Abstract
The assembly of large DNA constructs coding for entire pathways poses a major challenge in the field of synthetic biology. Here, we present AssemblX, a novel, user-friendly and highly efficient multi-gene assembly strategy. The software-assisted AssemblX process allows even unexperienced users to rapidly design, build and test DNA constructs with currently up to 25 functional units, from 75 or more subunits. At the gene level, AssemblX uses scar-free, overlap-based and sequence-independent methods, allowing the unrestricted design of transcriptional units without laborious parts domestication. The assembly into multi-gene modules is enabled via a standardized, highly efficient, polymerase chain reaction-free and virtually sequence-independent scheme, which relies on rare cutting restriction enzymes and optimized adapter sequences. Selection and marker switching strategies render the whole process reliable, rapid and very effective. The assembly product can be easily transferred to any desired expression host, making AssemblX useful for researchers from various fields.
INTRODUCTION
Synthetic biology is a relatively young, but fast developing, scientific discipline. The field combines aspects of life sciences, chemistry and engineering to design, build and test new biological systems. Many projects in synthetic biology require sophisticated cloning methods, suitable for the modular assembly of complex DNA constructs, for example for the programmable production of compounds of medical or technical interest, construction of synthetic genomes or the design of regulatory circuits (1–4 and https://doi.org/10.1101/041871).
While cloning of single DNA parts into target vectors has become a routine technique in laboratories long ago, the assembly of multi-gene constructs or even genomes by de novo assembly of DNA fragments from different sources still remains a challenge (5). To enable and standardize such assembly projects, different methods have been reported. These can be roughly separated into two groups: namely sequence-dependent and sequence-independent methods. One of the first multi-part assembly strategies described is the sequence-dependent BioBrick (6). It is based on iterative restriction and ligation steps, using a combination of four restriction endonucleases (REs) with 6 bp-long recognition sites and compatible overhangs. BioBrick allows only step-by-step assemblies precluding the rapid assembly of large multi-gene constructs. Furthermore, recognition sites of six-base cutters statistically occur every 4 kilo base pairs (kb), making it necessary to ‘domesticate’ assembly parts by eliminating undesired cutting sites. This is a drawback many sequence-dependent methods, like Golden Gate cloning, MoClo and Golden Braid, have in common (7–9).
To overcome the drawbacks of RE-based cloning methods, sequence-independent cloning methods have been developed. Such cloning methods avoid REs and are often based on short overlapping sequences shared by the assembly parts, thereby allowing the directed assembly of multiple DNA fragments. These methods include the in vitro Gibson assembly (10), the bacterial cell extract-based SLiCE (11) and in vivo transformation-associated recombination (TAR) in Saccharomyces cerevisiae (12,13). The overlapping regions for each assembly reaction have to be designed individually, depending on the sequence of neighboring assembly parts. This results in a lack of modularity and an increased design and planning effort. Nevertheless, these methods are widely used to assemble scar-free constructs.
An example for a modular, but still sequence-independent assembly strategy is MODAL, which in particular focuses on high modularity for easy shuffling of single parts (14). Universal prefix and suffix adapters for upstream and downstream primer binding are fused to individual assembly parts, which are then kept in storage plasmids. Specific linker sequences, computationally optimized for the overlap-guided assembly with neighboring fragments, are then fused to these adapters via a polymerase chain reaction (PCR) step. While MODAL allows the assembly of parts in different orders and orientations, the user has to accept large scar sequences, the risk of internal recombination (as every part contains the same adapter sequence), and, due to the necessary amplification steps, the risk of PCR-born errors with every new assembly reaction.
There are many more highly efficient assembly methods available, for example BASIC, yeast Golden Gate, VEGAS or MoClo (8,15–17). However, in most cases the user has to decide between scar-free, sequence-independent but non-modular methods or modular approaches, which generate scar sequences and require laborious parts domestication. We believe that this decision hinders many biology labs from outside the synthetic biology community to explore new grounds.
With these problems in mind, we have developed a new DNA assembly strategy, which makes use of several highly efficient cloning methods and combines the advantages in a streamlined process. The user-oriented AssemblX workflow allows scar-less, sequence-independent de novo assemblies from multiple parts at the single gene level and subsequent standardized, modular and PCR-free assemblies into multi-gene constructs. The whole process was designed for use with cost efficient cloning methods, enables multi parallel assemblies to speed up the process and allows the user to freely choose the final host organism for gene expression. The user is supported in all steps by the AssemblX web tool and gets a protocol including all primers, PCR conditions, digestions and assembly reactions. Taken together, AssemblX can be used in any lab and by any user to assemble complex genetic constructs for various purposes including pathway engineering.
MATERIALS AND METHODS
Strains, growth conditions, reagents and plasmids
Escherichia coli strains NEB5α, NEB10β (New England Biolabs, Frankfurt am Main, Germany) and DH5α were used for cloning purposes. All strains were grown in Luria-Bertani medium at 37°C, with kanamycin or ampicillin at 50 mg/ml as selection markers for transformed plasmids. For all yeast experiments, S. cerevisiae strains YPH500 (ATCC® 76626™) or BY4741 (ATCC® 201388™) were used and grown at 30°C in yeast extract peptone dextrose adenine (YPDA)-rich medium or in appropriate synthetic dextrose (SD) media lacking one or more amino acids to allow selection for transformed cells. Dominant selection markers were used in YPDA medium in the indicated final concentrations: G418 (200 μg/ml), hygromycin B (200 μg/ml), phleomycin (20 μg/ml) and nourseothricin (100 μg/ml). PCR purification and gel elution were done with NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Dueren, Germany) and plasmid purification was done with NucleoSpin Plasmid EasyPure kit (Macherey-Nagel). Gel elution of large fragments (>10 kb) was done with NucleoTrap kit (Macherey-Nagel). Plasmids pLOG1, pTPG1 and pUOGB are gifts from Tom Ellis, Imperial College London (18). Plasmids pCEV-G1-Ph and pCEV-G2-Km are gifts from Lars Nielsen and Claudia Vickers (Addgene plasmid # 46814 and 46815) (19). pAG36 is a gift from John McCusker (Addgene plasmid # 35126) (20). pHIS3p:mRuby2-Tub1+3΄UTR::HPH is a gift from Wei-Lih Lee (Addgene plasmid # 50633). EBFP2-pBAD is a gift from Michael Davidson (Addgene plasmid # 54542). pJC104_violacein is a gift from Jeff Boeke, NYU School of Medicine (17). p426-SNR52p-gRNA.CAN1.Y-SUP4t is a gift from George Church (Addgene plasmid # 43803) (21). p2GWL7,0 and pKGWFS7 were provided by VIB-Gent University (https://gateway.psb.ugent.be) (22,23). pYC6Lys-TRP1URA3 is a gift from Francisco Malagon (Addgene plasmid # 11010). pdCas9-bacteria is a gift from Stanley Qi (Addgene plasmid # 44249) (24). Sequences of plasmids generated in this work are included in the Supplementary Data of this manuscript. The sequences were also deposited at NCBI GenBank (Accession numbers KY131987–KY132089) and are available via www.assemblx.org.
PCR methods
PCR amplifications were done with Phusion (Thermo Fisher Scientific, Waltham, MA, USA) or Q5 DNA Polymerases (New England Biolabs), following the manufacturer's recommendations. Sequences of all primers and oligonucleotides used for cloning are listed in Supplementary Tables S3 and 4. Amplified DNA parts were analyzed by agarose gel electrophoresis and column- or gel-purified prior to further use.
In vitro DNA assembly and cell transformation protocols
For overlap-based in vitro DNA assembly methods like SLiCE, Gibson and NEBuilder HiFi DNA Assembly, DNA fragments containing 25- to 36-bp-long overlap regions were generated by PCR amplification and were assembled with a linearized plasmid. SLiCE assembly was done as previously reported (11,25). Gibson assembly and NEBuilder HiFi DNA Assembly (both New England Biolabs) were done according to the manufacturer's recommendations. Commercially available chemically competent E. coli NEB5α or NEB10β cells (New England Biolabs) or homemade competent DH5α cells were transformed with 2 μl of the assembly reaction. Transformations were performed following the High Efficiency Transformation Protocol from New England Biolabs. Assemblies were verified by sequencing.
In vivo DNA assembly and cell transformation protocols
Saccharomyces cerevisiae transformations were done according to the LiAc/SS carrier DNA/PEG method by Gietz and Schiestl (26). For the assembly of multiple fragments, 100 ng of each DNA part were mixed with water to a final volume of 34 μl and transformed as described. Plasmid rescue from yeast colonies was done with the Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research Corporation, Freiburg, Germany) and the entire plasmid DNA was subsequently transformed into chemically competent NEB10β cells. Level 2 constructs were transformed into electrocompetent NEB10β cells.
Assembly of Level 0 vector sets
To generate the Level 0 vector backbone, the pUC19 replication origin and the nptII kanamycin resistance gene from plasmid pHis2.1 (Takara Bio, Saint-Germain-en-Laye, France) were amplified by PCR using primer combinations P094/P095 and P096/P097, respectively. Primers contain overlaps for SLiCE cloning and introduce a SmaI site between the pUC19 origin and the nptII gene. The PCR products were combined in a SLiCE reaction to give plasmid pFM011. The vector pFM011 was linearized with SmaI and double-stranded oligonucleotides P251/P252 and P253/P254 were used in a SLiCE reaction to introduce two regions containing multiple 8-base cutter restriction sites, resulting in pFM012. Primers P288/P289 were used to amplify the 2-micron origin and the URA3 marker gene from pYES2/CT (Thermo Fisher Scientific). The resulting PCR product and AflIII-linearized pFM012 were used in a SLiCE reaction to give the final Level 0 backbone pFM032. To create individual Level 0 vectors (pL0 vectors) with different pairs of homology regions, pFM032 was cut by SmaI in between the 8-base cutter regions. Subsequently, two single-stranded oligonucleotides, each containing a different homology region and suitable overlaps for SLiCE cloning, were assembled into pFM032 together with a HindIII-flanked ccdB expression cassette, PCR-amplified from plasmid pDEST22 (Thermo Fisher Scientific) with primers P1165/P1166. The resulting Level 0 vectors contain both homology regions, separated by the HindIII-flanked ccdB cassette. For homology regions AR, BR, CR, DR and ER, PCR products containing the respective homology regions, coding for the first 17 codons of an auxotrophic marker gene, preceded by a promoter region for later control of the Level 1 marker gene, were generated (primer pairs P1173/P1174, P1207/P1208, P1240/P1241, P1274/P1275 or P1307/P1308). These PCR products were assembled together with the HindIII-flanked ccdB cassette and double-stranded oligonucleotides containing the other homology region in SmaI-linearized pFM032.
Assembly of Level 1 vectors
Two types of Level 1 vector backbones were constructed: a low-copy version with CEN/ARS region and a high-copy version with 2-micron origin. For the high-copy backbone, primers P104/P105 and P106/P107 were used to amplify the pBR322 bacterial origin of replication, the ampicillin resistance gene and the 2-micron origin from plasmid pYES2/CT (Thermo Fisher Scientific). Primers P108/P109 and P110/P111 were used to PCR-amplify stuffer fragments from pYES2/CT and to introduce PacI and NotI restriction sites. All four PCR fragments were combined in a SLiCE reaction to give plasmid pFM013. To create the low-copy plasmid pFM014, the PCR product obtained with primers P106/P107 was exchanged for a PCR product generated with primers P112/P113, containing the CEN/ARS origin from pDEST22 (Thermo Fisher Scientific). We further implemented a single-guide RNA (sgRNA) expression cassette for potential future CRISPR/Cas9 applications. Vectors pFM013 and pFM014 were linearized by NotI digestion. Primers P114/P115 and P199/P200 were used to PCR-amplify the crRNA expression cassette from plasmid p426-SNR52p-gRNA.CAN1.Y-SUP4t. The primer pairs were designed to amplify the S. cerevisiae SNR52 promoter and the structural crRNA. Instead of the 20 bp sgRNA target region, a NotI site was introduced via the primer sequence, to allow later integration of any desired sgRNA target sequence. Linearized pFM013 or pFM014 were combined with the above PCR products in a SLiCE reaction to give plasmids pFM015 and pFM016, respectively. Vectors pFM015 and pFM016 were PCR-amplified with primer pair P402/P403, thereby adding I-SceI restriction sites on both termini. Level 1 vectors (pL1 vectors) with different combinations of homology regions and promoter-less marker genes were generated combining the PCR product P402/P403 with different PCR products (primers P1124–P1140, Supplementary Table S3) containing promoter-less auxotrophic marker genes equipped with 50-bp homology regions and a PacI site in a SLiCE reaction.
Assembly of Level 2 vectors
The basic Level 2 backbone is identical to the Level 1 backbone. To assemble different Level 2 vectors (pL2 vectors), either pFM015 or pFM016 were amplified with primer pair P402/P403 and subsequently used in a SLiCE reaction with two oligonucleotides, containing the desired homology regions separated by an EcoRI site and equipped with appropriate overlaps.
Promoter library
To generate a yeast promoter library, different native promoter and terminator sequences were PCR-amplified from S. cerevisiae BY4741. The primers used are detailed in Supplementary Table S4. Each corresponding promoter and terminator pair was individually cloned into the same Level 0 vector pL0A_0R (with A0-AR homology regions), separated by a NotI-flanked stuffer fragment. The stuffer contains the first and the last 36 bp of the yEGFP CDS, separated by an AsiSI restriction site. Resulting plasmids were linearized with AsiSI to allow insertion of PCR-amplified yEGFP. The plasmids were cut with PmeI or AscI to release the complete reporter gene with the regulatory sequences and flanking homology regions. The resulting fragments were cotransformed with the corresponding PacI-linearized Level 1 CEN/ARS vector pL1A-lc into S. cerevisiae YPH500 to give single-gene Level 1 vectors. Promoter activity for each construct was evaluated by flow cytometry-based yEGFP fluorescence measurements.
Flow cytometry
Cells were inoculated in 500 μl drop-out medium, containing either glucose or galactose as carbon source, in 48-well deep-well plates and incubated with shaking for 24 h (30°C, 240 rpm). A main culture in 500 μl fresh medium (supplemented with 20 mM IPTG or 1 μg/ml ATc, if applicable) was then inoculated to OD600 ≈ 0.1 and incubated for another 16 h, before fluorescence was determined via flow cytometry. Fluorescence output of single cells was measured with a BD FACSCalibur flow cytometer (BD Biosciences, Heidelberg, Germany). S. cerevisiae cells grown as described above were diluted in water and passed through the cytometer with less than 3000 counts per second. Yeast cells were identified and gated in a forward/sideward scatter dot plot. Within this gate, 20 000 cells were counted per measurement. Fluorescence was analyzed in a histogram and the geometrical mean fluorescence per cell was calculated for each measurement, using Flowing Software 2 (version 2.5.1, www.flowingsoftware.com). All results shown are mean values calculated from the indicated number of samples. The error bars indicate the standard deviation.
Twenty-five units demo assembly
All Level 0 assemblies were performed via SLiCE or HiFi DNA Assembly in a preliminary AssemblX version, which used non-optimized homology regions. All individual parts and their source plasmids or organisms are given in Supplementary Table S2. To make these constructs compatible with the final AssemblX platform, all 25 Level 0 units were PCR-amplified with primers that exchanged the non-optimized homology regions for the R2o-optimized versions thereof. Subsequently, from these 25 PCR products, five Level 1 modules were assembled using TAR with PacI-linearized Level 1 vector backbones (CEN/ARS versions). Positive candidates were identified via yeast colony PCR and plasmid DNA of selected clones was isolated with the Zymo Yeast Plasmid II kit (Zymo Research). Plasmid DNA was then transformed into E. coli NEB10β cells. For further verification, plasmids were isolated and analysed by restriction digestion and verified by sequencing of assembly junctions. Positive plasmids were processed following the AssemblX protocol and assembled in the EcoRI-digested CEN/ARS Level 2 vector pL2_AE_lc. After verification of successful assembly by colony PCR the expression of incorporated genes was analyzed via different reporter assays. The final construct was subsequently isolated from yeast following a protocol for isolation of circular YACs (27). For further analysis by sequencing and restriction digestion, the construct was transformed into E. coli NEB10β cells by electroporation and isolated using the QIAGEN Large-Construct Kit (Qiagen, Hilden, Germany). Sequencing was done by LGC Genomics (Berlin, Germany).
Pulsed-field gel electrophoresis (PFGE)
Gel electrophoretic analysis of the full-length 25-gene construct was done with a CHEF MAPPER XA System (Bio-Rad Laboratories, Muenchen, Germany) on a 1% pulsed-field gel electrophoresis (PFGE)-grade agarose gel in 0.5 × TBE at 14°C. Instrument settings were: 6 V/cm, 120° pulse angle, initial switch time 0.22 s, final switch time 7.67 s with linear ramp, run time 15:16 h.
β-Galactosidase and β-glucuronidase overlay assays
Plate overlay assays to detect β-galactosidase (LacZ) or β-glucuronidase (GUS) were done according to the Dual Bait Hybrid Hunter Yeast Two-Hybrid System manual (Version C, Invitrogen). Results shown are from a 45 min incubation at 37°C and subsequent incubation at 4°C overnight.
Dual luciferase assay
Dual luciferase assays were done as described using the dual luciferase reporter assay system (Promega GmbH, Mannheim, Germany) and a GlowMax 20/20 luminometer (Promega) (28). For easy comparison, FLuc values are normalized by corresponding RLuc values and vice versa. Results shown are mean values with standard deviation from three biologically independent samples, each measured in triplicate during a single experiment.
Online tool
The web tool was implemented with the web application framework Struts 2 (Apache Software Foundation, Los Angeles, CA, USA) using the integrated development environment MyEclipse (Genuitec, Flower Mound, TX, USA). As a servlet container we use Tomcat (Apache Software Foundation). The communication with the web service j5 works by using an XML-RPC interface (29). The AssemblX web tool will be made available for a minimum full 2 years following publication, via www.assemblx.org and via GitHub (https://github.com/AssemblX/AssemblXWeb) under a GNU General Public License as published by the Free Software Foundation.
RESULTS
The AssemblX strategy
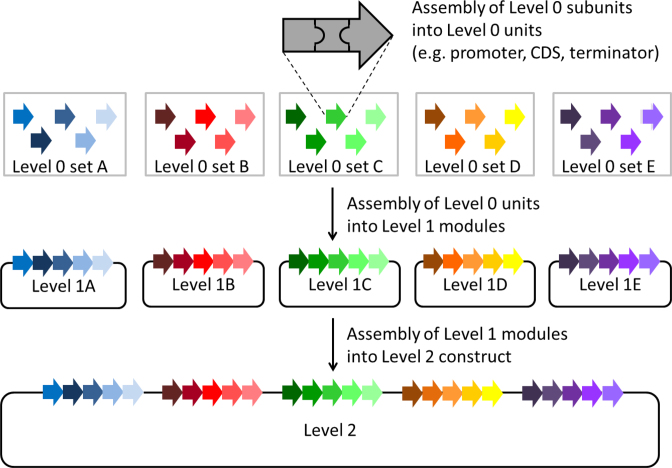
The assembly strategy presented here allows the de novo assembly of up to 25 (transcriptional) units from 75 or even more subunits into a single construct in a predefined order. AssemblX relies on overlap-based cloning methods, like in vitro SLiCE and HiFi DNA assembly (NEB) or in vivo TAR (10,11,13). These methods utilize overlapping homology regions of various lengths flanking the DNA parts to allow the scar-less assembly of multiple parts into a single DNA construct. To facilitate a fast assembly process, AssemblX is designed in three successive cloning Levels, each Level allowing multiple parallel assemblies: entry Level 0 for assembly of units from individual parts like promoter, terminator and coding sequence (CDS), or any other type of DNA sequence; Level 1 for the assembly of up to five Level 0 units into Level 1 modules, and Level 2 for the final assembly of up to five Level 1 modules into a single construct (Figure 1). Furthermore, AssemblX provides a j5-based web tool (www.assemblX.org), which assists the user with all design and assembly steps and therefore greatly reduces the time required to complete complex assemblies (29).
Figure 1.
The AssemblX workflow for the assembly of 25 units. Level 0 subunits, here with three DNA parts each, are assembled into five different sets of Level 0 vectors resulting in 25 Level 0 units. In the next step, Level 0 units from one Level 0 set are assembled in one corresponding Level 1 module. The five Level 1 modules, each containing five Level 0 units, are then assembled into one single Level 2 vector.
Assembly Level 0
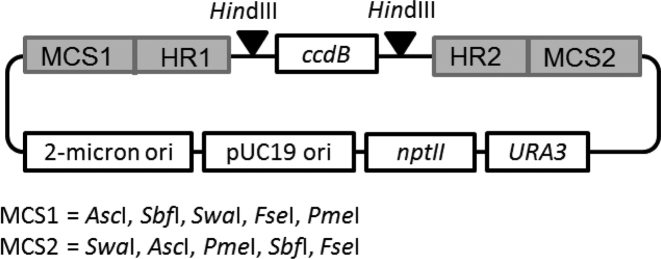
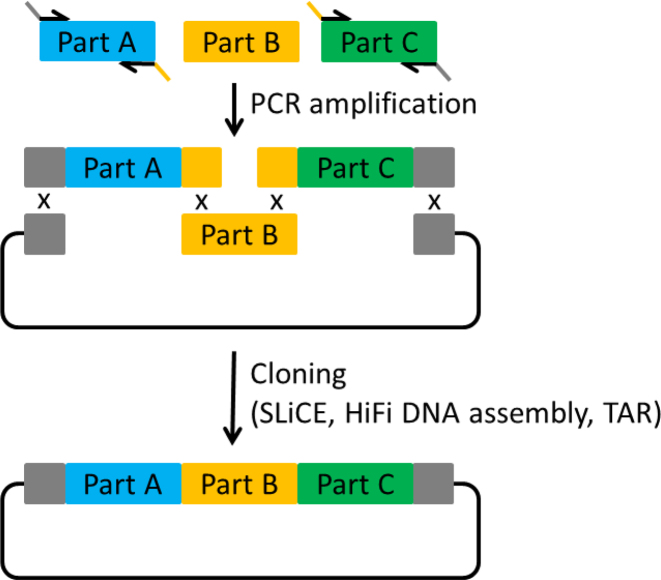
Level 0 assemblies allow the construction of assembly units from several subunits, e.g. promoter, CDS, terminator. For this purpose, we designed a vector backbone, which is similar for all Level 0 vectors and just differs in two variable homology regions that will later define the position of the assembly unit within the final construct (Figure 2). The Level 0 backbone consists of the E. coli pUC19 replication origin, the kanamycin resistance gene nptII, the 2-micron origin for replication in yeast and the yeast URA3 marker gene. Each Level 0 vector corresponds to a given position (position 1–25) in the final Level 2 assembly product and therefore acts as a positioning vector. The upstream (left) homology region of a vector defines the upstream neighbor, whereas the downstream (right) homology region defines the downstream neighbor. To this end, each downstream homology region is used as an upstream region in the neighboring downstream Level 0 vector. The homology regions are separated by a HindIII-flanked ccdB expression cassette. For the Level 0 assembly of a given assembly unit from multiple subunits, the ccdB cassette is released by HindIII digestion, to allow the overlap-based cloning in-between the homology regions, e.g. by well-established methods like SLiCE, TAR or HiFi DNA assembly. To this end, all subunits need to be equipped with customized overlaps to their neighboring subunits or to the vector sequence, respectively. This is usually achieved by introducing the overlaps with primers during PCR amplification (Figure 3). The primer design can be done manually or, for more convenience, with the help of the AssemblX web tool (see below). All PCR products necessary for the assembly are then combined with the linearized Level 0 backbone in an appropriate assembly reaction. The size, number of subunits and overall design of a Level 0 unit is completely user defined and only limited by the chosen assembly method. During the Level 0 cloning process, the ccdB gene reduces empty vector background and obviates purification of the linearized Level 0 backbone. A successfully assembled Level 0 unit can be released from the backbone for subsequent Level 1 assembly by using one of five rare 8-base cutter recognition sites (AscI, SbfI, SwaI, FseI, PmeI) flanking the homology regions. This makes it unnecessary to PCR-amplify the assembly units prior to Level 1 assemblies and eliminates the risk of PCR-born errors.
Figure 2.
Design of Level 0 vectors. The backbone is a yeast shuttle vector with a pUC19 replication origin, the kanamycin resistance gene nptII for selection in Escherichia coli, a high-copy 2-micron origin for replication and the URA3 marker gene for selection in Saccharomyces cerevisiae. Each Level 0 vector possesses a different combination of homology regions (HR). HR1 gives homology to the Level 1 vector backbone or to the HR2 of the adjacent Level 0 vector. HR2 gives homology to the next Level 0 vector or to the Level 1 vector backbone. These regions are separated by a HindIII-flanked ccdB expression cassette. The HindIII sites are used to release the ccdB cassette and to allow the subsequent insertion of the individual subunits in-between the homology regions by scar-free, overlap-based cloning. The homology regions are flanked by two multiple cloning sites (MCS), containing the five 8-base cutters AscI, SbfI, SwaI, FseI, PmeI to release Level 0 constructs for further cloning into Level 1.
Figure 3.
Level 0 assembly with overlap-based cloning methods. The overlaps between the individual subunits can be added via PCR, using primers overlapping with the neighboring fragment in their 5΄ region. After amplification of Part A and Part C with appropriate primers the three parts can be assembled into a vector backbone.
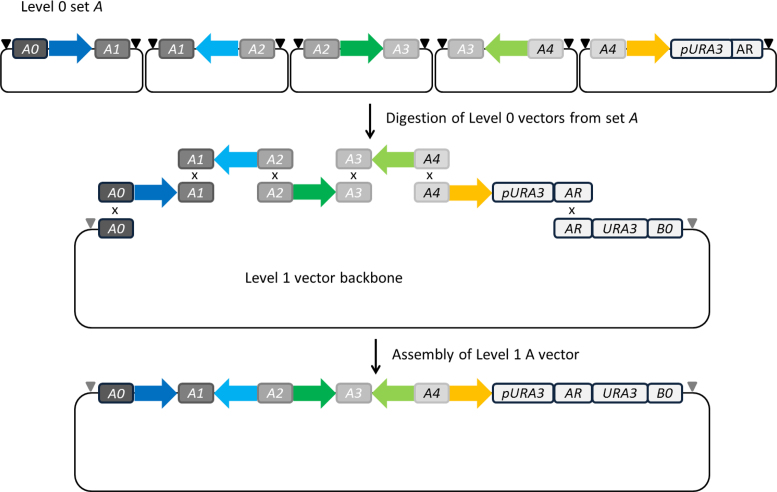
Level 0 vectors are grouped into five sets (set A, B, C, D and E), each containing five positioning vectors. All units, cloned in one set of Level 0 vectors, are assembled into one specific Level 1 vector, e.g. pL1A for Level 0 set A, or pL1B for Level 0 set B. The resulting Level 1 module holds up to five Level 0 units in the predefined order (Figure 4). The first (or left) homology region of a Level 0 vector set is designated as type X0 (X = A, B, C, D or E). These regions are 50 bp long and provide the left-arm homology to a specific Level 1 vector. The inlying homology regions of type X1 to X4 are 36 bp long each and will later provide the necessary overlaps for a directed assembly of Level 0 units into a single Level 1 module. The last homology region of a vector set, designated type XR, is 51 bp long and provides right arm homology to a Level 1 vector. The longer overlaps to the Level 1 vector support the TAR mediated assembly of large fragments in the later Level 2 assembly step. All homology regions of type XR were defined manually: each represents the first 17 codons from one out of five different auxotrophic yeast marker genes and is preceded by the corresponding promoter region and followed by a stop codon. Despite providing overlap to a Level 1 vector via XR, this region also drives the expression of an auxotrophic marker, in successfully assembled Level 1 constructs.
Figure 4.
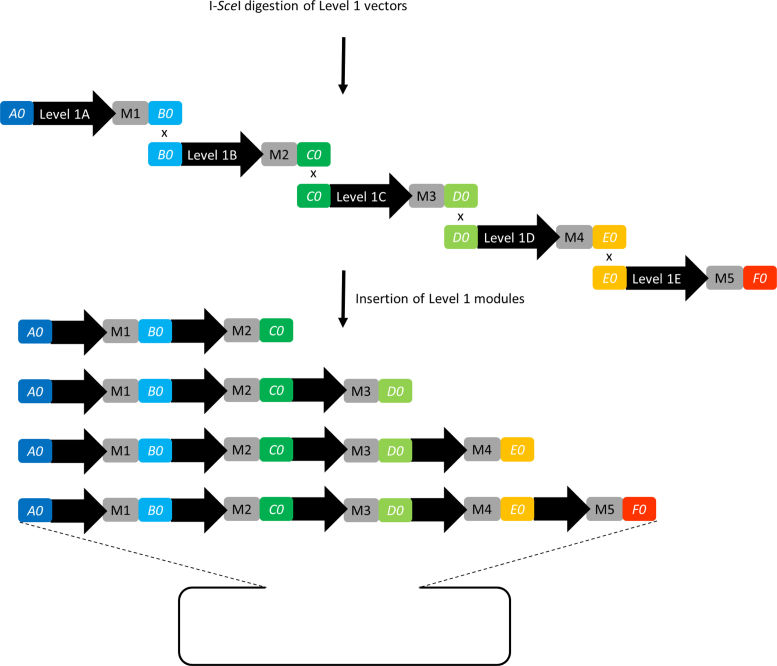
Level 1 assembly scheme. Five Level 0 units are released from Level 0 vectors by digestion with one of the designated 8-base cutters (black triangle). During in vivo or in vitro assembly, overlaps between the different Level 0 units and the Level 1 destination vector ensure a directed assembly into a single circular DNA construct. Located in front of the homology region AR is the URA3 promoter. AR itself represents the first 50 nucleotides of the URA3 CDS. The complete URA3 CDS and the appropriate terminator are located on the Level 1 vector backbone. In this way a functional selection marker is created only by a successful Level 1 assembly. Complete Level 1 modules are flanked by I-SceI sites (gray triangle).
To design the sequences for homology regions of type X0 and type X1 to X4 for each vector set, we employed the web tool R2o designer (30). The tool generates orthogonal sequences, intended to be biologically neutral and well suited for isothermal overlap-based assembly methods (40% GC content, Supplementary Table S1) (14).
As the X0 and XR regions of a given Level 0 set provide overlap to the Level 1 vector, they are strictly required for the next assembly level. Therefore, the vector collection described so far would only allow the cloning of exactly five Level 0 units per Level 1 module. To allow the assembly of less than five units, we created additional Level 0 vectors that contain the XR homology region of a vector set, in combination with X0, X1, X2 or X3. For example A1-AR vectors allow Level 1 assembly of two Level 0 units by using A0-A1 and A1-AR vectors from vector set A. Table 1 shows a complete list of all Level 0 vectors.
Table 1. Complete list of available Level 0 and Level 1 vectors.
| Level 0 vector set | Vector name | HR1 | HR2 | Corresponding Level 1 vectors (HR1/HR2) |
|---|---|---|---|---|
| A | pL0A_0-1 | A0 | A1 | pL1A-hc/pL1A-lc (A0/AR) |
| pL0A_1-2 | A1 | A2 | ||
| pL0A_2-3 | A2 | A3 | ||
| pL0A_3-4 | A3 | A4 | ||
| pL0A_0-R | A0 | AR | ||
| pL0A_1-R | A1 | AR | ||
| pL0A_2-R | A2 | AR | ||
| pL0A_3-R | A3 | AR | ||
| pL0A_4-R | A4 | AR | ||
| B | pL0B_0-1 | B0 | B1 | pL1B-hc/pL1B-lc (B0/BR) |
| pL0B_1-2 | B1 | B2 | ||
| pL0B_2-3 | B2 | B3 | ||
| pL0B_3-4 | B3 | B4 | ||
| pL0B_0-R | B0 | BR | ||
| pL0B_1-R | B1 | BR | ||
| pL0B_2-R | B2 | BR | ||
| pL0B_3-R | B3 | BR | ||
| pL0B_4-R | B4 | BR | ||
| C | pL0C_0-1 | C0 | C1 | pL1C-hc/pL1C-lc (C0/CR) |
| pL0C_1-2 | C1 | C2 | ||
| pL0C_2-3 | C2 | C3 | ||
| pL0C_3-4 | C3 | C4 | ||
| pL0C_0-R | C0 | CR | ||
| pL0C_1-R | C1 | CR | ||
| pL0C_2-R | C2 | CR | ||
| pL0C_3-R | C3 | CR | ||
| pL0C_4-R | C4 | CR | ||
| D | pL0D_0-1 | D0 | D1 | pL1D-hc/pL1D-lc (D0/DR) |
| pL0D_1-2 | D1 | D2 | ||
| pL0D_2-3 | D2 | D3 | ||
| pL0D_3-4 | D3 | D4 | ||
| pL0D_0-R | D0 | DR | ||
| pL0D_1-R | D1 | DR | ||
| pL0D_2-R | D2 | DR | ||
| pL0D_3-R | D3 | DR | ||
| pL0D_4-R | D4 | DR | ||
| E | pL0E_0-1 | E0 | E1 | pL1E-hc/pL1E-lc (E0/ER) |
| pL0E_1-2 | E1 | E2 | ||
| pL0E_2-3 | E2 | E3 | ||
| pL0E_3-4 | E3 | E4 | ||
| pL0E_0-R | E0 | ER | ||
| pL0E_1-R | E1 | ER | ||
| pL0E_2-R | E2 | ER | ||
| pL0E_3-R | E3 | ER | ||
| pL0E_4-R | E4 | ER |
The Level 0 vectors are grouped into five sets. Each set is compatible with a distinct Level 1 vector, which is available in either high- or low-copy format (hc/lc).
Design of Level 1 vectors
Level 1 vectors allow the assembly of up to five Level 0 units, released from their Level 0 backbone by digestion, into one Level 1 module. All Level 0 fragments for one module are combined with the appropriate, linearized Level 1 vector in an in vitro or in vivo overlap-based assembly reaction. Each Level 1 vector backbone is available in two versions, with either a high-copy 2-micron origin or a low-copy CEN/ARS region for replication in yeast. This boosts the flexibility for the direct expression of small assembly projects consisting of up to five units in a single Level 1 module. To create Level 1 vectors compatible with the different Level 0 vector sets, we inserted homology regions X0 and XR. The left homology region of type X0 is followed by a PacI site for linearization, and a promoterless auxotrophic marker gene, which contains a XR type homology region, defined as the first 17 codons of the respective gene. The resulting vectors are listed in Table 1. The marker gene is preceded by a third homology region Y0 (first homology region of the next Level 1 vector) downstream of the terminator, e.g. B0 for the pL1A vector. Homology regions X0 and Y0 are flanked by 18 bp long recognition sites for the homing endonuclease I-SceI (31,32). Digestion with I-SceI allows the release of Level 1 modules from the backbone and their further cloning into Level 2 without PCR amplification. During this step the auxotrophic marker genes integrated in the Level 1 constructs are also transferred into the Level 2 construct, which allows stringent selection of successful assemblies. Level 1 assembly may be done by in vitro overlap-based methods or by in vivo assembly in yeast. In contrast to the Level 0 vectors, the Level 1 destination vectors confer ampicillin resistance, allowing selection against undigested Level 0 backbones in E. coli.
Design of Level 2 vectors
After I-SceI mediated release from the Level 1 backbone, up to five Level 1 modules can be combined into a single Level 2 destination vector, preferably by using TAR. Level 2 vectors can be used directly to maintain and express the assembled units in S. cerevisiae either in high or low copy number. The basic Level 2 vector backbones are identical to the Level 1 backbones and differ only in the homology regions inserted between the two I-SceI sites. The different Level 2 vectors are designed to hold different numbers of Level 1 modules. A Level 2 vector for only one Level 1 module is not necessary, as it would be identical to the corresponding Level 1 construct. All Level 2 vectors contain homology region A0 in combination with a second region, providing overlap to the homology region Y0 from the last Level 1 module that is part of the assembly (Figure 5 and Table 2). The successful assembly can be monitored by selection of transformed yeast on SD dropout medium selecting for the presence of all incorporated Level 1 modules.
Figure 5.
Level 2 assembly scheme. Level 1 modules are released from Level 1 vectors by digestion with I-SceI. During assembly, overlaps between the different modules and the Level 2 destination vector ensure a directed assembly into a single circular DNA construct. Depending on the number of Level 1 modules to be assembled, different Level 2 vectors for the assembly of two to five Level 1 modules are used. M1: URA3, M2: LEU2, M3: HIS3, M4: LYS2, M5: TRP1.
Table 2. Available Level 2 vectors.
| Level 1 modules used for assembly | Corresponding Level 2 vectors (HR1/HR2) | Auxotrophic markers for selection |
|---|---|---|
| A, B | pL2_AB_hc/pl2_AB_lc (A0_C0) | URA3, LEU2 |
| A, B, C | pL2_AC_hc/pL2_AC_lc (A0_D0) | URA3, LEU2, HIS3 |
| A, B, C, D | pL2_AD_hc/pL2_AD_lc (A0_E0) | URA3, LEU2, HIS3, LYS2 |
| A, B, C, D, E | pL2_AE_hc/pl2_AE_lc (A0_F0) | URA3, LEU2, HIS3, LYS2, TRP1 |
Depending on the number of Level 1 modules to assemble, different Level 2 vectors must be used. Each vector is available in high- and low-copy format (hc/lc). Depending on the assembled Level 1 modules, the resulting Level 2 construct can be selected with up to five auxotrophic markers.
If the final construct is intended for use in host organisms different from yeast, any desired (expression) vector, e.g. a T-DNA vector for plant transformation, can be converted into an AssemblX compatible vector (Supplementary Figure S1). To this end, the target vector is linearized either with a suitable restriction enzyme, or by PCR amplification. Thereafter, the user has the following three options: (i) insertion of a vector conversion cassette, amplified from a pL2 vector containing the required homology regions. This cassette contains the homology regions and a yeast replication origin. The newly created vector can be used as backbone in the Level 2 assembly reaction in yeast. The resulting construct is suitable for transfer to the desired host organism. (ii) Insertion of an appropriate homology region cassette into the target vector for overlap-based subcloning of I-SceI digested Level 1 or Level 2 constructs. This cassette can be amplified from a Level 2 AssemblX vector containing the required homology regions. (iii) The insertion of a single I-SceI site or an I-SceI –ccdB- I-SceI cassette into the target vector for the subcloning of an I-SceI digested Level 1 or Level 2 construct by ligation.
The AssemblX web tool
The assembly of up to 25 genes with AssemblX requires a thorough design of primer sequences for Level 0 assemblies. To facilitate planning and design we established a web tool, which supports the user in every step of the assembly. The web tool is accessible via www.assemblx.org. The help function guides the user through every step of the intuitive planning and design workflow: beginning with the number of units to be assembled, organization of the individual assembly units into distinct modules, planning the variable parts of the final assembly, detailed primer design, assembly suggestions for Level 0 and ending with individualized protocols for higher-level assemblies. After the user has defined a schematic overview of the final assembly product, the Level 0 assemblies are addressed: the user provides DNA sequences for all individual subunits, in either GenBank or plain text format. The sequence files are then used to design primers for Level 0 assemblies. To this end, the user's input is converted into input files for the online tool j5, which is an automated design software for DNA assemblies (29). The web tool hands over the data to the j5 server and integrates the j5 results into the further process. Basically, the j5 output contains suggestions for PCR primers for amplification and attachment of homology regions to Level 0 subunits. From these suggestions, individual protocols for the assembly of Level 0 units are compiled by the AssemblX web tool and combined into a written bench protocol for all steps of the AssemblX process leading the user to the final product.
Highly efficient assembly of 25 expression cassettes using AssemblX
For the successful construction and expression of multi-gene pathways it is necessary to control the expression level of many individual genes. To provide a first starting point for researchers working with S. cerevisiae, we cloned and characterized a panel of 40 constitutive yeast promoters, each in combination with the terminator region from the same gene (Supplementary Results 1 and Supplementary Figure S2). To make use of this promoter collection and to demonstrate the capability of AssemblX, we designed and successfully cloned a panel of 25 individually regulated genes, consisting of 69 different DNA parts in total, into a single Level 2 construct, in about 4 weeks. Besides several fluorescent and enzymatic reporter genes (yEGFP, AsRed2, EBFP2, lacZ, uidA, FLuc, RLuc), a set of dominant yeast selection markers (G418, hygromycin, phleomycin and nourseothricin resistance genes) for easy and direct validation of successful assemblies in yeast has been incorporated. LacI and TetR genes and the corresponding modified GAL1 UAS-based promoters enable the IPTG- or ATc-controlled expression of yEGFP and AsRed2, respectively. Furthermore, the complete Level 2 assembly product contains the biosynthetic pathway for the purple pigment violacein consisting of five yeast codon optimized genes (vioA, vioB, vioC, vioD and vioE) from Chromobacterium violaceum, and five rpo genes for the production of E. coli RNA polymerase subunits (alpha, beta, beta’, omega and sigma70 factor) in yeast. Finally, two E. coli resistance markers (spectinomycin and chloramphenicol resistance) for easy monitoring of plasmid integrity, after transfer to E. coli, are included. Supplementary Table S2 shows a complete list of all genes incorporated into the assembly, the subunits from which the Level 0 units have been assembled, and their position in the final construct. All Level 0 assemblies have been done with a preliminary version of AssemblX, using a set of Level 0 vectors with non-optimized homology regions and lacking the ccdB cassette. Overall, the cloning efficiency for Level 0 assemblies greatly varied with complexity and choice of the assembly method (efficiencies between 2 and 100%, data not shown). Therefore, a meaningful general efficiency cannot be calculated. To increase the cloning efficiency and reduce empty vector background, we redesigned the Level 0 vectors by optimizing the homology regions using R2o designer (30) and inserting the ccdB cassette, as described above. To assess the efficiency of these changes, the four Level 0 assemblies that performed worst with the previous non-optimized AssemblX version (cloning efficiencies below 5%) were reassembled in the optimized pL0 vectors. The cloning efficiencies determined by colony PCR and restriction digestion increased to 33% for one construct and 100% for the remaining three constructs. This demonstrates the positive effect of ccdB counterselection and optimized homology regions. To change the old homology regions to the optimized ones in the remaining constructs, the complete Level 0 units were amplified with primers containing the new R2o-optimized homology regions. Subsequently, the PCR products were directly used in Level 1 assemblies. All Level 1 assemblies were done with TAR cloning in YPH500 yeast cells. Yeast colonies were pre-screened by colony PCR and potential positive plasmids were transferred to E. coli for subsequent analysis by restriction digestion and sequencing of assembly junctions (Table 3). For Level 1 modules A, B, C and D, 100% of clones analyzed by colony PCR were identified as potential positives and all clones further investigated by restriction digestion showed the expected fragment pattern. Sequencing of selected clones further confirmed the expected assembly junctions. Assembly of Level 1E was less efficient, as only 15/24 clones (62%) were identified as potential positives by colony PCR. Four of these clones were further analyzed by digestion and all were confirmed to be positive by sequencing. The R2o-optimized homology regions are probably not the reason for the lower assembly efficiency in Level 1E, as we experienced a much lower cloning efficiency for this module with non-optimized homology regions (4/47 clones, data not shown). Most likely, random internal homologies between the vioE CDS and the ACT1 and TDH1 promoters, which are parts of Level 1 module E, are the cause for the reduced cloning efficiency. Subsequent to the Level 1 assemblies, the individual Level 1 modules were assembled into the pL2AE-lc backbone using TAR. Analysis by multiplex colony PCR (24/24 clones positive), assays for the reporter genes lacZ and uidA (42/42 clones positive) and growth assays on plates containing G418, hygromycin, phleomycin and nourseothricin (42/42 clones positive) indicate a cloning efficiency of 100% and confirm the expression of six reporter genes (Table 3 and Supplementary Figure S3a–d). For further analysis, a single clone was randomly selected. The DNA was isolated and successfully transformed into E. coli. Selection of transformants was done with spectinomycin and chloramphenicol, indicating the presence of the respective resistance genes, which are part of the assembly. The DNA was isolated from E. coli and integrity was confirmed via gel electrophoresis and restriction mapping (Supplementary Figure S4). The size of the final assembly product was determined to be between 70 and 75 kb, which fits the expected size of 72 kb, and restriction with I-SceI releases a 68.5 kb fragment, which was confirmed with PFGE. Also, all expected restriction fragments from a SalI/PacI digestion were observed. Sequencing of the assembly junctions between all Level 1 modules and the Level 2 vector backbone gave further proof for a complete and error-free assembly. Functional analysis of the reporter genes FLuc and RLuc was done by dual luciferase measurements, confirming the expression of these genes in yeast (Supplementary Figure S5a). Similarly, flow cytometry based measurements for yEGFP or AsRed2 fluorescence with or without the inducers IPTG or ATc indicate the functionality of LacI and TetR, regulating the expression of yEGFP and AsRed2 in yeast, respectively (Supplementary Figure S5b). The above mentioned experiments confirm the function of 14 (reporter) genes. To investigate the successful expression of the five-gene violacein pathway, the construct was retransformed into BY4741 yeast cells. This was done, because the original assembly host YPH500 is auxotroph for tryptophane, which is converted to violacein by the vioABCDE pathway. Transformed BY4741 cells are purple, indicating the successful production of the purple pigment violacein by the five biosynthetic genes vioA, vioB, vioC, vioD and vioE (Supplementary Figure S3e and f). The five assembly units, containing E. coli rpo genes for expression in yeast, were verified by complete sequencing. Equally, the EBFP2 gene was confirmed by sequencing, because, for unknown reasons, the fluorescence signal in microscopic analyses was very weak. Taken together, the results from the demo assembly, which was completed in about 4 weeks, confirm the effectiveness of the assembly strategy presented here. Several other, but not yet published assembly projects, have already been completed using AssemblX. The testimonial section available at www.assemblX.org provides an overview on the different topics addressed so far.
Table 3. Efficiency of Level 1 and 2 assemblies during the 25-gene demo assembly.
| Assembly step | Positive in colony PCR | Confirmed by restriction analysis | Confirmed by sequencing |
|---|---|---|---|
| 1A | 24/24 | 4/4 | 2/2 |
| 1B | 24/24 | 4/4 | 2/2 |
| 1C | 24/24 | 4/4 | 2/2 |
| 1D | 24/24 | 4/4 | 2/2 |
| 1E | 15/24 | 4/4 | 2/2 |
| 2 | 24/24 | 1/1 | 1/1 |
DISCUSSION
The AssemblX toolkit provides a user-friendly strategy for reliable de novo multi-part assemblies in S. cerevisiae and the subsequent expression in yeast or any other host organism. The toolkit does not require parts domestication, allows performing all necessary Level 0 assemblies in parallel to save time and avoids PCR amplification for higher assembly levels. Furthermore, AssemblX enables the functional analysis and reuse of individual Level 0 units or complete Level 1 modules, is highly efficient, uses inexpensive methods, is flexible with regard to the user's preference for different overlap based cloning methods, and is accompanied by an online tool. The latter provides easy access for users regardless of their scientific background and minimizes experimental errors. In this way, undergraduate students and experienced researchers alike will be able to create complex genetic constructs, benefiting from the easy-to-follow three-step workflow.
The novelty of AssemblX lies in an elaborate combination of different methods to facilitate and optimize cloning workflow and expression of multi-gene constructs, rather than an optimization or modification of already functional cloning methods. The overall AssemblX strategy incorporates many useful tweaks that include reduced background through ccdB counter selection, E. coli marker switching between Level 0 and Level 1, and selection of successful in vivo assemblies in yeast through marker gene reconstitution. The homology regions used in AssemblX are R2o-optimized, supporting highly efficient assemblies (up to 100% correct clones in Level 1 and 2 assemblies) and can be used with most overlap-based assembly methods. AssemblX combines the advantages of overlap-based, scar-free and sequence-independent assemblies in Level 0 vectors and subsequent standardized adapter-based assemblies in Levels 1 and 2. This flexibility allows an unrestricted architecture for every Level 0 unit and minimizes the risk of running into intractable conflicts during cloning.
The sequence independency of AssemblX relies on the release of Level 0 units from the positioning vectors by restriction digestion with a set of five rare-cutting 8-base cutters for further Level 1 assemblies. Laborious deletion of unintended cut sites, or PCR amplifications which bear the risk of introducing sequence errors, thereby become unnecessary. This is also true for I-SceI-based release of Level 1 modules from their backbones, as the I-SceI recognition site is 18 bp long, extremely rare and does for example not cut within the entire yeast genome (33). The avoidance of PCR amplification facilitates Level 1 and 2 construct verification, as diagnostic restriction digestion or sequencing of assembly junctions is usually sufficient.
The features, uniquely combined in AssemblX, allow completing a 25-parts assembly in about a month, starting from scratch, as demonstrated in the present study by generating a 25-gene assembly resulting in a 72 kb plasmid. An even faster multi-gene assembly is possible when pre-existing modules are employed. While several cloning strategies enabling multi-gene cloning have been published, none of them aims at constructs of this complexity and simultaneously allows sequence-independent, scar-free gene-level assemblies in a user oriented workflow (8,14–17,34).
We chose S. cerevisiae as host for the in vivo cloning steps, as homologous recombination in yeast is an efficient and cost-effective alternative to other recombination-based cloning methods. Furthermore, S. cerevisiae is a well characterized organism with reported success in the production of important natural substances like taxol and artemisinic acid (35–37). The library of constitutive promoters that we established and characterized here will assist users in establishing expression constructs for complex biosynthetic pathways in yeast. Researchers working with a different host or even ‘non-model’ organisms can easily transfer every Level 1 or Level 2 AssemblX construct into a new host by either including the necessary markers and/or replication origins into the assembly itself or by I-SceI- or homology region-based subcloning into appropriate vectors. We believe that this will encourage researchers from various fields to make use of the AssemblX toolkit and to explore new grounds in synthetic biology.
Supplementary Material
ACKNOWLEDGEMENTS
B.M.-R. thanks the Federal Ministry of Education and Research of Germany for funding. The authors greatly acknowledge Dr Pamela Holzlöhner (University of Potsdam, Germany) for introduction to the flow cytometer, Nathan Hillson (Joint BioEnergy Institute, Emeryville, CA, USA) for support with the implementation of j5 into the AssemblX web tool, and Jessica Behrend and Christoph Alt for testing the web tool.
Author contributions: L.H. and F.M. designed and developed the overall AssemblX strategy, conducted the experiments, analyzed the data and wrote the paper. J.G. programmed the online tool. K.S. constructed the different vector sets. B.M.-R. initiated the project and the overall research strategy, and edited the paper. K.M. and B.M.-R. jointly supervised the work. All authors take full responsibility for the content of the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Federal Ministry of Education and Research of Germany (BMBF) [031A172 to B.M.-R.]. Funding for open access charge: BMBF [031A172].
Conflict of interest statement. The authors declare competing financial interests, due to a patent application for the AssemblX cloning strategy, filed by the University of Potsdam with L.H., F.M., K.M. and B.M.-R. as inventors (Patent number EP16188155.2).
REFERENCES
- 1. Si T., Luo Y., Xiao H., Zhao H.. Utilizing an endogenous pathway for 1-butanol production in Saccharomyces cerevisiae. Metab. Eng. 2014; 22:60–68. [DOI] [PubMed] [Google Scholar]
- 2. Runguphan W., Keasling J.D.. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 2014; 21:103–113. [DOI] [PubMed] [Google Scholar]
- 3. Annaluru N., Muller H., Mitchell L.A., Ramalingam S., Stracquadanio G., Richardson S.M., Dymond J.S., Kuang Z., Scheifele L.Z., Cooper E.M. et al. . Total synthesis of a functional designer eukaryotic chromosome. Science. 2014; 344:55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kiani S., Beal J., Ebrahimkhani M.R., Huh J., Hall R.N., Xie Z., Li Y., Weiss R.. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods. 2014; 11:723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen S.N., Chang A.C., Boyer H.W., Helling R.B.. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. U.S.A. 1973; 70:3240–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knight T. Idempotent vector design for standard assembly of biobricks. 2003; Cambridge, MA: DSpace. MIT Artificial Intelligence Laboratory; MIT Synthetic Biology Working Group, Massachusetts Institute of Technology. [Google Scholar]
- 7. Engler C., Kandzia R., Marillonnet S.. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008; 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S.. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011; 6:e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sarrion-Perdigones A., Falconi E.E., Zandalinas S.I., Juárez P., Fernández-del-Carmen A., Granell A., Orzaez D.. GoldenBraid: An iterative cloning system for standardized assembly of reusable genetic modules. PLoS One. 2011; 6:e21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gibson D.G. Voigt C. Enzymatic assembly of overlapping DNA fragments. Methods in Enzymology. 2011; 498:San Diego, CA: Academic Press; 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y., Werling U., Edelmann W.. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012; 40:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibson D.G. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 2009; 37:6984–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kouprina N., Larionov V.. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 2008; 3:371–377. [DOI] [PubMed] [Google Scholar]
- 14. Casini A., MacDonald J.T., De Jonghe J., Christodoulou G., Freemont P.S., Baldwin G.S., Ellis T.. One-pot DNA construction for synthetic biology: the modular overlap-directed assembly with linkers (MODAL) strategy. Nucleic Acids Res. 2014; 42:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Storch M., Casini A., Mackrow B., Fleming T., Trewhitt H., Ellis T., Baldwin G.S.. BASIC: A new biopart assembly standard for idempotent cloning provides accurate, single-tier DNA assembly for synthetic biology. ACS Synth. Biol. 2015; 4:781–787. [DOI] [PubMed] [Google Scholar]
- 16. Agmon N., Mitchell L.A., Cai Y., Ikushima S., Chuang J., Zheng A., Choi W.-J., Martin J.A., Caravelli K., Stracquadanio G. et al. . Yeast Golden Gate (yGG) for the efficient assembly of S. cerevisiae transcription units. ACS Synth. Biol. 2015; 4:853–859. [DOI] [PubMed] [Google Scholar]
- 17. Mitchell L.A., Chuang J., Agmon N., Khunsriraksakul C., Phillips N.A., Cai Y., Truong D.M., Veerakumar A., Wang Y., Mayorga M. et al. . Versatile genetic assembly system (VEGAS) to assemble pathways for expression in S. cerevisiae. Nucleic Acids Res. 2015; 43:6620–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ellis T., Wang X., Collins J.J.. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechol. 2009; 27:465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vickers C.E., Bydder S.F., Zhou Y., Nielsen L.K.. Dual gene expression cassette vectors with antibiotic selection markers for engineering in Saccharomyces cerevisiae. Microb. Cell Fact. 2013; 12:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldstein A.L., McCusker J.H.. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999; 15:1541–1553. [DOI] [PubMed] [Google Scholar]
- 21. DiCarlo J.E., Norville J.E., Mali P., Rios X., Aach J., Church G.M.. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013; 41:4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Licausi F., Weits D., Pant B.. Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 2011; 190:442–456. [DOI] [PubMed] [Google Scholar]
- 23. Karimi M., Inzé D., Depicker A.. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002; 7:193–195. [DOI] [PubMed] [Google Scholar]
- 24. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Messerschmidt K., Hochrein L., Dehm D., Schulz K., Mueller-Roeber B.. Characterizing seamless ligation cloning extract for synthetic biological applications. Anal. Biochem. 2016; 509:24–32. [DOI] [PubMed] [Google Scholar]
- 26. Gietz R.D., Schiestl R.H.. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007; 2:31–34. [DOI] [PubMed] [Google Scholar]
- 27. Noskov V.N., Chuang R.Y., Gibson D.G., Leem S.H., Larionov V., Kouprina N.. Isolation of circular yeast artificial chromosomes for synthetic biology and functional genomics studies. Nat. Protoc. 2011; 6:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McNabb D.S., Reed R., Marciniak R.A.. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot. Cell. 2005; 4:1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hillson N.J., Rosengarten R.D., Keasling J.D.. j5 DNA assembly design automation software. ACS Synth. Biol. 2011; 1:14–21. [DOI] [PubMed] [Google Scholar]
- 30. Casini A., Christodoulou G., Freemont P.S., Baldwin G.S., Ellis T., MacDonald J.T.. R2oDNA Designer: computational design of biologically neutral synthetic DNA sequences. ACS Synth. Biol. 2014; 3:525–528. [DOI] [PubMed] [Google Scholar]
- 31. Colleaux L., D’Auriol L., Galibert F., Dujon B.. Recognition and cleavage site of the intron-encoded omega transposase. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:6022–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Monteilhet C., Perrin A., Thierry A., Colleaux L., Dujon B.. Purification and characterization of the in vitro activity of I-SceI, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res. 1990; 18:1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thierry A., Perrin A., Boyer J., Fairhead C., Dujon B., Frey B., Schmitz G.. Cleavage of yeast and bacteriophage T7 genomes at a single site using the rare cutter endonuclease I-SceI. Nucleic Acids Res. 1991; 19:189–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sarrion-Perdigones A., Vazquez-Vilar M., Palaci J., Castelijns B., Forment J., Ziarsolo P., Blanca J., Granell A., Orzaez D.. GoldenBraid 2.0: A comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013; 162:1618–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Engels B., Dahm P., Jennewein S.. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab. Eng. 2008; 10:201–206. [DOI] [PubMed] [Google Scholar]
- 36. DeJong J.M., Yule L., Bollon A.P., Long R.M., Jennewein S., Williams D., Croteau R.B.. Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2006; 93:212–224. [DOI] [PubMed] [Google Scholar]
- 37. Ro D.-K., Paradise E.M., Ouellet M., Fisher K.J., Newman K.L., Ndungu J.M., Ho K.A., Eachus R.A., Ham T.S., Kirby J.. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006; 440:940–943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.