Abstract
Threose nucleic acid (TNA) is an artificial genetic polymer capable of undergoing Darwinian evolution to produce aptamers with affinity to specific targets. This property, coupled with a backbone structure that is refractory to nuclease digestion, makes TNA an attractive biopolymer system for diagnostic and therapeutic applications. Expanding the chemical diversity of TNA beyond the natural bases would enable the development of functional TNA molecules with enhanced physiochemical properties. Here, we describe the synthesis and polymerase activity of a fluorescent cytidine TNA triphosphate analogue (1,3-diaza-2-oxo-phenothiazine, tCfTP) that maintains Watson-Crick base pairing with guanine. Polymerase-mediated primer-extension assays reveal that tCfTP is efficiently added to the growing end of a TNA primer. Detailed kinetic assays indicate that tCfTP and tCTP have comparable rates for the first nucleotide incorporation step (kobs1). However, addition of the second nucleotide (kobs2) is 700-fold faster for tCfTP than tCTP due the increased effects of base stacking. Last, we found that TNA replication using tCfTP in place of tCTP exhibits 98.4% overall fidelity for the combined process of TNA transcription and reverse transcription. Together, these results expand the chemical diversity of enzymatically generated TNA molecules to include a hydrophobic base analogue with strong fluorescent properties that is compatible with in vitro selection.
INTRODUCTION
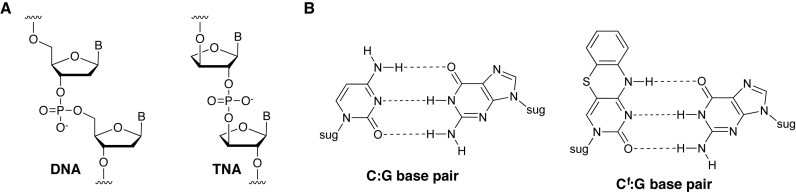
We are interested in advancing the development of threose nucleic acid (TNA) as an artificial genetic polymer for synthetic genetics (1). TNA is a particular type of XNA that has a backbone structure composed of repeating units of α-l-threose sugars that are vicinally linked by 2΄,3΄-phosphodiester bonds (Figure 1A) (2). Despite a backbone repeat unit that is one atom (or bond) shorter than that of DNA or RNA, TNA is capable of forming stable antiparallel Watson–Crick duplex structures with itself and with complementary strands of DNA and RNA (3–5). Solution NMR studies reveal that duplex formation in either the self-pairing mode (TNA/TNA) or cross-pairing modes (TNA/DNA or TNA/RNA) occurs through an A-like helical geometry that is templated by a rigid TNA backbone (5,6). More recently, stability assays performed under harsh biological conditions demonstrate that TNA is refractory to nuclease digestion, which makes TNA a strong candidate for diagnostic and therapeutic applications that require high biological stability (7).
Figure 1.
Molecular structures. (A) Constitutional structure for the linearized backbone of DNA (left) and α-l-threofuranosyl-(3΄-2΄) nucleic acid TNA (right). (B) Watson–Crick base pairs for C:G and Cf:G. Cf is the cytosine analogue, 1,3-diaza-2-oxo-phenothiazine.
The process of developing a polymerase-mediated replication system began with an effort to identify natural or engineered polymerases that could recognize TNA, either as a substrate in the template or as a nucleotide triphosphate. From these studies, several DNA polymerases were identified that could synthesize short segments of DNA on a TNA template and other polymerases that could copy limited stretches of TNA on a DNA template (8–11). Subsequent screening of Archaeal B-family thermophilic DNA polymerases led to the discovery of Therminator DNA polymerase as an enzyme that could copy DNA templates (∼50 nts) into full-length TNA products (12). Using a DNA display strategy that linked newly synthesized TNA strands to their encoding DNA templates (13), a TNA aptamer was evolved to bind human α-thrombin (14). The ability for TNA to fold into a structure with high ligand binding affinity (14), along with similar studies performed on hexitol nucleic acid (15), provided the first clear demonstration that the chemical problem of ligand binding was not unique to the natural ribofuranosyl polymers of DNA and RNA.
More recently, a microfluidics strategy called droplet-based optical polymerase sorting (DrOPS) was developed that allows engineered polymerases to be evolved in uniform water-in-oil microcompartments (16). This technique, coupled with the protein design strategy of scaffold sampling, led to the development of a new TNA polymerase called Kod-RI, which is an engineered version of a naturally occurring DNA polymerase isolated from the Archaean species Thermococcus kodakarensis that carries the TNA synthesis mutations A485R and E664I and the 3΄,5΄-exonuclease silencing mutations D141A and E143A (17). Kod-RI exhibits 5-fold faster primer-extension efficiency (3 hours versus 15 hours, respectively) and ∼20-fold higher fidelity (four errors per 1000 nucleotide incorporations versus 70 errors, respectively) as compared to Therminator DNA polymerase.
Although Kod-RI represents a substantial improvement over Therminator DNA polymerase, the enzyme stalls when it encounters sequential G-nucleotides in the DNA template. This problem is not unique to engineered polymerases, as many naturally occurring DNA polymerases are known to process G-rich sequences with reduced efficiency (18). However, relative to DNA synthesis, the problem is more acute for TNA synthesis as repeats as short as two G-nucleotides can lead to polymerase stalling. We postulated that polymerase stalling was due to poor substrate coordination in the enzyme active site, which is consistent with the structural differences between DNA and TNA nucleotide triphosphates. In an effort to solve this problem, we sought to develop a cytidine analogue that would introduce new physiochemical properties into the TNA strand, while at the same time help stabilize the tC:dG base pair during TNA synthesis. In this regard, the cytidine analogue, 1,3-diaza-2-oxo-phenothiazine (Figure 1B), provided an attractive candidate due to its expanded aromatic ring system that is both fluorescent and capable of Watson-Crick base pairing with guanine (19,20). Because TNA cytidine and the cytosine base analogue, 1,3-diaza-2-oxo-phenothiazine share the same abbreviation ‘tC’ in the prior literature, we chose to abbreviate the tricyclic cytidine ring system for a TNA nucleoside as ‘tCf’, which allowed us to distinguish it from TNA cytidine (tC). Previous studies have shown that the DNA version of Cf increases the thermal stability of model duplexes without perturbing the structure or dynamics of the DNA helix (21). In addition, the 5΄-O-triphosphate of 1,3-diaza-2-oxo-phenothiazine 2΄-deoxynucleoside is a known substrate for several DNA polymerases, demonstrating that DNA polymerases can accommodate the larger size of the Cf base (22–25). Moreover, tricyclic cytosine has also been investigated in RNA polymerase contexts (26).
Here, we report the chemical synthesis of 1,3-diaza-2-oxo-phenothiazine TNA nucleoside (tCf) derivatives and the in vitro replication of TNA polymers containing Cf as a fluorescent cytidine analogue. We show that Kod-RI can efficiently read-through sequential G-nucleotides in a DNA template when the reaction mixture contains tCfTP in place of tCTP. Competition assays performed at different ratios of tCTP and tCfTP indicate that tCf is preferentially incorporated onto the growing end of a primer strand. Pre-steady-state kinetics measurements reveal that the first nucleotide incorporation step (kobs1) is comparable for tCfTP and tCTP. However, incorporation of the second nucleotide (kobs2) is 700-fold faster for tCfTP than tCTP. Last, we found that a complete replication cycle of DNA into TNA and TNA back into DNA occurs with an overall fidelity of 98.4%, which is comparable to other xeno-nucleic acid polymers (15). Taken together, these results advance the field of synthetic genetics by enabling the synthesis of TNA polymers with new physiochemical properties.
MATERIALS AND METHODS
General methods and materials
Non-aqueous reactions were performed using oven-dried glassware under an atmosphere of argon or nitrogen. Chemicals and solvents were purchased from Sigma-Aldrich and used without further purification. Reactions were monitored by thin layer chromatography using UV-activated TLC plates with silica gel 60 F254 and aluminium backing (Sigma-Aldrich, St. Louis, US). Flash column chromatography was performed using SiliCycle 40–60 mesh silica gel (SiliCycle Inc., Quebec City, Canada). Yields are reported as isolated yields of pure compounds. 1H, 13C and 31P NMR spectra were obtained using a Bruker DRX400 or DRX500 NMR (Bruker, Billerica, USA). 1H and 13C NMR values are reported in ppm relative to Me4Si as internal standard. 31P NMR values are reported in ppm relative to an external standard of 85% H3PO4. Splitting patterns are designated as follows: s, singlet; br, broad; d, doublet; dd, doublet of doublets; t, triplet; q, quartet; m, multiplet. HPLC purification was performed with a C18 reverse phase 250 × 21.2 mm HPLC column (Thermo Scientific, US) using a mobile phase of 100 mM triethylammoniumacetate buffer (pH 7.0)–acetonitrile. Thermo Pol buffer and Bst DNA polymerase 2.0 were purchased from New England Biolabs (Ipswich, MA, USA). DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA) and the MALDI data were provided in Supplementary Information (Supplementary Table S1). The PBS7-TNA5-IR800 primer was obtained by solid-phase synthesis using chemically synthesized TNA phosphoramidites (27). Recombinant Kod-RI polymerase was expressed and purified from E. coli as described in the supplemental information section.
Chemical synthesis
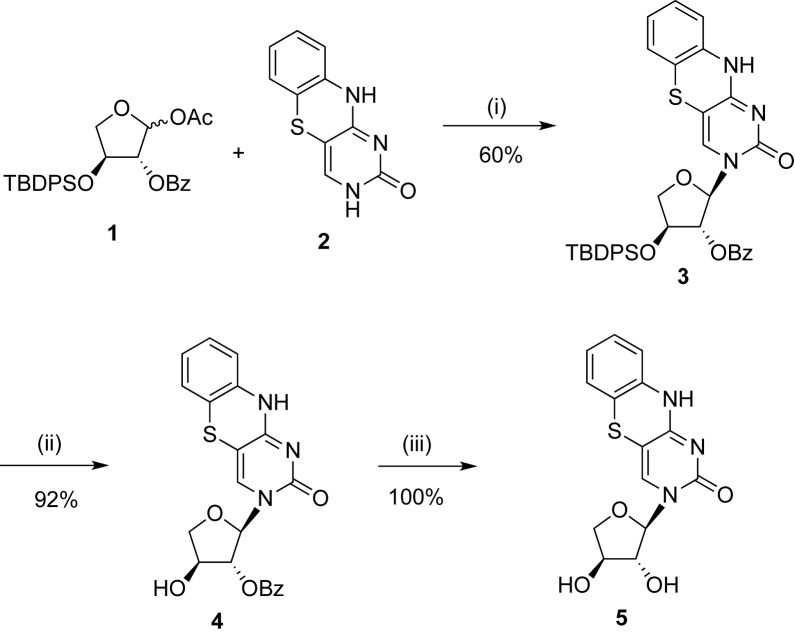
3-(2΄-O-Benzoyl-3΄-O-tert-butyldiphenylsilyl-α-L-threofuranosyl)-1,3-diaza-2-oxophenothiazine (3)
To a suspension of 1,3-diaza-2-oxophenothiazine (2) (0.66 g, 3.03 mmol) in 40 ml anhydrous acetonitrile was added N,O-bis(trimethylsilyl)acetamide (1.85 ml, 7.57 mmol) and the mixture was stirred for 2 h at 75°C. After cooling to 24°C, 1-O-acetyl-2-O-benzoyl-3-O-tert-butyldiphenylsilyl-l-threofuranose (1) (1.53 g, 3.03 mmol) in 10 ml anhydrous acetonitrile was added dropwise to the reaction mixture. TMSOTf (1.65 ml, 9.09 mmol) was then added and the mixture was heated for 5 h at 65°C. The mixture was cooled to 24°C, diluted with 100 ml of CH2Cl2, and poured into a stirring solution of sat aq. NaHCO3 (100 ml) to precipitate the unreacted nucleobase as a yellow solid. The precipitate was filtered and the organic layer was separated and washed with H2O (100 ml) and brine (100 ml), dried over MgSO4 and concentrated under reduced pressure. The residue was washed with 100 ml of hexane/EtOAc (1:1) and the crude nucleoside was obtained as yellow solid (1.2 g), which was used directly without further purification. A small amount of pure compound 3 was obtained by flash chromatography (silica gel, column 15 × 3 cm, hexane/CH2Cl2/EtOAc, 1:1:1). TLC (hexane/EtOAc, 1:1): Rf = 0.33. 1H NMR (500 MHz, CDCl3): δ 1.08 (s, 9H), 3.99–4.01 (dd, 1H, J = 4.0, 8.0 Hz), 4.11–4.13 (m, 1H), 4.35 (m, 1H), 5.60 (s, 1H), 6.17–6.18 (s, 1H), 6.87–6.93 (m, 2H), 7.03–7.05 (m, 2H), 7.20–7.22 (m, 1H), 7.38–7.65 (m, 14H), 7.94–7.96 (d, 2H, J = 4.0 Hz), 9.81 (s, 1H). 13C NMR (125.8 MHz, CDCl3): δ 19.2, 27.0, 75.9, 76.4, 82.3, 90.9, 116.6, 118.4, 124.5, 125.9, 127.8, 128.1, 128.1, 128.5, 129.3, 131.8, 132.8, 134.6, 135.8, 135.9, 136.0, 154.7, 161.1, 164.5. HRMS (ESI-TOF) calcd for C37H35O5N3SSiNa [M+Na]+ 684.1964; observed 684.1948.
3-(2΄-O-Benzoyl-α-L-threofuranosyl)-1,3-diaza-2-oxophenothiazine (4)
To a cold (0–5°C) solution containing crude 3 (1.54 g, 2.32 mmol) in THF (30 ml) was added dropwise tetrabutylammonium fluoride (1 M solution in THF (2.32 ml, 2.32 mmol) and the mixture was stirred for 1 h at 0°C. The solvent was evaporated under reduced pressure and the residue was purified by flash chromatography (SiO2, column 10 × 3 cm, CH2Cl2/MeOH, 20:1) to afford 4 (0.91 g, 92%) as yellow foam. TLC (DCM/MeOH, 10:1): Rf = 0.48. 1H NMR (500 MHz, CDCl3): δ 4.23–4.25 (m, 1H), 4.33–4.35 (m, 1H), 4.43 (m, 1H), 5.22 (brs, 1H), 5.90 (s, 1H), 6.80 (s, 2H), 6.86–6.89 (m, 2H), 7.37–7.43 (m, 3H), 7.55–7.58 (m, 2H), 7.95–7.96 (d, 2H, J = 4.0 Hz), 9.04 (brs, 1H). 13C NMR (125.8 MHz, CDCl3): δ 73.8,76.7, 82.1, 92.6, 96.8, 116.6, 117.7, 124.6, 125.9, 127.4, 128.6, 129.1, 130.0, 135.5, 135.6, 155.6, 160.5, 166.0. HRMS (ESI-TOF) calcd for C21H17O5N3SNa [M+Na]+ 446.0787; observed 446.0777.
3-α-l-Threofuranosyl-1,3-diaza-2-oxophenothiazine (5)
Compound 4 (208 mg, 0.49 mmol) was dissolved in a solution of NH3–MeOH (6 ml) and stirred for 3.5 h at 24°C. The solvent was removed under reduced pressure and crude 5 was purified by flash chromatography (SiO2, column 10 × 3 cm, CH2Cl2/MeOH, 10:1) to afford the free nucleoside 5 (157 mg, 100%) as a yellow solid. TLC (DCM/MeOH, 9:1): Rf = 0.35. UV λmax (MeOH)/nm (ε/dm3 mol−1 cm−1) 236 (26100), 260 (13300), 271 (12200), 368 (3900). 1H NMR (500 MHz, CD3OD): δ 4.13–4.16 (m, 2H), 4.20–4.26 (m, 2H), 5.68 (s, 1H), 6.82–6.83 (m, 1H), 6.90–6.97 (m, 2H), 7.03–7.06 (m, 1H), 7.54 (s, 1H). 13C NMR (125 MHz, CD3OD): δ 75.2, 76.3, 80.3, 93.9, 95.9, 116.5, 116.6, 124.2, 125.7, 127.1, 135.5, 136.1, 156.3, 160.7. HRMS (ESI-TOF) calcd for C14H13O4N3SNa [M+Na]+ 342.0525; observed 342.0522.
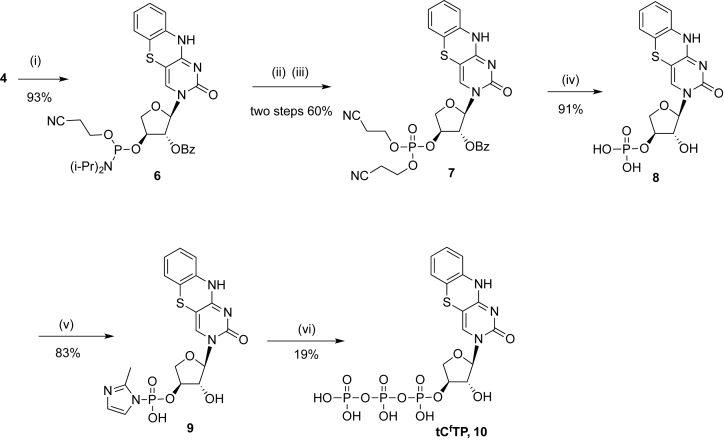
3-(2΄-O-Benzoyl-α-l-threofuranosyl)-1,3-diaza-2-oxophenothiazine-3΄-(2-cyanoethyl)-N,N’-diisopropyl phosphoramidite (6)
To a stirring solution of 4 (0.34 g, 0.80 mmol) and DMAP (20 mg, 0.16 mmol) in CH2Cl2 (10 ml) was added (i-Pr)2NEt (210 μl, 1.21 mmol) followed by the addition of 2-cyanoethyl-N,N-diisopropyl-chlorophosphine (215 μl, 0.96 mmol). After stirring for 40 min at 24°C, the solution was diluted with CH2Cl2 (40 ml) and extracted with 5% aq. NaHCO3 (40 ml). The organic layer was washed with brine, dried over Na2SO4 and evaporated. The residue was purified by flash chromatography (SiO2, column 10 × 3 cm, CH2Cl2/acetone, 6:1) affording 6 (0.47 g, 93%) as a yellow foam. Rf = 0.33 (CH2Cl2/acetone, 4:1). 31P NMR (162 MHz, CDCl3) δ 152.87, 150.85. HRMS (ESI-TOF) calcd for C30H34O6N5PSNa [M+Na]+ 646.1865; observed 646.1862.
3-(2΄-O-Benzoyl-α-l-threofuranosyl)-1,3-diaza-2-oxophenothiazine-3΄-bis(2-cyanoethyl)- phosphotriester (7)
To a stirring solution of 6 (0.47 g, 0.75 mmol) in acetonitrile (15 ml) was added 3-hydroxypropionitrile (82 μl, 1.2 mmol) followed by a solution of 0.45 M tetrazole in THF (2.67 ml, 1.2 mmol). After stirring for 3 h at 24°C, the solution was diluted with CH2Cl2 (50 ml) and washed with brine. The organic layer was dried over Na2SO4 and evaporated to afford a yellow foam. This material was directly used for the next step. The foam was dissolved in 6 ml of THF and H2O2 (30% in H2O) (153 μl, 1.5 mmol) was added to the solution. After stirring at 24°C for 20 min, the solution was diluted with CH2Cl2 (50 ml) and washed with brine. The organic layer was dried over Na2SO4 and evaporated. The residue was purified by flash chromatography (SiO2, column 10 × 3 cm, CH2Cl2/acetone, 1:1) to afford 7 (275 mg, 60%) as a yellow foam. Rf = 0.28 (CH2Cl2/acetone, 1:1). 1H NMR (500 MHz, CDCl3): δ 2.75–2.82 (m, 4H), 4.33–4.38 (m, 5H), 4.58–4.61 (d, J = 11.5 Hz, 1H), 5.02–5.04 (m, 1H), 5.73 (s, 1H), 6.14 (s, 1H), 6.84-6.85 (m, 2H), 6.99–7.00 (m, 1H), 7.09–7.11 (d, J = 7.5 Hz, 1H), 7.35 (s, 1H), 7.42–7.60 (m, 3H), 8.01 (d, 2H, J = 8.0 Hz), 9.71 (brs, 1H). 13C NMR (125.8 MHz, CDCl3) δ 19.6, 19.7, 63.1, 63.2, 74.5, 78.9, 79.9, 90.7, 96.6, 116.6, 118.2, 124.6, 125.8, 127.6, 128.4, 128.5, 130.0, 133.4, 133.9, 135.4, 154.5, 160.5, 164.9. 31P NMR (162 MHz, CDCl3): δ –3.80. HRMS (ESI-TOF) calcd for C27H24O8N5PSNa [M+Na]+ 632.0981; observed 632.0955.
3-α-l-Threofuranosyl-1,3-diaza-2-oxophenothiazine 3΄-monophosphate (8)
Compound 7 (185 mg, 0.30 mmol) was suspended in NH4OH 5mL and stirred for 16 h at 37°C. The solution was diluted with water and washed with CH2Cl2 (3 × 40 ml). The aqueous layer was lyophilized to afford the desired monophosphate 8 (119 mg, 91%) as a yellow solid. 31P NMR (162 MHz, D2O): δ 2.55. HRMS (ESI-TOF) calcd for C14H13O7N3PS [M−H]− 398.0212; observed 398.0219.
3΄-Phosphor-(2-methyl)imidazolide (9)
Monophosphate 8 (118 mg, 0.27 mmol) and 2-methylimidazole (200 mg, 2.44 mmol) were dissolved in DMSO under nitrogen. To this solution was added DMF (2 ml), triethylamine (0.10 ml), triphenylphosphine (140 mg, 0.53 mmol) and 2,2΄-dipyridyl disulfide (160 mg, 0.73 mmol), and the reaction was stirred for 6 h at 24°C. The product was precipitated as a sodium salt by dropwise adding the reaction mixture to a stirring solution of acetone (60 ml), diethyl ether (80 ml), triethylamine (6 ml), saturated NaClO4 in acetone (0.4 ml). The precipitate was collected by centrifugation, washed with (1:1) acetone/diethyl ether (3 × 10 ml), and dried over vacuum. 110 mg of product 9 (yield 83%) was obtained as sodium salt and used for next step directly without further purification. 31P NMR (162 MHz, DMSO-d6): δ −10.04. HRMS (ESI-TOF) calcd for C18H18O6N5PSNa [M+Na]+ 486.0613; observed 486.0602.
3-α-l-Threofuranosyl-1,3-diaza-2-oxophenothiazine 3΄-triphosphate (10)
To a DMF (2 ml) solution of compound 9 (49 mg, 0.10 mmol) was added tributylamine (47 μl, 0.20 mmol) and tributylammonium pyrophosphate (110 mg, 0.20 mmol), and the reaction was stirred for 6 h at 24°C under N2. The reaction mixture was diluted with water and washed with CH2Cl2 (3 × 40 ml). The aqueous layer was lyophilized to dryness and purified by reverse-phase HPLC. Fractions containing triphosphates was collected and lyophilized to afford the product as a triethylammonium salt. The yield of triphosphate 10 was measured by the UV spectrophotometry, assuming an extinction coefficient of 13.3 mM−1 cm−1, and found to be 19% (19 μmol). 31P NMR (162 MHz, D2O): δ –8.49 (d, J = 20.0 Hz), −12.50 (d, J = 20.0 Hz), −22.40 (t, J = 20.0 Hz). HRMS (ESI-TOF) calcd for C14H14O13N3P3SK [M–2H+ K]− 595.9097; observed 595.9072.
TNA synthesis
Primer-extension reactions were performed in a final volume of 10 μl using the PBS2-IR800 primer and PBS2-(1-3G) templates. Each reaction contained 10 pmol primer/template complex, 1× ThermoPol buffer [20 mM Tris–HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, pH 8.8], 1 μM Kod-RI, 100 μM of each tNTP and 1 mM MnCl2. Reactions were incubated for 30 min at 55°C, quenched with stop buffer (8 M urea, 45 mM EDTA) and analyzed by 20% denaturing urea PAGE.
Competition assay
Competition assays were performed as described above for the primer-extension assay using the PBS7-IR700 or PBS7-TNA5-IR800 primers and the PSB7-1G or PBS7-TNA5-1G templates. Reactions contained different molar ratios of tCTP and tCfTP (0–100%) poised at a final concentration of 100 μM cytidine triphosphate. Reactions were incubated for 5 min at 55°C, quenched with stop buffer and analyzed by 20% denaturing urea PAGE, and quantified using an ODYSSEY scanner (LI-COR, Lincoln, USA).
Pre-steady state kinetics
100 nM of PBS7-IR700 was annealed with 100 nM of PBS7-2G in 1 × ThermoPol buffer. The solution was combined with 400 nM Kod-RI and 100 μM of tNTP substrate. The reaction was incubated at 45°C for increasing amounts of time (0–15 min). At designated time points, a portion of the reaction was removed, quenched with stop buffer, analyzed by 20% denaturing PAGE, and quantified by ODYSSEY scanner. In the kinetics analysis (see SI section), kobs1 was measured from the decay profile of the starting primer (Eq. 1). The value for kobs1 was then introduced into Equation (4) to obtain kobs2, which describes the accumulation of the n + 2 product. Graphs were plotted using Prism (Lalla, CA, USA).
Fidelity of TNA replication
The fidelity assay was performed as previously described (17). Briefly, primer 1 μM PBS8_extra containing a single nucleotide mismatch was annealed with 1 μM of the 4NT9G template. The primer-extension reaction was performed for 1 h at 55°C in 1× Thermopol buffer supplemented with 1 mM MnCl2 and 100 μM of each tNTP. The polymerase Kod-RI was used at a final concentration of 0.5 mg/ml, which is in 4-fold excess to the primer–template concentration. The extension product was purified by denaturing PAGE and reverse transcribed into cDNA. The reverse transcription reaction was performed by incubating the TNA template–PBS7 primer complex (1 μM) with Bst DNA polymerase (1 μM) for 3 h at 50°C in a reaction mixture (50 μl) that contained 500 μM each dNTP, 3 mM MgCl2 and 1× Thermopol buffer. The cDNA product was PCR amplified with the extra/PBS7 primer pair, ligated into a TOPO vector (Thermo Fisher, Waltham, MA, USA), and cloned into Escherichia coli. Individual colonies were grown in liquid media and sequenced (Retrogene, San Diego, CA, USA). DNA sequences were aligned with the 4NT9G template and analyzed using DNASTAR (Madison, MI, USA).
RESULTS
Synthesis of α-l-threofuranosyl-1,3-diaza-2-oxo-phenothiazine nucleoside
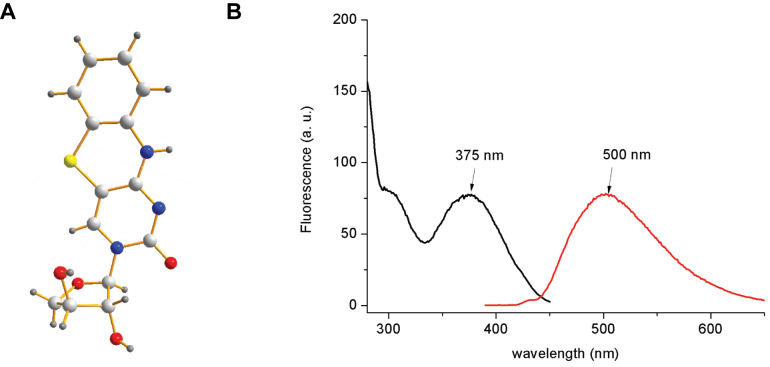
We began by synthesizing the α-L-threofuranosyl-1,3-diaza-2-oxo-phenothiazine nucleoside (5) from the universal glycosyl donor (1) and 1,3-diaza-2-oxo-phenothiazine (2). Compounds 1 and 2 were prepared from known literature procedures and conjugated together in a Vorbrüggen glycosylation reaction to produce nucleoside 3 in protected form (Scheme 1) (20,27). Compound 5 was generated with an overall yield of 55% by deprotection of the 3΄-O-TBDPS group of 3 with TBAF to produce 4, which was subsequently treated with ammonia in methanol to remove the 2΄-O-benzoyl group. The structure of α-l-threofuranose nucleoside 5 was confirmed by small molecule X-ray diffraction (Figure 2A). Torsion angles formed by the O–C(2΄)–C(3΄)–O and O–C(1΄)–N(3)–C(2) atoms (170.2° and –166.5°, respectively) are in close agreement with the bond angles previously observed for TNA nucleosides with natural bases (see Supplementary Data) (3,28).
Scheme 1.
aReagents and conditions. (i) N,O-bis(trimethylsilyl)-acetamide (BSA), trimethylsilyltriflate (TMSOTf), CH3CN, 65°C, 5 h; (ii) 1 M TBAF in THF, 0°C, 1 h; (iii) NH3–MeOH, rt, 3.5 h.
Figure 2.
(A) X-ray crystal structure of TNA nucleoside 5. (B) Fluorescence excitation (black line) and emission (red line) spectra of nucleoside 5 (5 μM in H2O containing 1% DMSO).
To confirm that tCf retains its fluorescent properties when constructed as a TNA nucleoside, we measured the absorption and emission spectra of the free nucleoside in water. As shown in Figure 2B, nucleoside 5 exhibits a significant Stokes shift with absorption and emission maxima of 375 and 500 nm, respectively. This result is in close agreement with a previously reported value for the DNA version of Cf (21,29), and highlights the value of extending the properties of modified bases to nucleic acid polymers with unnatural backbone structures.
Synthesis of the triphosphate derivative
TNA triphosphates are more difficult to synthesize than DNA and RNA triphosphates, because phosphorylation occurs at the sterically encumbered secondary hydroxyl group located at the 3΄ position of the sugar ring rather than the more accessible primary hydroxyl group located at the 5΄ position of pentose sugars (e.g. the sugar commonly found in DNA and RNA). For this reason, we have evaluated a number of different strategies for synthesizing TNA triphosphates (30). One of the more successful approaches developed to date follows a Hoard-Ott-like approach in which the TNA nucleoside is first converted to an activated monophosphate that is subsequently treated with pyrophosphate to afford the desired nucleoside triphosphate (31,32). As shown in Scheme 2, nucleoside 4 was converted to monophosphate 8 via the phosphoramidite and phosphotriester intermediates 6 and 7, respectively. Monophosphate 8 was then converted to a phosphorimidazolide (9), which was subsequently displaced with pyrophosphate to provide the desired nucleoside triphosphate (tCfTP, 10). Compounds were characterized by 1H, 13C, 31P NMR spectra as well as HRMS mass spectra (see Materials and Methods Section and Supplementary Data).
Scheme 2.
aReagents and conditions. (i) NC(CH2)2OP(Cl)N(i-Pr)2, (i-Pr)2EtN, DMAP, rt, 40 min; (ii) 3-hydroxypropionitrile, tetrazole, CH3CN, rt, 3h; (iii) H2O2, THF, rt. 20 min; (iv) NH4OH, 37°C, 16 h, ; (v) 2-methylimidazole, PPh3, 2,2΄-dipydiyl disulfide, triethylamine, DMF–DMSO, rt, 6 h; (vi) tributylammonium pyrophosphate, tributylamine, anhydrous DMF, rt, 6 h.
TNA synthesis with a modified base pair
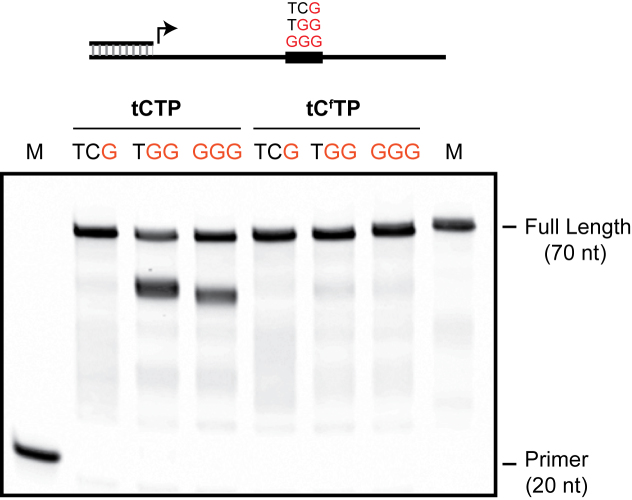
We evaluated the ability of tCfTP to be incorporated into a growing TNA strand using a standard primer-extension assay. For this experiment, a series of DNA primer–template complexes were designed to contain a central region of one to three sequential guanosine residues that were flanked on either side by an arbitrary sequence composed of A, C and T residues (Figure 3). Kod-RI was challenged to extend the DNA primer using chemically synthesized TNA triphosphates (tNTPs) that contained either tCTP or the fluorescent analogue tCfTP in the reaction mixture (30). In cases where the polymerase is able to read through the stretch of G nucleotides, the product will be an elongated strand in which 50 consecutive TNA residues have been added to the 3΄-end of a DNA primer. However, if the polymerase is unable to bypass the G repeat, it will produce truncated band that is easily detected by denaturing polyacrylamide gel electrophoresis (PAGE).
Figure 3.
TNA synthesis with a modified base pair. Primer-extension assay used to evaluate TNA synthesis on DNA templates with one to three internal G-nucleotides. TNA synthesis reactions were performed for 30 min at 55°C using Kod-RI TNA polymerase.
PAGE analysis of the primer-extension assay reveals a striking difference in the ability for tCTP and tCfTP residues to read through the G-nucleotide motifs (Figure 3). In the case of tCTP, Kod-RI can efficiently read through a single G-residue embedded in a DNA template. However, the enzyme struggles to read through DNA templates that contain two or more sequential G-nucleotides. In such cases, ∼50% of the elongated product is trapped at the pause site, while the remainder is extended to full-length product. By comparison, primer-extension reactions performed with tCfTP yield the desired full-length TNA product in all of the cases tested (Figure 3). This marked difference in TNA polymerization efficiency implies that the larger aromatic ring system of tCfTP may help stabilize the tC:dG base pair in the enzyme active site.
Competitive primer-extension assays
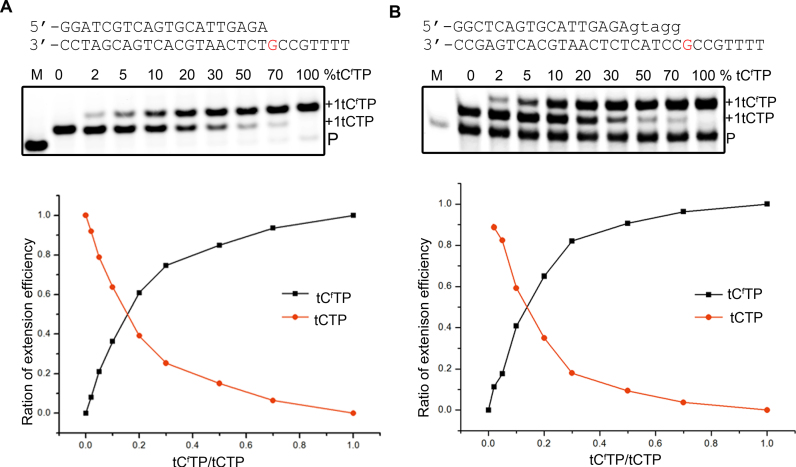
Recognizing the fact that DNA primers extended with tCf migrate more slowly on a denaturing polyacrylamide gel than primers extended with tC, we designed a competition assay to compare the relative preference of Kod-RI for tCfTP versus tCTP. Accordingly, a series of single-nucleotide primer-extension experiments were performed by varying the mole fraction of tCfTP in the tNTP mixture from 0 to 100%. If Kod-RI has an equal preference for both nucleotides, the primer will be extended equal amounts at a mole fraction of 0.5. However, if Kod-RI prefers one substrate over the other, then an imbalance will occur where the mole fraction will trend in the direction of the preferred substrate. As shown in Figure 4A, Kod-RI shows a clear preference for tCfTP over tCTP such that an equal distribution of the tCf and tC monomers occurs at a mole fraction of 0.15.
Figure 4.
Single-nucleotide competition assay comparing the primer-extension efficiency of tCTP versus tCfTP. Competition assays were performed using triphosphate solutions that contained defined ratios of tCTP and tCfTP ranging from 0 to 100% tCfTP. Primer-extension reactions were performed on primer–template complexes that were either all-DNA (A) or used a DNA/TNA chimeric primer that contained 5 TNA residues at the 3΄ end (B). Each dataset represents the average of two independent trials.
To show that this effect was not due to the regiochemistry of the DNA primer, we repeated the experiment using a chimeric DNA–TNA primer that carried 5 TNA residues at the 3΄ end. This new primer was designed to probe the downstream process of TNA synthesis where TNA monomers are added to the 2΄ end of a growing TNA strand to create 2΄-3΄ phosphodiester bonds. By contrast, strand elongation from a DNA primer produces a 3΄-3΄ phosphodiester bond between the DNA primer and first nucleotide adduct due to the strand polarity differences between TNA and DNA (3΄,2΄ versus 5΄,3΄, respectively). Similar to the all-DNA primer, an equal distribution of the tCfTP and tCTP monomers occurs at a mole fraction of 0.15 (Figure 4B). Together, these results indicate that the preference of Kod-RI for tCfTP is the same regardless of whether TNA extension occurs from a DNA primer or DNA/TNA chimeric primer.
Kinetic analysis of TNA synthesis
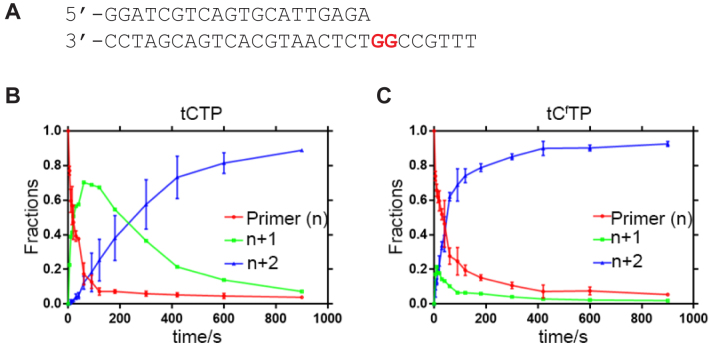
To examine the mechanism of TNA synthesis in greater detail, we used pre-steady-state kinetics to measure the rate of tCTP and tCfTP incorporation at the 3΄ end of a DNA primer. Although tCf provides an optical signal for gel analysis, we chose to use a 5΄-labeled dye for kinetic analysis so that the kinetic data from tCf could be compared directly to equivalent data obtained for tC. In brief, a pre-incubated solution of Kod-RI (400 nM) and 5΄-dye-labeled DNA primer–template complex (100 nM) in a reaction buffer at 45°C was rapidly mixed with either tCTP or tCfTP (100 μM). After brief incubations at various times, the reactions were quenched with EDTA and analyzed by denaturing PAGE. The data were fit to a single-exponential equation to yield kobs, which is the observed rate constant for a single-nucleotide incorporation event. Since the tCfTP substrate was found to insert opposite multiple G residues with higher efficiency than tCTP, we designed a DNA template that contained two sequential G-nucleotides. This template design allowed us to measure the first (kobs1) and second (kobs2) nucleotide incorporation events by monitoring the decay of the starting primer and the accumulation of the n + 2 product from the n + 1 product, respectively, in a single primer-extension reaction.
The complete set of kinetic profiles observed for primer-extension experiments performed with the tCTP and tCfTP substrates are provided in Figure 5. Accordingly, we observe that the rate of cytidine incorporation at the first nucleotide position (kobs1) is 2-fold faster for tCTP than tCf TP (0.031 ± 0.002 s−1 versus 0.017 ± 0.007 s−1, respectively). However, after the first TNA residue is added to the DNA primer, the n + 1 product accumulates in the tCTP reaction, while the tCfTP reaction rapidly progresses from the n + 1 product to the n + 2 product (Figure 5). Comparison of the incorporation rates for the second nucleotide extension step (kobs2) reveals that the modified tCfTP substrate is 700-fold faster than the tCTP substrate (2.6 ± 0.04 s−1 versus 0.0036 ± 0.001 s−1). We attribute the rate acceleration of the tCfTP substrate to favorable base stacking interactions that arise when two Cf bases are located adjacent to one another on the same strand, which could help stabilize the substrate in the enzyme active site. Alternatively, it is also possible that enhanced activity is due to increased Van der Waals interactions provided by aromatic side chains in the enzyme active site.
Figure 5.
Kinetic analysis of tCTP and tCfTP incorporation on a DNA primer–template complex. (A) Pre-steady state conditions were used to measure the rate of the first (kobs1) and second (kobs2) nucleotide incorporation events for tCTP (B) and tCfTP (C) on a DNA primer–template complex that contained two G-residues in the template region. The kinetic process is colored according to the decay of the primer strand (red), accumulation of the n+1 adduct (green), and accumulation of the n + 2 adduct (blue). Error bars represent the average of two or more independent trials.
Fidelity of TNA replication
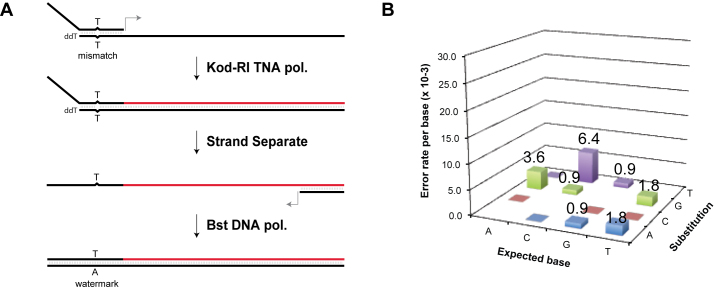
To ensure that Kod-RI faithfully incorporates the tCfTP substrate opposite dG residues in a DNA template, we measured the fidelity of TNA replication by sequencing the cDNA product. This fidelity assay measures the aggregate fidelity of a complete replication cycle (DNA→TNA→DNA), which is operationally different than the more restricted view of fidelity as the accuracy of a single-nucleotide incorporation event (Figure 6A). The fidelity determined by this assay is the actual accuracy by which full-length TNA is synthesized and reverse transcribed, and therefore reflects the combined effects of nucleotide misincorporation, insertions and deletions, and any mutations that occur during PCR amplification and cloning.
Figure 6.
Fidelity of TNA replication. (A) Schematic representation of the transcription and reverse transcription process used to evaluate the fidelity of TNA replication. DNA is shown in black, TNA is shown in red. The primer–template complex contains a T–T mismatch, which produces a T to A transversion in the cDNA strand. The transversion represents a watermark to ensure that the sequenced DNA was produced by TNA transcription and reverse transcription. The starting template contains a 3΄-deoxy residue to prevent extension across the unpaired region. (B) The mutation profile reveals that TNA transcription and reverse transcription proceeds with an error rate of 16 x 10-3 (98.4% fidelity).
Several controls were implemented to ensure that the sequencing results represented the true fidelity of TNA replication. First, to eliminate any possibility of contamination by the starting DNA template, the DNA primer–template complex used for TNA transcription was partially unpaired and contained a dideoxynucleotide at the 3΄ end of the template strand to prevent elongation across the unpaired region of the primer. This design also facilitates separation of the TNA product by denaturing PAGE as the product and template strands have different lengths. Second, all PCR amplification steps were performed using a negative control that contained the purified TNA template prior to the reverse transcription step. In no cases did we observe a DNA band, demonstrating that the purified TNA product contained minimal DNA template as a background contaminant. Third, to unambiguously demonstrate that each DNA sequence derived from a complete cycle of TNA replication, the DNA primer used for TNA transcription was engineered to contain a single-nucleotide mismatch that resulted in an A→T transversion in the sequenced product. Together, these controls allowed us to measure the fidelity of TNA replication with confidence.
We sequenced >1000 nucleotide positions from the cDNA isolated from a cycle of TNA transcription and reverse transcription. In this experiment, Kod-RI was used to facilitate TNA synthesis, while Bst DNA polymerase was used to reverse transcribe the TNA back into DNA (33). Analysis of the data yielded an overall replication fidelity of 98.4% (error rate of 1.6 × 10−2), as shown in Figure 6B. This level of fidelity is comparable to previous fidelity results for other XNA systems (15).
DISCUSSION
In the field of synthetic genetics, where engineered polymerases are used to synthesize artificial genetic polymers, there exist two main reasons for developing XNA polymers with modified bases. The first and most pressing reason is to overcome the limitations of engineered polymerase that function with inferior activities relative to natural polymerases. Analogues that compensate for problems with template sequence bias, primer-extension efficiency, or fidelity provide a chemical biology solution that can enhance the value of XNA polymers until new generations of XNA polymerases are produced by directed evolution. Diaminopurine, for example, was previously used as a base analogue of adenosine to increase the rate of TNA synthesis at a time when early TNA polymerases were unable to synthesize long stretches of TNA on DNA templates (11). A second reason for pursuing XNA with modified bases is to endow XNA polymers with functional properties not present in the four canonical bases of A, G, C and T (U). Chemically modified bases, for example, have long been used to expand the functional diversity of natural DNA and RNA (34). These include functional groups with amino acid side chains as well as unnatural chemical moieties with functionalities beyond those found in proteins (35,36).
In the current study, we sought to develop a cytidine analogue that would stabilize the tC:dG base pair in the active site of Kod-RI and expand the functional properties of TNA. A review of the literature identified 1,3-diaza-2-oxo-phenothiazine (Cf) as a base analogue that maintains Watson-Crick base pairing with guanine, but has an enhanced base stacking ability due to a large tricyclic aromatic ring system (19). In the context of a DNA helix, one Cf incorporation can increase the melting temperature of a DNA duplex by 2.7°C relative to an identical duplex containing natural cytosine (21). This level of stability, along with increased hydrophobic and fluorescence character, were viewed as favorable properties for a modified base analogue.
We show that Cf supports both reasons for generating XNA polymers with modified bases. Replacing cytosine with Cf enables Kod-RI to read through G-repeats in a DNA template that posed a strong barrier to TNA synthesis. The favorable kinetic properties and faithful replication profile make Cf a valuable base analogue for synthetic genetics, as this modification makes it possible to synthesize any reasonable TNA sequence that is accessible from a standard four letter genetic alphabet. In addition, the Cf ring system provides TNA with new chemical properties that are not found in the canonical bases (20,21). This includes strong fluorescent properties that are insensitive to environment as well as increased hydrophobic and base stacking character that may enhance the functional properties of TNA aptamers. Collectively, these benefits demonstrate the importance of pursuing base analogues in synthetic genetics, which is growing trend in the field of artificial genetic polymers (37).
CONCLUSION
In summary, our study identified Cf as a modified cytosine analogue that improves TNA synthesis by enabling an engineered TNA polymerase to read through sequential G-repeats in a DNA template. In addition to enhancing the sequence diversity of TNA polymers generated by enzyme-mediated synthesis, Cf introduces new physiochemical that are not present in TNA molecules composed of natural bases. This includes an expanded aromatic ring system with increased hydrophobic character and fluorescence activity that is insensitive to environmental conditions.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank members of the Chaput lab for helpful comments and suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DARPA Folded Non-Natural Polymers with Biological Function Fold F(x) Program [N66001-16-2-4061]; National Science Foundation [1615804 and 1607111]. Funding for open access charge: US federal grants.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chaput J.C., Yu H., Zhang S.. The emerging world of synthetic genetics. Chem. Biol. 2012; 19:1360–1371. [DOI] [PubMed] [Google Scholar]
- 2. Schöning K.U., Scholz P., Guntha S., Wu X., Krishnamurthy R., Eschenmoser A.. Chemical etiology of nucleic acid structure: the alpha-threofuranosyl-(3΄–>2΄) oligonucleotide system. Science. 2000; 290:1347–1351. [DOI] [PubMed] [Google Scholar]
- 3. Schöning K.-U., Scholz P.X.W., Guntha S., Delgado G., Krishnamurthy R., Eschenmoser A.. The a-L-Threofurnaosyl-(3΄-2΄)-oligonucleotide system ('TNA'): synthesis and pairing properties. Helv. Chim. Acta. 2002; 85:4111–4153. [Google Scholar]
- 4. Yang Y.-W., Zhang S., McCullum E.O., Chaput J.C.. Experimental evidence that GNA and TNA were not sequential polymers in the prebiotic evolution of RNA. J. Mol. Evol. 2007; 65:289–295. [DOI] [PubMed] [Google Scholar]
- 5. Anosova I., Kowal E.A., Sisco N.J., Sau S.P., Liao J.-Y., Bala S., Rozners E., Egli M., Chaput J.C., Van Horn W.D.. Structural insights into conformational differences betweeen DNA/TNA and RNA/TNA chimeric duplexes. ChemBioChem. 2016; 17:1705–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ebert M.-O., Mang C., Krishnamurthy R., Eschenmoser A., Jaun B.. The structure of a TNA-TNA complex in solution: NMR study of the octamer duplex derived from a-(L)-threofuranosyl-(3΄-2΄)-CGAATTCG. J. Am. Chem. Soc. 2008; 130:15105–15115. [DOI] [PubMed] [Google Scholar]
- 7. Culbertson M.C., Temburnikar K.W., Sau S.P., Liao J.-Y., Bala S., Chaput J.C.. Evaluating TNA stability under simulated physiological conditions. Bioorg. Med. Chem. Lett. 2016; 26:2418–2421. [DOI] [PubMed] [Google Scholar]
- 8. Chaput J.C., Ichida J.K., Szostak J.W.. DNA polymerase-mediated DNA synthesis on a TNA template. J. Am. Chem. Soc. 2003; 125:856–857. [DOI] [PubMed] [Google Scholar]
- 9. Chaput J.C., Szostak J.W.. TNA synthesis by DNA polymerases. J. Am. Chem. Soc. 2003; 125:9274–9275. [DOI] [PubMed] [Google Scholar]
- 10. Kempeneers V., Vastmans K., Rozenski J., Herdewijn P.. Recognition of threosyl nucleotides by DNA and RNA polymerases. Nucleic Acids Res. 2003; 31:6221–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horhota A., Zou K., Ichida J.K., Yu B., McLaughlin L.W., Szostak J.W., Chaput J.C.. Kinetic analysis of an efficient DNA-dependent TNA polymerase. J. Am. Chem. Soc. 2005; 127:7427–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ichida J.K., Horhota A., Zou K., McLaughlin L.W., Szostak J.W.. High fidelity TNA synthesis by therminator polymerase. Nucleic Acids Res. 2005; 33:5219–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ichida J.K., Zou K., Horhota A., Yu B., McLaughlin L.W., Szostak J.W.. An in vitro selection system for TNA. J. Am. Chem. Soc. 2005; 127:2802–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu H., Zhang S., Chaput J.C.. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 2012; 4:183–187. [DOI] [PubMed] [Google Scholar]
- 15. Pinheiro V.B., Taylor A.I., Cozens C., Abramov M., Renders M., Zhang S., Chaput J.C., Wengel J., Peak-Chew S.Y., McLaughlin S.H. et al. . Synthetic genetic polymers capable of heredity and evolution. Science. 2012; 336:341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Larsen A.C., Dunn M.R., Hatch A., Sau S.P., Youngbull C., Chaput J.C.. A general strategy for expanding polymerase function by droplet microfluidics. Nat. Commun. 2016; 7:11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dunn M.R., Otto C., Fenton K.E., Chaput J.C.. Improving polymerase activity with unnatural substrates by sampling mutations in homologous protein architectures. ACS Chem. Biol. 2016; 11:1210–1219. [DOI] [PubMed] [Google Scholar]
- 18. Skelly J.V., Edwards K.J., Jenkins T.C., Neidle S.. Crystal structure of an oligonucleotide duplex containing GG base pairs: influence of mispairing on DNA backbone conformation. Proc. Natl. Acad. Sci. USA. 1993; 90:804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin K.-Y., Jones R.J., Matteucci M.. Tricyclic 2-deoxycytidine analogs: syntheses and incorporation into oligodeoxynucleotides which have enhanced binding to complementary RNA. J. Am. Chem. Soc. 1995; 117:3873–3874. [Google Scholar]
- 20. Sandin P., Lincoln P., Brown T., Wilhelmsson L.M.. Synthesis and oligonucleotide incorporation of fluorescent cytosine analogue tC: a promising nucleic acid probe. Nat. Protoc. 2007; 2:615–623. [DOI] [PubMed] [Google Scholar]
- 21. Engman K.C., Sandin P., Osborne S., Brown T., Billeter M., Lincoln P., Norden B., Albinsson B., Wilhelmsson L.M.. DNA adopts normal B-form upon incorporation of highly fluorescent DNA base analogue tC: NMR structure and UV-vis spectroscopy characterization. Nucleic Acids Res. 2004; 32:5087–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stengel G., Purse B.W., Wilhelmsson L.M., Urban M., Kuchta R.D.. Ambivalent incorporation of the fluorescent cytosine analogues tC and tCo by human DNA polymerase a and Klenow fragment. Biochemistry. 2009; 48:7547–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sandin P., Stengel G., Ljungdahl T., Börjesson K., Macao B., Wilhelmsson L.M.. Highly efficient incorporation of the fluorescent nucleotide analogs tC and tCo by Kenow fragment. Nucleic Acids Res. 2009; 37:3924–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stengel G., Gill J.P., Sandin P., Wilhelmsson L.M., Albinsson B., Norden B., Millar D.. Conformational dynamics of DNA polymerase probed with a novel fluorescent DNA base analogue. Biochemistry. 2007; 46:12289–12297. [DOI] [PubMed] [Google Scholar]
- 25. Walsh J.M., Bouamaied I., Brown T., Wilhelmsson L.M., Beuning P.J.. Discrimination against the cytosine analog tC by Escherichia coli DNA polymerase IV DinB. J Mol Biol. 2011; 409:89–100. [DOI] [PubMed] [Google Scholar]
- 26. Stengel G., Urban M., Purse B.W., Kuchta R.D.. Incorporation of the fluorescent ribonucleotide analogue tCTP by T7 RNA polymerase. Anal Chem. 2010; 82:1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sau S.P., Fahmi N.E., Liao J.-Y., Bala S., Chaput J.C.. A scalable synthesis of α-L-threose nucleic acid monomers. J. Org. Chem. 2016; 81:2302–2307. [DOI] [PubMed] [Google Scholar]
- 28. Bain J.C., Ricardo A., Szostak J.W.. Synthesis and nonenzymatic template-directed polymerization of 2΄-amino-2΄-deoxythreose nucleotides. J. Am. Chem. Soc. 2014; 136:2033–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilhelmsson L.M., Sandin P., Holmén A., Albinsson B., Lincoln P., Nordén B.. Photophysical characterization of fluorescent DNA base analogue, tC. J. Phys. Chem. B. 2003; 107:9094–9101. [Google Scholar]
- 30. Sau S.P., Chaput J.C.. A one-pot synthesis of a-L-threofuranosyl nucleoside triphosphates. Bioorg. Med. Chem. Lett. 2016; 26:3271–3273. [DOI] [PubMed] [Google Scholar]
- 31. Burgess K., Cook D.. Syntheses of Nucleoside Triphosphates. Chem. Rev. 2000; 100:2047–2060. [DOI] [PubMed] [Google Scholar]
- 32. Chen Z., Meek K.N., Rangel A.E., Heemstra J.M.. Synthesis and polymerase incorporation of b,g-modified a-L-threofuranosyl thymine triphosphate mimics. Bioorg. Med. Chem. Lett. 2016; 26:3958–3962. [DOI] [PubMed] [Google Scholar]
- 33. Dunn M.R., Chaput J.C.. Reverse transcription of threose nucleic acid by a naturally occurring DNA polymerase. ChemBioChem. 2016; 17:1804–1808. [DOI] [PubMed] [Google Scholar]
- 34. Malyshev D.A., Romesberg F.E.. The expanded genetic alphabet. Angew. Chem. Int. Ed. Engl. 2015; 54:11930–11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vaught J.D., Bock C., Carter J., Fitzwater T., Otis M., Schneider D., Rolando J., Waugh S., Wilcox S.K., Eaton B.E.. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 2010; 132:4141–4151. [DOI] [PubMed] [Google Scholar]
- 36. Diafa S., Hollenstein M.. Generation of aptamers with an expanded chemical repertoire. Molecules. 2015; 20:16643–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bande O., El Asrar B.A., Braddick D., Dumbre S., Pezo V., Schepers G., Pinheiro V.B., Lescrinier E., Holliger P., Marliere P. et al. . Isoguanine and 5-methyl-isocytosine bases, in vitro and in vivo. Chem. A Eur. J. 2015; 21:5009–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.