, Carnazza KE, Klee GG, Polkinghorn W, Tindall DJ, Lewis JS, Sawyers CL. Androgen Receptor Upregulation Mediates Radioresistance after Ionizing Radiation. Cancer Res. 2015;75:4688–4696. doi: 10.1158/0008-5472.CAN-15-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, Arora VK, Yen WF, Cai L, Zheng D, Carver BS, Chen Y, Watson PA, Shah NP, Fujisawa S, Goglia AG, Gopalan A, Hieronymus H, Wongvipat J, Scardino PT, Zelefsky MJ, Jasin M, Chaudhuri J, Powell SN, Sawyers CL. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3:1245–1253. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hearn JW, AbuAli G, Reichard CA, Reddy CA, Magi-Galluzzi C, Chang KH, Carlson R, Rangel L, Reagan K, Davis BJ, Karnes RJ, Kohli M, Tindall D, Klein EA, Sharifi N. HSD3B1, resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. Lancet Oncol. 2016;17:1435–1444. doi: 10.1016/S1470-2045(16)30227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang KH, Li R, Kuri B, Lotan Y, Roehrborn CG, Liu J, Vessella R, Nelson PS, Kapur P, Guo X, Mirzaei H, Auchus RJ, Sharifi N. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell. 2013;154:1074–1084. doi: 10.1016/j.cell.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang KH, Li R, Papari-Zareei M, Watumull L, Zhao YD, Auchus RJ, Sharifi N. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2011;108:13728–13733. doi: 10.1073/pnas.1107898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharifi N, Auchus RJ. Steroid biosynthesis and prostate cancer. Steroids. 2012;77:719–726. doi: 10.1016/j.steroids.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 49.Ettinger SL, Sobel R, Whitmore TG, Akbari M, Bradley DR, Gleave ME, Nelson CC. Dysregulation of sterol response element-binding proteins and downstream effectors in prostate cancer during progression to androgen independence. Cancer Res. 2004;64:2212–2221. doi: 10.1158/0008-5472.can-2148-2. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 51.Tome-Garcia J, Li D, Ghazaryan S, Shu L, Wu L. ERBB2 increases metastatic potentials specifically in androgen-insensitive prostate cancer cells. PLoS One. 2014;9:e99525. doi: 10.1371/journal.pone.0099525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao S, Ye H, Gerrin S, Wang H, Sharma A, Chen S, Patnaik A, Sowalsky AG, Voznesensky O, Han W, Yu Z, Mostaghel EA, Nelson PS, Taplin ME, Balk SP, Cai C. ErbB2 Signaling Increases Androgen Receptor Expression in Abiraterone-Resistant Prostate Cancer. Clin Cancer Res. 2016;22:3672–3682. doi: 10.1158/1078-0432.CCR-15-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding CB, Yu WN, Feng JH, Luo JM. Structure and function of Gab2 and its role in cancer (Review) Mol Med Rep. 2015;12:4007–4014. doi: 10.3892/mmr.2015.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Z, Lutz J, Nevalainen MT. Transcription factor Stat5a/b as a therapeutic target protein for prostate cancer. Int J Biochem Cell Biol. 2010;42:186–192. doi: 10.1016/j.biocel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 56.Rae C, Haberkorn U, Babich JW, Mairs RJ. Inhibition of Fatty Acid Synthase Sensitizes Prostate Cancer Cells to Radiotherapy. Radiat Res. 2015;184:482–493. doi: 10.1667/RR14173.1. [DOI] [PubMed] [Google Scholar]
- 57.Menendez JA, Vellon L, Colomer R, Lupu R. Pharmacological and small interference RNA-mediated inhibition of breast cancer-associated fatty acid synthase (oncogenic antigen-519) synergistically enhances Taxol (paclitaxel)-induced cytotoxicity. Int J Cancer. 2005;115:19–35. doi: 10.1002/ijc.20754. [DOI] [PubMed] [Google Scholar]
- 58.Ventura R, Mordec K, Waszczuk J, Wang Z, Lai J, Fridlib M, Buckley D, Kemble G, Heuer TS. Inhibition of de novo Palmitate Synthesis by Fatty Acid Synthase Induces Apoptosis in Tumor Cells by Remodeling Cell Membranes, Inhibiting Signaling Pathways, and Reprogramming Gene Expression. EBioMedicine. 2015;2:808–824. doi: 10.1016/j.ebiom.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 60.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kast RE, Tucker SC, Killian K, Trexler M, Honn KV, Auner GW. Emerging technology: applications of Raman spectroscopy for prostate cancer. Cancer Metastasis Rev. 2014;33:673–693. doi: 10.1007/s10555-013-9489-6. [DOI] [PubMed] [Google Scholar]