Abstract
DNA library preparation for high-throughput sequencing of genomic DNA usually involves ligation of adapters to double-stranded DNA fragments. However, for highly degraded DNA, especially ancient DNA, library preparation has been found to be more efficient if each of the two DNA strands are converted into library molecules separately. We present a new method for single-stranded library preparation, ssDNA2.0, which is based on single-stranded DNA ligation with T4 DNA ligase utilizing a splinter oligonucleotide with a stretch of random bases hybridized to a 3΄ biotinylated donor oligonucleotide. A thorough evaluation of this ligation scheme shows that single-stranded DNA can be ligated to adapter oligonucleotides in higher concentration than with CircLigase (an RNA ligase that was previously chosen for end-to-end ligation in single-stranded library preparation) and that biases in ligation can be minimized when choosing splinters with 7 or 8 random nucleotides. We show that ssDNA2.0 tolerates higher quantities of input DNA than CircLigase-based library preparation, is less costly and better compatible with automation. We also provide an in-depth comparison of library preparation methods on degraded DNA from various sources. Most strikingly, we find that single-stranded library preparation increases library yields from tissues stored in formalin for many years by several orders of magnitude.
INTRODUCTION
High-throughput DNA sequencing has become deeply integrated with genetic research over the past years. As current technologies allow for sequencing millions or billions of DNA fragments in parallel at relatively low costs, the scope of data generation is often limited by difficulties in sample preparation rather than sequencing capacity. In spite of recent advances (1,2), nucleic acids cannot be efficiently sequenced in situ, thus requiring the extraction of nucleic acids from the material under study and their subsequent conversion into DNA libraries. This is achieved by attaching synthetic adapters to their ends, which provides a format that enables their amplification and the priming of the sequencing reaction. Losses of molecules occur during both steps of sample preparation and impose challenges on work with small quantities of nucleic acids. Furthermore, DNA molecules are sometimes present in a form that complicates their successful extraction and the preparation of DNA libraries, for example if they are very short.
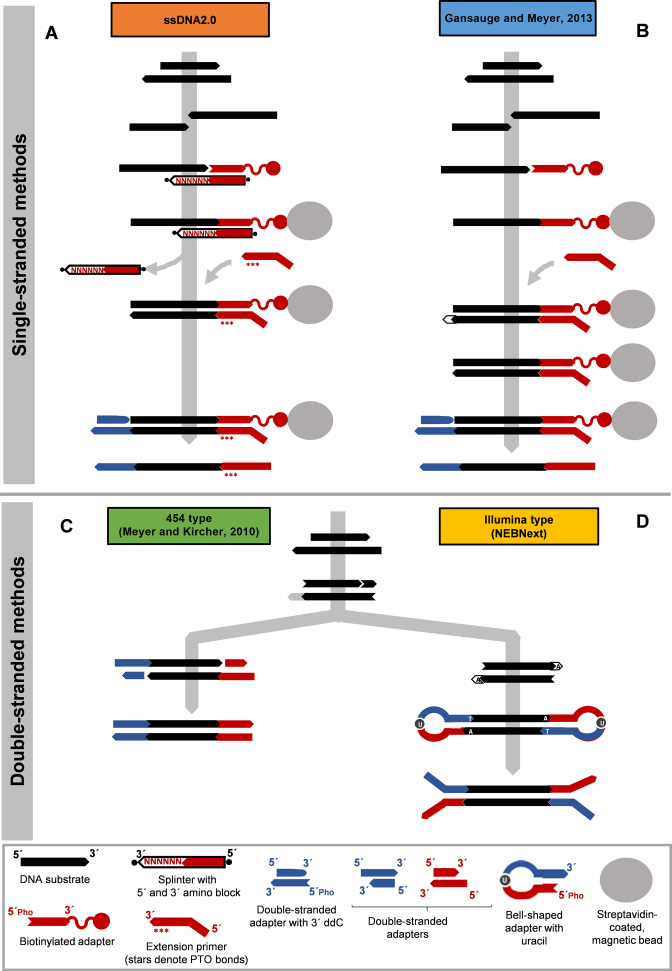
One type of material that is particularly difficult to work with is ancient DNA. The possibility to recover genomic sequences from organisms that died tens or even hundreds of thousand years ago has fascinated evolutionary biologists for decades and has spurred the development of methods to improve the recovery of DNA sequences from fossil remains. One significant leap forward came through the invention of a library preparation method that converts each strand of the DNA fragments separately into library molecules instead of attaching adapters to double-stranded DNA (3,4). A graphical outline of this method is presented in Figure 1B. Briefly, DNA fragments are dephosphorylated and denatured, after which the first adapter is joined to their 3΄ ends using CircLigase. Successfully ligated DNA strands are immobilized on streptavidin-coated magnetic beads. Subsequent reaction steps, which include copying the template strand with a DNA polymerase, the generation of blunt ends and the ligation of the second adapter, are carried out on beads, thereby minimizing losses of DNA in intermittent purification steps. As each strand of a double-stranded fragment can potentially be converted into a library molecule, chances are doubled that at least one strand of a given DNA fragment will be recovered. Moreover, DNA molecules with single-strand breaks, which are not consistently recovered with double-stranded methods, are disassembled and turned into suitable substrates for library preparation.
Figure 1.
Library preparation methods for highly degraded DNA. (A) In the single-stranded library preparation method described here (ssDNA2.0), DNA fragments (black) are 5΄ and 3΄ dephosphorylated and separated into single strands by heat denaturation. 3΄ biotinylated adapter molecules (red) are attached to the 3΄ ends of the DNA fragments via hybridization to a stretch of six random nucleotides (marked as ‘N’) belonging to a splinter oligonucleotide complementary to the adapter and nick closure with T4 DNA ligase. Following the immobilization of the ligation products on streptavidin-coated beads, the splinter oligonucleotide is removed by bead wash at an elevated temperature. Synthesis of the second strand is carried out using the Klenow fragment of Escherichia coli DNA polymerase I and a primer with phosphorothioate backbone modifications (red stars) to prevent exonucleolytic degradation. Unincorporated primers are removed through a bead wash at an elevated temperature, preventing the formation of adapter dimers in the subsequent blunt-end ligation reaction, which is again catalyzed by T4 DNA ligase. Adapter self-ligation is prevented through a 3΄ dideoxy modification in the adapter. The final library strand is released from the beads by heat denaturation. (B) In the single-stranded library preparation method originally described in Gansauge and Meyer, (4), the first adapter was attached through true single-stranded DNA ligation using CircLigase. The large fragment of Bst DNA polymerase was used to copy the template strand, leaving overhanging 3΄ nucleotides, which had to be removed in a blunt-end repair reaction using T4 DNA polymerase. (C) The ‘454’ method of double-stranded library preparation in the implementation of Meyer and Kircher, (23), is based on non-directional blunt-end ligation of a mixture of two adapters to blunt-end repaired DNA fragments using T4 DNA ligase. To prevent adapter self-ligation, no phosphate groups are present at the 5΄ ends of the adapters, resulting in the ligation of the adapter strands only and necessitating subsequent nick fill-in with a strand-displacing polymerase. Intermittent DNA purification steps are required in-between enzymatic reactions. (D) The ‘Illumina’ method of double-stranded library preparation, shown here as implemented in New England Biolabs’ NEBNext Ultra II kit, requires the addition of A-overhangs (marked as ‘A’) to blunt-end repaired DNA fragments using a 3΄-5΄ exonuclease deletion mutant of the Klenow fragment of E. coli DNA polymerase I. Both adapter sequences are combined into one bell-shaped structure, which carries a 3΄ T overhang to allow sticky end ligation with T4 DNA ligase. Following ligation, adapter strands are separated by excision of uracil. Excess adapters and adapter dimers are removed through size-selective purification.
The first application of the single-stranded library preparation method was the generation of a 30-fold coverage genome sequence from a tiny finger bone of a Denisovan individual, a type of extinct archaic human (3). This was followed by the generation of additional high-coverage genomes from other ancient hominins (5,6). More recently, single-stranded library preparation in combination with a DNA extraction method optimized for the recovery of extremely short DNA fragments (7) allowed the successful recovery of mitochondrial and nuclear DNA sequences from the hominin fossils of Sima de los Huesos, Spain (7,8), pushing back the temporal limits of DNA recovery from non-permafrost fossils to ∼430 000 years. Direct comparisons between single- and double-stranded library preparation methods confirmed that single-stranded library preparation greatly improves the yield of library molecules, especially those shorter than 50 bp (9,10). In addition, single-stranded library preparation increases the proportion of endogenous DNA in many highly degraded samples (9) and fully preserves the strand orientation of the sequenced fragments.
Beyond its application to ancient DNA, single-stranded library preparation provides a higher level of resolution in sequence data generated from circulating cell-free DNA from blood and urine (11,12) and dramatical increases in the yield of DNA sequences from formalin-fixed, paraffin-embedded (FFPE) tissues (13). Unfortunately, a more widespread use of the method is somewhat hindered by the fact that it is more time consuming than double-stranded methods and that it requires the use of an expensive enzyme, a thermostable RNA ligase from bacteriophage TS2126 (14) (branded ‘CircLigase’ by EpiCentre), for single-stranded DNA ligation. Variations of the method replace single-stranded ligation by polymerase-based adapter addition (15) or end-tailing with terminal transferases in order to create priming sites for downstream amplification and sequencing (16,17). However, no comparisons of efficiency have been made to the original method. Furthermore, the introduction of homopolymer stretches in the latter methods obscures the true ends of DNA fragments and may cause problems in paired-end sequencing. Currently, many studies of ancient DNA still rely on less expensive and simpler double-stranded methods (see Figure 1C and D for a description of the two most commonly used methods).
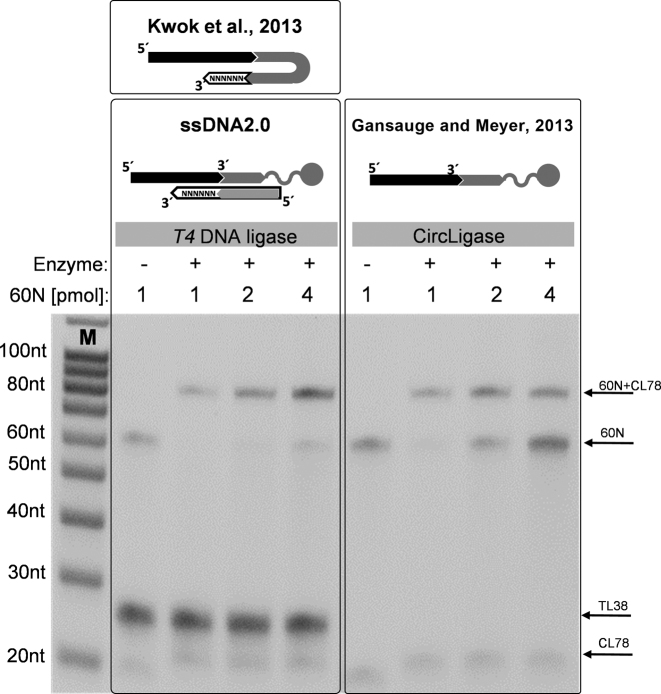
The most commonly used enzyme for joining double-stranded DNA fragments or sealing nicks in DNA is T4 DNA ligase. However, the enzyme shows little activity for ligating the ends of single-stranded DNA (18). In 2013, Kwok et al. described a scheme that enables the ligation of single-stranded acceptor DNA to a hairpin-shaped donor that carries a 3΄ tail of random nucleotides (19) (see Figure 2). Ligation occurs through nick sealing as acceptor molecules hybridize to the stretch of random nucleotides of the donor. The efficiency of this reaction was reported to vary substantially depending on the sequence of the donor, especially within its loop region. More recently, simpler schemes utilizing separate splinter and donor oligonucleotides were implemented in two methods for DNA replication analysis, emRiboSeq (20) and OK-seq (21), but the efficiencies of these reactions were not characterized. Here we provide an in-depth exploration of the suitability of splinted DNA ligation for single-stranded DNA library preparation. We show that this reaction scheme, among other modifications to the original approach (Figure 1A)—can be utilized for more robust and less costly single-stranded library preparation, a method we call ‘ssDNA2.0‘. We comprehensively compare the performance of ssDNA2.0 and CircLigase-based single-stranded library preparation on ancient DNA, cell-free DNA and DNA from formalin-fixed tissues. We also provide a more detailed comparison of single- and double-stranded methods than previous studies.
Figure 2.
Single-stranded DNA ligation with T4 DNA ligase and CircLigase. A pool of 60 nt acceptor oligonucleotides (‘60N’) were ligated to 10 pmol of a 3΄ biotinylated donor oligonucleotide (CL78) using either T4 DNA ligase in the presence of a splinter oligonucleotide (TL38) or CircLigase. Ligation products were visualized on a 10% denaturing polyacrylamide gel stained with SybrGold. Band shifts from 60 nt to 80 nt indicate successful ligation. Schematic overviews of the reaction schemes are shown on top. The scheme developed by Kwok et al. (19) is shown for comparison. M: Single-stranded DNA size marker.
MATERIALS AND METHODS
DNA extraction
DNA was extracted from between 50 and 68 mg of bone powder using the silica-based extraction technique by Dabney et al. (7) with small modifications described in Korlević et al. (22). Total extract volume was 50 μl. Specimens used in this study included Holocene and Late Pleistocene animal bones from three Eurasian cave sites, the North Sea and permafrost (see Supplementary Table S1 for details). DNA was extracted from 20 mg slices of horse and pig liver, which had been stored in buffered formalin for 5 and 11 years respectively, using Qiagen's DNeasy Blood &Tissue Kit. This kit was chosen with the aim of minimizing denaturation of double-stranded DNA during DNA extraction, as it uses relatively mild incubation temperatures that do not exceed 56°C. Briefly, each sample was squashed with a scalpel and transferred into a 1.5 ml tube. To ensure that the tissue was completely covered with digestion buffer, 260 μl ATL buffer and 40 μl Proteinase K were added, followed by a 24 h incubation at 56°C. Remaining tissue was pelleted by centrifugation at 14 000 g for 2 min and the supernatant was transferred to a fresh 1.5 ml tube. Subsequent steps of DNA extraction were performed according to the manufacturer's instructions (DNeasy Blood & Tissue Handbook, 07/2006; ‘Purification of Total DNA from Animal Tissues’). The purified DNA was eluted in 100 μl AE buffer.
Fragmented human genomic DNA was obtained by shearing 1 μg of human DNA (Promega, cat. no. G1471) in 130 μl of TE buffer with a Covaris S2 ultrasonicator using the following parameters: intensity 5, cycles/burst 200, duty cycles 10% and time 180 s. For the experiments with cell-free DNA, blood plasma was obtained by centrifugation of freshly drawn human blood (stored in ethylenediaminetetraacetic acid for <1.5 h) for 10 min at 800 g. The supernatant was transferred to a fresh tube and centrifuged for 10 min at 14 000 g. The centrifuge was cooled to 4°C in both steps. Aliquots of the final supernatant were stored at −80°C until DNA extraction. Circulating nucleic acids were isolated from 2 ml of plasma using the QIAAmp Circulating Nucleic Acid kit according to the manufacturer's instructions. To maximize the DNA yield, an additional elution step using 20 μl elution buffer was performed, leading to a total volume of 50 μl DNA extract. DNA concentration was estimated to be 0.33 ng/μl using the Qubit Fluorometer (ThermoFisher Scientific).
Single-stranded DNA ligation with T4 DNA ligase and CircLigase
To compare the efficiencies of splinter-mediated ligation with T4 DNA ligase and single-stranded DNA ligation with CircLigase, synthetic oligonucleotides were used as acceptor molecules in ligation and reaction products were visualized on denaturing polyacrylamide gels. Ligation with CircLigase was performed in 80 μl reactions containing 1 × CircLigase II reaction buffer (Epicentre), 2.5 mM MnCl2 (Epicentre), 20% PEG-4000 (Sigma-Aldrich), 10 picomole (pmol) donor oligonucleotide (CL78, TL128, TL130 or TL134; see Supplementary Table S2 for oligonucleotide sequences), 1, 2 or 4 pmol acceptor oligonucleotide (60N or HP8) and 400 U CircLigase II (Epicentre). Reactions were incubated at 60°C for 1 h. For ligation reactions with T4 DNA ligase, adapter and splinter oligonucleotides were first hybridized by combining 200 pmol adapter (CL78, TL128, TL130 or TL134) and 400 pmol splinter (TL38, TL129, TL131 or TL135, respectively) in a 20 μl reaction containing 1 × T4 RNA ligase buffer (50 mM Tris-HCl, 10 mM NaCl, 1 mM DTT, pH 7.5 at 25°C; New England Biolabs) and heated up to 95°C for 10 s in a thermal cycler, followed by a ramp to 10°C at 0.1°C/s. Ligation was performed in 80 μl reactions containing 1 × T4 RNA ligase buffer, 20% PEG-8000, 0.5 mM ATP, 10/20 pmol of adapter splinter mix CL78/TL38, 1, 2 or 4 pmol acceptor oligonucleotide and 30 U T4 DNA ligase (ThermoFisher Scientific). Incubation was carried out for 1 h at 37°C. All ligation products were purified using Qiagen's Nucleotide Removal Kit according to the manufacturer's instructions but using MinElute columns (Qiagen) instead of Qiagen's QiaQuick columns to enable a reduction of the elution volume to 10 μl. Eluates were combined with 10 μl 2 × TBE-Urea sample buffer (Bio-Rad), loaded onto a 10% TBE-UREA gel (Bio-Rad), separated for 35 min at 12.5 V/cm and stained with 1× SybrGold dye (ThermoFisher Scientific).
DNA library preparation
Single-stranded libraries using the CircLigase and ssDNA2.0 methods were prepared from between 0 and 27 μl DNA extract and 0.1 pmol of a positive control oligonucleotide (CL104) as described in detail in Supplementary Methods. In brief, CircLigase-based library preparation was performed as described previously (4) with the only major modification being a 3΄-5΄ exonuclease treatment of the single-stranded adapter oligonucleotide CL 78 using the Klenow fragment of Escherichia coli DNA polymerase I in the absence of nucleotides in order to remove synthesis artifacts and potential DNA contamination. ssDNA2.0 library preparation differed from this method in three aspects. First; single-stranded ligation of the first adapter oligonucleotide was carried out using T4 DNA ligase in the presence of a splinter oligonucleotide, which was used in two-fold excess over the adapter oligonucleotide (20 versus 10 pmol) to ensure that all adapters are hybridized to a splinter. Splinters with different end modifications and different numbers of degenerated nucleotides were used in successive experiments (Supplementary Table S3) with the aim of reducing artifact formation and biases in ligation. Second, an additional 45°C wash step was introduced to remove the splinter oligonucleotides after immobilization of the ligation products on streptavidin-coated beads. Third, copies of the template strands were created using an extension primer protected by phosphorothioate (PTO) linkages (CL130) in combination with Klenow fragment instead of Bst DNA polymerase, which avoided blunt-end repair and associated bead wash steps (steps 16–19 in Gansauge and Meyer, (4)).
In addition to Klenow fragment, the performance of three other polymerases was tested in ssDNA2.0. For fill-in with T4 DNA polymerase a 50 μl reaction mix was prepared containing 1× T4 DNA polymerase buffer (ThermoFisher Scientific), 0.05% Tween-20, 100 μM each dNTP, 100 pmol primer CL130 and 2 μl 5 U/μl T4 DNA polymerase (ThermoFisher Scientific). Before adding the enzyme as the final component, the beads were resuspended in the reaction mix, incubated for 2 min at 65°C and transferred to an ice-water bath. After enzyme addition, the bead suspension was incubated for 5 min at 25°C and 25 min at 37°C. Fill-in with Sulfolobus DNA polymerase IV (Dpo4) was performed using the same protocol except that 1× Thermopol buffer and 3 μl 2 U/μl Dpo4 (both New England Biolabs) were used as well as an extension primer that did not contain PTO linkages (CL9, Supplementary Table S2). Fill-in with Bst DNA polymerase and the associated blunt-end repair were performed as described for CircLigase-based library preparation in Supplementary Methods. Blunt-end repair was also carried out for the libraries prepared with Dpo4.
Double-stranded libraries were prepared using two methods: first, following the method described in Meyer and Kircher for highly degraded DNA (23) but omitting the final purification step after adapter fill-in to maximize recovery of library molecules; second, using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer's instructions. The final volume of all libraries was adjusted to 50 μl with water where necessary.
All libraries were quantified by quantitative polymerase chain reaction (PCR) as described elsewhere (4). For comparisons of library yields, molecule counts obtained for the libraries prepared with the method of Meyer and Kircher, (23), were divided by 2, as both strands of double-stranded library molecules generated with this method contribute to the molecule count while producing identical sequences. Library molecules carrying the same adapter sequences, which are also produced with this method, are not efficiently amplified (24) due to intramolecular hybridization of the adapters and do not contribute to the qPCR counts.
Ten microliter of each single-stranded and double-stranded library were amplified and double-indexed using AccuPrime Pfx DNA polymerase (ThermoFisher Scientific) (25,26) in 100 μl reaction mixes containing 1 × AccuPrime Pfx buffer, 1 μM each indexing primer and 1 μl (2.5 U) polymerase. PCR temperature profile included an activation step at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 20 s, annealing at 60°C for 30 s and elongation at 68°C for 1 min, with a final extension step at 68°C for 5 min. PCR products were then purified using the MinElute PCR purification kit (Qiagen). Amplification into PCR plateau inflates PCR biases but enabled subsequent pooling of the libraries in equal volumes while maintaining a relatively even sequence representation across libraries. Since library molecules prepared with the method of Meyer and Kircher, (23) may contain uracils, they were amplified and indexed in a 10-cycle PCR using AmpliTaq Gold (ThermoFisher Scientific) (25), purified using the MinElute PCR purification kit, amplified further in a 25-cycle PCR cycles using AccuPrime Pfx DNA polymerase and primers IS5 and IS6 (23), and purified again. Amplified libraries were pooled, and heteroduplexes that had formed in PCR plateau were removed by subjecting 500 ng of pooled PCR product to a single-cycle PCR with primers IS5 and IS6 using the same reaction conditions. The library pools were then purified, their concentration determined using a DNA1000 chip on the Bioanalyzed 2100 (Agilent Technologies) and sequenced on Illumina's MiSeq instrument using a recipe for 2 × 76 bp paired-end sequencing of double-indexed libraries (26).
Sequence data processing
Base calling was performed using Illumina's Bustard software. Reads were assigned to their original library based on perfect matches to one of the expected index combinations. Whenever possible, overlapping paired-end reads were merged into single sequences using leeHom (27) in order to reconstruct full-length molecule sequences. All sequences were aligned to the reference genome of a closely related species (see Supplementary Table S3) using Burrows-Wheeler Aligner (BWA) (28) with parameters optimized for ancient DNA (3). Summary statistics were computed using SAMtools (29) and custom perl scripts. Insert sizes of library molecules were either inferred directly from the length of overlap-merged sequences or indirectly from the length of the reference genome enclosed by mapped paired-end reads. The informative sequence content of each library was calculated as follows: [number of mapped sequences ≥35 bp (without duplicate removal)]/[number of raw sequences generated] x [qPCR molecule count].
RESULTS
Splinted end-to-end ligation of single-stranded DNA using T4 DNA ligase
To explore the efficiency of splinted end-to-end ligation of single stranded DNA with T4 DNA ligase in the absence of hair-pin structures, we designed a ligation scheme where the splinter oligonucleotide is hybridized to a biotinylated adapter oligonucleotide (the donor), allowing for subsequent immobilization of ligation products on beads and removal of the splinter by mild heat treatment.
We first determined the efficiency of the ligation reaction using a pool of 60 nt oligonucleotides with random sequences (‘60N’) as acceptors, which is analogous to the situation in library preparation where the input DNA consists of strands of unknown sequence. Under optimized conditions, T4 DNA ligase nearly completely turned over 1 pmol (∼6 × 1011 molecules) of the 60N pool to ligation products. Remarkable efficiency was also observed with higher 60N input amounts. In fact, ligation efficiency was even higher than that achieved with CircLigase (Figure 2). We obtained similar results when using other randomly designed adapter/splinter sequences (Supplementary Figure S1), underlining the robustness of the approach. Ligation efficiency decreased only when acceptor molecules of a defined sequence were used instead of a pool of oligonucleotides, as expected due to the limited availability of splinters with sufficient sequence complementarity to the acceptor (Supplementary Figure S2).
A simplified method of single-stranded library preparation
To evaluate if splinted single-stranded ligation with T4 DNA ligase can substitute CircLigase in single-stranded library preparation, we created libraries with both ligation strategies using a DNA extract of a >50 000 year-old cave bear. It is important to note that CircLigase shows an activity optimum of around 60°C (14) whereas splinted single-stranded DNA ligation with T4 DNA ligase is carried out at a lower temperature (37°C). Since reduced temperature could theoretically promote interactions between DNA strands that might interfere with ligation, we prepared libraries from four different volumes (1, 3, 9 and 27 μl) of DNA extract to determine the sensitivity of the reaction to varying concentrations of input DNA.
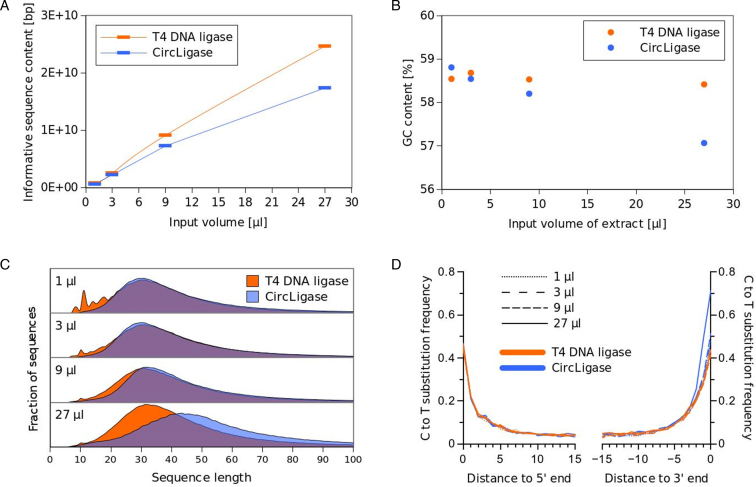
Based on library quantification by quantitative PCR and sequencing on Illumina's MiSeq system, we computed the informative sequence content in each library (Supplementary Table S3), i.e. the sum of nucleotides comprised in bear-like sequences of length 35 or greater. This measure disregards molecules that are too short to be surely mapped to the reference genome and compensates for the lower information content in short sequences. For small volumes of extract we observed the expected linear input/output relationship with both methods (Figure 3A). Interestingly however, we observed a decrease in library yield with the highest input volumes of extracts (9 and 27 μl) for CircLigase but not T4 DNA ligase libraries. CircLigase libraries with higher input volumes are also affected by other biases, including longer insert sizes, lower GC content and higher frequencies of C to T substitutions at their 3΄ end (Figure 3B–D) than the ones produced by T4 DNA ligase. The pronounced asymmetry in C to T substitutions, which result from deamination of cytosine to uracil in ancient DNA (30), points to a less efficient ligation of 3΄ cytosines compared to thymines by CircLigase in the presence of high concentrations of DNA. These biases could be due to saturation of the CircLigase reaction with DNA (31) or a lower sensitivity of the splinted ligation scheme toward inhibitory substances co-extracted from the bone.
Figure 3.
Effects of single-stranded ligation schemes on library characteristics. (A) Informative sequence content of libraries prepared with CircLigase and T4 DNA ligase as a function of the input volume of ancient DNA extract used for library preparation. (B) Average GC content of the sequences obtained with the two ligation schemes. Note that the average GC content exceeds that of a typical mammalian genome because most sequences derive from microbial DNA, which is the dominant source of DNA in most ancient bones. (C) Fragment size distribution in the libraries as inferred from overlap-merged paired-end reads. Short artifacts in the library prepared from extremely little input DNA (corresponding to ∼1 mg bone) are mainly due to the incorporation of splinter fragments. (D) Frequencies of damage-induced C to T substitutions near the 5΄ and 3΄ ends of sequences.
Another step in single-stranded library preparation with potential for improvement is the synthesis of the second strand, which is performed using Bst polymerase in the original protocol, thus requiring subsequent blunt-end repair to remove nucleotides added by the terminal transferase activity of the enzyme. Because it is desirable to perform both reactions in one step, we generated additional libraries from two ancient DNA extracts as well as a control oligonucleotide using Bst polymerase and two proof-reading enzymes: T4 DNA polymerase and the Klenow fragment of E. coli DNA polymerase I. The latter enzyme has already been used in single-stranded library preparation recently (22), but without presenting data comparing its performance to Bst polymerase. In addition, we included Sulfolobus DNA polymerase IV (Dpo4) in the experiment. Similar to Bst polymerase, Dpo4 lacks 3΄-5΄ exonuclease activity but incorporates nucleotides across a wide range of DNA lesions and may thus be particularly efficient in copying highly damaged ancient DNA molecules (32). To prevent exonucleolytic degradation we introduced three PTO linkages into the 3΄ terminus of the extension primer for its use with T4 DNA polymerase and Klenow fragment. The yield of library molecules varied relatively little among the polymerases, with slightly higher gains of molecules in the libraries prepared with polymerases lacking 3΄-5΄ exonuclease activity (Bst polymerase and Dpo4) (Supplementary Figure S3a and Supplementary Table S3). Investigation of the sequence alignments revealed no elevated substitution frequencies other than from C to T, including in the libraries prepared with Dpo4. We thus detected no signal that would suggest the presence of previously undetected types of damage in ancient DNA that may lead to false incorporations of nucleotides due to the lesion-bypass ability of this enzyme. However, we detected substantial differences in the frequencies of C to T substitutions near the 5΄ ends of sequence alignments, especially with T4 DNA polymerase, which led to a pronounced deprivation of C to T substitutions between the second and approximately the tenth alignment position. This suggests that the latter enzyme is less efficient in incorporating nucleotides across uracils located close to the 5΄ end of template strands (Supplementary Figure S3b). Since polymerase choice had only a small impact on library yields, we opted to replace Bst polymerase with the Klenow fragment in order to eliminate one reaction step from the protocol.
A comparison of library preparation methods on degraded DNA
To compare the performance of ssDNA2.0 to that of CircLigase-based library preparation, we prepared DNA libraries from multiple sources: several ancient bones, tissues stored in formalin for 5 to 11 years, circulating cell-free DNA and sheared genomic DNA. Additionally, we included in this comparison the two double-stranded methods that have been most commonly used in previous studies of ancient DNA. The first is a method that was originally developed for the sequencing of high-quality DNA by 454 Life Sciences (33) and later adapted for the use with highly degraded DNA (23,24). It is based on the ligation of an adapter mix to blunt-end repaired ancient DNA fragments using T4 DNA ligase (Figure 1C). Even though half of the molecules are left with identical adapters and are hence inaccessible to down-stream amplification and sequencing, the method is robust, relatively inexpensive and thus widely used in ancient DNA studies, especially those involving large numbers of samples (34,35). The second double-stranded method was originally proposed by Illumina (36) and relies on T/A-overhang ligation of a fork-like adapter, or, in the implementation of New England Biolabs’ NEBNext Ultra II DNA Library Prep Kit for Illumina used here, a bell-shaped adapter (Figure 1D). The Illumina method enables directional ligation and a greater degree of simplification, e.g. by combining some or all of the reactions without intermittent purification steps. It is widely used for library preparation from high-quality DNA, but represents a less ideal choice for highly degraded DNA, as it requires size-selective purification to remove adapter dimers. The method also bears the risk for formation of chimeric artifacts when used with short DNA fragments (37).
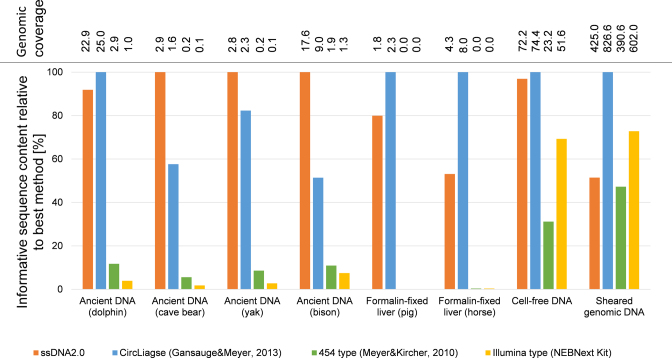
The content of informative nucleotides is similar in the libraries prepared with ssDNA2.0 and the CircLigase-based method for all samples (Figure 4). Both methods consistently outperform the best double-stranded method when used with highly degraded DNA. With ssDNA2.0, informative sequence yields are 11.6 times higher on average for the ancient DNA extracts and 1.4 times for cell-free DNA. In line with previous reports (9,10), single-stranded library preparation also increases the proportion of mapped sequences in the ancient DNA libraries (Supplementary Figure S5), thus reducing the burden of microbial contamination in sequencing. Strikingly, library yields from formalin-fixed DNA are ∼150- to ∼3100-fold higher with single-stranded library preparation, enabling multi-fold coverage genome sequencing from extracts that otherwise yield virtually no sequence information. Library yields from the degraded DNA samples are lowest with the Illumina method, which is at least partly due to a loss of short molecules (see Supplementary Figure S4 for sequence length distributions). It should also be noted that the specific implementation of Illumina-type library preparation used here reduces library yields from ancient DNA due to uracil-DNA-glycosylase treatment, which prevents amplification of DNA strands containing uracils, approximately half of which persist through the blunt-end repair step (30).
Figure 4.
Performance of single- and double-stranded library preparation methods using DNA from different sources. The informative sequence content of each library is provided in percent of that obtained with the best performing method for each sample. In addition, the number of nuclear genomes present in each library was calculated by dividing the informative sequence content by the size of the reference genome used for mapping.
All library preparation methods produced considerable numbers of library molecules even in the absence of input DNA (Supplementary Table S3). These artifacts are mainly caused by the incorporation of imperfectly synthesized adapter oligonucleotides into the libraries. ssDNA2.0 is more prone to the formation of artifacts than CircLigase-based library preparation as it uses an additional oligonucleotide to mediate single-stranded ligation. By changing the blocking modifications of the splinter oligonucleotide in the course of our experiments, we were able to successively reduce artifact formation in library preparation to ∼4 × 107 molecules (Supplementary Table S3). This number is slightly higher than the ∼2 × 107 molecules obtained with CircLigase-based library preparation and the ∼1 × 107 and ∼3 × 107 artifacts generated with the two double-stranded methods, but lower than the ∼1 × 108 molecules previously reported for the CircLigase method (4), indicating that not only the chemical structure of oligonucleotides but also batch-to-batch variation in oligonucleotide synthesis contributes to artifact formation. As many more library molecules are obtained from the same amount of sample DNA with single-stranded library preparation, artifacts are present in lower proportions in these libraries (Supplementary Table S3), further highlighting the suitability of single-stranded library preparation for work with small quantities of highly degraded DNA.
For the ancient DNA libraries we observed consistently lower deamination-induced C to T substitution frequencies at the 3΄ ends in the ssDNA2.0 compared to the CircLigase libraries (Supplementary Figure S6). While this signal may in part be due to preferential ligation of uracils by CircLigase as described above, we also observed an underrepresentation of thymines within the last six positions of ssDNA2.0 sequences (Supplementary Figure S7), matching in length exactly the hybridization site of the splinter oligonucleotide. This signal suggests that DNA strands with thymine-rich 3΄ ends—and possibly those with uracil-rich ends as well—are less efficiently joined by splinted ligation. To investigate whether the length of the degenerate sequence overhang of the splinter oligonucleotide contributes to ligation bias we prepared additional libraries from one of the ancient DNA extracts using splinters with seven and eight degenerate bases. While the overall yield of library molecules is similar with the three splinters (Supplementary Figure S8a), we find that longer splinters increase both the thymine content (Supplementary Figure S7) as well as the frequency of C to T substitution at the 3΄ end of sequences (Supplementary Figure S8b), suggesting that longer splinters reduce ligation biases associated with splinted DNA ligation.
DISCUSSION
The library preparation method described here offers several benefits over single-stranded DNA library preparation in its first implementation. The new method relies on the widely available and inexpensive T4 DNA ligase instead of CircLigase, which substantially reduces costs (see Supplementary Table S4 for a comparison of reagent costs) and makes the method more robust to larger quantities of input DNA. Even though ssDNA2.0 also comes with a small reduction in the number of reaction steps, single-stranded library preparation remains more time-consuming than double-stranded methods, limiting the number of samples that can be processed by manual pipetting. However, irrespective of the choice of protocols, high-throughput sample preparation is difficult to achieve without the use of laboratory automation systems. The most important advantage of ssDNA2.0 in our view thus lies in the removal of the extended high-temperature incubation step required by CircLigase, which makes the method compatible with automation on open liquid handling platforms and opens up the possibility for library preparation in microplate format.
We believe that the core of the method, single-stranded DNA ligation with splinter oligonucleotides carrying degenerate bases—as first described in proof-of-principle experiments by Kwok et al. (19)—bears unused potentials for methods development in molecular biology. We observed that highly efficient ligation can be achieved using randomly chosen adapter sequences and that the separation of the adapter and splinter oligonucleotide does not impair the efficacy of this ligation strategy. Further, analysis of sequence data indicates that ligation biases are minimized when using splinters carrying seven or eight degenerate bases. We also show that splinted DNA ligation is most efficient with acceptors of high sequence complexity, such as fragments of genomic DNA isolated from biological tissue. The sequencing of low complexity DNA using ssDNA2.0, for example for quality control of oligonucleotide synthesis, may therefore require the use of relatively low amounts of input DNA to ensure that enough splinters with sequence similarity to the acceptor strands are available for ligation.
Our study also provides a more comprehensive comparison of single- and double-stranded library preparation than previously made (9,10,13). Emphasis in these studies was placed mainly on the characteristics of the sequences obtained, for example their size distribution or the proportions of endogenous and contaminating microbial DNA sequences. However, one of the key parameters that determines the success of library preparation is the content of unique molecules in each library (often referred to as library complexity). With the exception of a single-study on FFPE samples (13), this parameter was either not assessed or merely extrapolated from the clonality of sequence reads based on very shallow sequence data. We insist that the number of library molecules should be determined prior to amplification and sequencing when evaluating the performance of library preparation methods. Quantitative PCR is well suited for this purpose (38), as it enables direct comparisons of library yield. By providing estimates of library complexity independent from sequencing, we conclusively show that single-stranded library preparation increases library yields from ancient DNA by approximately one order of magnitude and by a factor of ∼1.4 for cell-free DNA compared to the best double-stranded method. The difference is even more dramatic for formalin-fixed samples, where single-stranded library preparation recovers up to ∼3100 times more molecules. This observation is in line with a recent report where single-stranded library preparation was found to increase the number of library molecules from FFPE tissues by an average of 900-fold compared to double-stranded methods (13). However, in contrast to FFPE samples, which are typically fixed in formalin for hours or days, the samples used here had been stored in formalin for many years, suggesting that even material preserved under such extremely unfavorable conditions can be made accessible to genetic studies.
Lastly, our work illustrates to which extent details of library preparation, such as the choice of enzymes, influence the characteristics of sequences obtained from degraded DNA. For example, the elevation in frequency of deamination-induced substitutions near the end of DNA sequences has been used to infer the length of single-stranded overhangs in ancient DNA (30,39); however, the frequency of these substitutions at the terminal positions of sequence alignments and their decline toward the interior of the sequence depend substantially both on the ligase used to join adapters in library preparation and on the polymerase used for copying the template strands. The possibility of biases in DNA extraction and library preparation, which are generally still poorly understood, should thus be taken into consideration when inferring parameters of DNA damage from ancient DNA sequence data.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Svante Pääbo for his support throughout all stages of the project. We thank Viviane Slon, Manja Wachsmuth and Louisa Jauregui for comments on the manuscript, Elena Essel and Roland Schröder for help in the lab, Barbara Höber and Antje Weihmann for performing the sequencing runs, Gabriel Renaud and Udo Stenzel for help with raw sequence data processing. We are grateful to Klaas Post, Pavao Rudan, Michael Shunkov, Grant Zazula, Sahra Talamo and Kristin Müller for providing samples.
Author contributions. M.T.G., T.G. and M.M. developed the method. M.T.G., T.G., L.L., S. N. and A.S. performed laboratory experiments. I.G. and P.K. prepared ancient DNA extracts. L.R. prepared cell-free DNA. M.M. and M.T.G. analyzed the data and wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Society. Funding for open access charge: Max Planck Institute for Evolutionary Anthroplogy.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ke R., Mignardi M., Pacureanu A., Svedlund J., Botling J., Wahlby C., Nilsson M.. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013; 10:857–860. [DOI] [PubMed] [Google Scholar]
- 2. Lee J.H., Daugharthy E.R., Scheiman J., Kalhor R., Yang J.L., Ferrante T.C., Terry R., Jeanty S.S., Li C., Amamoto R. et al. . Highly multiplexed subcellular RNA sequencing in situ. Science. 2014; 343:1360–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prufer K., de Filippo C. et al. . A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012; 338:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gansauge M.T., Meyer M.. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 2013; 8:737–748. [DOI] [PubMed] [Google Scholar]
- 5. Prufer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. et al. . The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014; 505:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fu Q., Li H., Moorjani P., Jay F., Slepchenko S.M., Bondarev A.A., Johnson P.L., Aximu-Petri A., Prufer K., de Filippo C. et al. . Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014; 514:445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dabney J., Knapp M., Glocke I., Gansauge M.T., Weihmann A., Nickel B., Valdiosera C., Garcia N., Paabo S., Arsuaga J.L. et al. . Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15758–15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyer M., Arsuaga J.L., de Filippo C., Nagel S., Aximu-Petri A., Nickel B., Martinez I., Gracia A., Bermudez de Castro J.M., Carbonell E. et al. . Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature. 2016; 531:504–507. [DOI] [PubMed] [Google Scholar]
- 9. Bennett E.A., Massilani D., Lizzo G., Daligault J., Geigl E.M., Grange T.. Library construction for ancient genomics: single strand or double strand. Biotechniques. 2014; 56:289–300. [DOI] [PubMed] [Google Scholar]
- 10. Wales N., Caroe C., Sandoval-Velasco M., Gamba C., Barnett R., Samaniego J.A., Madrigal J.R., Orlando L., Gilbert M.T.. New insights on single-stranded versus double-stranded DNA library preparation for ancient DNA. Biotechniques. 2015; 59:368–371. [DOI] [PubMed] [Google Scholar]
- 11. Snyder M.W., Kircher M., Hill A.J., Daza R.M., Shendure J.. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016; 164:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burnham P., Kim M.S., Agbor-Enoh S., Luikart H., Valantine H.A., Khush K.K., De Vlaminck I.. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci. Rep. 2016; 6:27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stiller M., Sucker A., Griewank K., Aust D., Baretton G.B., Schadendorf D., Horn S.. Single-strand DNA library preparation improves sequencing of formalin-fixed and paraffin-embedded (FFPE) cancer DNA. Oncotarget. 2016; 7:59115–59128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blondal T., Thorisdottir A., Unnsteinsdottir U., Hjorleifsdottir S., Aevarsson A., Ernstsson S., Fridjonsson O.H., Skirnisdottir S., Wheat J.O., Hermannsdottir A.G. et al. . Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res. 2005; 33:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karlsson K., Sahlin E., Iwarsson E., Westgren M., Nordenskjold M., Linnarsson S.. Amplification-free sequencing of cell-free DNA for prenatal non-invasive diagnosis of chromosomal aberrations. Genomics. 2015; 105:150–158. [DOI] [PubMed] [Google Scholar]
- 16. Turchinovich A., Surowy H., Serva A., Zapatka M., Lichter P., Burwinkel B.. Capture and Amplification by Tailing and Switching (CATS). An ultrasensitive ligation-independent method for generation of DNA libraries for deep sequencing from picogram amounts of DNA and RNA. RNA Biol. 2014; 11:817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tin M.M., Economo E.P., Mikheyev A.S.. Sequencing degraded DNA from non-destructively sampled museum specimens for RAD-tagging and low-coverage shotgun phylogenetics. PLoS One. 2014; 9:e96793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuhn H., Frank-Kamenetskii M.D.. Template-independent ligation of single-stranded DNA by T4 DNA ligase. FEBS J. 2005; 272:5991–6000. [DOI] [PubMed] [Google Scholar]
- 19. Kwok C.K., Ding Y., Sherlock M.E., Assmann S.M., Bevilacqua P.C.. A hybridization-based approach for quantitative and low-bias single-stranded DNA ligation. Anal. Biochem. 2013; 435:181–186. [DOI] [PubMed] [Google Scholar]
- 20. Ding J., Taylor M.S., Jackson A.P., Reijns M.A.. Genome-wide mapping of embedded ribonucleotides and other noncanonical nucleotides using emRiboSeq and EndoSeq. Nat. Protoc. 2015; 10:1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petryk N., Kahli M., d’Aubenton-Carafa Y., Jaszczyszyn Y., Shen Y., Silvain M., Thermes C., Chen C.L., Hyrien O.. Replication landscape of the human genome. Nat. Commun. 2016; 7:10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korlevic P., Gerber T., Gansauge M.T., Hajdinjak M., Nagel S., Aximu-Petri A., Meyer M.. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques. 2015; 59:87–93. [DOI] [PubMed] [Google Scholar]
- 23. Meyer M., Kircher M.. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010; 2010, doi:10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 24. Maricic T., Paabo S.. Optimization of 454 sequencing library preparation from small amounts of DNA permits sequence determination of both DNA strands. Biotechniques. 2009; 46:51–57. [DOI] [PubMed] [Google Scholar]
- 25. Dabney J., Meyer M.. Length and GC-biases during sequencing library amplification: a comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. Biotechniques. 2012; 52:87–94. [DOI] [PubMed] [Google Scholar]
- 26. Kircher M., Sawyer S., Meyer M.. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012; 40:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Renaud G., Stenzel U., Kelso J.. leeHom: adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res. 2014; 42:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Briggs A.W., Stenzel U., Johnson P.L., Green R.E., Kelso J., Prufer K., Meyer M., Krause J., Ronan M.T., Lachmann M. et al. . Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:14616–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li T.W., Weeks K.M.. Structure-independent and quantitative ligation of single-stranded DNA. Anal. Biochem. 2006; 349:242–246. [DOI] [PubMed] [Google Scholar]
- 32. Yang W. Damage repair DNA polymerases Y. Curr. Opin. Struct. Biol. 2003; 13:23–30. [DOI] [PubMed] [Google Scholar]
- 33. Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z. et al. . Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005; 437:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K. et al. . Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015; 522:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allentoft M.E., Sikora M., Sjogren K.G., Rasmussen S., Rasmussen M., Stenderup J., Damgaard P.B., Schroeder H., Ahlstrom T., Vinner L. et al. . Population genomics of Bronze Age Eurasia. Nature. 2015; 522:167–172. [DOI] [PubMed] [Google Scholar]
- 36. Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G., Hall K.P., Evers D.J., Barnes C.L., Bignell H.R. et al. . Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008; 456:53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Star B., Nederbragt A.J., Hansen M.H., Skage M., Gilfillan G.D., Bradbury I.R., Pampoulie C., Stenseth N.C., Jakobsen K.S., Jentoft S.. Palindromic sequence artifacts generated during next generation sequencing library preparation from historic and ancient DNA. PLoS One. 2014; 9:e89676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meyer M., Briggs A.W., Maricic T., Hober B., Hoffner B., Krause J., Weihmann A., Paabo S., Hofreiter M.. From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res. 2008; 36:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jonsson H., Ginolhac A., Schubert M., Johnson P.L., Orlando L.. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013; 29:1682–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.