Abstract
Enhancers of yellow (e(y)) is a group of genetically and functionally related genes for proteins involved in transcriptional regulation. The e(y)3 gene of Drosophila considered here encodes a ubiquitous nuclear protein that has homologues in other metazoan species. The protein encoded by e(y)3, named Supporter of Activation of Yellow Protein (SAYP), contains an AT-hook, two PHD fingers, and a novel evolutionarily conserved domain with a transcriptional coactivator function. Mutants expressing a truncated SAYP devoid of the conserved domain die at a midembryonic stage, which suggests a crucial part for SAYP during early development. SAYP binds to numerous sites of transcriptionally active euchromatin on polytene chromosomes and coactivates transcription of euchromatin genes. Unexpectedly, SAYP is also abundant in the heterochromatin regions of the fourth chromosome and in the chromocenter, and represses the transcription of euchromatin genes translocated to heterochromatin; its PHD fingers are essential to heterochromatic silencing. Thus, SAYP plays a dual role in transcription regulation in euchromatic and heterochromatic regions.
Keywords: chromatin, PHD finger, position effect variegation, transcription activation
Introduction
Transcriptional regulation requires concerted action of a large number of proteins or protein complexes. Some of them modulate the local structure of chromatin, making it more or less accessible to other transcription factors, while others bind to the regulatory regions of the gene and recruit the general transcription factors to the promoter (for review, see George et al, 1995; Lee and Young, 1998; Bell and Tora, 1999).
Significant portions of the eukaryotic genome, in particular, the centromeric and telomeric regions of chromosomes, are packaged in constitutive heterochromatin, which is associated with condensed appearance and late replication in the S phase (Zhimulev, 1998). The introduction of euchromatic genes or transgenes to heterochromatin leads to mosaic expression known as position effect variegation (PEV) (Weiler and Wakimoto, 1995; Hwang et al, 2001). PEV is associated with transcriptional silencing of the gene in part of cells, caused by expansion of the heterochromatin conformation; this phenomenon appears to be general and to affect genes with different promoters (Schotta et al, 2003).
The mechanism of heterochromatin silencing is conserved in evolution and is believed to involve multiprotein complexes. Heterochromatin protein 1 (HP1) (James and Elgin, 1986) is supposed to play a key role in maintaining the heterochromatin structure (Clark and Elgin, 1992). It is mainly associated with pericentric heterochromatin (James et al, 1989), and a loss-of-function mutation of Drosophila HP1 (Su(var)2-5) acts as a dominant suppressor of PEV (Eissenberg et al, 1992; Cryderman et al, 1998; Eissenberg and Elgin, 2000). HP1 is thought to interact both with modified histones and with proteins instrumental in transcription silencing, such as histone H3 methyltransferase (SU(VAR)3-9), recruiting them to heterochromatin (Kellum, 2003). HP1 was also shown to directly interact with zinc-finger protein SU(VAR)3-7 (Cleard et al, 1997) and heterochromatin protein 2 (HP2), which has two AT-hook domains (Shaffer et al, 2002). These proteins colocalize with HP1 in heterochromatin regions on polytene chromosomes of Drosophila (chromocenter and the small fourth chromosome). Like Su(var)2-5, mutations of the corresponding genes are dominant suppressors of PEV, that is, these proteins are required for spreading of heterochromatin and establishment/maintenance of the silent state.
The group of enhancer of yellow (e(y)) genes has been isolated in a genetic screen aimed to find mutations influencing the activator-dependent transcription (Georgiev and Gerasimova, 1989). In our previous studies, we have shown that e(y)1 encodes TAF9, a subunit of both TFIID and the TFTC complexes (Soldatov et al, 1999), and that e(y)2 encodes a small nuclear protein highly conserved in metazoan evolution (Georgieva et al, 2001). E(y)2 was found to be present in a large multiprotein complex containing TAF9 and to potentiate transcription activation on chromatin templates (Georgieva et al, 2001). Recently, the yeast homologue of E(y)2, Sus1, was identified as a component of both the SAGA complex and the nuclear pore-associated mRNA transport machinery (Rodriguez-Navarro et al, 2004). Interestingly, the weak mutations of e(y) genes that do not influence the viability of flies proved to be lethal in compound, suggesting that these genes have overlapping and/or redundant functions (Georgiev, 1994).
The e(y)3 gene has been genetically shown to activate the transcription of the yellow gene: the e(y)3 mutation decreased the expression of the y2 allele (Georgiev and Gerasimova, 1989). Partial inactivation of the e(y)3 function impairs the expression of the white and cut genes, suggesting a general role in transcription for the e(y)3 protein product (Georgiev, 1994).
Here we identify and characterize the protein encoded by the e(y)3 gene. E(y)3, hereafter called Supporter of Activation of Yellow Protein (SAYP), is a large multidomain nuclear protein essential at early stages of embryonic development. It contains several nuclear localization signals, an AT-hook, a novel evolutionarily conserved domain, and two PHD fingers near the carboxy terminus. SAYP is present at numerous sites on polytene chromosomes and colocalizes with Pol II in transcriptionally active euchromatin. Its conserved domain is shown to be involved in transcription activation. On the other hand, SAYP is also found in heterochromatic regions of polytene chromosomes. It negatively regulates the expression of genes in heterochromatin, and its PHD fingers are essential to this function. Our results suggest a general role for SAYP/E(y)3 in regulation of transcription in both euchromatin and heterochromatin.
Results and discussion
Structure of the e(y)3 gene
Two mutant alleles of e(y)3 genetically mapped to 19C of the X chromosome have been isolated. Isolation of the viable e(y)3u1 allele, induced by insertion of a Stalker mobile element, was described before (Georgiev et al, 1990). The lethal allele e(y)3EMSl was later found in the progeny of ethylmethanesulfonate-treated (EMS) males (see Materials and methods).
To isolate the e(y)3 gene, the sequences surrounding the Stalker in e(y)3u1 flies were cloned. Sequencing demonstrated that Stalker insertion occurred in the genetic locus encoding a protein with two PHD fingers at the C terminus (FlyBase report CG12238). To prove that e(y)3 mutations really influence CG12238, the corresponding genomic region (Figure 1A) including the predicted promoter sequences was cloned in CaSpeR3 vector and used to rescue the e(y)3u1 and e(y)3EMSl mutants. Each of five independently obtained transgenes completely restored the wild-type phenotype, demonstrating that the isolated gene was really e(y)3 (CG12238).
Figure 1.
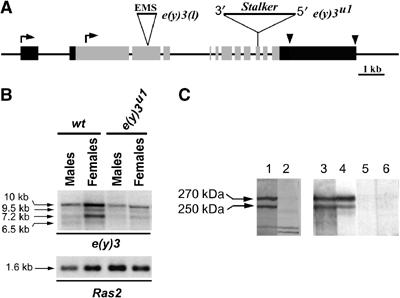
The structure of e(y)3 gene and the nature of e(y)3 mutations. (A) Molecular structure of e(y)3 gene and transcripts. Gray boxes indicate the coding regions. Black boxes indicate 5′- and 3′-untranslated regions. Two alternative transcription start sites are shown by bent arrows; the alternative polyadenylation sites are shown by arrowheads. The Stalker insertion (position 4956 from the beginning of the longest ORF) in the e(y)3u1 allele and the site of 11-nt deletion at position 3525 in the e(y)3EMSl allele are indicated (not to scale). Both mutations lead to stop codon formation. The probe corresponding to the second exon was used for Northern hybridization (panel B). (B) Transcription of e(y)3 in wild-type and e(y)3u1 flies. The level of e(y)3 transcription is decreased in mutant males and females. Ras2 was used for normalization. The e(y)3 transcripts did not change in length in mutated flies, because splicing between the 3′ end of exon 9 and the sequences of Stalker 5′LTR resulted in replacement of 24 nt of exon 10 by 23 nt of Stalker. This produced a stop codon 85 amino acids downstream of the place of Stalker insertion. (C) Western blot detection of SAYP in embryonic nuclear extract. The lanes were developed with (1) nonpurified antiserum 1, (2) antiserum 1 after 1-h incubation with the peptide used for immunization, (3, 4) affinity-purified Ab1 and Ab2, and (5, 6) preimmune serum. Ab1 were raised against residues 102–308. Ab2 were raised against residues 495–643.
Several cDNA clones corresponding to e(y)3 were isolated from a cDNA library prepared from strain Oregon R. This gene has 12 exons (Figure 1A), containing an open reading frame (ORF) for a protein of 2008 amino acids, that is, 165 residues longer than the one presented in FlyBase. The main e(y)3 mRNA detected by Northern blot hybridization was 10 kb long. However, three additional weaker transcripts of lower molecular weight were also found (Figure 1B). Analysis of the cDNA clones showed that three mRNAs of e(y)3 were identical in their coding sequences but different in their 3′-untranslated regions. The 6.5-kb transcript was found to have an alternative start of transcription at exon 2 and thus to contain the coding region, which is 163 amino acids shorter.
Next, polyclonal antibodies against two different peptides from the SAYP N-proximal region (Ab1 and Ab2; see Figure 2) were raised in rabbits. Antiserum 1 and affinity-purified Ab1 and Ab2 detected the same two closely migrating protein species (about 270 and 250 kDa) in a nuclear extract from Drosophila embryos (Figure 1C, lanes 1, 3, and 4) that were not recognized by preimmune sera (lanes 5 and 6). Moreover, the bands specifically disappeared if the antiserum was incubated with the peptides used for immunization before Western blotting (lane 2). The lower band seems to represent a version of SAYP lacking the N-terminal stretch and synthesized from the 6.5-kb transcript, as the difference between two bands (about 20 kDa) is quite close to the expected one (18 kDa). The molecular weight of SAYP proteins is higher than that calculated from the amino-acid sequence, which may be explained by post-translational modifications. Also, it cannot be excluded that the two bands detected on Western blot represent differently modified SAYP.
Figure 2.
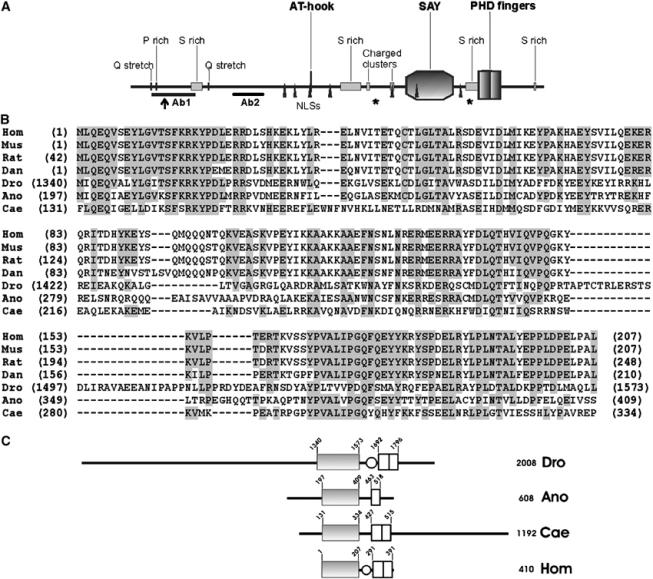
SAYP is a multidomain protein that has homologues in different species. (A) The structure of SAYP. AT-hook, SAY domain (amino-acid residues 1340–1573), PHD fingers (1692–1796), Ser-rich regions (293–345, 1018–1117, 1629–1693, 1960–1970), Pro-rich region (119–126), Gln stretches (93–97, 375–379), positively charged regions (1146–1160, 1297–1303), and nuclear localization signals (770–777, 805–811, 881–884, 991–997, 1297–1305, 1417–1420, 1610–1616) are indicated. The beginning of the short alternative form (residue 164) is indicated by an arrow. Asterisks mark the last amino acid of the protein in e(y)3u1 (1652) and e(y)3EMSl (1175) alleles. The positions of peptides used to raise antibodies are indicated (Ab1 and Ab2). (B) Sequence alignment of the SAY domains from proteins of different species. Hom, Homo sapiens, NP_060758 locus; Mus, M. musculus, NP_077212 locus; Rat, Rattus norvegicus, XP_214780 locus; Dan, D. rerio, AAH57492 locus; Dro, Drosophila melanogaster, SAYP; Ano, A. gambiae, XP_321190 locus; Cae, C. elegans, A88925 locus; amino acids identical to human are background-shaded when present in more than four species. (C) Comparison of SAYP domain structure with homologues from A. gambiae (Ano), C. elegans (Cae) and vertebrate (Hom). Solid box stands for the SAY domain, light box for the PHD finger, and oval for the Ser-rich region.
SAYP is a multidomain protein with PHD fingers near the C terminus
SAYP contains four serine-rich regions, a proline-rich region, two glutamine stretches, and two positively charged clusters. It also contains seven putative nuclear localization signals in different parts of the molecule (Figure 2).
An AT-hook motif is present in the central part of SAYP. The AT-hook, first described in HMGA proteins, is a small motif recognizing AT-rich sequences and binding to the minor groove of the DNA helix. Multiple or single AT-hook motifs have been detected in a wide range of nuclear proteins from different species, including multidomain chromatin-associated proteins involved in transcription activation or repression (for review, see Aravind and Landsman, 1998).
Near the carboxy terminus of SAYP, there are two PHD fingers. PHD is an orphan conserved Cys4-His-Cys3 Zn-finger domain found in many chromatin-associated proteins, including several transcription factors, for example, TrxG (Mazo et al, 1990) and PcG (DeCamillis et al, 1992); acetyltransferase CBP/p300 (Kalkhoven et al, 2002); Mi-2 protein, a component of the histone deacetylating complex (Wade et al, 1999); and the chromatin remodeling protein Acf1 (Ito et al, 1999).
The homologues of SAYP from different species contain a highly conserved domain followed by PHD fingers
A database search revealed sequences homologous to SAYP in human (hypothetical protein XAP135/PHF10) (Rebhan et al, 1997) and several other metazoan species including mouse Mus musculus, zebrafish Danio rerio, and nematode Caenorhabditis elegans as well as mosquito Anopheles gambiae (Figure 2B). The vertebrate proteins are very close in sequence (75% identity between human and fish) and length, representing the core of evolutionary conservation (Figure 2C). Both insect and nematode proteins are considerably longer, extending in both directions.
The comparison indicated that all homologues contained a highly conserved domain, hereafter referred to as the Supporter of Activation of Yellow (SAY) domain. The SAY domain (30% identity and 45% similarity between Drosophila and human proteins) is followed, after a short low-homology region, by PHD fingers. It is noteworthy that the two putative PHD fingers of SAYP share the conserved Cys residue, implying that only one PHD may function at a moment. The same is observed for SAYP homologues from other species, except for the mosquito protein, which has only one finger. The Drosophila protein, like the vertebrate homologues, has a serine-rich region in the spacer. The pronounced conservation of this domain arrangement from insects to mammals strongly suggests that both domains are essential to the function of these factors.
SAYP is a ubiquitous nuclear protein indispensable in oogenesis and early Drosophila development
The 10-kb e(y)3 mRNA was detected at all stages of insect development (Figure 3A) as well as were all weaker transcripts (data not shown). However, the most intense transcription was observed in adult females. The highest content of e(y)3 mRNA was detected in ovaries. It was present in the cytoplasm of nursing cells and growing oocytes at all stages of development and accumulated in mature oocytes (Figure 3B and C).
Figure 3.
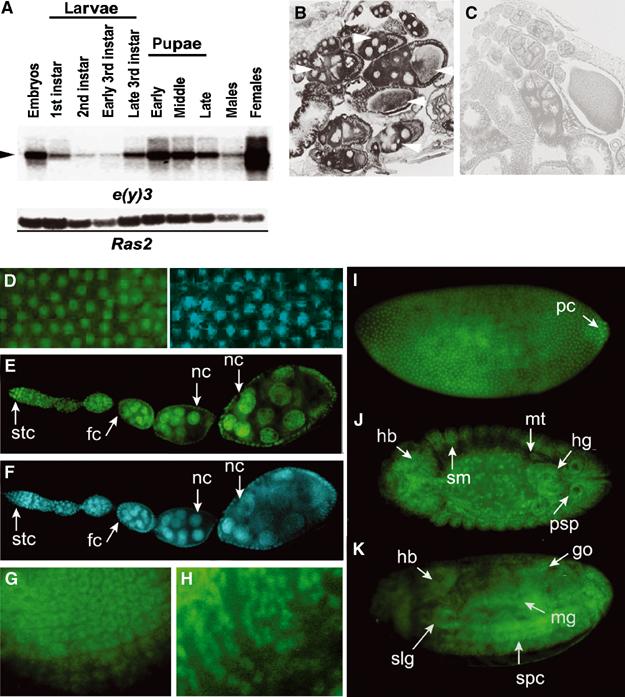
SAYP is a ubiquitous nuclear protein. (A) Northern blot hybridization of poly(A) RNA from different stages of Drosophila development. The 10-kb mRNA is indicated by an arrowhead. The lower panel shows the hybridization of the same membrane with the Ras2 probe. (B, C) In situ hybridization of a frontal tissue section of wt female abdomen with antisense (B) and sense (C) e(y)3 mRNA probes. Arrows indicate wt oocytes and accompanying nursing cells at different stages of development. (D) A field of stage-4 embryo stained with antibodies against SAYP (left) and DAPI (right). The localization of SAYP in the nuclei of syncytium blastoderm is well observed. (E) The distribution of SAYP in adult ovaries. (F) The same stained with DAPI. SAYP is detected in the nuclei of germarium stem cells (stc), follicular cells (fc), and nursing cells (nc). (G, H) SAYP is present in ommatidia precursors of eye-antennal imaginal disk (G) and in precursors of glial cells in the brain of third-instar larva (H). (I–K) SAYP is detected in different tissues of Drosophila embryo. Immunostaining of embryos at stage 4 (I); stage 14, dorsal view (J); and stage 16, ventrolateral view (K). hb, head brain; hg, hindgut; go, gonads; mt, malpigian tubes; mg, midgut; pc, polar cells; psp, posterior spiracles; slg, salivary glands; sm, somatic mesoderm; spc, spinal cord. All embryos are oriented to the left. Affinity-purified Ab1 and FITC-conjugated secondary antibodies were used.
Immunostaining also revealed SAYP in the nuclei of syncytium blastoderm of early embryos (Figure 3D and I) and in the nuclei of different tissues of late embryos (Figure 3J and K), larvae, and adults (Figure 3E–H and data not shown). According to the in situ hybridization data, SAYP is abundant in the nuclei of various ovary cells (Figure 3E and F). Thus, SAYP is a ubiquitous nuclear protein, expressed at all stages of development and in different tissues of flies. Interestingly, the cDNAs of XAP135/PHF10, the human counterpart of SAYP, were found by us in EST databases prepared from various tissues, suggesting that XAP135/PHF10 is also a ubiquitous protein (Rebhan et al, 1997).
In line with the essential role of SAYP in oogenesis shown above, the major phenotypic manifestation of the e(y)3u1 mutation was female sterility. The e(y)3u1 mutation also decreased fly viability—by 50% hemizygous males and by 20% in homozygous females—and caused disturbances in the development of femur, shortened body, and expanded wings. All these features were weak and were observed in 15–20% of flies.
Homozygous e(y)3EMSl females and hemizygous e(y)3EMSl males died at a midembryonic stage. Their survival at earlier stages appears to be due to the maternal effect of the e(y)3 gene. Examination of e(y)3EMSl embryos revealed multiple and variable disturbances in development including the formation of head, midgut, malpigian tubes, and embryonic gonads (data not shown). The mutations of genes encoding several different transcription factors have similar manifestations, suggesting that these genes probably interact with e(y)3 in development. Of particular interest is the cut locus, whose function is necessary for specification of a large number of cell types. Previously, we have reported the genetic interaction of e(y)3 and the cut locus (Melnikova et al, 1996). However, the interaction of e(y)3 with other genes needs further careful investigation. The manifestations of e(y)3 mutations suggest SAYP to be indispensable for oogenesis and early stages of development.
The molecular nature of e(y)3 mutations
The e(y)3u1 mutation is generated by Stalker insertion in exon 10 at position 4956 from the beginning of the longest ORF. It produces a stop codon close to the place of insertion (see legend to Figure 1B). Therefore, the mutant SAYP lacks the last 349 amino acids corresponding to PHD domains (Figure 2). Besides, e(y)3 transcription is considerably less intense in the e(y)3u1 flies (Figure 1B). Sequencing of the e(y)3EMSl allele revealed a stop codon produced upon an 11-nucleotide deletion at position 3525. Importantly, in this case, the SAYP is truncated close to the beginning of the conserved region, thus lacking most of the SAY domain and the two PHDs.
The truncated SAYP is detected in nuclear extracts of Drosophila embryos of e(y)3u1 and e(y)3/e(y)3EMSl strains by Western blot (Figure 4A), confirming the obtained data. In extracts from homozygous e(y)3u1 flies, the antibodies recognize two bands of 240 and 220 kDa corresponding to SAYP lacking PHDs. According to the transcription data, the level of the protein is decreased in e(y)3u1 flies (about 3–4 times). Two bands (180 and 160 kDa) in addition to the wild-type SAYP (270 and 250 kDa) are detected in heterozygous e(y)3/e(y)3EMSl flies. The presence of SAYP was also tested in embryos of the e(y)3EMSl/e(y)3EMSl strain containing the construct that expressed SAYP lacking PHDs (see below). The bands corresponding to protein lacking the SAY domain as well as two bands corresponding to transgenic SAYP (230 and 210 kDa) were also detected. It is noteworthy that, like the full-length SAYP, the truncated versions also exhibit abnormal mobility. However, the difference in electrophoretic mobility of wild-type and mutated or transgenic versions of SAYP coincides with the expected values. The same results were obtained with Ab2 raised against a different peptide of SAYP (Figure 4B).
Figure 4.
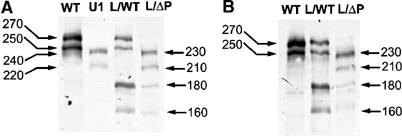
The molecular nature of the e(y)3u1 mutation. (A, B) The presence of SAYP in different strains of flies revealed by Western blotting with affinity-purified Ab1 (A) or Ab2 (B). The total protein from five embryos of Oregon R (WT), e(y)3u1/e(y)3u1 (U1), e(y)3/e(y)3EMSl (L/WT) strains and transgenic e(y)3EMSl/e(y)3EMSl; P{e(y)3ΔPHD} (L/ΔP) strain was resolved in 8% SDS–PAGE. The molecular weight is indicated in kDa.
SAYP is an abundant protein of euchromatin that also binds to heterochromatin regions
To assess the distribution of SAYP in chromatin, we undertook immunostaining of polytene chromosomes from Drosophila salivary glands. Affinity-purified antibodies Ab1 or Ab2 were used for immunostaining. Both antibodies were shown to be specific (Figure 4A and B): they selectively recognize on Western blots the wild-type and mutated versions of SAYP and do not recognize any unspecific bands. About 150 sites of SAYP binding were detected (Figure 5A); the two antibodies recognized the same sites in the arms of polytene chromosomes (Figure 5B). Most of them coincided with those containing Pol II (Figure 5A and B) and were localized in the less compact regions of chromatin poorly stained with DAPI (Figure 5B). On the other hand, Pol II was revealed at many more sites than SAYP, indicating that SAYP is only present at a certain fraction of the transcribed genes.
Figure 5.
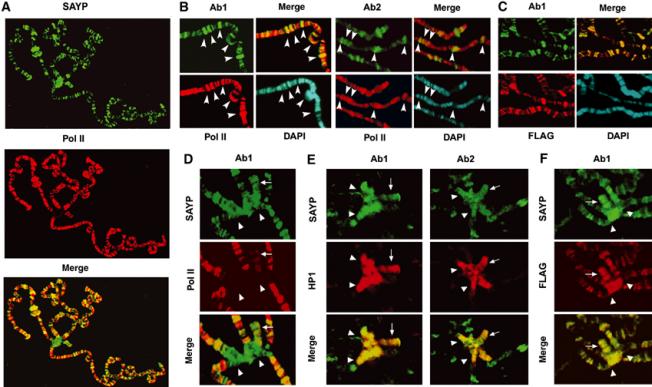
SAYP is abundant in euchromatin and binds to heterochromatin regions on Drosophila polytene chromosomes. (A) SAYP colocalizes with Pol II on polytene chromosomes of Drosophila: staining with antibodies (Ab1) against SAYP, Pol II, and the merged image. (B) Fragment of chromosome 2R stained with antibodies against SAYP (Ab1 or Ab2), Pol II, DAPI, and merged image. Arrowheads indicate sites 47A, 47C, 48B, 49E, and 50C (from left to right) strongly stained with both anti-SAYP antibodies. (C) Fragment of chromosome arms stained with antibodies against SAYP (Ab1), FLAG, DAPI, and merged image. (D) The chromocenter (marked with arrowheads in panels D–F) and the fourth chromosome (arrow) stained with antibodies against SAYP (Ab1), Pol II, and the merged image. (E) Chromocenter and the fourth chromosome stained with different antibodies against SAYP (either Ab1 or Ab2), HP1, and the merged images. (F) Chromocenter and the fourth chromosome from transgenic flies carrying FLAG-tagged SAYP stained with antibodies against SAYP (Ab1), FLAG, and merged image. Affinity-purified Ab1 and Ab2, monoclonal antibodies against CTD of Pol II, and monoclonal antibodies against HP1 and FLAG were used for staining.
Unexpectedly, immunostaining of polytene chromosomes also revealed SAYP in heterochromatic regions: in the chromocenter and at chromosome 4 (Figure 5D), most of which is represented by heterochromatin (Sun et al, 2000). Antibodies against SAYP strongly decorated these regions, while only weak staining of the fourth chromosome was observed with antibodies against Pol II. The heterochromatic nature of the sites of SAYP binding is further proved by comparison of the distribution of SAYP with that of heterochromatin protein 1 (HP1): their colocalization is apparent in Figure 5E.
To verify the observed pattern, transgenic flies bearing the construct expressing FLAG-tagged SAYP were obtained. The transgene was able to rescue the e(y)3u1 and e(y)3EMSl mutations, testifying that tagged SAYP is functional. The antibodies against FLAG stained euchromatic and heterochromatic regions on polytene chromosomes of the transgenic flies, demonstrating complete colocalization with SAYP both in the sites on the arms of chromosomes (Figure 5C) and on the fourth chromosome and chromocenter (Figure 5F).
The results obtained implicate SAYP in the organization of heterochromatin structure and hence in regulation of gene expression in heterochromatin (see below).
SAYP represses the expression of transgenes located in heterochromatin
To study the function of SAYP in heterochromatin, we investigated the effect of the e(y)3u1 mutation on expression of transgenes located in different heterochromatic regions of the fourth chromosome and in the chromocenter. The transgenic lines of flies bearing the P-element vector P[hsp26-pt, hsp70-w] have been described (Sun et al, 2000). This vector contained the white gene driven by the hsp70 promoter, which produced a convenient marker to monitor gene expression both visually and by quantitating the amount of pigment accumulated in eyes.
The transgenic flies with inserts in heterochromatic domains demonstrated a variegating eye phenotype due to silencing of the transgene. The effect of SAYP on transgene expression was investigated in females heterozygous for e(y)3u1 and P insertion and in males hemizygous for e(y)3u1 and heterozygous for P insertion. We observed significantly increased transgene expression in males of most of the tested lines (Figure 6A and B). Even in females, the effect, although less prominent, could be observed despite the presence of a wild-type e(y)3 allele. Increased expression was observed in lines in which the P insertion occurred in the fourth chromosome and in the centromeric region (line 118E-10). The extent of the observed effect of e(y)3u1 was similar to that shown previously for a missense mutation of HP1 (Su(var)2-5).
Figure 6.
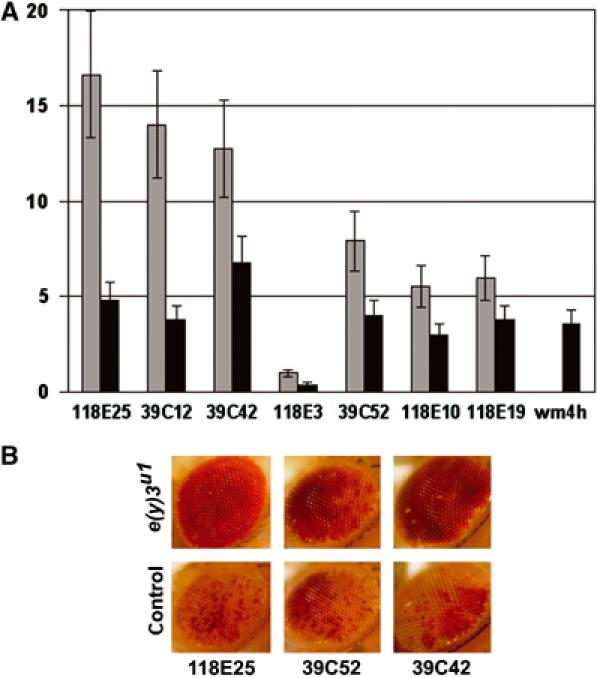
SAYP participates in heterochromatin silencing. (A) The influence of the e(y)3u1 mutation on transcription of genes situated in different heterochromatic regions of the fourth chromosome. The code numbers of transgenic strains are specified under paired bars along the horizontal axis. The ordinate is the ratio of the level of eye pigmentation in y2w1e(y)3u1/Y; P[hsp26-pt, hsp70-w]/+ males (dark gray bars) and y2w1e(y)3u1/X; P[hsp26-pt, hsp70-w]/+ females (light gray bars) to the level of pigmentation in control y1w1/Y; P[hsp26-pt, hsp70-w]/+ males and y1w1/y1w1; P[hsp26-pt, hsp70-w]/+ females, respectively. The rightmost bar shows the ratio of pigmentation level in heterozygous e(y)3u1wm4 to control heterozygous wm4 females. (B) The eye phenotypes of control +/Y; P[hsp26-pt, hsp70-w]/+ and e(y)3u1/Y; P[hsp26-pt, hsp70-w]/+ males.
We also tested the influence of e(y)3u1 on the natural white-mottled (wm4h) mutation, which leads to variegated color of eyes because of X-chromosome inversion bringing the white gene in proximity to the centromere. Introduction of the e(y)3u1 in wm4h flies tripled the white expression in females heterozygous for e(y)3u1 and wm4h mutations (Figure 6A).
Thus, like the known mutations of HP1 and other proteins shown to participate in heterochromatin silencing, the e(y)3u1 mutation is a dominant suppressor of PEV. It represses transcription of euchromatic genes brought into heterochromatin surroundings, affecting transgenes as well as a natural wm4h mutation. However, the influence of SAYP on the expression of genes originally residing in heterochromatin may be different. It may activate them, just as HP1 activates transcription of light and rolled genes of heterochromatin (Lu et al, 2000). Importantly, recent findings suggest that HP1 regulates both positively and negatively several genes of euchromatin (Li et al, 2003; Piacentini et al, 2003).
We further tested the influence of the mutation on expression of the same P-element construct located in 2L, 2R, and 3R telomeres, where SAYP binding was also detectable (data not shown). We did not find any significant changes in the level of white expression. The results obtained demonstrate that SAYP participates in repression of transcription in heterochromatin of the fourth chromosome and in pericentric heterochromatin, but not in telomeres.
The SAY domain is involved in transcription activation
In previous genetic experiments, the e(y)3 gene was shown to be required for activation of several genes (Georgiev and Gerasimova, 1989). Here we tested whether individual domains of SAYP would have transcriptional activation functions. To this end, several domains of SAYP were fused to the C terminus of LexA. In yeast cells, the fusion peptide containing the conserved region of SAYP (amino acids 1273–1629) efficiently activated the HIS3 and LacZ reporter genes containing LexA-binding sites upstream of their promoter regions (Table I), while other LexA-SAYP fusions, in particular those containing the PHDs, did not (data not shown). Cells expressing the LexA-SAYP(1273–1629) fusion grew efficiently on selective medium lacking histidine, as well as those expressing the LexA-GAL4 activation domain fusion used as the positive control. The rate of LacZ reporter gene activation by LexA-SAYP(1273–1629) was 10 times lower than that provided by LexA-GAL4. However, it was about 80 times higher than that provided by LexA-SAYP(1366–1629) that lacked the first 26 amino acids of the SAY domain. This fusion also resulted in a weaker growth. Deletion of 80 amino acids from the C terminus of the SAY domain in LexA-SAYP(1273–1493) completely abolished the activity of the fusion peptide (Table I). Thus, the whole conserved domain of SAYP (about 350 amino acids) is required for transcription activation in the yeast two-hybrid system, while the other domains do not possess this activity. These results testify that the SAY domain is a potent transcriptional activator that is responsible for the coactivator function of SAYP. However, in vivo, the other domains of SAYP may also be important for transcription activation.
Table 1.
The conserved domain of SAYP activates reporter genes HIS3 and LacZ in yeast
| LexA fusion | Growth without histidinea | β-Galactosidase activity (U)b |
|---|---|---|
| LexA-SAYP(1273–1629) | ++ | 31±2 |
| LexA-SAYP(1366–1629) | + | 0.4±0.1 |
| LexA-SAYP(1273–1493) | − | — |
| LexA-GAL4 activation domain | ++ | 317±30 |
| a‘++' potent growth, ‘+' weak growth, ‘−' no growth. | ||
| bβ-Galactosidase activity (U) was determined using the following formula: U=1000 × OD578/(t × 0.5 × OD600), where t is incubation time (in minutes). | ||
The SAY domain is essential for fly viability and transcription activation in vivo
Next, we investigated the function of the SAY domain and PHDs using the obtained mutations. The influence of the e(y)3EMSl mutation on fly viability is much more severe than that of e(y)3u1, suggesting that the SAY domain is indispensable for development of flies. To ascertain this, we constructed a transgene P{e(y)3ΔPHD} that expressed a protein truncated shortly after the SAY domain, retaining the Ser-rich stretch but having no PHDs. The expression of P{e(y)3ΔPHD} was confirmed by Western blot (Figure 4A and B). The P{e(y)3+} construct producing the full-length SAYP was used as a control.
Both constructs were first tested for the ability to rescue the lethal e(y)3EMSl allele. The results are schematically represented in Table II. Eight independent insertions of P{e(y)3+} and five independent insertions of P{e(y)3ΔPHD} were tested in rescue experiments. Just as P{e(y)3+}, the P{e(y)3ΔPHD} transgene restored the visible wild-type phenotype of the e(y)3EMSl mutants.
Table 2.
Overwiew of the results of experiments of rescue of e(y)3EMSl and e(y)3u1 mutations by different transgenes
| Genotype | Presence of SAY and PHDs | Female fertility | Viability | Expression of y2 and yInr alleles | Expression of transgene in heterochromatin |
|---|---|---|---|---|---|
| e(y)3+ | SAY+PP | Fertile | Normal | Normal | Repressed |
| e(y)3EMSl | — | ND | Lethal | ND | ND |
| e(y)3u1 | SAYa | Sterile | Reduced | Reduced | Activated |
| e(y)3EMSl | — | ||||
| P{e(y)3+} | SAY | Fertile | Normal | ND | ND |
| e(y)3EMSl | — | ||||
| P{e(y)3ΔPHD} | SAY | Fertile | Normal | ND | ND |
| e(y)3u1 | SAYa | ||||
| P{e(y)3+} | SAY+PP | Fertile | Normal | Normal | Repressed |
| e(y)3u1 | SAYa | ||||
| P{e(y)3ΔPHD} | SAY | Fertile | Normal | Normal | Activated |
| aThe level of SAYP expression is decreased in e(y)3u1 flies. | |||||
| Bold is used to highlight the transgenes. | |||||
We further tested the influence of the P{e(y)3ΔPHD} transgene on the phenotype of the e(y)3u1 mutation. As shown above, this mutation results in synthesis of the protein lacking PHDs. In addition, the amount of SAYP is decreased in the e(y)3u1 strain. Introduction of P{e(y)3ΔPHD} would increase the amount of truncated protein, making it possible to detect the consequences of mutation caused by the lack of PHDs. Unexpectedly, like the construct expressing the wild-type SAYP, P{e(y)3ΔPHD} was able to complement the main manifestations of the e(y)3u1 mutation, restoring female fertility and increasing the viability of the e(y)3u1 strain. Thus, a lower content of the SAY domain resulting from decreased e(y)3 expression, rather than the lack of PHDs, is the main cause of disturbances in e(y)3u1 flies.
We also assessed the influence of the SAY domain on gene expression in vivo. A significant manifestation of the e(y)3u1 mutation described previously was its ability to interfere with the expression of several genes (Georgiev, 1994). In particular, it affects the yellow gene, decreasing expression of the y2 allele in bristles (Georgiev and Gerasimova, 1989). The y2 allele is generated by insertion of the retrotransposon gypsy in the yellow regulatory region (Geyer et al, 1986). To exclude the role of gypsy in the e(y)3-mediated regulation of the yellow gene, we also used the yInr allele that is generated by mutation in the Initiator element of the yellow promoter (Morris et al, 1999). While the yInr allele displayed a wild-type phenotype, the e(y)3u1 mutation strongly reduced its expression in bristles.
We checked whether introduction of the P{e(y)3ΔPHD} construct in y2e(y)3u1 or yInre(y)3u1 flies would influence the expression of yellow, and observed complete restitution of the original y2 or yInr phenotype in transgenic y2e(y)3u1/Y; P{e(y)3ΔPHD} males.
Altogether, these results show the SAY domain to be crucial for the functioning of SAYP. A drop in its content to one-fourth in e(y)3u1 flies leads to disturbances in fly development, while deletion of the SAY domain appears to be lethal. It is involved in activation of transcription of the yellow gene in vivo, which confirms the results obtained in vitro in yeasts. At the same time, removal of PHD fingers seems to be not essential for these functions.
The PHD fingers of SAYP are specially needed for transcription repression in heterochromatin
As SAYP is involved in repression of transcription in heterochromatin, we investigated whether the P{e(y)3ΔPHD} transgene would interfere with the influence of the e(y)3u1 mutation on PEV. In the e(y)3u1 strain, SAYP mutation does not prevent the binding of mutated SAYP to polytene chromosomes (data not shown). Thus, either the weaker transcription of e(y)3 or the lack of PHDs, or both, suppresses PEV.
To discriminate between these possibilities, the P{e(y)3ΔPHD} and P{e(y)3+} constructs were introduced in e(y)3u1; P[hsp26-pt, hsp70-w] flies. Unlike the construct expressing the full-length protein, P{e(y)3ΔPHD} producing the truncated PHD-finger-less version failed to suppress the influence of e(y)3u1 on the expression of P[hsp26-pt, hsp70-w] transgenes in three tested lines. The transgenic females heterozygous for e(y)3u1 did not increase the expression of the reporter white gene after introduction of either one or two copies of P{e(y)3ΔPHD}. The same applied to males hemizygous for e(y)3u1. Hence, it is the PHD fingers of SAYP that are instrumental in repressing transcription in heterochromatin in vivo.
Concluding remarks
Our results demonstrate that SAYP is a chromatin-binding protein with a dual function that depends on chromatin surroundings. It operates positively or negatively in transcription regulation via different domains, which may interact with various transcription factors or protein complexes.
Previously, we observed strong genetic interaction between e(y)3 and e(y)1/taf9. This result suggests that SAYP may coactivate transcription by Pol II via interaction with TAF9-containing complexes, like TFIID or TFTC (Bell and Tora, 1999). This interaction may involve the SAY domain that was shown to possess an activator function; the high evolutionary conservation of SAY points to its possible interaction with general factors of transcription, while the variable N terminus may interact with some factors specific for particular promoters.
Our data demonstrate that the PHD domains are not important for SAYP functions in euchromatin. At the same time, PHD fingers are required for repression of the euchromatic genes inserted into the heterochromatin region. Thus, SAYP, and particularly its PHD fingers, may perform dissimilar functions in euchromatic and heterochromatic regions. The presence of PHD fingers in many chromatin-associated proteins suggests that PHD has chromatin-related function. Several PHDs were shown to participate in protein–protein interactions (Aasland et al, 1995; Fair et al, 2001; O'Connell et al, 2001; Schultz et al, 2001). However, the PHD fingers are very diverse in sequence, suggesting that their molecular function related to chromatin is also diverse. Recent studies have demonstrated that the bromodomain and PHD of transcriptional cofactor p300 cooperate in binding nucleosomes that have a high degree of histone acetylation (Ragvin et al, 2004), pointing to the possible function of PHD in histone code recognition. Deletion of the PHD domain from SAYP does not influence its ability to bind to polytene chromosome in euchromatin and heterochromatin regions. Thus, the PHD domains mediate some specific protein–protein interactions rather than recruit SAYP to chromatin.
Our results do not yet disclose the mechanisms of action of SAYP domains. Several models can be proposed to explain the dual activity of SAYP. It is possible that SAYP mutation suppresses PEV indirectly, decreasing the transcription level of genes responsible for transcription repression in heterochromatin. As the increase in the SAY domain content in transgenic flies does not influence PEV, this model implicates PHDs in transcription activation. We did not reveal the involvement of PHDs in transcription activation in yeast two-hybrid or in rescue experiments on Drosophila. The high concentration of SAYP in heterochromatin regions also suggests that SAYP is directly involved in repression.
The attractive possibility is that the SAYP-dependent silencing is realized via recruiting by the PHD domains of a protein or a protein complex involved in formation of pericentric heterochromatin. We did not find interaction between SAYP and HP1 in additional genetic experiments. We also observed no interaction between SAYP and Drosophila Mi-2 ATPase, a component of the NuRD complex that represses transcription through its remodeling and deacetylation activities (Brehm et al, 2000) (data not shown). However, these results do not exclude that the PHDs of SAYP may recruit to heterochromatin another complex responsible for transcription repression.
To explain the opposite activities of SAYP, we speculate that the SAY domain, once bound to euchromatin proteins, alters the PHD finger structure, thus blocking their interaction with a hypothetical transcription repressor (or repression complex). Conversely, in heterochromatin, there is no target for the SAY domain, and it is free or is blocked by heterochromatin proteins and thus does not prevent PHDs from binding with the repressor. It is also conceivable that SAYP enters into the composition of different multiprotein complexes having either coactivator or corepressor functions. Further studies should clarify the mechanism of action of the SAY domain and the PHD fingers of this versatile regulator protein.
Materials and methods
Genetic crosses and P-element-mediated constructs
Cultivation of flies, genetic crosses, and isolation of e(y)3u1 mutation were described previously (Georgiev and Gerasimova, 1989). The e(y)3u1 mutation in combination with zv77h (zeste null allele) had an inhibitory effect on expression of the white gene. The lethal allele e(y)3EMSl was found in the progeny of ethylmethanesulfonate (EMS)-treated males as a dominant suppressor of white expression in the presence of zv77h. The EMS treatment was performed as described by Kozitsina and Georgiev (1992). The e(y)3u1 and e(y)3EMSl mutations were maintained in y2w1e(y)3u1/FM4 and e(y)3EMSl/FM4 strains. The level of y2 expression was measured as described previously (Georgiev and Gerasimova, 1989). P{e(y)3} was obtained by inserting in the pCaSpeR 3 vector a genomic fragment flanked by BamHI sites at positions 1800 upstream of the beginning of the first exon and 1400 downstream of the stop codon. P{e(y)3ΔPHD} was obtained by deleting the 3′-terminal 2622 bp of the same genomic fragment (deletion comprises exons 10–12 and the 3′-untranslated region). P{FLAG-e(y)3+} construct expressing N-terminal FLAG-tagged SAYP was obtained by insertion of FLAG epitope after the first initiation codon of e(y)3 gene. The constructs were injected into y1w1 preblastoderm embryos as described elsewhere (Rubin and Spradling, 1982; Spradling and Rubin, 1982). The number of inserted copies was determined by Southern blot analysis using the P-element sequence as a probe.
Testing the effect of e(y)3u1 mutation on expression of transgenes
The y1w1e(y)3u1/FM4 females were crossed with the X/Y, P[hsp26-pt, hsp70-w]/P[hsp26-pt, hsp70-w] males from lines bearing insertions of the transgene into different heterochromatin regions of the fourth chromosome and chromocenter (provided by S Elgin); y1w1/FM4 females were used as controls. The extent of eye pigmentation in y2w1e(y)3u1/Y, P[hsp26-pt, hsp70-w/+ males and y2w1e(y)3u1/Õ, P[hsp26-pt, hsp70-w]/+ females was measured according to Sun et al (2000) as the transgenic/control OD485 ratio (mean of five independent samples). The photographs of eyes were taken on the fifth day after emergence.
Rescue experiments with P{e(y)3} and P{e(y)3ΔPHD} constructs
Three independently obtained insertions of P{e(y)3} and four of P{e(y)3ΔPHD} were used in each rescue experiment. Expression of yellow was evaluated in 3- to 5-day-old males developing at 25°C, ranked on a scale of 0 (pigmentation of y1 flies) to 5 (pigmentation of y+ flies). Viability was calculated as percentage of surviving transgenic males versus FM4 males. No less than 200 males were scored for each transgenic strain. To study the expression of the reporter transgene in heterochromatin, the e(y)3u1 mutation was introduced in three different strains (118E25 e(y)3u1, 39C52 e(y)3u1, 39C42 e(y)3u1).
Cloning of e(y)3 gene and Northern blot analysis
The preparation of genomic and cDNA libraries from wild-type Oregon R and e(y)3u1 flies was described by Georgieva et al (2000). Total cell RNA was isolated from Drosophila embryos, larvae, pupae, or imagoes according to Maes and Messens (1992). A 1.5 mg portion of poly(A)+ RNA was loaded per lane of agarose gel. Northern hybridization was performed as described in the same work. Membranes were exposed to a Storage Phosphor Screen and developed on a Cyclone Storage Phosphor System (Packard Instrument Company).
Preparation of nuclear extracts and immunoprecipitation
Nuclear extracts from Drosophila embryos were obtained as described previously (Sandaltzopoulos et al, 1995) by lysing the nuclei from 0- to 6-h embryos with 0.4 M ammonium sulfate. Affinity-purified polyclonal antibodies raised in rabbits against His-tagged SAYP peptides were used in Western blot analysis.
Immunostaining and in situ hybridization
Use was made of affinity-purified rabbit polyclonal antibodies against E(y)3 (dilution 1:300) and secondary FITC-conjugated and Cy-3 conjugated antibodies. Monoclonal antibodies against the CTD of the Pol II large subunit were a gift of Laszlo Tora; monoclonal antibodies against HP1 were a gift of Sarah Elgin. Antibodies against FLAG epitope (M2) were obtained from Sigma. Immunostaining of polytene chromosomes and in situ hybridization of tissue sections were performed as described previously (Soldatov et al, 1999). Embryos were collected, fixed, and stained as described by Rothwell and Sullivan (2000). Ovaries from wild-type flies were dissected in Ringer's solution (EBR: 130 mM NaCl, 5 mM KCl, 2 mM CaCl2, 10 mM Hepes pH 6.9), fixed, and stained as described by Lin and Spradling (1993). Immunostaining of whole mounted preparations of third-instar larval brain was carried out according to Donaldson et al (2001) with modifications. Final preparations of brain and ovaries were mounted in Vectashield (Vector Laboratories).
Tests in yeast two-hybrid system
The SAYP fragments were individually fused to the C terminus of LexA in pBTM117c vector. Transformed L40c yeast cells were plated on a selective medium without histidine (Wanker et al, 1997). Activation of the LacZ reporter gene was assayed using CPRG as a substrate.
Search for SAYP homologues and analysis of amino-acid sequences
Database search was performed with the BLAST (NCBI) program (Altschul et al, 1997). The multiple sequence alignment of proteins was performed using the MultAlign service (Corpet, 1988).
Acknowledgments
We are grateful to S Elgin and L Tora for the gift of antibodies and constructs, to P Becker, J Eisenberg, and J Kadonaga for helpful discussion, and to L Tora for critical reading of the manuscript. We are grateful to Tatiana Luchnik for help in preparation of the manuscript. Special thanks are due to AV Galkin for his invaluable help in bringing this work to publication. This work was supported by a Cellular and Molecular Biology grant from the Russian Academy of Sciences, INTAS #211, CRDF #RB1-2349-MO-02, RFBR #03-04-48502, and Scientific School grant 1792.2003.4. The work of AK, EN, and SG is supported by fellowships from the University of Oslo, Centre for Medical Studies, Moscow.
References
- Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20: 56–59 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Landsman D (1998) AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res 26: 4413–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B, Tora L (1999) Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp Cell Res 246: 11–19 [DOI] [PubMed] [Google Scholar]
- Brehm A, Langst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Muller J, Becker PB (2000) dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J 19: 4332–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RF, Elgin SC (1992) Heterochromatin protein 1, a known suppressor of position-effect variegation, is highly conserved in Drosophila. Nucleic Acids Res 20: 6067–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard F, Delattre M, Spierer P (1997) SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J 16: 5280–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman DE, Cuaycong MH, Elgin SC, Wallrath LL (1998) Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 107: 277–285 [DOI] [PubMed] [Google Scholar]
- DeCamillis M, Cheng NS, Pierre D, Brock HW (1992) The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome-binding sites with Polycomb. Genes Dev 6: 223–232 [DOI] [PubMed] [Google Scholar]
- Donaldson MM, Tavares AA, Ohkura H, Deak P, Glover DM (2001) Metaphase arrest with centromere separation in polo mutants of Drosophila. J Cell Biol 153: 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC (2000) The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev 10: 204–210 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Morris GD, Reuter G, Hartnett T (1992) The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair K, Anderson M, Bulanova E, Mi H, Tropschug M, Diaz MO (2001) Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol Cell Biol 21: 3589–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CP, Lira-DeVito LM, Wampler SL, Kadonaga JT (1995) A spectrum of mechanisms for the assembly of the RNA polymerase II transcription preinitiation complex. Mol Cell Biol 15: 1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva SG, Nabirochkina EN, Georgiev PG, Soldatov AV (2000) Gene enhancer of yellow 1 of Drosophila melanogaster codes for protein TAFII40. Dokl Biochem 375: 228–230 [DOI] [PubMed] [Google Scholar]
- Georgieva S, Nabirochkina E, Dilworth FJ, Eickhoff H, Becker P, Tora L, Georgiev P, Soldatov A (2001) The novel transcription factor e(y)2 interacts with TAF(II)40 and potentiates transcription activation on chromatin templates. Mol Cell Biol 21: 5223–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev PG (1994) Identification of mutations in three genes that interact with zeste in the control of white gene expression in Drosophila melanogaster. Genetics 138: 733–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev PG, Gerasimova TI (1989) Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol Gen Genet 220: 121–126 [DOI] [PubMed] [Google Scholar]
- Georgiev PG, Kiselev SL, Simonova OB, Gerasimova TI (1990) A novel transposition system in Drosophila melanogaster depending on the Stalker mobile genetic element. EMBO J 9: 2037–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Spana C, Corces VG (1986) On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J 5: 2657–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang KK, Eissenberg JC, Worman HJ (2001) Transcriptional repression of euchromatic genes by Drosophila heterochromatin protein 1 and histone modifiers. Proc Natl Acad Sci USA 98: 11423–11427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev 13: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC (1989) Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol 50: 170–180 [PubMed] [Google Scholar]
- James TC, Elgin SC (1986) Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol 6: 3862–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhoven E, Teunissen H, Houweling A, Verrijzer CP, Zantema A (2002) The PHD type zinc finger is an integral part of the CBP acetyltransferase domain. Mol Cell Biol 22: 1961–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R (2003) HP1 complexes and heterochromatin assembly. Curr Top Microbiol Immunol 274: 53–77 [DOI] [PubMed] [Google Scholar]
- Kozitsina MV, Georgiev PG (1992) [Preparation and analysis of new mutations of the mod(mdg4) gene in Drosophila melanogaster]. Genetika 28: 75–82 [PubMed] [Google Scholar]
- Lee TI, Young RA (1998) Regulation of gene expression by TBP-associated proteins. Genes Dev 12: 1398–1408 [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC (1993) Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev Biol 159: 140–152 [DOI] [PubMed] [Google Scholar]
- Li Y, Danzer JR, Alvarez P, Belmont AS, Wallrath LL (2003) Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 130: 1817–1824 [DOI] [PubMed] [Google Scholar]
- Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC (2000) Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M, Messens E (1992) Phenol as grinding material in RNA preparations. Nucleic Acids Res 20: 4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo AM, Huang DH, Mozer BA, Dawid IB (1990) The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA 87: 2112–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L, Kulikov A, Georgiev P (1996) Interactions between cut wing mutations and mutations in zeste, and the enhancer of yellow and Polycomb group genes of Drosophila melanogaster. Mol Gen Genet 252: 230–236 [DOI] [PubMed] [Google Scholar]
- Morris JR, Geyer PK, Wu CT (1999) Core promoter elements can regulate transcription on a separate chromosome in trans. Genes Dev 13: 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell S, Wang L, Robert S, Jones CA, Saint R, Jones RS (2001) Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J Biol Chem 276: 43065–43073 [DOI] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S (2003) Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol 161: 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland KR, Breen K, OYan AM, Eberharter A, Gibson TJ, Becker PB, Aasland R (2004) Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol 337: 773–788 [DOI] [PubMed] [Google Scholar]
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D (1997) GeneCards: encyclopedia for genes, proteins and diseases. Weizmann Institute of Science, Bioinformatics Unit and Genome Center (Rehovot, Israel). GeneCard for PHF10 (2003)
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E (2004) Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86 [DOI] [PubMed] [Google Scholar]
- Rothwell WF, Sullivan W (2000) Fluorescent analysis of Drosophila embryos. In Drosophila Protocols, Sullivan W, Ashburner M, Harwley RS (eds) pp 141–159. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Rubin GM, Spradling AC (1982) Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353 [DOI] [PubMed] [Google Scholar]
- Sandaltzopoulos R, Quivy JP, Becker PB (1995) Analysis of protein/DNA interactions by solid-phase footprinting. Methods Mol Cell Biol 5: 176–181 [Google Scholar]
- Schotta G, Ebert A, Dorn R, Reuter G (2003) Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol 14: 67–75 [DOI] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ III (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 15: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer CD, Stephens GE, Thompson BA, Funches L, Bernat JA, Craig CA, Elgin SC (2002) Heterochromatin protein 2 (HP2), a partner of HP1 in Drosophila heterochromatin. Proc Natl Acad Sci USA 99: 14332–14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov A, Nabirochkina E, Georgieva S, Belenkaja T, Georgiev P (1999) TAFII40 protein is encoded by the e(y)1 gene: biological consequences of mutations. Mol Cell Biol 19: 3769–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM (1982) Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347 [DOI] [PubMed] [Google Scholar]
- Sun FL, Cuaycong MH, Craig CA, Wallrath LL, Locke J, Elgin SC (2000) The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc Natl Acad Sci USA 97: 5340–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet 23: 62–66 [DOI] [PubMed] [Google Scholar]
- Wanker EE, Rovira C, Scherzinger E, Hasenbank R, Walter S, Tait D, Colicelli J, Lehrach H (1997) HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum Mol Genet 6: 487–495 [DOI] [PubMed] [Google Scholar]
- Weiler KS, Wakimoto BT (1995) Heterochromatin and gene expression in Drosophila. Annu Rev Genet 29: 577–605 [DOI] [PubMed] [Google Scholar]
- Zhimulev IF (1998) Polytene chromosomes, heterochromatin, and position effect variegation. Adv Genet 37: 1–566 [DOI] [PubMed] [Google Scholar]


