Abstract
The electron transport chain is the primary pathway by which a cell generates energy in the form of ATP. Byproducts of this process produce reactive oxygen species that can cause damage to mitochondrial DNA. If not properly repaired, the accumulation of DNA damage can lead to mitochondrial dysfunction linked to several human disorders including neurodegenerative diseases and cancer. Mitochondria are able to combat oxidative DNA damage via repair mechanisms that are analogous to those found in the nucleus. Of the repair pathways currently reported in the mitochondria, the base excision repair pathway is the most comprehensively described. Proteins that are involved with the maintenance of mtDNA are encoded by nuclear genes and translocate to the mitochondria making signaling between the nucleus and mitochondria imperative. In this review, we discuss the current understanding of mitochondrial DNA repair mechanisms and also highlight the sensors and signaling pathways that mediate crosstalk between the nucleus and mitochondria in the event of mitochondrial stress.
Keywords: Reactive oxygen species, mitochondrial DNA damage, mitochondrial DNA repair, mito-nuclear signaling, oxidative phosphorylation, mitochondrial dysfunction, REDOX signaling
Introduction
A few decades after the nucleus was discovered in the 1830s [1], another granular-looking organelle termed “bioblasts” by Richard Altmann was discovered [2]. In 1898, the term mitochondria was coined by Carl Benda who named this organelle after the Greek words “mitos” meaning thread and “chondros” meaning granule [2]. The discovery that mitochondria contained nucleic acids was made in the 1960s and the development of cloning and sequencing techniques aided in a much better understanding of these organelles [3, 4]. Despite their primary role in ATP generation, mitochondria also play critical roles in aspects of cellular signaling, fatty acid oxidation, calcium signaling, heme biosynthesis, and the assembly of iron-sulfur clusters in proteins [5–7]. These essential functions make mitochondria indispensible for cellular function; however, the first example of a eukaryotic microorganism that lacks mitochondria was recently reported [8]
The circular ~16.5 kb human mitochondrial DNA (mtDNA) is maternally inherited and encodes for 13 polypeptides, 22 tRNAs, and 2 rRNAs that participate in oxidative phosphorylation (OXPHOS) via the electron transport chain (ETC) [9]. Unlike nuclear DNA, which is packaged into nucleosomes, mtDNA molecules are tightly associated with the mitochondrial matrix and form compact structures called nucleoids [10]. Nucleoids are composed of mtDNA-protein complexes that include proteins involved in replication and transcription such as mitochondrial single-strand binding protein (mtSSB), DNA polymerase gamma (POLG), and mitochondrial transcription factor A (TFAM) [11].
Just like its nuclear counterpart, mtDNA is subjected to genotoxic assaults from exogenous sources such as exposure to chemotherapeutic drugs as well as from endogenous sources including reactive oxygen species (ROS) that form as byproducts of mitochondrial respiration [12]. While evidence suggests that mtDNA molecules are likely to be more susceptible to oxidized DNA damage than nuclear DNA owing to their proximity to sites of oxidative phosphorylation, our current knowledge on the extent of mtDNA damage is limited owing to the lack of experimental approaches to accurately detect oxidatively generated mtDNA damage [7, 12]. Several DNA repair mechanisms exist within a cell to restore DNA integrity and while these pathways have been extensively studied in the nucleus (reviewed in [13, 14]), the base excision repair (BER) pathway has been established as the primary repair pathway in the mitochondrion [15]. Evidence for DNA repair pathways occurring in the mitochondria has been presented where mtDNA repair enzymes are encoded by nuclear genes and translocate to the mitochondria [15, 16]. With the exception of the 13 mtDNA encoded polypeptides, the mitochondrial proteome comprises an estimated >1,500 proteins encoded by nuclear genes that are exported to the mitochondria for mtDNA maintenance [17, 18]. Multiple mitochondrial protein import pathways participate in protein translocation to the mitochondria [19]. For instance, many proteins possess a mitochondrial targeting signal (MTS) that enables the protein to be transported via translocases of the outer and inner membranes (TOM and TIM), while others rely on the redox-mediated MIA pathway for import into the mitochondria [18–20]. Web servers like MitoProt, MitoFates, and TPpred2 that predict mitochondrial localization by analysis of the primary amino acid sequence, are becoming useful tools to theoretically evaluate the probability of mitochondrial targeting of a protein [21–23].
Mitochondria are unique in their genome organization in that they can contain multiple copies of mtDNA molecules within a single mitochondrion. Therefore, mtDNA damage if left unrepaired can lead to mutations that could result in heteroplasmy, a condition where both undamaged and damaged mtDNA molecules co-exist within the same mitochondrion [24]. Mitochondrial mutations can cause coding errors in the 13 polypeptides involved in ATP generation via the ETC and in the 22 tRNA molecules encoded by the mitochondrial genome [25, 26]. Depending on the extent of DNA damage, heteroplasmy can lead to mitochondrial dysfunction and the fine threshold between normal mitochondria and those that develop into a disease state remains intriguing [27]. Several chronic human diseases including diabetes, aging, neurodegenerative disorders (such as Alzheimer’s and Parkinson’s disease), and cancer are believed to be associated with mitochondrial dysfunction [17, 26, 28–30]. Mitochondria signal and communicate with the rest of the cell through numerous signaling pathways in response to dysfunction caused by physiological stimuli, stresses, and biological events [31–33]. In this review, we discuss our current knowledge of mtDNA repair and highlight some of the signaling pathways that coordinate stress responses between the nucleus and mitochondria in the event of DNA damage.
Mitochondrial DNA genome maintenance
Mitochondrial antioxidant pathways present the first line of defense that protects mitochondrial genome integrity (reviewed in [34, 35]). These include superoxide dysmutases (SODs), and the glutathione (GSH) peroxidase, thioredoxin (Trx), and peroxiredoxin (Prx) pathways [36–39]. Manganese SOD, an example of a member of this family, is present in the mitochondrial matrix and converts superoxide (O2•−) to hydrogen peroxide (H2O2) [40]. The H2O2 generated can be neutralized to water by either GSH or the thiol-specific peroxidases, Prx [39]. If the H2O2 generated in the mitochondria is not neutralized to water, it can be readily converted to the hydroxyl radical (•OH), a strong oxidant, in the presence of Fe2+ via the Haber-Weiss Fenton reaction [41]. Since the mitochondria play a critical role in FeS cluster biogenesis and in iron homeostasis, these organelles are likely to be more susceptible to damage by H2O2 [12].
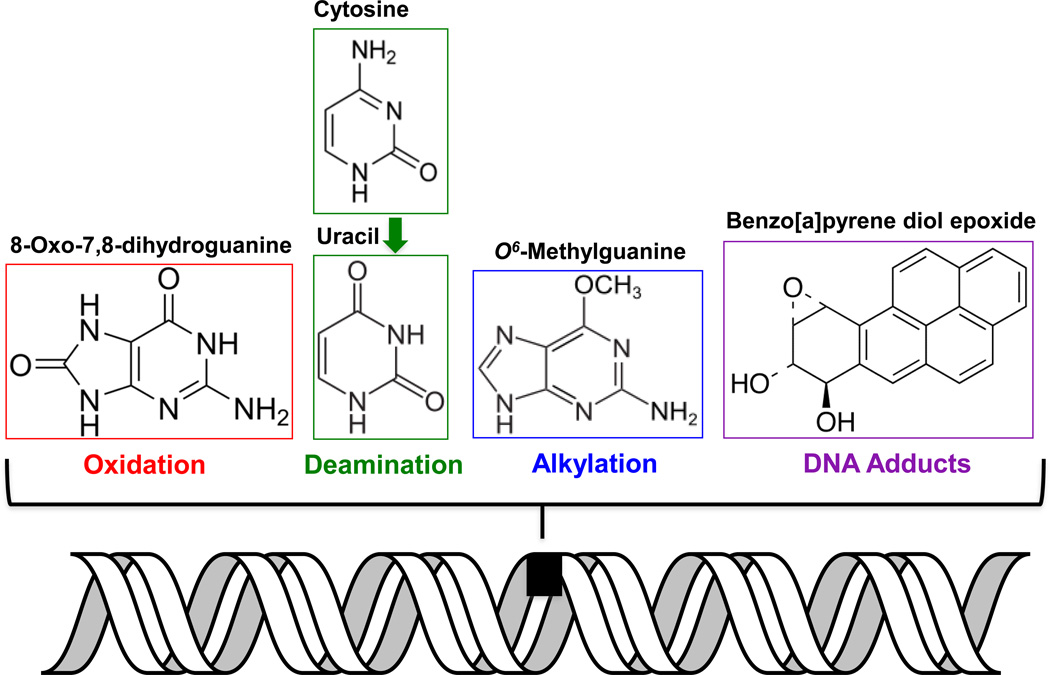
Oxidative stress results when the equilibrium between the antioxidant systems and ROS production is perturbed. Guanine has the lowest redox potential of the four DNA bases and is prone to oxidatively generated DNA damage produced by one-electron oxidants or singlet oxygen species [42–44]. The most common base modification that results from oxidation of guanine is 8-oxo-7,8-dihydroxyguanine (or 8-oxoG, Figure 1) [45, 46]. Thymine on the other hand is susceptible to •OH-mediated damage resulting in lesions such as 5,6-dihydroxy-5,6-dihydrothymine [43]. Other modifications including alkylation, bulky adducts, and deamination also occur within mtDNA (Figure 1). To combat DNA damage, like the nucleus, mammalian mitochondria also possess DNA repair pathways, which involve repair proteins encoded by nuclear genes that are transported to the mitochondria (see Table 1). Whether these repair enzymes pre-exist in mitochondria at the time of damage or translocate into the mitochondria in response to damage signals is still not clear. The DNA repair pathways in mammalian mitochondria include base excision repair (BER), direct reversal (DR), mismatch repair (MMR), translesion synthesis (TLS), and possibly double-strand break repair (DSBR) [16, 47–49]. Thus far, there is no evidence for the repair of helix-distorting bulky adducts and ultraviolet-induced photodimers by the nucleotide excision repair (NER) pathway in mitochondria [50]. However, in one study, XPD, an NER protein was seen to protect mtDNA from oxidative DNA damage [51]. These data need to be further clarified and substantiated [52]. Furthermore, bulky adducts such as benzo[a] pyrene diol epoxide that are also formed within mtDNA (Figure 1) have no known mechanism of repair within the mitochondria, and are susceptible to destruction by autophagy [17, 53]. Overall, of the DNA repair pathways available in the mitochondria, BER appears to be the major pathway for eliminating ROS-induced oxidative damage.
Figure 1. Types of DNA lesions found in the mitochondria.
Damage to mtDNA can occur in the form of alkylation, oxidation, spontaneous deamination, and bulky adducts. Examples of lesions from each of these categories are shown.
Table 1.
Mammalian proteins and enzymes involved in the repair of mtDNA
| Pathway | Protein/Enzyme | Function | Reference |
|---|---|---|---|
| BER | NEIL1 | DNA glycosylase (Bifunctional) | [241] |
| NEIL2 | DNA glycosylase (Bifunctional) | [69] | |
| OGG1 | DNA glycosylase (Bifunctional) | [242, 243] | |
| NTH1 | DNA glycosylase (Bifunctional) | [242] | |
| MUTYH | DNA glycosylase (Monofuctional) | [242] | |
| AAG | DNA glycosylase (Monofuctional) | [244] | |
| UNG | DNA glycosylase (Monofuctional) | [60, 245] | |
| TFAM | Mitochondrial transcription factor A stimulates glycosylase activity |
[88] | |
| TDP1 | DNA end processing | [87] | |
| PARP1 | Senses SSBs, PARylates itself and other proteins |
[82, 84] | |
| APE1 | DNA end processing endonuclease | [68] | |
| PNKP | DNA end processing | [67, 69] | |
| APTX | DNA end processing | [85] | |
| POLG | Mitochondrial DNA polymerase | [246] | |
| FEN1 | Flap endonuclease, flap processing | [75] | |
| EXOG | Flap processing | [77] | |
| LIGIII | DNA ligase | [133] | |
| DR | MGMT | Methytransferase | [98, 100] |
| MMR | YB-1 | Mismatch binding and repair | [104, 105] |
| MLH1 | Mismatch binding and recruitment of downstream proteins |
[106] | |
| TLS | PrimPol | DNA primase-polymerase | [117] |
| Polymerase zeta | Error-prone B family DNA polymerase | [116] | |
|
DSBR |
RAD51 | Central catalyst of HR | [247] |
| XRCC3 | Participates in HR and maintains chromosome stability |
[247] | |
| BRCA1 | DNA binding and protein interactions | [248] | |
| Ku70/80 | DSB binding during NHEJ | [249, 250] | |
| MRE11 | mtDNA binding and end processing | [251] | |
| NER | NONE to DATE | ||
Base excision repair (BER)
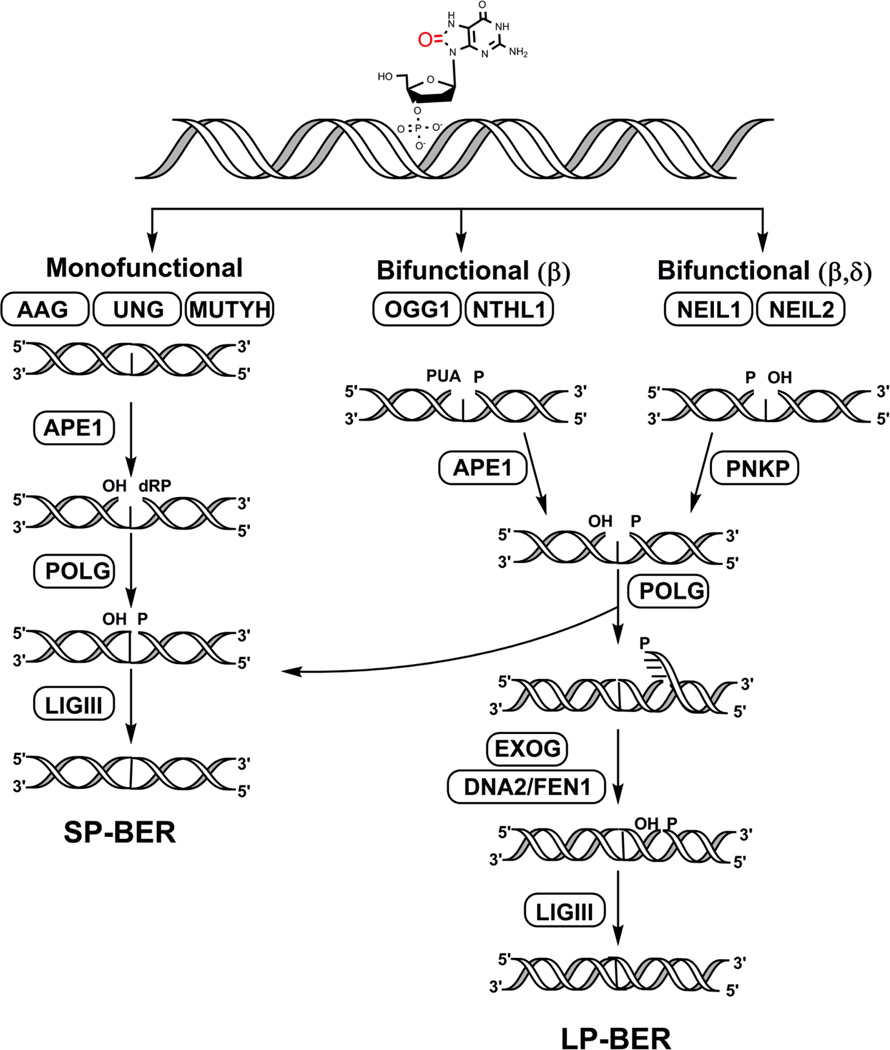
The BER pathway is a well-characterized, tightly-coordinated process that is carried out in a step-wise manner and includes: recognition and excision of the damaged DNA base, removal of the resulting abasic (apurinic/apyrimidinc or AP) site, end processing, gap filling, and ligation (summarized in Figure 2) [16, 48, 54]. BER can proceed via three sub-pathways, short-patch repair (SP-BER, 1-nt), long patch repair (LP-BER, 2 or more nt), and single-stranded break repair (SSBR) (extensively reviewed in [55–57]).
Figure 2. Overview of base excision repair in the mitochondria.
This figure displays the step-wise repair of DNA base damage via the BER pathway by enzymes identified in the mitochondria. Initially, the lesion is recognized by either mono or bifunctional DNA glycosylases depending on the type of damage. Of the eleven known mammalian DNA glycosylases, only seven have been identified in the mitochondria. These include: the monofunctional glycosylases AAG (alkyladenine DNA glycosylase), UNG (uracil N-glycosyalse), and MUTYH (MutY glycosylase homolog) as well as the bifunctional glycosylases OGG1 (8-oxoG DNA glycosylase 1), NTHL1 (Nth-Like 1), NEIL1 (Nei-like 1), and NEIL2 (Nei-like 2). The resulting AP sites are further processed either by APE1 in case of the monofunctional, OGG1, and NTHL1 glycosylases or by PNKP that processes the ends after the NEIL enzymes, thereby leaving suitable ends for gap-filling by POLG. Ligase III then seals the DNA nick and completes the process in SP-BER. In the long-patch (or LP, >2 or more nt) repair pathway, a 2–6 nt flap is generated by POLG that is further processed by DNA2/FEN1. Alternatively, EXOG may function as the major 5’ flap-processing enzyme in the mitochondria in both SP- and LP-BER. The final ligation step is carried out by LIGIII.
The initiation step of BER is carried out by DNA glycosylases, specialized enzymes, which recognize the damaged base and catalyze the cleavage of the N-glycosyl bond between the damaged base and its 2-deoxyribose resulting in an AP site [58, 59]. Glycosylases are either monofunctional or bifunctional, depending on whether they possess an intrinsic lyase activity. Seven of the eleven known mammalian DNA glycosylases contain an MTS that allows them to translocate to the mitochondria (summarized in Table 1 and Figure 2) [54]. The translocation of some DNA glycosylases to their respective nuclear and mitochondrial compartments is enabled by mechanisms of alternative splicing and different transcription start sites as observed with the uracil-DNA glycosylase (UDG), 8-oxoG DNA glycosylase 1 (OGG1), and MUTYH DNA glycosylase [60–62]. Monofunctional glycosylases excise non-oxidized damaged bases and rely on AP endonuclease (APE1) to complete the lyase elimination reaction whereas bifunctional DNA glycosylases are involved in the removal of oxidized DNA bases and nick the DNA backbone 3’ to the lesion [63, 64]. End processing following base excision and backbone cleavage by bifunctional glycosylases either involves APE1 as is seen in the case of OGG1 and NTHL1 (Nth-like 1) or polynucleotide kinase phosphate (PNKP) that processes the ends generated by the NEIL (Nei-like) enzymes [64, 65]. Both APE1 and PNKP have been identified in the mitochondria and play critical roles in mitochondrial BER [66–69]. DNA POLG, the primary polymerase in the mitochondrion, is responsible for the gap-filling synthesis step in the mitochondria [70, 71]. During SP-BER, POLGA, the catalytic subunit of heterotrimeric POLG, possesses lyase activity and is able to excise the 5’-phosphodeoxyribose (dRP) moiety generated by APE1 and fills the 1-nt gap [72]. During LP-BER, the strand displacement synthesis activity of POLGA is utilized, which generates flaps of 2–6 nt that are good substrates for the flap endonuclease 1 (FEN1) [73–75]. Although early reports suggested that SP-BER was the primary mitochondrial BER pathway, the choice between LP-BER and SP-BER is determined by several factors that are present in both the nucleus and mitochondria [64, 74]. For instance, certain types of oxidized abasic sites (such as 2-deoxyribonolactone) that are not suitable substrates for POLG undergo FEN1-dependent LP-BER [75]. Another enzyme DNA2, also found in mitochondrial extracts, assists in the FEN1-mediated cleavage of the DNA flaps by maintaining the length of single-stranded flap so that it is not immediately coated by mtSSB [76]. In contrast, another study demonstrates that 5’-exo/endonuclease (EXOG) removes 5’ flaps during mitochondrial LP-BER independently of FEN1 and DNA2 [77]. The final ligation step in the mitochondria, which is common in all BER sub-pathways, is carried out by DNA ligase III (LigIII) [78, 79].
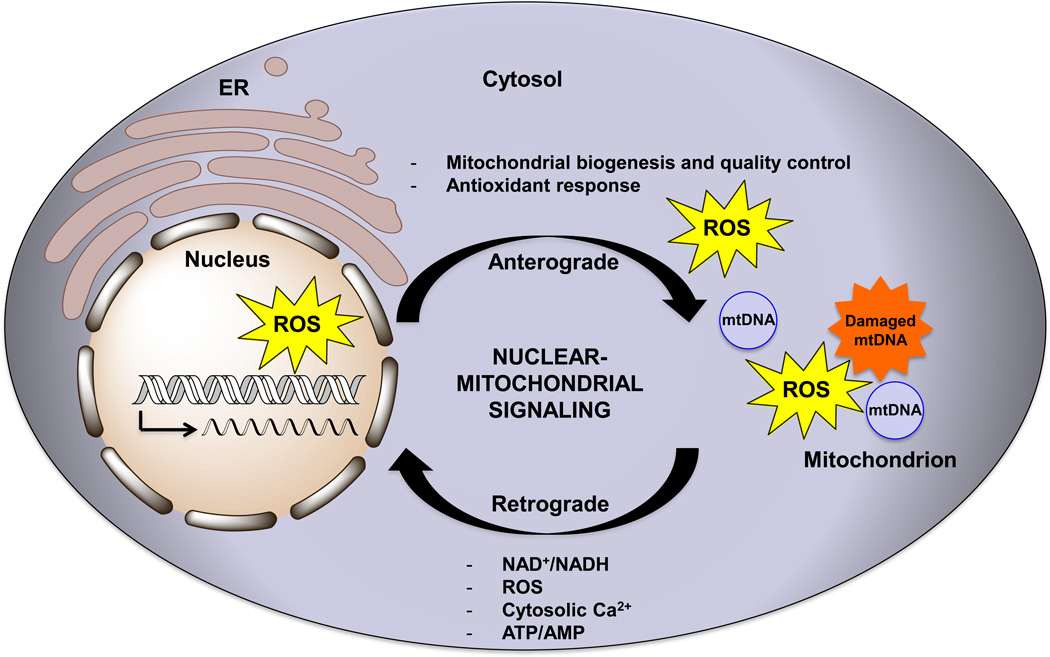
Figure 3. Signaling between the nucleus and mitochondria.
Crosstalk between the nucleus and mitochondria occurs not only to signal oxidative stress, DNA damage, and mitochondrial dysfunction but also occurs during normal cellular metabolism. The flow of information from the nucleus to the mitochondria (termed anterograde signaling) involves the transcription and translocation of genes involved with mitochondrial biogenesis. Anterograde regulation also includes responses to stressors that trigger an antioxidant response by regulating the expression of genes involved with Ca2+ metabolism and glycolysis. Mitochondria can signal to the nucleus (in a process called retrograde signaling) in times of stress via signals such as changes in the levels of NAD+/NADH, ROS, cytosolic Ca2+, and ATP/AMP as well as changes in membrane potential. ER stands for endoplasmic reticulum. The arrow within the nucleus signifies the transcriptional activation of nuclear genes either during normal conditions, or upon stress induced signaling.
The SSBR pathway is considered to be a form of SP-BER that involves detection of SSBs, end processing, gap filling, and ligation where the later two steps follow the same path as SP-BER [48, 80, 81]. The SSBR pathway in the nucleus is initiated by the detection of SSBs by enzymes of the poly (ADP ribose) polymerase family such as PARP1 that has been shown in some studies to localize to the mitochondria [82]. The exact role of PARP1 in mtDNA SSBR remains unknown partly due to conflicting reports describing the effects of PARP1 depletion on the accumulation and repair of mtDNA damage [82–84]. Although the BER enzymes APE1, and PNKP, discussed above, can perform end processing, tyrosyl-DNA phosphodiesterase 1 (TDP1), and aprataxin (APTX) have also been implicated in the end processing steps of mitochondrial SSBR (Table 1) [85–87]. TDP1 activity in the mitochondria is required to hydrolyze the bond between a tyrosyl moiety and a 3’ DNA end as well as for hydrolyzing other 3’-end alterations such as 3’-phosphoglycolate that are poor substrates for APE1 [87]. ATPX catalyzes the removal of chemically adducted 5’-AMP moieties generated by aborted DNA ligation reactions and its importance is highlighted in human neuroblastoma cells where depletion of APTX resulted in mitochondrial dysfunction and an increase in mtDNA damage [85, 86].
Factors other than the core BER enzymes also play a role in mitochondrial BER. For instance, mitochondrial TFAM possesses a greater affinity for oxidized lesions like 8-oxoG and inhibits the activity of OGG1, uracil-DNA glycosylase (UNG), and APE1 [88]. Furthermore, the tumor suppressor p53 is able to bind to TFAM and alter its DNA-binding so that the BER glycosylases are no longer inhibited. OGG1 also interacts with the Cockayne Syndrome A and B proteins (CSA and CSB), which may localize to the mitochondria under oxidative stress [89]. Given that BER is the major repair pathway in the mitochondria, disease states associated with mutations and deletions in the BER machinery result in aging-related neurodegenerative disorders, ataxia, diabetes, and cardiomyopathy reviewed elsewhere [13, 80, 90–92].
Direct reversal (DR)
The DR pathway is a simple one-step repair pathway that is responsible for the repair of lesions such as cyclobutane pyrimidine dimers and O6-alkylguanines. In bacteria, the enzyme photolyase is responsible for the removal of cyclobutane pyrimidine dimers and while this enzyme has been identified in plant and yeast mitochondria, no evidence for mammalian homologs has been presented [93, 94]. In mammalian cells, direct reversal of O6-methylguanine to guanine is carried out by methylguanine DNA methyltransferase (MGMT; Table 1) [95–97]. It has been suggested that a methyltransferase with a similar molecular weight as nuclear MGMT may exist in mitochondria, but these results have yet to be further substantiated [16, 95, 98–100].
Mismatch repair pathway (MMR)
The MMR pathway is a highly conserved repair process involved with the recognition and removal of mismatched bases and slippage errors caused by DNA polymerase during replication (reviewed in [101, 102]). The eukaryotic MSH (MSH2-6) or MutS homolog, and MUTL (MLH1,3) or MutL homolog proteins, are key components of this pathway in the nucleus where MMR ensues with the recognition of DNA mispairs by the MSH proteins followed by recruitment of the MLHs and activation of downstream molecules to repair the mismatch [102, 103]. Mitochondrial MMR appears to occur independently of the MSH and MLH proteins but relies instead on the Y-box binding protein (YB-1; Table 1) [104, 105]. While there is precedence for the presence of MLH1 in the mitochondria, further evidence is warranted to fully understand the role of these enzymes in the mitochondria [106].
Translesion synthesis (TLS)
TLS is an error-prone DNA damage tolerance pathway that utilizes a group of specialized DNA polymerases that can bypass DNA lesions and allow DNA replication to proceed [107, 108]. In eukaryotic nuclei, five polymerases perform TLS including Rev1, polymerases kappa, eta, and iota (Polκ, Polη, and Polι) that belong to the Y-family of polymerases and polymerase zeta (Polζ), which is a B-family polymerase [108]. The recruitment of these polymerases to the site of a lesion is described by two well-supported models, where one model proposes a “polymerase switching” mechanism whereas the other involves a “gap-filling” model (reviewed in [107–110]. Yet, how cells decide which TLS polymerase to use remains one of the most perplexing questions in this field.
In the mitochondria, POLG exhibits TLS activity and is able to bypass lesions such as cyclobutane pyrimidine dimers by incorporating purines opposite the lesion [111]. POLG is also able to bypass 8-oxoG and exocyclic DNA adducts that gives rise to increased mutagenicity [112, 113]. Both Polζ and Rev1 have been identified to have a role in yeast mitochondrial genome maintenance where overexpression of these enzymes led to a reduction in point mutations in the mtDNA [114, 115]. More recently, Polζ was detected in the mitochondria of mammalian cells and elevated expression of the enzyme in breast tumors was associated with cell migration and invasion [116]. PrimPol, a DNA primase-polymerase, was identified in the mammalian mitochondria [117] and evidence for its ability to bypass DNA lesions such as 8-oxoG was presented [118]. A conflicting report suggests that while PrimPol is unable to promote DNA damage bypass at a replication fork, its interactions with proteins involved in mtDNA replication, such as the Twinkle helicase, may be important for mtDNA metabolism [119].
Double-strand break repair (DSBR)
DNA double-strand breaks (DSBs) that occur in nuclear DNA are repaired by homologous recombination (HR), microhomology-mediated end-joining (MMEJ), and non-homologous end joining (NHEJ) pathways (reviewed elsewhere [48, 120]). While HR-mediated repair of mtDNA has been reported in plants and yeast, the occurrence of a bonafide HR repair mechanism in mammalian mitochondria has been debated and remains to be elucidated [121–125]. Some evidence for HR occurring within mammalian mitochondria was presented in studies using mitochondrial extracts where a recombinase RecA-mediated recombination event was observed [126]. The presence of recombination intermediates in mtDNA from human heart muscle were reported and suggest that an HR-like mechanism may occur in these cells [127]. Other studies revealed that if recombination events do occur, they do so at a low frequency, primarily under conditions where mtDNA is first depleted by treatment with either ethidium bromide or restriction endonucleases [128, 129].
NHEJ and MMEJ are relatively more error-prone when compared with the HR pathway and result in large deletions in the DNA. In yeast, direct-repeat-mediated deletions were seen to accumulate in mtDNA thereby implicating the existence of the DSB repair pathways in the mitochondria [130, 131]. Another report in yeast also indicated that NHEJ proteins such as the MRX (Mre11p, Rad50p, Xrs2p) and Ku70/80 (Ku70p, Ku80p) complexes were observed in the mitochondria at the site of induced DSBs [132]. End-joining activity of linearized mtDNA substrates was observed using mammalian mitochondrial protein extracts where most of the molecules were end joined accurately, but regions containing direct repeat sequences displayed deletions [133]. While some mammalian DSB proteins have been observed in the mitochondria (Table 1), overall, it appears as though DSB repair in mammalian mitochondria may exist as a minor pathway under certain conditions, and further evidence is required to support the existence of these pathways within this organelle.
Signaling between the nucleus and mitochondria
For efficient DNA repair to take place in the mitochondria, DNA repair proteins are first transcribed in the nucleus, synthesized as precursors in the cytosol, and then imported through the outer mitochondrial membrane aided by the TOM translocase complex [19]. Further transport into the inner membrane of the mitochondria is aided by the translocase of the inner membrane (or TIM23 complex) [19]. Since a majority of the mitochondrial proteome comprises genes synthesized by the nuclear machinery, the flow of information from the nucleus to the mitochondria can be described as a way for the nucleus to inform the mitochondria about changes in the cellular environment, energetic requirements, and to signal stress responses [134, 135]. The stream of information from the nucleus to the mitochondria is termed anterograde signaling and the reverse process is called retrograde signaling or mitochondrial stress signaling (summarized in Figure 3) [136–138]. Gene expression of mitochondrial-targeted proteins drives mtDNA replication, maintenance, and energy homeostasis [135, 137, 139]. In return, mitochondrial dysfunction resulting from loss of mtDNA, damage accumulation, and oxidative stress trigger signals (such as elevated Ca2+ and NAD+ levels) that then orchestrate a nuclear gene expression response to mitigate the defect [138]. In mammalian cells, the first report of inter-organellar signaling between the nucleus and the mitochondria was described by differential expression of respiratory chain complexes between wild-type and mtDNA depleted fibroblasts [139, 140]. Signaling between the two organelles is essential for deciding cellular fate and for the remainder of this review, we will discuss some of the damage sensors, metabolite signals, and pathway cascades that are triggered upon DNA damage.
Ca2+ signaling
The importance of calcium (Ca2+) uptake by mitochondria is known to have critical functions in metabolism, stress signaling, and cell survival (reviewed in [141, 142]). Stressors such as depletion of mtDNA and treatment with ionophores result in perturbation of membrane potential (ΔΨm) thereby causing a change in Ca2+ efflux [143, 144]. This was observed in work performed in mouse myocytes where elevated cytosolic Ca2+ levels prompted a nuclear response that included reduced levels of nuclear factor-κB (NF-κB) and activation of the calcineurin dependent nuclear factor of activated T-cells (NF-AT) [144]. These responses in turn stimulate other transcription factors that drive nuclear transcription of genes involved in Ca2+metabolism and glycolysis. Notably, restoration of mtDNA resulted in the reversal of these signals, thereby strengthening the relationship between mtDNA stress and nuclear transcription [144, 145]. Elevated calcium levels resulting from mtDNA damage can also directly influence pathway activation including protein kinase C (PKC), and c-Jun N-terminal kinase (JNK)/p38 as well as activation of Ca2+/calmodulin-dependent protein kinases (CAMKIV) that modulates phosphorylation of the cAMP-responsive element-binding protein (CREB) [146, 147]. Both NF-κB and the JNK signaling pathways have been linked to the DNA damage response and are involved with the post-translational modifications of DNA repair proteins [148, 149]. It still remains to be elucidated whether activation of these pathways influences import of DNA repair genes into the mitochondria.
Signaling mediated by NAD+/ NADH
Nicotinamide adenine dinucleotide (NAD) in its oxidized (NAD+, electron acceptor) and reduced (NADH, electron donor) forms is an essential metabolite and coenzyme that is involved in the regulation of pathways such as glycolysis, the ETC, and the tricarboxylic acid (TCA) cycle [150–152]. NAD+ content appears to be unevenly distributed throughout the cell with the lowest concentrations observed in the nucleus in comparison to the mitochondria and cytosol [151, 152]. NAD+ is a substrate for two families of enzymes that serve critical roles in the DNA repair response: the PARP family and the sirtuin family of NAD+-dependent deacetylases (SIRTs) [7, 153–155]. Three of the 17-membered PARP family of enzymes (PARPs1–3) have known roles in DNA repair where PARP1 and PARP2 play a role in BER, while PARP3 senses DSBs and is involved in DSBR [81, 156–159]. The PARP enzymes synthesize poly(ADP-ribose) or PAR by utilizing NAD+ as a substrate to transfer the ADP-ribose moiety of NAD+ to acceptor proteins. This type of post-translational modification occurs when nuclear DNA damage in the form of DNA strand breaks (SSBs and DSBs) is sensed. Both PARP1 and PARP2 auto PARylate themselves, interact with the DNA, and further recruit and PARylate other repair enzymes [157]. This consumption of NAD+ drives NAD+ synthesis presumably by either the de novo synthesis pathway or the salvage pathway to keep up with cellular demand [160]. Localization of PARP1 to the mitochondria has been reported but its function in this organelle is still a matter of debate [82]. PARP1 appears to have an inhibitory negative role in the mitochondria where it interacts with and PARylates POLG, and EXOG and inhibits BER, a function that is opposite to its roles in the nucleus [83].
The sirtuin family of proteins use NAD+ as a substrate to deacetylate proteins by the removal of acetyl groups from lysine residues [161]. Of the 7 sirtuin family members, SIRT1, SIRT6, and SIRT7 are localized in the nucleus, SIRT2 is mainly present in the cytoplasm and SIRT3, SIRT4, and SIRT5 are mitochondrial proteins [153, 161]. Despite this compartmental segregation, these enzymes can shuttle between the two organelles and the cytosol in response to cellular stimuli. SIRT1, the most studied sirtuin enzyme, plays a role in mitochondrial biogenesis, where it deacetylates key proteins such as peroxisome proliferator-activated receptor-γ co-activator 1α (PGC-1α), and in DNA repair via DSBR by deacetylating and activating proteins such as Ku70 [162–164]. Another target of SIRT1 deacetylation is the hypoxia inducible factor 1 alpha (HIF-1α), which belongs to the hypoxia-inducible transcription factor family of proteins that are regulators of cellular responses to oxygen deprivation [165, 166]. This SIRT1-mediated deacetylation event inactivates HIF-1α signaling under normal conditions, but allows for activation of this pathway when SIRT1 levels are low and under hypoxic conditions where NAD+ levels are also diminished [166]. Since both PARP1 and SIRT1 are involved in DNA repair and use NAD+ as a substrate for their respective activities, they compete for NAD+ in an interplay that is not completely understood. Furthermore, the rivalry between SIRT1 and PARP1 is also observed in NF-κB regulation where SIRT1 deacetylates the transcription factor and inhibits its activity whereas PARP1 activates and stimulates NF-κB activity [167]. Recent evidence presented in pulmonary artery endothelial cells indicates that an increase in Ca2+ levels in the mitochondria influences the NAD+/NADH ratio in the cytoplasm by causing an efflux of NADH from the mitochondria to the cytosol [168]. This change influences the NAD+ consuming SIRT family of enzymes, and provides a link between Ca2+ fluctuations, NAD+ levels, DNA damage, and mitochondrial dysfunction [168].
Energy homeostasis mediated by AMP-activated protein kinase signaling
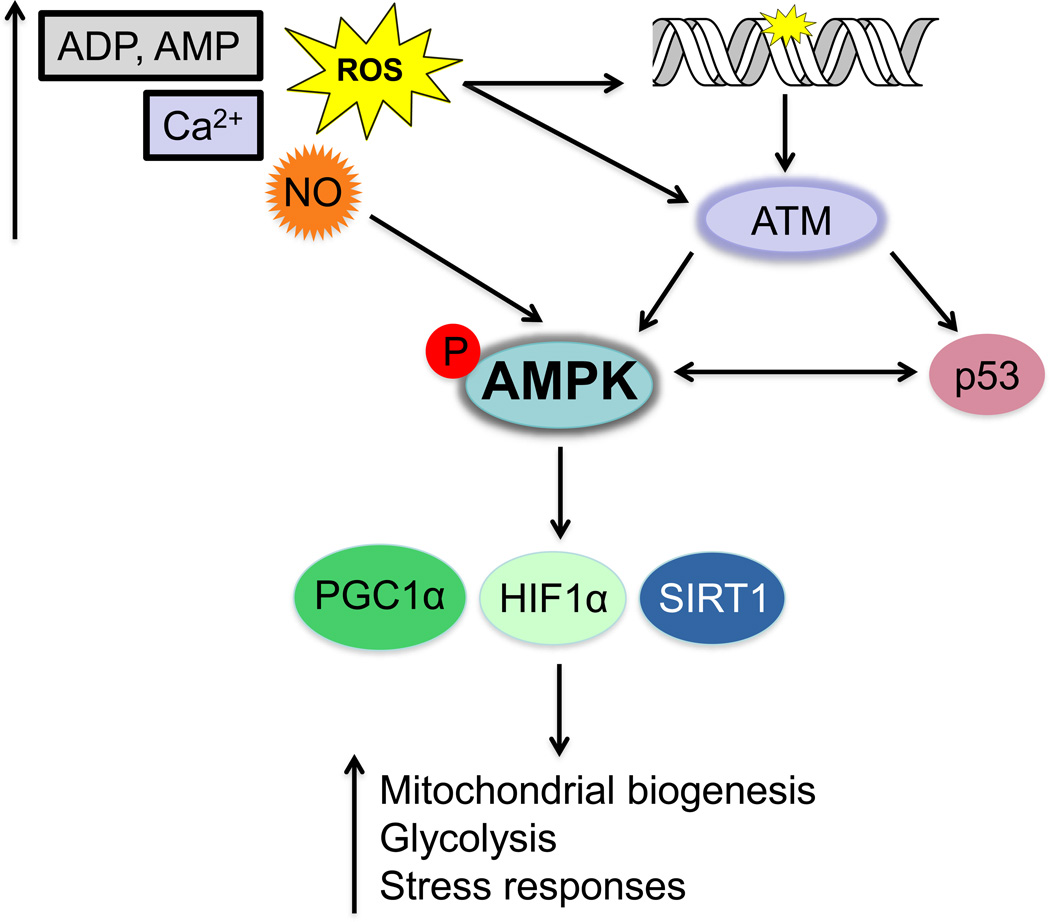
AMP-activated protein kinase, AMPK, is referred to as the energy sensor of the cell as it is highly sensitive to changes in intracellular energy levels (or the ATP:AMP ratio) [169]. AMPK is a serine/threonine kinase that is activated in response to increased levels of AMP and ADP and triggers a cascade of orchestrated responses that are able to restore ATP levels and maintain energy balance [169, 170]. To mention two examples, both p53 and the oxidative stress/ DNA damage sensor protein, ataxia telangiectasia mutated (ATM), can phosphorylate and activate AMPK (Figure 4) [171–173]. AMPK is activated by the Sestrin 1 and 2 proteins, which are targets of p53 activation upon genotoxic stress [172]. Incidentally, p53 is also a target of AMPK phosphorylation where this phosphorylation event triggers several outcomes including cell cycle-dependent checkpoint activation, and accumulation of mitochondrial p53 that promotes apoptosis via the Bak-Bcl-xL complex [174, 175]. ATM can phosphorylate AMPK in response to oxidative stress via the liver kinase B1 tumor suppressor protein [173, 176]. Phosphorylated AMPK in turn can phosphorylate a number of factors involved in nuclear-mitochondrial signaling responses including SIRT1, PGC-1α, and HIF-1α (Figure 4) [177–179]. Additionally, the mitochondrial SIRT4 protein regulates cellular ATP levels thereby contributing to mito-nuclear crosstalk via AMPK-mediated signaling [180]. These studies coupled with reports describing the role of redox molecules in triggering AMPK response mechanisms, further strengthens the importance of this signaling pathway in the maintenance of cellular homeostasis [169].
Figure 4. Schematic of AMP-activated kinase (AMPK) signaling triggered by changes in cellular metabolites and DNA damage.
AMPK is activated upon increased levels of AMP/ADP, ROS, and cytosolic calcium. Activation of AMPK can be triggered by the ataxia telangiectasia mutated (ATM) kinase that is activated upon oxidative stress or DNA damage. ATM triggers a DNA damage response that activates other factors such as p53 that can also directly activate AMPK. The activation of AMPK elicits downstream responses regulated by proteins including PGC-1α, SIRT1 and HIF-1α. These responses manifest in an upregulation of mitochondrial processes such as glycolysis, fatty acid oxidation, and responses to hypoxia.
Redox signaling: Reactive oxygen, nitrogen species, and APE1
Both ROS and reactive nitrogen species (RNS) are highly reactive and are generated during normal cellular metabolism as byproducts of processes such as respiration, and by enzymatic processes involving nitric oxide synthases (NOS), and NADPH oxidase [181]. Some forms of ROS including •OH and singlet oxygen species are highly reactive and are able to react with and damage DNA bases whereas H2O2 in the presence of metals (such as Fe2+) generates •OH via the Fenton reaction [43, 182]. Several antioxidant mechanisms (discussed above) exist to combat ROS including natural antioxidants such as vitamins A, E, and C obtained from the diet and endogenous enzymes like glutathione peroxidases and superoxide dismutases, which reduce free H2O2 to H2O and cause the dismutation of O2•− to H2O2 and O2, respectively [183, 184]. Increased levels of ROS can overwhelm the antioxidant responses and trigger the activation of transcription factors including NF-κB, HIF-1α, p53, and nuclear factor E2-related factor 2 (NRF2) [185–189]. An example of signaling between the mitochondria and nucleus under conditions of high levels of ROS is witnessed by the stimulation of over 100 target genes such as glutathione S-transferases mediated by the binding of NRF2 to antioxidant response elements [189]. Furthermore, the triggering of TFAM expression for driving mitochondrial transcription by NRF1 occurs in response to increased levels of ROS [190].
RNS such as nitric oxide (NO), which is biosynthesized by NOS readily diffuse across cell membranes and function as signaling molecules between the nucleus and mitochondria [191, 192]. Mitochondria also possess NOS (mtNOS) that is capable of generating NO in the inner mitochondrial membrane and modulates diffusion of NO from the mitochondria to the cytosol [193, 194]. NO can impart toxicity in a cell via its oxidation products that damage DNA as well as by the inhibition of mitochondrial respiration [195]. NO reacts with O2•−to produce peroxynitrite (ONOO−), which can lead to the nitrosative deamination of DNA bases, nitrated DNA bases (8-nitroguanine), and the nitration of tyrosine residues that blocks signaling cascades [195]. Mitochondrial NO binds to and inhibits cytochrome C oxidase (Complex IV) thereby modulating respiration by directly competing with intracellular O2 [196, 197]. One way that NO mediates mito-nuclear signaling under conditions of oxygen deprivation, is by destabilizing HIF-1α, which inhibits its transcriptional activity [198]. Furthermore, an increase in NO signaling correlates with an elevated number of mitochondria and is consistent with an upregulation in the levels of the mitochondrial biogenesis master regulator, PGC-1α that mediates downstream activation of factors such as TFAM [199, 200]. This increase in PGC-1α regulation by NO can also be influenced by the binding of NO to AMPK (Figure 4) [201] and is particularly useful for scrutinizing mitochondria-rich tumors such as thyroid oncocytoma [202]. While increased RNS levels can elicit harmful effects in a cell, their role in immune responses and as anti-tumor agents has been previously described [203].
Redox activities are not limited to ROS and RNS. Incidentally, the only known DNA repair protein that is able to regulate other proteins via a redox mechanism is APE1 [204, 205]. As a DNA repair protein, APE1 functions in the BER pathway (discussed above), and as a redox regulator, it plays roles in processes such as immune inflammatory responses, angiogenesis, and in tumor progression [206]. Cysteine residues 65, 93, and 99 have been implicated in mediating the redox activities of APE1 [207–209]. Cys 65 and 93 are not solvent exposed residues based on the crystal structure of APE1, implicating that a conformational change has to occur within APE1 in order for these cysteine residues to serve as nucleophiles during the redox process [210]. APE1 maintains downstream targets such as HIF-1α [211], NF-κB [212], and p53 [213] in their reduced forms to promote stabilization and enhanced DNA binding. APE1 can also negatively regulate the activity of the NRF2 protein that is involved in the oxidative stress response, thereby serving a critical role as a regulator in DNA repair, transcriptional control, and genome maintenance [214]. Thioredoxin, the antioxidant enzyme interacts with APE1 and mediates its redox activities by reducing the oxidized form of APE1 [215]. The importance of APE1 as an essential regulator of cellular processes is evidenced by the fact that knockdown of APE1 in mice leads to embryonic lethality [216]. Although the mitochondrial translocation of APE1 has been documented previously, recent evidence suggests that APE1 translocates to the mitochondria in a redox-dependent manner via interaction with Mia40, the mitochondrial import protein that is also used by p53 to translocate to the mitochondria [217, 218].
Mitochondrial unfolded protein response-like pathway
The maintenance of protein homeostasis via the unfolded protein response (or UPR) is tightly controlled in the cytosol, endoplasmic reticulum, as well as in the mitochondria and involves crosstalk between these organelles and the nucleus [219–221]. Mutations and deletions in mtDNA yield misfolded or mutant proteins that are unable to form stoichiometric complexes with the nuclear encoded proteins being transported to the mitochondria for OXPHOS. This can place a large amount of stress on the protein folding machinery in the mitochondria [220]. The UPR in mitochondria (UPRmt) has been studied extensively in yeast and in C. elegans and is reviewed in [219, 222, 223]. In C. elegans, UPRmt is triggered when ROS levels increase and involves expression of nuclear genes such as the heat shock proteins (hsp) hsp-6 and hsp-60, which encode for chaperone proteins [224]. In mammalian cells there are two modes of UPRmt that have been proposed in order to understand the key players involved in this process. The first pathway described by Hoogenraad and colleagues describes a UPRmt where unfolded proteins accumulate in the mitochondrial matrix and result in the expression of nuclear encoded proteins including HSP60 (chaperone 60), and the ClpP mitochondrial protease [225]. This process is mediated by activation of the CHOP (C/EBP homology protein) transcription factor [226]. The second CHOP-independent UPRmt model was described by Germain and colleagues in MCF-7 mammalian cells where protein aggregates accumulate in the inter-membrane space and trigger a response involving estrogen receptor α [227, 228].
Another level of regulation that occurs with the accumulation of misfolded proteins, depletion of mitochondrial chaperone proteins, or disruption of mitochondrial membrane potential, is mitochondrial autophagy (or mitophagy) [229]. In normal mitochondria, the multifunctional PINK1 kinase that regulates mitophagy is imported into the mitochondrial matrix via an MTS and is degraded by proteases [230, 231]. However, under conditions of inner membrane stress, PINK1 cannot cross the inner membrane and instead is integrated into the outer membrane where it recruits the ubiquitin ligase Parkin [232]. Parkin is phosphorylated and activated by PINK1 and is able to ubiquitilate multiple mitochondrial proteins that further leads to mitophagy [233]. However, while much is known about the unfolded protein response and mitophagy, the precise mechanisms of these processes still need to be resolved since many disease states have been attributed to a malfunction in UPRmt signaling and mitophagy including neurodegenerative disorders such as Parkinson’s disease, aging, and Friedreich’s ataxia [30, 222, 234].
Concluding remarks and unanswered questions
In this review, we have summarized the current knowledge of the DNA repair mechanisms that exist in the mitochondria and highlighted some of the signals that connect the two organelles during stress responses. Despite the vast body of literature on this subject, numerous questions still remain to be answered [7]. In terms of DNA repair, although we know that nuclear genes are responsible for the maintenance of the mitochondrial genome, how and when these proteins are targeted to the mitochondria remains unclear. Furthermore, the chaperone proteins responsible for the proper folding and further processing of these enzymes are still largely unknown. The role of post-translational modifications of DNA repair proteins either within the mitochondria, or in the cytosol that enables mitochondrial targeting, also remains an area of interest.
From our current understanding of mitochondrial biology, it is evident that numerous factors are responsible for the well-being and proper functioning of mitochondria. As presented here by us and in reports by other groups, the signaling cascades that occur between the mitochondria and the rest of the cellular milieu, represent a “web” of interactions that are interconnected and influence each other. When one of these pathways is disrupted, in the case of mitochondrial stress, we can easily fathom progression of a disease state or the triggering of pathways that lead to apoptosis. The recent development of mitochondrial-nuclear exchange (MNX) mice has helped us cultivate a better understanding of the contribution of damaged, mutated, and variant mtDNA to disease progression [235]. In these mice, the nuclear and mitochondrial genomes from different mice strains were interchanged, and from these animal models, and subsequent studies, it was evident that mtDNA contributes to increased levels of ROS, vulnerability to cardiovascular stress, and changes in lipid metabolism in chronic liver disease, independently of nuclear DNA [235, 236]. Furthermore, the importance of mtDNA repair is highlighted in studies that indicate that some repair enzymes are transported to the mitochondria not only to maintain mitochondrial genome integrity, but also to protect against tissue injury such as ventilator induced lung injury in mice [237–239]. In other animal models, a neuroprotective role was suggested for mitochondrial BER enzymes where increased levels these enzymes protected against ischemic brain injury [240]. It is therefore apparent that mtDNA serves a protective role in controlling cell fate in response to a variety of oxidative stresses and is essential for proper cellular maintenance and function. Unfortunately, in this review, we were unable to cover the all facets of mitochondrial signaling including mitophagy, apoptosis, mitochondrial fusion and fission, and mitochondrial diseases that result from dysfunction. In summary, despite the wealth of knowledge that exists in the fields of DNA repair and mito-nuclear signaling, much is left to be determined to deconvolute the complex array of interactions that maintain cellular homeostasis.
Acknowledgments
We would like to thank Dr. Peter Sykora, Dr. Ania Wilk, and Dr. Brian Eckenroth for critically reading this manuscript. We apologize for citing a number of review articles, not being able to cover all facets of the complex signaling between the nucleus and mitochondria, and acknowledge the valuable work of numerous groups in the rapidly growing areas of mtDNA repair and nuclear-mitochondrial signaling pathways. This work is supported by National Institutes of Health grant 4R00ES024417-03 awarded to AP by the National Institute of Environmental Health Sciences.
Footnotes
Conflict of Interest
None
References
- 1.Pederson T. The nucleus introduced. Cold Spring Harb Perspect Biol. 2011;3(5) doi: 10.1101/cshperspect.a000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernster L, Schatz G. Mitochondria: a historical review. J Cell Biol. 1981;91(3 Pt 2):227s–255s. doi: 10.1083/jcb.91.3.227s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4(9):a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson SG, Karlberg O, Canback B, Kurland CG. On the origin of mitochondria: a genomics perspective. Philos Trans R Soc Lond B Biol Sci. 2003;358(1429):165–177. doi: 10.1098/rstb.2002.1193. discussion 177-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowaltowski AJ. Alternative mitochondrial functions in cell physiopathology: beyond ATP production. Braz J Med Biol Res. 2000;33(2):241–250. doi: 10.1590/s0100-879x2000000200014. [DOI] [PubMed] [Google Scholar]
- 6.Stehling O, Lill R. The role of mitochondria in cellular iron-sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harbor perspectives in biology. 2013;5(8):a011312. doi: 10.1101/cshperspect.a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaughnessy DT, McAllister K, Worth L, Haugen AC, Meyer JN, Domann FE, Van Houten B, Mostoslavsky R, Bultman SJ, Baccarelli AA, Begley TJ, Sobol RW, Hirschey MD, Ideker T, Santos JH, Copeland WC, Tice RR, Balshaw DM, Tyson FL. Mitochondria, energetics, epigenetics, and cellular responses to stress. Environmental health perspectives. 2014;122(12):1271–1278. doi: 10.1289/ehp.1408418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karnkowska A, Vacek V, Zubacova Z, Treitli SC, Petrzelkova R, Eme L, Novak L, Zarsky V, Barlow LD, Herman EK, Soukal P, Hroudova M, Dolezal P, Stairs CW, Roger AJ, Elias M, Dacks JB, Vlcek C, Hampl V. A Eukaryote without a Mitochondrial Organelle. Curr Biol. 2016;26(10):1274–1284. doi: 10.1016/j.cub.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 9.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 10.Gilkerson R, Bravo L, Garcia I, Gaytan N, Herrera A, Maldonado A, Quintanilla B. The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harb Perspect Biol. 2013;5(5):a011080. doi: 10.1101/cshperspect.a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spelbrink JN. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. 2010;62(1):19–32. doi: 10.1002/iub.282. [DOI] [PubMed] [Google Scholar]
- 12.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94(2):514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang EF, Scheibye-Knudsen M, Chua KF, Mattson MP, Croteau DL, Bohr VA. Nuclear DNA damage signalling to mitochondria in ageing. Nat Rev Mol Cell Biol. 2016;17(5):308–321. doi: 10.1038/nrm.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalousi A, Soutoglou E. Nuclear compartmentalization of DNA repair. Current opinion in genetics & development. 2016;37:148–157. doi: 10.1016/j.gde.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, Lightowlers RN, Dietrich A. DNA repair in organelles: Pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta. 2011;1813(1):186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The maintenance of mitochondrial DNA integrity--critical analysis and update. Cold Spring Harb Perspect Biol. 2013;5(5):a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Houten B, Hunter SE, Meyer JN. Mitochondrial DNA damage induced autophagy, cell death, and disease. Front Biosci (Landmark Ed) 2016;21:42–54. doi: 10.2741/4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138(4):628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudek J, Rehling P, van der Laan M. Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta. 2013;1833(2):274–285. doi: 10.1016/j.bbamcr.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Mordas A, Tokatlidis K. The MIA pathway: a key regulator of mitochondrial oxidative protein folding and biogenesis. Accounts of chemical research. 2015;48(8):2191–2199. doi: 10.1021/acs.accounts.5b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claros MG. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci. 1995;11(4):441–447. doi: 10.1093/bioinformatics/11.4.441. [DOI] [PubMed] [Google Scholar]
- 22.Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14(4):1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savojardo C, Martelli PL, Fariselli P, Casadio R. TPpred2: improving the prediction of mitochondrial targeting peptide cleavage sites by exploiting sequence motifs. Bioinformatics. 2014;30(20):2973–2974. doi: 10.1093/bioinformatics/btu411. [DOI] [PubMed] [Google Scholar]
- 24.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5(11):a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yarham JW, Elson JL, Blakely EL, McFarland R, Taylor RW. Mitochondrial tRNA mutations and disease. Wiley Interdiscip Rev RNA. 2010;1(2):304–324. doi: 10.1002/wrna.27. [DOI] [PubMed] [Google Scholar]
- 26.Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2010;1797(2):113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. 2015;16(9):530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 28.Zong WX, Rabinowitz JD, White E. Mitochondria and Cancer. Mol Cell. 2016;61(5):667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace DC. A mitochondrial bioenergetic etiology of disease. J Clin Invest. 2013;123(4):1405–1412. doi: 10.1172/JCI61398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandel NS. Mitochondria as signaling organelles. BMC Biol. 2014;12:34. doi: 10.1186/1741-7007-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tait SW, Green DR. Mitochondria and cell signalling. J Cell Sci. 2012;125(Pt 4):807–815. doi: 10.1242/jcs.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldenthal MJ, Marin-Garcia J. Mitochondrial signaling pathways: a receiver/integrator organelle. Mol Cell Biochem. 2004;262(1–2):1–16. doi: 10.1023/b:mcbi.0000038228.85494.3b. [DOI] [PubMed] [Google Scholar]
- 34.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 35.Espinosa-Diez C, Miguel V, Mennerich D, Kietzmann T, Sanchez-Perez P, Cadenas S, Lamas S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015;6:183–197. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Che M, Wang R, Li X, Wang HY, Zheng XF. Expanding roles of superoxide dismutases in cell regulation and cancer. Drug Discov Today. 2016;21(1):143–149. doi: 10.1016/j.drudis.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brigelius-Flohe R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta. 2013;1830(5):3289–3303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 38.Miranda-Vizuete A, Damdimopoulos AE, Spyrou G. The mitochondrial thioredoxin system. Antioxid Redox Signal. 2000;2(4):801–810. doi: 10.1089/ars.2000.2.4-801. [DOI] [PubMed] [Google Scholar]
- 39.Rhee SG, Woo HA. Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H(2)O(2), and protein chaperones. Antioxid Redox Signal. 2011;15(3):781–794. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- 40.Miriyala S, Spasojevic I, Tovmasyan A, Salvemini D, Vujaskovic Z, St Clair D, Batinic-Haberle I. Manganese superoxide dismutase, MnSOD and its mimics. Biochim Biophys Acta. 2012;1822(5):794–814. doi: 10.1016/j.bbadis.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kehrer JP. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology. 2000;149(1):43–50. doi: 10.1016/s0300-483x(00)00231-6. [DOI] [PubMed] [Google Scholar]
- 42.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9(7):246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 43.Cadet J, Wagner JR. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol. 2013;5(2) doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cadet J, Wagner JR. Oxidatively generated base damage to cellular DNA by hydroxyl radical and one-electron oxidants: similarities and differences. Arch Biochem Biophys. 2014;557:47–54. doi: 10.1016/j.abb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 45.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17(10):1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 46.Valavanidis A, Vlachogianni T, Fiotakis C. 8-hydroxy-2' -deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2009;27(2):120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 47.Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 2005;5(2):89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Kazak L, Reyes A, Holt IJ. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat Rev Mol Cell Biol. 2012;13(10):659–671. doi: 10.1038/nrm3439. [DOI] [PubMed] [Google Scholar]
- 49.Cline SD. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim Biophys Acta. 2012;1819(9–10):979–991. doi: 10.1016/j.bbagrm.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15(7):465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, Fang H, Chi Z, Wu Z, Wei D, Mo D, Niu K, Balajee AS, Hei TK, Nie L, Zhao Y. XPD localizes in mitochondria and protects the mitochondrial genome from oxidative DNA damage. Nucleic Acids Res. 2015;43(11):5476–5488. doi: 10.1093/nar/gkv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Houten BV, Kuper J, Kisker C. Role of XPD in cellular functions: To TFIIH and beyond. DNA Repair (Amst) 2016;44:136–142. doi: 10.1016/j.dnarep.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 53.Bess AS, Ryde IT, Hinton DE, Meyer JN. UVC-induced mitochondrial degradation via autophagy correlates with mtDNA damage removal in primary human fibroblasts. J Biochem Mol Toxicol. 2013;27(1):28–41. doi: 10.1002/jbt.21440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prakash A, Doublie S. Base Excision Repair in the Mitochondria. J Cell Biochem. 2015;116(8):1490–1499. doi: 10.1002/jcb.25103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5(4):a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66(6):981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121(1):1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834(1):247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25(4):750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakabeppu Y. Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage. Prog Nucleic Acid Res Mol Biol. 2001;68:75–94. doi: 10.1016/s0079-6603(01)68091-7. [DOI] [PubMed] [Google Scholar]
- 62.Plotz G, Casper M, Raedle J, Hinrichsen I, Heckel V, Brieger A, Trojan J, Zeuzem S. MUTYH gene expression and alternative splicing in controls and polyposis patients. Hum Mutat. 2012;33(7):1067–1074. doi: 10.1002/humu.22059. [DOI] [PubMed] [Google Scholar]
- 63.Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci U S A. 2005;102(16):5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18(1):27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15(2):209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 66.Li M, Zhong Z, Zhu J, Xiang D, Dai N, Cao X, Qing Y, Yang Z, Xie J, Li Z, Baugh L, Wang G, Wang D. Identification and characterization of mitochondrial targeting sequence of human apurinic/apyrimidinic endonuclease 1. J Biol Chem. 2010;285(20):14871–14881. doi: 10.1074/jbc.M109.069591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tahbaz N, Subedi S, Weinfeld M. Role of polynucleotide kinase/phosphatase in mitochondrial DNA repair. Nucleic Acids Res. 2012;40(8):3484–3495. doi: 10.1093/nar/gkr1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitra S, Izumi T, Boldogh I, Bhakat KK, Chattopadhyay R, Szczesny B. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair (Amst) 2007;6(4):461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 69.Mandal SM, Hegde ML, Chatterjee A, Hegde PM, Szczesny B, Banerjee D, Boldogh I, Gao R, Falkenberg M, Gustafsson CM, Sarkar PS, Hazra TK. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3'-phosphatase in maintenance of mitochondrial genome. J Biol Chem. 2012;287(4):2819–2829. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bogenhagen DF, Pinz KG, Perez-Jannotti RM. Enzymology of mitochondrial base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:257–271. doi: 10.1016/s0079-6603(01)68105-4. [DOI] [PubMed] [Google Scholar]
- 71.Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem Rev. 2006;106(2):383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 72.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5'-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci U S A. 1998;95(21):12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7(4):605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 74.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283(39):26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28(16):4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Copeland WC, Longley MJ. DNA2 resolves expanding flap in mitochondrial base excision repair. Mol Cell. 2008;32(4):457–458. doi: 10.1016/j.molcel.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5'-EXO/endonuclease) in their repair. J Biol Chem. 2011;286(37):31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lakshmipathy U, Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 2000;28(20):3880–3886. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sykora P, Wilson DM, 3rd, Bohr VA. Base excision repair in the mammalian brain: implication for age related neurodegeneration. Mech Ageing Dev. 2013;134(10):440–448. doi: 10.1016/j.mad.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9(8):619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 82.Rossi MN, Carbone M, Mostocotto C, Mancone C, Tripodi M, Maione R, Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem. 2009;284(46):31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szczesny B, Brunyanszki A, Olah G, Mitra S, Szabo C. Opposing roles of mitochondrial and nuclear PARP1 in the regulation of mitochondrial and nuclear DNA integrity: implications for the regulation of mitochondrial function. Nucleic Acids Res. 2014;42(21):13161–13173. doi: 10.1093/nar/gku1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lapucci A, Pittelli M, Rapizzi E, Felici R, Moroni F, Chiarugi A. Poly(ADP-ribose) polymerase-1 is a nuclear epigenetic regulator of mitochondrial DNA repair and transcription. Molecular pharmacology. 2011;79(6):932–940. doi: 10.1124/mol.110.070110. [DOI] [PubMed] [Google Scholar]
- 85.Sykora P, Croteau DL, Bohr VA, Wilson DM., 3rd Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc Natl Acad Sci U S A. 2011;108(18):7437–7442. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Akbari M, Sykora P, Bohr VA. Slow mitochondrial repair of 5'-AMP renders mtDNA susceptible to damage in APTX deficient cells. Sci Rep. 2015;5:12876. doi: 10.1038/srep12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Das BB, Dexheimer TS, Maddali K, Pommier Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci U S A. 2010;107(46):19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Canugovi C, Maynard S, Bayne AC, Sykora P, Tian J, de Souza-Pinto NC, Croteau DL, Bohr VA. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 2010;9(10):1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dolle ME, Kuiper R, Majora M, Schaller M, van der Horst GT, van Steeg H, Rocken M, Rapaport D, Krutmann J, Mullenders LH, Berneburg M. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med. 2010;207(2):379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gredilla R. DNA damage and base excision repair in mitochondria and their role in aging. J Aging Res. 2010;2011:257093. doi: 10.4061/2011/257093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chan SS, Copeland WC. DNA polymerase gamma and mitochondrial disease: understanding the consequence of POLG mutations. Biochim Biophys Acta. 2009;1787(5):312–319. doi: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leandro GS, Sykora P, Bohr VA. The impact of base excision DNA repair in age-related neurodegenerative diseases. Mutat Res. 2015;776:31–39. doi: 10.1016/j.mrfmmm.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takahashi M, Teranishi M, Ishida H, Kawasaki J, Takeuchi A, Yamaya T, Watanabe M, Makino A, Hidema J. Cyclobutane pyrimidine dimer (CPD) photolyase repairs ultraviolet-B-induced CPDs in rice chloroplast and mitochondrial DNA. Plant. 2011;66(3):433–442. doi: 10.1111/j.1365-313X.2011.04500.x. [DOI] [PubMed] [Google Scholar]
- 94.Yasui A, Eker AP, Yasuhira S, Yajima H, Kobayashi T, Takao M, Oikawa A. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J. 1994;13(24):6143–6151. doi: 10.1002/j.1460-2075.1994.tb06961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brent TP, Dolan ME, Fraenkel-Conrat H, Hall J, Karran P, Laval L, Margison GP, Montesano R, Pegg AE, Potter PM, et al. Repair of O-alkylpyrimidines in mammalian cells: a present consensus. Proc Natl Acad Sci U S A. 1988;85(6):1759–1762. doi: 10.1073/pnas.85.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zak P, Kleibl K, Laval F. Repair of O6-methylguanine and O4-methylthymine by the human and rat O6-methylguanine-DNA methyltransferases. J Biol Chem. 1994;269(1):730–733. [PubMed] [Google Scholar]
- 97.Ragg S, Xu-Welliver M, Bailey J, D'Souza M, Cooper R, Chandra S, Seshadri R, Pegg AE, Williams DA. Direct reversal of DNA damage by mutant methyltransferase protein protects mice against dose-intensified chemotherapy and leads to in vivo selection of hematopoietic stem cells. Cancer Res. 2000;60(18):5187–5195. [PubMed] [Google Scholar]
- 98.Rasmussen AK, Rasmussen LJ. Targeting of O6-MeG DNA methyltransferase (MGMT) to mitochondria protects against alkylation induced cell death. Mitochondrion. 2005;5(6):411–417. doi: 10.1016/j.mito.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 99.Cai S, Xu Y, Cooper RJ, Ferkowicz MJ, Hartwell JR, Pollok KE, Kelley MR. Mitochondrial targeting of human O6-methylguanine DNA methyltransferase protects against cell killing by chemotherapeutic alkylating agents. Cancer Res. 2005;65(8):3319–3327. doi: 10.1158/0008-5472.CAN-04-3335. [DOI] [PubMed] [Google Scholar]
- 100.Myers KA, Saffhill R, O'Connor PJ. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis. 1988;9(2):285–292. doi: 10.1093/carcin/9.2.285. [DOI] [PubMed] [Google Scholar]
- 101.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 102.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 103.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281(41):30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 2009;8(6):704–719. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA. 2014;5(1):95–110. doi: 10.1002/wrna.1200. [DOI] [PubMed] [Google Scholar]
- 106.Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell. 2010;17(3):235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73(1):134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5(3):a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18(5):499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 110.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6(7):891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 111.Kasiviswanathan R, Gustafson MA, Copeland WC, Meyer JN. Human mitochondrial DNA polymerase gamma exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J Biol Chem. 2012;287(12):9222–9229. doi: 10.1074/jbc.M111.306852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kasiviswanathan R, Minko IG, Lloyd RS, Copeland WC. Translesion synthesis past acrolein-derived DNA adducts by human mitochondrial DNA polymerase gamma. J Biol Chem. 2013;288(20):14247–14255. doi: 10.1074/jbc.M113.458802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hanes JW, Thal DM, Johnson KA. Incorporation and replication of 8-oxo-deoxyguanosine by the human mitochondrial DNA polymerase. J Biol Chem. 2006;281(47):36241–36248. doi: 10.1074/jbc.M607965200. [DOI] [PubMed] [Google Scholar]
- 114.Zhang H, Chatterjee A, Singh KK. Saccharomyces cerevisiae polymerase zeta functions in mitochondria. Genetics. 2006;172(4):2683–2688. doi: 10.1534/genetics.105.051029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baruffini E, Serafini F, Ferrero I, Lodi T. Overexpression of DNA polymerase zeta reduces the mitochondrial mutability caused by pathological mutations in DNA polymerase gamma in yeast. PLoS One. 2012;7(3):e34322. doi: 10.1371/journal.pone.0034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Singh B, Li X, Owens KM, Vanniarajan A, Liang P, Singh KK. Human REV3 DNA Polymerase Zeta Localizes to Mitochondria and Protects the Mitochondrial Genome. PLoS One. 2015;10(10):e0140409. doi: 10.1371/journal.pone.0140409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Garcia-Gomez S, Reyes A, Martinez-Jimenez MI, Chocron ES, Mouron S, Terrados G, Powell C, Salido E, Mendez J, Holt IJ, Blanco L. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell. 2013;52(4):541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guilliam TA, Jozwiakowski SK, Ehlinger A, Barnes RP, Rudd SG, Bailey LJ, Skehel JM, Eckert KA, Chazin WJ, Doherty AJ. Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins. Nucleic Acids Res. 2015;43(2):1056–1068. doi: 10.1093/nar/gku1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stojkovic G, Makarova AV, Wanrooij PH, Forslund J, Burgers PM, Wanrooij S. Oxidative DNA damage stalls the human mitochondrial replisome. Sci Rep. 2016;6:28942. doi: 10.1038/srep28942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lockshon D, Zweifel SG, Freeman-Cook LL, Lorimer HE, Brewer BJ, Fangman WL. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell. 1995;81(6):947–955. doi: 10.1016/0092-8674(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 122.Stein A, Kalifa L, Sia EA. Members of the RAD52 Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in Saccharomyces cerevisiae. PLoS Genet. 2015;11(11):e1005664. doi: 10.1371/journal.pgen.1005664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fritsch ES, Chabbert CD, Klaus B, Steinmetz LM. A genome-wide map of mitochondrial DNA recombination in yeast. Genetics. 2014;198(2):755–771. doi: 10.1534/genetics.114.166637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Manchekar M, Scissum-Gunn K, Song D, Khazi F, McLean SL, Nielsen BL. DNA recombination activity in soybean mitochondria. J Mol Biol. 2006;356(2):288–299. doi: 10.1016/j.jmb.2005.11.070. [DOI] [PubMed] [Google Scholar]
- 125.Lieberman-Lazarovich M, Levy AA. Homologous recombination in plants: an antireview. Methods Mol Biol. 2011;701:51–65. doi: 10.1007/978-1-61737-957-4_3. [DOI] [PubMed] [Google Scholar]
- 126.Thyagarajan B, Padua RA, Campbell C. Mammalian mitochondria possess homologous DNA recombination activity. J Biol Chem. 1996;271(44):27536–27543. doi: 10.1074/jbc.271.44.27536. [DOI] [PubMed] [Google Scholar]
- 127.Kajander OA, Karhunen PJ, Holt IJ, Jacobs HT. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2001;2(11):1007–1012. doi: 10.1093/embo-reports/kve233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.D'Aurelio M, Gajewski CD, Lin MT, Mauck WM, Shao LZ, Lenaz G, Moraes CT, Manfredi G. Heterologous mitochondrial DNA recombination in human cells. Hum Mol Genet. 2004;13(24):3171–3179. doi: 10.1093/hmg/ddh326. [DOI] [PubMed] [Google Scholar]
- 129.Bacman SR, Williams SL, Moraes CT. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 2009;37(13):4218–4226. doi: 10.1093/nar/gkp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Phadnis N, Sia RA, Sia EA. Analysis of repeat-mediated deletions in the mitochondrial genome of Saccharomyces cerevisiae. Genetics. 2005;171(4):1549–1559. doi: 10.1534/genetics.105.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18(6):1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kalifa L, Quintana DF, Schiraldi LK, Phadnis N, Coles GL, Sia RA, Sia EA. Mitochondrial genome maintenance: roles for nuclear nonhomologous end-joining proteins in Saccharomyces cerevisiae. Genetics. 2012;190(3):951–964. doi: 10.1534/genetics.111.138214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lakshmipathy U, Campbell C. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res. 1999;27(4):1198–1204. doi: 10.1093/nar/27.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Monaghan RM, Whitmarsh AJ. Mitochondrial Proteins Moonlighting in the Nucleus. Trends Biochem Sci. 2015;40(12):728–735. doi: 10.1016/j.tibs.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 135.Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 136.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14(1):1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 137.Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17(4):213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 138.Jazwinski SM. The retrograde response: a conserved compensatory reaction to damage from within and from without. Prog Mol Biol Transl Sci. 2014;127:133–154. doi: 10.1016/B978-0-12-394625-6.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cagin U, Enriquez JA. The complex crosstalk between mitochondria and the nucleus: What goes in between? Int J Biochem Cell Biol. 2015;63:10–15. doi: 10.1016/j.biocel.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 140.Marusich MF, Robinson BH, Taanman JW, Kim SJ, Schillace R, Smith JL, Capaldi RA. Expression of mtDNA and nDNA encoded respiratory chain proteins in chemically and genetically-derived Rho0 human fibroblasts: a comparison of subunit proteins in normal fibroblasts treated with ethidium bromide and fibroblasts from a patient with mtDNA depletion syndrome. Biochim Biophys Acta. 1997;1362(2–3):145–159. doi: 10.1016/s0925-4439(97)00061-6. [DOI] [PubMed] [Google Scholar]
- 141.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13(9):566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 142.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1(1):11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 143.Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001;20(8):1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18(3):522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Biswas G, Guha M, Avadhani NG. Mitochondria-to-nucleus stress signaling in mammalian cells: nature of nuclear gene targets, transcription regulation, and induced resistance to apoptosis. Gene. 2005;354:132–139. doi: 10.1016/j.gene.2005.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim J, Sharma RP. Calcium-mediated activation of c-Jun NH2-terminal kinase (JNK) and apoptosis in response to cadmium in murine macrophages. Toxicological sciences : an official journal of the Society of Toxicology. 2004;81(2):518–527. doi: 10.1093/toxsci/kfh221. [DOI] [PubMed] [Google Scholar]
- 147.Arnould T, Vankoningsloo S, Renard P, Houbion A, Ninane N, Demazy C, Remacle J, Raes M. CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation. EMBO J. 2002;21(1–2):53–63. doi: 10.1093/emboj/21.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Prakash A, Cao VB, Doublie S. Phosphorylation Sites Identified in the NEIL1 DNA Glycosylase Are Potential Targets for the JNK1 Kinase. PLoS One. 2016;11(8):e0157860. doi: 10.1371/journal.pone.0157860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Picco V, Pages G. Linking JNK Activity to the DNA Damage Response. Genes Cancer. 2013;4(9–10):360–368. doi: 10.1177/1947601913486347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal. 2008;10(2):179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- 151.Stein LR, Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab. 2012;23(9):420–428. doi: 10.1016/j.tem.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Canto C, Menzies KJ, Auwerx J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015;22(1):31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Osborne B, Bentley NL, Montgomery MK, Turner N. The role of mitochondrial sirtuins in health and disease. Free Radic Biol Med. 2016 doi: 10.1016/j.freeradbiomed.2016.04.197. [DOI] [PubMed] [Google Scholar]
- 154.Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann N Y Acad Sci. 2009;1173(Suppl 1):E10–E19. doi: 10.1111/j.1749-6632.2009.04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Burkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ FEBS J. 2005;272(18):4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 156.Fouquerel E, Sobol RW. ARTD1 (PARP1) activation and NAD(+) in DNA repair and cell death. DNA Repair (Amst) 2014;23:27–32. doi: 10.1016/j.dnarep.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Talhaoui I, Lebedeva NA, Zarkovic G, Saint-Pierre C, Kutuzov MM, Sukhanova MV, Matkarimov BT, Gasparutto D, Saparbaev MK, Lavrik OI, Ishchenko AA. Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci U S A. 2011;108(7):2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41(1):33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 160.Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism, BioEssays : news and reviews in molecular. cellular and developmental biology. 2003;25(7):683–690. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- 161.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Molecular biology of the cell. 2005;16(10):4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 163.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Current opinion in lipidology. 2009;20(2):98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Experimental & molecular medicine. 2007;39(1):8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 165.Joo HY, Yun M, Jeong J, Park ER, Shin HJ, Woo SR, Jung JK, Kim YM, Park JJ, Kim J, Lee KH. SIRT1 deacetylates and stabilizes hypoxia-inducible factor-1alpha (HIF-1alpha) via direct interactions during hypoxia. Biochem Biophys Res Commun. 2015;462(4):294–300. doi: 10.1016/j.bbrc.2015.04.119. [DOI] [PubMed] [Google Scholar]
- 166.Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38(6):864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 167.Luna A, Aladjem MI, Kohn KW. SIRT1/PARP1 crosstalk: connecting DNA damage and metabolism. Genome integrity. 2013;4(1):6. doi: 10.1186/2041-9414-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Marcu R, Wiczer BM, Neeley CK, Hawkins BJ. Mitochondrial matrix Ca(2)(+) accumulation regulates cytosolic NAD(+)/NADH metabolism, protein acetylation, and sirtuin expression. Mol Cell Biol. 2014;34(15):2890–2902. doi: 10.1128/MCB.00068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shirwany NA, Zou MH. AMPK: a cellular metabolic and redox sensor. A minireview. Front Biosci (Landmark Ed) 2014;19:447–474. doi: 10.2741/4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Canto C, Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67(20):3407–3423. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Guo Z, Deshpande R, Paull TT. ATM activation in the presence of oxidative stress. Cell Cycle. 2010;9(24):4805–4811. doi: 10.4161/cc.9.24.14323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134(3):451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14(4):197–210. [PubMed] [Google Scholar]
- 174.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18(3):283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 175.Nieminen AI, Eskelinen VM, Haikala HM, Tervonen TA, Yan Y, Partanen JI, Klefstrom J. Myc-induced AMPK-phospho p53 pathway activates Bak to sensitize mitochondrial apoptosis. Proc Natl Acad Sci U S A. 2013;110(20):E1839–E1848. doi: 10.1073/pnas.1208530110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tripathi DN, Chowdhury R, Trudel LJ, Tee AR, Slack RS, Walker CL, Wogan GN. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2-mediated suppression of mTORC1. Proc Natl Acad Sci U S A. 2013;110(32):E2950–E2957. doi: 10.1073/pnas.1307736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y. AMPK and SIRT1: a long-standing partnership? Am J Physiol Endocrinol Metab. 2010;298(4):E751–E760. doi: 10.1152/ajpendo.00745.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Canto C, Auwerx J. Calorie restriction: is AMPK a key sensor and effector? Physiology (Bethesda) 2011;26(4):214–224. doi: 10.1152/physiol.00010.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104(29):12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ho L, Titus AS, Banerjee KK, George S, Lin W, Deota S, Saha AK, Nakamura K, Gut P, Verdin E, Kolthur-Seetharam U. SIRT4 regulates ATP homeostasis and mediates a retrograde signaling via AMPK. Aging (Albany NY) 2013;5(11):835–849. doi: 10.18632/aging.100616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Janssen-Heininger YM, Mossman BT, Heintz NH, Forman HJ, Kalyanaraman B, Finkel T, Stamler JS, Rhee SG, van der Vliet A. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic Biol Med. 2008;45(1):1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, Sauvaigo S. Hydroxyl radicals and DNA base damage. Mutat Res. 1999;424(1–2):9–21. doi: 10.1016/s0027-5107(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 183.Marengo B, Nitti M, Furfaro AL, Colla R, Ciucis CD, Marinari UM, Pronzato MA, Traverso N, Domenicotti C. Redox Homeostasis and Cellular Antioxidant Systems: Crucial Players in Cancer Growth and Therapy. Oxid Med Cell Longev. 2016;2016:6235641. doi: 10.1155/2016/6235641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Arthur JR. The glutathione peroxidases. Cell Mol Life Sci. 2000;57(13–14):1825–1835. doi: 10.1007/PL00000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Pantano C, Reynaert NL, van der Vliet A, Janssen-Heininger YM. Redox-sensitive kinases of the nuclear factor-kappaB signaling pathway. Antioxid Redox Signal. 2006;8(9–10):1791–1806. doi: 10.1089/ars.2006.8.1791. [DOI] [PubMed] [Google Scholar]
- 186.Janssen-Heininger YM, Poynter ME, Aesif SW, Pantano C, Ather JL, Reynaert NL, Ckless K, Anathy V, van der Velden J, Irvin CG, van der Vliet A. Nuclear factor kappaB, airway epithelium, and asthma: avenues for redox control. Proc Am Thorac Soc. 2009;6(3):249–255. doi: 10.1513/pats.200806-054RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med. 2008;44(8):1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Solaini G, Baracca A, Lenaz G, Sgarbi G. Hypoxia and mitochondrial oxidative metabolism. Biochim Biophys Acta. 2010;1797(6–7):1171–1177. doi: 10.1016/j.bbabio.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 189.Itoh K, Ye P, Matsumiya T, Tanji K, Ozaki T. Emerging functional cross-talk between the Keap1-Nrf2 system and mitochondria. J Clin Biochem Nutr. 2015;56(2):91–97. doi: 10.3164/jcbn.14-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Piantadosi CA, Suliman HB. Mitochondrial transcription factor A induction by redox activation of nuclear respiratory factor 1. J Biol Chem. 2006;281(1):324–333. doi: 10.1074/jbc.M508805200. [DOI] [PubMed] [Google Scholar]
- 191.Erusalimsky JD, Moncada S. Nitric oxide and mitochondrial signaling: from physiology to pathophysiology. Arterioscler Thromb Vasc Biol. 2007;27(12):2524–2531. doi: 10.1161/ATVBAHA.107.151167. [DOI] [PubMed] [Google Scholar]
- 192.Brown GC. Cell biology. NO says yes to mitochondria. Science. 2003;299(5608):838–839. doi: 10.1126/science.1082028. [DOI] [PubMed] [Google Scholar]
- 193.Ghafourifar P, Cadenas E. Mitochondrial nitric oxide synthase. Trends Pharmacol Sci. 2005;26(4):190–195. doi: 10.1016/j.tips.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 194.Valdez LB, Boveris A. Mitochondrial nitric oxide synthase, a voltage-dependent enzyme, is responsible for nitric oxide diffusion to cytosol. Front Biosci. 2007;12:1210–1219. doi: 10.2741/2139. [DOI] [PubMed] [Google Scholar]
- 195.Habib S, Ali A. Biochemistry of nitric oxide. Indian J Clin Biochem. 2011;26(1):3–17. doi: 10.1007/s12291-011-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sarti P, Arese M, Bacchi A, Barone MC, Forte E, Mastronicola D, Brunori M, Giuffre A. Nitric oxide and mitochondrial complex IV. IUBMB Life. 2003;55(10–11):605–611. doi: 10.1080/15216540310001628726. [DOI] [PubMed] [Google Scholar]
- 197.Brown GC. Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim Biophys Acta. 2001;1504(1):46–57. doi: 10.1016/s0005-2728(00)00238-3. [DOI] [PubMed] [Google Scholar]
- 198.Hagen T, Taylor CT, Lam F, Moncada S. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1alpha. Science. 2003;302(5652):1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 199.Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299(5608):896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 200.Bossy-Wetzel E, Lipton SA. Nitric oxide signaling regulates mitochondrial number and function. Cell Death Differ. 2003;10(7):757–760. doi: 10.1038/sj.cdd.4401244. [DOI] [PubMed] [Google Scholar]
- 201.Lira VA, Brown DL, Lira AK, Kavazis AN, Soltow QA, Zeanah EH, Criswell DS. Nitric oxide and AMPK cooperatively regulate PGC-1 in skeletal muscle cells. J Physiol. 2010;588(Pt 18):3551–3566. doi: 10.1113/jphysiol.2010.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Raharijaona M, Le Pennec S, Poirier J, Mirebeau-Prunier D, Rouxel C, Jacques C, Fontaine JF, Malthiery Y, Houlgatte R, Savagner F. PGC-1-related coactivator modulates mitochondrial-nuclear crosstalk through endogenous nitric oxide in a cellular model of oncocytic thyroid tumours. PLoS One. 2009;4(11):e7964. doi: 10.1371/journal.pone.0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fionda C, Abruzzese MP, Santoni A, Cippitelli M. Immunoregulatory and Effector Activities of Nitric Oxide and Reactive Nitrogen Species in Cancer. Curr Med Chem. 2016 doi: 10.2174/0929867323666160727105101. [DOI] [PubMed] [Google Scholar]
- 204.Kelley MR, Georgiadis MM, Fishel ML. APE1/Ref-1 role in redox signaling: translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Curr Mol Pharmacol. 2012;5(1):36–53. doi: 10.2174/1874467211205010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mikhed Y, Gorlach A, Knaus UG, Daiber A. Redox regulation of genome stability by effects on gene expression, epigenetic pathways and DNA damage/repair. Redox Biol. 2015;5:275–289. doi: 10.1016/j.redox.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Park MS, Kim CS, Joo HK, Lee YR, Kang G, Kim SJ, Choi S, Lee SD, Park JB, Jeon BH. Cytoplasmic localization and redox cysteine residue of APE1/Ref-1 are associated with its anti-inflammatory activity in cultured endothelial cells. Mol Cells. 2013;36(5):439–445. doi: 10.1007/s10059-013-0195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wang Z, Ayoub E, Mazouzi A, Grin I, Ishchenko AA, Fan J, Yang X, Harihar T, Saparbaev M, Ramotar D. Functional variants of human APE1 rescue the DNA repair defects of the yeast AP endonuclease/3'-diesterase-deficient strain. DNA Repair (Amst) 2014;22:53–66. doi: 10.1016/j.dnarep.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 208.Walker LJ, Robson CN, Black E, Gillespie D, Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol. 1993;13(9):5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res. 2008;643(1–2):54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403(6768):451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 211.Huang LE, Arany Z, Livingston DM, Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271(50):32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 212.Nishi T, Shimizu N, Hiramoto M, Sato I, Yamaguchi Y, Hasegawa M, Aizawa S, Tanaka H, Kataoka K, Watanabe H, Handa H. Spatial redox regulation of a critical cysteine residue of NF-kappa B in vivo. J Biol Chem. 2002;277(46):44548–44556. doi: 10.1074/jbc.M202970200. [DOI] [PubMed] [Google Scholar]
- 213.Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11(5):558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 214.Fishel ML, Wu X, Devlin CM, Logsdon DP, Jiang Y, Luo M, He Y, Yu Z, Tong Y, Lipking KP, Maitra A, Rajeshkumar NV, Scandura G, Kelley MR, Ivan M. Apurinic/apyrimidinic endonuclease/redox factor-1 (APE1/Ref-1) redox function negatively regulates NRF2. J Biol Chem. 2015;290(5):3057–3068. doi: 10.1074/jbc.M114.621995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal. 2009;11(3):601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci U S A. 1996;93(17):8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Barchiesi A, Wasilewski M, Chacinska A, Tell G, Vascotto C. Mitochondrial translocation of APE1 relies on the MIA pathway. Nucleic Acids Res. 2015;43(11):5451–5464. doi: 10.1093/nar/gkv433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zhuang J, Wang PY, Huang X, Chen X, Kang JG, Hwang PM. Mitochondrial disulfide relay mediates translocation of p53 and partitions its subcellular activity. Proc Natl Acad Sci U S A. 2013;110(43):17356–17361. doi: 10.1073/pnas.1310908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jovaisaite V, Mouchiroud L, Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol. 2014;217(Pt 1):137–143. doi: 10.1242/jeb.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochim Biophys Acta. 2013;1833(2):410–416. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123(Pt 22):3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 222.Arnould T, Michel S, Renard P. Mitochondria Retrograde Signaling and the UPR mt: Where Are We in Mammals? Int J Mol Sci. 2015;16(8):18224–18251. doi: 10.3390/ijms160818224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 224.Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117(Pt 18):4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 225.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21(17):4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Horibe T, Hoogenraad NJ. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS One. 2007;2(9):e835. doi: 10.1371/journal.pone.0000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J Cell Sci. 2011;124(Pt 9):1396–1402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol. 2014;34(4):699–710. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pellegrino MW, Haynes CM. Mitophagy and the mitochondrial unfolded protein response in neurodegeneration and bacterial infection. BMC Biol. 2015;13:22. doi: 10.1186/s12915-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Thomas RE, Andrews LA, Burman JL, Lin WY, Pallanck LJ. PINK1-Parkin pathway activity is regulated by degradation of PINK1 in the mitochondrial matrix. PLoS Genet. 2014;10(5):e1004279. doi: 10.1371/journal.pgen.1004279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Voigt A, Berlemann LA, Winklhofer KF. The mitochondrial kinase PINK1: functions beyond mitophagy. J Neurochem. 2016;139(Suppl 1):232–239. doi: 10.1111/jnc.13655. [DOI] [PubMed] [Google Scholar]
- 232.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9(11):1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Durcan TM, Fon EA. The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015;29(10):989–999. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85(2):257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Fetterman JL, Zelickson BR, Johnson LW, Moellering DR, Westbrook DG, Pompilius M, Sammy MJ, Johnson M, Dunham-Snary KJ, Cao X, Bradley WE, Zhang J, Wei CC, Chacko B, Schurr TG, Kesterson RA, Dell'italia LJ, Darley-Usmar VM, Welch DR, Ballinger SW. Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem J. 2013;455(2):157–167. doi: 10.1042/BJ20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Betancourt AM, King AL, Fetterman JL, Millender-Swain T, Finley RD, Oliva CR, Crowe DR, Ballinger SW, Bailey SM. Mitochondrial-nuclear genome interactions in non-alcoholic fatty liver disease in mice. Biochem J. 2014;461(2):223–232. doi: 10.1042/BJ20131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hashizume M, Mouner M, Chouteau JM, Gorodnya OM, Ruchko MV, Potter BJ, Wilson GL, Gillespie MN, Parker JC. Mitochondrial-targeted DNA repair enzyme 8-oxoguanine DNA glycosylase 1 protects against ventilator-induced lung injury in intact mice. Am J Physiol Lung Cell Mol Physiol. 2013;304(4):L287–L297. doi: 10.1152/ajplung.00071.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hashizume M, Mouner M, Chouteau JM, Gorodnya OM, Ruchko MV, Wilson GL, Gillespie MN, Parker JC. Mitochondrial Targeted Endonuclease III DNA Repair Enzyme Protects against Ventilator Induced Lung Injury in Mice. Pharmaceuticals (Basel) 2014;7(8):894–912. doi: 10.3390/ph7080894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gebb SA, Decoux A, Waggoner A, Wilson GL, Gillespie MN. Mitochondrial DNA damage mediates hyperoxic dysmorphogenesis in rat fetal lung explants. Neonatology. 2013;103(2):91–97. doi: 10.1159/000342632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Simon R. Post-conditioning and reperfusion injury in the treatment of stroke. Dose Response. 2014;12(4):590–599. doi: 10.2203/dose-response.14-026.Simon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280(49):40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 242.Takao M, Aburatani H, Kobayashi K, Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26(12):2917–2922. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61(14):5378–5381. [PubMed] [Google Scholar]
- 244.van Loon B, Samson LD. Alkyladenine DNA glycosylase (AAG) localizes to mitochondria and interacts with mitochondrial single-stranded binding protein (mtSSB) DNA Repair (Amst) 2013;12(3):177–187. doi: 10.1016/j.dnarep.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Krokan HE, Otterlei M, Nilsen H, Kavli B, Skorpen F, Andersen S, Skjelbred C, Akbari M, Aas PA, Slupphaug G. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog Nucleic Acid Res Mol Biol. 2001;68:365–386. doi: 10.1016/s0079-6603(01)68112-1. [DOI] [PubMed] [Google Scholar]
- 246.Bertazzoni U, Scovassi AI, Brun GM. Chick-embryo DNA polymerase gamma. Identity of gamma-polymerases purified from nuclei and mitochondria. Eur J Biochem. 1977;81(2):237–248. doi: 10.1111/j.1432-1033.1977.tb11945.x. [DOI] [PubMed] [Google Scholar]
- 247.Sage JM, Gildemeister OS, Knight KL. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J Biol Chem. 2010;285(25):18984–18990. doi: 10.1074/jbc.M109.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Coene ED, Hollinshead MS, Waeytens AA, Schelfhout VR, Eechaute WP, Shaw MK, Van Oostveldt PM, Vaux DJ. Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Molecular biology of the cell. 2005;16(2):997–1010. doi: 10.1091/mbc.E04-10-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Coffey G, Campbell C. An alternate form of Ku80 is required for DNA end-binding activity in mammalian mitochondria. Nucleic Acids Res. 2000;28(19):3793–3800. doi: 10.1093/nar/28.19.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Coffey G, Lakshmipathy U, Campbell C. Mammalian mitochondrial extracts possess DNA end-binding activity. Nucleic Acids Res. 1999;27(16):3348–3354. doi: 10.1093/nar/27.16.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Dmitrieva NI, Malide D, Burg MB. Mre11 is expressed in mammalian mitochondria where it binds to mitochondrial DNA, American journal of physiology. Regulatory. integrative and comparative physiology. 2011;301(3):R632–R640. doi: 10.1152/ajpregu.00853.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]