Abstract
In genomics, effectively identifying the biological effects of genetic variants is crucial. Current methods handle each variant independently, assuming that each variant acts in a context-free manner. However, variants within the same gene may interfere with each other, producing combinational (compound) rather than individual effects. In this work, we introduce COPE, a gene-centric variant annotation tool that integrates the entire sequential context in evaluating the functional effects of intra-genic variants. Applying COPE to the 1000 Genomes dataset, we identified numerous cases of multiple-variant compound effects that frequently led to false-positive and false-negative loss-of-function calls by conventional variant-centric tools. Specifically, 64 disease-causing mutations were identified to be rescued in a specific genomic context, thus potentially contributing to the buffering effects for highly penetrant deleterious mutations. COPE is freely available for academic use at http://cope.cbi.pku.edu.cn.
INTRODUCTION
Tremendous advances in high-throughput sequencing technologies have enabled several large-scale human genome sequencing projects, such as the 1000 Genomes Project (1) and the Personal Genome Project (2), to identify millions of genetic variants in thousands of individual genomes. Consequently, there is a great demand for effectively interpreting these variants (3–5). The majority of current variant annotation tools adopt a variant-centric approach that assesses the functional consequence of each variant independently by assuming that each variant acts in a context-free manner (6–8). Several reports have shown that such an assumption results in both false-positive and false-negative calls (9–11) when multiple variants affect the same gene.
A few recently released tools have applied additional filters to identify (and try to correct) the common false-positive calls caused by multiple Single Nucleotide Variants (SNVs) in the same codon (12), multiple indels in the same gene (11) and in-frame alternative acceptor sites (13). However, none of these tools integrate the entire sequential context in addition to their own particular configurations. For an accurate annotation algorithm, the entire sequential context within a gene should be considered together, because multiple variants in the same gene may interfere with each other, thus producing combinational rather than individual effects (e.g., the complementary rescue effect (9)). Here, we present a fully gene-centric variant annotation tool, COPE (Context-Oriented Predictor for variant Effect), for evaluating the effects of variants in a context-sensitive approach. Using each transcript as the basic annotation unit, COPE infers the ‘mutant peptide’ from the entire variant set input and reports the final amino acid alteration through comparison against the reference sequence. Incorporating the whole sequence context enables COPE to accurately annotate complex compound effects of multiple genetic variants like alternative isoforms caused by gain/loss of splicing sites, which are cannot be handled by previous tools (11–13) (Figure 1B, also see Supplementary Tables S1 and S5). The web server and source code of COPE are freely available for academic use at http://cope.cbi.pku.edu.cn/.
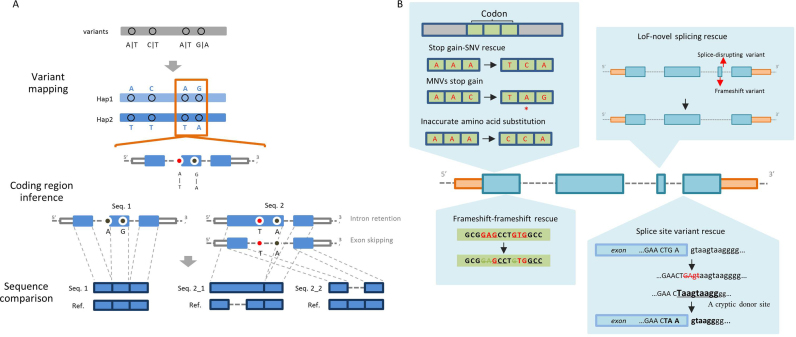
Figure 1.
(A) Overview of COPE. COPE uses each transcript as a basic annotation unit. The variant mapping step identifies variants within transcripts. The coding region inference step removes introns from each transcript; all possible splicing patterns are taken into consideration for splice-altering transcripts (in this case, the red dot indicates a splice acceptor site SNP, and intron retention and exon skipping are considered). The sequence comparison step compares a ‘mutant peptide’ against a reference protein sequence to obtain the final amino acid alteration. (B) Schematic diagram of typical types of annotation corrections implemented in COPE. A rescued stop-gained SNV indicates that another SNV (‘A’ to ‘C’) in the same codon rescues a variant-centric stop-gained SNV (‘A’ to ‘T’). Stop-gained MNV indicates that two or more SNVs result in a stop codon (‘A’ to ‘T’ and ‘C’ to ‘G’). A rescued frameshift indel indicates that another indel in the same haplotype recovers the original open reading frame. A splicing-rescued stop-gained/frameshift variant indicates that a stop-gained or frameshift variant is rescued by a novel splicing isoform. A rescued splice-disrupting variant indicates that a splice-disrupting variant is rescued by a nearby cryptic site (as shown in the figure) or a novel splice site. The asterisk in the figure indicates a stop codon.
MATERIALS AND METHODS
Overview of COPE
COPE is a framework for predicting the effects of variants through a context-sensitive, gene-centric approach (Figure 1A). Firstly, genetic variants are mapped to protein-coding genes derived from a user-supplied reference gene model such as RefSeq (14). Using the phase information, COPE handles two haplotypes separately. Then, COPE tries to reconstruct the ‘mutant peptide’ from the entire inputted variant set. Briefly, COPE attempts to identify splicing-changing variants (i.e. variants that disrupt existing splice sites or create novel splice sites), and, if a splicing-changing variant is found, new isoforms are inferred accordingly (Supplemental Figure S1). Finally, COPE translates all coding sequences into amino acid sequences and compares them against the reference sequence to obtain the final amino acid alterations.
Transcript inference
The accurate inference of a ‘mutant’ transcript is the most important step in the COPE pipeline. We used MaxEntScan (15), a commonly used splice sequence scoring tool, to identify splice site gain/loss events. Inspired by the results of Jian et al. (16), we used relative score variation to measure the scale of change caused by the variant and adopted the cut-off recommended in their paper (16).
We evaluated the performance of isoform inference in COPE by following the protocol from Jian et al. (16). Briefly, for splice site gain events, the positive set was derived from a publication by Stein et al. (17); the negative dataset was constructed from the 1000 Genomes Project Phase 3 genotype data through the following steps (16): (i) we kept only intronic variants within protein-coding genes of the Ensembl gene model that could be downloaded from ftp://ftp.ensembl.org/pub/release-83/gtf/homo_sapiens/Homo_sapiens.GRCh38.83.gtf.gz; (ii) we discarded variants within multi-transcript genes; (iii) we discarded variants within single-exon genes and (iv) we discarded variants with an allele frequency less than or equal to 0.1. For splice loss events, the positive set was derived from the publication by Jung et al. (18), and the negative dataset was derived from Jian et al. (16). The results showed that COPE is accurate for isoform inference, with a high sensitivity (85.4% for splice gain events and 94.7% for splice loss events) and a high specificity (89.8% for splice gain events and 92.9% for splice loss events) (Supplementary Figure S8).
Application of COPE
To provide a proof of concept, we applied COPE to the genotype data of a male Caucasian (NA12144), downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/.
We further applied COPE to a list of 1147 curated high-confidence LoF variants reported by MacArthur et al. (9). Because there was no phase information in the dataset reported in that paper, we used the phased dataset downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/. Ninety-three curated LoF variants were no longer existent in the current phased dataset (released in 2 May 2013) and were excluded from our analysis. Additionally, to exclude the effects resulting from dataset updates, we further assessed the context of 53 rescued (i.e. the putative damaging effect is neutralized by the specific genomic context) loss-of-function variant candidates in the original MacArthur et al. dataset (released in July 2010, downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/pilot_data/release/2010_07/) and discarded three variants with different sequential contexts (Supplementary Figure S11).
We then extended the analysis to the full 1000 Genomes Project Phase 3 SNVs and indels variant set (downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/). Inconsistent variant calls (SNVs/indels overlapping with indels) were removed. Then, the SNVs and indels were annotated with VEP by using the RefSeq gene model. Following previous studies (9,13), we removed putative LoF variants that result in less than 10% protein sequence alteration. Finally, 5559 splice-disrupting, 2092 frameshift and 9728 stop-gained variants were fed into COPE for reanalysis.
Validation of the rescued false-positive splice-disrupting variants
To validate the results on rescued false-positive splice-disrupting variants, we used RNA-seq data to confirm the novel splice junctions predicted by COPE. RNA-seq data on 445 individuals in the 1000 Genomes Project were obtained from the Geuvadis RNA Sequencing Project (19) (downloaded from EBI ArrayExpress, accession E-GEUV-1). For each rescued false-positive splice-disrupting variant, we extracted the inferred genomic sequence (with intron retained) of each transcript from each individual and then mapped the RNA-Seq reads to the inferred genomic sequence by HISAT2 (20). We used junction reads to validate the rescued false-positive splice-disrupting variants. Briefly, a given junction is called as ‘expressed’ if and only if ≥1 RNA-Seq reads are found to be spanning the junction. A transcript is considered as ‘expressed’ in a particular sample when all its reference junctions (i.e. junctions annotated in the reference gene model) are expressed in the given sample. And a putative novel junction identified by COPE in an expressed transcript will be classified as ‘True Positive’ if and only if the junction itself is called as ‘expressed’ too (Supplementary Figure S9).
Search for rescued pathogenic LoF variants in healthy individuals
We compiled a list of pathogenic LoF variants by merging 60,556 LoF variants tagged as ‘DM’ from the HGMD (21) database and 11,777 annotated with the label ‘(likely) pathogenic mutations’ from the ClinVar (22) database. In addition, we downloaded a total of 9,362 disease-associated genes from DisGeNET (23). Then, we searched for pathogenic variants that were rescued in at least one healthy individual from the 1000 Genomes Project and identified 64 pathogenic LoF variants, including 21 from the HGMD database and 43 from DisGeNET (Supplementary Table S4).
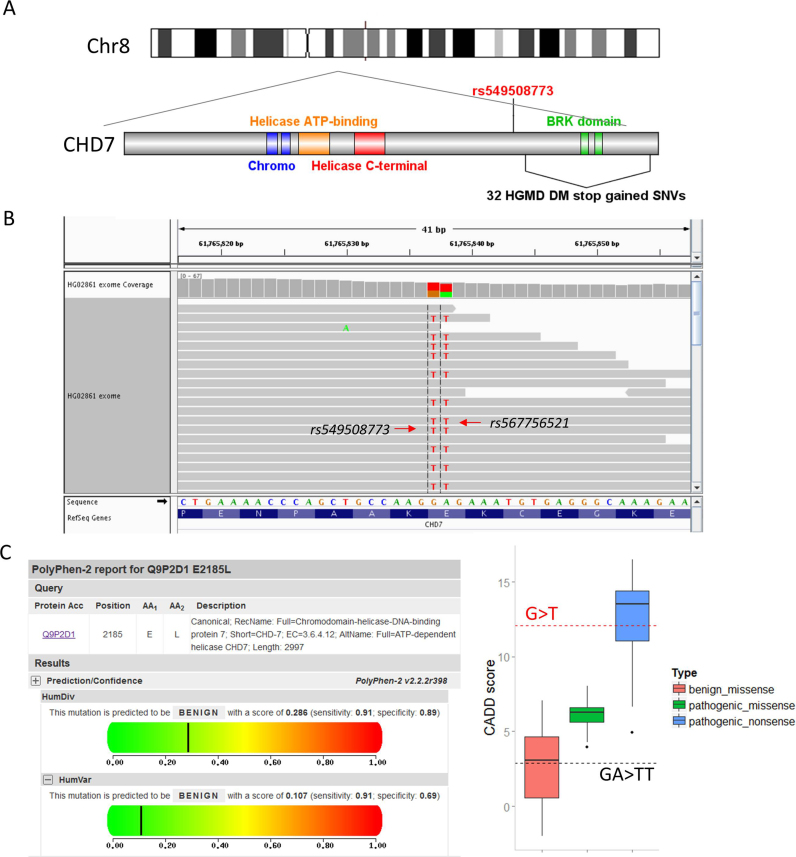
The variant rs549508773 is a stop-gained SNV within the gene CHD7, a driver gene of CHARGE syndrome. To demonstrate the disease-causing effect of SNP rs549508773, 175 disease-causing (CHARGE syndrome) stop-gained SNVs were collected from the HGMD database. In addition, we also collected CADD scores (24) of 43 pathogenic missense variants, 156 pathogenic stop-gained variants and 26 benign missense variants from the CHD7 database (25) to demonstrate the benign effect of the single amino acid substitution resulting from SNP rs549508773 together with SNP rs567756521.
RESULTS
COPE handles complex compound effects of multiple variants correctly
As a gene-centric annotation tool, COPE is able to handle complex compound effects of multiple variants correctly (Figure 1B, also see Supplementary Table S1). For proof-of-concept analysis, we applied COPE to the male Caucasian sample NA12144 from the 1000 Genomes Project. We compared the COPE results with the official variant annotation generated by Variant Effect Predictor (VEP) (6), a commonly used variant-centric annotation tool. COPE corrected two false-positive stop-gained calls, five false-positive frameshift calls, eight false-positive splice-disrupting calls and one false-negative stop-gained call (Supplementary Table S2). For example, the VEP called two indels (rs67712719 and rs67322929) in ZFPM1 ‘frameshift variants’ and suggested that both of them lead to a loss-of-function (LoF) event. In contrast, COPE correctly identified the combinational effect of these two variants as one amino acid deletion (Supplementary Figure S2). Similarly, COPE also accurately identified a cryptic splicing site 3 bp downstream of the VEP-reported splice acceptor variant rs1152522 within the C14orf105 gene, which can rescue the splicing at the cost of a single amino acid (glutamine) deletion; this finding was validated by both corresponding RNA-Seq data (Supplementary Figure S3) and an independent report (26).
To further assess the performance of COPE, we reanalyzed the manually curated high-confidence LoF variants listed in 1000 Genomes (9). After exclusion of 93 nonexistent variants in the current phased release, COPE identified 4.74% curated LoF variants (consisting of one stop-gained and 49 splice-disrupting variants) as potential false-positive calls in at least one sample from 1000 Genomes (Supplementary Figure S11 and Table S3). Further inspection showed that the stop-gained variant was rescued by another SNV in the same codon (Supplementary Figure S4) that was previously incorrectly handled, and all 49 variants were able to be correctly annotated only when the entire sequential context was considered, thus demonstrating the necessity of COPE even after manual variant reannotation.
Application to genotype data from the 1000 Genomes Project
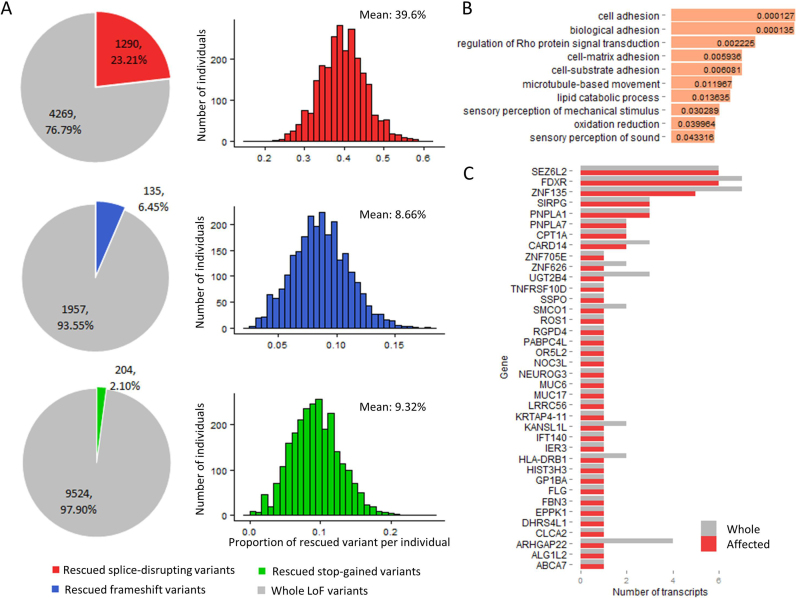
We then extended the analysis to the full 1000 Genomes Project Phase 3 SNVs set. All LoF variants reported by VEP, including splice-disrupting variants (in either donor or acceptor site), frameshift indels, and stop-gained SNVs, were extracted and reanalyzed by COPE. Unexpectedly, we found that a total of 1290 (23.21%) reported splice-disrupting variants were rescued in their particular sequential context and 1251 (97.0%) were rescued by in-frame cryptic splice sites within 100 bp. An average of 39.6% VEP-annotated splice-disrupting variants in each individual were rescued (Figure 2A). On the basis of the Geuvadis RNA-Seq data (19), we validated 78 (79.6%) out of 98 rescued false-positive splice-disrupting variants supported by 129 expressed transcripts. Additionally, COPE also identified 6.45% (135 out of 2092, Figure 2A) reported frameshift indels and 2.10% (204 out of 9728, Figure 2A) reported stop-gained SNVs as false-positive calls. Statistical analysis showed that 1398 genes containing these false-positive LoF calls were likely to be involved in several particular biological processes, including adhesion and signal transduction, thus suggesting a systematic bias in the variant-centric function calling tool (Figure 2B, even such bias did not significantly change the global functional spectrum of LoF genes (Supplementary Figure S12)). Notably, by incorporating the entire sequential context, COPE was also able to identify false-negative LoF variants, such as stop-gained MNV (i.e. multiple co-occurring SNVs in the same codon that jointly introduce a new STOP codon), which are usually neglected by variant-centric tools. We identified 38 stop-gained MNVs in 38 genes, including TNFRSF10D, encoding a member of the TNF-receptor superfamily, and NEUROG3, encoding a transcription factor involved in neurogenesis (Figure 2C). Unexpectedly, we found that 78% (1960 of 2504) of the individuals sequenced by the 1000 Genomes Project had at least one of the 38 identified stop-gained MNV-harboring genes (Supplementary Figure S5). In particular, the stop-gained MNV-harboring zinc finger protein ZNF705E (Supplementary Figure S6) was found in 64.5% (1616) of individuals.
Figure 2.
LoF variants in the 1000 Genomes Project rescued in a specific genomic context. (A) The number of rescued LoF variants. The pie charts show the proportion of rescued LoF variants, and the histograms show the proportion of rescued LoF variants in each individual. The ‘mean’ labels in the histograms indicate the average number. (B) Enrichment analysis of the rescued LoF transcripts. The numbers represent the corrected P-values. (C) The 38 genes affected by stop-gained MNVs. The red bar represents the number of transcripts affected by each stop-gained MNV, and the gray bar represents the total number of transcripts of the gene.
Pathogenic variants rescued by specific genomic context
We also found 64 rescued disease-causing mutations in the list (Supplementary Table S4). One highly intriguing case was the rescued stop-gained SNV rs549508773 within the CHD7, a confirmed disease-causing gene for CHARGE syndrome (OMIM 214800), a severe childhood autosomal dominant disease with a recognizable appearance of birth defects (27–31). The variant rs549508773 itself results in a nonsense mutation and, along with 32 downstream HGMD-reported stop-gained SNVs (Figure 3A), was identified as deleterious by variant-centric tools. However, in the individual HG02861 harboring this SNV, a co-occurring SNV rs567756521 was found in the same codon (Figure 3B), thus leading to a mild single amino acid substitution together with rs549508773 that was predicted to be neutral by both PolyPhen-2 (32) and CADD (24) (Figure 3C), thus suggesting a plausible mechanism for the buffering effects of highly penetrant, deleterious mutations (31) (also see another case in Supplementary Figure S7).
Figure 3.
A pathogenic SNV is rescued in its specific genomic context. (A) Compared with SNV rs549508773 (red), 32 disease-causing (DM) stop-gained SNVs (black) recorded in the HGMD database are located downstream of the gene CHD7. (B) The figure shows an IGV image of the whole exome sequencing data of HG02861, a healthy participant in the 1000 Genomes Project. As shown in the figure, SNV rs567756521 rescues the stop-gained mutation, and the combined effect is a single amino acid substitution (Glu>Leu). (C) Polyphen-2 predicted the single amino acid (Glu>Leu) as a benign mutation (Left). Boxplot of CADD scores for three different kinds of mutations (benign missense, pathogenic missense and pathogenic nonsense) collected from the CHD7 database (Right). The score for rs549508773 (G>T) is located within the range of pathogenic nonsense (red dotted line). The score for the MNV (GA>TT) is located within the range of benign missense (black dotted line).
Online web server and standalone package
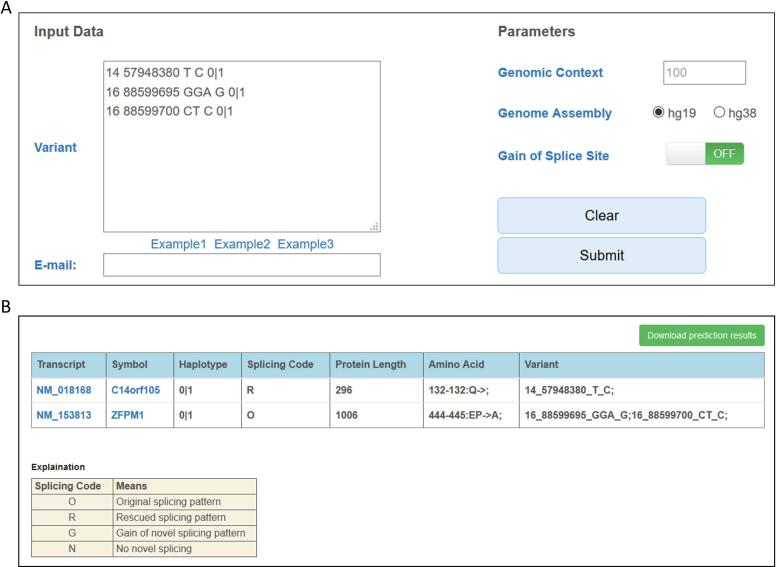
A web server is available at http://cope.cbi.pku.edu.cn/whole_PCG_Analysis.html for users to try COPE online (Figure 4). The input is a space-delimited file with five columns per line: chromosome, position, reference allele, alternative allele and haplotype information. The output on the website includes seven columns: transcript, symbol, splicing code, protein length, amino acid (including all amino acid alterations), and variant (including all variants in the transcript).
Figure 4.
Screenshot of the COPE web server. (A) An example of input. (B) Annotation by COPE.
For large-scale analysis, we also provide a standalone package, which can be downloaded freely for academic use. A detailed guideline for installation and setup is also available at http://cope.cbi.pku.edu.cn/PCG_manual.html.
DISCUSSION
During recent years, annotating each variant independently has been taken for granted and variant-centric annotation algorithms have widely been used in the downstream analysis of genome sequencing. The challenges of a variant-centric algorithm have been discussed previously (9). COPE aims to avoid the annotation errors caused by variant-centric methods by considering the genomic sequential context. COPE was designed as an isoform-oriented annotator, and all isoforms of a gene are analyzed simultaneously. By analyzing the genotype data from the 1000 Genomes Project, we demonstrated that COPE is able to correct numerous annotation errors, including both false-positive and false-negative LoF calls. To the best of our knowledge, COPE is the first fully gene-centric tool for annotating the effects of variants in a context-sensitive approach. Detailed comparison based on both typical sample and whole 1000 Genome dataset shows COPE's gene-centric strategy significantly improves the accuracy of variant annotation (Supplementary Tables S6 and S7).
Phase information is important for accurate annotation. Several algorithms have been proposed for inferring the haplotype from un-phased sequencing data (33,34). COPE makes full use of phasing information for accurately annotating the variant effect. We have made a script available for inferring the phase directly from short-read sequencing data (http://cope.cbi.pku.edu.cn/phase.html) when such information is not available. Our evaluation suggested that our pipeline achieves a rather high (>90%) haplotype recovery rate (i.e, the proportion of completely phased transcripts over transcripts with multiple variants) for un-phased data with reasonable coverage (>30x), and the rate kept increasing with higher coverage (Supplementary Figure S10). We also note that rapid development of the sequencing technology is effectively enabling haplotype resolved experimentally (35–37).
COPE accesses functional effects of genetic variants by comparing the inferred ‘mutant’ transcript with the ‘wild-type’ one based on user-specified reference gene models. The quality and completeness of reference gene models is critical to the accuracy of COPE annotation. Thus, while COPE is designed to be species-neutral, its performance may suffer when being applied to less-annotated genomes (e.g. genomes of non-model organism).
Epistasis is a phenomenon in which the functional influence of a variant at a genetic locus is affected by another variant at another locus (38). It leverages the SNP–SNP interaction between different genes to explain the lack of heritability of numerous types of complex human disease (39,40). COPE is a variant effect annotator that considers the compound effects of multiple variants within the same gene, in contrast to epistasis, to annotate their effects accurately.
Protein-coding genes are not the only player in the complex biological network; the current framework could also be readily extended to and adapted for other functional molecules other than protein-coding genes, such as miRNAs and long noncoding RNAs, when more functional and mechanistic information becomes available.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Liping Wei, Cheng Li, Zemin Zhang and Jian Lu at Peking University for their helpful comments and suggestions during the study.
Footnotes
Present address: Huan Liu, Novo Nordisk China Pharmaceuticals Co. Ltd, 1 W Dawang Rd, Chaoyang, Beijing 100026, People's Republic of China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program [2016YFC0901603]; China 863 Program [2015AA020108]; International Collaboration Program for Proteome Biological Big Data and Standard System [2014DFB30030]; State Key Laboratory of Protein and Plant Gene Research. The research of G.G. was supported in part by the National Program for Support of Top-notch Young Professionals. Part of the analysis was performed on the Computing Platform of the Center for Life Sciences of Peking University. Funding for open access charge: State Key Laboratory of Protein and Plant Gene Research, China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Genomes Project, C. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A. et al. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Church G.M. The personal genome project. Mol. Syst. Biol. 2005; 1, doi:10.1038/msb4100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shameer K., Tripathi L.P., Kalari K.R., Dudley J.T., Sowdhamini R.. Interpreting functional effects of coding variants: challenges in proteome-scale prediction, annotation and assessment. Brief. Bioinformatics. 2016; 17:841–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015; 17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ward L.D., Kellis M.. Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol. 2012; 30:1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F.. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010; 26:2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang K., Li M., Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramos A.H., Lichtenstein L., Gupta M., Lawrence M.S., Pugh T.J., Saksena G., Meyerson M., Getz G.. Oncotator: cancer variant annotation tool. Hum. Mutat. 2015; 36:E2423–E2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B. et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012; 335:823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arndt S., Hobbs A., Sinclaire I., Lane A.B.. Chitotriosidase deficiency: a mutation update in an african population. JIMD Rep. 2013; 10:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vergara I.A., Frech C., Chen N.. CooVar: co-occurring variant analyzer. BMC Res. Notes. 2012; 5:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wei L., Liu L.T., Conroy J.R., Hu Q., Conroy J.M., Morrison C.D., Johnson C.S., Wang J., Liu S.. MAC: identifying and correcting annotation for multi-nucleotide variations. BMC Genomics. 2015; 16:569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Narasimhan V.M., Hunt K.A., Mason D., Baker C.L., Karczewski K.J., Barnes M.R., Barnett A.H., Bates C., Bellary S., Bockett N.A. et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016; 352:474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yeo G., Burge C.B.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004; 11:377–394. [DOI] [PubMed] [Google Scholar]
- 16. Jian X., Boerwinkle E., Liu X.. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014; 42:13534–13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stein S., Lu Z.X., Bahrami-Samani E., Park J.W., Xing Y.. Discover hidden splicing variations by mapping personal transcriptomes to personal genomes. Nucleic Acids Res. 2015; 43:10612–10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jung H., Lee D., Lee J., Park D., Kim Y.J., Park W.Y., Hong D., Park P.J., Lee E.. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 2015; 47:1242–1248. [DOI] [PubMed] [Google Scholar]
- 19. Lappalainen T., Sammeth M., Friedlander M.R., t Hoen P.A., Monlong J., Rivas M.A., Gonzalez-Porta M., Kurbatova N., Griebel T., Ferreira P.G. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013; 501:506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cooper D.N., Stenson P.D., Chuzhanova N.A.. The Human Gene Mutation Database (HGMD) and its exploitation in the study of mutational mechanisms. Curr. Protoc. Bioinformatics. 2006; doi:10.1002/0471250953.bi0113s12. [DOI] [PubMed] [Google Scholar]
- 22. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pinero J., Queralt-Rosinach N., Bravo A., Deu-Pons J., Bauer-Mehren A., Baron M., Sanz F., Furlong L.I.. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database. 2015; bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van den Akker P.C., Jonkman M.F., Rengaw T., Bruckner-Tuderman L., Has C., Bauer J.W., Klausegger A., Zambruno G., Castiglia D., Mellerio J.E. et al. The international dystrophic epidermolysis bullosa patient registry: an online database of dystrophic epidermolysis bullosa patients and their COL7A1 mutations. Hum. Mutat. 2011; 32:1100–1107. [DOI] [PubMed] [Google Scholar]
- 26. Hiller M., Huse K., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M.. Single-nucleotide polymorphisms in NAGNAG acceptors are highly predictive for variations of alternative splicing. Am. J. Hum. Genet. 2006; 78:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C. et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004; 36:955–957. [DOI] [PubMed] [Google Scholar]
- 28. Lalani S.R., Safiullah A.M., Fernbach S.D., Harutyunyan K.G., Thaller C., Peterson L.E., McPherson J.D., Gibbs R.A., White L.D., Hefner M. et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am. J. Hum. Genet. 2006; 78:303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Janssen N., Bergman J.E., Swertz M.A., Tranebjaerg L., Lodahl M., Schoots J., Hofstra R.M., van Ravenswaaij-Arts C.M., Hoefsloot L.H.. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum. Mutat. 2012; 33:1149–1160. [DOI] [PubMed] [Google Scholar]
- 30. Zentner G.E., Layman W.S., Martin D.M., Scacheri P.C.. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am. J. Med. Genet. Part A. 2010; 152:674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen R., Shi L., Hakenberg J., Naughton B., Sklar P., Zhang J., Zhou H., Tian L., Prakash O., Lemire M. et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 2016; 34:531–538. [DOI] [PubMed] [Google Scholar]
- 32. Adzhubei I., Jordan D.M., Sunyaev S.R.. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013; doi:10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bansal V., Bafna V.. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics. 2008; 24:I153–I159. [DOI] [PubMed] [Google Scholar]
- 34. Delaneau O., Marchini J., Zagury J.F.. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2012; 9:179–181. [DOI] [PubMed] [Google Scholar]
- 35. Schadt E.E., Turner S., Kasarskis A.. A window into third-generation sequencing. Hum. Mol. Genet. 2010; 19:R227–R240. [DOI] [PubMed] [Google Scholar]
- 36. Snyder M.W., Adey A., Kitzman J.O., Shendure J.. Haplotype-resolved genome sequencing: experimental methods and applications. Nat. Rev. Genet. 2015; 16:344–358. [DOI] [PubMed] [Google Scholar]
- 37. Kitzman J.O., Mackenzie A.P., Adey A., Hiatt J.B., Patwardhan R.P., Sudmant P.H., Ng S.B., Alkan C., Qiu R., Eichler E.E. et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat. Biotechnol. 2011; 29:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cordell H.J. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002; 11:2463–2468. [DOI] [PubMed] [Google Scholar]
- 39. Murk W., Bracken M.B., DeWan A.T.. Confronting the missing epistasis problem: on the reproducibility of gene-gene interactions. Hum. Genet. 2015; 134:837–849. [DOI] [PubMed] [Google Scholar]
- 40. Hemani G., Knott S., Haley C.. An evolutionary perspective on epistasis and the missing heritability. Plos Genet. 2013; 9:e1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.