Abstract
Successful vaccination relies on driving the immune response towards high specificity, affinity and longevity. Germinal centers facilitate the evolution of antigen-specific B cells by iterative rounds of diversification, selection, and differentiation to memory and plasma cells. Experimental evidence points to B cell receptor affinity and amount of antigen presented to follicular helper T cells as main drivers of clonal evolution. Concurrent studies suggest that modifiers of cognate contact, temporal mechanisms, and stochastic factors can also shape diversity and influence differentiation to memory and plasma cells, but molecular pathways driving these selection decisions are unresolved. Due to rapid cycles of transcriptional change in the germinal center, single-cell resolution is imperative to dissect mechanisms dictating the mature antigen-specific repertoire. Future studies linking high-resolution analysis of this diverse evolving population with cellular outcome are needed to fully understand the complex mechanisms of selection driving antigen-specific humoral immunity.
Introduction
Vaccination remains an important public health tool to prevent infection and the spread of disease. By driving the evolution of antigen-specific B cell populations, vaccines elicit robust antibody-mediated immunity while bypassing infection. Affinity maturation through clonal selection in germinal centers (GCs) allows evolution of the B cell repertoire to generate antibodies against virtually any foreign antigen [1] (Figure 1). Though antigen affinity is a major driving force for selection, patterns of molecular signals drive B cells through this process, ensuring the production of not only antibody-producing plasma cells but also memory B cells that can respond and re-diversify to secondary challenge [2]. Understanding the regulation of this process in vivo is paramount to formulating novel vaccines to produce efficient and diverse immune responses.
Figure 1. Immunization-driven antigen-specific immunity.
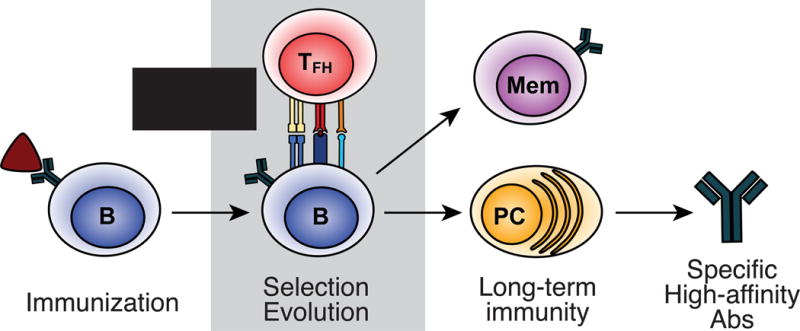
Immunization with protein antigen primes naïve antigen-specific B cells and T cells separately. Activated B cells uptake bound antigen, processing and presenting antigenic peptide on MHCII to TFH cells and a germinal center is formed. The population of germinal center (GC) B cells undergoes evolution toward higher antigenic affinity and specificity, marked by continual antigenic binding, processing, and presentation to cognate TFH cells, which deliver selection signals resulting in further diversification or exit to join the memory compartment (Mem) or differentiate to plasma cells (PC), which secrete specific, high-affinity antibodies (Abs).
This selection process is highly regulated by complex molecular signals at multiple stages. Following immunization, antigen-specific B cell precursors are activated, binding antigen and moving to the outer follicular zones. Here, they present antigenic peptide on MHCII to specialized subsets of separately-activated follicular helper T (TFH) cells to form GCs [3–6]. In this structure, B cells undergo cycles of Darwinian evolution through repeated rounds of expansion, diversification, and selection by limiting numbers of cognate TFH cells to form a both a diverse and highly-specific repertoire in both the memory and plasma cell compartments.
Central to understanding these concurrent processes of diversification, affinity maturation, and exit are spatial, temporal, and transcriptional dynamics in the GC. Robust model antigen systems and recent advances in genetic and imaging approaches currently allow access to this complex and ever-changing population of GC B cells. In this review, we will outline literature informing our present understanding of GC physical structure over time as it relates to transcriptional programs as well as the cellular and molecular mechanisms that regulate them in the primary and secondary response. Finally, we will discuss future directions of the field, with an eye on uncovering dynamics of evolutionary development by using the power of single-cell resolution.
Spatiotemporal control of GC B cell programs
The physical organization of the GC is reflective of and intimately tied to spatiotemporal function. Originally observed in histological sections of secondary lymphoid tissue, GC B cells were described to reside in two compartments that would be known as the “light zone” and “dark zone” (LZ and DZ, respectively) [7]. The LZ contains B cells that bind antigen trapped on the follicular dendritic cell network and interact with GC-associated TFH cells. The DZ contains large numbers of proliferating cells undergoing rapid division and somatic hypermutation. Early pulse-chase experiments using 3H-thymidine and BrdU [8,9] implied movement between the two zones that was later suggested to be controlled by CXCR4- and CXCR5-mediated chemotaxis [10]. In a series of seminal studies using two-photon microscopy, the real-time dynamics of cellular movement during early GC events [11] and dynamic cycling between the LZ and DZ [12–14] were directly visualized for the first time.
In more recent studies, Victora and colleagues utilized a fluorescent photoactivatable reporter to label DZ and LZ GC B cells in situ to provide direct confirmation of the connection between GC localization, cellular phenotype, and gene expression [15]. They found that DZ B cells were characterized by increased expression of CXCR4 protein and mRNA, along with upregulation of distinct patterns of expression for cell cycle and somatic hypermutation machinery. In contrast, CD86 mRNA and protein were upregulated in LZ cells, and these cells also displayed increased transcription of gene programs essential for antigen presentation [15]. Together, these studies revealed that distinct GC processes were dependent upon cell location within the GC structure, and that GC B cells undergo spatiotemporal cycling through progressive stages of transcription over the course of clonal evolution.
Characterization of DZ and LZ transcriptional programs informed further studies to dissect regulators of GC B cell movement and location-specific processes. The cell cycle regulator c-Myc, previously found to have low or completely absent activity in bulk GC B cells, was revisited and shown to be transiently expressed in selected subsets of GC B cells in the LZ [16,17]. The short-lived expression of Myc in the LZ was suggested to initiate downstream regulators, such as AP4, to mediate multiple rounds of division and somatic hypermutation in the DZ [18]. Additionally, high expression of the transcription factor FOXO1 in DZ B cells appeared to control DZ formation, organization, and gene expression [19,20], likely working in concert with Bcl6, an important regulator of GC B cell function [21–23]. Furthermore, the presence of FOXO1+ cells in the LZ suggested an initiation program for DZ entry established during cognate selection [20].
Paired analysis of repertoire, transcription, and protein
While visualizing the spatiotemporal organization and movement of GC B cells and TFH cells in real time has illuminated essential information regarding the cellular dynamics of GC clonal selection, many of these studies relied heavily on adoptive transfer systems of transgenic Ag-specific T and B cells. It remains important to consider the complex evolutionary dynamics within an intact polyclonal system, where there is a much broader range of binding affinities competing for selection. Additionally, while genetic experiments are informative for mechanistic analysis of GC processes, there is no one definitive marker for GC B cells, and many surface markers and transcription factors active in the GC also play a large role in its formation, making results from these experiments complicated to interpret. Finally, single-cell resolution is necessary to disentangle the complex, heterogeneous population of cells in the GC. Analyzing repertoire, mRNA, and protein levels from the same cell allow integrated analysis of antigen-specific affinity, molecular programming, and cellular phenotype to make conclusions about the mechanisms of GC selection and differentiation.
Utilizing this approach, Victora and colleagues used multicolor labeling to illustrate clonal selection and dominance within GCs of an intact polyclonal system [24]. To visualize clonal composition of individual GCs in real time, B cell clones were irreversibly labeled with a heritable color label after GC formation, and the GC was allowed to progress. Clonal dominance was indicated by the heterogeneity of colors visualized in situ. Microdissection and repertoire analysis of single cells within individual GCs linked clonal dominance with affinity maturation. Remarkably, lower-affinity B cells were able to maintain fitness even within GCs that contained higher-affinity competition, providing evidence that mechanisms independent of BCR affinity are important in GC clonal composition [24].
In recent studies, we have integrated surface phenotype and transcription with BCR repertoire analysis of single cells to detail how antigen-specific GC B cells progress through stages of selection [25]. Transcription of 96 genes for single cells was obtained using high-order quantitative PCR and paired with immunoglobulin repertoire to link gene expression with clonal selection. Multidimensional clustering using the machine learning algorithm, t-distributed stochastic neighbor embedding (t-SNE), was utilized to visualize patterns of similar gene expression within the heterogeneous GC population. Four clusters of similar gene expression emerged, with a clear pattern of progression defined most clearly by expression of Cd83 and Polh, two genes whose differential mRNA and protein expression had been previously established as part of the hallmark LZ and DZ programs, respectively, in bulk RNA-seq [15] (Figure 2). Expression of Polh, which encodes the DNA polymerase that introduces somatic hypermutations in the DZ [26], was highly expressed in the DZ program, while Cd83 expression characterized cognate interactions between TFH cells and GC B cells. Transitions between these four stages were correlated with genes important for cognate contact (Stage 1 to 2), DZ transition (Stage 2 to 3), proliferation and diversification (Stage 3 to 4), and LZ re-entry (Stage 4 to 1) [25].
Figure 2. Stage-specific regulation of genetic programs in the GC.
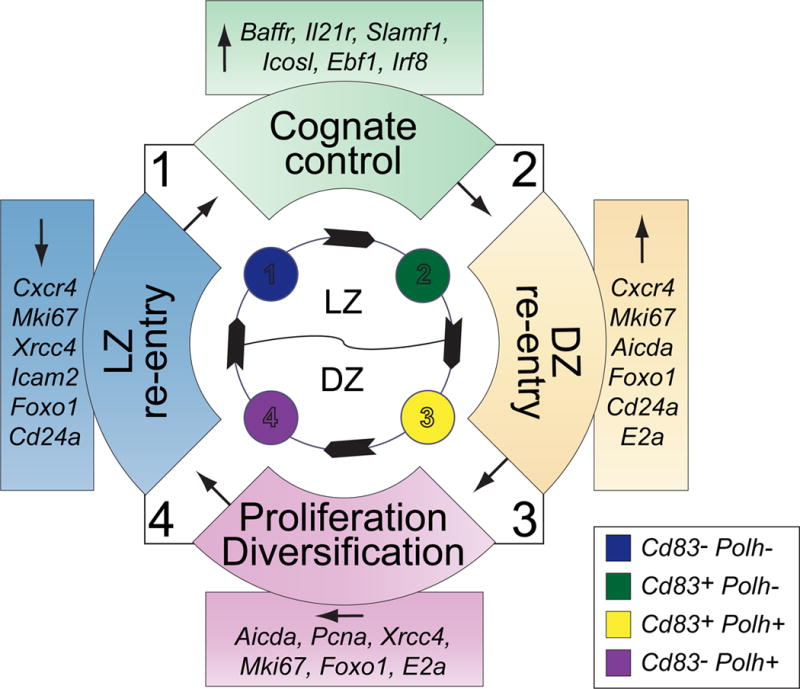
Gene expression of single cells in the GC assorted into four distinct patterns of gene expression (Stages 1–4). Gene clusters were determined according to patterns of similar expression using the machine learning algorithm t-distributed stochastic neighbor embedding (t-SNE), and order of cyclic progression computationally inferred by the trajectory detection algorithm Wanderlust. Genes in colored boxes change significantly between the stages marked in the colored semicircles in the direction indicated by the arrow. Stages 1–2 represent cognate control in the light zone (LZ), where germinal center (GC) B cells pick up antigen trapped on the follicular dendritic cell network, processing and presenting them to limiting numbers of cognate follicular helper T cells (Tfh), which select B cells based on levels of cognate antigenic peptide. Between Stages 2–3, selected B cells travel to the dark zone (DZ), upregulating genes associated with somatic hypermutation and proliferation, as well as the chemokine receptor Cxcr4, which enables spatial migration. From Stages 3–4, GC B cells diversify the B cell receptor and proliferate, maintaining high levels of Aicda and Mki67, among others. Finally, from Stages 4–1, GC B cells re-enter the LZ, downregulating genes necessary for affinity maturation and prepare to re-express the new B cell receptor to pick up and present antigen once again.
TFH cells as molecular controllers of clonal evolution
To drive clonal evolution and initiate progression through the GC, effective selection pressure is required. Two-photon imaging studies first illustrated contact between GC B cells and TFH cells in real time in vivo, lending appreciation for a possible role of TFH cells to provide selection pressure [12–14]. To further investigate the effect of this interaction on GC selection, Nussenzweig and colleagues manipulated peptide presentation levels on MHCII independently of BCR crosslinking via utilization of the endocytic receptor DEC205. The increased expansion of GC B cells that presented more antigen supported the model that TFH cells serve as the primary limiting selection pressure for GC B cells [15]. Indeed, further study using this system revealed that cell division and hypermutation by GC B cells was proportional to the amount of antigen presented to cognate TFH cells [27], and the cells presenting the highest levels of pMHCII to TFH cells had sustained increases in intracellular free calcium and IL-4 and IL-21 expression [28], suggesting an active mode of selection. Evidence linking increased antigen presentation on MHCII to rapid cell cycle progression in the DZ [29] provided a mechanism for clonal evolution. These results support a model in which TFH cell help in the LZ determines the timing of proliferation and diversification in the DZ to preferentially shape the GC repertoire [30]. In addition to controlling re-entry into the DZ, antigen affinity may also regulate GC exit by inducing memory or PC transcriptional regulators. Kurosaki’s group correlated Bach2 expression with antigen affinity to show induction of the post-GC memory program in lower-affinity, less mutated cells, which would preserve a wide range of repertoire diversity in the memory compartment [31].
Despite much evidence pointing to peptide presentation as a primary mode of selection, concurrent mechanisms also influence GC clonal evolution. Even though individual GCs may become dominated by a single clone, high-affinity GC clones can be found evolving alongside low-affinity clones, even within the same GC [24]. Heterogeneous populations of TFH cells may select multiple antigen-specific B cell clones binding different epitopes, and could contribute to the maintenance of clonal variety [32,33]. Sets of costimulatory molecules expressed on GC TFH cells and GC B cells can modify the signaling consequences of cognate contact and direct post-GC fate [32]. In a complete knockout of PD-1, a costimulatory molecule highly expressed on TFH cells, decreased Il4 and Il21 expression by TFH cells was accompanied by lowered long-lived antigen-specific PCs following immunization [34]. Furthermore, ICOS-ICOSL interactions between TFH cells and GC B cells were found to enhance cell-cell contact and initiate a feed-forward signal of positive selection events to enhance antigen-specific affinity selection in a competitive adoptive transfer system [35].
Differential selection may also rely on the timing of the immune response. Shlomchik and colleagues proposed that low-affinity, antigen-specific B cells with few mutations differentiate into memory B cells early on, and are followed by differentiation of high-affinity plasma cells later in the response [36]. While studying the humoral response to a Th2-type infection, Craft’s group attributed this switch to modulation of TFH cell phenotype over the course of infection, suggesting TFH cell-mediated temporal control by differential cytokine secretion [37]. These ongoing studies continue to emphasize the importance of TFH cells in shaping GC B cell populations and outcomes.
Negative selection in the GC
Because of the prevalence of DNA damage driving the rapid mutational processes in the GC, stringent negative selection mechanisms must be in place to prevent autoreactivity [38] and cancer [39]. Follicular regulatory T (Tfr) cells have emerged as negative regulators that dampen the GC response [40–42] and may play an integral role in preventing aberrant pathological outcomes; however, their mechanism of action is largely unresolved. Like TFH cells, they express the co-receptors PD-1 [43] and CTLA-4 [44,45], which modify the potency of their activity and may serve as modulators of negative selection. Furthermore, in vitro co-culture experiments have suggested that TFR-mediated suppression of B cell activation relies on the presence of TFH cells and may epigenetically restrict access to genes important to GC B cell differentiation, notably Aicda, Myc, and Pou2af1 [46]. TFR cells can be specific for both foreign and self-antigen [47]. Nevertheless, it is unclear how antigen-specificity and TCR affinity contribute to TFR cell differentiation, as they do for TFH cells [48], and how cognate contact might contribute to TFR cell function.
Another requirement for GC negative regulation may be to preserve diversity of BCR specificity across many antigen epitopes. Plasma cell differentiation itself within the GC may serve as a negative feedback loop. Our studies have shown that PCs, enabled by expression of MHCII, costimulatory molecules, and antigen presentation machinery, can negatively regulate the TFH cell program in an antigen-specific manner [49]. Another study by Toellner and colleagues provided evidence that antibody secreted by differentiated PCs can bind antigen in the GC, limiting access to antigen and driving BCR evolution by providing an advantage to specificities that have not already been produced [50].
Memory and the secondary response
The ability to produce memory B cells that can rapidly differentiate into memory-response PCs (mPCs) and re-diversify in memory-response GCs (mGCs) upon antigenic rechallenge is a hallmark trait of the GC [2,25]. Experimental evidence from several studies points to heterogeneity of memory cells with varying capacities for mPC versus mGC differentiation. Antibody class is suggested to be a major divergent trait induced in the primary response because it has been correlated to divergent differentiation pathways upon re-challenge. In a murine malaria model, Pepper and colleagues found that IgM+ memory B cells (MBCs) preferentially expanded and generated PCs upon secondary infection [51], while Jenkins’ group revealed that phycoerythrin (PE) immunization with complete Freund’s adjuvant (CFA) induced domination of the secondary response by class-switched MBCs [52].
Reynaud, Weill, and colleagues suggested that both IgM+ and class-switched cells participated in the secondary response in different ways. Immunization with sheep red blood cells induced antigen-specific IgG1+ MBCs that preferentially differentiate to mPC, while antigen-specific IgM+ MBCs induced secondary GCs [53]. Alternatively, a virus-like particle (VLP) prime-boost immunization strategy by Bachmann’s group drove a robust IgG+ and IgM+ mPC response with negligible mGCs forming [54]. Finally, Shlomchik and colleagues found that CD80 and PD-L2 could define MBC heterogeneity in a class-independent manner. After immunization with NP-CGG in alum, CD80+ PD-L2+ MBCs seeded mGCs, and CD80− PD-L1− MBCs became mPCs regardless of antibody class [55]. These seemingly conflicting results may indicate that memory-response programming may be dependent on immunization or infection conditions.
An additional important characteristic of memory B cells is longevity, which is crucial for lasting antigen-specific immunity. The enzyme activation-induced cytidine deaminase (AID), which is responsible for both class-switch recombination (CSR) and somatic hypermutation (SHM) [56], has been implicated as a key driver in MBC longevity. Jenkins and colleagues originally suggested class-switch recombination as the key factor for persistence, as PE-specific IgM+ MBCs survived longer than class-switched MBCs [52]. However, in a more recent study, Nussenszweig’s group used a complex genetic model to disentangle AID-driven mechanisms of CSR and SHM to conclude that levels of SHM were a better indicator of MBC longevity, independent of class [57]. Single-cell resolution is still needed to dissect divergent programming of populations arising post-GC to understand mechanisms of antigen-specific regulation. Regardless, antigen experience, independent of class, is most likely a strong component of memory B cell programming [58], which itself diverges in a class-specific manner [59].
Conclusions and future questions
While much information has been uncovered from bulk analysis, single-cell approaches are needed to uncover the ever-changing clonal and molecular dynamics of heterogeneous populations within the GC. The studies already performed will inform future work using techniques such as single-cell RNA-seq to detail more completely the molecular mechanisms of GC cyclic progression. Pairing these analyses with information on clonal selection and evolution within the GC, as well as its products memory B cells and PCs, will enlighten our understanding of these changing dynamics. Finally, studies pairing GC B cell evolution with active programs of T cell-mediated selection will help to design novel vaccine formulations that induce efficient immune responses against antigen.
Highlights.
Germinal centers (GCs) facilitate antigen-specific evolution of B cell repertoire.
Follicular helper T cells regulate GC evolution, memory, and plasma cell formation.
Complex molecular signals drive rapid transcriptional changes in GC B cells.
Single-cell analysis details regulation of the diverse, evolving GC population.
Acknowledgments
This work was supported by grants from the National Institute of Health (AI047231, AI040215 to M.G.M-W.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mesin L, Ersching J, Victora GD. Germinal center B cell dynamics. Immunity. 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McHeyzer-Williams M, Okitsu S, Wang N, McHeyzer-Williams L. Molecular programming of B cell memory. Nat Rev Immunol. 2012;12:24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nutt SL, Tarlinton DM. Germinal center B and follicular helper T cells: siblings, cousins or just good friends? Nat Immunol. 2011;12:472–477. doi: 10.1038/ni.2019. [DOI] [PubMed] [Google Scholar]
- 4.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinuesa CG, Linterman MA, Yu D, MacLennan ICM. Follicular helper T cells. Annu Rev Immunol. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- 6.Qi H. T follicular helper cells in space-time. Nat Rev Immunol. 2016;16:612–625. doi: 10.1038/nri.2016.94. [DOI] [PubMed] [Google Scholar]
- 7.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–39. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 8.Hanna MG. An autoradiographic study of the germinal center in spleen white pulp during early intervals of the immune response. Lab Investig J Tech Methods Pathol. 1964;13:95–104. [PubMed] [Google Scholar]
- 9.Liu Y-J, Zhang J, Lane PJL, Chan EY-T, Maclennan ICM. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991;21:2951–2962. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- 10.Allen CDC, Ansel KM, Low C, Lesley R, Tamamura H, Fujii N, Cyster JG. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol. 2004;5:943–952. doi: 10.1038/ni1100. [DOI] [PubMed] [Google Scholar]
- 11.Okada T, Miller MJ, Parker I, Krummel MF, Neighbors M, Hartley SB, O’Garra A, Cahalan MD, Cyster JG. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3:e150. doi: 10.1371/journal.pbio.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen CDC, Okada T, Tang HL, Cyster JG. Imaging of Germinal Center Selection Events During Affinity Maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 13.Schwickert TA, Lindquist RL, Shakhar G, Livshits G, Skokos D, Kosco-Vilbois MH, Dustin ML, Nussenzweig MC. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007;446:83–87. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 14.Hauser AE, Junt T, Mempel TR, Sneddon MW, Kleinstein SH, Henrickson SE, von Andrian UH, Shlomchik MJ, Haberman AM. Definition of germinal-center B cell migration in vivo reveals predominant intrazonal circulation patterns. Immunity. 2007;26:655–667. doi: 10.1016/j.immuni.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calado DP, Sasaki Y, Godinho SA, Pellerin A, Köchert K, Sleckman BP, de Alborán IM, Janz M, Rodig S, Rajewsky K. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012;13:1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, Dalla-Favera R. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol. 2012;13:1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou C, Verbaro DJ, Tonc E, Holmgren M, Cella M, Colonna M, Bhattacharya D, Egawa T. The transcription factor AP4 mediates resolution of chronic viral infection through amplification of germinal center B cell responses. Immunity. 2016;45:570–582. doi: 10.1016/j.immuni.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Dominguez-Sola D, Kung J, Holmes AB, Wells VA, Mo T, Basso K, Dalla-Favera R. The FOXO1 transcription factor instructs the germinal center dark zone program. Immunity. 2015;43:1064–1074. doi: 10.1016/j.immuni.2015.10.015. With Ref. [20], one of two genetic ablation studies investigating the relationship between transcriptional regulation, GC organization, and GC clonal evolution, in particular showing the importance of FOXO1 for DZ formation, DZ transcriptional programs, and GC affinity selection. [DOI] [PubMed] [Google Scholar]
- 20*.Sander S, Chu VT, Yasuda T, Franklin A, Graf R, Calado DP, Li S, Imami K, Selbach M, Di Virgilio M, et al. PI3 kinase and FOXO1 transcription factor activity differentially control B cells in the germinal center light and dark zones. Immunity. 2015;43:1075–1086. doi: 10.1016/j.immuni.2015.10.021. See Ref. [19] [DOI] [PubMed] [Google Scholar]
- 21.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang Y-H, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 24**.Tas JMJ, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill J-C, Reynaud C-A, Browne EP, et al. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. A temporally-driven, B cell-specific, Cre-mediated coloring system in “confetti mice” allows for clear visualization of B cell clones at various stages of the GC response. High-affinity clones could be found evolving alongside low-affinity clones, even within the same GCs, providing evidence that factors other than affinity are responsible for driving antigen-specific selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.McHeyzer-Williams LJ, Milpied PJ, Okitsu SL, McHeyzer-Williams MG. Class-switched memory B cells remodel BCRs within secondary germinal centers Nat. Immunol. 2015;16:296–305. doi: 10.1038/ni.3095. Using a single-cell approach, our study utilized flow cytometry, repertoire sequencing, and high-order 96-gene RT-qPCR to obtain multidimensional information of each B cell in a GC. Using machine learning algorithms allowed the visualization of cellular programs in a two-dimensional plot, while trajectory-detection methods allowed us to infer progression of GC B cell programs using these highly-detailed transcriptional snapshots. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delbos F, Aoufouchi S, Faili A, Weill J-C, Reynaud C-A. DNA polymerase η is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28**.Shulman Z, Gitlin AD, Weinstein JS, Lainez B, Esplugues E, Flavell RA, Craft JE, Nussenzweig MC. Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science. 2014;345:1058–1062. doi: 10.1126/science.1257861. This study utilized two-photon microscopy to characterize interactions between GC B cells and TFH cells in the germinal center. Visualization of sustained intracellular Ca2+ and expression of IL-4 and IL-21 was correlated with cognate interactions, and TFH cells were shown to be highly motile and capable of scanning many GC B cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Gitlin AD, Mayer CT, Oliveira TY, Shulman Z, Jones MJK, Koren A, Nussenzweig MC. T cell help controls the speed of the cell cycle in germinal center B cells. Science. 2015;349:643–646. doi: 10.1126/science.aac4919. Using DEC-205 to manipulate antigenic peptide presentation on B cells without interfering with BCR signaling, this study links the strength of cognate help by TFH cells to provide proliferative advantages to high-affinity GC B cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bannard O, Horton RM, Allen CDC, An J, Nagasawa T, Cyster JG. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 2013;39:912–924. doi: 10.1016/j.immuni.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Shinnakasu R, Inoue T, Kometani K, Moriyama S, Adachi Y, Nakayama M, Takahashi Y, Fukuyama H, Okada T, Kurosaki T. Regulated selection of germinal-center cells into the memory B cell compartment. Nat Immunol. 2016;17:861–869. doi: 10.1038/ni.3460. This study investigated the regulation of post-GC fate, seeking to disentangle stochastic versus instructed differentiation to memory B cells, and finding that lower-affinity GC B cells were found to preferentially differentiate into the memory compartment and correlate with high expression of Bach2. [DOI] [PubMed] [Google Scholar]
- 32.Fazilleau N, Mark L, McHeyzer-Williams LJ, McHeyzer-Williams MG. Follicular helper T cells: lineage and location. Immunity. 2009;30:324–335. doi: 10.1016/j.immuni.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shulman Z, Gitlin AD, Targ S, Jankovic M, Pasqual G, Nussenzweig MC, Victora GD. T follicular helper cell dynamics in germinal centers. Science. 2013;341:673–677. doi: 10.1126/science.1241680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, Shlomchik MJ. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol. 2010;11:535–542. doi: 10.1038/ni.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Liu D, Xu H, Shih C, Wan Z, Ma X, Ma W, Luo D, Qi H. T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 2015;517:214–218. doi: 10.1038/nature13803. Two-photon analysis of the effects of ICOS-ICOSL interactions as they pertain to physical contact between GC B cells and TFH cells in vivo, and their implications for GC clonal evolution. Results revealed the propensity of costimulatory actions to trigger molecular feed-forward mechanisms that amplify small differences in cognate help and shape affinity maturation in the GC. [DOI] [PubMed] [Google Scholar]
- 36*.Weisel FJ, Zuccarino-Catania GV, Chikina M, Shlomchik MJ. A temporal switch in the germinal center determines differential output of memory b and plasma cells. Immunity. 2016;44:116–130. doi: 10.1016/j.immuni.2015.12.004. Using BrdU pulse-chase experiments, this study addressed temporal control of post-GC differentiation to memory and plasma cells, concluding that GC output favors memory cell output early in the response, followed by a preference for PCs output later on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Weinstein JS, Herman EI, Lainez B, Licona-Limón P, Esplugues E, Flavell R, Craft J. TFH cells progressively differentiate to regulate the germinal center response. Nat Immunol. 2016;17:1197–1205. doi: 10.1038/ni.3554. The authors investigate progression of the TFH cell response over time from an IL-21-producing population to an IL-4-producing population, addressing heterogeneity within the TFH cell compartment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vinuesa CG, Sanz I, Cook MC. Dysregulation of germinal centres in autoimmune disease. Nat Rev Immunol. 2009;9:845–857. doi: 10.1038/nri2637. [DOI] [PubMed] [Google Scholar]
- 39.Basso K, Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat Rev Immunol. 2015;15:172–184. doi: 10.1038/nri3814. [DOI] [PubMed] [Google Scholar]
- 40.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang Y-H, Lim H, Reynolds JM, Zhou X, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wollenberg I, Agua-Doce A, Hernández A, Almeida C, Oliveira VG, Faro J, Graca L. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J Immunol. 2011;187:4553–4560. doi: 10.4049/jimmunol.1101328. [DOI] [PubMed] [Google Scholar]
- 43.Sage PT, Francisco LM, Carman CV, Sharpe AH. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol. 2013;14:152–161. doi: 10.1038/ni.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sage PT, Paterson AM, Lovitch SB, Sharpe AH. The coinhibitory receptor CTLA-4 controls B cell responses by modulating T follicular helper, T follicular regulatory, and T regulatory cells. Immunity. 2014;41:1026–1039. doi: 10.1016/j.immuni.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wing JB, Ise W, Kurosaki T, Sakaguchi S. Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity. 2014;41:1013–1025. doi: 10.1016/j.immuni.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 46*.Sage PT, Ron-Harel N, Juneja VR, Sen DR, Maleri S, Sungnak W, Kuchroo VK, Haining WN, Chevrier N, Haigis M, et al. Suppression by TFR cells leads to durable and selective inhibition of B cell effector function. Nat Immunol. 2016;17:1436–1446. doi: 10.1038/ni.3578. This in vitro investigation using co-cultures of TFH cells, TFR cells, and B cells reveals programming and epigenetic changes that occur in the presence of TFR cells, offering insight into mechanisms of TFR negative regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Aloulou M, Carr EJ, Gador M, Bignon A, Liblau RS, Fazilleau N, Linterman MA. Follicular regulatory T cells can be specific for the immunizing antigen and derive from naive T cells. Nat Commun. 2016;7:10579. doi: 10.1038/ncomms10579. This study offers evidence that TFR cells can be specific for both foreign and self-antigen and is a key study in unraveling the mechanisms of TFR cell-mediated negative GC regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat Immunol. 2009;10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelletier N, McHeyzer-Williams LJ, Wong KA, Urich E, Fazilleau N, McHeyzer-Williams MG. Plasma cells negatively regulate the follicular helper T cell program. Nat Immunol. 2010;11:1110–1118. doi: 10.1038/ni.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Meyer-Hermann M, George LA, Figge MT, Khan M, Goodall M, Young SP, Reynolds A, Falciani F, Waisman A, et al. Germinal center B cells govern their own fate via antibody feedback. J Exp Med. 2013;210:457–464. doi: 10.1084/jem.20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Krishnamurty AT, Thouvenel CD, Portugal S, Keitany GJ, Kim KS, Holder A, Crompton PD, Rawlings DJ, Pepper M. Somatically hypermutated plasmodium-specific IgM+ memory B cells are rapid, plastic, early responders upon malaria rechallenge. Immunity. 2016;45:402–414. doi: 10.1016/j.immuni.2016.06.014. This study interrogates the importance of IgM+ memory B cells and their involvement in the secondary response in a murine model of malaria, highlighting the importance of IgM+ MBCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pape KA, Taylor JJ, Maul RW, Gearhart PJ, Jenkins MK. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331:1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dogan I, Bertocci B, Vilmont V, Delbos F, Mégret J, Storck S, Reynaud C-A, Weill J-C. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 54.Zabel F, Mohanan D, Bessa J, Link A, Fettelschoss A, Saudan P, Kündig TM, Bachmann MF. Viral particles drive rapid differentiation of memory B cells into secondary plasma cells producing increased levels of antibodies. J Immunol. 2014;192:5499–5508. doi: 10.4049/jimmunol.1400065. [DOI] [PubMed] [Google Scholar]
- 55.Zuccarino-Catania GV, Sadanand S, Weisel FJ, Tomayko MM, Meng H, Kleinstein SH, Good-Jacobson KL, Shlomchik MJ. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat Immunol. 2014;15:631–7. doi: 10.1038/ni.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 57*.Gitlin AD, von Boehmer L, Gazumyan A, Shulman Z, Oliveira TY, Nussenzweig MC. Independent roles of switching and hypermutation in the development and persistence of B lymphocyte memory. Immunity. 2016;44:769–81. doi: 10.1016/j.immuni.2016.01.011. This study attempts to disentangle the two distinct events driven by AID: CSR and SHM. Utilizing a complex genetic system whereby CSR in B cells is driven by the Aicda promoter and mimicked by Cre recombination, the authors find that in this system, SHM is likely to play a bigger role than CSR in the longevity of the memory compartment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kometani K, Nakagawa R, Shinnakasu R, Kaji T, Rybouchkin A, Moriyama S, Furukawa K, Koseki H, Takemori T, Kurosaki T. Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory b cells toward plasma cell differentiation. Immunity. 2013;39:136–147. doi: 10.1016/j.immuni.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Wang NS, McHeyzer-Williams LJ, Okitsu SL, Burris TP, Reiner SL, McHeyzer-Williams MG. Divergent transcriptional programming of class-specific B cell memory by T-bet and RORα. Nat Immunol. 2012;13:604–611. doi: 10.1038/ni.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]


