Abstract
Spatial and temporal brain transcriptomics has recently emerged as an invaluable data source for molecular neuroscience. The complexity of such data poses considerable challenges for analysis and visualization. We present BrainScope: a web portal for fast, interactive visual exploration of the Allen Atlases of the adult and developing human brain transcriptome. Through a novel methodology to explore high-dimensional data (dual t-SNE), BrainScope enables the linked, all-in-one visualization of genes and samples across the whole brain and genome, and across developmental stages. We show that densities in t-SNE scatter plots of the spatial samples coincide with anatomical regions, and that densities in t-SNE scatter plots of the genes represent gene co-expression modules that are significantly enriched for biological functions. We also show that the topography of the gene t-SNE maps reflect brain region-specific gene functions, enabling hypothesis and data driven research. We demonstrate the discovery potential of BrainScope through three examples: (i) analysis of cell type specific gene sets, (ii) analysis of a set of stable gene co-expression modules across the adult human donors and (iii) analysis of the evolution of co-expression of oligodendrocyte specific genes over developmental stages. BrainScope is publicly accessible at www.brainscope.nl.
INTRODUCTION
The field of molecular neuroscience has seen a sharp rise in the availability of spatially mapped molecular data, accessible through public databases. General databases such as GTEx (1) and Encode (2), but also brain-specific databases like PsychENCODE (3), contain anatomically annotated gene expression and epigenetic data across the brain. Where some projects focus on specific diseases (such as Huntington's disease (4) and autism spectrum disorder (5)), others aim to capture general patterns in the healthy brain. A strong example of the latter are the efforts of the Allen Institute for Brain Science (6) to measure spatially mapped gene expression in mouse, macaque and human brain, both in the healthy adult individual and throughout brain development. These genome-wide studies of the transcriptome aim to elucidate relationships between brain structure and brain function, and identify genes that play a role in this.
Understanding brain transcriptome data is challenging, since it encompasses RNA expression over all genes, across many spatial coordinates of the brain, and through development in time. A powerful way to obtain insight into such complex multi-way data sets is by visually exploring the data using principles of presenting, browsing, and selecting. Currently available tools for analyzing gene expression in the brain that incorporate visualization include the Allen Institute's AGEA (7) and Neuroblast (8). These two portals represent two distinct views on the data. With AGEA, researchers can explore the interplay between anatomical connections and the gene expression similarities of brain areas. It shows sample-sample similarities and provides a parcellation of the brain entirely based on transcriptome data. A different view on the same data is offered by Neuroblast. Here, the focus lies on gene-gene comparisons: it shows which genes have similar spatial expression patterns in the healthy brain. Both AGEA and Neuroblast are valuable tools that have been used to study, for instance, bipolar disorder (9). However, these tools focus either on relationships between genes, or on the relationships between brain regions, while the interplay between these two is an essential part of the data. A suitable representation of brain transcriptome data that links a gene-centric and a sample-centric view is currently lacking.
The relationships between genes or samples can intuitively be represented in plots, where these elements are shown as points. The closeness of the points then represents their similarity. However, with a large number of samples and thousands of genes, a plot that reflects similarities needs to capture a high-dimensional space in a two-dimensional map. Common ways to reduce this dimensionality are multi-dimensional scaling (MDS) (10) and principle component analysis (PCA) (11). A more recently introduced non-linear dimension reduction method is t-distributed stochastic neighborhood embedding (t-SNE) (12). The power of t-SNE comes from the fact that it tries to accurately represent the local neighborhoods of points, so neighbors in the plot match those in the original high dimensional data. In return, the distances between non-similar points are less well-preserved. This is in marked contrast to, for example, PCA where the important components capture the direction of the largest variance across the points, which is generally reflected in distant (non-similar) points. t-SNE has been used to produce transcriptional maps of brain regions in the Allen Brain Atlas (ABA) (13,14), and it is popular in the analysis of single-cell molecular data (15–18).
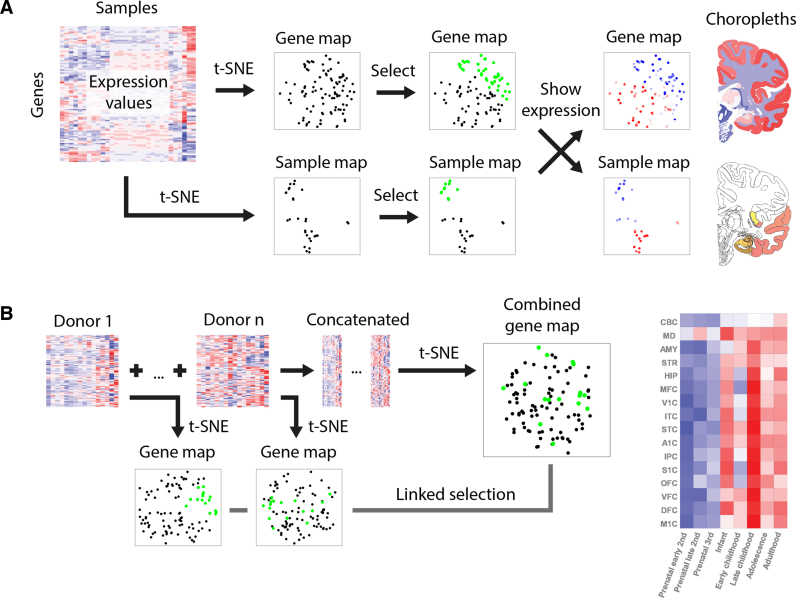
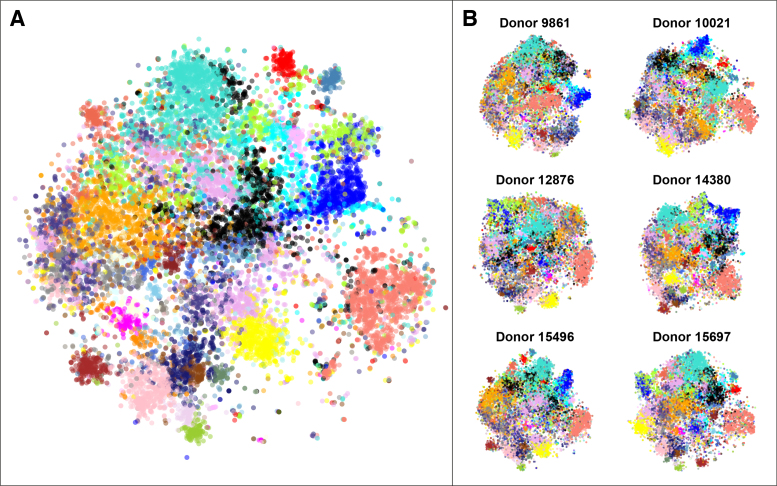
Here, we present BrainScope, a portal that uses t-SNE maps of both samples and genes in an interactive visualization of the transcriptional landscape of the brain. It gives a brain- and transcriptome-wide view of gene co-expression and transcriptional similarity of brain regions, based on the human brain data of the Allen Institute (19,20). It allows for interactive analysis of gene expression in the human brain, in an intuitive visual way. To connect the gene-centered and the sample-centered views, we make use of linked maps: t-SNE plots where a selection of points is rendered as a visual change in the linked plots. The first instance of this is the dual explorer (see Figure 1A), which has a single transcriptome-wide gene map and a brain-wide sample map. Users can select genes or samples and show their mean expression patterns in the other map. In addition, this part of the portal contains brain choropleths: user-selected slices of the human brain that are used to localize samples and illustrate spatial expression patterns. In addition to the dual explorer, the portal contains the comparative explorer (see Figure 1B), which focuses on the comparison of several gene maps, representing distinct donor brains. Therefore, the comparative explorer reveals inter-donor similarities in co-expression. Using the adult human data it shows robustness of co-expression modules, while for the developmental human data it shows changes in co-expression through time. Each part of the portal contains a direct link to enrichment tools Enrichr (21) and ToppGene (22), to provide a functional interpretation of selected gene sets.
Figure 1.
BrainScope views. (A) In the dual explorer, the gene expression data is visualized in two directions: a map for genes and a map for samples. Points that are close in the map have a high similarity. The portal allows for selection of points in either of the maps and shows the expression in the other map (red is high, blue is low): a set of genes has a profile across samples, which is averaged; and a set of samples has expression values in all genes, which are also averaged. When genes are selected, their average expression is also shown on brain slice choropleths; and when samples are selected, their location is shown on the same choropleths. (B) In the comparative explorer, only gene t-SNE maps are shown, but it contains data for multiple donor brains (replicates, or developmental stages). A t-SNE map is made for each donor and, in addition, one map is made for the combined data sets. When a selection of genes is made in either of the maps, this selection is carried over to the other maps and the average regional expression of these genes is shown in a heatmap.
The linked t-SNE maps of the BrainScope can conceptually be used in several ways. Selection of a single point reveals the corresponding expression pattern, either of a gene throughout the spatially mapped samples, or of a sample across all genes. Selection of points in the sample map reveals gradients of expression in the gene map, which elucidate gene-gene relationships. In addition to single point selection, a set of points can be selected to study the relationships between these points (co-expression or transcriptional similarity) and characterize sub-clusters by their mean expression patterns. In the comparative explorer, any selection of genes is carried over to all gene maps, showing differences in co-expression between brains. We demonstrate the usefulness of BrainScope by exploring the major patterns of gene expression in the adult human brain, and the way these reflect gene function and cell type composition of brain samples. In addition, user-supplied gene sets can be examined for structure. With the comparative explorer, we highlight the stability of the gene t-SNE maps over the six donor brains of the Allen Brain Atlas, in line with recently published consensus modules (23). Finally, the spatio-temporal transcriptome shows that the changes in expression of oligodendrocyte marker genes reflect the development of the brain. Combined, these applications enable a unique view of the rich gene expression data of the Allen Human Brain Atlases.
MATERIALS AND METHODS
Gene expression data
Gene expression data was obtained from the Allen Institute for Brain Science. The Adult Human Brain Atlas (19) contains gene expression measurements of six healthy adult donors. Samples were taken using macro- and microdissection of anatomically annotated regions. The number of samples differs per donor, from 363 to 946, with a total of 3702 samples. The expression values in each sample were determined with a customized micro-array chip, measuring 58 692 probes. Initial data processing was performed by the Allen Institute, and the data were made available on their website (http://human.brain-map.org/static/download).
The Developing Human Brain Atlas (20) has a lower spatial resolution, but samples were taken from human donors of a broad range of ages. In total 42 brains were sampled, ranging in age from 8 weeks post-conception to 40 years old. The number of samples per brain ranges from 1 to 16, with a total of 524 anatomically annotated samples. Gene expression was determined using RNA sequencing, and RPKM values are available online for 52 376 genes (http://www.brainspan.org/static/download).
Data preprocessing
In the adult human brain data the 58 692 probes were mapped to 19 992 genes, using their Entrez identifiers. For genes that have two probes, the probe with the highest variance was selected. For genes with more than two probes, we picked the probe with the highest connectivity to all other probes (defined as the sum of Pearson correlations). The number of samples differs per donor brain. To enable combination of the data for dual t-SNE, all expression sets were reduced to have 105 values per gene, corresponding to the annotated regions that were sampled in each brain. Finally, to obtain a single gene and sample map in the dual explorer, the expression values for each combination of sample and gene were averaged over the six donors. The comparative explorer of the adult brain instead uses processed data for all brain samples (23). To enable a direct comparison between densities in the gene t-SNE maps and previously defined WCGNA-based gene modules (23), both were computed from identical gene-sample data matrices.
In the developing human dataset, samples were pooled into eight age windows, to obtain subsets with higher sample sizes. Supplementary Table S1 shows which donor brains were combined into each age window, with sample sizes and donor characteristics. Anatomical regions were required to have at least one sample for each age group, giving 16 regions with eight measurements each. From the 52 376 genes, only the 18 233 genes were selected that had an RPKM-value above 1 in at least 20% of all samples.
The dimension reduction results of t-SNE are dependent on scale and location of the data. For the gene maps, all genes were z-score normalized, to have zero mean and a standard deviation of 1. For the sample maps the values for each sample were instead z-scored.
Dimension reduction
Dimension reduction was performed with t-distributed stochastic neighborhood embedding (t-SNE), a non-linear embedding technique (12). It creates a low-dimensional map of high-dimensional data, while preserving as much of the local structure as possible. The method has one main parameter, the perplexity value, which determines the variances of the Gaussian kernels that are used to calculate similarities in the high-dimensional space. The higher the perplexity value, the larger the number of neighboring points to which similarities are preserved. Because t-SNE only aims to preserve neighborhoods, the rotation of the maps is arbitrary. In the comparative explorers, the maps were rotated to be as similar as possible (defined by the mean Euclidean distance of all points). In many applications a PCA reduction to a somewhat lower dimensional space is performed, for computational and noise reduction reasons. In our analyses we did not perform this step, in order to retain the original neighborhoods. The gene t-SNE maps were made with the default perplexity value of 30. The sample t-SNE maps were made with a lower perplexity value of 10, due to the lower number of points.
Gene set clustering and analyses
We characterized the 3000 genes in the regions of highest density of the gene map for the adult human data. These were identified in a Gaussian density estimate of the map, with an identity covariance matrix. The 3000 genes with highest local densities were then hierarchically clustered with Euclidean distance and complete linkage. The optimal number of clusters (27) was determined by maximizing the silhouette score, and the 23 clusters with more than 30 genes were characterized by their expression patterns and enrichments in ToppGene (22).
To define cell-type marker genes, we made use of a database of gene expressions from fluorescence-activated sorted cells of the mouse cerebral cortex (24). Genes were selected as markers when they had a 20-fold higher expression in the cell type of interest than the geometric mean of the other cell types. Mouse gene identifiers were matched to human orthologs using builds GRCm38.p4 and GRCh38.p3 in BioMart (25).
Clustering of post synaptic density related genes (19,26) in the gene t-SNE map was performed with a Gaussian mixture model, where the number of clusters was optimized using the Bayesian information criterion. The reported gene set enrichment analyses were performed in ToppGene (22).
RESULTS
Dimension reduction of gene expression in the brain
BrainScope aims to visualize gene expression data of the brain, in an interactive and intuitive way (see Supplementary Video S1-3). It is built on spatially resolved gene expression data in the adult human brain, and the Brainspan atlas of the developing human brain, both provided by the Allen Institute for Brain Science. The adult human brain atlas contains genome-wide expression values of six donors, five males and one female, aged 24–57 years old. The in total 3702 samples cover a wide range of anatomical regions, with 105 distinct regions that are sampled at least once in every donor. For the dual explorer, we averaged the expression values to these 105 regions for 19 992 genes (see the Materials and Methods section). For the comparative explorer of the developing human atlas, we grouped the measurements of 16 brain regions in eight developmental stages (see Supplementary Table S1). To produce 2D maps of the expression data, we made use of t-SNE (12). A comparative analysis between t-SNE and PCA is provided in the Supplementary Text and Supplementary Figure S2.
The dual explorer shows localized transcriptional similarity in human neuro-anatomy
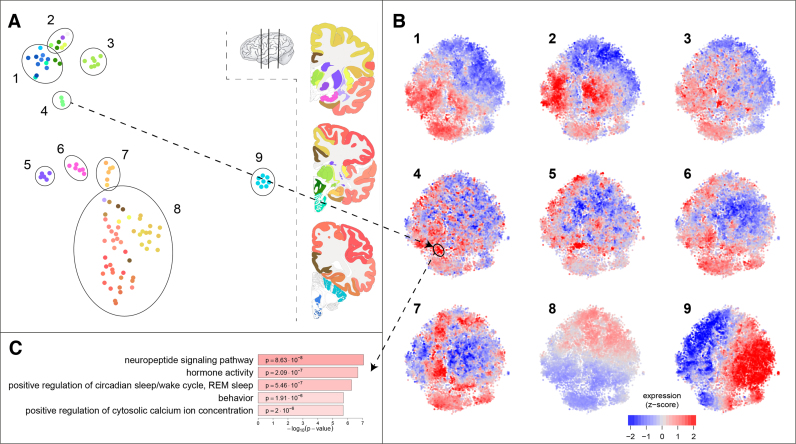
The dual explorer contains two maps: a sample map and a gene map. In the sample map, samples are close together if they are similar in their gene expression profiles. By coloring samples with anatomical annotation colors, this map shows both anatomical relationships between samples, and their transcriptional similarity. For the adult human data, Figure 2A shows that samples of close spatial proximity are more likely to have similar expressions, so the sample t-SNE map reflects the anatomy of the brain. Note that this map was produced using only the transcriptional profiles of the samples, not their locations. All samples of the cerebral cortex are co-located in the map (cluster 8), while sub-clusters can be recognized for example for the frontal lobe and the hippocampal formation (cluster 7). The six regions of the amygdala cluster together (cluster 6), as do the five striatum regions (cluster 5), the three hypothalamus regions (cluster 4), and the seven dorsal thalamus regions (cluster 3). The ventral thalamus, on the other hand, is more similar to the anatomically adjacent globus pallidus and midbrain. The samples of the cerebellar cortex form a distinct cluster in the map (cluster 9), whereas the cerebellar nuclei (represented by the dentate nucleus) are most similar to samples from medulla and pons, the structures that anatomically connect the cerebellum to the midbrain.
Figure 2.
Characterization of the gene and sample maps in the dual explorer. (A) The samples show a clustered pattern, matching the anatomical annotation shown in the color coding of the brain slices on the right. Nine groups of samples are highlighted in the map. (B) The mean expression of the nine groups of samples shown on the gene map. Each cluster of samples has its own distinct expression pattern, where red is high expression and blue is low expression. The dual explorer facilitates exploration of gene-sample relationships. The hypothalamus samples that are selected in A4 have a high expression in the gene cluster highlighted in B4. (C) The five strongest GO-term enrichments in the hypothalamus related gene cluster, which point to well-known hypothalamus functions.
The sample map reflects the similarities between samples, but the same transcriptome data can be used to infer gene-gene similarities. Figure 2B shows the transcriptional activity of 19 992 genes in the nine sets of samples that are selected in the sample map. The positions of the points (genes) in this t-SNE map capture brain-wide expression profiles, so the co-expression over the 105 selected regions. The colors of the points, on the other hand, show the activity of the genes in a selected subset of samples. For example, Figure 2B9 shows the average expression in cerebellar cortex samples, where we see a strong expression gradient from left-to right. These patterns of expression reflect how the gene map was made: genes with a similar expression across brain regions should be nearby in the t-SNE map.
The dual use of sample and gene maps (dual t-SNE) can give valuable insights. The differences between anatomical annotation and the sample map highlight the importance of exploring similarities in the characterization of brain regions. For example, the cerebellar nuclei samples (which are part of cluster 1 in Figure 2) are very different from those of the cerebellar cortex (all part of cluster 9). If one were to look at the average expression pattern of the full cerebellum, this would be a mixture of two distinct expression patterns (that of Figure 2B1 and 2B9). The interplay between gene and sample map allows for quick exploration of brain region specific expression. For example, one group of spatially co-expressed genes are highly expressed in the hypothalamus samples (of cluster 4). When these genes are analyzed for GO-term enrichment, we find they comprise several genes with hormone activity (Figure 2C). In addition to these specific analyses, one can also directly see large-scale patterns in the maps, such as the fact that few genes are highly expressed in both brain-stem and cerebral cortex (Figure 2B1 and 2B8).
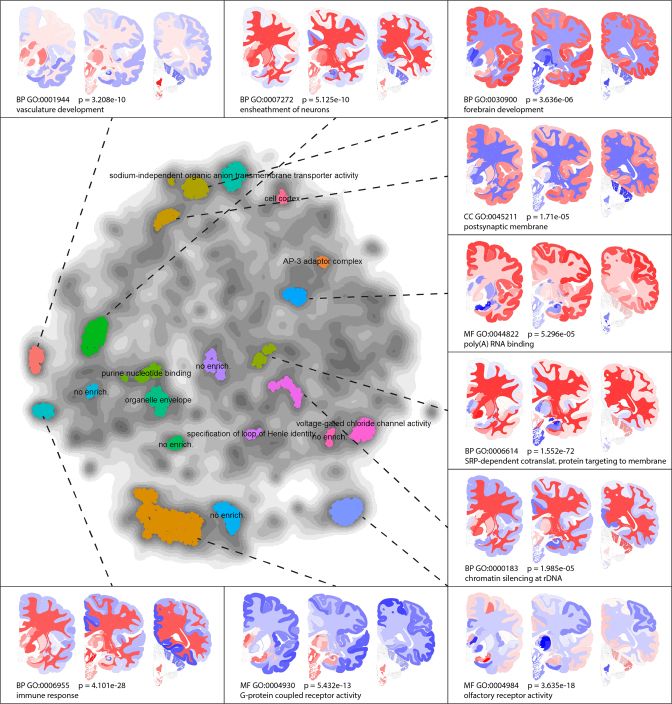
As we have seen in the hypothalamus example, similarities in gene expression may point to similarities in function. The gene map captures gene co-expression networks, which may consist of functionally related genes. To test this, we considered parts of the gene map with large numbers of genes, so with a high density. Figure 3 shows the gene ontology enrichments and spatial expression patterns of 3000 genes with the highest density values in the density map. Where Figure 2A shows similarities between neighboring brain regions, Figure 3 captures spatial co-expression networks. The results confirm the hypothesis that co-expression is related to shared functions, and it provides a global annotation of the gene map. The link between co-expression and function can also be used to characterize a gene by its neighbors in the gene map, as is illustrated for the APOE gene in the Supplementary Text and Supplementary Figure S1.
Figure 3.
Functional characterization of the gene map: 3000 genes within the highest densities were clustered, and clusters containing over 30 genes were characterized using ToppGene. Only the most significant GO-term is shown for each cluster, while the 10 clusters with strongest enrichments are provided with spatial expression choropleths. Most high-density areas in the gene map contain genes with common functions. All P-values are Bonferroni corrected in ToppGene, and the gene modules are provided in Supplementary Table S2.
Gene-expression reflects cell type composition
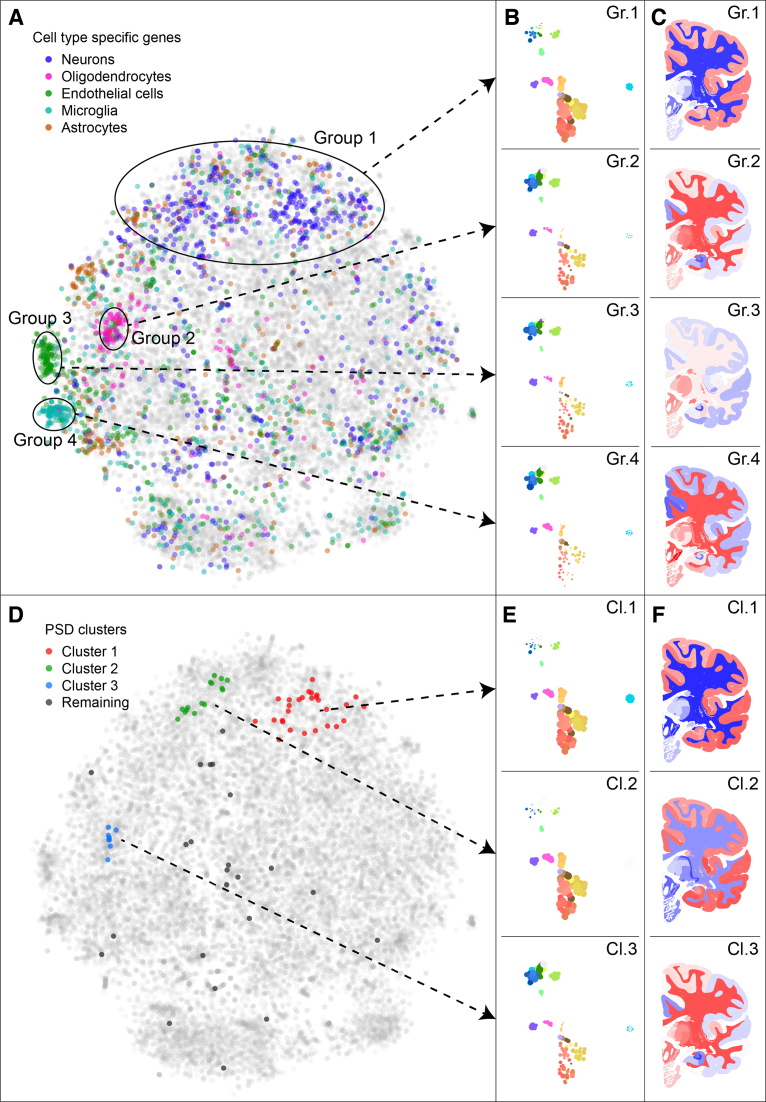
Gene expression measurements in the brain are partly determined by cell type composition of the samples (27). Therefore, the similarities between genes in their expression patterns may reflect cell type specific expression. As a result, cell type specific genes are likely to be co-located in the t-SNE map of the genes. To test this hypothesis, we obtained expression data from a cell-sort experiment of mouse cerebral cortex samples (24). We selected expression data of five major cell types present in the brain: astrocytes, endothelial cells, microglia, neurons and oligodendrocytes. Genes are labeled as cell type specific if they have at least a 20-fold expression in a specific cell type compared to the geometric mean of the other cell types.
The cell type specific genes (or cell type ‘markers’) are co-located in the gene t-SNE map. Figure 4A shows the location of these genes, where neuronal markers are found at the top of the map, which contains genes with high expression in cerebral cortex (Figure 4B1 and C1). The endothelial cell markers are also strongly co-expressed, with high average expression in thalamus, striatum and medulla (Figure 4B/C3). The microglia and oligodendrocyte markers form distinct clusters that share a high expression in the white matter (Figure 4B/C2 and 4B/C4). Microglia are known to be prevalent in the corpus callosum, which contains the ‘fountain of microglia’, from which these cells migrate to other parts of the brain (28). Compared to the microglia markers, the oligodendrocyte markers have a somewhat higher expression in cerebral cortex and thalamus, but lower in hippocampal formation and amygdala. Oligodendrocytes are responsible for myelination in the central nervous system, so they are prevalent in white matter of the brain. Combined, these results show that the maps in the BrainScope portal pick up the detailed patterns of cell type specific expression that partly underlie the transcriptome of the brain.
Figure 4.
Gene sets show cell type specificity of spatial gene expression and clusters of post synaptic density related genes. (A) The adult human gene t-SNE map, with highlighted cell type markers. The cell type markers were picked based on data from fluorescence-activated cell sorted brain cells from mouse (24), as genes with 20-fold expression in one of the types compared to the geometric mean in the other types. In the map, four groups of genes are highlighted, which correspond to areas of the map with high numbers of cell type markers. (B) The mean expression of all genes in the four groups in the gene map, shown by point sizes in the sample maps. (C) The mean expression of all genes in the same four groups, shown on a brain slice. (D) The adult human gene t-SNE map with highlighted post synaptic density (PSD) related genes (19). Three clusters of co-expressed PSD related genes are highlighted. (E and F) The expression patterns of the three clusters, shown in the sample t-SNE map and choropleths. Clusters 1 and 2 contain genes mostly expressed in the cerebral cortex, where cluster 2 is distinct from cluster 1 because of its stronger expression in cerebellar cortex. Genes in cluster 3 are expressed most strongly in subcortical regions such as thalamus, hypothalamus, and brain stem.
Dual explorer is an instrument for visual exploration of a set of genes of interest
The dual explorer captures robust patterns of spatial co-expression in the brain. This allows for characterization of sets of genes with respect to their shared expression, and therefore potentially shared brain specific functions. To illustrate this, we selected the 74 genes that were identified to have strong regional expression in the brain and presence in post synaptic density (PSD) (19), using data from a proteomic profiling of human neocortex (26). Post-synaptic densities connect neuronal cells and are essential to signal transmission in the brain. The 74 genes may all be specific to the PSD, but they do not all have identical spatial expression patterns in the brain. Figure 4D shows the PSD related genes in the gene map, where they can be separated into three clusters (and a remainder of unclustered genes). Cluster 1 contains 28 genes that are preferentially expressed in the cerebral cortex, and compared to all genes in the genome are enriched for the GO-term synapse part (GO:0044456, P = 4.52 × 10−23). Cluster 2 contains 15 genes that are similar in expression pattern, but have lower expression in the cerebellar cortex. They have the strongest GO-enrichment for synapse (GO:0045202, P = 8.82 × 10−8). The eight genes in cluster 3 have low expression in the cerebral cortex and cerebellum and high expression in subcortical regions, such as the thalamus and brainstem. This cluster is enriched for GO-term myelin sheath (GO:0043209, P = 6.94 × 10−9). In fact, 5 out of the eight genes share this annotation, which is surprising for genes that have been identified as being PSD related. Hence, the dual explorer gives a clear view of the clusteredness of this gene set of interest and the number of discernible clusters. In general, it allows for rapid interactive exploration of spatial expression patterns and gene function.
Comparative explorer shows expression stability across donors in the adult human brain
The Allen Brain Atlas contains gene expression measurements for six adult brains. We compared the gene t-SNE maps of these donors, using the comparative explorer. The explorer contains a consensus gene map, made by concatenating all samples before dimension reduction, and six per-donor gene maps that use all samples taken from a single donor. Details on data processing can be found in the Materials and Methods section. Figure 5 shows all seven gene maps side-by-side. To allow for visual comparison, the genes are labeled using gene modules that have been found to be consistently co-expressed in each of the six donor brains (23). These previously published modules were created by first assessing each gene for stability, defined as the correlation between the expression vectors for each pair of donor brains. The 50% of genes with the highest differential stability were then selected for an initial clustering of genes. Subsequently, weighted gene co-expression analysis (WGCNA) (29) was used to obtain 32 modules, which were characterized by module eigengenes. To obtain genome-wide gene modules, the remaining genes (with lower differential stability) were then linked to their most similar modules, defined on the correlation with the module eigengenes. Figure 5 shows that many of the previously reported WCGNA modules consistently form clusters in the consensus t-SNE map, as well as in the per-donor maps, pointing to the robustness of these maps. The t-SNE method, with only one main parameter, offers a visual representation of the data that is strongly in line with the results of the more parameter sensitive WGCNA algorithm. The relative positions of the modules in these maps vary to some extent, which is a result of the limited importance of large distances in t-SNE. In addition, the differences in brain region sampling may account for variability between donors.
Figure 5.
Gene t-SNE maps are robust and reproducible across donors. (A) The combined gene t-SNE map, showing previously reported stable gene modules (23). The map separates the 32 modules and shows their relationships. (B) The gene maps for each of the six donor brains. The maps are made using independent data sets, so they reflect the robustness of spatial gene expression patterns in the human brain. Data was pre-processed as in the original publication (23) to enable direct comparison of the WGCNA modules to the gene t-SNE maps.
Developmental comparative explorer captures spatio-temporal co-expression patterns
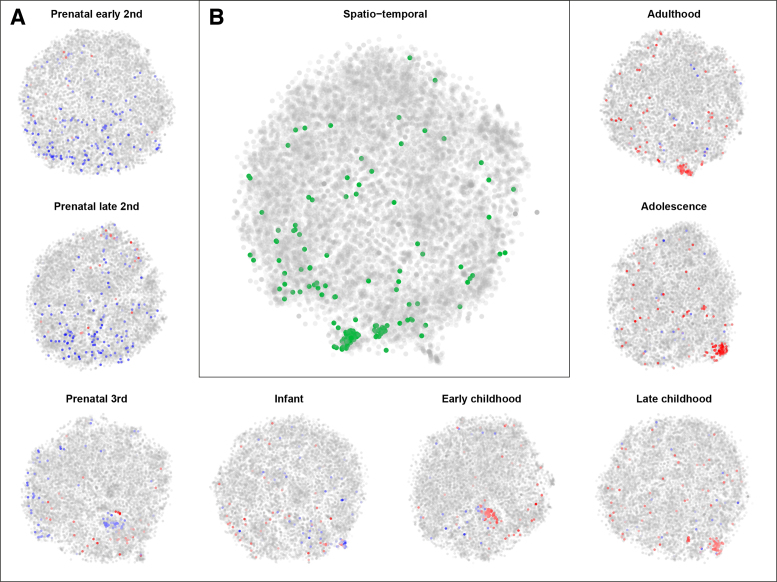
Thus far, we have only considered spatial gene expression patterns in the adult human brain. The Brainspan atlas of the developing human brain contains spatially and temporally resolved transcriptome data. To visualize this atlas, we developed the Brainspan comparative explorer (Figure 6). The Brainspan human developmental atlas contains gene expression data for 42 brains, ranging in donor age from 8 weeks post-conception to 40 years after birth. From each brain, up to 16 anatomical regions were sampled. We summarized the data to contain mean expression values for each of the 16 anatomical regions, for 8 developmental stages: early second trimester of pregnancy, late second trimester, third trimester, infancy, early childhood, late childhood, adolescence and adulthood.
Figure 6.
Developmental gene expression patterns show oligodendrocyte activity after birth. (A) The gene t-SNE maps per developmental stage. These maps reflect spatial co-expression of genes at each stage of development from early second trimester to adulthood. Oligodendrocyte marker genes are highlighted by their average expression across the brain at that stage of development. The oligodendrocyte marker genes have a low pre-natal expression (blue) and, as a result, a weak co-expression. After birth these genes become more active (red), and more co-expressed, which reflects the formation of white matter after birth. (B) The spatio-temporal gene t-SNE map of gene expression. Genes cluster together if they have a similar expression pattern through developmental time and anatomical space. The highlighted genes (green) are those that are oligodendrocyte specific.
This summarized data set is visualized with the comparative explorer in Figure 6B, i.e. genes are close together in the map if they behave similarly through time and anatomical regions simultaneously. It also shows gene maps for each developmental stage individually (Figure 6A), i.e. genes are close together in a map when they behave similarly across anatomical regions within that developmental stage. The comparative explorer gives insight in the transcriptional background of development. For example, Figure 6 shows that oligodendrocyte marker genes are spatio-temporally co-expressed, but before birth these genes are not co-expressed. In fact, these marker genes have a very low expression before birth, which reflects the fact that myelination is largely a post-natal process. The rise in expression of myelination related genes after birth has been observed before (30), and BrainScope's comparative explorer shows that this is also reflected in changes in co-expression over time.
DISCUSSION
We present the BrainScope portal for interactive visual analysis of gene expression in the brain. Through the use of linked t-SNE maps both global and local patterns in the data can be elucidated. Specific cell types give rise to expression patterns, which can be explored in both the sample and gene map using cell type marker genes. Users can upload their own gene sets of interest to find the spatial expression and co-expression patterns in the healthy human brain. The fact that neighboring genes in the gene map reflect co-expression, and therefore possible functional links, means that genes can be studied in their co-expressional context. In addition, the comparative explorer for the adult brain allows for the assessment of inter-donor stability of co-expression. The maps show transcriptomic robustness over donors, in a similar manner as the widely-used WGCNA algorithm. Finally, the developmental comparative explorer captures transcriptional patterns through development and age. Taken together, BrainScope gives an instant overview of similarities of all genes and of all brain regions.
The non-linearity and focus on local neighborhood structure of t-SNE make it well-suited for the visualization of similarities between samples and between genes in 2D plots. A practical advantage of t-SNE is that it has only one main parameter, the perplexity value, which controls the relative size of the neighborhood that is taken into account. It performs better in separating co-expression gene modules than PCA.
The gene and sample maps in BrainScope are based on all samples, and hence are affected by the anatomical distribution and sampling density. In addition, the portal is genome-wide. This means users are likely to find their brain regions and genes of interest represented in the portal. A filtering of genes could, however, give a stronger signal for specific applications, and a selected gene set may provide tailored sample-sample relationships. In addition, the current maps are affected by the strong difference between cerebellar cortex and cerebral samples. Therefore, an extension to the portal would be the option to recalculate the t-SNE maps on a subset of samples or genes, in an interactive manner (31). A user could select points based on prior knowledge or visual inspection of expression and update the maps. This would require more investment in server-side calculations.
Currently the portal contains only the gene expression data for the Allen Atlases of the adult human and developing human brain. However, the Allen Institute also provides spatial transcriptomic data for mouse (32), developing mouse (33), macaque (34), and developing macaque (35). In addition to these large scale datasets by the Allen Institute, spatially resolved epigenetic data of the brain is now available from the PsychENCODE project (3). The concepts of the BrainScope portal are applicable to these datasets as well. To illustrate this, we have applied the methodology to the spatial gene expression data of the UK Brain Expression Consortium (36) (see Supplementary Text and Supplementary Figure S3).
The amount of data available to molecular neuroscientists is rapidly growing. The availability of increasingly high-dimensional data, even on a single-cell level, calls for visualization tools that can offer both a birds-eye view of the full data, and an entry point to formulating specific questions. Consequently, BrainScope is a valuable tool for neurologists to gain a deeper understanding of the interactions between brain anatomy and molecular function.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Mike Hawrylycz of the Allen Institute for Brain Science.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Dutch Technology Foundation STW, as part of the STW project [12721: ‘Genes in Space’ and 12720: ‘VANPIRE’], under the IMAGENE STW Perspective program. Funding for open access charge: Nederlandse Organisatie voor Wetenschappelijk Onderzoek—Stichting voor de Technische Wetenschappen Project [12721].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N. et al. . The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. The ENCODE Project Consortium Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.a., Doyle F., Epstein C.B., Frietze S., Harrow J. et al. . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akbarian S., Liu C., Knowles J.A., Vaccarino F.M., Farnham P.J., Crawford G.E., Jaffe A.E., Pinto D., Dracheva S., Geschwind D.H. et al. . The PsychENCODE project. Nat. Neurosci. 2015; 18:1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neueder A., Bates G.P.. A common gene expression signature in Huntington's disease patient brain regions. BMC Med. Genomics. 2014; 7:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., Geschwind D.H.. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011; 474:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sunkin S.M., Ng L., Lau C., Dolbeare T., Gilbert T.L., Thompson C.L., Hawrylycz M.J., Dang C.. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013; 41:D996–D1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ng L., Bernard A., Lau C., Overly C.C., Dong H.W., Kuan C., Pathak S., Sunkin S.M., Dang C., Bohland J.W. et al. . An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 2009; 12:356–362. [DOI] [PubMed] [Google Scholar]
- 8. Hawrylycz M.J., Ng L., Page D., Morris J., Lau C., Faber S., Faber V., Sunkin S., Menon V., Lein E.S. et al. . Multi-scale correlation structure of gene expression in the brain. Neural Networks. 2011; 24:933–942. [DOI] [PubMed] [Google Scholar]
- 9. McCarthy M.J., Liang S., Spadoni A.D., Kelsoe J.R., Simmons A.N.. Whole brain expression of bipolar disorder associated genes: Structural and genetic analyses. PLoS ONE. 2014; 9:e100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tzeng J., Lu H.H., Li W.H.. Multidimensional scaling for large genomic data sets. BMC Bioinf. 2008; 9:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma S., Dai Y.. Principal component analysis based methods in bioinformatics studies. Brief. Bioinf. 2011; 12:714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maaten L.V.D., Hinton G.. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008; 9:2579–2605. [Google Scholar]
- 13. Ji S. Computational genetic neuroanatomy of the developing mouse brain: dimensionality reduction, visualization, and clustering. BMC Bioinf. 2013; 14:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahfouz A., Giessen M.V.D., Maaten L.V.D., Huisman S.M.H., Reinders M., Hawrylycz M.J., Lelieveldt B.P.F.. Visualizing the spatial gene expression organization in the brain through non-linear similarity embeddings. Methods. 2015; 73:79–89. [DOI] [PubMed] [Google Scholar]
- 15. Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. et al. . Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015; 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shekhar K., Brodin P., Davis M.M., Chakraborty A.K.. Automatic classification of cellular expression by nonlinear stochastic embedding (ACCENSE). Proc. Natl. Acad. Sci. U.S.A. 2014; 111:202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wong M.T., Chen J., Narayanan S., Lin W., Anicete R., Kiaang H.T.K., De Lafaille M.A.C., Poidinger M., Newell E.W.. Mapping the diversity of follicular helper T cells in human blood and tonsils using high-dimensional mass cytometry analysis. Cell Rep. 2015; 11:1822–1833. [DOI] [PubMed] [Google Scholar]
- 18. van Unen V., Li N., Molendijk I., Temurhan M., Höllt T., van der Meulen-de Jong A.E., Verspaget H.W., Mearin M.L., Mulder C.J., van Bergen J. et al. . Mass cytometry of the human mucosal immune system identifies tissue- and disease-associated immune subsets. Immunity. 2016; 44:1227–1239. [DOI] [PubMed] [Google Scholar]
- 19. Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.a., Ebbert A., Riley Z.L. et al. . An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012; 489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Ebbert A., Riley Z.L., Royall J.J., Aiona K. et al. . Transcriptional landscape of the prenatal human brain. Nature. 2014; 508:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A.. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013; 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J., Bardes E.E., Aronow B.J., Jegga A.G.. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009; 37:305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hawrylycz M.J., Miller J.A., Menon V., Feng D., Dolbeare T., Guillozet-Bongaarts A.L., Jegga A.G., Aronow B.J., Lee C.K., Bernard A. et al. . Canonical genetic signatures of the adult human brain. Nat. Neurosci. 2015; 18:1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y., Chen K., Sloan S.a., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. et al. . An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014; 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smedley D., Haider S., Durinck S., Pandini L., Provero P., Allen J., Arnaiz O., Awedh M.H., Baldock R., Barbiera G. et al. . The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015; 43:W589–W598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bayés A., van de Lagemaat L.N., Collins M.O., Croning M.D.R., Whittle I.R., Choudhary J.S., Grant S.G.N.. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 2011; 14:19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grange P., Bohland J.W., Okaty B.W., Sugino K., Bokil H., Nelson S.B., Ng L., Hawrylycz M.J., Mitra P.P.. Cell-type-based model explaining coexpression patterns of genes in the brain. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5397–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gehrmann J., Matsumoto Y., Kreutzberg G.W.. Microglia: Intrinsic immuneffector cell of the brain. Brain Res. Rev. 1995; 20:269–287. [DOI] [PubMed] [Google Scholar]
- 29. Zhang B., Horvath S.. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005; 4, doi:10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 30. Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M.M., Pletikos M., Meyer K.A., Sedmak G. et al. . Spatio-temporal transcriptome of the human brain. Nature. 2011; 478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pezzotti N., Lelieveldt B., van der Maaten L., Hollt T., Eisemann E., Vilanova A.. Approximated and user steerable tSNE for progressive visual analytics. IEEE Trans. Visual. Comput. Graphics. 2016; 2016, doi:10.1109/TVCG.2016.2570755. [DOI] [PubMed] [Google Scholar]
- 32. Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J. et al. . Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007; 445:168–176. [DOI] [PubMed] [Google Scholar]
- 33. Thompson C.L., Ng L., Menon V., Martinez S., Lee C.K., Glattfelder K., Sunkin S.M., Henry A., Lau C., Dang C. et al. . A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron. 2014; 83:309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bernard A., Lubbers L.S., Tanis K.Q., Luo R., Podtelezhnikov A.A., Finney E.M., McWhorter M.M., Serikawa K., Lemon T., Morgan R. et al. . Transcriptional architecture of the primate neocortex. Neuron. 2012; 73:1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bakken T.E., Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Dalley R.A., Royall J.J., Lemon T. et al. . Comprehensive transcriptional map of primate brain development. Nature. 2016; 535:367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramasamy A., Trabzuni D., Guelfi S., Varghese V., Smith C., Walker R., De T., Hardy J., Ryten M., Trabzuni D. et al. . Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 2014; 17:1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.