Abstract
DNA double-strand breaks (DSBs) are repaired by either the non-homologous end joining (NHEJ) or homologous recombination (HR) pathway. Pathway choice is determined by the generation of 3΄ single-strand DNA overhangs at the break that are initiated by the action of the Mre11–Rad50–Xrs2 (MRX) complex to direct repair toward HR. DSB repair occurs in the context of chromatin, and multiple chromatin regulators have been shown to play important roles in the repair process. We have investigated the role of the SWI/SNF ATP-dependent nucleosome-remodeling complex in the repair of a defined DNA DSB. SWI/SNF was previously shown to regulate presynaptic events in HR, but its function in these events is unknown. We find that in the absence of functional SWI/SNF, the initiation of DNA end resection is significantly delayed. The delay in resection initiation is accompanied by impaired recruitment of MRX to the DSB, and other functions of MRX in HR including the recruitment of long-range resection factors and activation of the DNA damage response are also diminished. These phenotypes are correlated with a delay in the eviction of nucleosomes surrounding the DSB. We propose that SWI/SNF orchestrates the recruitment of a pool of MRX that is specifically dedicated to HR.
INTRODUCTION
DNA double-strand breaks (DSBs) are potent cytotoxic lesions that must be accurately repaired to prevent cellular senescence, apoptosis or oncogenic transformation (1). Cells encounter a barrage of genomic insults that can lead to the formation of DSBs, including exogenous sources such as ionizing radiation, environmental toxins, and chemotherapeutic agents, as well as endogenous sources such as reactive oxygen species (ROS) and DNA replication stress (2). There are two major pathways to correct DSBs: non-homologous end joining (NHEJ) and homologous recombination (HR). NHEJ is a cell-cycle independent process that involves the direct rejoining of broken ends. While it can be error-free, especially with DSB ends that are complementary and free of base damage, when the DSB ends are non-complementary or chemically altered, such as in breaks generated by ionizing radiation, end processing to make them ligatable can result in insertions and deletions (3). In contrast, HR is a highly accurate but cell-cycle dependent process that requires a homologous template such as a sister chromatid for copying in order to replace the damaged segment of DNA (4). It is imperative that cells utilize the correct DSB repair pathway depending on cell cycle stage and lesion context (5,6) in order to prevent or limit genome instability and ensure organism survival.
A key repair intermediate that drives pathway choice is 3΄ single-strand DNA (ssDNA) generated at break ends (7,8). After a DSB is generated, both the NHEJ-promoting Ku70/Ku80 (KU) complex and the multifunctional Mre11/Rad50/Xrs2 (MRX in yeast; Mre11/Rad50/Nbs1, MRN in mammals) complex rapidly associate with DSB ends. When KU binding predominates, generally during G1 phase, accessory NHEJ proteins are recruited to bridge, process, and ligate the break ends (9,10). In contrast, during S-G2 phases, the MRX/MRN complex associates with Sae2/CtIP to initiate the process of DNA end resection, which generates 3΄ ssDNA overhangs that antagonize KU association and channel the DSB ends into the HR pathway (11,12).
During HR, DNA end resection occurs in two distinct phases. The initial phase is carried out by MRX/MRN in conjunction with Sae2/CtIP, creating short 100–300 bp 3΄ ssDNA overhangs (13,14). In the next phase, known as long-range resection, there are two resection activities—exonuclease 1 (Exo1) and the nuclease/helicase complex Dna2-Sgs1/Top3/Rmi1 (Dna2–STR; BLM-Topo IIIα/RMI1/RMI2, BTR in mammals)—that extend the ssDNA tracts for many kilobases on either side of the break (14–17). The ssDNA is first coated by Replication Protein A (RPA), which is then actively replaced by the Rad51 recombinase to form a nucleoprotein filament that mediates homology search and strand invasion (18).
The initiation of end resection is tightly regulated in cells to prevent inappropriate recombination, for example during G1 or early S phase when a sister chromatid is not available (19). In G1 phase cells, KU and mammalian 53BP1, as well as the yeast 53BP1 ortholog Rad9, accumulate on DSB ends where they antagonize the initiation of end resection (20–23). The nuclease activity of MRX/MRN is also impaired in G1 phase cells due to the low CDK-dependent phosphorylation of Sae2/CtIP (24,25), whereas in S and G2 phases cells, multiple resection factors, including Sae2/CtIP, Dna2, and Exo1, have increased expression and/or phosphorylation that enhance their participation in resection (26). Furthermore, KU is reported to be modified by phosphorylation, ubiquitylation, and neddylation, promoting its removal from DSB ends in S and G2 phase cells (22,27,28). BRCA1 replaces 53BP1 in mammals and CDK-phosphorylated Fun30 antagonizes yeast Rad9, thereby removing additional barriers to resection and enhancing the recombinogenic environment in late S and G2 phases (29–31). Thus, it is apparent that the initiation of DNA end resection is controlled by multiple mechanisms because this is a key step in determining repair pathway choice.
Since DNA end resection and other steps in DSB repair occur in the context of chromatin, chromatin regulators play influential roles in repair outcomes. While some chromatin regulators deposit covalent modifications on histone tails to facilitate DNA damage signaling and repair factor recruitment (32), others alter the structure of chromatin either by replacing canonical histones with histone variants or by moving or evicting nucleosomes. These latter functions are carried about by a class of enzymes known as ATP-dependent nucleosome remodelers, which utilize the energy of ATP hydrolysis to modify histone–DNA interactions (33). In yeast, multiple ATP-dependent nucleosome remodelers, all of which are conserved and diversified in higher eukaryotes, are recruited to DNA DSBs (34–36). For example, the RSC complex is recruited almost immediately after a DSB is formed, inducing a rapid shift of nucleosomes next to a DSB (37,38). This activity is posited to facilitate both KU and MRX recruitment to break ends and to promote the formation of cohesion to tether the broken region to the donor locus (38,39). The INO80 complex is recruited to a DSB later and participates in the sequential eviction of nucleosomes on either side of a DSB, an activity that facilitates Rad51 nucleoprotein filament formation (40). While there are conflicting reports about the role of RSC or INO80 in DNA end resection (36), two other remodelers have been demonstrated to facilitate long-range resection. The SWR-C complex replaces canonical H2A with the variant H2A.Z to promote long-range resection by the Exo1 pathway (41), whereas the Fun30 nucleosome remodeler travels with the long-range resection machinery to facilitate resection through nucleosomes by removing the Rad9 checkpoint protein from chromatin (42–44). Finally, the prototypical ATP-dependent nucleosome-remodeling complex, SWI/SNF, is recruited to DNA DSBs, where it plays a critical but unknown role in pre-synaptic events during HR (45).
In this study, we have investigated the role of SWI/SNF in DSB repair in budding yeast. We provide the first evidence that SWI/SNF is required for the timely initiation of DNA end resection during HR. This role appears to be mediated through the action of SWI/SNF in the recruitment and/or stabilization of the MRX complex at DSB ends. We also observed that nucleosome eviction at a DSB is delayed in a SWI/SNF mutant, suggesting that the chromatin remodeling activity of SWI/SNF may contribute to its role in MRX recruitment and resection initiation. Together our results reveal critical early roles for SWI/SNF in orchestrating successful DSB repair by HR.
MATERIALS AND METHODS
Strains and growth conditions
Saccharomyces cerevisiae strains are listed in Supplementary Table 1. Gene knockouts and epitope tagging were performed by genomic recombination with PCR-amplified cassettes (46). Strains were pre-grown to O.D.600 nm 0.4–0.6 in YPD medium (1% yeast extract, 2% peptone, 2% dextrose). Cells were then diluted into pre-induction GLGYP medium (3% glycerol, 2% d-lactate, 0.05% dextrose, 1% yeast extract, 2% peptone, pH 5.5) and grown for 12–15 h until mid-log phase. To induce a MAT DSB, galactose was added to 2% and cells were harvested at time points after addition.
Cell cycle analysis
Fluorescence activated cell sorting (FACS) was performed as previously described (47). Briefly, ∼0.6 O.D.600 nm units of mid-log phase cells were fixed in three volumes of ethanol, followed by storage at −20°C for up to 1 month. Before staining, ∼0.2 O.D.600 nm units of cells were removed and suspended in TE (10 mM Tris, 1 mM EDTA, pH 8.0) with 100 μg/ml RNAse A and incubated at 37°C overnight, followed by incubation in pepsin solution (0.48% HCl, 5% pepsin in TE) for 30 min. After suspension in Sybr Green solution (0.25% NP-40, 0.02% SYBR-Green I in TE) for 24–36 h at 4°C, cells were briefly sonicated before sorting on a FACScalibur machine using CellQuest Pro software (BD Biosciences). At least 30,000 events were collected for each sample. Data were exported to FlowJo software for gating, visualization and analysis.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously described, with minor modifications (48). Briefly, 30–50 O.D.600 nm units of mid-log phase cells were fixed with 1% formaldehyde for 30 min, quenched with 125 mM glycine for 5 min, washed with PBS, and stored at −80°C. Pellets were suspended in FA lysis buffer (50 mM HEPES–KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) supplemented with 1× protease inhibitor cocktail (Sigma P2714) and 1 mM phenylmethylsulfonyl fluoride (PMSF). 0.45 g of 425–600 μm glass beads (Sigma G8772) were added and cells were disrupted by vortexing for 17 min at 4°C (Scientific Industries SI-D248). Lysates were centrifuged and the soluble fraction was discarded. Chromatin was solubilized by sonicating the pellet on ice in FA lysis buffer supplemented with protease inhibitors using a Branson 250 sonifier with a microtip probe for six 10-s cycles on output 3, with a minimum one minute break between pulses. Sonicated lysates were centrifuged, and the clarified, sonicated chromatin fractions were removed, quantitated by Bradford assay with BSA standards (49), and stored at −80°C.
A specified amount of sonicated chromatin was diluted to 1 ml in FA lysis buffer containing protease inhibitors. A 5% input (INP) sample was removed, and then antibody was added before overnight incubation at 4°C (see Supplemental Table S2 for ChIP conditions). Protein A/G beads (40 μl) were then added and incubated for 2 h at 4°C (Santa Cruz Biotechnology sc-2003), followed by sequential washes with FA lysis buffer, FA lysis buffer plus high salt (500 mM NaCl), LiCl buffer (0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, in TE) and TE. Immunocomplexes (IP) were eluted from beads with 1% SDS in TE by incubating at 65°C for 15 min. IP and INP samples were incubated in Pronase solution (2 mg/ml pronase + 10 mM CaCl2) at 42°C for 2 h. Samples were then incubated at 65°C overnight to reverse crosslinks, and DNAs were purified using a QIAquick PCR Purification kit (Qiagen).
Quantitative real-time PCR
DNAs isolated for ChIP and end resection assays were analyzed by quantitative real-time PCR using a Step One Plus instrument (Bio-Rad). For each reaction, 5 μl of DNA (diluted 1:10 for IP and 1:50 for INP) was added to a 20 μl reaction mixture consisting of 0.75× Maxima Sybr Green Master Mix (Thermo Fisher Scientific) and 0.15 μM forward and reverse primers in nuclease-free water (see Supplementary Table S3 for primer sequences). Samples were performed in triplicate and relative quantitations were obtained by plotting cycle numbers against a standard curve generated from six serial 1:10 dilutions of genomic DNA (gDNA) prepared in the same manner as a ChIP INP sample, with the first dilution containing 2 ng/μl of gDNA as measured by a NanoDrop spectrophotometer (Thermo Fisher Scientific).
In general, signals at MAT loci were first normalized to the POL5 INP signal to adjust for DNA quantity, and then normalized for the fraction of cells containing a MAT DSB using primers that span the MAT HO cut site. For both ChIP and end resection experiments, signals at time points represent the fraction of the signal at T0 (pre-induction). There were two exceptions to this general rule. First, for Exo1-Myc ChIPs, which have low signal and are highly affected by background, the signals were further normalized to the POL5 IP signal to account for background fluctuations (50) (S.E. Lee, personal communication). Second, for the FLAG-H2B ChIPs, the POL5 IP signal was used to normalize for DNA quantity.
DNA end resection and long-range kinetics analysis
For end resection assays, either INP DNA from corresponding ChIP experiments was utilized or 15 μg of sonicated chromatin was freshly prepared. Resection kinetics were analyzed as previously described (42). Briefly, the time required for 25% resection to occur (0.75 fraction intact) at locations to the right of the MAT DSB was interpolated by assuming a linear relationship between time points from the resection graphs. The times to 25% resection were then plotted against the distances from the MAT DSB, with distance on the y-axis and time on the x-axis, and the slope of the line was obtained by linear regression, yielding the resection rate (Δy/Δx = Δdistance/Δtime, or kb/h). A minimum of three loci were used for regression analysis.
Western blot analysis
Trichloroacetic acid (TCA) lysates were prepared as previously described (48). Briefly, 10 O.D.600 nm units of mid-log phase cells were collected and washed with 20% TCA, and pellets were stored at −80°C. Pellets were suspended in 20% TCA, 0.5 g of 425–600 μm glass beads (Sigma G8772) were added, and lysates were prepared by vortexing with a Turbo Vortexer (Scientific Industries SI-D248) for 15 min at 4°C. Lysates were incubated for 10 min on ice and precipitated proteins were collected by centrifugation. Pellets were suspended in Laemmli Buffer (5% SDS, 10% glycerol, 0.25 M unbuffered Tris, 0.01% Bromophenol Blue), boiled for 5 min, centrifuged, and the soluble fraction was removed for standard SDS-PAGE. Primary antibodies used were Rabbit α-Rad53 1:2000 (Abcam ab104232), Rabbit α-Mre11 1:5000 (Patrick Sung), Rabbit α-Hdf1 1:5000 (Alan Tomkinson) and Mouse α-beta Actin 1:20 000 (Abcam ab8224). Immunoblots were analyzed by probing with HRP-conjugated secondary antibodies (Bio-Rad), incubating with Clarity ECL substrate reagent per manufacturers instructions (Bio-Rad), and exposing to X-ray film (Phoenix Research Products), which was then developed with a Konica SRC-101 developer. Films were digitized, and band densitometry was performed using Quantity One software (Bio-Rad) with one pixel local background subtraction.
RNA expression analysis
Total RNA was harvested from exponentially growing cells in YPD using a hot phenol method as previously described (51). For cDNA synthesis, RNA was first treated with RQ1 RNase-free DNase to remove residual genomic DNA (Promega), and cDNA was synthesized using the SuperScript III First-Strand Synthesis System with Oligo(dT)20 primers (Invitrogen). Quantitative real-time PCR was performed as described above with gene specific primers, and signals were normalized to ACT1 to adjust for RNA quantity (see Supplemental Table S3 for primer sequences).
Statistical analysis
Data were expressed as mean ± one standard deviation. For comparison of one variable, a two-tailed unequal variance t-test was performed. For comparison of two variables, a two-way unequal variance ANOVA with Holm–Sidak post-hoc analysis was performed. All statistical analyses were performed using SigmaPlot software.
RESULTS
SWI/SNF facilitates the initiation of DNA end resection
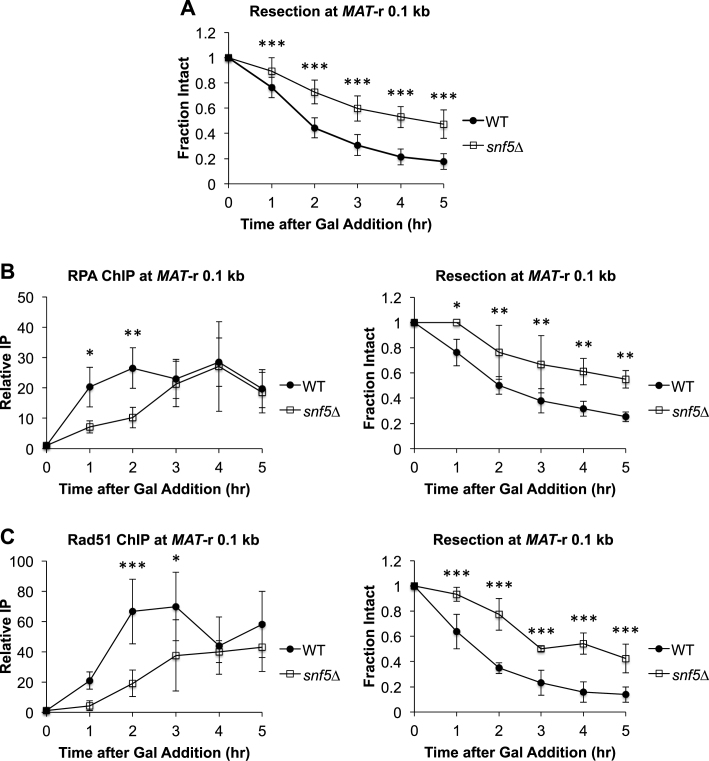
To investigate repair protein and end resection dynamics at DSBs, we utilized an established site-specific DSB assay (52). In this assay, addition of galactose to the medium leads to the rapid induction of the HO endonuclease, which introduces a single DSB within the yeast mating-type (MAT) locus. We used a strain in which the MAT homology regions HMLα and HMRa were also deleted, leading to a MAT DSB that can initiate but not complete HR (53). However, the initial events of recombinational repair, including repair factor recruitment and 5΄ to 3΄ DNA end resection, as well as DSB repair by NHEJ, can be monitored spatiotemporally (54–56). As previously shown, end resection initiated rapidly after DSB induction at MAT in wild-type (WT) cells (Figure 1A) (13,54,57). To address the role of the SWI/SNF complex in HR, we deleted SNF5, a core subunit that is required to form a fully functional and correctly targeted SWI/SNF complex (58–60) and found that there was a 1–2 h delay in the initiation of resection at MAT in the snf5Δ mutant strain, as well as a reduction in the extent of resection (Figure 1A; Supplementary Figure S1A–C). Consistent with the defect in the initiation of resection the snf5Δ mutant strain, there was also a delay in the recruitment of both the RPA single-stranded DNA binding protein (Figure 1B) and Rad51 recombinase (Figure 1C) to the DSB in snf5Δ cells, and diminished recruitment of these factors distal to the break (Supplementary Figure S1A–C).
Figure 1.
Initiation of DNA end resection at MAT is impaired in snf5Δ cells. OK (A) Asynchronous WT (n = 9) and snf5Δ (n = 10) cells were harvested at 1 h intervals after addition of galactose to induce a MAT DSB. Resection was monitored by qPCR with primers that anneal 0.1 kb to the right of the MAT DSB. (B) Recruitment of RPA 0.1 kb to the right of the MAT DSB was monitored by ChIP (left) and DNA end resection was simultaneously monitored (right) in WT (n = 3) and snf5Δ (n = 3) asynchronous cells after addition of galactose to induce a MAT DSB. (C) Recruitment of Rad51 0.1 kb to the right of the MAT DSB was monitored by ChIP (left) and DNA end resection was simultaneously monitored (right) in WT (n = 3) and snf5Δ (n = 3) asynchronous cell populations as described in B. Error bars denote one standard deviation. Statistical differences between WT and snf5Δ at time points were assessed by two-way ANOVA with Holm–Sidak post-hoc analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
While the results suggested a previously uncharacterized role for SWI/SNF in the initiation of DNA end resection, it was possible that this was an indirect effect. This prompted us to examine the impact of deleting SNF5 on cell cycle distribution, as this influences the expression and activity of multiple components of the resection machinery (26) and the efficiency by which the MAT DSB itself is formed. We found that an asynchronous population of snf5Δ cells contained ∼20% more G2/M phase cells than WT cells, and that both WT and snf5Δ cell populations demonstrated a shift from G1 phase to G2/M phase after addition of galactose to the pre-induction medium (Supplementary Figure S2A and B). Thus, the resection impairment in snf5Δ is not due to an accumulation of cells in G1 phase. While there was rapid cleavage at MAT in WT cells after addition of galactose, MAT cleavage was less efficient in snf5Δ cells (Supplementary Figure S3A). Because either reduced transcription of GAL-HO in snf5Δ cells or increased nucleosome occupancy at the MAT HO cut site could account for this phenotype, we explored both possibilities (56). In agreement with the known role of SWI/SNF in facilitating the transcription of inducible genes, including GAL10, SUC2 and HO (61,62), transcription of GAL-HO was modestly impaired in snf5Δ cells after addition of galactose to the medium (Supplementary Figure S3B). In contrast, no difference was observed in nucleosome occupancy at the MAT HO cut site, consistent with previous nucleosome mapping around the MAT DSB in snf5Δ cells (37) (Supplementary Figure S3C). To account for the decrease in MAT cleavage in snf5Δ cells, all data presented, including end resection and repair factor recruitment, have been adjusted for the fraction of cells containing a MAT DSB as described in Materials and Methods.
SWI/SNF controls MRX recruitment to DSB ends
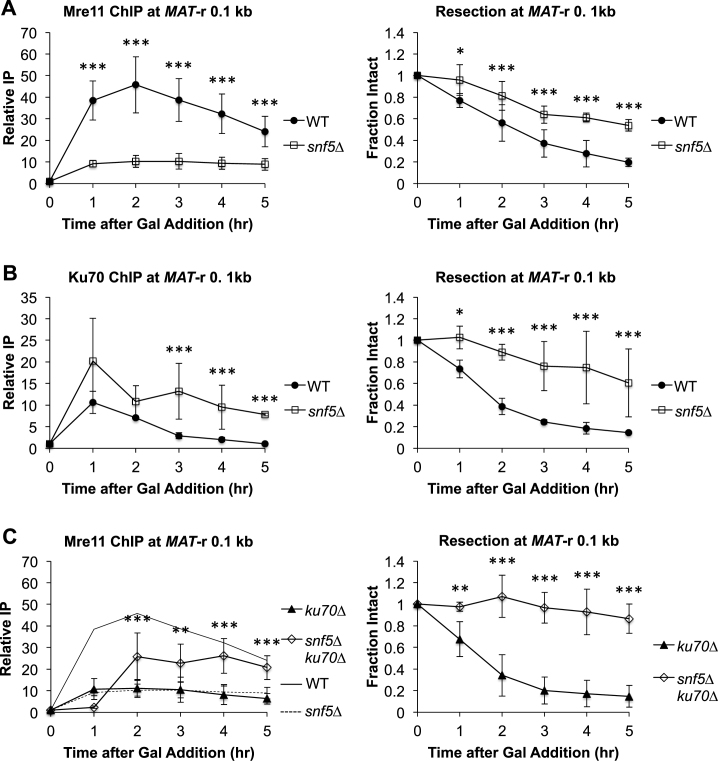
The observation that snf5Δ cells had a defect in the initiation of DNA end resection led us to investigate the upstream events in DSB repair that determine pathway choice between HR and NHEJ. Two complexes compete for DSB ends: Mre11/Rad50/Xrs2 (MRX; Mre11/Rad50/Nbs1, MRN in mammals) and Ku70/Ku80 (KU). When MRX binding predominates, generally in G2/M phase, Sae2 (CtIP in mammals) stimulates the endonucleolytic activity of MRX to initiate the first phase of DNA end resection (63). Furthermore, the activity of MRX contributes to KU dissociation from the DSB, facilitating further end resection (55). When KU binding predominates, generally in G1 phase, it blocks DNA end resection and recruits accessory NHEJ factors, including the MRX complex, which in this context acts as an end-bridging factor rather than as a nuclease (10,64,65).
We examined the recruitment of these pathway-regulating complexes to DSB ends in both WT and snf5Δ cells after DSB induction. While there was an ∼4.5-fold decrease in Mre11 binding to the MAT DSB in snf5Δ cells (Figure 2A), Ku70 was recruited at higher levels and retained longer at the break in the mutant (Figure 2B). Complementing the snf5Δ strain with a plasmid bearing a wild-type SNF5 gene completely rescued the DSB induction, MRX binding and DNA end resection phenotypes of snf5Δ, demonstrating that the results were not due to the presence of another mutation in this strain (Supplementary Figure S4A–C). Furthermore, the snf5Δ mutation did not alter the expression of genes encoding DSB repair factors or MRX or KU components (66) (Supplementary Figure S5A and B). Together, these data argue that SWI/SNF has a direct role in recruiting MRX to DSB ends.
Figure 2.
SWI/SNF regulates recruitment of MRX to a MAT DSB. (A) Asynchronous WT (n = 7) and snf5Δ (n = 6) cells were harvested at 1 h intervals after addition of galactose to induce a MAT DSB. Recruitment of Mre11 0.1 kb to the right of the MAT DSB was monitored by ChIP (left) and DNA end resection was simultaneously monitored (right). (B) Asynchronous WT (n = 3) and snf5Δ (n = 3) cells were harvested after addition of galactose. Recruitment of Ku70 (left) and DNA end resection (right) were monitored as in A. (C) Asynchronous ku70Δ (n = 5) and snf5Δku70Δ (n = 3) cells were harvested after addition of galactose. Recruitment of Mre11 (left) and DNA end resection (right) were monitored as in A. WT (solid line) and snf5Δ (dashed line) Mre11 ChIP recruitment data from A are overlaid on the graph. Error bars denote one standard deviation. Statistical differences between strains at time points were assessed by two-way ANOVA with Holm–Sidak post-hoc analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
We initially considered a model in which MRX and KU directly compete for binding to DSB ends and that SWI/SNF promotes the binding of MRX over KU. In support of this model, deletion of KU70 in snf5Δ cells partially rescued MRX recruitment to the MAT DSB (Figure 2C). However, we noted that snf5Δku70Δ cells exhibited reduced growth, increased accumulation in G1 phase, and a failure to enter S phase after DSB induction (Supplementary Figure S6A). These phenotypes were accompanied by a lack of end resection (Figure 2C, right panel) and increased impairment in DSB induction compared to the snf5Δ single mutant (Supplemental Figure S6B). Thus, we speculate that the partial rescue of MRX recruitment in the snf5Δku70Δ double mutant may represent the presence of stalled, inactive MRX that is unable to initiate end resection. Moreover, MRX recruitment was significantly decreased in ku70Δ cells (Figure 2C, ku70Δ), as others have also shown (55,67). Thus, as discussed in more detail below, a paradigm of simple competition for ends does not account for the known roles of MRX in both HR and NHEJ. Instead, our data are consistent with an alternative model in which there are HR-active and NHEJ-active pools of MRX at DNA ends (55), with SWI/SNF promoting the association of the pool of HR-active MRX.
Activation of the DNA damage response is impaired in snf5Δ cells
The DNA damage response (DDR) can be initiated by both the yeast ATR homolog Mec1, which is activated by pathologic ssDNA generation, and the yeast ATM homolog Tel1, which associates with damage-recruited MRX (68). As we observed both impaired DNA end resection and reduced MRX recruitment to a MAT DSB in snf5Δ cells, we investigated whether the DDR response was also altered in this mutant. We found that DDR activation, as measured by phosphorylation of Rad53, was abolished in snf5Δ (Supplementary Figure 7). Interestingly, the magnitude of the defect in Rad53 phosphorylation was greater than the magnitude of the defects in initiation of end resection and MRX recruitment (compare Figures 1A and 2A to Supplementary Figure S7). This could be due to the recently discovered non-chromatin role for the SWI/SNF ATPase subunit, Snf2, in activating the Mec1 kinase (69). Thus, in snf5Δ cells, both the signaling events for DDR activation and Mec1-mediated signal transduction are impaired.
Long-range resection is delayed in snf5Δ cells and relies upon Exo1
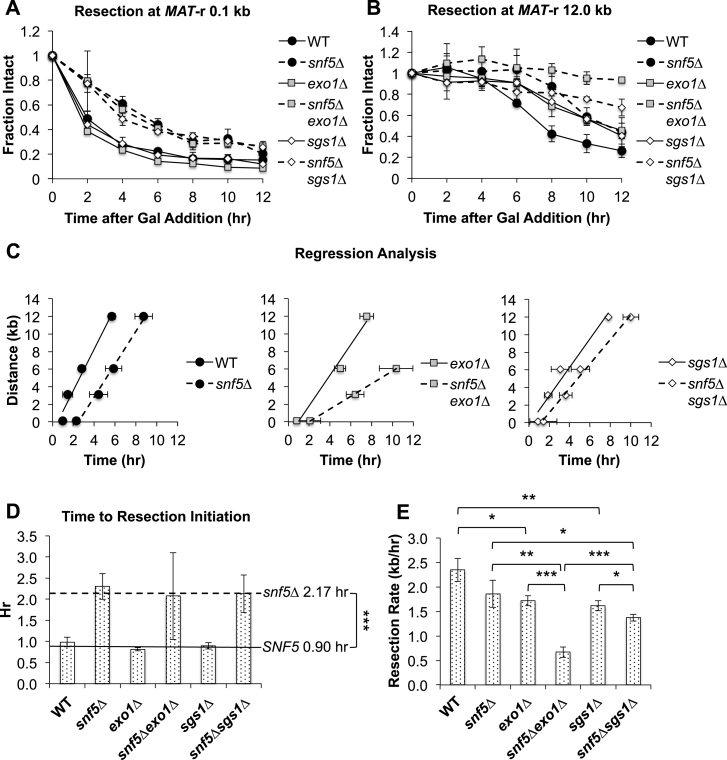
DNA resection during HR is a multistep process consisting of an initial resection phase mediated by MRX-Sae2 (MRN-CtIP in mammals) that generates short, 3΄ ssDNA overhangs, and a subsequent long-range resection phase mediated by either Exo1 or the nuclease/helicase complex Dna2-Sgs1/Top3/Rmi1 (Dna2–STR) (11,13–15). Our observation that snf5Δ cells had defects in both initial DNA resection and MRX recruitment to the MAT DSB led to the prediction that long-range resection would also be impaired. To address the role of SWI/SNF in the long-range resection pathways, we measured resection at distal locations from the MAT DSB in WT and snf5Δ cells and examined the effect of exo1Δ and sgs1Δ mutations in both WT and snf5Δ backgrounds on long-range resection after MAT DSB induction. While initiation of resection was delayed in snf5Δ compared to WT cells (Figure 3A; Supplementary Figure S8A, left panel), resection initiated at the same time in exo1Δ and sgs1Δ single mutants as in WT cells (Figure 3A). Furthermore, there was no further delay in the initiation of resection in a snf5Δ exo1Δ or snf5Δsgs1Δ double mutant compared to snf5Δ cells (Figure 3A; Supplementary Figure S8B, C, left panels), confirming that Exo1 and Dna2–STR do not participate in the initiation of DNA end resection (13).
Figure 3.
Long-range resection is delayed in snf5Δ cells and relies upon Exo1. Asynchronous WT, snf5Δ, exo1Δ, snf5Δexo1Δ, sgs1Δ and snf5Δsgs1Δ cells (n = 3) were harvested at 2 h intervals after addition of galactose to induce a MAT DSB. Resection was monitored by qPCR with primers annealing either 0.1 kb (A) or 12.0 kb (B) to the right of the MAT DSB. (C) The time for 25% resection to occur (0.75 fraction intact) at positions to the right of the MAT DSB. (D) The time for 25% resection to occur immediately adjacent to the MAT was designated as the time to resection initiation, and (E) resection rates were calculated by determining the slopes of the graphs by linear regression analysis. Error bars denote one standard deviation. Statistical comparisons between strains were assessed by Student's t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
In contrast to the results proximal to the MAT DSB, both exo1Δ and sgs1Δ mutants demonstrated impaired long-range resection to the right of the break (Figure 3B; Supplementary Figure S8B and C, middle and right panels). Although long-range resection was delayed in snf5Δ cells, presumably because of the defect in resection initiation, the pattern of resection was otherwise similar to that seen in WT cells (Figure 3B; Supplementary Figure S8A, middle and right panels). Interestingly, long-range resection was only modestly decreased in snf5Δ sgs1Δ cells, but was completely abolished 12 kb to the right of the break in a snf5Δexo1Δ double mutant (Figure 3B; Supplementary Figure S8B and C).
To gain further insight into the characteristics of long-range resection in these strains, we interpolated the time to 25% resection (0.75 fraction intact by qPCR) at various distances to the right of the MAT DSB (42) (Figure 3C). By using the time to 25% resection immediately next to the break as a measure of resection initiation, we found that initiation was delayed by ∼1.3 h in all snf5Δ strains compared to SNF5 cells, and that timely initiation depended only on SWI/SNF and not Exo1 or Dna2–STR (Figure 3D). Using linear regression analysis to obtain resection rates (42) (Materials and Methods), we found that there was no significant difference in the resection rate between WT and snf5Δ cells (Figure 3E). Thus, these data provide strong evidence that the resection phenotype of snf5Δ is characterized by a delay in initiation but does not involve a significant reduction in resection velocity. The rate of resection was reduced by ∼25% in either exo1Δ or sgs1Δ cells, demonstrating that either pathway can largely compensate for the loss of the other (Figure 3E). There was also a ∼25% decrease in resection rate in a snf5Δsgs1Δ double mutant compared to a snf5Δ single mutant, suggesting that Dna2–STR has a relatively minor role in long-range resection in the absence of SWI/SNF and/or the Exo1 pathway can still compensate for the loss of Dna2–STR in the absence of SNF5 (Figure 3E). In contrast, there was a ∼65% decrease in resection rate in a snf5Δexo1Δ double mutant compared to a snf5Δ mutant (Figure 3E). These observations suggested that SWI/SNF is critical for regulating the association of Dna2–STR with the MAT DSB, as snf5Δ cells were disproportionately reliant upon Exo1 to accomplish long-range resection in the absence of functioning SWI/SNF.
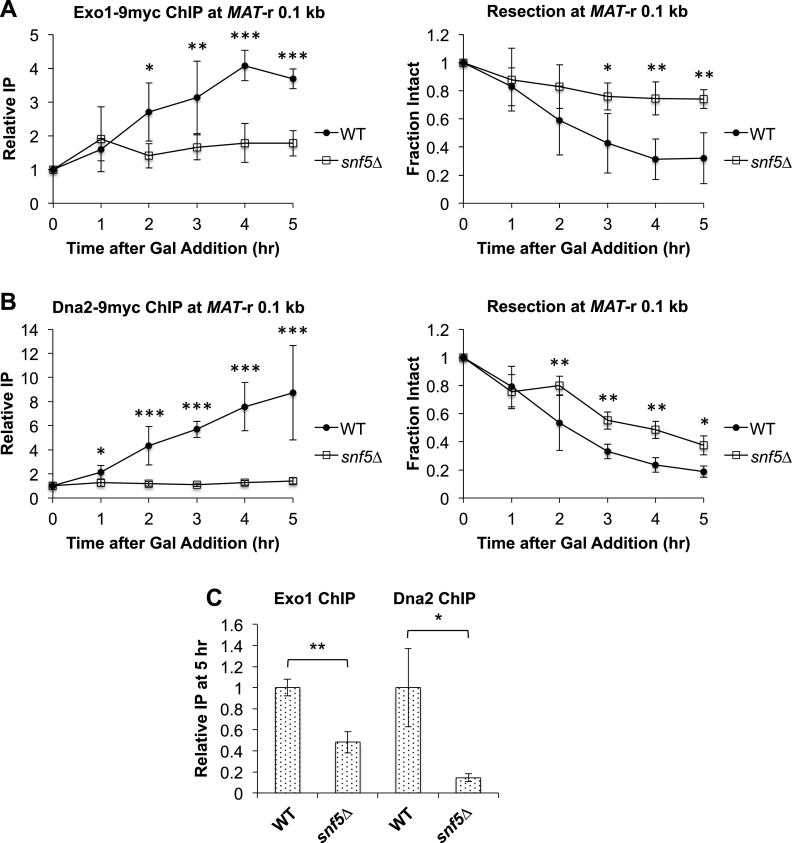
To address the relationship between SWI/SNF and the Exo1 and Dna2–STR pathways, we examined the effect of snf5Δ on the recruitment of Exo1-Myc and Dna2-Myc to the MAT DSB (Figure 4), and found that the recruitment of both factors was impaired in snf5Δ cells compared to WT (Figure 4A and B). However, while maximal Exo1 recruitment was decreased by ∼50%, maximal Dna2 recruitment was decreased by ∼90% in snf5Δ cells (Figure 4C). This suggested that Exo1 has a decreased reliance on SWI/SNF to load onto breaks compared to Dna2–STR (Figure 4C). Together, these results demonstrated that SWI/SNF acts early in the long-range resection process to coordinate the timely initiation of resection and subsequent loading of long-range resection factors, and that in the absence of functional SWI/SNF, Exo1-mediated resection constitutes the primary pathway.
Figure 4.
Recruitment of Exo1 and Dna2 to a MAT DSB is impaired in snf5Δ cells. Asynchronous WT (n = 3) and snf5Δ (n = 3) strains containing Exo1-Myc or Dna2-Myc were harvested at 1 h intervals after addition of galactose to induce a MAT DSB. Recruitment of (A) Exo1-Myc or (B) Dna2-Myc 0.1 kb to the right of the MAT DSB was monitored by ChIP (left) and DNA end resection was simultaneously monitored (right). Error bars denote one standard deviation. Statistical differences between strains at time points were assessed by two-way ANOVA with Holm–Sidak post-hoc analysis. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Recruitment of Exo1-Myc and Dna2-Myc at 5 h in snf5Δ relative to recruitment in WT, which was set as 1. Error bars denote one standard deviation. Statistical differences between recruitment at 5 h in WT and snf5Δ cells were assessed by Student's t-test. *P < 0.05, **P < 0.01.
Nucleosome eviction is delayed in snf5Δ cells
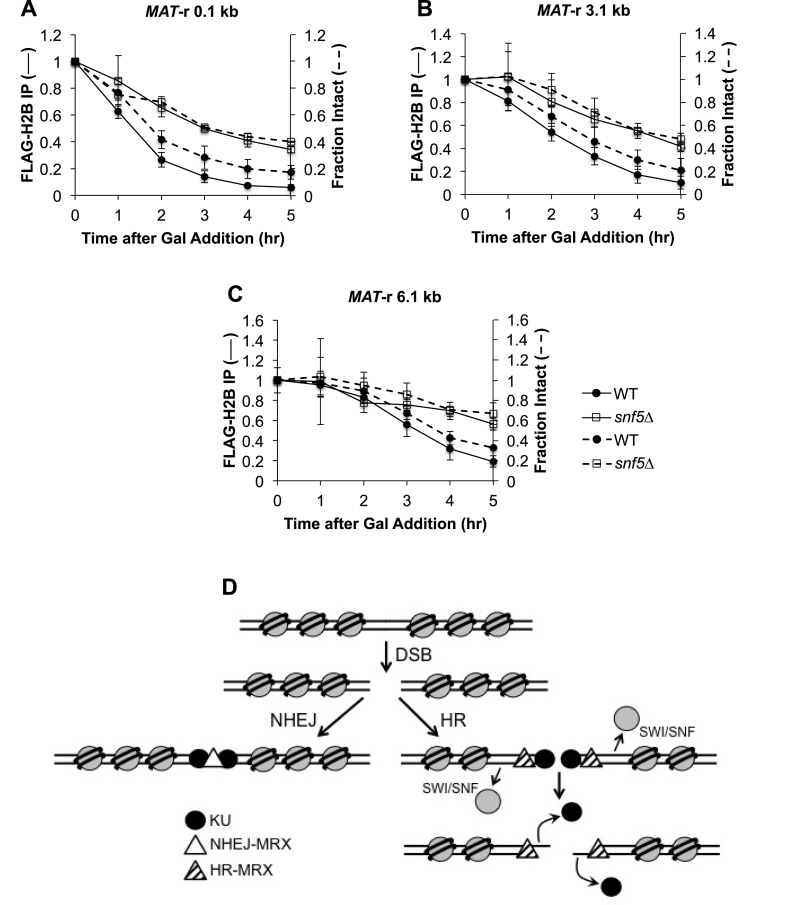
After a DSB is formed at MAT, nucleosomes are rapidly evicted for many kilobases on both sides of the break, a process postulated to facilitate the recruitment of the Rad51 recombinase and therefore later steps in HR (40). As SWI/SNF has nucleosome eviction activity and is recruited to a MAT DSB (45,70,71), we examined whether nucleosome displacement was impaired at a MAT DSB in snf5Δ cells, thereby accounting for the reduced recruitment of MRX and the delay in end resection and Rad51 recruitment at the MAT DSB. We monitored nucleosome eviction by FLAG-H2B ChIP at several positions near the MAT locus after DSB induction in WT and snf5Δ cells, and compared nucleosome occupancy to end resection (Figure 5A–C). In WT cells, both FLAG-H2B eviction and DNA end resection initiated within 1 h after formation of the MAT DSB and extended rapidly to more distal positions, whereas FLAG-H2B eviction and end resection were concurrently delayed in snf5Δ cells by 1–2 h. The close relationship between nucleosome eviction and end resection in both WT and snf5Δ cells strongly suggested that the two processes are linked. Interestingly, nucleosome eviction appeared to slightly precede end resection in WT but not snf5Δ cells (Figure 5A–C), although the difference was trending and not significant (Supplementary Figure S9). Taken together, our results argue that nucleosome eviction and DNA end resection during HR are tightly linked, and that SWI/SNF contributes a significant role to the timely initiation of both processes.
Figure 5.
Nucleosome eviction at MAT is delayed in snf5Δ cells. Asynchronous WT (n = 4) and snf5Δ (n = 3) cells containing FLAG-H2B were harvested at 1 h intervals after addition of galactose to induce a MAT DSB. H2B eviction was monitored by ChIP (solid lines) using qPCR with primers that anneal (A) 0.1 kb; (B) 3.1 kb and (C) 6.1 kb to the right of the DSB, and resection was simultaneously monitored (dashed lines). Error bars denote one standard deviation. (D) Model for role of SWI/SNF in the initiation of HR repair. Only initial events in the repair of a DSB by NHEJ or HR are shown. After a DSB is formed, KU rapidly associates with broken ends and recruits Dnl4-Lif1, leading to the recruitment of a pool of NHEJ-MRX that tethers broken ends and stimulates end ligation that is essential for repair by NHEJ (left panel). Recruitment of SWI/SNF to a DSB promotes nucleosome eviction in the vicinity of the break, leading to the recruitment or stabilization of a distinct pool of HR-active MRX (right panel). The nuclease activity of MRX promotes the initiation of end resection, which leads to the displacement of KU. Long-range resection factors, Exo1 and Dna2–STR, are then recruited, the ssDNA overhang is coated with RPA, and the DNA damage checkpoint is activated. RPA is replaced with the Rad51 recombinase and the nucleoprotein filament initiates homology search for HR repair.
DISCUSSION
In this study, we have identified a novel role for the SWI/SNF ATP-dependent nucleosome remodeler in facilitating early events during DNA DSB repair by HR in S. cerevisiae. Previous research demonstrated that SWI/SNF is recruited to a MAT DSB in yeast where it is functions at or just preceding strand invasion during mating-type switching, a prototypical gene conversion event (45). In the absence of functional SWI/SNF, single-strand annealing (SSA), an HR subtype in which two homologous sequences are annealed after extensive end resection at a DSB (72), is also reduced (45). These phenotypes suggested a possible role for the remodeling complex in DNA end resection, a prerequisite for HR/SSA. The present study demonstrates that the initiation of DNA end resection is significantly delayed in a snf5Δ mutant, thereby identifying an early role for the SWI/SNF complex in HR. This role appears to be mediated through the recruitment and/or stabilization of a distinct pool of MRX to DSBs to promote HR, and is related to the function of SWI/SNF in nucleosome eviction, as outlined in the model in Figure 5D.
In the absence of functional SWI/SNF, there was an ∼1.3 h delay in the resection of DNA just proximal to a MAT DSB, but once initiated, long-range resection occurred with similar kinetics as in WT cells. While SWI/SNF is recruited late to a MAT DSB and is detectable only ∼0.75 h after DSB induction (45), our results demonstrated that resection initiated at ∼0.9 h post-break induction in WT cells. Thus, DNA end resection begins shortly after detectable SWI/SNF recruitment. In contrast, the initiation of long-range resection in snf5Δ cells did not occur until >2 h after DSB induction. This suggests that SWI/SNF acts rapidly upon its recruitment to orchestrate the successful initiation of end resection. In contrast, the Fun30 nucleosome remodeler is not required for efficient resection initiation but plays an important role in long-range resection by removing the inhibitory Rad9 checkpoint protein from nucleosomes (42–44).
An intricate choreography of events occurs at DSBs to determine the choice between the NHEJ and HR pathways, including a complex relationship between the pathway-regulating MRX/MRN and KU complexes. KU binds to dsDNA with high affinity (73) and recruits Dnl4-Lif1 to form a DNA–protein complex that recruits MRX (67). The NHEJ-specific functions of MRX include tethering broken ends to maintain their intermolecular proximity, stimulating end ligation by Dnl4-Lif1, and providing a key interaction between Xrs2 and Lif1 that is essential for NHEJ (65,74). In contrast, during G2/M phases, the MRX complex is able to evict KU from DSB ends and initiate 5΄ to 3΄ end resection in conjunction with Sae2/CtIP, creating a ssDNA substrate that is not amenable to classical end joining and requires HR for repair (14,55,75). Thus, MRX has both NHEJ-active and HR-active roles with non-overlapping and opposing activities, leading to the proposal that there are two different modes of MRX recruitment to DNA DSBs that lead to its distinct activities (55,67,76).
Our study demonstrated that SWI/SNF is required for the efficient recruitment of MRX to a MAT DSB. In contrast, the association of KU with the MAT DSB was not affected in the snf5Δ mutant, and the retention of KU over time was significantly increased. Previous research found that NHEJ, as measured by a plasmid-based end-joining assay, is intact in both snf5Δ and snf2Δ mutants, while HR/SSA is defective (45). We therefore suggest that SWI/SNF is specifically required for the recruitment or stabilization of a distinct pool of MRX that is active in HR. MRX has multiple roles in HR, including initiation of DNA end resection (14,15), recruitment of long-range resection factors (50), and activation of the DNA damage response (36,74). In support of a role for SWI/SNF in regulating HR-active MRX functions, our data showed that in a snf5Δ mutant the initiation of DNA end resection was delayed, recruitment of long-range resection factors was reduced, and the DNA damage response was significantly impaired. Thus, all the known functions of HR-active MRX at a DSB are either lost or greatly diminished in the absence of SWI/SNF.
Another nucleosome remodeler, the RSC complex, has also been demonstrated to regulate the association of MRX with a MAT DSB. However, unlike SWI/SNF, RSC acts very early after break induction and facilitates the recruitment of both KU and MRX (38). RSC catalyzes the sliding of nucleosomes proximal to the break to create a stretch of nucleosome-free DNA that facilitates the recruitment of factors for both HR and NHEJ (37,38,77). Also, unlike SWI/SNF mutants, RSC mutants have defects in both NHEJ and HR (38,45,77,78). Moreover, there appears to be functional differences between RSC isoforms, and contrasting results have been obtained with plasmid versus chromosomal end joining assays in RSC mutants (38,45,77,78). A general picture that emerges from these combined studies supports a model in which RSC works rapidly after DSB induction to create a chromatin microenvironment that is generally conducive for DSB repair, while SWI/SNF acts more specifically to promote HR through recruiting or stabilizing an HR-active pool of MRX.
Although the absence of functional SWI/SNF does not affect long-range resection kinetics, a surprising finding was that snf5Δ cells depend on Exo1 rather than Dna2–STR for long-range resection. Previous in vitro studies demonstrated that Dna2–STR more readily processes nucleosomal templates than Exo1 because of the ability of Sgs1 to unwind DNA from nucleosomes (41). In addition, Dna2–STR can also compensate for the loss of the nuclease activity of MRX in DSB repair (79), thus making it a logical candidate to substitute for the loss of HR-active MRX in the snf5Δ strain. However, this phenotype may be explained by the observation that Dna2 recruitment is almost completely abolished in snf5Δ cells, similar to the loss of Dna2 recruitment to a MAT DSB in rad50Δ and mre11Δ strains (50). It is likely, therefore, that Exo1 has a greater ability to load onto a DSB in the absence of MRX than Dna2–STR.
The increased binding of KU to DSB ends and the reliance on Exo1 for long-range resection in snf5Δ cells is paradoxical since KU binding is known to block Exo1 activity (20,50). Previous data have demonstrated that KU association with DSBs in vivo is dynamic (67). Together with the recent discovery that phosphorylation of KU reduces its affinity for DNA ends in vivo and increases accessibility of DSB ends to Exo1 in vitro, it appears that Exo1 could initiate processing of DSB ends when KU transiently dissociates (22). Alternatively, the successive cycles of end joining and HO cleavage at MAT may create the opportunity for Exo1 to occasionally process the breaks before KU can associate. Either way, once the minimum amount of resection has occurred to recruit RPA and nucleate a Rad51 nucleofilament, KU binding will be repelled (80) and multiple chromatin remodelers will be recruited by Rad51 (71) to assist in processing the chromatin landscape to allow Exo1 to proceed with resection.
Nucleosome eviction is a conserved activity during the repair of DSBs in both yeast and mammals (40,81,82). An open question in the field is the identity of the factors that evict nucleosomes from DNA during HR in vivo. Previous research demonstrated that both MRX and the INO80 ATP-dependent nucleosome remodeler contribute to the removal of nucleosomes near a MAT DSB, although eviction eventually occurs after a delay (40,83). Our data showed that nucleosome eviction was delayed in snf5Δ cells in a manner that temporally paralleled the delay in resection initiation in this mutant, suggesting that these events are coupled. In both WT and snf5Δ cells, FLAG-H2B eviction and DNA end resection occurred at approximately the same time. This supports the view that that SWI/SNF increases the efficiency of nucleosome eviction, and that eviction is a prerequisite for the efficient recruitment of HR dependent MRX and resection initiation. Thus, there may be redundant nucleosome eviction pathways during HR that are mediated by different factors, including ATP-dependent nucleosome remodelers as well as helicases such as Sgs1, all of which co-operate to ensure the generation of nucleosome-free, recombinogenic ssDNA that can be efficiently coated by Rad51.
It is important to note that while the data were normalized for MAT DSB efficiency, the late induction of DSBs in snf5Δ cells may partially account for the observed resection delay at the MAT DSB. However, the combination of phenotypes that supersede the moderate delay in DSB induction, including highly reduced MRX recruitment, greatly impaired DDR activation, and near abolishment of Dna2 recruitment in snf5Δ cells, all support a role for SWI/SNF in DNA end resection and recruitment of MRX for its activities in HR. Additionally, we noted that when observing resection over a 12-h time course, a highly significant difference in resection between WT and snf5Δ cells was maintained for at least 10 h, which is far longer than the delay in induction, further supporting a role for SWI/SNF in the initiation of DNA end resection. Future studies utilizing an inducible DSB system that is independent of SWI/SNF for induction could assist in clarifying the exact temporal sequence of events in SWI/SNF mutants.
In conclusion, we have demonstrated that SWI/SNF plays a role in the initiation of end resection at the MAT DSB in yeast and is critical for the robust recruitment of MRX to broken ends. The small pool of MRX that is recruited in snf5Δ cells appears to lack the well-known functions of MRX in HR, including resection initiation, recruitment of the long-range resection machinery, and activation of the DDR, suggesting that SWI/SNF orchestrates the recruitment and/or stabilization of an HR-active pool of MRX to DNA DSBs that has distinct activities compared to NHEJ-active MRX. Furthermore, we suggest that this role of SWI/SNF is mediated through its activity in nucleosome eviction at a DSB. SWI/SNF is an important tumor suppressor that is mutated in ∼20% of human malignancies (84,85), and recent studies have shown that mammalian SWI/SNF is recruited to DSBs, where it contributes important roles in activating the DNA damage response and recruiting repair proteins to damaged DNA (86–88). Furthermore, SWI/SNF contributes to end resection and HR in mammalian cells (89). Thus, we believe that our study sheds light on important and conserved roles for the SWI/SNF complex in maintaining genomic and epigenomic stability during DNA double-strand break repair.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Wolf-Dietrich Heyer, Michael Lichten, Philippe Prochasson, and Patrick Sung for their generous gifts of antibodies, plasmids and strains, and Cory Hillyer for her expert technical assistance. Toyoko Tsukuda is gratefully acknowledged for her important insights and advice during this project.
Footnotes
Present addresses:
Scott Houghtaling, Center for Developmental Biology and Regenerative Medicine, Seattle Children's Research Institute, Seattle, WA 98101, USA.
Joseph Sanchez, Molecular and Cellular Biology Program, University of Washington, Seattle, WA 98195, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM40118 to M.A.O. and GM47251 to A.T.]; UNM Cancer Center core grant from the National Cancer Institute [NIH/NCI 2 P30 CA118100]; National Institutes of Health Initiative to Support Student Development grant [GM060201]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mladenov E., Magin S., Soni A., Iliakis G.. Seminars in cancer biology. Semin. Cancer Biol. 2016; 37-38:51–64. [DOI] [PubMed] [Google Scholar]
- 2. Mehta A., Haber J.E.. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harbor Perspect. Biol. 2014; 6:a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bétermier M., Bertrand P., Lopez B.S.. Is non-homologous end-joining really an inherently error-prone process?. PLoS Genet. 2014; 10:e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jasin M., Rothstein R.. Repair of strand breaks by homologous recombination. Cold Spring Harbor Perspect. Biol. 2013; 5:a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sonoda E., Sasaki M.S., Buerstedde J.M., Bezzubova O., Shinohara A., Ogawa H., Takata M., Yamaguchi-Iwai Y., Takeda S.. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998; 17:598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saleh-Gohari N., Bryant H.E., Schultz N., Parker K.M., Cassel T.N., Helleday T.. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 2005; 25:7158–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Symington L.S., Gautier J.. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011; 45:247–271. [DOI] [PubMed] [Google Scholar]
- 8. Cejka P. DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem. 2015; 290:22931–22938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lieber M.R. The Mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis A.J., Chen D.J.. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res. 2013; 2:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clerici M., Mantiero D., Lucchini G., Longhese M.P.. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005; 280:38631–38638. [DOI] [PubMed] [Google Scholar]
- 12. Rathmell W.K., Chu G.. A DNA end-binding factor involved in double-strand break repair and V(D)J recombination. Mol. Cell. Biol. 1994; 14:4741–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu Z., Chung W.-H., Shim E.Y., Lee S.E., Ira G.. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008; 134:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcia V., Phelps S.E.L., Gray S., Neale M.J.. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2012; 479:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daley J.M., Niu H., Miller A.S., Sung P.. Biochemical mechanism of DSB end resection and its regulation. DNA Repair. 2015; 32:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhong S., Chen X., Zhu X., Dziegielewska B., Bachman K.E., Ellenberger T., Ballin J.D., Wilson G.M., Tomkinson A.E., MacKerell A.D.. Identification and validation of human DNA ligase inhibitors using computer-aided drug design. J. Med. Chem. 2008; 51:4553–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daley J.M., Chiba T., Xue X., Niu H., Sung P.. Multifaceted role of the Topo III -RMI1-RMI2 complex and DNA2 in the BLM-dependent pathway of DNA break end resection. Nucleic Acids Res. 2014; 42:11083–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X., Haber J.E.. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004; 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hustedt N., Durocher D.. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2017; 19:1–9. [DOI] [PubMed] [Google Scholar]
- 20. Mimitou E.P., Symington L.S.. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010; 29:3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bunting S.F., Callén E., Wong N., Chen H.-T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010; 141:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee K.-J., Saha J., Sun J., Fattah K.R., Wang S.-C., Jakob B., Chi L., Wang S.-Y., Taucher-Scholz G., Davis A.J. et al. Phosphorylation of Ku dictates DNA double-strand break (DSB) repair pathway choice in S phase. Nucleic Acids Res. 2015; doi:10.1093/nar/gkv1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferrari M., Dibitetto D., De Gregorio G., Eapen V.V., Rawal C.C., Lazzaro F., Tsabar M., Marini F., Haber J.E., Pellicioli A.. Functional interplay between the 53BP1-Ortholog Rad9 and the Mre11 complex regulates resection, end-tethering and repair of a double-strand break. PLoS Genet. 2015; 11:e1004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huertas P., Cortés-Ledesma F., Sartori A.A., Aguilera A., Jackson S.P.. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008; 455:689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cannavo E., Cejka P.. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature. 2014; 514:122–125. [DOI] [PubMed] [Google Scholar]
- 26. Symington L.S. Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2016; 51:195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Postow L., Ghenoiu C., Woo E.M., Krutchinsky A.N., Chait B.T., Funabiki H.. Ku80 removal from DNA through double strand break–induced ubiquitylation. J. Cell Biol. 2008; 182:467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown J.S., Lukashchuk N., Sczaniecka-Clift M., Britton S., le Sage C., Calsou P., Beli P., Galanty Y., Jackson S.P.. Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites. Cell Rep. 2015; 11:704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Panier S., Boulton S.J.. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2013; 15:7–18. [DOI] [PubMed] [Google Scholar]
- 30. Chen H., Lisby M., Symington L.S.. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell. 2013; 50:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen X., Niu H., Yu Y., Wang J., Zhu S., Zhou J., Papusha A., Cui D., Pan X., Kwon Y. et al. Enrichment of Cdk1-cyclins at DNA double-strand breaks stimulates Fun30 phosphorylation and DNA end resection. Nucleic Acids Res. 2016; doi:10.1093/nar/gkv1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hunt C.R., Ramnarain D., Horikoshi N., Iyengar P., Pandita R.K., Shay J.W., Pandita T.K.. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat. Res. 2013; 179:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou C.Y., Johnson S.L., Gamarra N.I., Narlikar G.J.. Mechanisms of ATP-dependent chromatin remodeling motors. Annu. Rev. Biophys. 2016; 45:153–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Osley M.A., Tsukuda T., Nickoloff J.A.. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2007; 618:65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hargreaves D.C., Crabtree G.R.. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011; 21:396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsabar M., Haber J.E.. Chromatin modifications and chromatin remodeling during DNA repair in budding yeast. Curr. Opin. Genet. Dev. 2013; 23:166–173. [DOI] [PubMed] [Google Scholar]
- 37. Kent N.A., Chambers A.L., Downs J.A.. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J. Biol. Chem. 2007; 282:27693–27701. [DOI] [PubMed] [Google Scholar]
- 38. Shim E.Y., Hong S.J., Oum J.H., Yanez Y., Zhang Y., Lee S.E.. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 2007; 27:1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oum J.H., Seong C., Kwon Y., Ji J.H., Sid A., Ramakrishnan S., Ira G., Malkova A., Sung P., Lee S.E. et al. RSC facilitates Rad59-dependent homologous recombination between sister chromatids by promoting cohesin loading at DNA double-strand breaks. Mol. Cell. Biol. 2011; 31:3924–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsukuda T., Fleming A.B., Nickoloff J.A., Osley M.A.. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005; 438:379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adkins N.L., Niu H., Sung P., Peterson C.L.. Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 2013; 20:836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eapen V.V., Sugawara N., Tsabar M., Wu W.H., Haber J.E.. The saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol. Cell. Biol. 2012; 32:4727–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen X., Cui D., Papusha A., Zhang X., Chu C.-D., Tang J., Chen K., Pan X., Ira G.. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2013; 489:576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Costelloe T., Louge R., Tomimatsu N., Mukherjee B., Martini E., Khadaroo B., Dubois K., Wiegant W.W., Thierry A., Burma S. et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2013; 489:581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chai B. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005; 19:1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004; 21:947–962. [DOI] [PubMed] [Google Scholar]
- 47. Trujillo K.M., Osley M.A.. A role for H2B ubiquitylation in DNA replication. Mol. Cell. 2012; 48:734–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trujillo K.M., Tyler R.K., Ye C., Berger S.L., Osley M.A.. A genetic and molecular toolbox for analyzing histone ubiquitylation and sumoylation in yeast. Methods. 2011; 54:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976; 72:248–254. [DOI] [PubMed] [Google Scholar]
- 50. Shim E.Y., Chung W.-H., Nicolette M.L., Zhang Y., Davis M., Zhu Z., Paull T.T., Ira G., Lee S.E.. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010; 29:3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Collart M.A., Oliviero S.. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. 2001; doi:10.1002/0471142727.mb1312s23. [DOI] [PubMed] [Google Scholar]
- 52. Jensen R.E., Herskowitz I.. Directionality and regulation of cassette substitution in yeast. Cold Spring Harb. Symp. Quant. Biol. 1984; 49:97–104. [DOI] [PubMed] [Google Scholar]
- 53. Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E.. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998; 94:399–409. [DOI] [PubMed] [Google Scholar]
- 54. White C.I., Haber J.E.. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990; 9:663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu D., Topper L.M., Wilson T.E.. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in saccharomyces cerevisiae. Genetics. 2008; 178:1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haber J.E. Mating-type genes and MAT switching in saccharomyces cerevisiae. Genetics. 2012; 191:33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Llorente B., Symington L.S.. The Mre11 nuclease is not required for 5΄ to 3΄ resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 2004; 24:9682–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peterson C.L., Dingwall A., Scott M.P.. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:2905–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dutta A., Sardiu M., Gogol M., Gilmore J., Zhang D., Florens L., Abmayr S.M., Washburn M.P., Workman J.L.. Composition and function of mutant Swi/Snf complexes. Cell Rep. 2017; 18:2124–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sen P., Luo J., Hada A., Hailu S.G., Dechassa M.L., Persinger J., Brahma S., Paul S., Ranish J., Bartholomew B.. Loss of Snf5 induces formation of an aberrant SWI/ SNF complex. Cell Rep. 2017; 18:2135–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stern M., Jensen R., Herskowitz I.. Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol. 1984; 178:853–868. [DOI] [PubMed] [Google Scholar]
- 62. Peterson C.L., Herskowitz I.. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992; 68:573–583. [DOI] [PubMed] [Google Scholar]
- 63. Mimitou E.P., Symington L.S.. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008; 455:770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shao Z., Davis A.J., Fattah K.R., So S., Sun J., Lee K.-J., Harrison L., Yang J., Chen D.J.. Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair. 2012; 11:310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen L., Trujillo K., Ramos W., Sung P., Tomkinson A.E.. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell. 2001; 8:1105–1115. [DOI] [PubMed] [Google Scholar]
- 66. Sudarsanam P., Iyer V.R., Brown P.O., Winston F.. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Y., Hefferin M.L., Chen L., Shim E.Y., Tseng H.-M., Kwon Y., Sung P., Lee S.E., Tomkinson A.E.. Role of Dnl4–Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 2007; 14:639–646. [DOI] [PubMed] [Google Scholar]
- 68. Gobbini E., Cesena D., Galbiati A., Lockhart A., Longhese M.P.. Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks. DNA Repair. 2013; 12:791–799. [DOI] [PubMed] [Google Scholar]
- 69. Kapoor P., Bao Y., Xiao J., Luo J., Shen J., Persinger J., Peng G., Ranish J., Bartholomew B., Shen X.. Regulation of Mec1 kinase activity by the SWI/SNF chromatin remodeling complex. Genes Dev. 2015; 29:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kwon H., Imbalzano A.N., Khavari P.A., Kingston R.E., Green M.R.. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994; 370:477–481. [DOI] [PubMed] [Google Scholar]
- 71. Bennett G., Papamichos-Chronakis M., Peterson C.L.. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat. Commun. 2013; 4:2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bhargava R., Onyango D.O., Stark J.M.. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 2016; 32:566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Blier P.R., Griffith A.J., Craft J., Hardin J.A.. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J. Biol. Chem. 1993; 268:7594–7601. [PubMed] [Google Scholar]
- 74. Oh J., Al-Zain A., Cannavo E., Cejka P., Symington L.S.. Xrs2 dependent and independent functions of the Mre11-Rad50 complex. Mol. Cell. 2016; doi:10.1016/j.molcel.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M. et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004; 431:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Emerson C.H., Bertuch A.A.. Consider the workhorse: Nonhomologous end-joining in budding yeast 1. Biochem. Cell Biol. 2016; 94:396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chambers A.L., Brownlee P.M., Durley S.C., Beacham T., Kent N.A., Downs J.A.. The two different isoforms of the RSC chromatin remodeling complex play distinct roles in DNA damage responses. PLoS ONE. 2012; 7:e32016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shim E.Y., Ma J.-L., Oum J.-H., Yanez Y., Lee S.E.. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 2005; 25:3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Budd M.E., Campbell J.L.. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS ONE. 2009; 4:e4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Krasner D.S., Daley J.M., Sung P., Niu H.. Interplay between Ku and replication protein A in the restriction of Exo1-mediated DNA break end resection. J. Biol. Chem. 2015; 290:18806–18816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Berkovich E., Monnat R.J., Kastan M.B.. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007; 9:683–690. [DOI] [PubMed] [Google Scholar]
- 82. Li X., Tyler J.K.. Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. Elife. 2016; 5:e15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tsukuda T., Trujillo K.M., Martini E., Osley M.A.. Analysis of chromatin remodeling during formation of a DNA double-strand break at the yeast mating type locus. Methods. 2009; 48:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kadoch C., Hargreaves D.C., Hodges C., Elias L., Ho L., Ranish J., Crabtree G.R.. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013; 45:592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wilson B.G., Roberts C.W.M.. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer. 2011; 11:481–492. [DOI] [PubMed] [Google Scholar]
- 86. Park J.-H., Park E.-J., Lee H.-S., Kim S.-J., Hur S.-K., Imbalzano A.N., Kwon J.. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006; 25:3986–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Smith-Roe S.L., Nakamura J., Holley D., Chastain P.D., Rosson G.B., Simpson D.A., Ridpath J.R., Kaufman D.G., Kaufmann W.K., Bultman S.J.. SWI/SNF complexes are required for full activation of the DNA-damage response. Oncotarget. 2015; 6:732–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kwon S.-J., Lee S.-K., Na J., Lee S.-A., Lee H.-S., Park J.-H., Chung J.-K., Youn H., Kwon J.. Targeting BRG1 chromatin remodeler via its bromodomain for enhanced tumor cell radiosensitivity in vitro and in vivo. Mol. Cancer Ther. 2015; 14:597–607. [DOI] [PubMed] [Google Scholar]
- 89. Vélez-Cruz R., Manickavinayaham S., Biswas A.K., Clary R.W., Premkumar T., Cole F., Johnson D.G.. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev. 2016; 30:2500–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.