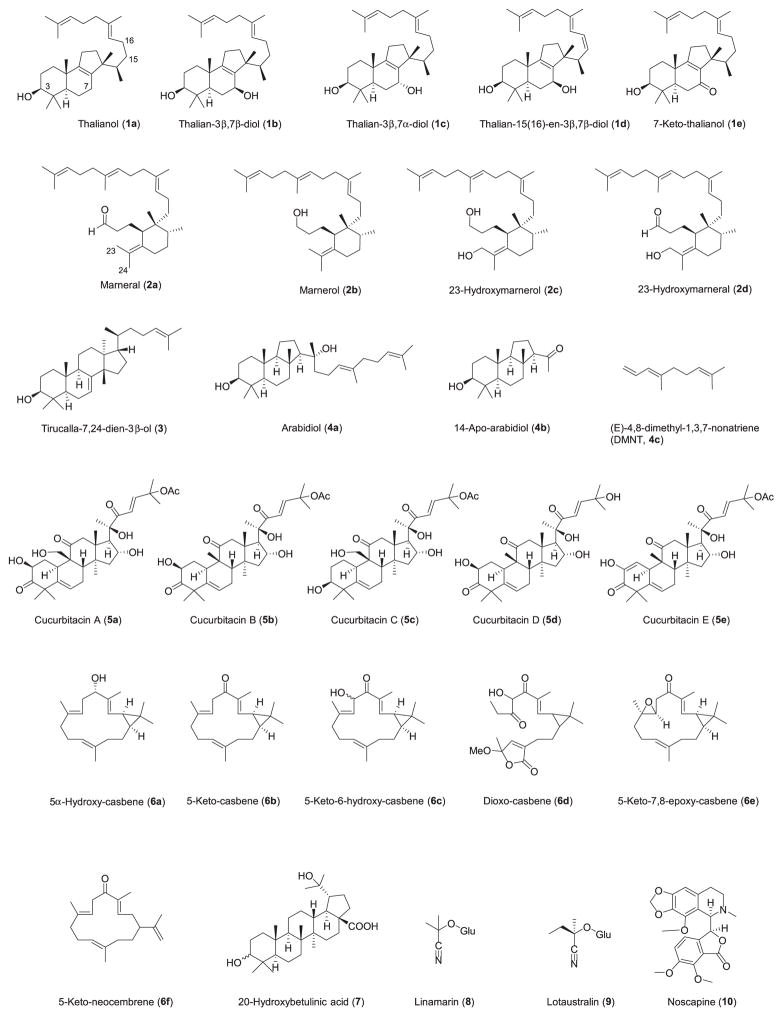
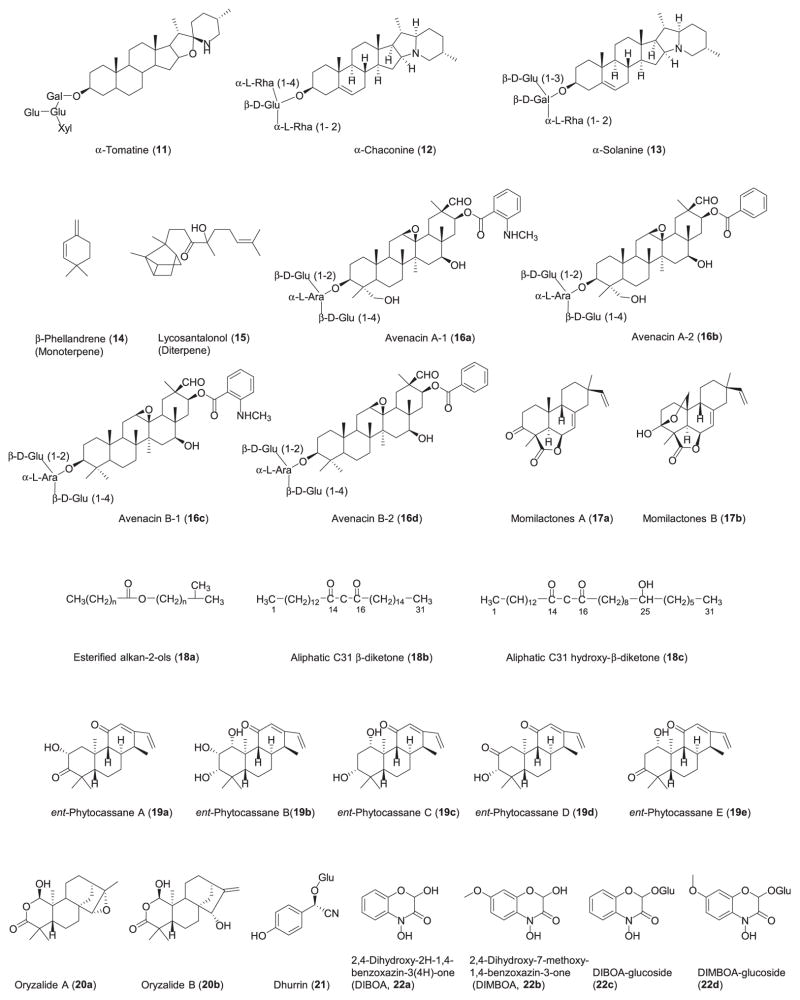
Summary
Plant natural products are of great value for agriculture, medicine and a wide range of other industrial applications. The discovery of new plant natural product pathways is currently being revolutionized by two key developments. First, breakthroughs in sequencing technology and reduced cost of sequencing are accelerating the ability to find enzymes and pathways for the biosynthesis of new natural products by identifying the underlying genes. Second, there are now multiple examples in which the genes encoding certain natural product pathways have been found to be grouped together in biosynthetic gene clusters within plant genomes. These advances are now making it possible to develop strategies for systematically mining multiple plant genomes for the discovery of new enzymes, pathways and chemistries. Increased knowledge of the features of plant metabolic gene clusters – architecture, regulation and assembly – will be instrumental in expediting natural product discovery. This review summarizes progress in this area.
Keywords: biosynthetic gene clusters, chromatin, genome mining, natural products, operons
I. Introduction
Plant natural products have been used by humans for hundreds, and in some cases thousands, of years as medicines, drugs, dyes, pigments, flavourings, fragrances, nutraceuticals, cosmetics and agrochemicals. The plant kingdom is a fertile source of biologically active molecules. These molecules, in contrast to randomly constructed compound libraries, have been prescreened and honed by nature for stability, bioavailability and ‘useful’ biological activities. The vast majority of the chemical potential of the plant kingdom awaits discovery and is hidden away in the dark matter of plant genomes. One of the great challenges for plant biology, and for evolutionary biology more widely, lies in understanding how and why different plants make different kinds of chemicals, and how new natural product pathways are formed.
The discovery of new plant natural product pathways and chemistries is now being revolutionized by two key developments. First, breakthroughs in sequencing technology and reduced cost of sequencing are accelerating the ability to discover new natural product pathways by identifying the underlying genes, as genome sequences of numerous different plant species are becoming available. Transcriptomics in combination with metabolite analysis of different plant tissues, developmental stages and/or elicitor-treated material is also being used to great effect to identify candidate genes for the synthesis of natural products of interest and to piece together pathways (e.g. Geu-Flores et al., 2012; Lau & Sattely, 2015). Second, it has emerged that the genes encoding certain natural product pathways are organized in biosynthetic gene clusters within plant genomes (Boycheva et al., 2014; King et al., 2014; Nützmann & Osbourn, 2014; Shang et al., 2014; Boutanaev et al., 2015; Schneider et al., 2016). These examples include pathways for diverse classes of chemicals from a wide variety of different plant species, including eudicots and monocots (both annuals and perennials) (Table 1; Fig. 1). This clustering phenomenon is interesting because it challenges the assumption that gene ordering on plant chromosomes is more or less random. It is also beginning to allow the straightforward prediction and subsequent characterization of biosynthetic pathways from genome sequences, an approach that has proved highly successful in bacteria and fungi (Cimermancic et al., 2014; Doroghazi et al., 2014; Medema et al., 2014; Ziemert et al., 2014; Li et al., 2016; Van der Lee & Medema, 2016). Until recently, much of what is known about pathways for the synthesis of natural products in plants has stemmed from examination of individual pathways. It is now becoming possible to mine multiple plant genomes for the discovery of a huge number of predicted genes and enzymes of biosynthetic pathways (Boutanaev et al., 2015). The challenges now lie in developing refined search methods to streamline this discovery platform, and in making sense of and exploiting the outputs. Understanding the features of the metabolic clusters that have been reported so far will be instrumental in enabling such methods to be developed.
Table 1.
Clustered pathways for the biosynthesis of plant natural products
| Pathway | Class of compound | Plant species | Annual/perennial | Method of cluster discovery | Reference |
|---|---|---|---|---|---|
| Eudicotsa | |||||
| Thalianol (1a–e) | Triterpene | Arabidopsis thaliana | Annual | Cluster mining | Field & Osbourn (2008) |
| Marneral (2a–d) | Triterpene | Arabidopsis thaliana | Cluster mining | Field et al. (2011) | |
| Tirucalla-7,24-dien-3β-ol (3) | Triterpene | Arabidopsis thaliana | Cluster mining | Boutanaev et al. (2015) | |
| Arabidiol (4a–c) | Triterpene | Arabidopsis thaliana | Cluster mining | Castillo et al. (2013); Sohrabi et al. (2015) | |
| Cucurbitacins (5a–e) | Triterpene | Cucumis sativus | Annual | Cluster mining; trait-based discovery (characterization of the Bi locus for bitterness) | Boutanaev et al. (2015); Shang et al. (2014) |
| Casbene diterpenoids (6a–b) | Diterpene | Euphorbia peplus | Annual | Characterized biosynthetic genes to clusteringc | King et al. (2014) |
| Casbene diterpenoids | Diterpene | Jatropha curcas | Perennial | Genome mining; genetics | King et al. (2014) |
| 20-Hydroxybetulinic acid (7) | Triterpene | Lotus japonicus | Annual | Cluster mining | Krokida et al. (2013) |
| Linamarin (8)/lotaustralin (9) | Cyanogenic glycoside | Lotus japonicus | Annual | Isolation of cyanogenesis deficient mutants; genomics | Takos et al. (2011) |
| Linamarin (8)/lotaustralin (9) | Cyanogenic glycoside | Manihot esculenta | Perennial | Characterized biosynthetic genes to cluster | Takos et al. (2011) |
| Noscapine (10) | Benzylisoquinoline alkaloid | Papaver somniferumb | Annual | Trait-based discovery (characterization of High Noscapine 1 locus) | Winzer et al. (2012) |
| Casbene diterpenoids (6a–f) | Diterpene | Ricinus communis | Perennial with annual forms | Cluster mining; characterized biosynthetic genes to cluster | Boutanaev et al. (2015); King et al. (2014) |
| α-Tomatine (11) | Steroidal alkaloid | Solanum lycopersicum | Perennial with annual forms | Characterized biosynthetic genes to cluster | Itkin et al. (2013) |
| α-Chaconine (12)/α-solanine (13) | Steroidal alkaloid | Solanum tuberosum | Perennial | Characterized biosynthetic genes to cluster; synteny with Solanum lycopersicon | Itkin et al. (2013) |
| Monoterpenes (14)/diterpenes (15) | Terpenes | Solanum lycopersicum | Perennial with annual forms | Characterized biosynthetic genes to cluster | Falara et al. (2011); Matsuba et al. (2013, 2015) |
| Monocots | |||||
| Avenacins (16a–d) | Triterpene | Avena strigosab | Annual | Trait-based discovery (forward screen for avenacin-deficient mutants) | Qi et al. (2004, 2006); Mugford et al. (2009, 2013) |
| β-Diketones (18a–c) | Polyketide | Hordeum vulgare | Annual | Trait-based discovery (characterization of the Cer-cqu leaf wax locus) | Schneider et al. (2016) |
| Momilactones (17a, b) | Diterpene | Oryza sativa | Annual | Characterized biosynthetic genes to cluster | Wilderman et al. (2004); Shimura et al. (2007) |
| Phytocassanes (19a–e), oryzalides (20a, b) | Diterpene | Oryza sativa | Annual | Characterized biosynthetic genes to cluster | Swaminathan et al. (2009) |
| Dhurrin (21) | Cyanogenic glycoside | Sorghum bicolor | Annual | Characterized biosynthetic genes to cluster | Takos et al. (2011) |
| 2,4-dihydroxy-7-methoxy-1, 4-benzoxazin-3-one (DIMBOA, 22a–d) | Hydroxamic acid | Zea mays | Annual | Trait-based discovery (screen for bx1 mutants) | Frey et al. (1997); von Rad et al. (2001); Frey et al. (2003); Jonczyk et al. (2008) |
The products of the pathways are shown in Fig. 1.
Species for which genome sequences are not publicly available. Clusters were therefore defined by assembling and sequencing bacterial artificial chromosome (BAC) contigs (Mugford et al., 2009; Mugford et al., 2013; Qi et al., 2006; Winzer et al., 2012).
For casbene diterpene biosynthesis in E. peplus, only two adjacent genes have been discovered so far (a casbene synthase and CYP72A19). Although this region does not contain genes encoding a minimum of three different types of enzymes, it has been included in the table since it emerged from a wider investigation of production of casbene diterpenoids in the wider Euphorbiaceae (see entries for castor (Ricinus communis) and jatropha (Jatropha curcas)) (King et al., 2014).
Fig. 1.
Structures of the products of characterized clustered plant metabolic pathways. The structures are numbered according to Table 1.
II. Plant metabolic gene clusters – some rules and anomalies
The natural product biosynthetic gene clusters so far reported from plants range from ~ 35 kb to several hundred kb in size and consist of three to 10 genes. They include genes for a minimum of at least three (and sometimes six or more) different types of biosynthetic enzymes (King et al., 2014; Nützmann & Osbourn, 2014; Shang et al., 2014; Boutanaev et al., 2015). The salient features of these plant metabolic clusters will be considered here.
1. Scaffolds and networks – what constitutes the first step in a clustered pathway?
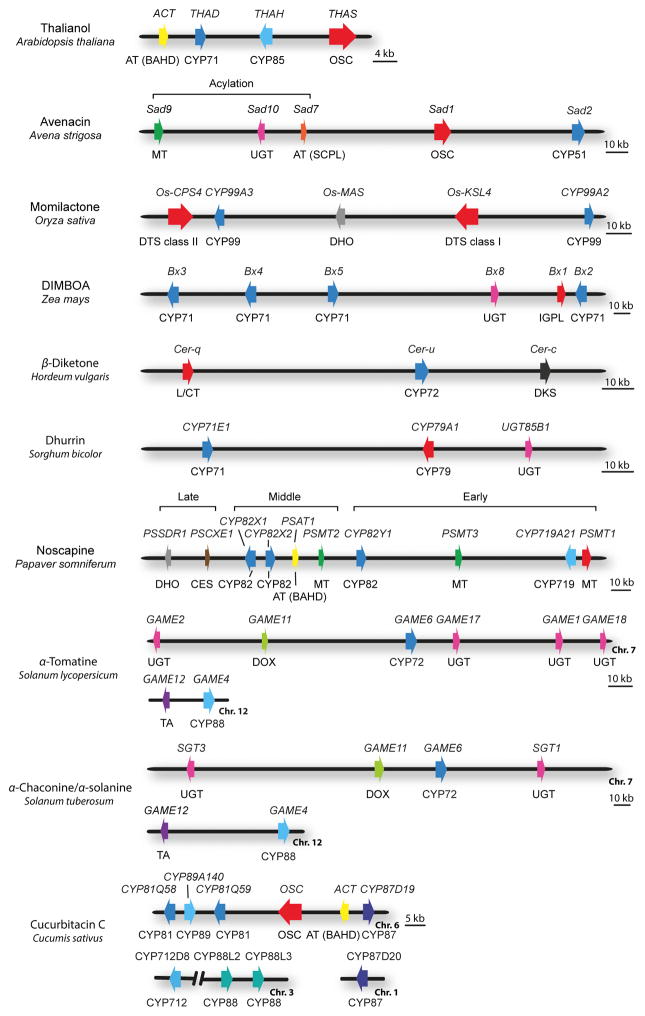
Plant metabolic clusters typically (but not always) contain the gene encoding the first committed pathway step and two or more genes encoding other downstream pathway enzymes (Fig. 2). Examples of enzymes that catalyse the first committed step include oxidosqualene cyclases (OSCs) (for the synthesis of triterpenes) (Thimmappa et al., 2014), class I and II diterpene synthases (DTSs) (which together make diterpene scaffolds) (Chen et al., 2011; Zi et al., 2014), and the tryptophan synthase α homologue BX1 (which diverts indole-3-glycerol phosphate from tryptophan biosynthesis into indole for the synthesis of cyclic hydroxamic acids) (Frey et al., 1997) (Fig. 2). These enzymes draw metabolites from primary metabolism into specialized metabolism. A type III chalcone synthase-like enzyme required for the synthesis of diketone wax aliphatics in barley has also been reported (Schneider et al., 2016). While in most cases the nature of the ‘signature’ enzyme, and hence the type of scaffold, is evident based on cursory examination of these clusters, this may not always be the case. For example, the first committed step in cyanogenic glycoside biosynthesis (the conversion of amino acids to oximes) is catalysed by cytochrome P450 (CYP) enzymes belonging to the CYP79 family (Koch et al., 1995; Bak et al., 1998; Andersen et al., 2000; Forslund et al., 2004; Olsen et al., 2008; Takos et al., 2011; Saito et al., 2012) (e.g. the dhurrin pathway from sorghum – Fig. 2). At first glance the scaffold for the cyanogenic glycoside clusters may not be immediately obvious. However, CYP79 enzymes are unusual amongst the CYP superfamily in having amino acids as their substrates (Hamberger & Bak, 2013). These CYPs convert amino acids to oximes, thereby siphoning them into specialized metabolism. Of note, CYP79 enzymes also catalyse the first committed step in the biosynthesis of noncyanogenic plant defence compounds such as glucosinolates and camalexin (Hull et al., 2000; Mikkelsen et al., 2000; Forslund et al., 2004; Glawischnig et al., 2004; Hamberger & Bak, 2013), pathways for which the genes are not clustered.
Fig. 2.
Genomic organization of plant metabolic gene clusters. Examples that exemplify different cluster types and biosynthetic pathways are shown. The genes are indicated by arrows. The gene(s) for the first committed pathway step are indicated in red. Gene names are indicated above the clusters, and class of biosynthetic enzyme below. Abbreviations: OSC, oxidosqualene cyclase; DTS, diterpene synthase (class I and class II shown); IGPL, indole 3-glycerol phosphate lyase; DKS, diketone synthase; AT (BAHD), BAHD-acyltransferase; AT (SCPL), SCPL-acyltransferase; MT, methyltransferase; UGT, UDP-dependent sugar transferase, DHO, dehydrogenase/reductase; L/CT, lipase/carboxyltransferase; CES, carboxylesterase; DOX, dioxygenase; TA, transaminase; CYP, cytochrome P450. The maize DIMBOA pathway includes three genes that are not shown in the figure: Bx7, which is separated from the core cluster by an intervening region of 15 Mb; the sugar transferase gene Bx9, which is located on a different chromosome; and finally Bx6, which is not shown because its genomic location has not yet been established. [Correction added after online publication 26 April 2016; β-Diketone Hordeum vulgaris cluster has been corrected.]
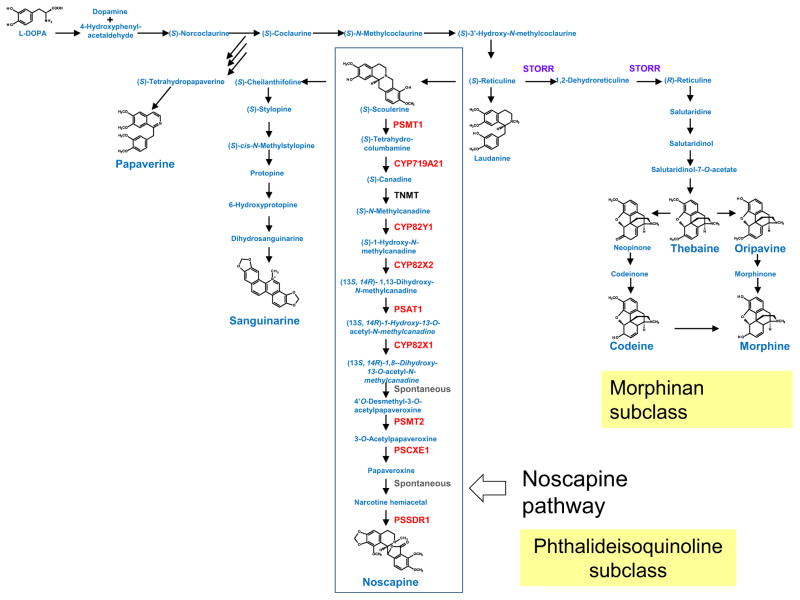
Opium poppy (Papaver somniferum) produces a variety of different types of benzylisoquinoline alkaloids (BIAs), including the potential anticancer drug noscapine (Beaudoin & Facchini, 2014; Chen et al., 2015) (Fig. 3). A gene cluster for the synthesis of noscapine has recently been discovered in a high noscapine-producing poppy variety (Winzer et al., 2012) (Fig. 2). This cluster contains 10 genes encoding five distinct enzyme classes and includes the gene for the first committed step in the noscapine pathway – the conversion of scoulerine to tetrahydrocolumbamine by the O-methyltransferase PSMT1 (Fig. 3). One of the steps in the pathway has previously been shown to be carried out by the enzyme tetrahydroprotoberberine cis-N-methyltransferase (TNMT) (Liscombe & Facchini, 2007). The TNMT gene is not represented within the noscapine cluster, although open reading frames with TNMT homology are present in the flanking regions. Unlike the rest of the pathway genes, TNMT is also present and expressed in nonnoscapine-synthesizing poppy varieties and is likely to have additional functions (Winzer et al., 2012).
Fig. 3.
Benzylisoquinoline alkaloid biosynthesis in poppy. Poppy synthesizes a variety of different benzylisoquinoline alkaloids, including noscapine, codeine and morphine. The first committed step of the noscapine pathway (boxed) is the conversion of (S)-scoulerine to (S)-tetrahydrocolumbamine by PSMT1. The enzymes encoded by the genes within the noscapine cluster are indicated in red. The gene for one of the noscapine pathway steps (tetrahydroprotoberberine cis-N-methyltransferase; TNMT) is not represented within the cluster. Unlike the rest of the pathway genes, TNMT is present and expressed in HN1, HT1 and HM1 poppy varieties and so is likely to have other functions in addition to its role in noscapine biosynthesis (Winzer et al., 2012). The P450 oxidoreductase fusion protein STORR, which catalyses the first step in morphinan biosynthesis in poppy – the conversion of (S)-reticuline to (R)-reticuline – is indicated in purple.
The step that provides the branch point between primary metabolism and BIA biosynthesis lies several steps upstream of scoulerine (Fig. 3). The genome sequence of P. somniferum is not yet available. It therefore remains to be seen whether the noscapine cluster is a discrete group of 10 genes dedicated to this particular branch of BIA biosynthesis or whether it forms part of a larger cluster that contains additional genes required for the synthesis of the scoulerine precursor. Similarly, it is not known whether the genes for other branches of the BIA biosynthetic network are clustered or unclustered. It has recently been shown, however, that the first committed step in morphinan biosynthesis in poppy is catalysed by a P450 oxidoreductase fusion protein (STORR: (S)- to (R)-reticuline) (Winzer et al., 2015) (Fig. 3). The existence of this chimaeric protein implies that extreme selection pressure has been at work to drive functional modularity in other parts of BIA biosynthesis in addition to the noscapine pathway.
Analysis of a draft genome sequence for Madagascan periwinkle (Catharanthus roseus) has provided evidence for partial clustering of genes for the biosynthesis of the monoterpene indole alkaloids (MIAs) vinblastine and vincristine (Kellner et al., 2015). MIA biosynthesis is highly complex, involving condensation of the indole tryptamine with the iridoid secaloganin to form strictosidine. Strictosidine is a common intermediate in the biosynthesis of thousands of MIAs, including those of vinblastine and vincristine (De Bernonville et al., 2015). Thus, as for noscapine, the vinblastine/vincristine pathway is part of a much larger and more complex biosynthetic network that gives rise to a wealth of other diverse products. The MIA pathway also has one of the most complex and elaborate forms of compartmentalization known for a plant specialized metabolic pathway (Verma et al., 2012; De Bernonville et al., 2015). The N50 scaffold size for the draft C. roseus genome sequence was in the region of 26–27 kb. Although not optimal for discovery of linked genes in plant genomes because of the large intergenic distances, Kellner et al. (2015), aided by bacterial artificial chromosome (BAC) sequencing, were able to identify seven small clusters each of two to three genes that contained genes encoding enzymes for vinblastine/vincristine biosynthesis and also other genes that may encode missing steps in the pathway, along with a gene for a predicted multi-antimicrobial extrusion protein (MATE) transporter that could potentially be involved in the transport of pathway intermediates. The development of a higher quality genome sequence will establish whether these small clusters are dispersed throughout the genome or whether they form a larger cluster, and how these genes are distributed relative to those required for the synthesis of other types of MIAs produced by C. roseus. A simplistic assumption may be that clustering is a feature of dedicated linear pathways that are insulated from the rest of metabolism and that take place in the same cell types. Analysis of complex networks such as those for alkaloid biosynthesis in P. somniferum and C. roseus should help to test this hypothesis and to understand what precisely constitutes a metabolic gene cluster in the context of wider metabolism.
In summary, most of the plant metabolic gene clusters that have been reported so far contain the genes encoding the enzymes for the first committed steps in the respective pathways. These may be obvious scaffold-generating enzymes (e.g. oxidosqualene cyclases; class I and class II diterpene synthases); other enzymes that divert metabolites from primary metabolism into specialized metabolism (e.g. the tryptophan synthase α homologue BX1, which catalyses the first step in DIBOA/DIMBOA biosynthesis; CYP79 P450s, which convert amino acids into oximes); or enzymes that siphon intermediates from complex specialized metabolic networks into dedicated pathways for specific end products (e.g. the O-methyltransferase PSMT1, which converts scoulerine to tetrahydrocolumbamine – the first committed step in the noscapine pathway). As more metabolic gene clusters are characterized from plants, the inventory of ‘signature’ enzymes that are diagnostic of particular types of biosynthetic pathways will grow, so making pathway prediction easier. In some cases, such genes do not appear to be present. For example, the tomato steroidal glycoalkaloid α-tomatine is predicted to be synthesized from cholesterol (Eich, 2008). The genes for α-tomatine biosynthesis are located in one major cluster of six genes on chromosome 7, with a further two pathway genes adjacent to each other on chromosome 12 (Fig. 2). The CYP72 gene GAME7, which is predicted to catalyse the first step in this pathway (the conversion of cholesterol to 22-hydroxycholesterol) (Itkin et al., 2013), is also located on chromosome 7 but is 7880 kb away from the nearest cluster gene (Sato et al., 2012; A. Aharoni, pers. comm.).
2. Tailoring enzymes, modularity, collinearity and moonlighting
The tailoring enzymes encoded within metabolic clusters may variously include CYPs, sugar transferases, methyltransferases, acyltransferases, dioxygenases, carboxylesterases, dehydrogenases/reductases and transaminases (King et al., 2014; Nützmann & Osbourn, 2014; Shang et al., 2014; Boutanaev et al., 2015; Schneider et al., 2016) (Fig. 2). It is likely that this list will continue to grow as more natural product clusters are discovered. The arbitrary definition of a metabolic gene cluster requires that it should contain genes for at least three different types of enzymes (Nützmann & Osbourn, 2014). In this regard, it is important to note that the CYP superfamily is divided into families based on sequence similarity (family members having > 40% amino acid sequence identity) (Nelson & Werck-Reichhart, 2011), and that for the purposes of defining clusters each CYP family should be regarded as a different type of biosynthetic enzyme.
The Arabidopsis thaliana thalianol and marneral clusters are the smallest plant metabolic clusters characterized so far. These are compact clusters that are 35–38 kb in size (Field & Osbourn, 2008; Field et al., 2011). The largest clusters are several hundred kb (Nützmann & Osbourn, 2014). In some cases, the genes in the pathway are contiguous, with no other obvious intervening genes (e.g. the thalianol cluster in A. thaliana and the avenacin cluster in diploid oat) (Qi et al., 2004, 2006; Field & Osbourn, 2008; Mugford et al., 2009, 2013) (Fig. 2). In other cases, some of the pathway genes are less tightly linked and there may be intervening genes with no obvious function in specialized metabolism between these ‘peripheral’ genes and the core cluster. For example, the maize Bx7 gene, which encodes an O-methyltransferase required for DIMBOA biosynthesis (Jonczyk et al., 2008), lies within 15 Mb of the core cluster. Another gene, Bx9, which encodes a sugar transferase that is active towards DIBOA/DIMBOA, is located on a different chromosome (von Rad et al., 2001). In diploid oat, the Sad3 and Sad4 loci have been shown by mutation to be required for avenacin glucosylation (Papadopoulou et al., 1999; Qi et al., 2004; Mylona et al., 2008) but not yet cloned. Sad3 is essential for avenacin glucosylation but is only loosely linked to the core cluster (genetic linkage distance 3.6 cM) (Qi et al., 2004). Full characterization of the wider avenacin cluster region will reveal the nature of Sad3 and the physical distance between this locus and the core cluster. Sad4 is unlinked to the cluster, and unlike Sad3 has only a partial effect on avenacin glucosylation when mutated and also glucosylates other oat metabolites (Papadopoulou et al., 1999; Mylona et al., 2008). Sad4 is therefore not a dedicated pathway component and appears to be ‘moonlighting’.
While it is important to emphasize that plant metabolic gene clusters contain at least three nonhomologous genes encoding different types of enzymes in order to distinguish them from regions of the genome that contain only tandem arrays of highly homologous genes, some of the characterized clusters do contain duplicated genes that, in some cases, have evolved different functions. A noteworthy example is the maize DIMBOA cluster, which contains genes for four CYPs belonging to the CYP71C subfamily (Frey et al., 1997) (Fig. 2). These closely related CYP genes (Bx2-5) have almost certainly arisen from a common ancestor by tandem gene duplication (Dutartre et al., 2012). However, functional analysis following expression in yeast has revealed that each carries out a highly specific and dedicated step in the conversion of indole (the product of the signature enzyme, BX1) to DIBOA (Frey et al., 1997). This group of four genes therefore provides some intriguing insights into the evolution of sequential CYP-mediated steps in this pathway.
A further intriguing finding is the observation that the genes required for a particular modification can be grouped within a cluster as a module. This is exemplified by a set of three genes within the oat avenacin cluster that are required for triterpene acylation (Fig. 2). The major avenacin, A-1, is acylated with an N-methyl anthranilate group. This acyl group is introduced by the acyltransferase SAD7, which is a member of the SCPL family of acyltransferases (Mugford et al., 2009). Unlike the BAHD acyltransferases, which use CoA-thioesters as the acyl donor, SCPL-acyltransferases use acyl sugar donors (Milkowski & Strack, 2004). The acyl glucose donor used by SAD7 is N-methyl anthranilate glucose, which is generated by methylation of the readily available precursor anthranilate (a product of primary metabolism) by the methyl transferase SAD9 followed by glucosylation by the glucosyl transferase SAD10 (Mugford et al., 2013; Owatworakit et al., 2013). The genes encoding SAD7, SAD9 and SAD10 are adjacent in the avenacin cluster, so forming an ‘acylation module’ (Mugford et al., 2013).
Interestingly, the genes within the noscapine cluster in poppy appear to show collinearity in that they are organized in groups that roughly correspond to the early, middle and late steps of the pathway (Winzer et al., 2012) (Fig. 2). Occasional examples of apparent collinearity of metabolic gene clusters have been noted in bacteria and filamentous fungi (Donadio et al., 1991; Cary et al., 2005; Erlich et al., 2005; Osbourn, 2010). Collinearity appears to be the exception rather than the rule in plants. Nevertheless, the fact that it occurs at all is noteworthy and suggests that, at least for the noscapine pathway, gene order (and by extrapolation temporal control of gene expression) may be significant for pathway function and/or potentially also for pathway assembly.
3. Core clusters and satellite subgroups
Several examples of pathways that have core metabolic clusters along with one or two peripheral pathway genes have been mentioned earlier (e.g. the O-methyltransferase gene Bx7 and the sugar transferase gene Bx9 in maize, and the Sad3 locus required for glucosylation of avenacins in oat). In some cases, the pathways are more fragmented. For example, as already mentioned, most of the genes for the synthesis of the steroidal glycoalkaloid α-tomatine in tomato are clustered on chromosome 7. These include two early pathway genes – the dioxygenase gene GAME11, and the CYP72 gene GAME6 – along with the four sugar transferases, GAME1, GAME17, GAME18 and GAME2. Two further pathway genes encoding the transaminase GAME12 and the CYP88D GAME4 are adjacent to each other on a different chromosome (Itkin et al., 2013) (Fig. 2). Note that, of these regions, only the first contains genes for at least three different types of biosynthetic enzyme, i.e. fulfils the requirements to be classified as a metabolic cluster. The genes for the synthesis of the related steroidal glycoalkaloids α-solanine and α-chaconine are similarly organized in potato on chromosomes 7 and 12 in regions that are syntenic to those in tomato (Itkin et al., 2013). Thus it appears that these pathways share a common evolutionary origin but have acquired different functions in tomatoes and potatoes.
By the same token, in cucumber (Cucumis sativus) five genes encoding enzymes required for the biosynthesis of cucurbitacins (triterpenoids that confer bitterness) are clustered on chromosome 6 – the gene for the oxidosqualene cyclase that synthesizes the triterpenoid scaffold cucurbitadienol, along with three different types of CYP genes (CYP81, CYP87, CYP89) and an acyltransferase gene. Four other CYP genes that are also required for cucurbitacin biosynthesis are located elsewhere (a CYP71 gene and two clustered CYP88 genes on chromosome 3; and a CYP87 gene on chromosome 1) (Shang et al., 2014) (Fig. 2). By extending such analyses out across the wider Solanaceae (for the steroidal glycoalkaloids) and the Cucurbitaceae (for the cucurbitacins), it should be possible to unearth other related pathways and enzymes that represent further variations on these themes. Such investigations will also help to establish whether these pathways may represent evolutionary snapshots of clusters in the process of assembling/disassembling/diversifying.
In maize, the five genes that are necessary and sufficient for the synthesis of the cyclic hydroxamic acid DIBOA are located in a single core cluster, the Bx cluster (Frey et al., 1997, 2003; von Rad et al., 2001; Jonczyk et al., 2008). The Bx cluster is believed to be of ancient origin. However, in wheat and rye, which also make cyclic hydroxamic acids, the genes for DIBOA biosynthesis are found as two subclusters located on different chromosomes. The organization of these genes in these two other cereals has been attributed to a reciprocal translocation in a common ancestor of wheat and rye (Nomura et al., 2002; Sue et al., 2011).
4. Summary – the architectures of plant metabolic clusters
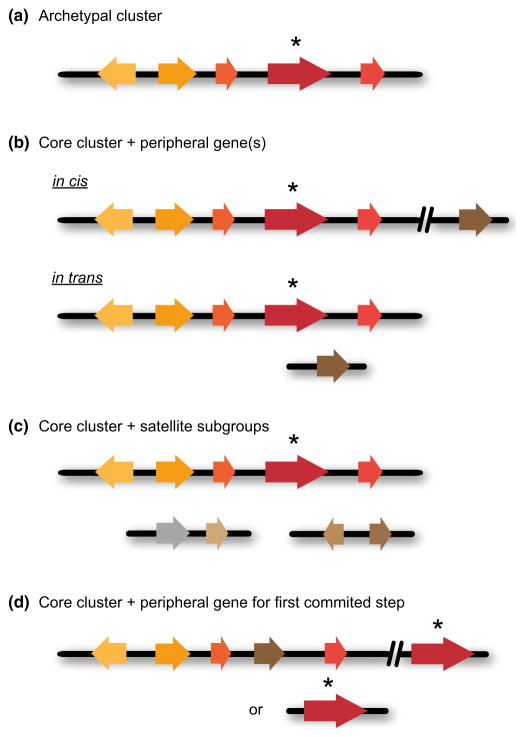
The archetypal plant metabolic cluster (Fig. 4a) is compact and contains contiguous pathway genes, including the gene encoding the enzyme for the first committed step in the pathway. In other cases, the genes for most of the metabolic pathway steps (including the gene for the first committed step) are clustered and contiguous, but there may be one or more peripheral genes encoding other pathway steps. These peripheral genes may be loosely linked to the core cluster (in cis) or unlinked (in trans) (Fig. 4b). As more plant metabolic clusters are characterized, the relative importance of in cis and in trans peripheral genes should become clearer. It will also be interesting to establish whether these peripheral genes tend to encode particular types of enzymes (the earlier examples suggest that sugar transferase genes may have a tendency to be peripheral), although it is too early to make any definitive generalizations. In other cases, the pathways are more fragmented. In these situations, the bulk of the pathway genes, including the gene for the first committed step, are organized in a core cluster with other genes present as satellite subgroups elsewhere in the genome (Fig. 4c). For the examples reported so far, these satellite subgroups contain genes for one or two different types of tailoring enzyme. In a fourth scenario the gene for the first committed step may be peripheral, as is the case for the α-tomatine cluster (Fig. 4d). These peripheral genes would, however, be expected to be strongly coexpressed with the clustered genes encoding the other pathway steps.
Fig. 4.
Types of cluster organization. (a) Archetypal cluster. The archetypal plant metabolic cluster is compact and contains contiguous pathway genes, including the gene encoding the enzyme for the first committed step in the pathway. (b) Core cluster plus peripheral gene(s). The genes for most of the metabolic pathway steps (including the gene for the first committed step) are clustered and contiguous, but there may be one or more peripheral genes encoding other pathway steps. These peripheral genes may be loosely linked to the core cluster (in cis) or may be unlinked (in trans). (c) Core cluster plus satellite subgroups. The bulk of the pathway genes, including the gene for the first committed step, are organized in a core cluster with small groups of two to three other genes present as satellite subgroups elsewhere in the genome. For the examples reported so far, these satellite subgroups contain genes for one or two different types of tailoring enzymes. (d) Core cluster plus peripheral gene for first step. In this scenario the core cluster encodes three or more different types of tailoring enzyme and the gene encoding the first committed step is elsewhere in the genome (either in cis or in trans). In each case the gene encoding the first committed pathway step is indicated in dark red and marked with an asterisk.
III. Cluster regulation
Clustered pathways for natural product biosynthesis are common in bacteria and fungi, and often contain genes encoding pathway-specific transcription factors that regulate the biosynthetic genes within the cluster (Chater & Bibb, 1997; Brakhage, 2013). Loss-of-function mutations in these transcription factors result in loss of expression of pathway genes, while overexpression can lead to coordinate up-regulation and enhanced metabolite production (Uguru et al., 2005; Bergmann et al., 2007). Such transcription factors are therefore very useful tools, both for pathway delineation and for metabolic engineering. By contrast, the clusters that have been reported so far in plants do not appear to contain genes for pathway-specific transcription factors. However, recent advances are beginning to yield insights into how plant metabolic gene clusters are regulated.
In some cases, quite a bit is known about the temporal and spatial expression patterns of particular clusters. For example, the major product of the avenacin pathway for the synthesis of antimicrobial triterpene glycosides in oats, avenacin A-1, has strong autofluorescence under ultraviolet illumination and can be readily localized to the epidermal cells of the root tips and lateral roots, consistent with its role in defence against soilborne pathogens (Osbourn et al., 1994). mRNA in situ hybridization indicates that the characterized avenacin pathway genes, including the first and last steps in the pathway, are all expressed specifically in these cell types (Qi et al., 2004, 2006; Mugford et al., 2009, 2013; Wegel et al., 2009). Regulation of the avenacin pathway is therefore under strict developmental control and does not appear to be altered in response to pathogen attack or abiotic stress treatment. The avenacin pathway has evolved relatively recently in evolutionary time (since the divergence of oats from other cereals some 25 million yr ago) and the ability to synthesize avenacins is restricted to oat. Interestingly, however, the promoters of the avenacin pathway genes retain very similar expression patterns when introduced into A. thaliana, rice and other plant species as reporter fusion constructs, i.e. they are expressed in the epidermal cells of the tips of the primary and lateral roots, suggesting that this pathway has hooked into an ancient root development process that is conserved across monocots and eudicots (Kemen et al., 2014). The class IV homeodomain leucine zipper transcription factor AsHDZ1 has been implicated in regulation of the avenacin gene cluster, but its function has not yet been validated in oat (Kemen et al., 2014).
The momilactone and phytocassane/oryzalide clusters in rice make diterpenes that are implicated in plant defence. Momilactones, oryzalexins and phytocassanes are phytoalexins (i.e. are produced in the leaves in response to pathogen attack or elicitor treatment), while oryzalides are present in unchallenged plants and so are regarded as phytoanticipins (Schmelz et al., 2014). Importantly, momilactones are also produced constitutively in the roots as part of normal growth and development, where they have been implicated in allelopathy (Xu et al., 2011; Kato-Noguchi & Peters, 2013). These rice diterpene clusters are therefore subject to different types of regulation, both developmental and inducible. A chitin oligosaccharide elicitor-inducible basic leucine zipper (bZIP) transcription factor (OsTGAP1) coordinately regulates the gene clusters for the synthesis of momilactone and phytocassane/oryzalide diterpenes in rice (Okada et al., 2009; Miyamoto et al., 2014). However, this effect is not direct, as the promoters of the diterpene cluster genes are not targets for OsTGAP1 binding (Miyamoto et al., 2014). A conserved APETALA2/ethylene response factor (AP2/ERF) transcription factor (GAME9) is involved in regulation of synthesis of the cholesterol-derived steroidal glycoalkaloids α-tomatine in tomato, and α-solanine and α-chaconine in potato (Cárdenas et al., 2016). GAME9 control of steroidal glycoalkaloid biosynthesis is not restricted to the clustered pathway genes and also encompasses upstream genes of the cholesterol pathway. Although concerted changes in cluster expression are observed in GAME9 knockdown and overexpression lines, it is not yet known whether GAME9 directly regulates each cluster gene. Initial data suggest that GAME9 binds to the promoter sequences of some of the cluster genes (GAME4 and GAME7) cooperatively with an MYC2 transcription factor (Cárdenas et al., 2016).
In cucumber, two transcription factors that regulate cucurbitacin C synthesis in leaves and fruit, respectively, have been identified – Bl (Bitter leaf) and Bt (Bitter fruit) (Shang et al., 2014). Bl encodes a basic helix–loop–helix (bHLH) transcription factor required for cluster regulation. Yeast one-hybrid analysis, tobacco transient reporter (luciferase) activation assays, chromatin immunoprecipitation analysis and electrophoretic mobility-shift assays (EMSAs) have collectively shown that Bl can selectively bind to cis elements within the promoter of the gene encoding the first step in the cucurbitacin C pathway Bi. Bt is also a bHLH transcription factor and lies within 8.5 kb of Bl. The bt allele confers the nonbitterness trait of cultivated cucumber and has been selected during domestication. Bl and Bt can specifically bind to the promoters of nine different cucurbitacin C pathway genes, including those of the core cluster and satellite subgroup. Bl and Bt are on a different chromosome to the cucurbitacin C core biosynthetic cluster and satellite pathway genes.
In some cases, the genes within reported clusters do not show consistent coexpression. For example, the cyanogenic glycoside cluster in Lotus japonicus and the terpene cluster in tomato deviate from the general coregulation pattern of metabolic gene clusters. The former is characterized by overlapping expression of all clustered genes in young leaf tissues. The transcript abundances of CYP79D3 and CYP736 are reduced in older leaf tissue but remain constant for the third pathway gene, UGT85K3 (Takos et al., 2011). The terpene cluster in tomato shows coregulation of subsets of genes. Expression of Sl-CPT1, Sl-TPS19, Sl-TPS20 and Sl-TPS41 is primarily restricted to the stem and leaf trichomes, while Sl-CPT2, Sl-CYP71BN1 and Sl-TPS21 show highest expression in the petiole and Sl-CPT18 is expressed in the roots (Matsuba et al., 2013, 2015). Other examples of inconsistencies in expression patterns of cluster genes have also been reported (von Rad et al., 2001; Swaminathan et al., 2009).
In filamentous fungi there is good evidence that gene clusters for natural product pathways are subject to regulation by chromatin-based mechanisms (Schwab et al., 2007; Bok et al., 2009; Brakhage, 2013). There is a growing body of evidence to indicate that this is also the case in plants. High-resolution DNA fluorescence in situ hybridization (FISH) analysis indicates that expression of the oat avenacin cluster is associated with chromatin decondensation (Wegel et al., 2009). Metabolic gene clusters in A. thaliana, maize, oat and rice show pronounced trimethylation of histone H3 lysine 27 (H3K27me3) and the levels of this chromatin mark are inversely correlated with cluster expression (Yu et al., 2016). H3K27me3 is a conserved histone modification in eukaryotes and is generally associated with gene silencing. Consistent with this, the thalianol and marneral clusters in A. thaliana show increased expression in Polycomb mutants that are compromised in H3K27 trimethylation, and reduced expression in mutants for PICKLE, a positive regulator that counteracts H3K27me3 silencing (Yu et al., 2016). These two A. thaliana clusters are also positively regulated (either directly or indirectly) by the SWR1 chromatin remodelling complex, which is required for the deposition of the histone 2 variant H2A.Z into nucleosomes, suggesting that H2A.Z is required for normal cluster expression. A model is emerging based on analysis of chromatin modifications and mutants in which H3K27me3 is implicated in cluster silencing and H2A.Z in cluster activation (Nützmann & Osbourn, 2015; Yu et al., 2016). Importantly, these features can be used in genome-wide analyses to mine for new metabolic gene clusters (Yu et al., 2016). Although mutations in chromatin regulators have been reported to affect the biosynthesis of phenylpropanoids and glucosinolates, compounds that are made by unclustered pathways (Rider et al., 2004; Bennett et al., 2005; Walley et al., 2008), it is not clear whether these effects are direct or indirect. RNAseq analysis of H2A.Z mutant lines did not support a role for this chromatin mark in the regulation of nonclustered glucosinolate and flavonoid pathways in A. thaliana (Yu et al., 2016).
IV. Why are the genes clustered?
The amount of metabolic diversity represented within the plant kingdom is huge. Very little is known by comparison about the locations of the genes encoding specialized metabolic pathways in plant genomes. Indeed, only a handful (< 50) of plant specialized metabolic pathways have been fully characterized in terms of both their biochemistry and the genomic locations of the pathway genes. As this inventory grows and more plant genome sequences become available, it will be possible to gain a wider overview of the organization of specialized metabolism in plants. The genes for well characterized pathways such as the flavonoid, carotenoid and glucosinolate pathways are patently unlinked (Sonderby et al., 2010; Saito et al., 2013; Nisar et al., 2015). Why, then, should the genes for the natural product biosynthetic pathways shown in Table 1 be clustered? It is clear that for the clusters that have been reported so far there is no evidence of horizontal gene transfer from microbes (Nützmann & Osbourn, 2014). This raises intriguing questions about why some pathways are clustered and others are not.
1. Coinheritance, toxic/bioactive intermediates, coexpression and metabolons
Various theories have been put forward to explain clustering. These theories are not mutually exclusive – neither are they likely to be exhaustive. The overriding matter is the benefit of clustering for the plant, not for the pathway. This brings in the issue of population genetics and the environment, as the benefit to the plant will depend on its ability to compete and survive in nature. Another key point is that these clusters are likely to be dynamic and so in their current manifestation represent a moment in evolutionary time. Thus the clustering phenomenon should be considered in the wider context of the birth, life and death of clusters.
The coinheritance argument
Genetic linkage of genes encoding complex traits that confer a selective advantage will reduce the risk of disruption of these favourable gene sets by recombination. Metabolic diversification in plants is likely to be a reflection of adaptation to survival in particular ecological niches. Many of the compounds shown in Table 1 and Fig. 1 have been variously reported to provide protection against pathogens and herbivores (Arneson & Durbin, 1968; Da Costa & Jones, 1971; Niemeyer, 1988; Fewell & Roddick, 1997; Papadopoulou et al., 1999; Tattersall et al., 2001; Balkema-Boomstra et al., 2003; Toyomasu et al., 2014; Sohrabi et al., 2015), to confer competitiveness against competing plant species (allelopathy) (Nicollier et al., 1983; Xu et al., 2011; Kato-Noguchi & Peters, 2013; Toyomasu et al., 2014), to form protective leaf waxes (Schneider et al., 2016) and to determine bitterness or antinutritional properties (Friedman, 2002, 2006; Shang et al., 2014). Many of the plant metabolic gene clusters reported so far appear to have arisen relatively recently in evolutionary time and are restricted to narrow taxonomic subgroups (e.g. Qi et al., 2004; Field et al., 2011; Matsuba et al., 2013). This ability to assemble new metabolic pathways is quite remarkable, and suggests that plant genomes are highly plastic and versatile. Once a favourable combination of alleles of different pathway genes is found, it will presumably be beneficial to inherit these genes together. Clustering may therefore be a feature of extreme selection for coinheritance and optimization of the best combinations of genes for new metabolic pathways that confer selective advantages. The arguments for cluster formation and maintenance are complex and sit at the interface of molecular biology, population and evolutionary genetics and genomics (Yeaman & Whitlock, 2011; Takos & Rook, 2012).
Toxic/bioactive intermediates
Clusters that are not optimal owing to the presence of maladapted alleles of pathway genes, or because of deletion, insertion or mutation, will be compromised in their ability to make protective chemicals. Importantly, there is growing evidence to indicate that interference with clustered pathways in plants through mutation, RNAi-mediated silencing or overexpression of pathway genes can result in accumulation of toxic intermediates (von Rad et al., 2001; Kristensen et al., 2005; Field & Osbourn, 2008; Mylona et al., 2008; Field et al., 2011; Itkin et al., 2011; Xu et al., 2011). Interestingly, in some cases, accumulation of pathway intermediates can have more subtle effects, such as increased root length in A. thaliana (Field & Osbourn, 2008) and altered cell specification, leading to super-hairy roots in oat (Kemen et al., 2014). Clustering may therefore not only enable coinheritance of genes for complete metabolic pathways, but may also protect against ectopic production of toxic/bioactive intermediates. The cause of various human disorders has been attributed to the accumulation of toxic intermediates, which occurs when the product of an enzyme is not used as a substrate by a downstream neighbour in the metabolic network. It has been proposed by analogy that clustering of metabolic pathway genes in filamentous fungi may be an adaptation against the accumulation of toxic intermediates (McGary et al., 2013). Further research is required to establish whether or not this may be the case in plants. Nevertheless, the various phenotypes observed in plants when metabolic pathways are disrupted provide some tantalizing glimpses into possible scenarios in pathway evolution and the likely consequences for plant growth, development and survival.
Coexpression
Physical clustering may provide opportunities for pathway regulation at the levels of localized chromatin modifications and nuclear organization, as discussed earlier. This could be important in order to avoid expression of incomplete pathways with consequent reduced product formation and accumulation of bioactive intermediates. Colocalization of pairs or groups of genes is known to increase the likelihood of coregulation of genes. Bidirectional promoters, locally spreading gene silencing mechanisms and formation of local chromatin domains have all been shown to be important for concerted transcription of colocalized genes in eukaryotes (Hurst et al., 2004). Large-scale expression analysis also indicates a tendency towards coregulation of neighbouring genes in different eukaryotic species (Cohen et al., 2000; Kruglyak & Tang, 2000; Boutanaev et al., 2002; Lercher et al., 2002; Spellman & Rubin, 2002; Kosak & Groudine, 2004). Underlining this pattern, a recent study reports a significant correlation of the expression profiles of neighbouring genes during evolution (Ghanbarian & Hurst, 2015). Furthermore, a number of examples throughout the eukaryotes provide evidence for local regulation patterns associated with clusters of functionally related genes.
Localized chromatin modification has been implicated in the stringent regulation of other types of eukaryotic gene clusters, including the major histocompatibility (MHC) locus, the β-globin locus and the homeodomain (Hox) transcription factor cluster in animals and a catabolic gene cluster in yeast (the DAL cluster) (Meneghini et al., 2003; Gialitakis et al., 2006; Garrick et al., 2008; Osbourn & Field, 2009; Montavon & Duboule, 2013). Like the A. thaliana thalianol and marneral clusters, the DAL cluster has pronounced H2A.Z marking that readily distinguishes it at the genome-wide level (Meneghini et al., 2003; Guillemette et al., 2005; Wong & Wolfe, 2005). Metabolic clusters in filamentous fungi are also demarcated by chromatin marks that are associated with activation and silencing (Bok et al., 2009; Reyes-Dominguez et al., 2010; Nützmann et al., 2011; Connolly et al., 2013). DNA-FISH and chromosome conformation capture analysis have shown that local three-dimensional chromatin domains are formed at animal gene clusters (Tolhuis et al., 2002; Simonis et al., 2006; Noordermeer et al., 2011; Tena et al., 2011; Vieux-Rochas et al., 2015). It has been suggested that these domains may separate cluster regions from the surrounding chromosomal areas and facilitate efficient and concerted silencing and activation of the clustered genes.
Work on a second catabolic cluster from yeast, the GAL gene cluster for galactose assimilation, indicates that changing the conformation of this three-gene cluster by placing one of the genes in trans results in reduced coregulation but does not incur a fitness cost, leading to the conclusion that genetic linkage is probably the selective force driving the origin and maintenance of GAL gene clusters (Lang & Botstein, 2011). By contrast, disruption of gene order in the DAL gene cluster, which is required for allantoin degradation under nitrogen limitation conditions, leads to both changes in expression of cluster genes and reduced fitness in starvation conditions (Naseeb & Delneri, 2012).
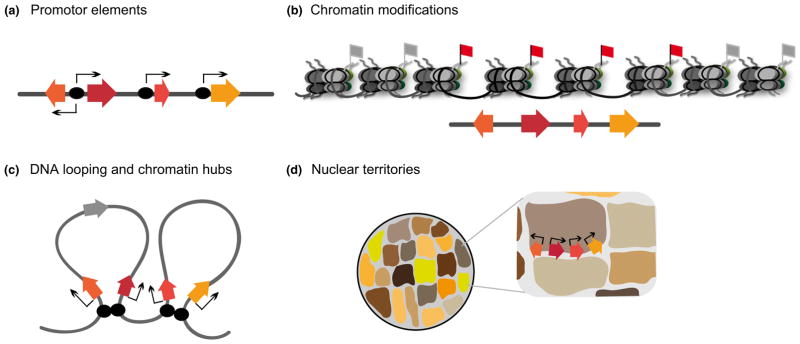
Investigation of the regulation of plant metabolic gene clusters at all levels – from analysis of promoters and transcription factors through to chromatin modifications, DNA looping and higher-level nuclear organization – is likely to reveal important new insights into the regulation of specialized metabolism and should also shed light on how new coregulated clustered pathways become established in plants (Fig. 5).
Fig. 5.
Schematic representation of potential levels of gene cluster regulation. (a) Coregulated promotor motifs (black ovals) are located upstream of each cluster gene. (b) Distinct chromatin modifications facilitate the formation of readily accessible open chromatin structures (red flag) and closed condensed chromatin regions (grey flag). An open chromatin structure will allow transcription, whereas a closed structure will suppress it. (c) At the tertiary level, cis-acting regulatory elements (black ovals) associate to form chromatin hubs, looping out the intervening DNA and bringing the cluster genes together in close proximity. The genes close to the regulatory elements are expressed (coloured), whereas genes located further away are silent (grey). (d) Gene clusters may be located in discrete nuclear territories that are characterized by distinct chromosomal conformations.
Metabolic channelling/metabolons
The formation of multienzyme complexes (metabolons) in which pathway components are held together by noncovalent interactions has been proposed as a mechanism for enhancing local accumulation of substrates and avoiding the release of toxic intermediates through metabolic channelling (Jorgensen et al., 2005). There is evidence to suggest that the pathway for the biosynthesis of the cyanogenic glycoside dhurrin in sorghum is highly channelled and that the pathway enzymes are organized as a metabolon (Møller & Conn, 1980; Kristensen et al., 2005; Nielsen et al., 2008; Møller, 2010). It has also been suggested that the STORR fusion protein that has recently been shown to catalyse the first committed step in morphinan biosynthesis may facilitate channelling of unstable or reactive pathway intermediates (Winzer et al., 2015). By contrast, for several other clustered pathways including the maize DIBOA/DIMBOA and oat avenacin pathways, the pathway enzymes are not colocalized and are distributed across several subcellular compartments (Frey et al., 1997, 2009; Jonczyk et al., 2008; Mugford et al., 2009, 2013). Thus metabolic channelling does not appear to be a general feature of clustered metabolic pathways. Furthermore, it is not clear why physical clustering of pathway genes may be expected to be beneficial for the formation of multienzyme complexes, although conceivably coordinate transcription of genes in distinct nuclear areas may facilitate cotranslation and subsequent colocalization of the cognate protein products.
In summary, the above arguments cannot readily be untangled and are unlikely to represent the full story. Further work is needed to shed light on the selective forces and mechanisms that drive the assembly and maintenance of plant metabolic gene clusters and on the significance of clustering for pathway function. It will also be important to explore the exceptions to the rule – where clusters are split (e.g. the DIBOA/DIMBOA pathway in wheat and rye) or have additional peripheral or satellite genes – as these examples are likely to be helpful when investigating the above possibilities.
2. Assembly of clustered pathways
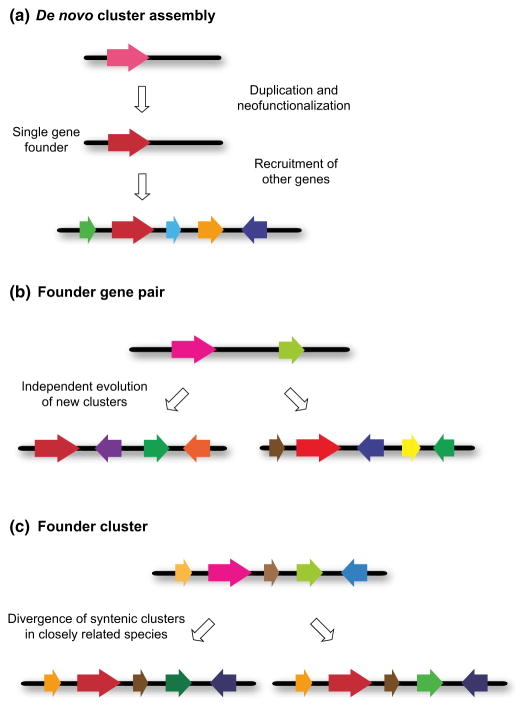
Based on knowledge of the plant metabolic gene clusters reported so far, several scenarios for the assembly of clustered pathways can be envisaged. Although these are illustrated separately here (Fig. 6), based on existing evidence, these different scenarios are in reality likely to represent snapshots in an evolutionary continuum that, once fully understood, will reveal the events underlying the full life cycle – the birth, life and death – of metabolic clusters. In some cases the gene for the first committed step in plant metabolic clusters is likely to have been recruited either directly or indirectly from primary metabolism and to have ‘seeded’ the formation of a multigene cluster (Fig. 6a). An example of this is the avenacin cluster in oat. The first step in the avenacin pathway is the cyclization of 2,3-oxidosqualene to the triterpene scaffold β-amyrin, catalysed by the oxidosqualene cyclase enzyme β-amyrin synthase (SAD1) (Haralampidis et al., 2001). The oat β-amyrin synthase gene has been recruited from the sterol pathway and is a divergent relative of cycloartenol synthase (Haralampidis et al., 2001; Qi et al., 2004). The avenacin pathway is restricted to oats (Avena species) and has arisen some 25 million yr ago, after the divergence of oats from other cereals and grasses (Qi et al., 2004). This duplicated gene then appears to have served as the founder gene for the de novo formation of the avenacin cluster. The second step in the avenacin pathway (the divergent CYP51 enzyme SAD2) has also been recruited from sterol biosynthesis, from obtusifoliol 14α-demethylase. However, the cycloartenol synthase and obtusifoliol 14α-demethylase genes are not linked to the avenacin cluster or to each other in oat (Qi et al., 2004, 2006) and there is no evidence for linkage of these highly conserved sterol biosynthetic genes in A. thaliana or rice.
Fig. 6.
Origins of plant metabolic clusters. The scenarios shown are based on current knowledge of characterized plant metabolic clusters (see text). (a) De novo cluster assembly. The first step in this process is the recruitment of the gene for the first committed step in the pathway by gene duplication and neofunctionalization. This gene then ‘seeds’ the formation of a multigene cluster for a new metabolic pathway by an as yet unknown mechanism. (b) Formation of new metabolic clusters from a founder gene pair template. In this scenario, an ancestral gene pair (e.g. encoding a terpene synthase and a cytochrome P450 (CYP) enzyme) may give rise to different metabolic clusters for new natural product pathways as a result of independent events involving gene rearrangement and recruitment of new genes. This is exemplified by the Arabidopsis thaliana thalianol and marneral clusters (Field et al., 2011). These are both triterpene pathways, but the scaffolds are not the same and the subsequent modifications are different. For both (a) and (b) the process of cluster formation will be accompanied by refinement and optimization of individual pathway components and establishment of a functional gene neighbourhood that allows for coordinate pathway control. (c) Diversification of an ancestral founder cluster in syntenic regions of closely related taxonomic lineages. In these cases, these pathways may have a common scaffold but make slightly different compounds.
In other cases, there is evidence that cluster founding events may involve pairs of genes rather than single ones (Fig. 6b). For example, the A. thaliana thalianol and marneral clusters, both of which make triterpenes, may have been founded by the duplication of an ancestral gene pair consisting of an oxidosqualene cyclase gene and a CYP705 gene. Independent events involving gene rearrangement and recruitment of additional genes would then have led to the formation of the present-day clusters (Field et al., 2011). These clusters formed relatively recently in evolutionary time after the α-duplication event within the Brassicales 4–20 million yr ago. Phylogenetic analysis of the DIBOA/DIMBOA gene cluster in maize suggests a similar cluster assembly path to that proposed by Field et al. (2011) for the thalianol and marneral clusters. A cluster founding event involving colocalization of the Bx1 and Bx2 genes, which together are required for the first two steps in the pathway, is believed to have been followed by recruitment of additional biosynthesis genes and cluster extension (Frey et al., 2009; Dutartre et al., 2012). In contrast to the thalianol and marneral clusters, the DIBOA/DIMBOA cluster is thought to be of ancient origin. A complex terpene cluster in tomato only partially follows this pattern. Here, after the colocalization of an original pair of biosynthesis genes (TPS41 and diTPS1), local duplications, rearrangements, gene recruitment and neofunctionalization have led to the entanglement of two functionally active clusters with different products and expression patterns (Matsuba et al., 2013).
A recent study of terpene diversification and the evolution of terpene biosynthetic genes across multiple plant species has provided interesting insights into mechanisms of metabolic cluster formation (Boutanaev et al., 2015). Systematic analysis of 17 sequenced plant genomes was carried out to determine the organization and evolution of terpene pathway genes. This study revealed that terpene synthase (TS) and CYP genes are clustered far more often than would be expected by chance, indicating nonrandom association of TS and CYP gene pairs within the surveyed genomes. All previously characterized terpene metabolic clusters were identified in this analysis along with other new candidate clusters, several of which were then experimentally validated (for Ricinus communis, A. thaliana and C. sativus). These experiments further demonstrated that certain TS/CYP pairings predominate, so providing signals for key events that are likely to have shaped terpene diversity. Importantly, comparison of the TS/CYP pairs from this analysis has provided evidence for different mechanisms of terpene cluster assembly in eudicots and monocots. In the eudicots, microsyntenic blocks of TS/CYP gene pairs duplicate and provide templates for the evolution of new clustered pathways. By contrast, in the monocots, new terpene clusters arise by mixing and matching of TS and CYP genes through dynamic genome rearrangements. These findings are consistent with the proposed model for the origin of the thalianol and marneral clusters from a common ancestral gene pair in A. thaliana (Field et al., 2011) and also with the single founder gene scenario proposed for the oat avenacin cluster (Qi et al., 2004, 2006; Mugford et al., 2009, 2013).
The thalianol and marneral clusters are located in regions of the A. thaliana genome that are significantly enriched in transposable elements and are located in dynamic chromosome regions (Field et al., 2011; Field & Osbourn, 2012). Other plant metabolic clusters also contain transposable elements that may have some function in gene rearrangement for cluster formation (e.g. Qi et al., 2004; Wilderman et al., 2004; Shimura et al., 2007; Takos et al., 2011; Winzer et al., 2012; Krokida et al., 2013). The flanking regions of the monocot TS/CYP gene pairs identified by Boutanaev et al. (2015) were significantly enriched in transposable elements relative to the norm for all TS and CYP genes across the monocot genomes analysed. A small but significant enrichment in transposable elements was also observed for eudicots when clustered and dispersed genes were compared (Boutanaev et al., 2015). Thus transposable element-mediated recombination may contribute to cluster formation in both monocots and eudicots.
A third scenario for cluster formation is the apparent diversification of a founder cluster in the syntenic regions of closely related taxonomic lineages (Fig. 6c). Here there is clear synteny between the metabolic clusters within the related species but each species makes slightly different compounds (Table 1; Fig. 1). This is exemplified by the steroidal glycoalkaloid clusters in tomato and potato. Interestingly, these pathways are the most fragmented of the clustered pathways reported so far. It is not yet clear whether the satellite/peripheral genes may have formed part of an ancestral progenitor cluster or whether they represent more recent acquisitions.
V. Conclusions and outlook
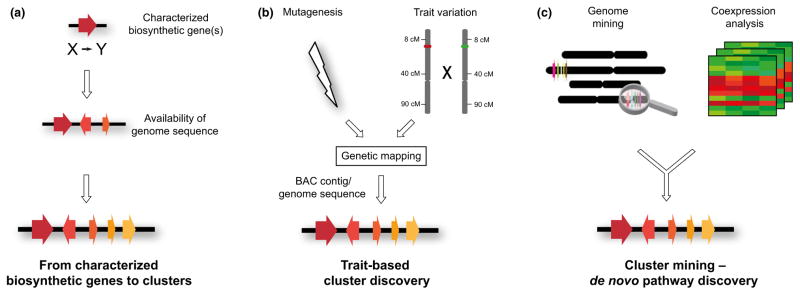
Technological advances in sequencing and bioinformatics coupled with the growing body of knowledge about the organization of metabolic pathways in plant genomes now hold great promise for accelerating the discovery and elucidation of new natural product pathways. Genetics-driven, trait-based approaches have proved to be very powerful and have led to the discovery of clustered pathways that determine crop traits that are important for food security, food quality and medicine (Fig. 7). In the cases of the oat avenacin cluster and the poppy noscapine cluster, this was achieved using BAC contigs in the absence of publicly available genome sequence information (Qi et al., 2006; Osbourn et al., 2012; Winzer et al., 2012; Mugford et al., 2013). Strategies for pathway discovery are now beginning to shift from characterization of individual pathway steps to genomics-based prediction and validation of entire pathways (Fig. 8). Increased knowledge of plant metabolic gene clusters and the systematic analysis, curation and standardization of these clusters and their parts (Medema et al., 2015; Patron et al., 2015) will feed back into future genome mining efforts, so enhancing the genome to natural product discovery pipeline. It will also establish how common this clustering phenomenon is in plants and shed further light on the raison d’être for metabolic clusters. Last but by no means least, it is intriguing to consider whether plant genomes might harbour other types of nonhomologous gene clusters that have functions other than in secondary metabolism.
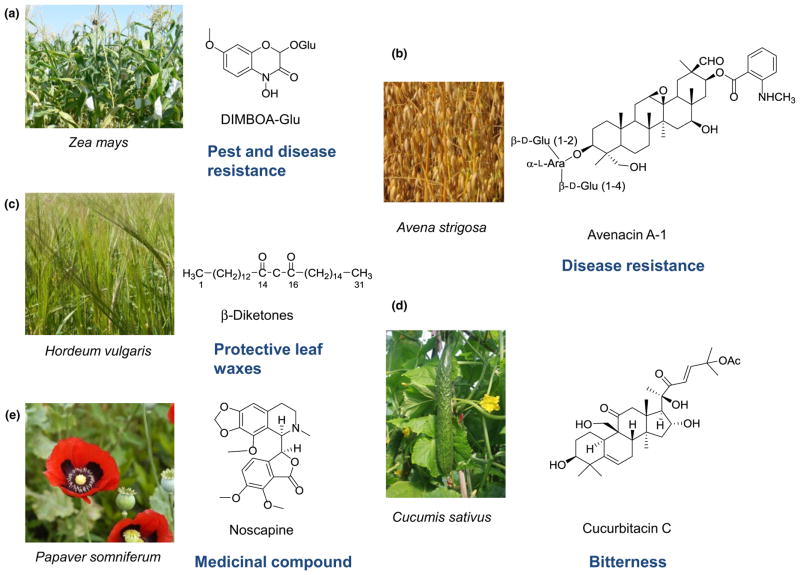
Fig. 7.
Trait-based cluster discovery. Examples of compounds of agronomic/medicinal importance for which the underlying metabolic pathways were characterized bytrait-based cluster discovery are shown. The images of plants are reproduced with the kind permission of: (a) Paul Cristou, Institució Catalanade Recerca I Estudis, Lleida, Spain; (b) Anthony Pugh, Institute for Biological, Environmental and Rural Sciences, Aberystwyth, UK; (c) John Innes Centre Photography; (d) Sanwen Huang, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences; (e) Tanja Niggendijker/Creative Commons.
Fig. 8.
Strategies used for identification of plant metabolic clusters. (a) Biochemical characterization of one or more pathway steps in the pregenome era. The existence of clustering may then become apparent once genome sequence data for the species of interest is available. (b) Investigation of induced or natural variation in a trait provides genetic evidence for a multigene locus. The relevant region is then further defined by mapping. The underlying cluster is identified by assembly and sequencing of bacterial artificial chromosome (BAC) contigs spanning the region (in the absence of genome sequence information) or by exploiting available genome sequence data in the region of the locus. (c) Discovery of metabolic clusters for new pathways by genome mining. Physical clustering of genes implicated in a specialized metabolic pathway in combination with transcriptomics data enables the delineation of new candidate metabolic gene clusters.
Acknowledgments
We thank Mike Ambrose and Thu-Thuy Dang (John Innes Centre, UK), Eran Pichersky (University of Michigan, USA) and Peter Facchini (University of Calgary, Canada) for helpful discussions and Ian Graham (University of York) for material for inclusion in Fig. 3. A.O.’s laboratory is supported by the UK Biotechnological and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Grant ‘Understanding and Exploiting Plant and Microbial Metabolism’ (BB/J004561/1) and the John Innes Foundation. We also acknowledge the joint Engineering and Physical Sciences Research Council/BBSRC-funded OpenPlant Synthetic Biology Research Centre grant BB/L014130/1 (A.O., H-W.N.) and National Institutes of Health Genome to Natural Products Network award U101GM110699 (A.O., A.H.).
References
- Andersen MD, Busk PK, Svendsen I, Møller BL. Cytochromes P450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin. Journal of Biological Chemistry. 2000;275:1966–1975. doi: 10.1074/jbc.275.3.1966. [DOI] [PubMed] [Google Scholar]
- Arneson PA, Durbin RD. Sensitivity of fungi to alpha-tomatine. Phytopathology. 1968;58:536–537. [Google Scholar]
- Bak S, Nielsen HL, Halkier BA. The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to oxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Molecular Biology. 1998;38:725–734. doi: 10.1023/a:1006064202774. [DOI] [PubMed] [Google Scholar]
- Balkema-Boomstra AG, Zijlstra S, Verstappen FWA, Inggamer H, Mercke PE, Jongsma MA, Bouwmeester HJ. Role of cucurbitacin C in resistance to spider mite (Tetranychus urticae) in cucumber (Cucumis sativus L.) Journal of Chemical Ecology. 2003;29:225–235. doi: 10.1023/a:1021945101308. [DOI] [PubMed] [Google Scholar]
- Beaudoin GAW, Facchini PJ. Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta. 2014;240:19–32. doi: 10.1007/s00425-014-2056-8. [DOI] [PubMed] [Google Scholar]
- Bennett RN, Wenke T, Freudenberg B, Mellon FA, Ludwig-Muller J. The tu8 mutation of Arabidopsis thaliana encoding a heterochromatin protein 1 homolog causes defects in the induction of secondary metabolite biosynthesis. Plant Biology. 2005;7:348–357. doi: 10.1055/s-2005-837634. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nature Chemical Biology. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, et al. Chromatin-level regulation of biosynthetic gene clusters. Nature Chemical Biology. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- Boutanaev A, Zi J, Nelson DR, Mugford ST, Peters RJ, Osbourn A. Investigation of terpene diversification across multiple sequenced plant genomes. PNAS. 2015;112:E81–E88. doi: 10.1073/pnas.1419547112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycheva S, Daviet L, Wolfender JL, Fitzpatrick TB. The rise of operon-like gene clusters in plants. Trends Plant Science. 2014;19:447–459. doi: 10.1016/j.tplants.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Brakhage AA. Regulation of fungal secondary metabolism. Nature Reviews: Microbiology. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- Cárdenas PD, Sonawane PD, Pollier J, Vanden Bossche R, Dewangan V, Weithorn E, Tal L, Meir S, Rogachev I, Malitsky S, et al. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nature Communications. 2016;7:10654. doi: 10.1038/ncomms10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary JW, Klich MA, Beltz SB. Characterization of aflatoxin-producing fungi outside of Aspergillus section Flavi. Mycologia. 2005;97:425–432. doi: 10.3852/mycologia.97.2.425. [DOI] [PubMed] [Google Scholar]
- Castillo DA, Kolesnikova MD, Matsuda SP. An effective strategy for exploring unknown metabolic pathways by genome mining. Journal of the American Chemical Society. 2013;135:5885–5894. doi: 10.1021/ja401535g. [DOI] [PubMed] [Google Scholar]
- Chater KF, Bibb MJ. Regulation of bacterial antibiotic production. In: Kleinkauf H, Von Dohren H, editors. Products of secondary metabolism. Vol. 7. Weinheim, Germany: VCH; 1997. pp. 57–105. [Google Scholar]
- Chen X, Dang T-TT, Facchini PJ. Noscapine comes of age. Phytochemistry. 2015;111:7–13. doi: 10.1016/j.phytochem.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant Journal. 2011;66:212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- Cimermancic P, Medema MH, Claesen J, Kurita K, Brown LCW, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nature Genetics. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- Connolly LR, Smith KM, Freitag M. The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genetics. 2013;9:e1003916. doi: 10.1371/journal.pgen.1003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa CP, Jones CM. Cucumber beetle resistance and mite susceptibility controlled by the bitter gene in Cucumis sativus L. Science. 1971;172:1145–1146. doi: 10.1126/science.172.3988.1145. [DOI] [PubMed] [Google Scholar]
- De Bernonville TD, Clastre M, Besseau S, Oudin A, Burlat V, Glévarec G, Lanoue A, Papon N, Gigliolo-Guivarc’h N, St-Pierre B, et al. Phytochemical genomics of the Madagascar periwinkle: unravelling the last twists of the alkaloid engine. Phytochemistry. 2015;113:9–23. doi: 10.1016/j.phytochem.2014.07.023. [DOI] [PubMed] [Google Scholar]
- Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nature Chemical Biology. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutartre L, Hilliou F, Feyereisen R. Phylogenomics of the benzoxazinoid biosynthetic pathway of Poaceae: gene duplications and origin of the Bx cluster. BMC Evolutionary Biology. 2012;12:64–64. doi: 10.1186/1471-2148-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eich E. Solanaceae and convolvulaceae: secondary metabolites: biosynthesis, chemotaxonomy, biological and economic significance. Berlin, Germany: Springer; 2008. [Google Scholar]
- Erlich KC, Yu J, Cotty PJ. Aflatoxin biosynthesis gene clusters and flanking regions. Journal of Applied Microbiology. 2005;99:518–527. doi: 10.1111/j.1365-2672.2005.02637.x. [DOI] [PubMed] [Google Scholar]
- Falara V, Akhtar TA, Nguyen TTH, Spyropoulou EA, Bleeker PM, Schauvinhold I, Matsuba Y, Bonini ME, Schilmiller AL, Last RL, et al. The tomato terpene synthase gene family. Plant Physiology. 2011;157:770–789. doi: 10.1104/pp.111.179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell AM, Roddick JG. Potato glycoalkaloid impairment of fungal development. Mycological Research. 1997;101:597–603. [Google Scholar]
- Field B, Fiston-Lavier A-S, Kemen A, Geisler K, Quesneville H, Osbourn AE. Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proceedings of the National Academy of Sciences, USA. 2011;108:16116–16121. doi: 10.1073/pnas.1109273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Osbourn AE. Metabolic diversification – independent assembly of operon-like gene clusters in different plants. Science. 2008;320:543–547. doi: 10.1126/science.1154990. [DOI] [PubMed] [Google Scholar]
- Field B, Osbourn A. Order in the playground. Mobile Genetic Elements. 2012;2:46–50. doi: 10.4161/mge.19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Morant M, Jørgensen B, Olsen CE, Asamizu E, Sato S, Tabata S, Bak S. Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiology. 2004;135:71–84. doi: 10.1104/pp.103.038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grun S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- Frey M, Huber K, Park WJ, Sicker D, Lindberg P, Meeley RB, Simmons CR, Yalpani N, Gierl A. A 2-oxoglutarate-dependent dioxygenase is integrated in DIMBOA-biosynthesis. Phytochemistry. 2003;62:371–376. doi: 10.1016/s0031-9422(02)00556-3. [DOI] [PubMed] [Google Scholar]
- Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A. Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry. 2009;70:1645–1651. doi: 10.1016/j.phytochem.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Friedman M. Tomato glycoalkaloids: role in the plant and in the diet. Journal of Agricultural and Food Chemistry. 2002;50:5751–5780. doi: 10.1021/jf020560c. [DOI] [PubMed] [Google Scholar]
- Friedman M. Potato glycoalkaloids: role in the plant and in the diet. Journal of Agricultural and Food Chemistry. 2006;54:8655–8681. doi: 10.1021/jf061471t. [DOI] [PubMed] [Google Scholar]
- Garrick D, De Gobbi M, Samara V, Rugless M, Holland M, Ayyub H, Lower K, Sloane-Stanley J, Gray N, Koch C, et al. The role of the polycomb complex in silencing alpha-globin gene expression in nonerythroid cells. Blood. 2008;112:3889–3899. doi: 10.1182/blood-2008-06-161901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, Nims E, Cui Y, O’Connor SE. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature. 2012;492:138–142. doi: 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]
- Ghanbarian AT, Hurst LD. Neighbouring genes show correlated evolution in gene expression. Molecular Biology and Evolution. 2015;32:1748–1766. doi: 10.1093/molbev/msv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gialitakis M, Kretsovali A, Spilianakis C, Kravariti L, Mages J, Hoffmann R, Hatzopoulos AK, Papamatheakis J. Coordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by Trichostatin A. Nucleic Acids Research. 2006;34:765–772. doi: 10.1093/nar/gkj462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:8245–8250. doi: 10.1073/pnas.0305876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLOS Biology. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Bak S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 2013;368:20120426. doi: 10.1098/rstb.2012.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralampidis K, Bryan G, Qi X, Papadopoulou K, Bakht S, Melton R, Osbourn AE. A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proceedings of the National Academy of Sciences, USA. 2001;98:13431–13436. doi: 10.1073/pnas.231324698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenze JL. Arabidopsis cytochrome P450s that catalyse the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proceedings of the National Academy of Sciences, USA. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nature Reviews Genetics. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science. 2013;341:175–179. doi: 10.1126/science.1240230. [DOI] [PubMed] [Google Scholar]
- Itkin M, Rogachev I, Alkan N, Rosenberg T, Malitsky S, Masini L, Meir S, Iijima Y, Aoki K, de Vos R, et al. GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell. 2011;23:4507–4525. doi: 10.1105/tpc.111.088732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonczyk R, Schmidt H, Osterrieder A, Fiesselmann A, Schullehner K, Haslbeck M, Sicker D, Hofmann D, Yalpani N, Simmons C, et al. Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiology. 2008;146:1053–1063. doi: 10.1104/pp.107.111237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen K, Rasmussen AV, Morant M, Nielsen AH, Bjarnholt N, Zagrobelny M, Bak S, Moller BL. Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Current Opinion in Plant Biology. 2005;8:280–291. doi: 10.1016/j.pbi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Kato-Noguchi H, Peters RJ. The roll of momilactones in rice allelopathy. Journal of Chemical Ecology. 2013;39:175–185. doi: 10.1007/s10886-013-0236-9. [DOI] [PubMed] [Google Scholar]
- Kellner F, Kim J, Clavijo BJ, Hamilton JP, Childs KL, Vaillancourt B, Cepela J, Habermann M, Steuernagel B, Clissold L, et al. Genome-guided investigation of plant natural product biosynthesis. Plant Journal. 2015;82:680–692. doi: 10.1111/tpj.12827. [DOI] [PubMed] [Google Scholar]
- Kemen AC, Honkanen S, Melton R, Findlay K, Mugford ST, Hayashi K, Haralampidis K, Rosser S, Osbourn A. Investigation of triterpene synthesis and regulation in oats reveals a role for β-amyrin in determining root epidermal cell patterning. Proceedings of the National Academy of Sciences, USA. 2014;111:8679–8684. doi: 10.1073/pnas.1401553111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AJ, Brown GD, Gilday AD, Larson TR, Graham IA. Production of bioactive diterpenoids in the Euphorbiaceae depends on evolutionarily conserved gene clusters. Plant Cell. 2014;26:3286–3298. doi: 10.1105/tpc.114.129668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch BM, Sibbesen O, Halkier BA, Svendsen I, Møller BL. The primary sequence of cytochrome P450tyr, the multifunctional N-hydroxylase catalyzing the conversion of L-tyrosine to p-hydroxyphenylacetaldhyde oxime in the biosynthesis of the cyanogenic glycoside dhurrin in Sorghum bicolor (L.) Moench. Archives of Biochemistry and Biophysics. 1995;323:177–186. doi: 10.1006/abbi.1995.0024. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Groudine M. Form follows function: the genomic organization of cellular differentiation. Genes & Development. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- Kristensen C, Morant M, Olsen CE, Ekstrøm CT, Galbraith DW, Møller BL, Bak S. Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proceedings of the National Academy of Sciences, USA. 2005;102:1779–1784. doi: 10.1073/pnas.0409233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokida A, Delis C, Geisler K, Garagounis C, Tsikou D, Pena-Rodriguez LM, Katsarou D, Field B, Osbourn AE, Papadopoulou KK. A metabolic gene cluster in Lotus japonicus discloses novel enzyme functions and products in triterpene biosynthesis. New Phytologist. 2013;200:675–690. doi: 10.1111/nph.12414. [DOI] [PubMed] [Google Scholar]
- Kruglyak S, Tang H. Regulation of adjacent yeast genes. Trends in Genetics. 2000;16:109–111. doi: 10.1016/s0168-9525(99)01941-1. [DOI] [PubMed] [Google Scholar]
- Lang GI, Botstein D. A test of the coordinated expression hypothesis for the origin and maintenance of the GAL cluster in yeast. PLoS ONE. 2011;6:e25290. doi: 10.1371/journal.pone.0025290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau W, Sattely ES. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015;349:1224–1228. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nature Genetics. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- Li YF, Tsai KJS, Harvey CJB, Li JJ, Ary BE, Berlew EE, Boehman BL, Findley DM, Friant AG, Gardner CA, et al. Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genetics and Biology. 2016;89:18–28. doi: 10.1016/j.fgb.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe DK, Facchini PJ. Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. Journal of Biological Chemistry. 2007;282:14741–14751. doi: 10.1074/jbc.M611908200. [DOI] [PubMed] [Google Scholar]
- Matsuba Y, Nguyen TT, Wiegert K, Falara V, Gonzales-Vigil E, Leong B, Schafer P, Kudrna D, Wing RA, Bolger AM, et al. Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell. 2013;25:2022–2036. doi: 10.1105/tpc.113.111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba Y, Zi J, Jones AD, Peters RJ, Pichersky E. Biosynthesis of the diterpenoid lycosantalonol via nerylnerly diphosphate in Solanum lycopersicum. PLoS ONE. 2015;10:e0119302. doi: 10.1371/journal.pone.0119302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary KL, Slot JC, Rokas A. Physical linkage of metabolic genes in fungi in an adaptation against the accumulation of toxic intermediate compounds. Proceedings of the National Academy of Sciences, USA. 2013;110:11481–11486. doi: 10.1073/pnas.1304461110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Computational Biology. 2014;10:e1004016. doi: 10.1371/journal.pcbi.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Choo YH, Claesen J, Coates RC, et al. Minimum information about a biosynthetic gene cluster. Nature Chemical Biology. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. Journal of Biological Chemistry. 2000;275:33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- Milkowski C, Strack D. Serine carboxypeptidase-like acyltransferases. Phytochemistry. 2004;66:517–524. doi: 10.1016/j.phytochem.2003.12.018. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Matsumoto T, Okada A, Komiyama K, Chujo T, Yoshikawa H, Nojiri H, Yamane H, Okada K. Identification of target genes of the bZIP transcription factor OsTGAP1, whose overexpression causes elicitor-induced hyperaccumulation of diterpenoid phytoalexins in rice cells. PLoS ONE. 2014;9:e105823. doi: 10.1371/journal.pone.0105823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller BL. Dynamic metabolons. Science. 2010;330:1328–1329. doi: 10.1126/science.1194971. [DOI] [PubMed] [Google Scholar]
- Møller BL, Conn EE. The biosynthesis of cyanogenic glucosides in higher plants – channelling of intermediates in dhurrin biosynthesis by a microsomal system from Sorghum bicolor (Linn.) Moench. Journal of Biological Chemistry. 1980;255:3049–3056. [PubMed] [Google Scholar]
- Montavon T, Duboule D. Chromatin organization and global regulation of Hox gene clusters. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 2013;368:20120367. doi: 10.1098/rstb.2012.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford ST, Louveau T, Melton R, Qi X, Bakht S, Hill L, Tsurushima T, Honkanen S, Rosser SJ, Lomonossoff GP, et al. Modularity of plant metabolic gene clusters: a trio of linked genes that are collectively required for acylation of triterpenes in oat. Plant Cell. 2013;25:1078–1092. doi: 10.1105/tpc.113.110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford ST, Qi X, Bakht S, Hill L, Wegel E, Hughes RK, Papadopoulou K, Melton R, Philo M, Sainsbury F, et al. A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell. 2009;21:2473–2484. doi: 10.1105/tpc.109.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona P, Owatworakit A, Papadopoulou K, Jenner H, Qin B, Findlay K, Hill L, Qi X, Bakht S, Melton R, et al. Sad3 and Sad4 are required for saponin biosynthesis and root development in oat. Plant Cell. 2008;20:201–212. doi: 10.1105/tpc.107.056531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseeb S, Delneri D. Impact of chromosomal inversions on the yeast DAL cluster. PLoS ONE. 2012;7:e42022. doi: 10.1371/journal.pone.0042022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D, Werck-Reichhart D. A P450-centric view of plant evolution. Plant Journal. 2011;66:194–211. doi: 10.1111/j.1365-313X.2011.04529.x. [DOI] [PubMed] [Google Scholar]
- Nicollier GF, Pope DF, Thompson AC. Biological activity of dhurrin and other compounds from Johnson Grass (Sorghum halepense) Journal of Agricultural and Food Chemistry. 1983;31:744–748. [Google Scholar]
- Neilsen KA, Tattersall DB, Jones PR, Møller BL. Metabolon formation in dhurrin biosynthesis. Phytochemistry. 2008;69:88–98. doi: 10.1016/j.phytochem.2007.06.033. [DOI] [PubMed] [Google Scholar]
- Niemeyer HM. Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones), defense chemicals in the Gramineae. Phytochemistry. 1988;27:3349–3358. [Google Scholar]
- Nisar N, Li L, Lu S, Khin NC, Pogson BJ. Carotenoid metabolism in plants. Molecular Plant. 2015;8:68–82. doi: 10.1016/j.molp.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Nomura T, Ishihara A, Imaishi H, Endo T, Ohkawa H, Iwamura H. Molecular characterization and chromosomal localization of cytochrome P450 genes in the biosynthesis of cyclic hydroxamic acids in hexaploid wheat. Molecular Genetics and Genomics. 2002;267:210–217. doi: 10.1007/s00438-002-0653-x. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- Nützmann H-W, Osbourn A. Gene clustering in plant specialized metabolism. Current Opinion in Biotechnology. 2014;26:91–99. doi: 10.1016/j.copbio.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Nützmann H-W, Osbourn A. Regulation of metabolic gene clusters in Arabidopsis thaliana. New Phytologist. 2015;205:503–510. doi: 10.1111/nph.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann HW, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schumann J, Hertweck C, Strauss J, Brakhage AA. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proceedings of the National Academy of Sciences, USA. 2011;108:14282–14287. doi: 10.1073/pnas.1103523108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A, Okada K, Miyamoto K, Koga J, Shibuya N, Nojiri H, Yamane H. OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. Journal of Biological Chemistry. 2009;284:26510–26518. doi: 10.1074/jbc.M109.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Hsu S-C, Small LL. Evidence on the molecular basis of the Ac/ac adaptive cyanogenesis polymorphism in white clover (Trifolium repens L.) Genetics. 2008;179:517–526. doi: 10.1534/genetics.107.080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends in Genetics. 2010;26:449–457. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Osbourn AE, Clarke BR, Lunness P, Scott PR, Daniels MJ. An oat species lacking avenacin is susceptible to infection by Gaeumannomyces graminis var. tritici. Physiological and Molecular Plant Pathology. 1994;45:457–467. [Google Scholar]
- Osbourn AE, Field B. Operons. Cellular and Molecular Life Sciences. 2009;66:3755–3775. doi: 10.1007/s00018-009-0114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A, Papadopoulou KK, Qi X, Field B, Wegel E. Finding and analyzing plant metabolic gene clusters. Methods in Enzymology. 2012;517:113–138. doi: 10.1016/B978-0-12-404634-4.00006-1. [DOI] [PubMed] [Google Scholar]
- Owatworakit A, Townsend B, Louveau T, Jenner J, Rejzek M, Hughes RK, Saalbach G, Qi X, Bakht S, Deb Roy A, et al. Glycosyltransferases from oat (Avena) implicated in the acylation of avenacins. Journal of Biological Chemistry. 2013;288:3696–3704. doi: 10.1074/jbc.M112.426155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn AE. Compromised disease resistance in saponin-deficient plants. Proceedings of the National Academy of Sciences, USA. 1999;96:12923–12928. doi: 10.1073/pnas.96.22.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron NJ, Orzaez D, Marillonnet S, Warzecha H, Matthewman C, Youles M, Raitskin O, Leveau A, Farré G, Rogers C, et al. Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytologist. 2015;208:13–19. doi: 10.1111/nph.13532. [DOI] [PubMed] [Google Scholar]
- Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A. A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proceedings of the National Academy of Sciences, USA. 2004;101:8233–8238. doi: 10.1073/pnas.0401301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bakht S, Qin B, Leggett M, Hemmings A, Mellon F, Eagles J, Werck-Reichhart D, Schaller H, Lesot A, et al. A new function for the most ancient and conserved cytochrome P450 family – from primary sterol biosynthesis to plant defence. Proceedings of the National Academy of Sciences, USA. 2006;103:18848–18853. doi: 10.1073/pnas.0607849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rad U, Huttl R, Lottspeich F, Gierl A, Frey M. Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant Journal. 2001;28:633–642. doi: 10.1046/j.1365-313x.2001.01161.x. [DOI] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Molecular Microbiology. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider SD, Jr, Hemm MR, Hostetler HA, Li HC, Chapple C, Ogas J. Metabolic profiling of the Arabidopsis pkl mutant reveals selective derepression of embryonic traits. Planta. 2004;219:489–499. doi: 10.1007/s00425-004-1254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Motawia MS, Olsen CE, Møller BL, Bak S. Biosynthesis of rhodiocyanosides in Lotus japonicus: rhodiocyanoside A is synthesized from (Z)-2-methylbutanaloxime via 2-methyl-2-butenenitrile. Phytochemistry. 2012;77:260–267. doi: 10.1016/j.phytochem.2012.01.020. [DOI] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiology and Biochemistry. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, Kaneko T, Nakamura Y, Shibata D, Aoki K, et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K, Peters RJ. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant Journal. 2014;79:659–678. doi: 10.1111/tpj.12436. [DOI] [PubMed] [Google Scholar]
- Schneider LM, Adamski NM, Christensen CE, Stuart DB, Hansson M, Uauy C, von Wettstein-Knowles P. The Cer-cqu gene cluster determines three key players in a β-diketone synthase polyketide pathway synthesizing aliphatics in epicuticular waxes. Journal of Experimental Botany. 2016 doi: 10.1093/jxb/erw105. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab EK, Bok J, Tribus M, Galehr J, Graessle S, Keller NP. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryotic Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Ma Y, Zhou Y, Zhang H, Duan L, Chen H, Zeng J, Zhou Q, Wang S, Gu W, et al. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science. 2014;346:1084–1088. doi: 10.1126/science.1259215. [DOI] [PubMed] [Google Scholar]
- Shimura K, Okada A, Okada K, Jikumaru Y, Ko KW, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N, et al. Identification of a biosynthetic gene cluster in rice for momilactones. Journal of Biological Chemistry. 2007;282:34013–34018. doi: 10.1074/jbc.M703344200. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nature Genetics. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Sohrabi R, Huh J-H, Badieyan S, Rakotondraibe LH, Kliebenstein DJ, Sobrado P, Tholl D. In planta variation of volatile biosynthesis: an alternative biosynthetic route to the formation of the pathogen-induced volatile homoterpene DMNT via triterpene degradation in Arabidopsis roots. Plant Cell. 2015;27:874–890. doi: 10.1105/tpc.114.132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates – gene discovery and beyond. Trends in Plant Science. 2010;15:283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. Journal of Biology. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue M, Nakamura C, Nomura T. Dispersed benzoxazinone gene cluster: molecular characterization and chromosomal localization of glycosyltransferase and glucosidase genes in wheat and rye. Plant Physiology. 2011;157:985–997. doi: 10.1104/pp.111.182378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell. 2009;21:3315–3325. doi: 10.1105/tpc.108.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos AM, Knudsen C, Lai D, Kannangara R, Mikkelsen L, Motawia MS, Olsen CE, Sato S, Tabata S, Jorgensen K, et al. Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant Journal. 2011;68:273–286. doi: 10.1111/j.1365-313X.2011.04685.x. [DOI] [PubMed] [Google Scholar]
- Takos AM, Rook F. Why biosynthetic genes for chemical defense compounds cluster. Trends in Plant Science. 2012;17:383–388. doi: 10.1016/j.tplants.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Tattersall DB, Bak S, Jones PR, Olsen CE, Nielsen JK, Hansen ML, Hoj PB, Moller BL. Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science. 2001;293:1826–1828. doi: 10.1126/science.1062249. [DOI] [PubMed] [Google Scholar]
- Tena JJ, Alonso ME, de la Calle-Mustienes E, Splinter E, de Laat W, Manzanares M, Gomez-Skarmeta JL. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nature Communications. 2011;2:310. doi: 10.1038/ncomms1301. [DOI] [PubMed] [Google Scholar]
- Thimmappa R, Geisler K, Louveau T, O’Maille P, Osbourn A. Triterpene biosynthesis in plants. Annual Review of Plant Biology. 2014;65:225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Molecular Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Usui M, Sugawara C, Otomo K, Hirose Y, Miyao A, Hirochika H, Okada K, Shimizu T, Koga J, et al. Reverse-genetic approach to verify physiological roles of rice phytoalexins: characterization of a knockdown mutant of OsCPS4 phytoalexin biosynthetic gene in rice. Physiologia Plantarum. 2014;150:55–62. doi: 10.1111/ppl.12066. [DOI] [PubMed] [Google Scholar]
- Uguru GC, Stephens KE, Stead JA, Towle JE, Baumberg S, McDowall KJ. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Molecular Microbiology. 2005;58:131–150. doi: 10.1111/j.1365-2958.2005.04817.x. [DOI] [PubMed] [Google Scholar]
- Van der Lee TAJ, Medema MH. Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genetics and Biology. 2016;89:29–36. doi: 10.1016/j.fgb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Verma P, Mathur AK, Srivastava A, Mathur A. Emerging trends in research on spatial and temporal organization of terpenoid indole alkaloid pathway in Catharanthus roseus: a literature update. Protoplasma. 2012;249:255–268. doi: 10.1007/s00709-011-0291-4. [DOI] [PubMed] [Google Scholar]
- Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proceedings of the National Academy of Sciences, USA. 2015;112:4672–4677. doi: 10.1073/pnas.1504783112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ, Wagner D, Dehesh K. The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathogens. 2008;4:e1000237. doi: 10.1371/journal.ppat.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegel E, Koumproglou R, Shaw P, Osbourn A. Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell. 2009;21:3926–3936. doi: 10.1105/tpc.109.072124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiology. 2004;135:2098–2105. doi: 10.1104/pp.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- Winzer T, Kern M, King AJ, Larson TR, Teodor RI, Donninger SL, Li Y, Dowle AA, Cartwright J, Bates R, et al. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science. 2015;349:309–312. doi: 10.1126/science.aab1852. [DOI] [PubMed] [Google Scholar]
- Wong S, Wolfe KH. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nature Genetics. 2005;37:777–782. doi: 10.1038/ng1584. [DOI] [PubMed] [Google Scholar]
- Xu Y, Galhano R, Wiemann P, Bueno E, Tiernan M, Wu W, Chung I-M, Hershenzon J, Tudzynski B, Sesma A, et al. Genetic evidence for natural product-mediated plant-plant allelopathy in rice (Oryza sativa) New Phytologist. 2011;193:570–575. doi: 10.1111/j.1469-8137.2011.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S, Whitlock MC. The genetic architecture of adaptation under migration-selection balance. Evolution. 2011;65:1897–1911. doi: 10.1111/j.1558-5646.2011.01269.x. [DOI] [PubMed] [Google Scholar]
- Yu N, Nützmann H-W, MacDonald JT, Moore B, Field B, Berriri S, Trick M, Rosser SJ, Kumar SV, Freemont P, et al. Delineation of metabolic gene clusters in plant genomes by chromatin signatures. Nucleic Acids Research. 2016;44:2255–2265. doi: 10.1093/nar/gkw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ. To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annual Review of Plant Biology. 2014;65:259–286. doi: 10.1146/annurev-arplant-050213-035705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Lechner A, Wietz M, Millán-Aguiñaga N, Chavarria KL, Jensen PR. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinospora. Proceedings of the National Academy of Sciences, USA. 2014;111:E1130–E1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]