Abstract
Although it is accepted that the environment within the granuloma profoundly affects Mycobacterium tuberculosis (Mtb) and infection outcome, our ability to understand Mtb gene expression in these niches has been limited. We determined intragranulomatous gene expression in human-like lung lesions derived from nonhuman primates with both active tuberculosis (ATB) and latent TB infection (LTBI). We employed a non–laser-based approach to microdissect individual lung lesions and interrogate the global transcriptome of Mtb within granulomas. Mtb genes expressed in classical granulomas with central, caseous necrosis, as well as within the caseum itself, were identified and compared with other Mtb lesions in animals with ATB (n = 7) or LTBI (n = 7). Results were validated using both an oligonucleotide approach and RT-PCR on macaque samples and by using human TB samples. We detected approximately 2,900 and 1,850 statistically significant genes in ATB and LTBI lesions, respectively (linear models for microarray analysis, Bonferroni corrected, P < 0.05). Of these genes, the expression of approximately 1,300 (ATB) and 900 (LTBI) was positively induced. We identified the induction of key regulons and compared our results to genes previously determined to be required for Mtb growth. Our results indicate pathways that Mtb uses to ensure its survival in a highly stressful environment in vivo. A large number of genes is commonly expressed in granulomas with ATB and LTBI. In addition, the enhanced expression of the dormancy survival regulon was a key feature of lesions in animals with LTBI, stressing its importance in the persistence of Mtb during the chronic phase of infection.
Keywords: Mycobacterium tuberculosis, macaque, granuloma, microdissection, hypoxia
Clinical Relevance
Our results conclusively show that the hypoxia-sensing dormancy survival regulon is expressed in tuberculosis (TB) lesions in vivo. In addition, we describe, in detail, hundreds of other Mycobacterium tuberculosis genes, the expression of which was detected in the natural niche. This will inform future drug and vaccine development efforts.
Mycobacterium tuberculosis (Mtb) is constantly subjected to stress in vivo and must successfully adapt to survive its ever-changing extracellular milieu (1). The study of the mycobacterial state in vivo is further complicated by variability in granuloma pathology, physiology, and morphology. Currently, we do not fully understand the drivers of Mtb reactivation in the lung (2), and disease progression can be highly variable given the differences in host genetics, environment, microbiota, and the presence of comorbidities. A comparative systems biology approach that incorporates the pathological complexities of Mtb infection would allow us to better understand the physiology of the pathogen.
Various approaches have been used to attempt to understand the importance of Mtb genes and therefore better elucidate tuberculosis (TB) pathogenesis. Initially, the TB field focused on a gene-by-gene approach and, with time and technological advancements, began shifting to whole-genome–based approaches. Leveraging transposon site hybridization (TraSH), genes required for Mtb growth have been identified in mice, murine macrophages, and computationally. Another mutant-based technique, designer arrays for defined mutant analysis, have been used in mice, guinea pigs, and nonhuman primates (NHPs). In addition, several groups have attempted to model the granuloma environment, whereas others have detected the expression of a small number of specific Mtb genes in human macrophages. Some have conducted massively parallel RNA-PCR of Mtb transcriptome using the mouse model. Yet others have identified whole-genome Mtb gene expression in various mouse models and used in vitro granuloma models. These studies have provided insight into genes required for Mtb growth in survival, but no studies to date have profiled in vivo gene profiles within its natural microenvironment and characteristic pulmonary lesion, the granuloma, while also using a model that most closely recapitulates the human disease spectrum. As such, failure to effectively control Mtb infections and TB disease via devising new vaccinations and therapeutic strategies has been due to the lack of effective disease models and fundamental knowledge of the pathogen in its natural niches.
Here, we change the way that we identify Mtb treatment and vaccine strategies by switching from traditionally used peripheral responses to a localized approach by assessing Mtb gene induction within the pulmonary granulomatous environment. Due to the many similarities between infected NHPs and humans, NHPs are the ideal model for addressing these questions (3). NHPs, such as rhesus (Macaca mulatta) (2–18) and cynomolgus (Macaca fascicularis) (19, 20) macaques, recapitulate the wide spectrum of human TB pathology and outcomes upon experimental infection with Mtb. We currently lack an understanding of the physiology and the metabolic state of Mtb in this granulomatous environment during different states of infection. Understanding the physiology and metabolism of the intragranulomatous environment is critical, because effective vaccines and drugs must target this state to eradicate the bacteria and subsequently control the infection. We therefore propose to study and compare the intragranulomatous gene expression of Mtb during ATB, latent TB infection (LTBI), and reactivation.
We hypothesize that the Mtb transcriptional profiles, with respect to metabolism and physiology, exhibit changes over time and upon interaction with both a variety of environmental cues and host immune responses. Consequently, we hypothesize that different infective stages, as well as different areas of the granuloma, lead to differential bacillary expression profiles. Furthermore, and more importantly, we propose that these changes can be used to understand the physiology of the pathogen as well as its virulence.
The purpose of evaluating the entire granulomatous pathology (i.e., the combination of all lesion and granuloma types on the formalin-fixed, paraffin-embedded [FFPE] lung slides) is to generate a bacterial transcriptome profile that is representative of the entire infective state. In addition, the purpose of specifically evaluating the classical, caseating-type granuloma is to evaluate the bacterial transcripts specific to this lesion type. Overall, evaluation of the mycobacterial transcriptome in granulomatous tissue is likely to further our understanding of the mechanisms involved in their formation and maintenance, as well as those genes that are expressed in each state of the infection. Analysis of Mtb gene expression within this environment in vivo will further our understanding of the environmental stressors that Mtb encounters within this specific niche, and allow us to identify genetic programs that are critical for the transition between ATB and LTBI. This information can be used to facilitate development of Mtb vaccines, diagnostics, and therapeutics, and consequently generate a more targeted approach to prevent, identify, and treat TB infection.
Materials and Methods
Study Design and Statistical Analysis
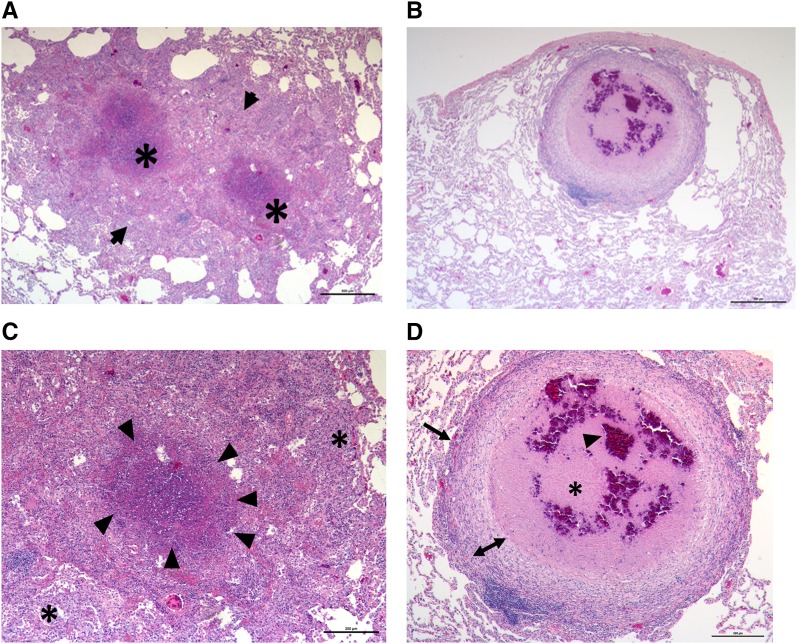
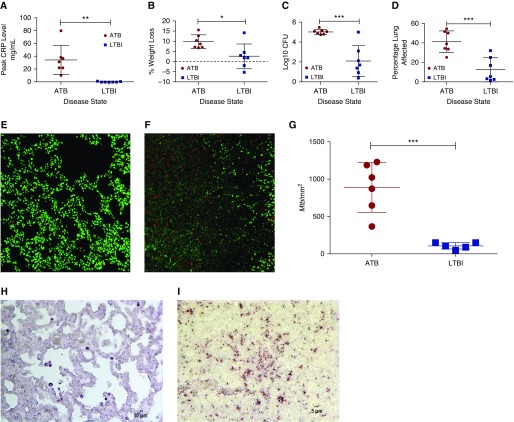
The goal of this study was to assess the in vivo Mtb transcriptome in defined microanatomic compartments typical of ATB (Figures 1A and 1B, see Figure E1 in the online supplement) and LTBI (Figures 1C and 1D, Figure E2). We observed that LTBI correlated with solitary lesions with well defined central necrosis (caseum) and defined cellular layers including fibrosis. These were referred to as “classical granulomata” or “classical granulomas.” In contrast, pulmonary pathology in ATB was characterized by the presence of a wide variety of granulomata, including coalescing of classical granulomas with caseum as well as other less-organized TB-related lesions, which include, but are not limited to: (1) granulomatous inflammation characterized by poorly organized cellular infiltration, predominately consisting of macrophages, but also consisting of other inflammatory cells; and (2) nonnecrotizing granulomas with a thin layer of lymphocytes surrounding a central core of predominantly epithelioid macrophages. In addition, due to the increased diversity of lesions in ATB, Mtb transcriptome was examined in the amalgamation of ATB-induced lesions by dissecting an area, referred to as “representative pathology,” consisting of all of the aforementioned lesion types evident on the section. Sections of FFPE lung tissue obtained from two groups of previously infected Mtb-infected NHPs (ATB [n = 7] or LTBI [n = 7]; Table E1) (4, 18) were microdissected, tissue digested, and RNA extracted, as described previously (21). Animals with ATB had higher serum C-reactive protein levels (Figure 2A; P < 0.01), significantly greater percent weight loss (Figure 2B; P < 0.05), and significantly higher lung bacterial burden (Figure 2C; P < 0.005) and lung pathology (Figure 2D; P < 0.005) as compared with animals with LTBI. These results are further supported by the Mtb-specific staining of lung sections using multilabel confocal microscopy (Figures 2E–2G) and chromogen staining (Figures 2H and 2I). In each of these instances, significantly higher Mtb burden was detected by staining fixed, random lung tissue sections with Mtb-specific antibodies in the animals with ATB relative to LTBI. In the case of confocal microscopy, we were able to quantify these signals, and found that the quantity of Mtb present per square millimeter was significantly higher in the lungs of animals with ATB than in those with LTBI (P < 0.005). Because the extracted heterogeneous RNA samples contain predominately host-specific transcripts, RNA was amplified (Ovation FFPE WTA System; Nugen, San Carlos, CA) and purified (QIAquick PCR Purification Kit; Qiagen, Germantown, MD). For microarray analysis, 3 μg cDNA samples (Alexa Fluor 5) and 3 μg CDC1551 gDNA (Alexa Fluor 3; BEI Resources, Manassas, VA) were labeled. Mtb microarrays were used to compare samples to control (4). Statistically significant genes were determined using linear models for microarray analysis (22). Multiple hypothesis error was corrected using a Bonferroni correction (P < 0.05) that has been previously described (14, 16).
Figure 1.
(A and B) Representative hematoxylin and eosin (H&E) image of active tuberculosis (ATB; DF30) lung. (A) A ×25 magnification image of two coalescing granulomata, with extensive intervening and surrounding regions of inflammation. We termed this presentation as “representative pathology,” which includes classical granulomata and associated caseum. Arrows define the two coalescing lung granulomas. (B) A ×50 magnification image of an animal with ATB. (C and D) Representative H&E image of latent tuberculosis infection (LTBI) (FJ05) lung. (C) A ×25 magnification image of a solitary granuloma surrounded by regions of normal lung. (D) A ×50 magnification image of the same granuloma clearly describes the presence of a central necrotic region (the caseum). We termed such lesions as “classical granulomata.” (A,C, and D) Asterisks correspond to necrotic-hypoxic centers within granulomas. (C) The arrowheads define the boundary between the necrotic center and the cellular layer to the outside. (D) The arrows define the boundary between the granuloma and the normal lung as well as between the myeloid and the lymphocytic layers. Scale bars: 500 μm.
Figure 2.
Clinical correlates of disease state and associated lung bacterial burdens and pathology. (A) Two groups of seven macaques were classified with either ATB (red) or LTBI (blue). Peak serum C-reactive protein (CRP; mg/ml), (B) changes in body weight, (C) Mycobacterium tuberculosis (Mtb) bacterial burden per gram of lung tissue after killing, (D) percentage lung affected by tuberculosis pathology at necropsy and over course of infection, representative confocal staining of a section of lung with anti-Mtb antibody for an animal with LTBI (E) and with ATB (F). Mtb, red; To-Pro-3 iodide, green; quantification of Mtb-positive signal over area (mm2) (G); representative chromogen staining of a section of lung with anti-Mtb antibody for an animal with LTBI (H) and with ATB (I). Scale bar in panel H is 10 μm, and scale bar in panel I is 5 μm. *P < 0.05, **P < 0.01 and ***P < 0.001 using unpaired Student’s t test.
Host-specific hypoxia was assessed using Agilent Rhesus Macaque microarrays (Agilent Technologies, Santa Clara, CA) relative to normal lung tissue derived from uninfected macaques as baseline. Lesion hypoxia was assessed in tissues of animals injected with pimonidazole hydrochloride (PIMO; Hypoxyprobe, Burlington, MA) coupled to daylight red (16). A subset of 86 genes (five housekeeping) was used to validate microarray profiles using nCounter Analysis (23). In addition, FFPE human lung blocks from patients with ATB were obtained from R.L.H. and S.-A.H. for validation of NHP gene expression profiles. A subset of bacillary genes with clearly defined expression profiles were used for real-time RT-PCR, performed as described previously (4, 21, 24–30), for an additional level of validation of results. For this purpose, RNA isolated from in vitro–grown, log-phase (0.4 optical density) Mtb cultures was used for comparison, and data were normalized using 16S gene as internal reference.
Results
Mesodissected samples from ATB and LTBI were used to identify Mtb genes that were expressed in a statistically significant manner in vivo. Significance was defined as those genes the expression level of which differed from that generated by the use of a discrete, constant amount of Mtb genomic DNA (control). The expression of approximately three-fourths of the entire Mtb genome could be detected in a statistically significant manner in all lesion types derived from NHPs with ATB, with 2,909, 2,848, and 2,910 genes being detected in representative pathology, classical granuloma, and caseum of the classical granuloma, respectively (Tables E2–E4). A similar analysis in lesions derived from animals with LTBI showed that 1,874 genes exhibited expression in classical granulomas and 1,872 in the caseum of the classical granuloma (Tables E5 and E6).
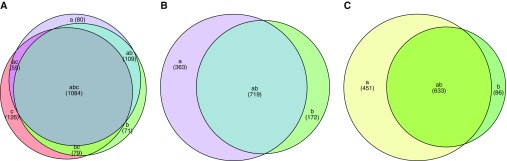
Microarray sample inputs consisted of 3 μg of either amplified, lung-derived mixed sample or Mtb control. Therefore, genes detected with a positive fold change in the mixed sample compared with the pure Mtb control reflect much larger actual fold changes. Given these limitations, we subsequently focused on statistically significant genes with a positive fold change. In ATB samples, 1,344, 1,328, and 1,343 genes were detected in a positive manner in representative pathology, classical granuloma, and caseum of the classical granuloma, respectively (Figure 3A, Tables E2–E4). In LTBI samples, the expression of 1,082 and 891 genes with a positive fold change could be detected in classical granuloma and associated caseum (Figure 3B, Tables E5 and E6). The expression of a core group of 633 genes was induced in ATB, LTBI, and among all lesion types (Figure 3C, Table E7).
Figure 3.
Transcriptome profiles. Statistically significant genes whose fold change expression is positively induced within nonhuman primate (NHP) lung in (A) ATB (a) classical granuloma, (b) caseum of classical granuloma, (c) representative pathology; and (B) LTBI (a) classical granuloma, (b) caseum of classical granuloma. (C) Corresponding genetic similarity among genes with induced expression in (a) all ATB granulomatous pathology, (b) all LTBI granulomatous pathology and (ab) genes with induced expression in all NHP-derived samples. P < 0.05 using linear models for microarray analysis and Bonferroni correction.
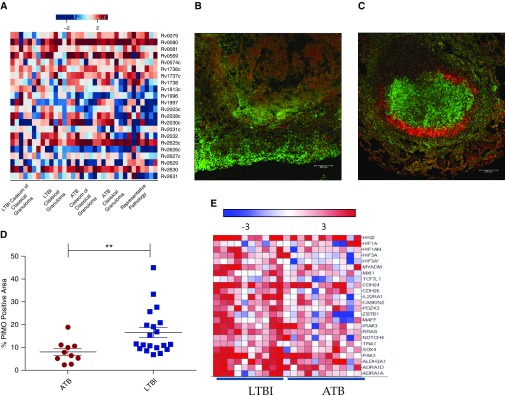
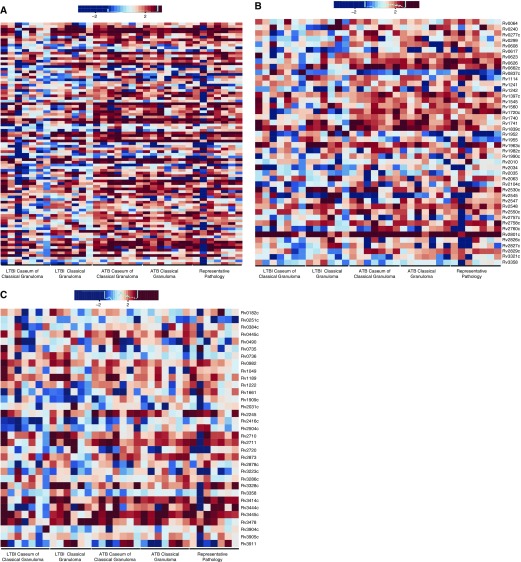
The dormancy survival regulon (dosR) consists of 48 genes up-regulated during a multitude of in vitro stress conditions that mimic the environment faced by Mtb in vivo, including hypoxia (31). This regulon has generally been considered essential for Mtb dormancy, although conclusive evidence in this regard was lacking until recently (16). In ATB lesions, the expression of approximately 26, 34, and 32% of the genes within this regulon were found to be induced in a statistically significant manner in representative pathology, classical granuloma, and caseum of the classical granuloma, respectively (Table E8). In LTBI samples, approximately 30% of the genes within the regulon could be similarly detected in both classical granuloma and caseum of the classical granuloma. In addition, eight dosR regulon members (Rv0080, Rv0081, Rv1736c, Rv1737c, Rv2032, Rv2625c, and Rv2630) were present in the core group common to all ATB and LTBI samples. We predicted that the expression of members of this regulon would occur at higher levels in animals progressing to LTBI. The greatest induction of genes of the dosR regulon occurred in the most hypoxic areas of the granuloma (e.g., the caseum derived from LTBI granulomas, followed by entire LTBI classical granuloma and the caseum derived from ATB lesions; Figure 4A).
Figure 4.
Expression of Mtb dormancy survival regulon (dosR regulon) and detection of hypoxia in ATB and LTBI lung granulomas. (A) Comparison of genes within the dosR regulon with induced expression in all NHPs with either LTBI or ATB in each granuloma sample subset. For all heat maps included here, genes within the regulon or gene family of interest were included based upon published association and statistically significant positively induced expression within at least one granuloma category. The least induction is seen in ATB representative pathology, and the most in LTBI. (B) Confocal image of pimonidazole hydrochloride (PIMO) conjugated with Daylight-546 (red) with To-Pro-3 iodide staining all nuclei (green). Signal in a representative ATB animal was much less than signal in a representative LTBI animal (C). Scale bars: 100 μm. In addition, PIMO signal within the lung as a percentage of total microscopic field was significantly greater in LTBI (blue squares) than ATB (red circles) (Student’s t test, **P < 0.005) (D). Finally, heat map of genes associated with host hypoxic response showed greater response during LTBI as compared with ATB within NHP granulomas (E).
Next, to further gauge the degree of hypoxia within the granuloma, we surveyed the amount of hypoxia within ATB and LTBI using PIMO (16) (Figures 4B and 4C). TB lung lesions in both ATB- and LTBI-derived granulomas were positive for PIMO, indicating hypoxia. During ATB, PIMO was evident throughout granulomatous pathology (Figure 4B). In contrast, in LTBI samples, PIMO signal was predominately localized to the inner rim of the granuloma surrounding the caseum (Figure 4C). PIMO signal was calculated as a percentage of lung and granuloma area within various microscopic fields (Figure 4D). The highest levels of PIMO were observed in the classical granulomas of NHPs infected with LTBI (Figure 4D). To determine if the expression of host genes known to be induced by hypoxia was correspondingly increased in LTBI compared with ATB, we screened a macaque transcriptome dataset of granuloma lesions isolated from 21 animals (LTBI [n = 10] or ATB [n = 11]) relative to normal lung tissue (S.M., A.A.L., and D.K., unpublished data), specifically focusing on host genes induced by hypoxia and regulated by hypoxia-inducible factor 1 (HIF-1) (32) (Tables E9 and E10). These genes exhibited higher expression in lesions derived from both groups (Figures 4B and 4E) with relatively higher expression of numerous hypoxia-sensitive genes in LTBI rather than ATB samples. These data suggest that the extent of the hypoxic environment is greater in lesions derived from animals with LTBI, strongly supporting our bacterial transcriptome data.
We detected the induced expression of a large number of genes that belong to the proline-glutamate (PE) or proline-proline-glutamate (PPE) family of genes (Table E11). We further assessed the expression of PE/PPE in vivo by supervised clustering and observed increased induction in ATB samples (Figure 5A). The PE/PPE family consists of more than 160 members unique to mycobacteria, which have been implicated in antigenicity and associated with persister formation (33). We also investigated six known gene families and regulons for their intragranulomatous expression, including sigma factors, toxin–antitoxin (TA) systems, lipid metabolism, Esx1, enduring hypoxic response, and persisters. The expression of several TA genes, which potentially aid in the survival and persistence of Mtb, was highly induced (Table E12). Although some differences existed between lesion types, which is expected given the widely accepted concept of lesion heterogeneity, we were able to detect approximately 26% of these genes in each of the granuloma types derived from animals with ATB (34). In animals with LTBI, we detected approximately 21% in the classical granuloma and 16% in the caseum. TAs depicted greater induction in ATB than LTBI (Figure 5B). The majority of the TAs expressed belonged to the largest TA family in the genome, the vapBC family. In total, we found the induction of 15 toxins and 14 antitoxins belonging to this family, including 4 pairs: vapBC19, vapBC21, vapBC33, and vapBC34, within granulomas.
Figure 5.
Mechanisms of persistence and validation. Comparison of genes within the (A) proline-glutamate/proline-proline-glutamate gene family, (B) toxin–antitoxin complexes, and (C) sigma factor family with induced expression in all NHPs with either LTBI or ATB in each granuloma sample subset.
To better assess the mechanisms of modulation of Mtb gene expression in vivo, we assessed known sigma (sig) factors and their associated genes (Table E13). We detected the following five in all samples: sigB, sigD, sigI, sigJ, and sigF. We also detected the following factors within specific environments and disease states: LTBI—sigL and sigM (caseum), as well as sigK and sigG (granuloma); ATB—sigM, sigK, sigG (caseum), sigK (granuloma), sigG (representative pathology), and sigH (caseum). Overall, greater expression was detected in ATB, similarly to many of the aforementioned pathways (Figure 5C). These results may suggest specific roles for each of these factors in facilitating the transition from ATB to LTBI. Thus, sigF and sigD have been shown to be required in the adaptation to stationary phase (35), whereas the induction of sigJ, sigI, sigB, sigK, and sigH supports the importance of the oxidative stress on various bacterial components within the granuloma (35). These results point to a battery of alternate sigma factors being critically important for the survival of Mtb in vivo by modulating gene expression in response to changing milieu.
We detected the up-regulation of 539 genes in a statistically significant manner common to all six granulomatous samples derived from patients with ATB (Table E14). In addition, 391 of the aforementioned genes were also detected in all NHP-derived samples. Using a subset of 86 genes, we validated gene expression profiles via a microfluidic approach (Figure E4, supplemental Materials and Methods, Table E15).
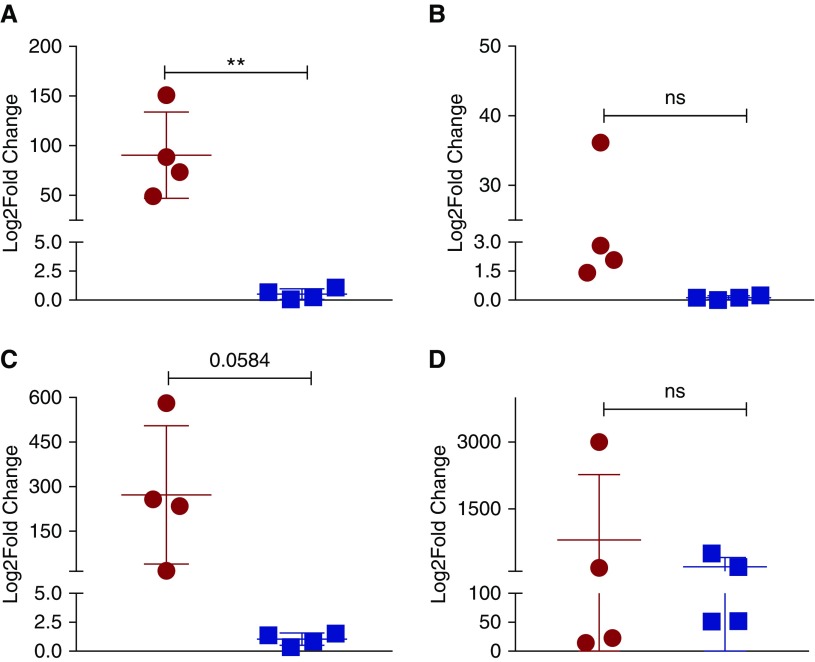
In addition, a small subset of genes with specific and interesting expression profiles were cherry picked and transcriptome profiles validated by real-time RT-PCR in comparable classical granulomata derived from animals with ATB and LTBI (Figure 6). The expression of vapB21, an antitoxin of the TA system, was validated to be induced approximately 100-fold in the classical lesions derived from animals with ATB, significantly higher than the expression in classical granulomata samples from animals with LTBI, where no induction was observed (Figure 6A). The expression of oxidative stress response factors, sigE (12- versus 0.1-fold) and sigH (∼300-fold versus 1.2-fold), was highly induced in lesions derived from ATB relative to LTBI animals (Figures 6B and 6C). In the case of sigH, the difference approached statistical significance (P = 0.0584). These results were comparable to those derived from whole-genome microarray analysis. The expression of the dosR gene was robustly detected in lesions derived from animals with both clinical outcomes, as was suggested by transcriptome and NanoString data (Figure 6D). The dosR levels were not statistically significantly different, but the overall expression was slightly higher in granulomata from animals with LTBI relative to ATB.
Figure 6.
Real-time RT-PCR to validate macaque Mtb transcriptome results. Using cDNA derived from in vitro, log-phase–grown Mtb, and using the 16S gene as an internal reference, the fold changes of expression of vapB21 (A), sigE (B), sigH (C), and dosR (D) was assessed in classical granulomata derived from animals with ATB (red circles) or LTBI (blue squares) (Student’s t test, **P < 0.005). ns, not significant.
We compared the genes expressed within granulomas to those previously essential for survival of the bacilli (Figure E5A) (4, 36–39). We found the greatest degree of similarity between intragranulomatous data described here to our previously conducted NHP mutant experiment, where we identified 108 genes unable to survive in macaque lungs during active TB, implying that the genes whose expression was interrupted in these mutants were important for pathogenesis and likely expressed in vivo (4) (Table E16). An overwhelming number of genes overlapped in each disease state with 27% belonging to all ATB derived lesions, 18% to all LTBI derived lesions, and 17% to all NHP-derived lesions (see the online supplement). The next greatest degree of similarity was found with a computational prediction model (39), followed by the in vitro/mouse TraSH (36), murine macrophage TraSH (38), and a mouse designer arrays for defined mutant analysis study (37) (Tables E17–E20). We detected more essential genes in ATB, presumably due to a higher burden of actively replicating bacteria. Pathway analysis of ATB and LTBI shows a high degree of known connection between essential genes defined by DeJesus and colleagues (39) (Figures E5B and E5C). Interestingly, we detected 115 essential genes common to all ATB and LTBI NHP samples (Figure E5D) (39), approximately 50% of which function in intermediary metabolism and respiration, representing essential mechanisms that Mtb employs to transition between active and latent disease. These results suggest the reprogramming of Mtb metabolism within lung lesions due to the varied availability of nutrients and metabolites.
Discussion
We assessed Mtb genes induced within defined microanatomic compartments of the TB lung in NHPs with ATB and LTBI. Due to route of infection, pathologic, and disease spectrum similarities between NHPs and humans, especially in the context of TB, the NHP model is ideally suited to assess in vivo Mtb gene expression. Lung samples used here reflect human TB disease, because, as with human infection, NHPs were infected via aerosol. In addition, NHP produce a spectrum of lesions most similar to those found in human patients with TB as compared with other animal models; therefore, this is the best model for assessing specific granuloma microenvironments. This work extends beyond just assessing active disease transcriptome profiles, but also assesses the transition between LTBI and ATB. As in humans, NHPs also develop LTBI. Unlike humans, it is possible to ethically obtain pulmonary endpoint samples from latently infected NHPs. Here, we identified transcriptome profiles of both ATB and LTBI. In addition, profiles were validated with lesions derived from human samples with active disease and comparable results obtained. Our results suggest that certain Mtb pathways are critical for the transition from LTBI to ATB disease.
Here, we identify regulons associated with ATB, LTBI, and the combination thereof in representative ATB pathology, ATB and LTBI classical granulomas, and their subsequent caseum. We identify a core group of genes common to all lesions, indicating an underlying shared bacterial program to respond to the granulomatous environment, regardless of the clinical disease status. Our results conclusively demonstrate that the response of the bacillus to hypoxia is critical for the transition to latency. By finding the enhanced expression of hypoxia-responsive regulons, we provide the conclusive in vivo evidence of its importance in maintaining Mtb in the chronic state of infection with the granuloma. By identifying genes and pathways involved in Mtb persistence, maintenance, and growth in vivo, and coupling these findings to pre-existing mutant-based studies, this study provides critical information on genes that can be used in a potentially novel, targeted vaccine and therapeutic approaches.
Unsurprisingly, more genes were detected in a statistically significant manner in ATB than LTBI, as the latter is characterized by a greater bacterial burden as well as a higher magnitude of host response (5). The induced expression of a core group of 633 genes was identified in all intragranulomatous lesions, representing a core group of genes essential for Mtb survival and persistence within the granuloma, suggesting that these genes may be necessary for the transition from ATB to LTBI.
Stress is vitally important for Mtb gene regulation (40); consequently, we studied regulons known to respond to specific validated conditions, with a primary focus on the expression of the hypoxia-sensing dosR gene and its regulon (31). While not required for initial infection, the dosR regulon is essential for the persistence of Mtb in human-like caseous granulomas (16). Furthermore, the lack of dosR in Mtb allows the recruitment of stronger adaptive immunity (16). A large number of dosR-dependent genes was expressed in every sample, disease state, and pathologic lesion studied. Importantly, intragranulomatous expression patterns of dosR-dependent genes circumvented our selection bias and strongly correlated with oxygenation patterns. Thus, the expression of the dosR regulon was at the lowest level in the least hypoxic lesion type (ATB-representative pathology), whereas it was the greatest in the most hypoxic samples (LTBI, particularly caseum). This pattern was best exemplified by Rv1813c, which is coregulated by both dosR and mprAB two-component regulatory systems. It exhibited increased expression in LTBI, especially in the hypoxic caseum of the classical granuloma, demonstrating the ability of the bacillus to use multiple regulatory pathways concurrently to recognize, respond, and persist in specific environmental conditions.
At least two of the genes identified in this screen, Rv1996 and Rv2028c, are implicated in inducing greater T cell responses in patients with LTBI than ATB (41). Finally, Rv2031c, which exhibited the highest induction in expression in LTBI and ATB caseum-derived samples, is up-regulated during latency. This gene has recently been implicated in silico as an important regulator in cellular hypoxic stress response via its regulation of the Rv2028-Rv2031 operon. The higher induction of the dosR regulon in samples derived from macaques with LTBI was reinforced by increased hypoxia (PIMO) signal, as well as increased host HIF-1 expression, which is a known regulator of hypoxic responses (32). The expression of the downstream HIF-regulated host genes also occurred at much higher levels in LTBI- relative to ATB-derived samples. Our study shows the critical importance of such mechanisms that allow the bacteria to alter its metabolism to favor survival in hostile conditions.
The expression of some dosR-dependent genes was high throughout the granuloma types and disease states. This profile was exemplified by Rv0569, which encodes a signal transducer during hypoxia and by Rv2625c (rip3), which encodes a mitochondrial reactive oxygen species induced in a TNF-associated pathway (42). In conditions of excess TNF, ROS induces macrophage necrosis, allowing bacterial release into the extracellular environment, increasing host susceptibility (42). Because hypoxia was detected in all samples tested, our data strongly suggest that the induction of dormancy is critical in all granuloma stages, a result supported by observations that lesions of macaques with both ATB and LTBI contain hypoxia (Figures 4B–4D) (16). Our results are also supported by the recent evidence of dosR expression in the lungs of humans with TB (43). Overall, the pathogen’s response to hypoxia is a critical component of its intragranulomatous physiology. Although long suspected (44), we are only beginning to appreciate the importance of dosR and hypoxia in governing the transition from active to latent TB in classical lesions, and its impact on immunity.
Due to sustained Mtb replication, granulomata of different maturation levels can be observed in the same animal with ATB. These granulomata of varying pathologies provide an array of diverse environments to which the bacilli must respond (45). Some of these lesions, especially the less mature ones, have not yet evolved to contain a fibrotic cuff on the exterior (Figure 1). As a result, these lesions are able to “breathe,” and don’t experience radically reduced oxygen tensions. On the contrary, virtually every LTBI lesion is highly ordered (Figure 1). Hence, it is not surprising that the latter are more hypoxic and invoke a greater hypoxic response from both the host and the pathogen. It is believed that hypoxia within the LTBI lesions is the trigger that alters the physiology of the pathogen in a manner such that bacilli acquire a dormant (or persistent) phenotype. There is support for our contention in the published literature. Thus, metronidazole is a drug that is only effective in hypoxic conditions, presumably against persisters. Treatment of LTBI, but not ATB, with metronidazole in this system prevents conversion to ATB (46).
Recent studies have in particular underlined the importance of the dosR regulon. T cells from humans with LTBI recognize dosR-regulated antigens, suggesting that these proteins are expressed and presented in vivo during latency (47). Infection of macaques with Mtb mutants in the dosR regulon causes nonpathogenic infection with enhanced adaptive immune responses recruited to the lung (16). Hence, the dosR response appears integral to Mtb pathogenesis, and helps the pathogen manipulate immunity. Finally, an unbiased, system-wide proteomic approach found that, upon in vitro hypoxic stress, 20% of all Mtb protein mass is contributed by the fewer than 50 dosR-regulated genes (48). Moreover, the expression of this regulon was recently reported to be strongly induced in human TB and to significantly lower levels in patients with HIV (43). This suggests that the expression of DosR may promote more robust granulomas, a contention supported by data that expression of DosR by WhiB6 promotes granuloma stability in an environment of chronic oxidative/nitrosative stress (49). Thus, the notion that dosR is critical to the reprogramming of Mtb physiology in hypoxic conditions in vivo is increasingly supported by experimental data (50). We propose that, in macaque as well as human lesions, progressively increasing hypoxia elicits the expression of the DosR regulon. This results in the blockade of the highly bactericidal Th1 response from accessing the bacilli within the lesion through the expression of DosR-regulated antigens. The lesions characterized by high DosR expression are therefore likely robust granulomas that do not allow the bacilli to spread. The current study cements the role of the dosR regulon in Mtb persistence within human-like caseous lung granulomas by providing conclusive evidence for its deployment in this important niche (16). Because it is likely that dosR-regulated antigens are expressed intragranulomatously, cognate Mtb-specific T cells could be effective in controlling or eradicating infection. By extension, our data suggest that the induction of dosR (or dormancy) results in the expression of numerous specific antigenic epitopes. Our results provide a compelling rationale to study responses specific to dosR-expressed antigens, and suggest that vaccination approaches that induce CD4 and CD8 responses to these may be successful.
Acknowledgments
Acknowledgments
This article is dedicated to the memory of Andrew A. Lackner, D.V.M., Ph.D., D.A.C.V.P., a co-author on this study and the long-serving Director of the Tulane National Primate Research Center, who passed away after this article had been accepted for publication.
Footnotes
This work was supported by Public Health Service grants AI089323, HL106790, AI091457, OD011104, OD011124, AI058609, and GM103424, the Louisiana Board of Regents, the Tulane National Primate Research Center Office of the Director, the Tulane Research Enhancement Fund, the Tulane Office of Vice President for Research, and the Wetmore Foundation of Louisiana.
Author Contributions: Research—T.A.H., T.W.F., U.S.G., A.V.V., D.N.L., K.M.G., A.C., M.M., and X.A.; analysis—T.A.H., T.W.F., N.B., U.S.G., A.V.V., D.N.L., N.A.G., S.-A.H., R.L.H., X.A., A.A.L., J.S.B., S.M., and D.K.; provided clinical tuberculosis samples—S.-A.H. and R.L.H.; writing—T.A.H., T.W.F., A.A.L., S.M., and D.K.; funding—A.A.L., S.M., and D.K.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0239OC on January 30, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Russell DG, Barry CE, III, Flynn JL. Tuberculosis: what we don’t know can, and does, hurt us. Science. 2010;328:852–856. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foreman TW, Mehra S, LoBato DN, Malek A, Alvarez X, Golden NA, Bucşan AN, Didier PJ, Doyle-Meyers LA, Russell-Lodrigue KE, et al. CD4+ T-cell-independent mechanisms suppress reactivation of latent tuberculosis in a macaque model of HIV coinfection. Proc Natl Acad Sci USA. 2016;113(38):E5636–E5644. doi: 10.1073/pnas.1611987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehra S, Golden NA, Stuckey K, Didier PJ, Doyle LA, Russell-Lodrigue KE, Sugimoto C, Hasegawa A, Sivasubramani SK, Roy CJ, et al. The Mycobacterium tuberculosis stress response factor SigH is required for bacterial burden as well as immunopathology in primate lungs. J Infect Dis. 2012;205:1203–1213. doi: 10.1093/infdis/jis102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutta NK, Mehra S, Didier PJ, Roy CJ, Doyle LA, Alvarez X, Ratterree M, Be NA, Lamichhane G, Jain SK, et al. Genetic requirements for the survival of tubercle bacilli in primates. J Infect Dis. 2010;201:1743–1752. doi: 10.1086/652497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehra S, Pahar B, Dutta NK, Conerly CN, Philippi-Falkenstein K, Alvarez X, Kaushal D. Transcriptional reprogramming in nonhuman primate (rhesus macaque) tuberculosis granulomas. PLoS One. 2010;5:e12266. doi: 10.1371/journal.pone.0012266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehra S, Golden NA, Dutta NK, Midkiff CC, Alvarez X, Doyle LA, Asher M, Russell-Lodrigue K, Monjure C, Roy CJ, et al. Reactivation of latent tuberculosis in rhesus macaques by coinfection with simian immunodeficiency virus. J Med Primatol. 2011;40:233–243. doi: 10.1111/j.1600-0684.2011.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta NK, Mehra S, Martinez AN, Alvarez X, Renner NA, Morici LA, Pahar B, Maclean AG, Lackner AA, Kaushal D. The stress-response factor SigH modulates the interaction between Mycobacterium tuberculosis and host phagocytes. PLoS One. 2012;7:e28958. doi: 10.1371/journal.pone.0028958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushal D, Mehra S. Faithful experimental models of human Mycobacterium tuberculosis infection. Mycobact Dis. 2012;2:e108. doi: 10.4172/2161-1068.1000e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaushal D, Mehra S, Didier PJ, Lackner AA. The non-human primate model of tuberculosis. J Med Primatol. 2012;41:191–201. doi: 10.1111/j.1600-0684.2012.00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gopal R, Monin L, Torres D, Slight S, Mehra S, McKenna KC, Fallert Junecko BA, Reinhart TA, Kolls J, Báez-Saldaña R, et al. S100A8/A9 proteins mediate neutrophilic inflammation and lung pathology during tuberculosis. Am J Respir Crit Care Med. 2013;188:1137–1146. doi: 10.1164/rccm.201304-0803OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slight SR, Rangel-Moreno J, Gopal R, Lin Y, Fallert Junecko BA, Mehra S, Selman M, Becerril-Villanueva E, Baquera-Heredia J, Pavon L, et al. CXCR5+ T helper cells mediate protective immunity against tuberculosis. J Clin Invest. 2013;123:712–726. doi: 10.1172/JCI65728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darrah PA, Bolton DL, Lackner AA, Kaushal D, Aye PP, Mehra S, Blanchard JL, Didier PJ, Roy CJ, Rao SS, et al. Aerosol vaccination with AERAS-402 elicits robust cellular immune responses in the lungs of rhesus macaques but fails to protect against high-dose Mycobacterium tuberculosis challenge. J Immunol. 2014;193:1799–1811. doi: 10.4049/jimmunol.1400676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo Q, Mehra S, Golden NA, Kaushal D, Lacey MR. Identification of biomarkers for tuberculosis susceptibility via integrated analysis of gene expression and longitudinal clinical data. Front Genet. 2014;5:240. doi: 10.3389/fgene.2014.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaushal D, Foreman TW, Gautam US, Alvarez X, Adekambi T, Rangel-Moreno J, Golden NA, Johnson AM, Phillips BL, Ahsan MH, et al. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat Commun. 2015;6:8533. doi: 10.1038/ncomms9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine DM, Dutta NK, Eckels J, Scanga C, Stein C, Mehra S, Kaushal D, Karakousis PC, Salamon H. A tuberculosis ontology for host systems biology. Tuberculosis (Edinb) 2015;95:570–574. doi: 10.1016/j.tube.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehra S, Foreman TW, Didier PJ, Ahsan MH, Hudock TA, Kissee R, Golden NA, Gautam US, Johnson AM, Alvarez X, et al. The DosR regulon modulates adaptive immunity and is essential for M. tuberculosis persistence. Am J Respir Crit Care Med. 2015;191:1185–1196. doi: 10.1164/rccm.201408-1502OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mothé BR, Lindestam Arlehamn CS, Dow C, Dillon MB, Wiseman RW, Bohn P, Karl J, Golden NA, Gilpin T, Foreman TW, et al. The TB-specific CD4 T cell immune repertoire in both cynomolgus and rhesus macaques largely overlap with humans. Tuberculosis (Edinb) 2015;95:722–735. doi: 10.1016/j.tube.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips BL, Mehra S, Ahsan MH, Selman M, Khader SA, Kaushal D. LAG3 expression in active Mycobacterium tuberculosis infections. Am J Pathol. 2015;185:820–833. doi: 10.1016/j.ajpath.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehra S, Alvarez X, Didier PJ, Doyle LA, Blanchard JL, Lackner AA, Kaushal D. Granuloma correlates of protection against tuberculosis and mechanisms of immune modulation by Mycobacterium tuberculosis. J Infect Dis. 2013;207:1115–1127. doi: 10.1093/infdis/jis778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutta NK, McLachlan J, Mehra S, Kaushal D. Humoral and lung immune responses to Mycobacterium tuberculosis infection in a primate model of protection. Trials Vaccinol. 2014;3:47–51. doi: 10.1016/j.trivac.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudock TA, Kaushal D. A novel microdissection approach to recovering Mycobacterium tuberculosis specific transcripts from formalin fixed paraffin embedded lung granulomas. J Vis Exp. 2014;88:51693. doi: 10.3791/51693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 23.Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol. 2011;Chapter 25:Unit25B.10. doi: 10.1002/0471142727.mb25b10s94. [DOI] [PubMed] [Google Scholar]
- 24.Mehra S, Kaushal D. Functional genomics reveals extended roles of the Mycobacterium tuberculosis stress response factor sigmaH. J Bacteriol. 2009;191:3965–3980. doi: 10.1128/JB.00064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutta NK, Mehra S, Kaushal D. A Mycobacterium tuberculosis sigma factor network responds to cell-envelope damage by the promising anti-mycobacterial thioridazine. PLoS One. 2010;5:e10069. doi: 10.1371/journal.pone.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehra S, Dutta NK, Mollenkopf HJ, Kaushal D. Mycobacterium tuberculosis MT2816 encodes a key stress-response regulator. J Infect Dis. 2010;202:943–953. doi: 10.1086/654820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gautam US, McGillivray A, Mehra S, Didier PJ, Midkiff CC, Kissee RS, Golden NA, Alvarez X, Niu T, Rengarajan J, et al. DosS is required for the complete virulence of Mycobacterium tuberculosis in mice with classical granulomatous lesions. Am J Respir Cell Mol Biol. 2015;52:708–716. doi: 10.1165/rcmb.2014-0230OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGillivray A, Golden NA, Gautam US, Mehra S, Kaushal D. The Mycobacterium tuberculosis Rv2745c plays an important role in responding to redox stress. PLoS One. 2014;9:e93604. doi: 10.1371/journal.pone.0093604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautam US, Mehra S, Kaushal D. In-vivo gene signatures of Mycobacterium tuberculosis in C3HeB/FeJ mice. PLoS One. 2015;10:e0135208. doi: 10.1371/journal.pone.0135208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGillivray A, Golden NA, Kaushal D. The Mycobacterium tuberculosis Clp gene regulator is required for in vitro reactivation from hypoxia-induced dormancy. J Biol Chem. 2015;290:2351–2367. doi: 10.1074/jbc.M114.615534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boon C, Dick T. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J Bacteriol. 2002;184:6760–6767. doi: 10.1128/JB.184.24.6760-6767.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fishbein S, van Wyk N, Warren RM, Sampson SL. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol. 2015;96:901–916. doi: 10.1111/mmi.12981. [DOI] [PubMed] [Google Scholar]
- 34.Lin PL, Ford CB, Coleman MT, Myers AJ, Gawande R, Ioerger T, Sacchettini J, Fortune SM, Flynn JL. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med. 2014;20:75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sachdeva P, Misra R, Tyagi AK, Singh Y. The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS J. 2010;277:605–626. doi: 10.1111/j.1742-4658.2009.07479.x. [DOI] [PubMed] [Google Scholar]
- 36.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamichhane G, Tyagi S, Bishai WR. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect Immun. 2005;73:2533–2540. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci USA. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeJesus MA, Zhang YJ, Sassetti CM, Rubin EJ, Sacchettini JC, Ioerger TR. Bayesian analysis of gene essentiality based on sequencing of transposon insertion libraries. Bioinformatics. 2013;29:695–703. doi: 10.1093/bioinformatics/btt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flentie K, Garner AL, Stallings CL. Mycobacterium tuberculosis transcription machinery: ready to respond to host attacks. J Bacteriol. 2016;198:1360–1373. doi: 10.1128/JB.00935-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hozumi H, Tsujimura K, Yamamura Y, Seto S, Uchijima M, Nagata T, Miwa S, Hayakawa H, Fujisawa T, Hashimoto D, et al. Immunogenicity of dormancy-related antigens in individuals infected with Mycobacterium tuberculosis in Japan. Int J Tuberc Lung Dis. 2013;17:818–824. doi: 10.5588/ijtld.12.0695. [DOI] [PubMed] [Google Scholar]
- 42.Roca FJ, Ramakrishnan L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell. 2013;153:521–534. doi: 10.1016/j.cell.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter ND, de Jong BC, Garcia BJ, Dolganov GM, Worodria W, Byanyima P, Musisi E, Huang L, Chan ED, Van TT, et al. Adaptation of Mycobacterium tuberculosis to impaired host immunity in HIV-infected patients. J Infect Dis. 2016;214:1205–1211. doi: 10.1093/infdis/jiw364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boon C, Dick T. How Mycobacterium tuberculosis goes to sleep: the dormancy survival regulator DosR a decade later. Future Microbiol. 2012;7:513–518. doi: 10.2217/fmb.12.14. [DOI] [PubMed] [Google Scholar]
- 45.Gengenbacher M, Kaufmann SH. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev. 2012;36:514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin PL, Dartois V, Johnston PJ, Janssen C, Via L, Goodwin MB, Klein E, Barry CE, III, Flynn JL. Metronidazole prevents reactivation of latent Mycobacterium tuberculosis infection in macaques. Proc Natl Acad Sci USA. 2012;109:14188–14193. doi: 10.1073/pnas.1121497109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, Kim Y, Sidney J, James EA, Taplitz R, et al. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog. 2013;9:e1003130. doi: 10.1371/journal.ppat.1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubert OT, Ludwig C, Kogadeeva M, Zimmermann M, Rosenberger G, Gengenbacher M, Gillet LC, Collins BC, Röst HL, Kaufmann SH, et al. Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microbe. 2015;18:96–108. doi: 10.1016/j.chom.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Chen Z, Hu Y, Cumming BM, Lu P, Feng L, Deng J, Steyn AJ, Chen S. Mycobacterial WhiB6 differentially regulates ESX-1 and the Dos regulon to modulate granuloma formation and virulence in zebrafish. Cell Reports. 2016;16:2512–2524. doi: 10.1016/j.celrep.2016.07.080. [DOI] [PubMed] [Google Scholar]
- 50.Lipworth S, Hammond RJ, Baron VO, Hu Y, Coates A, Gillespie SH. Defining dormancy in mycobacterial disease. Tuberculosis (Edinb) 2016;99:131–142. doi: 10.1016/j.tube.2016.05.006. [DOI] [PubMed] [Google Scholar]