Abstract
The membrane-water interface forms a uniquely heterogeneous and geometrically constrained environment for enzymatic catalysis. Integral membrane enzymes sample three environments – the uniformly hydrophobic interior of the membrane, the aqueous extramembrane region, and the fuzzy, amphipathic interfacial region formed by the tightly packed headgroups of the components of the lipid bilayer. Depending on the nature of the substrates and the location of the site of chemical modification, catalysis may occur in each of these environments. The availability of structural information for alpha-helical enzyme families from each of these classes, as well as several beta-barrel enzymes from the bacterial outer membrane, has allowed us to review here the different ways in which each enzyme fold has adapted to the nature of the substrates, products, and the unique environment of the membrane. Our focus here is on enzymes that process lipidic substrates.
1. Introduction
Despite the rapid advances in structural studies of many classes of membrane proteins, most notably channels, receptors and transporters, progress has been markedly slower for others, such as of integral membrane enzymes involved in formation and modification of the lipidic constituents of the membrane itself.
There are several biophysical issues that are shared by all membrane proteins involved in lipid biosynthesis and modification of lipidic ligands that are not well understood. First, how is recognition specificity generated for hydrophobic substrates, which typically interact with proteins through often-interchangeable non-polar contacts? Second, how does a protein compete with or interact with the membrane for the recruitment of specific lipid components, which themselves are best suited to reside in the hydrophobic environment provided by the bilayer? Third, specific to enzymes in the membrane, and the central theme of this review, how does the protein environment reconcile the requirement of charged groups in enzyme-catalyzed chemical reactions with the apparently incompatible hydrophobic nature of lipidic ligands and hydrophobic interior of the membrane bilayer? If the active site is outside the membrane, how are both soluble and insoluble substrates brought into apposition for catalysis to occur? If catalysis occurs within the bilayer, how do hydrophilic substrates enter?
The principles governing recognition, specificity, and function of membrane proteins that interact with hydrophobic substrates are best investigated by high-resolution structures. An atomic level structure is unique in providing detailed insight into these processes at a molecular level, revealing the precise protein residues involved in substrate recognition, catalysis or translocation, and, overall, providing us with hypotheses, which can then be tested by functional assays and biophysical techniques. However, our knowledge of the molecular determinants of lipid-enzyme and lipid-transporter interactions is scarce at best, reflecting the persistent technical difficulties associated with structural studies of membrane proteins. For example, amongst a total of 643 unique membrane protein entries, only very few are of polytopic transmembrane (TM) enzymes that process lipid substrates.
This review focuses on the current knowledge of enzymes requiring lipid substrates, the architecture of their active sites and how they accommodate lipid and hydrophilic substrates or cofactors. We have thus excluded respiratory and photosynthetic complexes, as we believe they are out of the scope of this review. We have divided the alpha-helical transmembrane enzymes into two main groups: those whose reaction occurs where the aqueous environment and the membrane meet (interfacial; Fig. 1) and those with an active site outside the membrane (extramembrane; Fig. 1). We also examine a unique example of an enzyme with an active site inside the borders of the membrane (intramembrane; Fig. 1). Finally, we also take a look at the known structures of beta-barrel lipid modifying enzymes and how they function uniquely from their alpha-helical counterparts. Table 1 summarizes information about available structures, architecture and biology of all enzymes that are being discussed in this review.
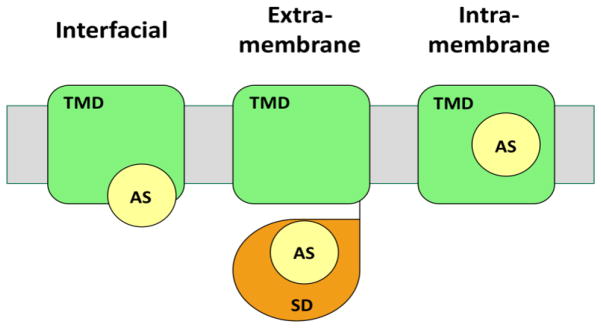
Figure 1. Schematic representation of the different catalytic modes at the membrane-water interface.
The grey region represents the membrane. The green rectangle represents the transmembrane domain of an enzyme (TMD), the orange teardrop represents a soluble domain (SD), and the yellow circle represents the location of the active site (AS).
Table 1.
Summary of enzymes discussed.
| INTERFACIAL CATALYSIS | |||
|---|---|---|---|
| Enzyme | Architecture | Structures and substrates | Function |
| PIP-synthase from Renibacterium salmoninarum | 6 TM α-helix dimer | Apo (PDB 5D91) CDP-DAG (PDB 5D92; Fig. 2A) [5] |
Catalyzes the synthesis of phosphatidylinositol-phosphate from inositol-phosphate and CDP- DAG |
| ICMT from Methanosarcina acetivorans | 5 TM α-helix | SAH (PDB 4A2N; Fig. 2B) [15] | Methylates the carboxy terminus of a lipid-linked protein |
| Cds from Thermatoga maritima | 9 TM α-helix dimer | Apo (PDB 4Q2E; Fig. 2C) [19] | Catalyzes the synthesis of CDP- DAG from phosphatidic acid and CTP. |
| DgkA from Escherichia coli | 3 TM α-helix trimer | 7.8 MAG (PDB 3ZE4) [21, 22] ACP and 9.9 MAG (PDB 4UXX; Fig. 2D) [21, 22] |
Phosphorylates diacylglycerol to produce phosphatidic acid |
| VKOR from Synechococcus sp. | 5 TM α-helix | Ubiquinone (PDB 3KP9; Fig. 2E) [31] | Disulfide bond formation in proteins couple to the reduction of vitamin K epoxide to vitamin K hydroquinone |
| DsbA/DsbB complex from Escherichia coli | 4 TM α-helix | Apo (PDB 2HI7) [115] | Catalyzes disulfide bond formation in proteins |
| PgpB from Escherichia coli | 6TM, α-helix | Apo (PDB 4PX7; Fig. 2F) [37] | Dephosphorylates lipid head groups |
| MraY from Aquifex aeolicus | 10 TM α-helix | Apo (PDB 4J72; Fig. 3A) [44] Inhibitor Muramycin D2 (PDB 5CKR) [47] | Transfers a peptidoglycan precursor to the lipid carrier UndP. |
| ArnT from Culpriavidus metallidurans | 13 TM α-helical | Apo (PDB 5EZM) UndP (PDB 5F15; Fig. 3B) [50] |
Transfers L-Ara4N sugar from UndP carrier to Lipid A. |
| Lgt from Escherichia coli | 7 TMα-helix | Phosphatidylglycerol (PG) (PDB 5AZC; Fig. 3C) Inhibitor palmitic acid (PDB 5AZB) [57] |
Catalyzes first step in conjugation of a lipobox protein to a lipid |
| UbiA from Aeropyrum pernix | 9 TM α-helix | Apo (PDB 4OD4) GSPP, PHB (PDB 4OD5; Fig. 3D) [58–60] |
Catalyzes the transfer of a prenyl group from donor to acceptor |
| UBIAD1 homolog from Archaeoglobus fulgidus | 9 TM α-helix | GPP (PDB 4TQ3) DMAPP (PDB 4TQ4) [58, 63] |
Catalyzes the transfer of a prenyl group from donor to acceptor |
| LTC4S from Homo sapiens | 4TM α-helix trimer | Apo (PDB 2UUI) GSH (PDB 2UUH) [67] LT analog I (PDB 4JCZ) LT analog II (PDB 4J7T; Fig. 3E) LT analog III (PDB 4J7Y) [68] |
Catalyzes the conjugation of fatty acid LTA4 with tripeptide GSH to form LTC4 |
| PGES1 from Homo sapiens | 4TM α-helix trimer | GSH (PDB 3DWW) [66] GSH analog (PDB 4AL1) Apo (PDB 4AL0) [65] |
Catalyzes the isomerization of prostaglanding H2 to prostaglanding E2 |
| EXTRAMEMBRANE CATALYSIS | |||
| Enzyme | Architecture | Structures and substrates | Function |
| GtrB from Synechocystis sp. | 2TM α-helix tetramer | UDP (PDB 5EKP; Fig. 4A) [72] | Transfers a glucose from UDP- glucose to UndP |
| STS (also known as ES) from Homo sapiens | 2 TM α-helix | Apo (PDB 1P49; Fig. 4B) [77] | Removes sulfate group from steroid-sulfate conjugates |
| MGT from Staphylococcus aureus | 1 TM α-helix | Apo (PDB 3VMQ) Lipid II analog 3 (PDB 3VMT; Fig. 4C) Moenomycin (PDB 3VMR) NBD-Lipid II (PDB 3VMS) [81, 82] |
Transglycosylase (TG) that catalyzes the formation of Lipid IV by transferring a disaccharide peptide from one Lipid II to another. |
| PBP1b from Escherichia coli | 1 TM α-helix | Apo (PDB 3VMA) [79] | TG that catalyzes the formation of Lipid IV by transferring a disaccharide peptide from one Lipid II to another. Also has transpeptidase activity, which covalently links peptides from different Lipid IV molecules together. |
| SCD1 from Mus musculus | 4 TMα-helix | Stearoyl-CoA (PDB 4YMK; Fig. 4D) [85, 86] | Introduces double bond into saturated fatty acyl-CoA |
| SCD1 from Homo sapiens | 4 TM α-helix | Stearoyl-CoA (PDB 4ZYO) [85, 86] | Introduces double bond into saturated fatty acyl-CoA |
| Scs7p from Saccharomyces cerevisiae | 4 TM α-helix | Apo (PDB 4ZR0) [89] | Carries out a hydroxylation reaction on a sphingolipid |
| PglB from Campylobacter lari | 13 TM α-helix | Acceptor peptide (PDB 3RCE; Fig. 4E) [51] | Transfers an oligosaccharide from a lipid carrier (UndP) to a protein |
| AglB from Archaeoglobus fulgidus | 13 TM α-helix | Apo (PDB 3WAJ) [52] | Transfers an oligosaccharide from a lipid carrier (UndP) to a protein |
| INTRAMEMBRANE CATALYSIS | |||
| Enzyme | Architecture | Structures and substrates | Function |
| SR1 from Methylomicrobium alcaliphilum | 10 TM α-helix | NADPH (PDB 4QUV; Fig. 5) [98] | Reduces double bonds of a sterol using a reducing cofactor. |
| CATALYSIS BY β-BARREL ENZYMES | |||
| Enzyme | Architecture | Structures and substrates | Function |
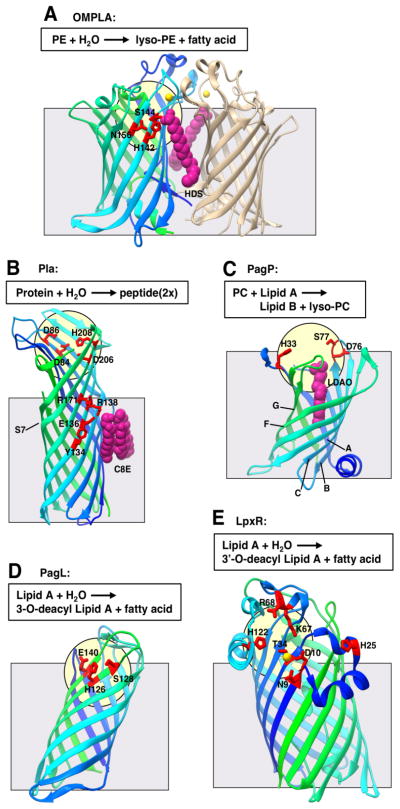
| OMPLA from Escherichia coli | 12 stranded β-barrel | Monomer (PDB 1QD5) Dimer (PDB 1QD6; Fig. 6A) [100] |
Hydrolyzes the acyl-ester bonds of phospholipids |
| OmpT from Escherichia coli | 10 stranded β-barrel | Apo (PDB 1I78) [105] | Catalyzes LPS dependent proteolysis |
| Pla from Yersinia pestis | 10 stranded β-barrel | Apo (PDB 2X55; Fig. 6B) [106] | Catalyzes LPS dependent proteolysis |
| PagP from Escherichia coli | 8 stranded β-barrel | Apo (PDB 1THQ; Fig. 6C) [110] SDS/Cosolvent (PDB 3GP6) [111] | Catalyzes the transfer of a palmitate moiety from a lipid donor to Lipid A |
| PagL from Pseudomonas aeruginosa | 8 stranded β-barrel | Apo (PDB 2ERV; Fig. 6D) [112] | Catalyzes the removal of a hydromyristate moiety from Lipid A |
| LpxR from Salmonella enterica | 12 stranded β-barrel | Apo (PDB 3FID; Fig. 6E) [114] | Catalyzes the removal of an acyloxyacyl moiety from Lipid A |
2. Structural basis of interfacial lipid modification
The most abundant category of alpha-helical transmembrane lipid modifying enzymes for which we have structural information – predominantly obtained by X-ray crystallography – are those whose active site is located at the membrane boundary. The abundance may be because the basic mechanism is, in fact, more frequently found in nature, or that enzymes with these requisites are more prone to crystallization, or simply that they have received thus far the greatest degree of attention from the structural biology community. Though these enzymes can be grouped together by active site location, they show wide diversity in biological function and substrate. Here we present the current knowledge of these enzymes, based on the structures available.
2.1 CDP-APs, a model for glycerophospholipid biosynthesis
The identity of a glycerophospholipid – whether it is a phosphatidyl-choline, or -inositol or -serine – is defined, across all kingdoms of life, by the enzymatic transfer of a substituted phosphate group from a CDP-linked donor to an acceptor alcohol [1, 2]. This reaction is catalyzed by CDP-alcohol phosphotransferases (CDP-APs). CDP-AP family members have unusually broad substrate specificity. The acceptor or donor substrate may carry a diacyl tail, both substrates may be lipids, as in cardiolipin synthase [3], or neither, with both substrates being polar small molecules [1]. CDP-APs are integral membrane enzymes that vary in number of predicted TM segments from 6 to 10, according to the nature of the acceptor substrate. CDP-APs are characterized by an absolutely conserved signature sequence of 8 amino acids, essential for function (D1xxD2G1xxAR…G2xxxD3xxxD4; [4]).
Information derived from the crystal structures of three enzymes [5–7] has elucidated how the active site of CDP-APs adapts to accommodate both lipidic and soluble substrates, thus addressing the question of specificity for lipidic molecules and shedding light at a molecular level on the chemistry of this reaction.
The structure of the CDP-AP Af2299 from Archaeoglobus fulgidus, which was determined from crystals grown in LCP with bound CDP, CMP/Apo and donor substrate CDP-glycerol to 2, 1.9 and 3.1 Å resolution respectively [5–7], shows molecular details of a 6 TM segment architecture, which represents the conserved core of all CDP-APs. The active site is harbored between TMs 2, 3 and 5, spanning from the water-membrane interface to within the transmembrane region, explaining the versatility of CDP-APs to accommodate a combination of soluble and membrane-embedded substrates. The universally conserved CDP-AP signature motif defines most of the active site, split between TM helices 2 & 3. A precise role for each one of the eight residues of the signature sequence has been assigned, and a mechanism proposed for the chemical reaction, universal for all CDP-APs. These results are in great part shared with those reported concomitantly on di-myo-inositol-1,3′-phosphate-1′-phosphate synthase (DIPPS), a CDP-AP related to Af2299 and also from Archaeoglobus fulgidus [6]. Interestingly, both Af2299 and DIPPS carry a soluble N-terminal cytidyltransferase-like domain [8], which provided the essential contacts in the crystal lattice between membrane layers of the lipidic crystallization matrix (lipidic cubic phase; LCP) [6, 7].
Phosphatidylinositol-phosphate (PIP) synthases catalyze the key step in the biosynthesis of phosphatidylinositol in prokaryotes, utilizing CDP-diacylglycerol (CDP-DAG) and inositol-phosphate as substrates to yield PIP [9]. The structure of PIP synthase from Renibacterium salmoninarum (RsPIPS) was determined without and with bound CDP-DAG to 2.5 and 3.6 Å resolution respectively, again from crystals grown in LCP following an engineering approach in which the soluble domain from Af2299 was used to generate a chimera to provide useful crystal lattice contacts [5]. The structure of RsPIPS (Fig. 2A) shows a similar architecture to the one observed for Af2299. The most notable difference is a large interfacial hydrophobic cavity between TM2 & TM5, which is the binding site for the diacyl chains of CDP-DAG. The diacylglycerol fits in a crevice between the TM helices, allowing the CDP to slot into the otherwise conserved active site. This work provides us with a first snapshot of how membrane and enzyme may interplay to allow catalysis of this lipidic substrate [5].
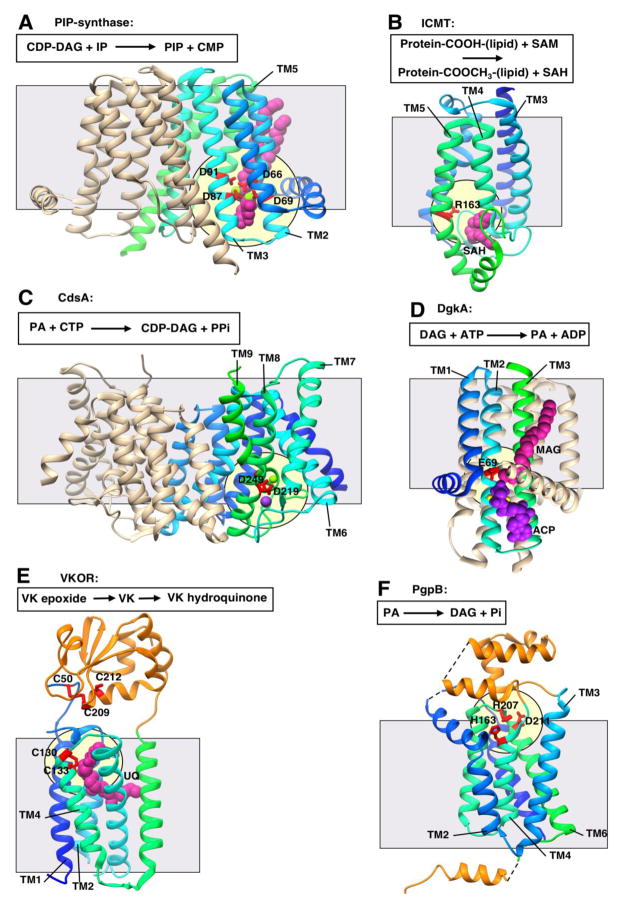
Figure 2. Representative structures showing Interfacial catalysis (I).
All structures are shown in ribbon representation with bound ligands shown in CPK representation. Transmembrane domains are shown in gradient coloring from the N-terminus to the C-terminus (blue to cyan to green) while extramembrane soluble domains are shown in orange. Grey square shows approximate location of the membrane border. Pale yellow circle indicates location of the putative active site. All structures are oriented with the cytosolic face of the membrane on the bottom. A schematic of the reaction catalyzed is shown above each structure. A. Phosphatidylinositol-phosphate synthase (PIP-synthase; PDB 5D92) with four aspartate residues from the CDP-AP signature sequence in red responsible for metal coordination and catalysis. Magnesium ion in light green and lipid CDP-diacylglycerol (CDP-DAG) in magenta. CDP-DAG, cytidine diphosphate diacylglycerol; IP, inositol-phosophate; PIP, phosphatidylinositol-phosphate; CMP, cytidine monophosphate. B. Isoprenylcysteine carboxyl methyltransferase (ICMT; PDB 4A2N) with putative catalytic residue in red. Methyl donor product S-Adenosyl-homocysteine (SAH) in magenta. SAM, S-Adenosyl methionine C. CDP-DAG synthetase (CdsA; PDB 4Q2E) with putative metal binding and catalytic residues in red. Magnesium ion in light green. Potassium ion in purple. PA, phosphatidic acid; CTP, cytidine triphosphate. D. Diacylglycerol kinase (DgkA; PDB 4UXX) with putative catalytic residue in red. Monoacylglycerol (MAG) in magenta, adenylmethylenediphosphonate (ACP) in purple, two zinc ion in yellow (partially obscured by ACP). E. Vitamin K epoxide reductase (VKOR; PDB 3KP9) cysteines proposed in electron transfer in red. Ubiquinone (UQ) in magenta. VK, vitamin K. F. Phosphatidylglycerolphosphate phosphatase B (PgpB; PDB 4PX7) with putative catalytic residues in red.
The structure of RsPIPS shows several conserved residues, which appear important for substrate recognition and catalysis. It was shown that, by mediating the access to the active site, hydrophobic residues determine substrate specificity for PIP synthases, and the functional importance of these residues was confirmed experimentally [5]. Results pertaining to PIP synthase will likely apply to all CDP-APs utilizing CDP-DAG as a substrate.
2.2 Methylation by Isoprenylcysteine Carboxyl Methyltransferase (ICMT)
Post-translational modification of C-terminal CAAX motifs (C, cysteine; A, aliphatic amino acid; X, any amino acid) is important for proper cellular trafficking, localization and function of many proteins within the mammalian cell including Ras, many Rho GTPases and G protein γ subunits. The CAAX motif is a signal for the prenylation of the protein containing the domain to be tethered to the membrane [10].
This posttranslational modification occurs in three steps in the endoplasmic reticulum (ER) [11, 12]. First, an isoprenyl lipid is attached to the cysteine of the CAAX motif. The cysteine is either farnesylated or geranylgeranylated by the respective transferase. Second, The -AAX tripeptide is cleaved by an integral membrane endoprotease. Third, the remaining C-terminus of the prenylated protein is methylated by isoprenylcysteine carboxyl methyltransferase (ICMT) using S-adenosyl-L-methionine (SAM) as a methyl donor. The carboxy methylation of these CAAX proteins is thought to help stabilize their interaction with the membrane environment. Inhibition of ICMT shows an anti-proliferative effect on cancer cells due to altered localization of Ras and disruption of Rho-mediated cell migration and adhesion. Therefore, ICMT is a promising anticancer drug target and ICMT inhibitors are being actively pursued for the treatment of cancers [13, 14].
The structure of ICMT ortholog from Methanosarcina acetivorans (MaICMT) has been solved using x-ray crystallography to 3.4 Å resolution [15]. MaICMT was crystallized as a monomer and shows a five transmembrane alpha helical topology (Fig. 2B). It also has a short C-terminal helix on the cytosolic side, which interacts with a long transmembrane helix protruding from the membrane (TM5). The enzyme was co-crystallized with S-adenosyl-L-homocysteine (SAH), the reaction product of SAM. This elucidated the SAM binding site and a putative catalytic site for the enzyme. The SAM binding site is enclosed on the cytoplasmic side by the L3 loop, which connects TM 3 and 4. The observation is made that in order for the SAH to exit the structure, there must be some rearrangement of the enzyme, most likely in the L3 loop. The binding site for SAM leads into a hydrophobic tunnel connected to the membrane environment, which is thus considered the putative lipid access point. Furthermore, in the structure, this hydrophobic tunnel is occupied by a long, snakelike density, which was interpreted as a prenyl chain. Though the identified sites for substrate binding differ greatly with those of other methyltransferases, the catalytic mechanism seems to remain the same. A highly conserved arginine residue (R163) sits between the SAM binding site and the hydrophobic tunnel, thought to coordinate the carboxyl group of the prenylated protein. The neutralization of the carboxyl negative charge is thought to be enough to coordinate a direct nucleophilic attack on the highly reactive SAM.
MaICMT presents an interesting example of catalysis at the interface of different substrate environments. MaICMT carries out catalysis at the point of protein prenylation, where the cytosolic protein and lipid meet. The enzyme must accommodate a lipid chain entering from the membrane, whilst restrained by a bulky protein partner. As a result, in order for all components to be favorably received, the prenylation site between protein and lipid has to dip into MaICMT laterally, in a unique way.
2.3 Cytidinediphosphate Diacylglycerol synthase (CdsA)
Glycerophospholipids are the main structural components of the lipid bilayer and their populations determine important characteristics such as membrane permeability and electrostatics [2]. They can also mediate cellular signaling, i.e. role of phosphoinositides (phosphatidylinositol derivatives) in membrane trafficking, and phosphatidylserine in apoptosis [16, 17]. As such central components of the cell, it is clear that the maintenance of glycerophospholipid homeostasis is vital in healthy cell function. Cytidinediphosphate diacylglycerol (CDP-DAG) is the precursor molecule for the biosynthesis of many glycerophospholipid species with variable polar head groups, such as phosphatidylinositol, phosphatidylserine, and phosphatidyl glycerol. CDP-DAG synthase (CdsA) is the integral membrane enzyme responsible for the production of CDP-DAG from nucleotide cytidine triphosphate (CTP) and phosphatidic acid (PA) [18].
The structure of CdsA from Thermotoga maritima (TmCdsA) was solved to 3.4 Å resolution by x-ray crystallography (Fig. 2C) [19]. The structure presented was a double cysteine mutant engineered for stability and crystallizability. The structure reveals a homo-dimer–corroborated with cross-linking and light scattering experiments–with each monomer containing nine transmembrane helices, organized into three distinct domains. TM 1 makes up the N-terminal domain (NTD), TM 2–5 make up the middle domain (MD), and TM 6–9 make up the C-terminal domain (CTD). Each domain forms a discrete entity, coming together to form a cone-shape cavity, extending into the transmembrane assembly. This cavity has two plausible substrate entry points, one directly from the cytosol and the other laterally from the membrane environment. Though no substrates were co-crystallized, based on metal coordination sites, the substrate-binding and catalytic sites are inferred. In particular, conserved motifs on two loops (‘SPXKXXEG’ on Loop A, between TM 6 and 7; ‘HGGXXDRXD’ on Loop B, between TM 8 and 9) line the cone cavity and are proposed to be important for substrate binding. TM 6 and 7 create a hydrophobic groove, which is implicated in the binding of PA.
Conserved residues (Asp219, Asp249, and Glu222 as numbered in TmCdsA) within the cone shaped cavity coordinate a Mg2+ and a K+ ion in what the authors call a “hetero-di-metal center.” Both metals have been shown to be important for catalysis and based on functional data the authors propose a two metal ion catalytic mechanism, in which Mg2+ activates the phosphate group of PA and promotes nucleophilic attack on CTP, while K+ coordinates the β- and γ-phosphates of CTP to aid in dissociation of the pyrophosphate after catalysis.
CdsA repeats a scheme for reactions between hydrophobic and hydrophilic substrates that is seen often in the structures discussed in this Review. An internal cone-shaped pocket extending into the membrane from the cytosol or periplasm bearing a lateral entrance site for a lipidic substrate is seen here as well as in PIP synthase and MraY, which we discuss next.
2.4 Diacylglycerol kinase (DgkA)
Phosphatidic acid is an essential precursor for phospholipid biosynthesis, a structural component of the membrane influencing membrane curvature, and a vital molecule for cellular signaling. Phosphatidic acid is produced, in part, when diacylglycerol is phosphorylated by diacylglycerol kinase (DgkA), using adenosine triphosphate (ATP) as a phosphate donor [20].
The structure of DgkA from Escherichia coli (EcDgkA), was solved without and with bound adenylylmethylenediphosphonate (ACP; non-hydrolyzable ATP analogue) to 2.05 Å and 2.7 Å resolution respectively by x-ray crystallography from crystals grown in LCP [21, 22]. Both structures contained bound monoacylglycerol (MAG) lipids of different lengths from the LCP formulation (7.8 MAG in the apo structure; 9.9 MAG in the ACP bound structure). A preceding structure, solved by NMR [23], was found to be distorted, hence is less informative than the more recent crystal structures [24]. EcDgkA exists in the membrane as a homo-trimer with each monomer having 3 transmembrane helices of varying lengths and one amphipathic surface helix lying parallel to and interacting with the membrane on the cytosolic side (Fig. 2D). TM 2 and 3 extend out of the membrane and are connected by a cytosolic loop (CL). In one of the three protomers, ACP appears bound to the CL. Each of the three putative active sites is formed by the three transmembrane helices of one protomer, and the amphipathic surface helix of an adjacent one. Two zinc ions were identified in each protomer, which are coordinated by conserved residues E28 and E76 and mediate the binding of ACP phosphates, when present. ACP is bound on the cytosolic side between TM 2 and 3, with the cytosolic loop between the two transmembrane helices interacting with the adenosine of ACP. The phosphates of ACP are pointed toward the membrane and the head of the lipid substrate. The head of a MAG molecule in the structure meets the phosphate tail of ACP in proximity to the putative catalytic residue E69. The acyl chain of the lipid substrate binds in a hydrophobic groove made up of the three TMs of one subunit and the surface helix hugs around the active site, enclosing the lipid substrate. According to the proposed mechanism, E69 strips a proton from the lipid hydroxyl, facilitating an attack on the γ-phosphate of the zinc-stabilized ATP.
The structure shows how DgkA brings together the polar head of a lipidic substrate, the acyl tail of which is bound to the transmembrane stalk of the protein, and a polar substrate (ATP), which is bound to the cytosolic protrusion of the protein. The active site, indicated by coordinated zinc ions, lies at the interface of the cytosol and membrane environment, where the soluble ATP meets the polar head group of the lipid to be phosphorylated.
2.5 Vitamin K epoxide reductase (VKOR)
VKOR catalyzes a step in the vitamin K cycle that is fundamental for the production of functional coagulation factors. Specifically, it is responsible for the regeneration of vitamin K hydroquinone from vitamin K epoxide [25]. Vitamin K hydroquinone is essential for the function of γ-carboxylase, which makes crucial modifications to glutamate residues of coagulation factors. Without these modifications, coagulation factors are not successfully activated by calcium at sites of injury [25]. In the vitamin K cycle, VKOR reduces vitamin K epoxide in two steps, first to vitamin K quinone and then to vitamin K hydroquinone [26, 27]. Mutations in human VKOR can cause resistance to warfarin, an anticoagulant commonly prescribed to prevent the formation of blood clots in patients [28].
VKOR has also been identified as a contributing enzyme to the formation of disulfide bonds in proteins within the endoplasmic reticulum (ER). VKOR homologs in bacteria have been shown to catalyze the formation of disulfide bonds, important for the stability of secreted proteins [29]. Moreover, a VKOR homolog from Mycobacterium tuberculosis is inhibited by warfarin, and the warfarin-resistant strains show mutations proximal to those in the human VKOR in patients resistant to the anticoagulant [30].
The structure of a homolog of VKOR from Synechococcus sp (ssVKOR) was solved, co-crystallized with endogenous redox partner ubiquinone (UQ; identified as mostly ubiquinone by mass spectrometry and the density was modeled as ubiquinone in the structure) to 3.6 Å resolution [31]. The enzyme is made up of five transmembrane helices (TM1-5), of which the first 4 are homologous to the mammalian VKOR, and a periplasmic thioredoxin (Trx)-like domain (Fig. 2E). In many other species, this Trx-like domain is a separate protein, but still participates in catalysis. This is exemplified in the bacterial homolog complex DsbA/DsbB, where DsbB is a transmembrane protein and DsbA is soluble, acting as a redox partner [32, 33].
There is a conserved active site CXXC motif, located in the structure near the periplasmic-membrane interface on TM4. The Trx-like domain also contains a CXXC domain. Both of these domains are thought to cycle between disulfide bonds and free-sulfides depending on the redox state. The five TM bundle is capped by a “half helix” or “horizontal helix” domain, existing on a loop between TM1 and TM2, which also contains a disulfide bond. The ubiquinone species is cradled in a space between the first four transmembrane helices, coordinated by hydrophobic residues within this cavity. The UQ is closer to the periplasmic side of the membrane and is proximal to the active site motif. It is thought that the disulfide bond-forming function of this protein is carried out in an electron transfer pathway. Three different cysteine mutants gave snapshots of redox states undergone by the enzyme [34]. These structures support the proposed progression of electron transfer in the formation of disulfide bonds. The proposed mechanism starts with the reduction of the CXXC motif of the Trx-like domain by a newly synthesized protein, causing reduction of the disulfide bond on the horizontal helix domain, and in turn reducing the CXXC disulfide bond on TM4. The disulfide bond on TM4 is reformed by the reduction of UQ. Dynamics of the horizontal helix are thought to be instrumental in the progression of the electron transfer as well as isolation of the reactive ubiquinone species.
VKOR is an example of a protein that holds the hydrophobic substrate steady within a hydrophobic protein environment. Stimulus from outside of the membrane promotes catalysis of the substrate at the interface through a chain reaction.
2.6 Phosphatidylglycerophosphate phosphatase B (PgpB)
Phosphorylation states of lipids influence their reactivity as carriers for sugars and proteins. The effectiveness of glycerophospholipids in cellular roles is dependent on the machinery that maintains different phosphorylation states, such as phosphatases. For example, phosphatidylglycerophosphate phosphatase B (PgpB) is responsible for the dephosphorylation of diacylglycerol pyrophosphate (DGPP) to phosphatidic acid (PA), PA to diacylglycerol (DAG), and undecaprenyl-pyrophosphate (UndPP) to undecaprenyl-phosphate (UndP) [35, 36].
Phosphatidylglycerophosphate phosphatase B (PgpB) is a member of the PAP2 (type II phosphatidic acid phosphatase) family. Type II enzymes differ from Type I in that they function in the absence of Mg2+, and they do not require any other cofactors. PAP2 enzymes process different substrates, but share a signature sequence (KX6RPX12-54PSGHX31-54SRX5HX3D), which is thought to be important for catalysis. A previously established two-step mechanism suggests that the second histidine and the aspartate residue of the signature sequence (KX6RPX12-54PSGHX31-54SRX5HX3D) are responsible for the initial nucleophilic attack and the formation of an enzyme-phosphate transition state. Then, the first histidine of the signature sequence (KX6RPX12-54PSGHX31-54 SRX5HX3D) steadies the intermediate and catalyzes the final cleavage of the phosphate from the substrate [37].
The structure of PgpB from E. coli (EcPgpB) was solved to 3.2 Å resolution [37]. The structure reveals six transmembrane helices, a soluble perisplasmic domain consisting of four helical domains, and a C-terminal helix on the cytosolic side of the membrane (Fig. 2F). TM 2 and 3 form a v-shaped cleft opening to the periplamic side of the membrane, which contains the putative active site. This active site is made up of the signature sequence, which localizes on the periplasmic side of the TM 3 to 6. In EcPgpB, a molybdate group is within the putative active site, plausibly mimicking a bound phosphate. The relative orientation of the active site and molybdate suggested that the UndP lipid substrate could bind within the groove created between TM 2 and 3, with the phosphate group oriented toward the periplasmic side of the membrane in the catalytic site of the enzyme. Mutagenesis designed to occlude the lipid entrance to the active site showed decreased enzyme activity, consistent with the proposed lipid-binding scheme. However, a recently published structure, co-crystallized with phosphatidylethanolamine (PE), shows PE binding in a hydrophobic tunnel formed by TMs 3, 4, and 6, on the other side of the enzyme [38]. PE is not a catalytic substrate of PgpB, but it does seem to act as a competitive inhibitor, suggesting that this binding site may also be utilized by physiological substrates such as UndPP and PA. Though it shares structural similarity to previously solved soluble PAP2 enzymes within a catalytic core (made up by TM 4 to 6 in EcPgpB) [39, 40], the substrate-binding site is, as could be expected, different in PgpB, given the presence of transmembrane segments, required to stabilize the enzyme in the membrane environment as well as to accommodate its lipidic substrate.
PgpB is a unique example of an enzyme coordinating a lipidic substrate, functioning without metal cofactors or other soluble substrates. It coordinates a lipidic substrate adjacent to a positively charged pocket, to catalyze a dephosphorylation reaction. The coordination of the acyl chain of the lipid is similar to other examples in this category, such as PIPS and CdsA.
2.7 Phospho-MurNAc-pentapeptide translocase (MraY)
The cell wall of almost all bacteria (gram-negative and gram-positive) is composed of peptidoglycan, a mesh of carbohydrates cross-linked by small peptides, which is essential for growth, division and viability of the organism. Any disruption of the biosynthesis of peptidoglycan, whether by genetic mutation, inhibition with antibiotics or degradation by lysozyme, results in bacterial cell lysis. Peptidoglycan helps maintain cell shape and serves as an anchor for accessory proteins and other cell wall components. As essential components of the cell wall, enzymes contributing to the peptidoglycan biosynthetic pathway can be exploited as antibiotic targets [41, 42].
After a hydrophilic peptidoglycan precursor (UDP-MurNAc-pentapeptide) is synthesized in the cytosol, it is attached to the lipid carrier undecaprenyl phosphate (UndP). The lipid-linked precursor (undecaprenyl-pyrophosphoryl-MurNAc-pentapeptide or Lipid I) is modified further to undecaprenyl-pyrophosphoryl-MurNAc-(pentapeptide)-GlcNAc (Lipid II) by addition of a GlcNAc moiety. Lipid II is then flipped across the membrane to the periplasm where its sugars are polymerized to form the glycan strands of the peptidoglycan mesh. MraY is the enzyme catalyzing the Lipid I synthesis [42]. MraY belongs to the polyprenylphosphate N-acetyl hexosamine 1-phosphate transferase (PNPT) superfamily of enzymes, which includes other potential antibiotic targets, WecA and TarO, enzymes responsible for bacterial cell wall synthesis. The PNPT superfamily also includes eukaryotic enzymes involved in protein N-linked glycosylation [43]. A high-resolution structure of MraY has provided a context for the accommodation of both hydrophobic and hydrophilic substrates, contributing to the understanding of the PNPT superfamily.
MraY from Aquifex aeolicus (AaMraY) was solved to 3.3 Å resolution by x-ray crystallography [44]. MraY is a dimer with each monomer containing ten transmembrane helices. The structure also reveals an interfacial helix at the N-terminus, a periplasmic β-hairpin between TM 4 and 5, a periplasmic helix between TM 6 and 7, and five cytoplasmic loops (Fig. 3A). TM 9 is split into two helical segments (TM 9a and 9b) with TM 9b bent away from the main helical bundle structure of the monomer, jutting into the membrane environment. This protruding helix helps to form the putative active site on the cytoplasmic side of the membrane. The active site coordinates a metal ion and connects to a hydrophobic groove leading to the membrane environment.
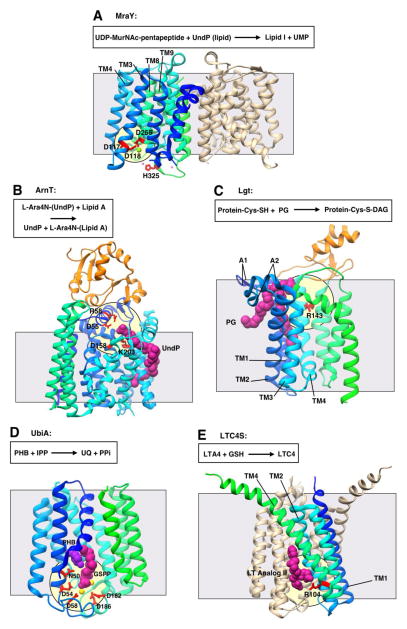
Figure 3. Representative structures showing Interfacial catalysis (II).
All structures are shown in ribbon representation with bound ligands shown in CPK representation. Transmembrane domains are shown in gradient coloring from the N-terminus to the C-terminus (blue to cyan to green), while extramembrane soluble domains are shown in orange. The grey square shows the approximate location of the membrane border. The pale yellow circle indicates the location of the putative active site. All structures oriented with the cytosolic face of the membrane on the bottom. A. Phospho-MurNAc-pentapeptide translocase (MraY; PDB 4J72) with putative catalytic residues in red. Nickel ion in pink, magnesium ion in light green. UDP, uridine diphosphate; UMP uridine monophosphate; UndP, undecaprenyl phosphate. B. A4-amino-4-deoxy-L-arabinose transferase (ArnT; PDB 5F15) with putative catalytic residues in red. UndP in magenta. C. Lipoprotein diacylglycerol transferase (Lgt; PDB 5A2C) with putative catalytic residues in red. Phosphatidylglycerol (PG) in magneta. D. Prenyltransferase UbiA (PDB 4OD5) with putative catalytic residues in red. Geranyl thiolopyrophosphate (GSPP) in magenta, p-hydroxybenzoate (PHB) in purple, magnesium ions in yellow. A1 and A2, arm 1 and arm 2; IPP, isoprenylpyrophosphate; UQ, ubiquinone. E. Leukotriene C4 Synthase (LTC4S; PDB 4J7T) with putative catalytic residue in red. Leukotriene (LT) analog in magenta. LTA4, (5S)-trans-5,6-oxido-7,9-trans-11,14-cis-eicosatetraenoic acid; LTC4, (6R)-S-glutathionyl-7,9-trans-11,14-cis-eicosatetraenoic acid ; GSH, glutathione.
Based on sequence homology analysis and mutagenesis coupled to a functional assay, the authors identified a substrate binding and catalytic cleft formed on the cytosolic side of the membrane flanked by TM3, 4, 8 and 9b. Three conserved aspartate residues (Asp117, Asp118, and Asp265) and one (of two) conserved histidine residues (His325) are implicated as essential for catalysis based on mutagenesis experiments. Asp117 and Asp118 were thought to coordinate a magnesium ion because it is similar to the magnesium coordinating sequence from prenyl transferases (DDXXD/N). However, manganese-soaking experiments revealed that Mn2+ only interacts with Asp265 directly. So, it was hypothesized that Asp117 may serve as the binding site for the phosphate group of UndP. An inverted U-shaped groove near TM9b is thought to accommodate the polyisoprenyl tail of UndP. Loop E, connecting TM 9b and 10, contains the conserved sequence (PXHHHXEXXG; also termed the ‘HHH motif’) made up of a short helix coordinating two Ni2+ ions. This conserved region is thought to contribute to the recognition of sugar groups [45, 46]. In MraY, this loop region and TM 9b help form the active site cleft and the HHH motif lies close to the coordinated Mg2+, forming the putative UDP-MurNAc-pentapeptide binding site.
A recent MraY structure in complex with a natural soluble inhibitor, muramycin D2 (MD2), was solved to 2.95 Å resolution [47]. This structure revealed that MD2 binding creates a large conformational change in the MraY and, though elements of MD2 mimic the natural soluble substrate, UDP-MurNAc-pentapeptide, it does not interact directly with any of the key acidic residues previously identified. The structure reveals the structural plasticity of MraY, leaving the potential for a wide range of inhibiting molecules. This structure has provided insight into the targeted inhibition of MraY and other members of the PNPT enzyme family.
MraY is another example of an enzyme, which seems to accommodate lipidic and soluble substrates in an intracellular pocket with cytosolic and membrane-embedded regions.
2.8 Lipid-to-lipid glycosyltransferase ArnT
The cell wall is an essential component of bacteria, providing structural integrity and defense against threats. In Gram-negative bacteria, the cell wall has an additional outer membrane lipopolysaccharide (LPS) component that is necessary for virulence and confers bacterial serotype. LPS is attached to the outer membrane through a complex lipid, called lipid A. Lipid A is modified by covalent addition of glycosides by different enzymes. ArnT catalyzes the attachment of an aminoarabinose moiety (L-Ara4N) from 4-Amino-4-deoxy-α-L-undecaprenyl-phosphate (UndP-α-L-Ara4N) to phosphates on lipid A [48]. This charge neutralization reaction leads to bacterial resistance to polymyxin class antibiotics by preventing binding of cationic antimicrobial peptides to their outer membrane [49]. ArnT is located in the inner membrane of Gram-negative bacteria [48]. Both its substrates are lipids and the reaction takes place on the outer leaflet of the inner membrane, raising the question of how the structure of this enzyme has been adapted to reconcile the hydrophobicity of two substrates with a chemical reaction requiring an aqueous environment.
The structure of ArnT from Cupriavidus metallidurans (CmArnT) was solved by x-ray crystallography without and with bound partial substrate undecaprenyl phosphate (UndP) to 2.8 Å and 3.2 Å resolution respectively [50]. The CmArnT structure shows a monomer consisting of two domains, a TM domain with 13 TM helices and 3 juxtamembrane (JM) helices, and a periplasmic domain with 7 helices and 5 beta sheets (Fig. 3B). The overall fold is reminiscent of protein oligosaccharyl-transferases PglB [51] and AglB [52].
The CmArnT structures revealed three cavities on the protein that are likely used to accommodate the lipidic substrates of the catalyzed reaction. The largest cavity was hypothesized to accommodate lipid A, based in most part on the need to receive the very bulky lipid A molecule. The UndP-bound structure of CmArnT provided further details on the utilization of the other two cavities. The one closer to the membrane interface is used to coordinate the headgroup of UndP (and thus the transferred sugar as well, which is not present in the structure), whereas the third cavity, the smallest of the three, receives the tail (last three prenyl groups) of UndP. The functional significance of the latter is uncertain, as mutagenesis of residues that may participate in hydrophobic interactions with the lipid tail was not successful in disrupting the function of ArnT. Contrary to that, residues that seem to coordinate the phosphate group of UndP (based on the bound-structure) and the aminoarabinose sugar (based on molecular docking of L-Ara4N) were necessary for function based on the results of structure-based mutagenesis experiments on CmArnT, and on ArnT from Salmonella typhimurium, coupled to functional assays. Interestingly, the two cavities that reside close to the membrane-water interface converge, showing a likely location for the active site of the enzyme.
Binding of UndP to CmArnT triggers a conformational change, resulting in the release of a bound Zn2+, extension of one of the JM helices and formation of the active site at the membrane-periplasm interface. This observation led to the hypothesis that the catalytic reaction by ArnT may be taking place in two steps: first the donor substrate carrying the L-Ara4N sugar enter the active site and triggers a conformational transition that locks it into the active site, the enzyme is then primed for catalysis and can bind the lipid A acceptor.
The likely catalytic residues have been identified through sequence analysis, location on the structure and mutagenesis experiments. D55 and D158 are located at the converging point of the two interfacial cavities (Fig. 3B). They are coordinated by R58 and K203 respectively. Mutations of this proposed catalytic quad leads to loss of function of ArnT. It was proposed that the two aspartates coordinate the acceptor phosphate of lipid A and enable it to perform a direct nucleophilic attack on C1 carbon of the arabinose ring, thus enabling direct transfer of sugar from the donor to the acceptor.
The structure of CmArnT shows how two amphipathic substrates can be coordinated within different parts of the same enzyme. Matching the local environment encountered by different parts of the amphipathic substrates allows the polar moieties to react within the aqueous environment of an active site, while the hydrophobic parts are efficiently shielded from this environment by hydrophobic interactions towards other parts of the enzyme.
2.9 Lipoprotein Diacylglycerol transferase (Lgt)
Proteins tethered to the membrane by covalent linkage to lipid have diverse roles associated with the bacterial membrane, including but not limited to facilitating uptake of nutrients and mediating host immune response to infection [53, 54]. Phosphatidylglycerol:prolipoprotein Diacylglyceryl transferase (Lgt) is an enzyme involved in the transfer of a protein containing a lipobox sequence to a lipid anchor. More specifically, Lgt catalyzes the transfer of diacylglycerol (from phosphatidylglycerol) to the cysteine thiol group within the lipobox consensus sequence recognized by the enzyme [55, 56]. After this, the membrane-anchoring signal peptide will be cleaved off the lipoprotein by LspA and then Lnt will complete the lipid anchoring reaction (only in Gram-negative bacteria). The lipoprotein synthesis pathway is present in both Gram-negative and Gram-positive bacteria, however an Lgt knockout is lethal in Gram-negative bacteria [53].
Lgt from E. coli (EcLgt) was solved co-crystallized with substrate phosphatidylglycerol (PG) and inhibitor palmitic acid to 1.9 and 1.6 Å resolution, respectively [57]. The enzyme contains 7 transmembrane (TM) helices divided into major and minor transmembrane domains. There is also a periplasmic domain containing a “head” and two amphipathic “arm” domains (arm1 and arm2). The head domain is made up of a 4-strand β-sheet and two α-helices. Arm1 is an amphipathic β-hairpin located N-terminally to TM1, and arm2, located between TM 2 and 3, is made up of two short α-helices (Fig. 3C). The central cavity surrounded by the 7 TM helices is the location of the active site and essential residues for catalysis were mapped to the periplasmic end of the central cavity. The putative critical residue (R143) of the catalytic center is located on TM 4 within the major TM domain near the periplasmic side of the central cavity. There are two clefts at the interface between the major and minor TM domains, which are thought to be lipid/prolipoprotein access points to the active site.
The structure of EcLgt is unique, revealing two separate binding sites for the lipid substrate. Two mechanisms were proposed. In the first hypothesis, one lipid may undergo catalysis in the second site, then upon release of the products, a lipid occupying the first site shuttles into the second one, which will then be poised for another lipoprotein-linkage reaction. In the second hypothesis, a lipid waits in the first binding site. When the lipobox protein binds, it causes a conformational change in a loop (between TM 6 and 7), which acts as a gate. This conformational change allows the lipid to move into the second binding site, for catalysis to occur with the lipobox protein.
2.10 UbiA family of enzymes, a model for ubiquinone biosynthesis
The UbiA family of enzymes consists of intramembrane prenyltransferases responsible for the biosynthesis of such molecules as ubiquinones, menaquinones, phylloquinones, chlorophyll, heme, Vitamin E and Vitamin K [58–60]. These molecules play key roles in electron and proton transport and respiration, photosynthesis, lipid metabolism, cell signaling, and cell wall formation [59]. They can also act as antioxidants and cofactors [61]. Mutations in human members of the UbiA family of enzymes have been shown to contribute to diseases such as Schnyder crystalline corneal dystrophy, which causes blindness, cardiovascular degeneration, Parkinson’s disease, mitochondrial diseases and urologic cancers [62].
Reactions catalyzed by this enzyme family are diverse as are the substrates utilized, however all UbiAs catalyze the transfer of a prenyl group from a donor to acceptor. In most cases, this acceptor is an aromatic molecule. Members of the UbiA family are known for their substrate promiscuity. For example, UbiA recognizes isoprenyldiphosphates of various lengths with similar specificity yielding ubiquinones of different lengths and function. In addition, the eukaryotic enzyme UBIAD1 is expressed in different subcellular locations determining the predominant ubiquinone species synthesized by the enzyme. For example, UBIAD1 transfers an isoprenyl chain from geranylgeranyl pyrophosphate to menadione phylloquinone yielding menaquinone-4 (the major storage form of vitamin K) important in mitochondria, while in the Golgi, it participates in the synthesis of ubiquinone UQ10 by a similar mechanism. Exceptions to this observed substrate promiscuity exist within the family, such as PHB geranyltransferase from Lithospermum erythrorhizon, which can only utilize geranylpyrophosphate (GPP) as a prenyl donor [59]. The promiscuity or fidelity of enzymes of the same family for lipidic substrates presents an intriguing problem from a structural perspective.
The structures of two members of the UbiA family – a UbiA homolog from Aeropyrum pernix (ApUbiA; Fig. 3D) and a UBIAD1 homolog from Archeoglobus fulgidus (AfUbiA) – have been solved by X-ray crystallography to 3.3 Å and 3.2 Å resolution respectively [58, 63]. Both structures reveal a 9 transmembrane topology with an extramembrane cap domain made of three short helical segments. This cap domain contains a majority of the conserved residues, including the characteristic aspartate-rich motifs (NDXXDXXXD and DXXXD), responsible for divalent-cation binding essential for activity.
ApUbiA presents a U-shaped architecture of the transmembrane helices containing a large central cavity (Fig. 3D). This cavity contains a polar pocket,capped by the aspartate-rich motifs, which in this structure coordinate the pyrophosphate of the non-cleavable analog geranyl thiolodiphosphate (GSPP, the prenyl donor) through magnesium intermediates. The cavity also shows a basic pocket binding PHB, and a hydrophobic wall to coordinate the isoprenyl chains. This cavity opens laterally to the membrane, providing a passage for prenyl donors of variable length to reach the active site.
AfUbiA differs in its binding pocket domains. One of the cap helices (L2-3) is disordered in the structure solved without ligand (GPP, DMAPP). The authors hypothesize that this could allow for access of soluble prenyl acceptors, including short chain polyprenyl diphosphates. The authors also suggest a conformational change for the protein to accept long-chain prenyl donors from the membrane. If TM9 were removed, there would be clear access of the substrate tunnel to the membrane environment. Though the exact substrates of AfUbiA are unknown, it may accept two prenylated substrates (short chain and long chain) as does DGGGPS, another member of the UbiA family.
Two structures from the UbiA family illustrate how similar enzymes can facilitate prenylation of different molecular species. The UbiA family of enzymes also presents a fascinating example of lipidic substrate promiscuity, as one enzyme is known to utilize several different prenyl donors. Substrate promiscuity highlights the complexities of enzyme specificity and is an important biological phenomenon when considering regulatory mechanisms.
2.10 Prostaglandin E synthase (PGES1) and LTC4 synthase (LTC4S)
Prostaglandins and leukotrienes are lipid signaling molecules acting to mediate the inflammatory response [64]. Prostaglandin E synthase (PGES1) and Leukotriene C4 synthase (LTC4S) are both members of the MAPEG (Membrane-Associated Proteins Involved in Eicosanoid and Glutathione metabolism) superfamily of enzymes. Structures of these two enzymes present similar models for lipid substrate binding, even though they share a low percentage identity (around 15%) and exhibit different catalytic mechanisms [65].
Prostaglandin E2 synthase 1 (PGES1) catalyzes the isomerization of prostaglandin H2 (PGH2) to prostaglandin E2 (PGE2) in the presence of soluble cofactor glutathione (GSH). The structure of human PGES1 (hPGES1) with bound GSH and a GSH analog inhibitor (bis-phenyl-GSH) has been solved to 1.2 and 1.95 Å resolution, respectively [65]. Another structure had been previously determined to 3.5 Å resolution [66]. Leukotriene C4 synthase (LTC4S) catalyzes the reaction between fatty acid LTA4 ((5S)-tans-5,6-oxido-7,9-trans-11,14-cis-eicosatetraenoic acid) and glutathione (GSH) to produce LTC4 ((5S)-hydroxy-(6R)-S-glutathionyl-7,9-trans-11,14-cis-eicosatetraenoic acid). The structure of LTC4S was first solved with and without bound GSH to 2.15 and 2 Å resolution, respectively [67]. It was also solved co-crystallized with three different leukotriene analogs (I, II, and III) to 2.75, 3.2, and 2.9 Å, respectively (Fig. 3E) [68].
PGES1 and LTC4S structures show a trimeric assembly, with each monomer consisting of four transmembrane helices. The interface between two monomers creates a hydrophobic groove acting as a putative binding site for lipidic substrate. This groove is made up of TM 1 from one monomer and TM 2 and 4 from another monomer. The GSH binding sites of PGES1 and LTC4S are highly conserved and both structures show GSH binding in a U-shaped conformation. The structure of PGES1 also reveals a cone-shaped entry point from the cytosolic side acting as the access point for soluble GSH. The structures of PGES1 with GSH and bis-prenyl-GSH clearly identify the residues involved in GSH binding. Based on this, the authors propose that the catalytic site is within the cavity of GSH binding, and hypothesize that the aspartate residue, Asp49, acts as a base in the proton transfer of the isomerization reaction. The structure of LTC4S reveals a tryptophan residue (W116) in the lipid-binding site. Though not important for catalysis, W116 seems to play a key role in lipid binding to the hydrophobic groove and product release after catalysis. Its role was examined in the context of binding of different product analogs in the lipid binding site of LTC4S. The catalytic arginine (R104) was also identified in the structure in proximity to the binding sites for the two substrates, thus defining the active site.
The structure of PGES1 shows a much shallower hydrophobic groove for lipid binding than that observed for LTC4S. This is likely due to the different function of the two enzymes. PGES1 acts as an isomerase and uses GSH as a cofactor. LTC4S on the other hand catalyzes the covalent joining of two substrates, one being GSH, which requires a deeper hydrophobic groove in order to bring them into apposition for the reaction to occur.
3. Structural basis of extramembrane lipid modification
Here we present examples of enzymes whose active site lies outside of the lipid environment, in a soluble protein domain. There are many examples and structures of such enzymes, but we have decided here to focus only on those for which the transmembrane domains were included in the structural analysis, which are rather few. The examples presented here illustrate distinctive solutions to the problem of bringing polar and hydrophobic substrates together for catalysis.
3.1 GtrB, a model for polyisoprenyl-glycosyltransferases
For a protein to be glycosylated or attached to the membrane via a glycosylated lipid, or for cell wall lipids to be modified, saccharides need to be exported from the cytoplasm to outer membrane compartments [69]. This is achieved across all kingdoms of life by enzymatic attachment of the sugar to specific polyisoprenyl-phosphates (PI-Ps), acting as lipid carrier molecules [70]. The most commonly used PI-Ps are undecaprenyl-phosphate (UndP) in bacteria and dolichol-phosphate (DolP) in eukaryotes [70]. The enzymes responsible for the attachment are 35–40kDa membrane-bound GTs called polyisoprenyl-glycosyltransferases (PI-GTs) [70]. From a structural biologist’s perspective, the most intriguing feature of PI-GTs is the fact that the catalytic domain is hydrophilic and separate from the TM domain, posing the immediate question of how attachment of a sugar to an extremely hydrophobic substrate may occur.
The PI-GT, GtrB, catalyzes the conversion of UndP to UndP-glucose using an activated sugar in the form of UDP-glucose, a reaction required to modify bacterial O-antigen (for a review see [71]). The crystal structure of GtrB from Synechocystis sp. at 3.0 Å resolution [72] shows a tetramer, with each subunit consisting of an N-terminal cytosolic glycosyltransferase domain (GT) of the GT-A fold (Fig. 4A) [73], two TM helices (TM1 & TM2) which form a compact stalk anchoring the intracellular domains to the membrane and two amphipathic JM helices (JM1 & JM2) that lie at the surface of the membrane, and connect the GT and TM domains.
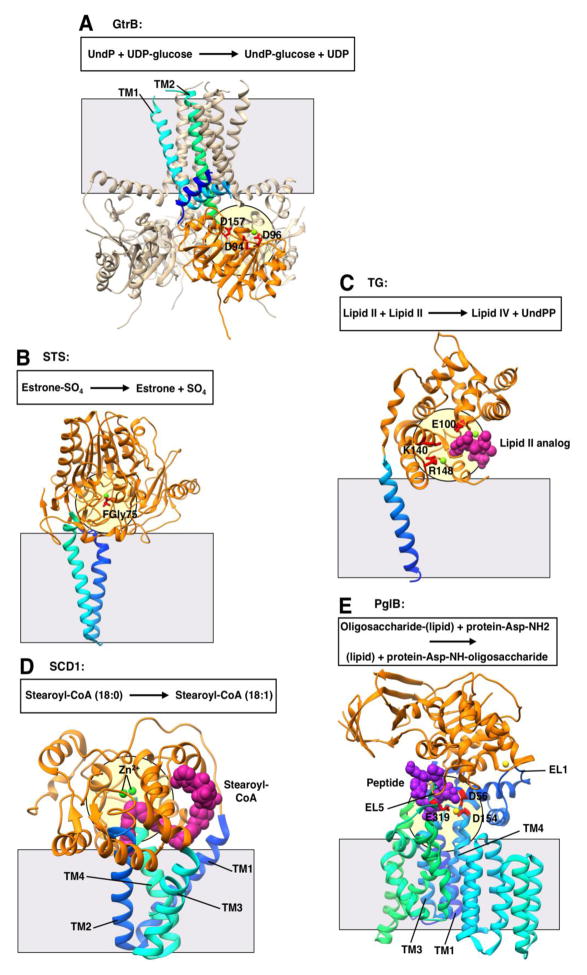
Figure 4. Representative structures showing extramembrane catalysis.
All structures are shown in ribbon representation with bound ligands shown in CPK representation. Transmembrane domains are shown in gradient coloring from the N-terminus to the C-terminus (blue to cyan to green), while extramembrane soluble domains are shown in orange. The grey square shows the approximate location of the membrane border. The pale yellow circle indicates the location of the putative active site. All structures oriented with the cytosolic face of the membrane on the bottom. A. Polyisoprenyl glycosyltransferase GtrB (PDB 5EKP) with putative metal biding and catalytic residues in red. Magnesium ion in light green. UndP, undecaprenyl phosphate; UDP, uridine diphosphate. B. Steroid sulfatase (STS; PDB 1P49) with formylglycine-sulfate (FGly; modified cysteine residue) in red. Calcium ion in light green. C. Transglycosylase (TG; PDB 3VMT) with putative catalytic and metal binding residues in red. Lipid II analog in magenta and magnesium ions in light green. UndPP, undecaprenyl pyrophosphate. D. Stearoyl-CoA desaturase (SCD1; PDB 4YMK) with zinc ions (indicating the di-iron center) in green. Stearoyl-CoA in magenta. E. Oligosaccharyltransferase PglB (PDB 3RCE) putative metal binding and catalytic residues in red. Oligosaccharide acceptor peptide in purple, magnesium ions in yellow. EL, external loop.
The active site of GtrB, which was identified by a combination of structure-based sequence alignments, co-crystallization experiments with substrate analogues and mutagenesis coupled to functional assays, is located in the cytosol approximately 15 Å from the membrane. Therefore, for catalysis to occur, either the GT domain must move to the membrane, or the lipid-phosphate acceptor must travel to the active site, shielded from the solvent. Analysis of the structure, coupled to functional experiments shifts favor to the latter hypothesis. First, ~900 Å2 of predominantly hydrophobic surface is buried between each pair of GT domains in the cytosol, with ten inter-subunit hydrogen bonds formed at each interface, suggesting a stable structure that is unlikely to disassemble for the GT domain to move to the membrane. Second, the enzyme is highly specific for acceptors built from isoprenyl units, but the lack of conservation in the TM region suggests that specificity for the lipid resides elsewhere. Third, several absolutely conserved hydrophobic and basic residues were identified in the JM region that could form a translocation pathway for the substrate from the membrane to the active site while shielded from the solvent. Thus, the structure of GtrB has provided a testable model for enzyme function. Given the high degree of sequence conservation within the family, these results from this system are expected to be applicable to all PI-GTs.
GtrB solves the problem of hydrophobic/hydrophilic substrate mismatch by translocating the hydrophobic molecule from the membrane to the active site via a hydrophobic path. This mechanism could prove to be generally applicable to numerous membrane protein families in which hydrophobic and hydrophilic ligands react in an active site outside the membrane.
3.2 Steroid Sulfatase (STS)
The cycling between sulfate-conjugated and unconjugated forms is an important regulatory mechanism for steroid hormone function. Steroid Sulfatase (STS) is an enzyme responsible for the conversion of estrone-sulfate (E1S) to estrone (E1), which is then reduced to 17β-estradiol (E2) (the active form of estrogen) by a separate enzyme. STS also hydrolyzes dehydroepiandrosterone-sulfate (DHEAS) to dehydroepiandrosterone (DHEA), cholesterol-sulfate to cholesterol, and pregnenolone-sulfate to pregnenolone [74]. X-linked ichthyosis, a disease characterized by the scaling of the skin, has been associated with STS gene deletion or the presence of non-functional point mutants [75]. Higher STS activity has been observed in gynecological diseases such as endometrial cancer and ovarian cancer [76].
The structure of STS purified from human placenta was determined by x-ray crystallography to 2.6 Å resolution [77]. STS has two transmembrane helices and a catalytic cap domain. The catalytic cap domain is made up of subdomains SD1 and SD2 with a α/β-sandwich fold (Fig. 4B). SD1 contains the active site, which lies on the underside of the cap close to the membrane. SD1 is formed by an 11-stranded mixed β-sheet surrounded by 13 α-helical structures. SD2 is made up of a four-stranded antiparallel β-sheet and one α-helix. SD2 packs against the β-sheet of SD1. The soluble cap domain closely resembles the fold of other soluble sulfatases with known structure. With the same fold and a high level of conservation at the catalytic site, it follows that all these sulfatases may function by the same mechanism. However, the sulfatase cap domain differs at the C-terminal end (SD2), which is thought to mediate interactions with the surface of the membrane [74, 77].
Within SD1, conserved residues coordinate a divalent cation (Ca2+ in the structure). Proximal to the Ca2+ is a conserved cysteine (C75), which is modified posttranslationally to formylglycine (FG75) in order for the enzyme to be functional. This is a key catalytic residue as it covalently accepts the sulfate group that is removed from the steroid substrate. In the structure, FG75 was found as a hydroxyformylglycine covalently linked to a sulfate group, suggesting that this is the resting state of the enzyme. It has been proposed that the catalytic cycle of STS starts with the activation of FG75 by a water molecule, which, facilitated by Ca2+, leads to a nucleophilic attack on the steroid-sulfate substrate. After the steroid product is released, the FG75-sulfate can be returned to FG75 for the next reaction to occur. It has been proposed that a water molecule, carried in by a sulfate-bearing substrate, acts in a nucleophilic attack of FG75-sulfate to clear the covalently bound sulfate group [74].
A “hydrophobic tunnel” leads from the two transmembrane helices to the active site. Assuming that estrone-sulfate exists in the membrane with the sulfate moiety interacting with the polar face of the membrane, the estrone-sulfate could first interact with the transmembrane portion of STS and enter the active site through the described “hydrophobic tunnel.” It was also proposed that loops in the cap domain could play a role in the binding of substrate and/or release of product. However, whether the product is released directly into the membrane to be trafficked by vesicular transport or into the lumen by interacting with binding proteins is unclear.
In summary, STS describes a protein in which a lipid substrate is taken from the membrane and moved into the catalytic domain, similar to what has been proposed for the GtrB enzyme [72], though the movement required for catalysis to occur appears to be less dramatic.
3.3 Transglycosylase (TG)
Transglycosylase (TG) is an essential enzyme in the biosynthesis of the peptidoglycan mesh of the bacterial membrane. Lipid I, synthesized by MraY as previously discussed, is further modified to Lipid II by the addition of a GlcNAc moiety. Lipid II is then flipped across the membrane to the periplasm where an enzyme with transglycosylase activity transfers sugars from donor Lipid II molecules to a growing glycan chain (on a Lipid II starter molecule) and the peptides are linked by an enzyme with transpeptidase activity to form the final peptidoglycan mesh [42]. TG, in the simple case, catalyzes the transfer of the disaccharide peptide unit from a donor Lipid II to an acceptor Lipid II, forming Lipid IV. In subsequent steps, the enzyme continues to transfer a growing glycan chain onto a Lipid II acceptor.
Many structures have elucidated transglycosylase activity and substrate binding. The structure of a truncated PBP2 enzyme from Staphylococcus aureus was determined with and without inhibiting antibiotic moenomycin to 2.8 and 2.9 Å resolution, respectively [78]. The structure of PBP1b from Escherichia coli was also solved co-crystallized with bound moenomycin to 2.16 Å resolution [79]. Both PBP2 and PBP1b are bifunctional enzymes with transglycosylase and transpeptidase activity. The Aquifex aeolicus PBP1a glycosyltransferase domain bound to moenomycin was also solved to 2.3 Å resolution [80]. Furthermore, the structure of the soluble domain from monofunctional glycosyltransferase (MGT) from Staphylococcus aureus (SaMGT) bound to moenomycin was solved to 2.1 Å resolution and the structure of the full-length SaMGT bound to a Lipid II analog to 2.3 Å resolution (Fig. 4C) [81, 82].
Bifunctional and monofunctional transglycosylases contain one anchoring transmembrane helix. This helix is truncated in many of the structures, however it has been shown to be essential for enzyme function. The majority of the enzyme consists of a GT cap domain of unconventional fold with 10 alpha helical segments (H1-H10). The cap domain is made up of a “head” and a “jaw.” The jaw domain interacts with the membrane and helps anchor the enzyme close to the surface. Structures of TGs with inhibitor moenomycin and Lipid II analog revealed putative binding sites for the glycosyl acceptor (occupied by the Lipid II analog) and glycosyl donor (occupied by moenomycin).
Though the active site lies outside of the membrane environment, the acyl chain of lipid II does not seem to be interacting with the protein and is not drawn out of the membrane as has been hypothesized for GtrB [72] or SCD1 [9, 84]. Here, only the sugar-peptide head groups of the lipids seem to be interacting with the active site, presenting yet another distinct mode of interaction between a lipid-modifying enzyme and a membrane embedded substrate.
3.4 Stearoyl-CoA desaturase (SCD1)
Stearoyl-CoA desaturase (SCD1) is responsible for introducing a cis-double bond in the saturated acyl chain of stearoyl- or palmitoyl-CoA, forming precursors for important lipid species such as phospholipids and triglycerides. The reaction is activated by a di-iron center in the presence of oxygen [83]. Disruption of normal SCD1 function has been associated with cancer, diabetes and other metabolic syndromes and is a popular target for inhibitor development in search of novel treatment of such disorders [84].
Two structures of mammalian SCD1 have been determined: one from mouse, solved to 2.6 Å resolution (Fig. 4D) and the other from human solved to 3.25 Å [85, 86]. Both structures contain bound stearoyl-CoA and two zinc ions and are strikingly similar. Although previous in vivo studies concluded that the protein acts as a dimer, it was purified with detergent and crystallized as a monomer. This may be an artifact of the removal of the enzyme from the membrane environment, although it seems unlikely given that both human and mouse structures were crystallized using different detergents and both solved as monomers.
SCD1 consists of four transmembrane helices (TM 1–4). TM2 and 4 protrude from the membrane to meet the cytosolic domain, which consists of ten helical segments interconnected with loop regions. The cytosolic cap domain accommodates the acyl chain of stearoyl-CoA to be desaturated in a distinctive hydrophobic tunnel. The hydrophobic tunnel is characterized by a kink centered on carbons 9 and 10 of the bound substrate tail. There is also a di-metal center proximal to this kink (occupied by zinc in the structure), which is coordinated by three conserved histidine residues. This di-iron center differs from the common coordination in a four-helix bundle by two EXXH motifs, as observed in soluble epoxidase BoxB [87] and Δ9 stearoyl-ACP desaturase [88]. SCD1 is generally permissive to a range of acyl chain lengths, however they always desaturate in the same position – between carbons 9 and 10 – seemingly due to the active site arrangement. It appears unlikely that the fatty acyl-CoA substrate can simply diffuse in and out of this hydrophobic tunnel given the wide network of interactions with the protein. However, if key hydrogen bonds are broken, the fatty acyl-CoA may be able to move back to the membrane environment. This would likely require a conformational change.
SCD1 also interacts with cytochrome b5 (which functions to restore the di-iron center) however a binding site was not definitively identified. The structures were docked together in a likely interaction state close enough for the cytochrome b5 to affect the di-metal center.
This structure provides a unique view of how internalized acyl chains of variable length can be uniformly desaturated based solely on the enzyme environment and how it binds the lipid.
SCD1 was compared to a recently solved structure of a sphingolipid α-hydroxylase from Saccharomyces cerevisiae (Scs7p), which shows a similar overall fold to SCD1 [89]. Scs7p, which was solved to 2.6 Å resolution, also contains four transmembrane helices with a di-iron center (occupied by zinc ions in the structure) localized to a soluble cap domain. However, the enzyme seems to have a tilted conformation in the membrane, moving the catalytic site inferred at the di-iron center much closer to the membrane. This would categorize the mechanism of Scs7p more as an interfacial than extramembrane reaction. Scs7p was not co-crystallized with a lipid substrate, though ceramide B was modeled into the structure based on densities of detergent and unidentified lipid [89].
3.5 Protein oligosaccharyltransferases PglB and AglB
N-linked glycosylation is a common posttranslational modification of proteins seen across all levels of life. It can act, for example, in mediating recognition events such as in pathogen-host interaction, modulating protein function and can even be important for maintaining proper structure of proteins [90]. Oligosaccharyltransferases catalyze the transfer of a sugar group from a lipid carrier to an asparagine residue on a protein. The glycosylation most commonly occurs on the protein at a NXS/T motif, where X is any amino acid except proline [91]. Well-studied examples of integral membrane protein glycosyltransferases include protein glycosylation B (PglB) and archaeal glycosylation B (AglB).
Isolated soluble domains of PglB and AglB were crystallized previously [92–94], but these enzymes were found to be non-functional without the transmembrane region. The structure of full length PglB from Campylobacter lari (ClPglB) bound to an acceptor peptide was solved by X-ray crystallography to 3.4 Å resolution (Fig. 4E) [51]. In addition, the structure of full length AglB from Archaeoglobus fulgidus (AfAglB) was solved also by X-ray crystallography with and without bound sulfate to 2.5 Å and 3.4 Å resolution, respectively [52]. Both PglB and AglB show similar enzyme architecture with thirteen transmembrane helices and a pronounced C-terminal periplasmic cap domain. The largest structural differences between the enzymes seem to be in the periplasmic domain and the organization of the catalytic site.
The catalytic pocket of ClPglB is located above the membrane boundary, indicated by notable metal coordinating residues lying on periplasmic loop regions (D56, D154, E319). The co-crystallized peptide showed coordination of the glycosylation target asparagine in the active site by residues D56 and E319. Protein binding to ClPglB, illustrated by the bound peptide, occurs where TMs 1, 3 and 4 meet external loops (EL) 1 and 5 of the perisplasmic domain. This catalytic pocket also opens to the other side of the ClPglB, where it is thought that the oligosaccharide lipid donor will bind. The proposed mechanism for substrate binding and product release involves a rearrangement of EL5.
The catalytic site of AfAglB was revealed by the coordination of a zinc ion and the binding of a sulfate in the structure, and is very similar to that in ClPglB. The zinc ion identified in the structure is coordinated by residues (D47, D161, H163) on EL1 and 2. The location of the sulfate was identified as a putative binding site for the phosphate group of the dolichol-phoshate, the oligosaccharide carrier for the reaction. The AfAglB structure with sulfate shows a completely disordered EL5, while the sulfate bound structure shows a structured EL5. Taken together with the ClPglB structure, it was proposed that in the apo state of the protein, EL5 is structured and that binding to a peptide causes EL5 to become partially unstructured, allowing glycosylated-lipid binding. After the oligosaccharide transfer reaction, the glycoprotein exits the enzyme, leaving a completely disordered EL5.
Though neither structure was solved with a bound lipid substrate, these oligosaccaryltransferases illustrate a unique mechanism involving soluble protein and lipid substrates. Though similar in structure to the enzyme ArnT, they differ in location of their active sites due to the different character of the substrates they process.
4. Structural basis of intramembrane lipid modification
In addition to the previous examples with active sites at the membrane-water interface or clearly external to the membrane, here we present a unique case where the site of catalysis resides within the membrane boundary. This enzyme requires separate consideration and description as it solves the problem of catalysis at the membrane perimeter in its own unique way.
4.1 Sterol reductase
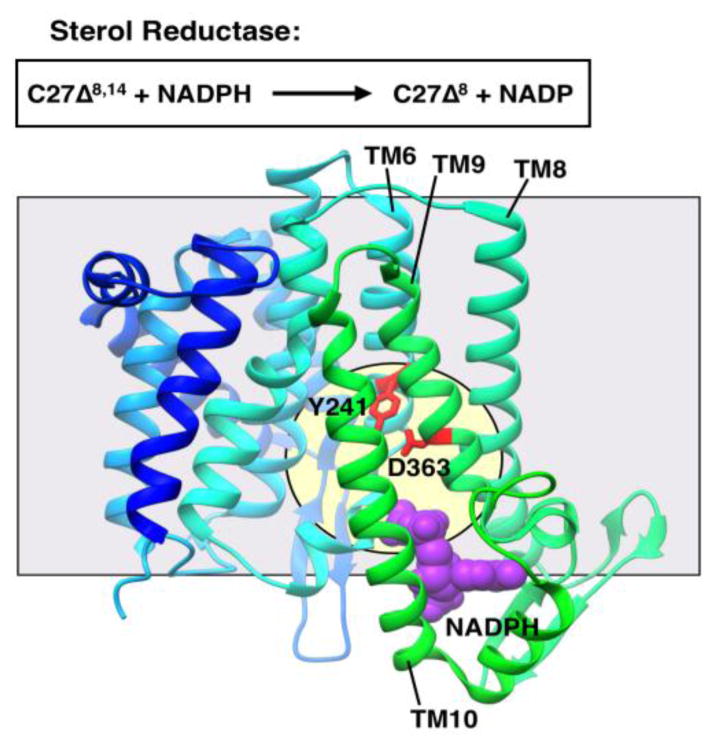
Largely hydrophobic sterols include such molecules as hopanoids, cholesterol, phytosterol, and ergosterol, which are found in animals, plants, fungi and some prokaryotes. Cholesterol is an abundant and essential sterol in animals and an important factor in cell membrane rigidity [95]. Modification of cholesterol is also the basis of steroid hormone biosynthesis [96]. One of the enzyme families that modifies sterols is the sterol reductase family. This family includes Δ14-sterol reductase (C14SR or TM7SF2), 7-dehydrocholesterol reductase (DHCR7) and 24-dehydrocholesterol reductase (DHCR24). All these enzymes catalyze the reduction of a specific carbon-carbon bond of a sterol using a reducing cofactor such as NADPH [97].
The structure of a Δ14-sterol reductase from Methylomicrobium alcaliphilum 20Z (MaSR1) was solved with bound NADPH to 2.7 Å resolution by x-ray crystallography [98]. MaSR1 contains 10 transmembrane helices, two anti-parallel β-sheets on cytosolic loops and two short α-helical domains (one at the cytoplasmic side and the other on the periplasmic side; Fig. 5). MaSR1 is known to catalyze the reaction of 5α-cholesta-8,14-dien-3β-ol (C27Δ8,14) to 5α-cholesta-8-en-3β-ol (C27Δ8) in the presence of NADPH. Two cavities are presented, which facilitate this reaction. One is a polar cavity, housing the co-crystallized NADPH, which is surrounded by the C-terminal domain (TM 6, 8–10). The other was identified housing density of an unknown molecule, flanked by TM 7 and 10. This second cavity was considered to be the putative sterol binding site because of 1) its lateral access to the membrane environment, 2) its similarity to the progesterone binding site of the soluble steroid-5β reductase (AKR1D1), and 3) its proximity to the bound NADPH. The two cavities meet where the putative active site resides, potentially allowing the hydrophilic and lipophilic substrates to be brought into apposition for catalysis to occur. Mutations at the NADPH binding site ablated activity of the enzyme. No mechanism of catalysis was proposed for MaSR1.
Figure 5. Representative structure showing intramembrane catalysis.
Sterol reductase (SR1; PDB 4QUV) with putative catalytic residues in red. Cofactor NADPH in purple in CPK representation. Structure is shown in ribbon representation colored in a gradient from the N-terminus to the C-terminus (blue to cyan to green). The grey square shows the approximate location of the membrane border. The pale yellow circle indicates the location of the putative active site. The protein is oriented with the cytoplasmic face of the membrane on the bottom.
A search for structural homologs of MaSR1 returned ICMT as the closest entry for the C-terminus (TM 6–10). The similarity seems to be most predominant in the binding of the soluble cofactor (SAH in ICMT and NADPH in MaSR1). ICMT also has a cavity accessible laterally from the membrane environment, however it accommodates an acyl chain and a bulky protein hanging off of the site important for catalysis, whereas MaSR1 only has to facilitate the binding of a small amphipathic sterol. Though these transmembrane enzymes have similar predicted structure, they tackle different biophysical problems unique to their substrates.
5. β-barrel enzymes
β-barrel membrane proteins are encountered primarily in Gram-negative bacteria, yet some mitochondrial and chloroplast membrane proteins are also thought to adopt a β-barrel fold [99]. They reside exclusively in the outer membrane of Gram-negative bacteria, a property that may be linked with their ability to efficiently dissipate proton gradients, leading to negative selection towards an inner membrane localization [100]. The βbarrel fold is often encountered in transport and structural proteins of the outer membrane, but it has also been adapted for enzymatic activity. β-barrel outer membrane proteins have 8–22 strands and typically exhibit a hydrophobic surface that comes in contact with the membrane environment, and a more polar barrel interior. βbarrel membrane proteins commonly have clusters of aromatic residues that demarcate the boundaries of the membrane bilayer on the protein, perhaps to stabilize the fold in the membrane environment. All β-barrel membrane protein structures solved so far contain an even number of strands and have a periplasmic localization of both their termini.
Even though there are many crystal structures of β-barrel membrane proteins, the majority corresponds to transport or structural proteins of the outer membrane. The number of β-barrel enzymes that have been structurally characterized is much smaller. Four of them have been reviewed previously [100], and will be presented briefly here, together with two new structures that have surfaced since.
5.1 Outer Membrane Phospholipase A (OMPLA)
OMPLA is a glycerophospholipid phospholipase that hydrolyzes the acyl-ester bonds of phospholipids in the outer leaflet of the outer membrane of Gram-negative bacteria [101]. It has been proposed to maintain the asymmetry of the outer membrane, by hydrolyzing phospholipids that have flipped to the outer leaflet [100, 101]. It is also important for virulence in several Gram-negative bacterial species [100].
Crystal structures of OMPLA from E. coli (EcOMPLA) in monomeric and dimeric forms (solved at 2.2 and 2.1 Å, respectively) revealed a β-barrel with 12 antiparallel strands [102]. The presence of phospholipids promotes dimerization that enables the enzymatic activity of EcOMPLA. The dimeric structure was solved with the competitive inhibitor hexadecane sulphonyl fluoride in place of the regular phospholipid substrate, thus enabling visualization of the likely mode of phospholipid coordination at the dimer interface of EcOMPLA, with two inhibitor molecules coordinated between the two EcOMPLA monomers [102, 103] (Fig. 6A). Calcium is an essential cofactor for activity and dimerization with binding sites located at the dimer interface (one in each monomer) [102]. A secondary binding site, different from the one observed on the dimer, was located in the monomer structure between two external loops (L3/L4) by soaking monomer crystals with calcium [103]. The monomer calcium-binding site was proposed to facilitate interactions with LPS and act as Ca2+ ion sink to feed the dimer site [103]. The active site residues His142, Ser144 and Asn156 are organized in a triad at the boundary of the outer leaflet of the outer membrane, and participate in a mechanism similar to that of esterases [102] (Fig. 6A). OMPLA is able to target the acyl chains at both sn-1 and sn-2 positions of phospholipids and is relatively unspecific towards the polar head group of its substrates [100].
Figure 6. Representative structures of integral membrane beta-barrel enzymes.
All structures are shown in ribbon representation with bound ligands in CPK representation. The proteins are colored with a gradient from the N-terminus to the C-terminus (blue to cyan to green). The grey square shows the approximate location of the membrane border. The pale yellow circle indicates the location of the putative active site. All structures oriented with the periplamic face of the membrane on the bottom. A. Outer membrane phospholipase (OMPLA; PDB 1QD6). The inhibitor hexadecanesulphonyl fluoride (HDS) is covalently bound to the nucleophilic serine residue (S144; shown in red). The catalytic calcium ion is shown in yellow. PE, phosphatidylethanolamine. B. Plasminogen activator protease (Pla; PDB 2X55). Observed bound acyl chains represented as C8E4 detergent chains potentially correspond to bound Lipid A with partial occupancy (shown in magenta). Putative catalytic and LPS-binding residues shown in red. C. Palmitoyltransferase (PagP; PDB 1THQ) with bound LDAO detergent molecule in the hydrocarbon ruler pocket. LDAO shown in magenta. PC, phosphatidylcholine; Lipid B, hepta-acylated Lipid A. D. Deacylase PagL (PDB 2ERV). Putative catalytic residues are shown in red. E. Deacylase LpxR (PDB 3FID). Putative catalytic and Kdo sugar binding residues are shown in red. Zinc ion observed in the structure is shown in yellow.
5.2 Proteases OmpT and Pla (Omptins)
OmpT and Pla are members of the omptin family of outer membrane proteases, widely present in Enterobacteriaceae. Omptins are proteases that cleave polycationic peptides, with the target site consisted of two adjacent basic amino acids [100, 104]. LPS is known to act as a cofactor for the catalytic activity of OmpT and other omptins, prompting us to include them in this review despite not utilizing any lipidic substrates during catalysis.
OmpT is expressed in E. coli K12 in a growth temperature-dependent manner [100]. The main substrate of OmpT is Proteamine P1, a highly basic antimicrobial peptide that is secreted by epithelial cells of the urinary tract [104, 105]. Omptins are believed to act generally as a defense mechanism towards antimicrobial peptides that disrupt the LPS network and reach the protease during uptake [100]. Pla is expressed in Yersinia pestis, an important pathogen that causes plague in humans [104, 106]. Inactivation of Pla greatly reduces virulence of Y. pestis, linked to the ability of Pla to cleave human plasminogen and generate plasmin, a blood protease that targets several blood proteins, most significantly fibrin, a protein involved in blood clotting [104, 106]. A secondary function that operates independently of protease activity is adhesion to host tissues, exemplified by Pla of Y. pestis, but also observed with OmpT of E. coli and PgtE of Salmonella enterica [104].
The crystal structure of OmpT from E. coli (EcOmpT) solved at 2.6 Å resolution showed a β-barrel fold consisted of 10 antiparallel strands [105]. EcOmpT protrudes ~40 Å above the lipid bilayer, with the outermost loops located just above the rim of the lipopolysaccharide (LPS) core region. The five external loops connecting the β-barrel strands create a crevice with a negatively-charged bottom, thought to be the site of coordination for the signature dibasic motif targeted by OmpT and other omptins. The active site of the enzyme is presented within the external crevice and consists of paired residues Asp83/Asp85 on one face and Asp210/His212 on the other. Catalysis has been proposed to proceed through a water molecule, coordinated between the two residue pairs, that acts as a nucleophile to attack the scissile bond [105]. While OmpT was originally classified as a serine protease, it was later reclassified as an aspartate protease based on weak inhibition by serine protease inhibitors and structural evidence. Yet the proposed mechanism, involving both an Asp-Asp pair similar to aspartate proteases and a Asp-His pair similar to serine carboxyl proteases, underscores the unique nature of the catalytic scheme employed by omptin proteases [104, 105]. By comparing the EcOmpT structure to transporter FhuA that has been solved together with LPS, residues Glu136, Arg138 and Arg174 were identified as potential LPS-interacting residues, yet in the absence of an OmpT-LPS complex structure no further inferences regarding the LPS effect on catalysis could be made.
Further insight about the LPS interaction and support for the proposed catalytic scheme came from a crystal structure of Pla from Y. pestis (YpPla) solved at 1.9 Å resolution [106]. The Pla structure consists of a β-barrel with 10 antiparallel strands (Fig. 6B) and high overall similarity to the EcOmpT structure (RMSD 1.1 Å). Importantly, the improved resolution allowed visualization of a network of water molecules situated within the active site, with one in particular (W1) located between the two-pair catalytic quad (D84/D86 and D206/H208 in YpPla), and thus likely corresponding to the nucleophilic water proposed to participate in catalysis. Also differences in purification techniques may have led to partial occupancy of the LPS-bound state. Densities corresponding to partially ordered acyl chains were present near the LPS-binding residues of YpPla (Y134, E136, R138 and R171). These densities may correspond to LPS acyl chains or detergent molecules occupying a similar space to the former. By comparing this partially “LPS-liganded” state of Pla with the apo state of EcOmpT, it was proposed that LPS binding may slightly alter active site geometry, pushing strand 7 inward, thus leading to activation of omptins [106] (Fig. 6B).
5.3 Palmitoyltransferase PagP
PagP is a lipid A modification enzyme located in the outer membrane of many Gram-negative bacteria. It transfers a palmitate moiety from the sn-1 position of a phospholipid donor to the N-linked hydroxymyristate of the proximal glucosamine unit of lipid A, the lipidic anchor of bacterial lipopolysaccharide (LPS) [107]. The resulting hepta-acylated species of lipid A endows bacteria expressing PagP with dual protection against host defenses: 1) it attenuates innate immune responses acting as a competitive antagonist of TLR4 activation, and 2) it enables resistance to host cationic antimicrobial peptides, likely by changing the properties of the bacterial outer membrane [100, 108]. Given that phospholipids are not typically found in the outer leaflet of the outer membrane (OM) due to its well-known asymmetry, PagP may be effectively activated by perturbations of the asymmetric nature of the outer membrane [100, 108].
A solution structure of PagP from E. coli (EcPagP) by NMR spectroscopy showed that EcPagP adopts a βbarrel fold with 8 antiparallel strands [109]. Further insights related to substrate recognition were gained through a structure obtained by x-ray crystallography, which was solved to 1.9 Å resolution [110]. The axis of the barrel appears to be tilted by roughly 25° at the plane of the membrane (Fig. 6C). A hydrophobic pocket, known as the hydrocarbon ruler, is situated at the extracellular face of the molecule, and is believed to be responsible for selection of acyl chains of appropriate size for transfer to lipid A. In fact, EcPagP displays a narrow selectivity for palmitate with activity being significantly attenuated even with one methylene group deviation from the optimal sixteen carbon long palmitate [110]. Gly88, located near the bottom of the hydrocarbon ruler pocket appears to control the selectivity of PagP towards palmitate, because mutation of this residue leads to significant attenuation of PagP selectivity. An LDAO detergent molecule was observed inside the pocket and is believed to approximate the binding mode of palmitate. The LDAO molecule was oriented appropriately into the pocket, with the polar headgroup located near the extracellular phase of the membrane and the hydrophobic tail embedded within the hydrophobic pocket. Discontinuities in the hydrogen bond network due to conserved prolines at key positions between strands A/B and F/G are hypothesized to provide pathways for lateral entry and/or exit of the lipidic substrate from the hydrophobic pocket [110]. Additionally, the extracellular loops display significant flexibility based on NMR and crystal structure evidence [109, 111], and may undergo rearrangements to enable access to the pocket and productive orientation of the catalytic residues. Molecular dynamics simulations have shown that entry of lipidic substrate to the pocket between strands F and G (F/G route) is likely the preferred route, whereas exit may also occur between strands B and C [111]. Moreover, a tyrosine residue (Y147) may function as a “gate” for the F/G route entry, since mutations to an alanine or phenylalanine increase the rate of enzymatic activity. Three residues that have been determined to be critical for catalysis (His33, Asp76, Ser77) are located in the first and second extracellular loop (Fig. 6C) [109]. Finally, PagP also has weak phospholipase activity, essentially slowly transferring the palmitate moiety to water. Mutations on L1 loop specifically affect the palmitoyltransferase activity of the enzyme, leaving the phospholipase activity intact, may thus be involved in Lipid A recognition [110].
5.4 Deacylase PagL
PagL is an outer membrane 3-O deacylase that removes a hydromyristate moiety from the 3 position of Lipid A and its precursors. The enzyme was initially identified and characterized in Salmonella enterica [48]. Similarly to PagP, modification of Lipid A by this enzyme leads to reduced Lipid A endotoxicity and increased resistance to antimicrobial peptides [100].
The crystal structure of PagL from Pseudomonas aeruginosa (PaPagL) was solved at 2.0 Å resolution and revealed an 8-stranded antiparallel β-barrel that resembles the fold seen in the PagP enzyme [112]. As is the case with most outer membrane β-barrels, PaPagL has aromatic residue belts, delineating the boundaries of the membrane. Based on a hydrophobicity profile, it is likely that the axis of the protein is tilted by ~30° relative to the plane of the membrane. The active site of PaPagL is very similar to the OMPLA active site. In PaPagL, a catalytic triad formed by His126, Ser128 and Glu140 is located on the beta-barrel exterior (Fig. 6D), and is superimposable with residues His142, Ser144 and Asn156 of OMPLA that form the catalytic triad. Moreover, the two active sites are located at similar heights of the β-barrel fold. PagL shows much lower selectivity towards the length of the targeted acyl chain compared to the PagP enzyme. Docking of the minimal substrate, Lipid X, onto the structure provides a reason for the broader selectivity towards acyl chains, as Lipid X is coordinated on the exterior of the β-barrel, and a hydrocarbon ruler pocket is not utilized in PagL, as it is in PagP. The positioning and orientation of two PaPagL molecules in the asymmetric unit raises the possibility that it can form a dimer. Indeed it was shown that a mutant of the active site serine into a cysteine can support dimer formation under non-reducing conditions. The authors of this study hypothesized that dimerization may represent an inactivation mechanism for PagL, contrary to the role of dimerization in OMPLA activation, because of occlusion of the active site [112].
5.5 Deacylase LpxR
The latest structure of a β-barrel enzyme to be solved is that of LpxR, an outer membrane enzyme that is again responsible for a Lipid A modification. Similarly to PagL, LpxR was identified in Salmonella enterica, and catalyzes the removal of the complete acyloxyacyl moiety from the 3’ position of Salmonella lipid A [113]. The reaction appears to be dependent on the presence of calcium (or other divalents). The resulting tetra-acylated lipid A has lower immunological activity (endotoxicity), thus facilitating evasion from the innate immune response of the host. Interestingly, in H. pylori LpxR is constantly activated leading to conversion of the entire lipid A population into the tetra-acylated form [113, 114].
LpxR from Salmonella enterica (SeLpxR) was solved by x-ray crystallography to 1.9 Å resolution [114]. The structure revealed a 12-stranded antiparallel β-barrel with an unusually long periplasmic turn 4 that forms a loop inside the β-barrel effectively sealing off the fold from the periplasmic side. The active site is located between the barrel wall and an alpha helix that is formed by the extracellular loop 3. Based on location of conserved residues, a catalytic mechanism similar to that of PLA2 has been proposed. Residues N9, D10, T34 and H122 are all essential for activity and are located in a cleft just above the interface of the membrane (Fig. 6E). A Ca2+ ion (zinc in the structure) is proposed to form interactions with both SeLpxR and the lipid A substrate and appears to be involved in the oxyanion hole of the enzyme necessary for catalysis, thus explaining the observed dependence of catalytic activity on the presence of divalent cations. Molecular docking of the complete lipid A molecule (Kdo2-lipid A) into the structure showed that only the polar headgroups are efficiently coordinated on the fold, whereas the acyl chains largely seem to remain embedded in the membrane environment. The Kdo sugars of lipid A are coordinated by positively charged residues H25, K67 and R68, located just above the active site of SeLpxR, by means of electrostatic interactions. This observation may help explain why lipid IV(A) (Kdo-less lipid A) acts as a poor substrate for LpxR-mediated catalysis.
6. Conclusions
Integral membrane enzymes represent an essential part of the biosynthetic capabilities of a cell, accommodating the need for catalysis involving lipidic substrates. The relevant catalytic reactions must take place at or near the membrane-water interface given the unique nature of their substrates. Thus, membrane enzymes have evolved folds and features that are appropriate to support this requirement. In this review, we have attempted to illustrate the structural diversity of integral membrane enzymes that has been captured by high-resolution structural biology techniques so far.
Alpha-helical integral membrane enzymes appear to have developed a wide array of folds, which can be subdivided into three categories, reflecting in large extent the character of the catalytic reactions they accommodate. Interfacial and extramembrane enzymes provide an obvious, yet elegant, solution to the problem of accommodating amphipathic substrates that contain both polar and hydrophobic elements. Intramembrane enzymes are also an obvious solution to the problem of catalyzing reactions that involve purely hydrophobic substrates or that target hydrophobic parts of amphipathic substrates. Further structural diversity is encapsulated by the number of alpha-helical transmembrane segments that each enzyme has been equipped with. Single transmembrane segment enzymes (or slightly more elaborated like GtrB) are well-suited to accommodate reactions that may have one or more soluble substrates, but still need to occur near the membrane-water interface (i.e. the second substrate is lipidic, or the product must be localized close to the membrane for utilization by another membrane enzyme). Thus, the fold(s) included in the extramembrane category likely represent an adaptation of soluble enzymes for catalysis at the membrane-water interface. Interfacial and intramembrane enzymes need a greater number of transmembrane segments to efficiently bind increasingly complex lipidic substrates, with the exact number of transmembrane segments possibly relating to the degree of coordination of the lipidic substrate necessary to enable efficient catalysis.
β-barrel enzymes seem to have followed a different approach by essentially adapting the same fold to accommodate diverse enzymatic reactions involving either soluble substrates (i.e. OmpT) or lipidic substrates. The special environment of the outer membrane of Gram-negative bacteria may be largely responsible for the lack of structural diversity of β-barrel enzymes. Moreover, β-barrel enzymes appear to utilize rather minimal interaction of the protein with its substrates especially when compared with the more elaborated themes of substrate recognition and coordination observed in alpha-helical membrane proteins. The need to adapt the same fold for many different functions could be a potential explanation for this fact.
In the coming years, we expect the field of integral membrane enzyme structure to continue to grow and provide further information about this important subset of cellular proteins. It will be interesting to see how the field will respond to the issue of sparse or incomplete substrate information that often hinders full interpretation of catalytic mechanisms even for enzyme classes that have been successfully solved. It is an inherent issue that we face in attempting to incorporate highly hydrophobic lipidic substrates into our protein crystals. Perhaps the answer to that will come from the rapidly rising use of cryo-electron microscopy for structural studies that has seen a renaissance after the advent of direct electron detectors. Further improvements in resolution and size-limitations in the coming years will make it an attractive choice for the study of enzyme-substrate interactions, particularly for cases that are resistant to co-crystallization attempts.
Highlights.
Here we review the current structural landscape of membrane enzymes utilizing lipidic substrates in diverse chemical reactions.
Within this class of integral membrane proteins, alpha-helical enzymes exhibit diverse structural folds, enabling three main modes of catalysis depending on the location of the catalytic site: intramembrane, extramembrane, and interfacial.
Beta-barrel enzymes on the other hand seem to lack diversity in fold, however they accommodate a variety of chemical reactions and substrates.
Acknowledgments
This work was supported in part by NIH-NIGMS grants R01 GM111980 (Mancia), R21 AI119672 (Mancia), U54 GM095315 (Hendrickson) and P41 GM116799 (Hendrickson).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vance JE, Vance DE. Phospholipid biosynthesis in mammalian cells. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2004;82:113–128. doi: 10.1139/o03-073. [DOI] [PubMed] [Google Scholar]
- 2.Cronan JE. Bacterial membrane lipids: where do we stand? Annu Rev Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- 3.Houtkooper RH, Akbari H, van Lenthe H, Kulik W, Wanders RJ, Frentzen M, Vaz FM. Identification and characterization of human cardiolipin synthase. FEBS letters. 2006;580:3059–3064. doi: 10.1016/j.febslet.2006.04.054. [DOI] [PubMed] [Google Scholar]
- 4.Williams JG, McMaster CR. Scanning alanine mutagenesis of the CDP-alcohol phosphotransferase motif of Saccharomyces cerevisiae cholinephosphotransferase. The Journal of biological chemistry. 1998;273:13482–13487. doi: 10.1074/jbc.273.22.13482. [DOI] [PubMed] [Google Scholar]
- 5.Clarke OB, Tomasek D, Jorge CD, Dufrisne MB, Kim M, Banerjee S, Rajashankar KR, Shapiro L, Hendrickson WA, Santos H, Mancia F. Structural basis for phosphatidylinositol-phosphate biosynthesis. Nat Commun. 2015;6:8505. doi: 10.1038/ncomms9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nogly P, Gushchin I, Remeeva A, Esteves AM, Borges N, Ma P, Ishchenko A, Grudinin S, Round E, Moraes I, Borshchevskiy V, Santos H, Gordeliy V, Archer M. X-ray structure of a CDP-alcohol phosphatidyltransferase membrane enzyme and insights into its catalytic mechanism. Nat Commun. 2014;5:4169. doi: 10.1038/ncomms5169. [DOI] [PubMed] [Google Scholar]
- 7.Sciara G, Clarke OB, Tomasek D, Kloss B, Tabuso S, Byfield R, Cohn R, Banerjee S, Rajashankar KR, Slavkovic V, Graziano JH, Shapiro L, Mancia F. Structural basis for catalysis in a CDP-alcohol phosphotransferase. Nat Commun. 2014;5:4068. doi: 10.1038/ncomms5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brito JA, Borges N, Vonrhein C, Santos H, Archer M. Crystal structure of Archaeoglobus fulgidus CTP:inositol-1-phosphate cytidylyltransferase, a key enzyme for di-myo-inositol-phosphate synthesis in (hyper)thermophiles. Journal of bacteriology. 2011;193:2177–2185. doi: 10.1128/JB.01543-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morii H, Ogawa M, Fukuda K, Taniguchi H, Koga Y. A revised biosynthetic pathway for phosphatidylinositol in Mycobacteria. Journal of biochemistry. 2010;148:593–602. doi: 10.1093/jb/mvq093. [DOI] [PubMed] [Google Scholar]
- 10.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 11.Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. The Journal of biological chemistry. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- 12.Romano JD, Schmidt WK, Michaelis S. The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:2231–2247. doi: 10.1091/mbc.9.8.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5:405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- 14.Lau HY, Ramanujulu PM, Guo D, Yang T, Wirawan M, Casey PJ, Go ML, Wang M. An improved isoprenylcysteine carboxylmethyltransferase inhibitor induces cancer cell death and attenuates tumor growth in vivo. Cancer Biol Ther. 2014;15:1280–1291. doi: 10.4161/cbt.29692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Kulkarni K, Manolaridis I, Zhang Z, Dodd RB, Mas-Droux C, Barford D. Mechanism of isoprenylcysteine carboxyl methylation from the crystal structure of the integral membrane methyltransferase ICMT. Mol Cell. 2011;44:997–1004. doi: 10.1016/j.molcel.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 17.Chaurio RA, Janko C, Munoz LE, Frey B, Herrmann M, Gaipl US. Phospholipids: key players in apoptosis and immune regulation. Molecules. 2009;14:4892–4914. doi: 10.3390/molecules14124892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sparrow CP, Raetz CR. Purification and properties of the membrane-bound CDP-diglyceride synthetase from Escherichia coli. The Journal of biological chemistry. 1985;260:12084–12091. [PubMed] [Google Scholar]
- 19.Liu X, Yin Y, Wu J, Liu Z. Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis. Nat Commun. 2014;5:4244. doi: 10.1038/ncomms5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bullen HE, Soldati-Favre D. A central role for phosphatidic acid as a lipid mediator of regulated exocytosis in apicomplexa. FEBS Lett. 2016;590:2469–2481. doi: 10.1002/1873-3468.12296. [DOI] [PubMed] [Google Scholar]
- 21.Li D, Lyons JA, Pye VE, Vogeley L, Aragao D, Kenyon CP, Shah ST, Doherty C, Aherne M, Caffrey M. Crystal structure of the integral membrane diacylglycerol kinase. Nature. 2013;497:521–524. doi: 10.1038/nature12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Stansfeld PJ, Sansom MS, Keogh A, Vogeley L, Howe N, Lyons JA, Aragao D, Fromme P, Fromme R, Basu S, Grotjohann I, Kupitz C, Rendek K, Weierstall U, Zatsepin NA, Cherezov V, Liu W, Bandaru S, English NJ, Gati C, Barty A, Yefanov O, Chapman HN, Diederichs K, Messerschmidt M, Boutet S, Williams GJ, Marvin Seibert M, Caffrey M. Ternary structure reveals mechanism of a membrane diacylglycerol kinase. Nat Commun. 2015;6:10140. doi: 10.1038/ncomms10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, Karra MD, Tian C, Sonnichsen FD, Sanders CR. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science. 2009;324:1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray DT, Li C, Gao FP, Qin H, Cross TA. Membrane protein structural validation by oriented sample solid-state NMR: diacylglycerol kinase. Biophys J. 2014;106:1559–1569. doi: 10.1016/j.bpj.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tie JK, Stafford DW. Structure and function of vitamin K epoxide reductase. Vitam Horm. 2008;78:103–130. doi: 10.1016/S0083-6729(07)00006-4. [DOI] [PubMed] [Google Scholar]
- 26.Oldenburg J, Marinova M, Muller-Reible C, Watzka M. The vitamin K cycle. Vitam Horm. 2008;78:35–62. doi: 10.1016/S0083-6729(07)00003-9. [DOI] [PubMed] [Google Scholar]
- 27.Goodstadt L, Ponting CP. Vitamin K epoxide reductase: homology, active site and catalytic mechanism. Trends Biochem Sci. 2004;29:289–292. doi: 10.1016/j.tibs.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Czogalla KJ, Watzka M, Oldenburg J. Structural Modeling Insights into Human VKORC1 Phenotypes. Nutrients. 2015;7:6837–6851. doi: 10.3390/nu7085313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wajih N, Hutson SM, Wallin R. Disulfide-dependent protein folding is linked to operation of the vitamin K cycle in the endoplasmic reticulum. A protein disulfide isomerase-VKORC1 redox enzyme complex appears to be responsible for vitamin K1 2,3-epoxide reduction. The Journal of biological chemistry. 2007;282:2626–2635. doi: 10.1074/jbc.M608954200. [DOI] [PubMed] [Google Scholar]
- 30.Dutton RJ, Wayman A, Wei JR, Rubin EJ, Beckwith J, Boyd D. Inhibition of bacterial disulfide bond formation by the anticoagulant warfarin. Proc Natl Acad Sci U S A. 2010;107:297–301. doi: 10.1073/pnas.0912952107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Schulman S, Dutton RJ, Boyd D, Beckwith J, Rapoport TA. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh AK, Bhattacharyya-Pakrasi M, Pakrasi HB. Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. The Journal of biological chemistry. 2008;283:15762–15770. doi: 10.1074/jbc.M800982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutton RJ, Boyd D, Berkmen M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, Cheng W, Fowle Grider R, Shen G, Li W. Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer. Nat Commun. 2014;5:3110. doi: 10.1038/ncomms4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dillon DA, Wu WI, Riedel B, Wissing JB, Dowhan W, Carman GM. The Escherichia coli pgpB gene encodes for a diacylglycerol pyrophosphate phosphatase activity. The Journal of biological chemistry. 1996;271:30548–30553. doi: 10.1074/jbc.271.48.30548. [DOI] [PubMed] [Google Scholar]
- 36.Touze T, Blanot D, Mengin-Lecreulx D. Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. The Journal of biological chemistry. 2008;283:16573–16583. doi: 10.1074/jbc.M800394200. [DOI] [PubMed] [Google Scholar]
- 37.Fan J, Jiang D, Zhao Y, Liu J, Zhang XC. Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B. Proc Natl Acad Sci U S A. 2014;111:7636–7640. doi: 10.1073/pnas.1403097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong S, Lin Y, Lu S, Wang M, Bogdanov M, Zheng L. Structural Insight into Substrate Selection and Catalysis of Lipid Phosphate Phosphatase PgpB in the Cell Membrane. The Journal of biological chemistry. 2016;291:18342–18352. doi: 10.1074/jbc.M116.737874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Messerschmidt A, Wever R. X-ray structure of a vanadium-containing enzyme: chloroperoxidase from the fungus Curvularia inaequalis. Proc Natl Acad Sci U S A. 1996;93:392–396. doi: 10.1073/pnas.93.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishikawa K, Mihara Y, Gondoh K, Suzuki E, Asano Y. X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate. EMBO J. 2000;19:2412–2423. doi: 10.1093/emboj/19.11.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Breukink E. The Membrane Steps of Bacterial Cell Wall Synthesis as Antibiotic Targets. Antibiotics (Basel) 2016;5 doi: 10.3390/antibiotics5030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 44.Chung BC, Zhao J, Gillespie RA, Kwon DY, Guan Z, Hong J, Zhou P, Lee SY. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–1016. doi: 10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amer AO, Valvano MA. Conserved amino acid residues found in a predicted cytosolic domain of the lipopolysaccharide biosynthetic protein WecA are implicated in the recognition of UDP-N-acetylglucosamine. Microbiology. 2001;147:3015–3025. doi: 10.1099/00221287-147-11-3015. [DOI] [PubMed] [Google Scholar]
- 46.Anderson MS, Eveland SS, Price NP. Conserved cytoplasmic motifs that distinguish sub-groups of the polyprenol phosphate:N-acetylhexosamine-1-phosphate transferase family. FEMS Microbiol Lett. 2000;191:169–175. doi: 10.1111/j.1574-6968.2000.tb09335.x. [DOI] [PubMed] [Google Scholar]
- 47.Chung BC, Mashalidis EH, Tanino T, Kim M, Matsuda A, Hong J, Ichikawa S, Lee SY. Structural insights into inhibition of lipid I production in bacterial cell wall synthesis. Nature. 2016;533:557–560. doi: 10.1038/nature17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 49.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrou VI, Herrera CM, Schultz KM, Clarke OB, Vendome J, Tomasek D, Banerjee S, Rajashankar KR, Belcher Dufrisne M, Kloss B, Kloppmann E, Rost B, Klug CS, Trent MS, Shapiro L, Mancia F. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science. 2016;351:608–612. doi: 10.1126/science.aad1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lizak C, Gerber S, Numao S, Aebi M, Locher KP. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 2011;474:350–355. doi: 10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- 52.Matsumoto S, Shimada A, Nyirenda J, Igura M, Kawano Y, Kohda D. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc Natl Acad Sci U S A. 2013;110:17868–17873. doi: 10.1073/pnas.1309777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LoVullo ED, Wright LF, Isabella V, Huntley JF, Pavelka MS., Jr Revisiting the Gram-negative lipoprotein paradigm. Journal of bacteriology. 2015;197:1705–1715. doi: 10.1128/JB.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovacs-Simon A, Titball RW, Michell SL. Lipoproteins of bacterial pathogens. Infect Immun. 2011;79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sankaran K, Wu HC. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. The Journal of biological chemistry. 1994;269:19701–19706. [PubMed] [Google Scholar]
- 56.Qi HY, Sankaran K, Gan K, Wu HC. Structure-function relationship of bacterial prolipoprotein diacylglyceryl transferase: functionally significant conserved regions. Journal of bacteriology. 1995;177:6820–6824. doi: 10.1128/jb.177.23.6820-6824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao G, Zhao Y, Kang X, Li Z, Zhang Y, Wang X, Sun F, Sankaran K, Zhang XC. Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat Commun. 2016;7:10198. doi: 10.1038/ncomms10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng W, Li W. Structural insights into ubiquinone biosynthesis in membranes. Science. 2014;343:878–881. doi: 10.1126/science.1246774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W. Bringing Bioactive Compounds into Membranes: The UbiA Superfamily of Intramembrane Aromatic Prenyltransferases. Trends Biochem Sci. 2016;41:356–370. doi: 10.1016/j.tibs.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakagawa K, Hirota Y, Sawada N, Yuge N, Watanabe M, Uchino Y, Okuda N, Shimomura Y, Suhara Y, Okano T. Identification of UBIAD1 as a novel human menaquinone-4 biosynthetic enzyme. Nature. 2010;468:117–121. doi: 10.1038/nature09464. [DOI] [PubMed] [Google Scholar]
- 61.Mugoni V, Postel R, Catanzaro V, De Luca E, Turco E, Digilio G, Silengo L, Murphy MP, Medana C, Stainier DY, Bakkers J, Santoro MM. Ubiad1 is an antioxidant enzyme that regulates eNOS activity by CoQ10 synthesis. Cell. 2013;152:504–518. doi: 10.1016/j.cell.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orr A, Dube MP, Marcadier J, Jiang H, Federico A, George S, Seamone C, Andrews D, Dubord P, Holland S, Provost S, Mongrain V, Evans S, Higgins B, Bowman S, Guernsey D, Samuels M. Mutations in the UBIAD1 gene, encoding a potential prenyltransferase, are causal for Schnyder crystalline corneal dystrophy. PLoS One. 2007;2:e685. doi: 10.1371/journal.pone.0000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang H, Levin EJ, Liu S, Bai Y, Lockless SW, Zhou M. Structure of a membrane-embedded prenyltransferase homologous to UBIAD1. PLoS Biol. 2014;12:e1001911. doi: 10.1371/journal.pbio.1001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Serhan CN, Chiang N, Dalli J, Levy BD. Lipid mediators in the resolution of inflammation. Cold Spring Harb Perspect Biol. 2015;7:a016311. doi: 10.1101/cshperspect.a016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sjogren T, Nord J, Ek M, Johansson P, Liu G, Geschwindner S. Crystal structure of microsomal prostaglandin E2 synthase provides insight into diversity in the MAPEG superfamily. Proc Natl Acad Sci U S A. 2013;110:3806–3811. doi: 10.1073/pnas.1218504110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jegerschold C, Pawelzik SC, Purhonen P, Bhakat P, Gheorghe KR, Gyobu N, Mitsuoka K, Morgenstern R, Jakobsson PJ, Hebert H. Structural basis for induced formation of the inflammatory mediator prostaglandin E2. Proc Natl Acad Sci U S A. 2008;105:11110–11115. doi: 10.1073/pnas.0802894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez Molina D, Wetterholm A, Kohl A, McCarthy AA, Niegowski D, Ohlson E, Hammarberg T, Eshaghi S, Haeggstrom JZ, Nordlund P. Structural basis for synthesis of inflammatory mediators by human leukotriene C4 synthase. Nature. 2007;448:613–616. doi: 10.1038/nature06009. [DOI] [PubMed] [Google Scholar]
- 68.Niegowski D, Kleinschmidt T, Olsson U, Ahmad S, Rinaldo-Matthis A, Haeggstrom JZ. Crystal structures of leukotriene C4 synthase in complex with product analogs: implications for the enzyme mechanism. The Journal of biological chemistry. 2014;289:5199–5207. doi: 10.1074/jbc.M113.534628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orlean P. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:5796–5805. doi: 10.1128/mcb.10.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bugg TD, Brandish PE. From peptidoglycan to glycoproteins: common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol Lett. 1994;119:255–262. doi: 10.1111/j.1574-6968.1994.tb06898.x. [DOI] [PubMed] [Google Scholar]
- 71.Allison GE, Verma NK. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8:17–23. doi: 10.1016/s0966-842x(99)01646-7. [DOI] [PubMed] [Google Scholar]
- 72.Ardiccioni C, Clarke OB, Tomasek D, Issa HA, von Alpen DC, Pond HL, Banerjee S, Rajashankar KR, Liu Q, Guan Z, Li C, Kloss B, Bruni R, Kloppmann E, Rost B, Manzini MC, Shapiro L, Mancia F. Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis. Nat Commun. 2016;7:10175. doi: 10.1038/ncomms10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 74.Ghosh D. Human sulfatases: a structural perspective to catalysis. Cell Mol Life Sci. 2007;64:2013–2022. doi: 10.1007/s00018-007-7175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hernandez-Martin A, Gonzalez-Sarmiento R, De Unamuno P. X-linked ichthyosis: an update. Br J Dermatol. 1999;141:617–627. doi: 10.1046/j.1365-2133.1999.03098.x. [DOI] [PubMed] [Google Scholar]
- 76.Rizner TL. The Important Roles of Steroid Sulfatase and Sulfotransferases in Gynecological Diseases. Front Pharmacol. 2016;7:30. doi: 10.3389/fphar.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hernandez-Guzman FG, Higashiyama T, Pangborn W, Osawa Y, Ghosh D. Structure of human estrone sulfatase suggests functional roles of membrane association. The Journal of biological chemistry. 2003;278:22989–22997. doi: 10.1074/jbc.M211497200. [DOI] [PubMed] [Google Scholar]
- 78.Lovering AL, de Castro LH, Lim D, Strynadka NC. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science. 2007;315:1402–1405. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- 79.Sung MT, Lai YT, Huang CY, Chou LY, Shih HW, Cheng WC, Wong CH, Ma C. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:8824–8829. doi: 10.1073/pnas.0904030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan Y, Fuse S, Ostash B, Sliz P, Kahne D, Walker S. Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design. ACS Chem Biol. 2008;3:429–436. doi: 10.1021/cb800078a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heaslet H, Shaw B, Mistry A, Miller AA. Characterization of the active site of S. aureus monofunctional glycosyltransferase (Mtg) by site-directed mutation and structural analysis of the protein complexed with moenomycin. J Struct Biol. 2009;167:129–135. doi: 10.1016/j.jsb.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 82.Huang CY, Shih HW, Lin LY, Tien YW, Cheng TJ, Cheng WC, Wong CH, Ma C. Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism. Proc Natl Acad Sci U S A. 2012;109:6496–6501. doi: 10.1073/pnas.1203900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab. 2009;297:E28–37. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Z, Dales NA, Winther MD. Opportunities and challenges in developing stearoyl-coenzyme A desaturase-1 inhibitors as novel therapeutics for human disease. J Med Chem. 2014;57:5039–5056. doi: 10.1021/jm401516c. [DOI] [PubMed] [Google Scholar]
- 85.Bai Y, McCoy JG, Levin EJ, Sobrado P, Rajashankar KR, Fox BG, Zhou M. X-ray structure of a mammalian stearoyl-CoA desaturase. Nature. 2015;524:252–256. doi: 10.1038/nature14549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Klein MG, Zou H, Lane W, Snell G, Levin I, Li K, Sang BC. Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate. Nat Struct Mol Biol. 2015;22:581–585. doi: 10.1038/nsmb.3049. [DOI] [PubMed] [Google Scholar]
- 87.Rather LJ, Weinert T, Demmer U, Bill E, Ismail W, Fuchs G, Ermler U. Structure and mechanism of the diiron benzoyl-coenzyme A epoxidase BoxB. The Journal of biological chemistry. 2011;286:29241–29248. doi: 10.1074/jbc.M111.236893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lindqvist Y, Huang W, Schneider G, Shanklin J. Crystal structure of delta9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu G, Koszelak-Rosenblum M, Connelly SM, Dumont ME, Malkowski MG. The Crystal Structure of an Integral Membrane Fatty Acid alpha-Hydroxylase. The Journal of biological chemistry. 2015;290:29820–29833. doi: 10.1074/jbc.M115.680124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21:576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 92.Matsumoto S, Igura M, Nyirenda J, Matsumoto M, Yuzawa S, Noda N, Inagaki F, Kohda D. Crystal structure of the C-terminal globular domain of oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 A resolution. Biochemistry. 2012;51:4157–4166. doi: 10.1021/bi300076u. [DOI] [PubMed] [Google Scholar]
- 93.Matsumoto S, Shimada A, Kohda D. Crystal structure of the C-terminal globular domain of the third paralog of the Archaeoglobus fulgidus oligosaccharyltransferases. BMC Struct Biol. 2013;13:11. doi: 10.1186/1472-6807-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maita N, Nyirenda J, Igura M, Kamishikiryo J, Kohda D. Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. The Journal of biological chemistry. 2010;285:4941–4950. doi: 10.1074/jbc.M109.081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miller WL, Bose HS. Early steps in steroidogenesis: intracellular cholesterol trafficking. J Lipid Res. 2011;52:2111–2135. doi: 10.1194/jlr.R016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller WL. Steroid hormone synthesis in mitochondria. Mol Cell Endocrinol. 2013;379:62–73. doi: 10.1016/j.mce.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 97.Porter FD, Herman GE. Malformation syndromes caused by disorders of cholesterol synthesis. J Lipid Res. 2011;52:6–34. doi: 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li X, Roberti R, Blobel G. Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum. Nature. 2015;517:104–107. doi: 10.1038/nature13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 100.Bishop RE. Structural biology of membrane-intrinsic beta-barrel enzymes: sentinels of the bacterial outer membrane. Biochim Biophys Acta. 2008;1778:1881–1896. doi: 10.1016/j.bbamem.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dekker N. Outer-membrane phospholipase A: known structure, unknown biological function. Molecular microbiology. 2000;35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 102.Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, Dijkstra BW. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 103.Snijder HJ, Kingma RL, Kalk KH, Dekker N, Egmond MR, Dijkstra BW. Structural investigations of calcium binding and its role in activity and activation of outer membrane phospholipase A from Escherichia coli. J Mol Biol. 2001;309:477–489. doi: 10.1006/jmbi.2001.4675. [DOI] [PubMed] [Google Scholar]
- 104.Hritonenko V, Stathopoulos C. Omptin proteins: an expanding family of outer membrane proteases in Gram-negative Enterobacteriaceae. Mol Membr Biol. 2007;24:395–406. doi: 10.1080/09687680701443822. [DOI] [PubMed] [Google Scholar]
- 105.Vandeputte-Rutten L, Kramer RA, Kroon J, Dekker N, Egmond MR, Gros P. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J. 2001;20:5033–5039. doi: 10.1093/emboj/20.18.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eren E, Murphy M, Goguen J, van den Berg B. An active site water network in the plasminogen activator pla from Yersinia pestis. Structure. 2010;18:809–818. doi: 10.1016/j.str.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 107.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bishop RE. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Molecular microbiology. 2005;57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 109.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Prive GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Natl Acad Sci U S A. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ahn VE, Lo EI, Engel CK, Chen L, Hwang PM, Kay LE, Bishop RE, Prive GG. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cuesta-Seijo JA, Neale C, Khan MA, Moktar J, Tran CD, Bishop RE, Pomes R, Prive GG. PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler. Structure. 2010;18:1210–1219. doi: 10.1016/j.str.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rutten L, Geurtsen J, Lambert W, Smolenaers JJ, Bonvin AM, de Haan A, van der Ley P, Egmond MR, Gros P, Tommassen J. Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2006;103:7071–7076. doi: 10.1073/pnas.0509392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CR, Trent MS. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3'-acyloxyacyl moiety of lipid A. The Journal of biological chemistry. 2006;281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rutten L, Mannie JP, Stead CM, Raetz CR, Reynolds CM, Bonvin AM, Tommassen JP, Egmond MR, Trent MS, Gros P. Active-site architecture and catalytic mechanism of the lipid A deacylase LpxR of Salmonella typhimurium. Proc Natl Acad Sci U S A. 2009;106:1960–1964. doi: 10.1073/pnas.0813064106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Inaba K, Murakami S, Suzuki M, Nakagawa A, Yamashita E, Okada K, Ito K. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]