Abstract
Circadian rhythm exerts its influence on animal physiology and behavior by regulating gene expression at various levels. Here we systematically explored circadian long non-coding RNAs (lncRNAs) in mouse liver and examined their circadian regulation. We found that a significant proportion of circadian lncRNAs are expressed at enhancer regions, mostly bound by two key circadian transcription factors, BMAL1 and REV-ERBα. These circadian lncRNAs showed similar circadian phases with their nearby genes. The extent of their nuclear localization is higher than protein coding genes but less than enhancer RNAs. The association between enhancer and circadian lncRNAs is also observed in tissues other than liver. Comparative analysis between mouse and rat circadian liver transcriptomes showed that circadian transcription at lncRNA loci tends to be conserved despite of low sequence conservation of lncRNAs. One such circadian lncRNA termed lnc-Crot led us to identify a super-enhancer region interacting with a cluster of genes involved in circadian regulation of metabolism through long-range interactions. Further experiments showed that lnc-Crot locus has enhancer function independent of lnc-Crot's transcription. Our results suggest that the enhancer-associated circadian lncRNAs mark the genomic loci modulating long-range circadian gene regulation and shed new lights on the evolutionary origin of lncRNAs.
INTRODUCTION
Circadian rhythm is an intrinsic 24 h oscillation of various physiological processes and behaviors synchronized with daily light/dark cycle in a wide-range of species. In mammals, suprachiasmatic nucleus (SCN) in the brain is known as the control center of circadian rhythm that orchestrates rhythms of peripheral tissues. Circadian control of many aspects of physiology and behavior is manifested by the widespread circadian regulation of gene expression. It is now known that the central molecular clock is formed by two main feedback loops consisting of core clock genes (1–4). In the first loop, BMAL1 and CLOCK form heterodimers to initiate the expression of Period (Per1/2/3) and Cryptochrome (Cry1/2) genes that in return repress BMAL1/CLOCK activities. In the second loop, BMAL1 and CLOCK activate the expression of Rev-erbα/β (Nr1d1/2) that in return repress the expression of Bmal1 and Clock. The studies of genome-wide circadian transcription have mainly focused on the protein coding genes (5–8) and small non-coding RNAs such as microRNAs (9,10).
In recent years, a new class of non-coding RNAs referred to as long non-coding RNAs (lncRNAs) has gained much attention. It was shown that lncRNAs are pervasively transcribed from mammalian genomes (11) and tens of thousands of lncRNAs have been detected in a variety of tissues. In contrast to the protein coding mRNAs, lncRNAs are considered to have lower expression level, stronger tissue specificity and lower sequence conservation (12–16) and have been found to play important roles in many biological processes (17,18).
Recently, lncRNAs showing circadian expression have been reported in several species. In Neurospora, qrf, a lncRNA residing on the anti-sense strand of a core clock gene, frequency (frq), oscillates in anti-phase to frq and inhibits the expression of frq through histone modification and premature termination of transcription (19). In mouse, a lncRNA anti-sense to the core circadian gene, Per2, has also been found oscillating anti-phase to Per2 but its exact function is still unknown (20). A recent study of mouse circadian transcriptome (21) suggested that at least 1000 known lncRNAs showed circadian expression in multiple mouse tissues. However, the roles of these lncRNAs in circadian rhythm remain unclear. Also another class of short and non-spliced non-coding RNAs transcribed from enhancers, called enhancer RNAs (eRNAs), were found with circadian expression in mouse liver (22). However how these eRNAs function in circadian gene regulatory network was unclear and their relationship with lncRNAs is yet to be discovered (23).
In this study, we compiled a comprehensive list of circadian lncRNAs in mouse liver. Among them, we identified a class of circadian lncRNAs that are transcribed from enhancer loci in mouse liver and regulated by circadian transcription factors (TFs) including BMAL1 and REV-ERBα. These lncRNAs were found to show close circadian phases and similar responses upon BMAL1 or REV-ERBα knockout in mouse liver with their nearby circadian protein coding genes. We hypothesized that these lncRNA loci may harbor enhancers to form chromatin loops to facilitate BMAL1 or REV-ERBα regulation on nearby genes through long-range interaction. Comparing nascent-seq with RNA-seq data, circadian lncRNAs were found more nucleus-enriched than circadian protein coding mRNAs but less than previously defined circadian eRNAs. Furthermore, strong association between enhancers and circadian lncRNAs were also discovered in non-hepatic mouse tissues. Our analysis of circadian lncRNAs from RNA-seq data in rat liver showed that circadian transcription of lncRNA locus is more conserved than lncRNAs themselves. This suggests that circadian lncRNAs may result from clock-controlled transcription at the enhancers, some of which are shared across tissues and even conserved between species. We found that a circadian lncRNA, termed lnc-Crot, is expressed at a super-enhancer upstream of the circadian oscillating gene Crot and is regulated by BMAL1 and REV-ERBα. With circular chromosome conformation capture sequencing (4C-seq), we showed that lnc-Crot locus interacts with the circadian oscillating genes that tend to oscillate with similar phases through long-range interaction while these interactions appeared invariant throughout the day. The deletion of the enhancer region of lnc-Crot locus disrupted the regulation of REV-ERBα upon lnc-Crot interacted genes. However, the expression of these genes did not show significant changes after in cis over-expression of lnc-Crot. This suggests that lnc-Crot locus functions as an enhancer independent of lnc-Crot's transcription. Our study suggests that a class of lncRNAs with circadian expression mark active enhancers that facilitate circadian TFs’ regulation on nearby genes through long-range interaction.
MATERIALS AND METHODS
Animal preparation and sample collection
All mice were male, C57BL/6J strain and aged around 8 weeks. All animals were provided with food and water available ad libitum under 12/12 h light-dark (LD) conditions. After the entrainment under light-dark condition for at least 7 days, animals were then transferred to constant darkness (DD) condition and sacrificed after 24 h of DD. Livers from three mice at each time point every 4 h for 48 h were collected, quickly frozen in liquid nitrogen and then stored at −80°C. Liver samples of two pairs of REV-ERBα knock-out (KO) and wild-type (WT) mice were collected at CT0 and CT12 respectively. All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee of Shanghai Institutes for Biological Sciences and conformed to institutional guidelines of vertebrates study.
RNA sequencing
Total RNA was extracted from tissue samples by Trizol (Invitrogen). Liver RNA samples of three liver-specific BMAL1 KO mice and three WT control mice at CT0 and CT12 respectively were provided by Prof. Yi Liu (Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences). Each of the RNA samples was then used for strand-specific library construction and sequencing with single-ended 100 bp reads and the WT samples were used for lncRNA de novo assembly. The RNAs of two pairs of REV-ERBα KO and control mouse livers were used for un-stranded RNA sequencing. RNA sequencing was performed using the Illumina HiSeq 2000. All sequencing raw data were submitted to Gene Expression Omnibus (GEO) with accession number GSE87299.
qPCR analysis
For quantitative polymerase chain reaction (qPCR) analysis, 500 ng of total RNA was reverse transcribed using random hexamer and the Superscript II reverse transcriptase (Invitrogen). A total of 4 μl of reverse transcribed (RT) product (1:10 diluted) and SYBR Green I Master Mix (Roche) were used in qPCR on a LightCycler 480 (Roche). Expression of all genes and lncRNAs were normalized by control gene, Actb. Primers used were listed in Supplementary Table S7.
Assembly of lncRNAs in mouse
The lncRNAs can be viewed on UCSC genome browser by visiting this address (http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=lumos21&hgS_otherUserSessionName=liver_lncRNAs). We assembled lncRNAs de novo from mouse liver RNA-seq data and combined them with annotated lncRNAs from public databases of Ensembl, Refseq and UCSC. Two mouse liver RNA-seq datasets were used in this study: (i) strand-specific RNA-seq dataset of livers from three above-mentioned WT mice collected at CT0 and CT12 respectively. (ii) un-stranded and pair-ended RNA-seq dataset of mouse liver (24). The discovery pipeline of lncRNAs was basically adapted from the method discussed in the study of Cabili et al. (13). The details were described in Supplementary Data. LncRNA assembly and identification from mouse pancreas and rat liver followed the similar method as in liver.
Analysis of circadian expression of lncRNAs and protein coding genes
Circadian RNA-seq data in mouse liver from dataset 1 (20) and dataset 2 (21) sampled every 4 and 6 h respectively for 48 h were downloaded from GEO with accession numbers: GSE39860 and GSE54651. LncRNAs and protein coding genes were quantified by RNA-seq with reads per kilobase of transcript per million mapped reads (RPKM) and fitted by cosine functions with 24 h period and shifting phases as described in the study of Yan et al. (25) in two datasets separately. The circadian transcripts shared by the two datasets were selected as summed –log2-transformed fitting P-value > 10 and phase difference < = 4 between the two datasets. A lncRNA cluster was defined as circadian if at least one of its lncRNA transcript was circadian and the same applied for protein coding genes. The details of false discovery rate (FDR) calculation were described in Supplementary Data.
To calculate the correlations of time-course data between lncRNAs and neighboring protein coding genes, for each lncRNA cluster, Pearson's correlation coefficients between all lncRNA transcripts within the cluster and all protein coding genes that were within 50 kb region of this cluster were calculated. The median value of these pairwise correlation coefficients was used as the final correlation coefficient between this lncRNA cluster with its nearby protein coding genes. While calculating the correlations between circadian lncRNAs with circadian protein coding genes, we required that both lncRNA clusters and their nearby protein coding genes were circadian.
Analysis of nascent-seq data
Circadian nascent-seq data of mouse liver were downloaded from GEO database: GSE36916. To measure nucleus/cytoplasm expression ratios, lncRNA and protein-coding gene expression was quantified by the exonic reads from stranded nascent-seq as well as from RNA-seq of dataset 2. Average RPKM of all time points of nascent-seq was taken as nuclear gene expression and average RPKM of all time points of RNA-seq was taken as cytoplasmic gene expression. Circadian expression of lncRNAs and protein coding genes at the primary RNA level was quantified with the unstranded nascent-seq, which has two replicates every 4 h and selected by cosine fitting P-value < 0.05.
ChIP-seq data analysis
Two sets of BMAL1 chromatin immunoprecipitation sequencing (ChIP-seq) data with GEO accession numbers: GSE26602 and GSE39860 were downloaded from GEO database (26,20). We obtained processed BMAL1 binding sites directly from these studies combining all six time points in 24 h. ChIP-seq data of CLOCK, PER1, PER2, CRY1 and CRY2 shown in Supplementary Figure S2A were directly downloaded from GEO database with accession number: GSE39860 (20). We analyzed other ChIP-seq datasets from raw read data. Two sets of REV-ERBα ChIP-seq data were from studies of Cho et al. (27) and Feng et al. (28) (GEO accession numbers: GSE34020 and GSE26345) sampled at ZT10 and ZT8 respectively. H3K4me3 and H3K27ac ChIP-seq in mouse liver were from ENCODE project (29) (GEO accession numbers: GSE29184). Cap analysis gene expression (CAGE) data of mouse liver were from the study of Forrest et al. (30). PPARa and HNF4a ChIP-seq datasets were from studies of Rakhshandehroo et al. (31) (GEO accession number: GSE8292) and Schmidt et al. (32) (GEO accession number: GSE22078). The ChIP-seq data analysis details were described in Supplementary Data.
Identification of enhancers and super-enhancers
Enhancer regions in this study were defined as the regions that contained both H3K27ac and H3K4me1 enriched marks but were away from the promoter regions flanking 1 kb of transcription start sites (TSS) of protein coding genes. Super-enhancers were identified based on the method from the study of Whyte et al. (33). First, H3K27ac binding intensity on each enhancer was quantified after subtracting the input intensity both measured in reads per million mapped reads per base pair. Then all enhancers were plotted with x-axis as the rank of its H3K27ac intensity and y-axis as its intensity value scaled to the same length of x-axis (Supplementary Figure S1D). The turning point above which intensity increased rapidly and thus separated super-enhancers from conventional enhancers was identified as the point where the tangent line of slope equaled to 1. All enhancers above this point were defined as super-enhancers.
4C-Seq analysis
Liver samples of three mice collected from CT6 and CT18 respectively were used for 4C assays as described in studies of Zhao et al. (34) and Xu et al. (35). The data around bait regions were displayed on Intergrative Genomics View (IGV) (36) in Supplementary Figure S5A. The PCR-amplified library was purified and sequenced with a 100 bp read length using the Illumina HiSeq 2000. Raw reads of 4C-seq were first de-multiplexed according to the viewpoint primers and restrict enzyme sites before mapping to mouse genome. All reads were sequenced in the direction from the first cutting enzyme HindIII (AAGCTT) to the second enzyme DpnII (GATC). Sequence A from primer to HindIII RE (restrict enzyme) site at 5΄ end: GGGCCTTGTGAGTCGCACAACTTTCAGAACTCCTAGAACCTCAAAGCTT as well as sequence B from DpnII RE site to primer at 3΄ end: GATCTGACCCCAGGCCAATCTGATTGAGGCCTTTTGTCAATTGTCTTCAAAACATCTGATTATGGCTGTATTATCACTGAGGTACCC have been removed. For a sequence read qualified for downstream analysis, sequence A must be present on the 5΄ end while sequence B may not be present or partially present on the 3΄ end. The sequence reads without sequence A were discarded. Then sequence A and B were trimmed from each read and then reads were aligned to mouse genome mm9 by Bowtie (37). As inter-chromosomal interactions captured by 4C assays may more likely be random-ligations and have low coverage, here we only focused on cis-chromosome interactions. We downloaded all HindIII restriction sites on the cis-chromosome from UCSC genome browser and obtained the fragments flanked by any two neighboring restriction sites on cis-chromosome. The sequence reads located on each fragment were counted. As ligation may happen between both ends of a fragment when constructing 4C library, our counting scheme ensured that the reads on both end of a fragment can be assigned to this fragment. Peak selection was based on the method in a previous study (38). First, the fragments without any read count were removed and read counts of the remaining fragments were smoothed by applying a running window with a median window size of 10 fragments. Second, the smoothed counts were log2-transformed. Then background count distribution along the cis-chromosome was generated by applying Loess regression to the log2-transformed count values, where Loess regression was implemented by R (39) with parameter α = 0.7. Then a fragment with log2-transformed value higher than regression fitted value was defined as interacted fragment. The fragment present in at least two samples was selected as the overall interacted fragments. Finally, a gene was defined as an interacted gene if this gene, including its gene body and its upstream 2 kb region, overlapped with any interacted fragments. The GEO accession number for the 4C datasets is: GSE73271.
Rapid amplification of cDNA ends (RACE)
5΄ rapid amplification of cDNA ends (RACE) experiment was performed according to instruction manual (version 2.0) (Catalog no. 18374-058, Invitrogen) and 3΄ RACE experiment was performed according to instruction manual (Catalog no. 18373-019, Invitrogen). Experiment details were described in Supplementary Data.
Analysis of subcellular localization
Mouse liver cells were quickly dispersed and filtered through the 40 μm cell strainer to make a single-cell suspension. Cells were then lysed in cold lysis buffer (10 mM Tris, 150 mM Nacl, 0.5% NP-40 and 1 mM Ribonucleoside vanadyl complexes) for 5 min on ice, followed by centrifugation for 3 min at 1000 g at 4°C. After collecting the supernatant and extracting cytosolic RNA with acid-phenol:chloroform (pH 4.5) and the pellet was lysed by Trizol to extract nuclear RNA. Entire RNA from nucleus was resolved to 5 μl and RNA from cytoplasm was resolved to 10 μl at final concentration of 200 ng/10 μl, followed by reverse transcription and then qPCR.
lncRNA assembly in rat and comparative analysis between mouse, rat and human
Rat liver samples were collected every 4 h for 24 h after the rats were entrained in light-dark (LD) cycles for at least 7 days using the same procedure as for mice and were used for RNA-seq (GEO accession: GSE77572). Rat circadian lncRNAs were defined as those with cosine fitting P-value < 0.01.
We utilized synteny (40) to define orthologous lncRNA regions between mouse and rat. For each intergenic lncRNA, its two flanking protein-coding genes were identified on the mouse genome (mm9) by closestBed of Bedtools (41). Then we searched for the orthologs of these two flanking genes in rat (rn5) using BioMart (42). If the two orthologs in rat were still neighboring genes, we defined this region as orthologous intergenic regions (OIRs).
Sequence alignments between mouse and rat were conducted by basic local alignmentsearch tool (BLAST) program (2.2.26). Sequences of mouse lncRNAs and rat lncRNAs were extracted from mouse genome (mm9) and rat genome (rn5) respectively. BLAST was conducted with mouse lncRNAs as query sequences and rat lncRNAs as search database and then vice versa. For each query sequence, top three best alignments with E-value < 1 × 10−10 were retained as target sequences. The sequence identity is obtained from the result of BLAST output.
Human lncRNAs annotations were first downloaded from a database called NONCODE (43) and the expression of these lncRNAs was quantified by a set of human liver RNA-seq data (ENCODE, GSE78569) (44) and only those lncRNA transcripts expressed (reads ≥ 10) in liver were retained as human liver-expressed lncRNAs.
Non-radioactive RNAin situ hybridization (ISH)
We performed RNA in situ hybridization (ISH) experiment by using digoxigenin-labeled RNA probes, a non-radioactive technique based on dual signal amplification, named ‘CARD’. RNA ISH was carried out on highly automated robotic equipment (TECAN GenePaint). The minimum amount of RNA probe used was 25 ng/slide. Each lncRNA and mRNA gene was analyzed on 24 sagittal sections of mouse embryonic E14.5 and 20 sagittal sections of P56 brain tissue sections. The entire protocol was performed according to method in the study of Eichele et al. (45).
Double RNA fluorescence in situ hybridization (FISH)
Double RNA fluorescence ISH (FISH) was performed on highly automated ISH robotic equipments and the protocol was performed according to method in the study of Eichele et al. (45) with additional modifications for FISH. In brief, using in vitro transcription we synthesized Crot mRNA labeled with fluorescein isothiocyanate (FITC) and lnc-Crot labeled with digoxigenin (DIG). The probes were detected using Anti-FITC and Anti-Cy3 respectively. FISH was performed on 20 μm adult mouse brain (P56) tissue sections. Following FISH, the slides were mounted with 4΄,6-diamidino-2-phenylindole (DAPI) (VectaShield mounting medium) and coverslipped. Confocal imaging was performed at 60× using oil immersion to visualize the expression of both Crot and lnc-Crot.
Genome editing by CRISPR-Cas9 to delete lnc-Crot locus
The pX330-mcherry plasmid used in this study was provided by Dr Jinsong Li's Lab (46). The sgRNAs targeting specific locations (L: caccgcaagatattaacagtagggc, aaacgccctactgttaatatcttgc, R: caccgcccagagctactgaggccgt, aaacacggcctcagtagctctgggc) were synthesized, annealed and ligated to pX330-mcherry plasmid that was digested with BbsI (New England Biolabs). The pX330-mcherry plasmids harboring corresponding sgRNAs were transfected into Hepa1-6 cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instruction. About 24 h after transfection, the Hepa1-6 cells expressing red fluorescence protein were sorted with flow cytometry (MoFlo™ XDP cell sorter, Beckman Coulter) and seeded into 96-well plates. Three weeks after plating, single colony was picked and genotyped with PCR and sequencing. REV-ERBα siRNAs were obtained from GenePharma (Shanghai). All siRNA experiments were conducted at a final concentration of 50 nM. Transfections were conducted using Lipofectamine RNAiMAX (Invitrogen).
Lnc-Crot over-expression in Hepa1-6 cells by CRISPR-Cas9
The method of genome editing in cells by CRISPR-Cas9 was based on the study of Xu et al. (35). CRISPR-Cas9 method (47) was used to insert a strong promoter (PGK promoter) to over-express lnc-Crot in Hepa1-6 cells. The sgRNA sequence (gCACAGCTACAGCCAAGGTACNGG) was designed by E-CRISP program (48). The primers for two homologous arms for donor plasmids were as follows: left-arm forward (ATGGCTACCGGCTTGTGCAATGTT); left-arm reverse (CTTGGCTGTAGCTGTGGT); right-arm forward (CACTCGCTGGTACAGGCAT); right-arm reverse (CCATCCCTGATTTGCCCTCCTGTTG). The regulatory module (hPGK promoter/PuroR) was amplified from commercially available expression vector pLKO.1. Homologous arms and PGK/puroR were assembled into pGEM-T Easy vector (Promega). Hepa1-6 cells were cultured with 10% fetal bovine serum (FBS) in Dulbecco's modified eagle's medium (Life technology) and co-transfected with two gRNA/Cas9 vectors and linearized donor DNA. Then the cells were screened with 3 μg/ml puromycin (Merck/ millipore) for 2 weeks. Expression of lnc-Crot was measured by qPCR for WT cells and CRISPR-Cas9 treated cells. Similar method was used for lnc-Rere over-expression. The sgRNA sequence was CTTCTGCACGCATCCCCAGAGGG, and the primers for two homologous arms were as follows: left-arm forward (TTGGCCGAGGATTCTTGTTGC); left-arm reverse (CATTAGCTGGATGGACGGGTG); right-arm forward (TTCTCTGGCCTCACATACTGC); right-arm reverse (CTCCTTCCTGGCTGCTTGACA).
Statistical analysis
The comparisons in Figures 1E, 2B, 2C, 2E and 3A were performed with Kolmogorov–Smirnov (KS) Test. The enrichment analyses in Figures 2A and 2D, 3E and 3F, and 5B were performed with Fisher's Exact Test. The comparisons in Figure 5F and 5E were performed with Student's t-test. All statistical analyses were performed in R environment (39).
Figure 1.
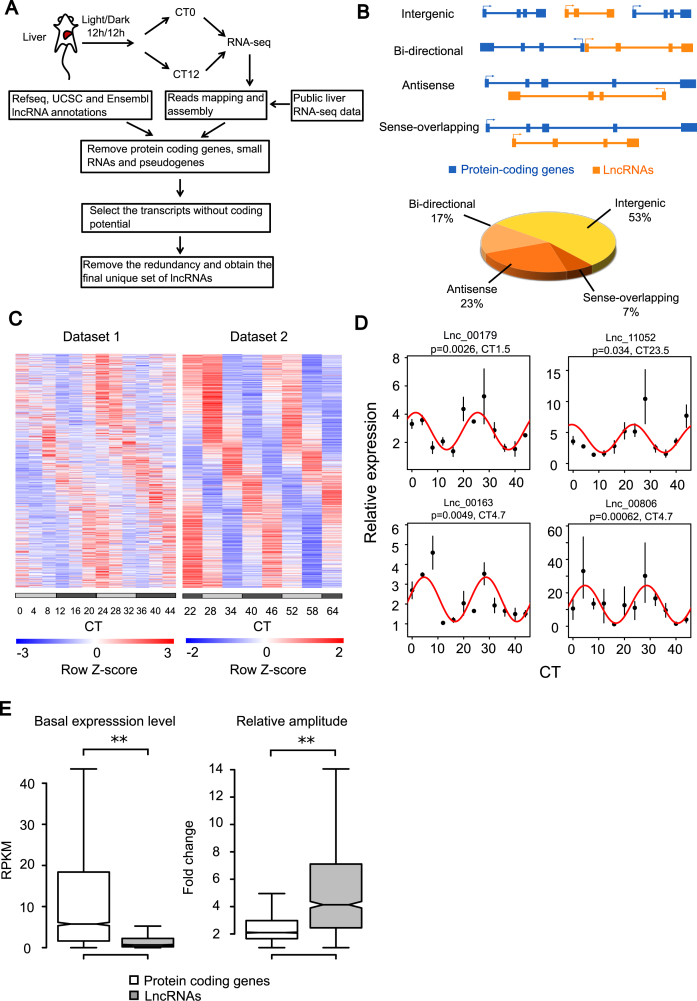
Identification of circadian lncRNAs in mouse liver. (A) The schematic work flow of de novo lncRNA assembly from RNA-seq data of mouse liver and integration with public annotation of lncRNAs. (B) Classification of lncRNAs based on their genomic locations relative to protein coding genes and the percentage of each category. (C) Heatmap of circadian expression of all circadian lncRNAs identified by the meta-analysis of dataset 1 (20) and dataset 2 (21). (D) Four selected circadian lncRNAs validated by quantitative polymerase chain reaction (qPCR) (n = 2–3). Lines were the best fitted cosine functions with fitted P-values and phases. All expression values in qPCR were quantified relative to expression of Actb. (E) Comparison of basal expression levels and relative amplitudes between circadian lncRNAs and circadian protein coding genes revealed lower expression levels but higher relative amplitudes of circadian lncRNAs than circadian protein coding genes. Relative amplitude was calculated by the ratio between peak and trough of circadian expression. Kolmogorov–Smirnov (KS) test was performed for the comparisons of both basal expression levels (P-value < 2.2 × 10−16) and relative amplitudes (P-value < 2.2 × 10−16), where the significance level was determined by ** P-value < 0.01 and * P-value < 0.05.
Figure 2.
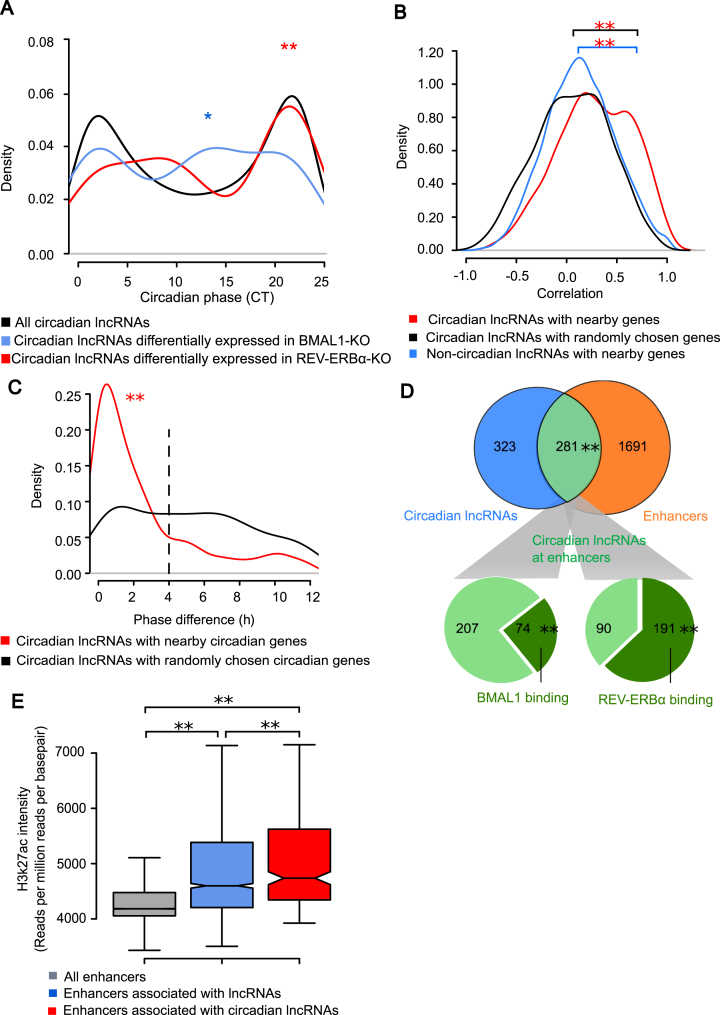
Circadian lncRNAs were enriched in enhancer regions. (A) Phase distribution of all circadian lncRNAs and those differentially expressed in BMAL1 KO or REV-ERBα KO mice. Phases of circadian lncRNAs differentially expressed in BMAL1 KO and REV-ERBα KO were enriched around CT12 (CT10–CT14, Fisher's Exact Test, *P-value = 0.025) and CT0 (CT21–CT23, Fisher's Exact Test, P-value = 0.0002) respectively, compared with all circadian lncRNAs. (B) Distribution of Pearson's correlation coefficients of lncRNAs with nearby protein coding genes in dataset 2. Comparisons of distribution were performed using KS test, where **P-value = 8.0 × 10−12 (circadian lncRNAs with nearby genes versus circadian lncRNAs with random genes) and **P-value = 3.6 × 10−15 (non-circadian lncRNAs with nearby genes vs. circadian lncRNAs with random genes). (C) Distribution of phase differences between circadian lncRNAs and nearby circadian protein coding genes. Comparison between two distributions of phase differences was performed using KS test, with **P-value = 7.5 × 10−12. (D) Enhancers, BMAL1 binding sites and REV-ERBα binding sites on circadian lncRNA loci. Significance of **P-value < 2.2 × 10−16 was determined by Fisher's Exact Test for the overlap between circadian lncRNAs and enhancers. The binding sites are over-represented on circadian lncRNA loci at enhancer regions compared to those at non-enhancer regions, where significance of **P-value < 2.2 × 10−16 and **P-value < 2.2 × 10−16 were calculated by Fisher's exact test for BMAL1 binding and REV-ERBα binding respectively. (E) Distribution of H3K27ac ChIP intensities on enhancer regions. Comparisons were performed by KS test between pairs, where **P-value < 2.2 × 10−16 (all enhancers vs. enhancers associated with lncRNAs), **P-value < 2.2 × 10−16 (all enhancers vs. enhancers associated with circadian lncRNAs) and **P-value = 0.0017 (enhancers associated with lncRNAs vs. enhancers associated with circadian lncRNAs) suggested they were all significantly different.
Figure 3.
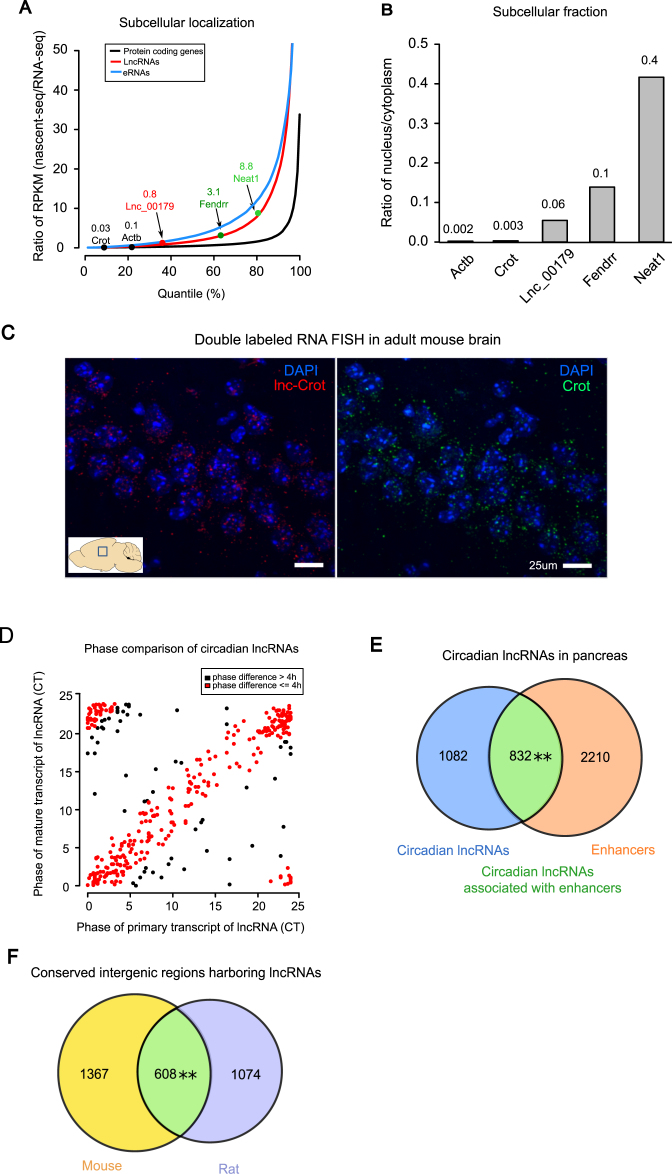
Subcellular localization of circadian lncRNAs in liver and circadian lncRNAs in non-hepatic tissues. (A) Distributions of relative nucleus to cytoplasm ratios for protein coding genes, lncRNAs and enhancer RNAs (eRNAs). Protein coding genes Actb and Crot, lncRNAs Fendrr and Neat1, and one selected circadian lncRNA were highlighted. Comparisons were performed using KS test, where **P-value < 2.2 × 10−16 (coding genes vs. lncRNAs) and **P-value = 2.2 × 10−16 (lncRNAs vs. eRNAs). (B) Relative nucleus to cytoplasm ratios assayed by qPCR of extracted RNAs separately from nucleus and cytoplasm in mouse liver. (C) Double RNA FISH confocal imaging of Crot mRNA labeled with FITC (right image, green), lnc-Crot labeled with DIG (left image, red) and nucleus labeled with DAPI (blue) in CA3 region of hippocampus in adult mouse brain (P56). Images were taken at 60× magnification. Scale bar: 25 μm. (D) Phases of circadian lncRNAs that also oscillated on nascent-seq. Phases of circadian lncRNAs from RNA-seq (Y-axis) and nascent-seq (X-axis) were plotted. Red ones are the lncRNAs with phase differences <4 h between RNA-seq and nascent-seq. (E) Circadian lncRNAs in pancreas also have significant overlap with the enhancers in pancreas. Pancreatic lncRNAs were assembled and identified by RNA-seq data of pancreas and enhancers were identified by H3K27ac ChIP-seq of pancreas (58). Significance level was determined by Fisher's exact test, where **P-value = 0.007. (F) Overlap of intergenic regions harboring lncRNAs in mouse and rat is significant (Fisher's exact test **P-value < 2.2 × 10−16).
Figure 5.
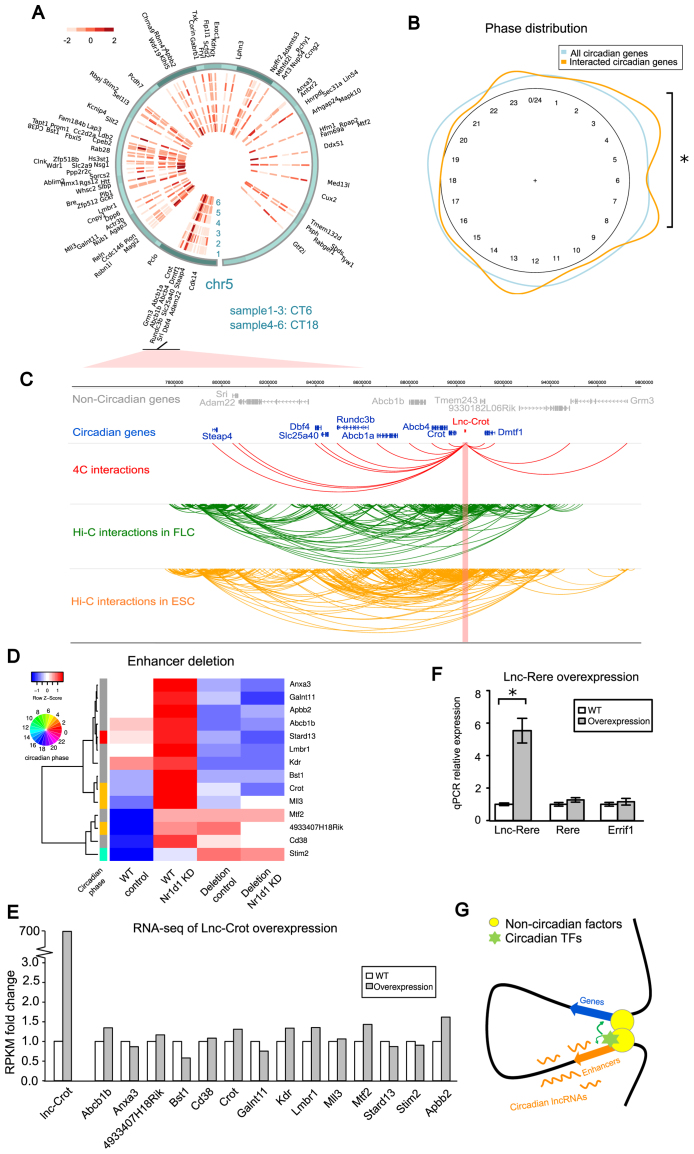
Interactome of lnc-Crot locus by 4C-seq and RNA-independent enhancer function of lnc-Crot. (A) Heatmap of interaction intensities of 4C-seq and interacted genes on chr5 (cis-chromosome) at CT6 and CT18. Interaction intensities were quantified by z-score (‘Materials and Methods’ section). Bait region was lnc-Crot locus. (B) Circadian phases of interacted circadian protein coding genes in 4C-seqand phases of all circadian genes. The phases of interacted genes were enriched around CT0 and CT8 (single-sided Fisher's exact test, P-value = 0.034) compared with all circadian genes. (C) 2Mb regions surrounding the lnc-Crot locus was displayed by WashU EpiGenome Browser (http://epigenomegateway.wustl.edu/browser/). Interacted genes were shown at top track as circadian and non-circadian. Interactions with lnc-Crot locus in liver from 4C-seq were shown below. Then interactions in fetal liver cells (FLC) and embryonic stem cells (ESC) from Hi-C study (64) in this region were shown. (D) Differentially expressed 4C-interacted genes in the RNA-seq of lnc-Crot enhancer deletion in Hepa1-6 cells. (E) Differentially expressed 4C-interacted genes did not show significant change in the RNA-seq upon lnc-Crot in cis over-expression. (F) In cis over-expression of lnc-Rere did not lead to significant changes of its neighboring gene expression assayed by qPCR. (G) Our hypothetical model that the lnc-Crot enhancer and target genes first form a scaffold through long-range interactions then histone modifications and circadian TF bindings take place to confer circadian gene expression.
RESULTS
Identification of circadian lncRNAs
In order to obtain a comprehensive list of lncRNAs expressed in mouse liver, we extracted total RNA and conducted single-ended strand-specific RNA sequencing (RNA-seq) in the livers of six WT mice collected at CT0 (n = 3) and CT12 (n = 3) respectively under DD after the entrainment under light/dark cycle. The gene models of lncRNAs were de novo assembled from our RNA-seq data based on the methods as described in the study of Cabili et al. (13). To reduce transcriptional noise and ensure high quality of transcripts, single-exonic lncRNAs were further selected by requiring the presence of 5΄ CAGE tags mapped within 500 bp upstream and 200 bp downstream of 5΄ ends of lncRNAs using high resolution CAGE data from Fantom5 (30). In order to achieve better annotation of mouse liver lncRNAs, we integrated another set of mouse liver lncRNAs assembled from a public RNA-seq data (24). After removing the redundancy, we obtained 9827 lncRNAs defined as RNA-seq derived liver lncRNAs. Among them, 4000 are multi-exonic transcripts. Furthermore, 3727 public annotated lncRNAs from databases including Ensembl, UCSC and Refseq were found to be expressed in mouse liver according to our RNA-seq data (Figure 1A). We found that a total of 8736 RNA-seq derived lncRNAs in our study had not been previously annotated in these public databases likely because of their liver-specific expression. After combining the RNA-seq derived and public annotated lncRNAs, we finally obtained a total of 12 463 putative lncRNA transcripts expressed in mouse liver (Supplementary Table S1). Based on their genomic locations relative to protein coding genes, these lncRNAs can be divided into four categories: sense-overlapping (7%), anti-sense (23%), bi-directional (17%) and intergenic (53%) lncRNAs (Figure 1B). As the lncRNA transcripts may contain various isoforms due to alternative splicing, 5΄ initiation and 3΄ termination, the overlapping transcripts from the same lncRNA locus were merged as one cluster, resulting in 8705 lncRNA clusters. Compared to protein coding genes, these lncRNAs have shorter lengths with median length of 668 bp and have less exons as half of them are single-exonic. This is consistent with the previously known characteristics of lncRNAs. All these liver lncRNA models can be visualized by UCSC genome browser (link address in Materials and Methods section).
Next, we searched for the lncRNAs that showed circadian expression using two mouse circadian liver RNA-seq datasets from the studies of Koike et al. (20) and Zhang et al. (21), referred as dataset 1 and dataset 2, respectively, in the downstream analysis. The mice were sampled with 4 h intervals in dataset 1, and 6 h intervals in dataset 2 for two consecutive days. Through a meta-analysis of the two datasets (sum of –log2-transformed fitting P-value > 10 and phase difference < 4 h), a total of 604 lncRNA clusters were identified to show circadian expression (Figure 1C) with an FDR = 0.17. Using the same criteria, 2245 circadian protein-coding genes were identified with a similar level of FDR (0.20). Selected circadian lncRNAs were validated by real-time qPCR (Figure 1D) in liver samples of entrained WT mice taken every 4 h in 48 h. All of them showed consistent circadian oscillations with RNA-seq. Because of the relatively low expression of lncRNAs, the median expression level of circadian lncRNAs was only about 11% of that of circadian protein coding genes (Figure 1E). However, the circadian lncRNAs had higher relative amplitude of oscillation, defined as the ratio between peak and trough, compared with circadian protein coding genes (Figure 1E). This may be caused by faster degradation of lncRNAs, as it is known that lower stability and shorter half-lives of transcripts may lead to lower baseline level but higher relative amplitude of circadian expression (10,49).
Circadian regulation of lncRNAs
Circadian TFs including BMAL1 and REV-ERBα have been considered as the master regulators in circadian clock. We wondered whether the circadian oscillating lncRNAs are regulated by these two TFs. We defined BMAL1 and REV-ERBα binding sites from two sets of BMAL1 ChIP-seq studies (26,20) and two sets of REV-ERBα ChIP-seq data (27,50) in mouse liver respectively, resulting in 8001 BMAL1 and 17545 REV-ERBα binding sites. A total of 259 (43%) circadian lncRNA clusters contain either BMAL1 or REV-ERBα binding sites. These binding sites are significantly enriched in circadian lncRNA clusters over the non-circadian lncRNA clusters (Fisher's exact test, P-value < 2.2 × 10−16). This indicates that these circadian lncRNAs are more likely regulated by these two circadian TFs. In order to investigate how genome-wide transcription of lncRNAs was functionally regulated by these two TFs, we utilized liver-specific BMAL1 knockout mice and full REV-ERBα knockout mice (2). We sequenced liver transcriptomes of six BMAL1 conditional knockout mice together with the above-mentioned WT mice at CT0 (n = 3) and CT12 (n = 3), respectively. Similarly, liver transcriptomes of four REV-ERBα knockout mice and WT mice at CT0 (n = 2) and CT12 (n = 2) were also sequenced.
For each lncRNA cluster, we quantified its expression in the BMAL1 knockout and WT mouse livers at CT0 and CT12 using our RNA-seq data. We used two-way analysis of variance (ANOVA) to evaluate the statistical significance of lncRNA expression changes. Taking both factors of genotype (BMAL1-KO versus WT) and circadian time (CT0 versus CT12) into consideration, a BMAL1-regulated lncRNA cluster was defined by genotype and circadian time interaction P-value < 0.01 in ANOVA. This resulted in 221 BMAL1-regulated lncRNA clusters and 53 (24%) of them showed circadian expression (Supplementary Table S2) (Supplementary Figure S1A). With similar method for REV-ERBα knockout data, it resulted in 447 REV-ERBα-regulated lncRNA clusters (ANOVA P-value < 0.1) and 51 (11.4%) of them showed circadian expression (Supplementary Table S3). Interestingly, we found the circadian phases of 53 BMAL1-regulated circadian lncRNA clusters were enriched around CT12 (CT10–CT14, Fisher's Exact Test, P-value = 0.025) (Figure 2A) and similar to BMAL1-regulated protein coding genes (CT10–CT14, Fisher's Exact Test, P-value = 6.3 × 10−12) (Supplementary Figure S1B). Meanwhile, the circadian phases of 51 REV-ERBα regulated circadian lncRNA clusters were enriched around CT0 (CT21–CT23, Fisher's Exact Test, P-value = 0.0002) (Figure 2A). This suggested that circadian lncRNAs were regulated by these two master circadian TFs in the similar way as circadian protein coding genes.
Circadian lncRNAs associated with enhancer
To address whether these circadian lncRNAs are involved in the regulation of other circadian protein-coding genes, we first investigated whether circadian lncRNAs co-express with their nearby protein coding genes across the genome. We defined protein-coding genes located within 50 kb distance to each lncRNA cluster as its nearby genes and calculated their Pearson's correlation coefficients of circadian expression in dataset 2 (Figure 2B) and dataset 1 (Supplementary Figure S1C). For each lncRNA cluster, correlations of all lncRNA transcripts with all nearby genes were calculated and the median value of all correlations were taken as the correlation of this lncRNA cluster with nearby genes.
We observed that the correlation coefficients of circadian lncRNA clusters with randomly chosen genes on the genome are distributed almost symmetrically around zero with a median value of 0.03 (black line in Figure 2B). In contrast, the correlations between circadian lncRNA clusters with their nearby protein coding genes showed significant bias toward more positive values with a median correlation coefficients value of 0.28 (KS test, P-value < 2.2 × 10−16) (red line in Figure 2B). The correlation coefficients of non-circadian lncRNA clusters with nearby genes did not show this bias and were symmetrically distributed around zero with a median value of 0.13 (blue line in Figure 2B). Therefore, circadian lncRNAs are more strongly correlated with nearby protein-coding genes than non-circadian lncRNAs. Furthermore, circadian lncRNA clusters also tended to show closer phases of circadian oscillations to the nearby circadian protein-coding genes (Figure 2C). The phase differences between 77% circadian lncRNA clusters and nearby protein-coding genes were within 4 h while the phase differences were more evenly distributed among circadian protein-coding genes randomly selected across the genome (Figure 2C, KS test P-value = 7.5 × 10−12). Taken together, circadian lncRNAs and nearby circadian protein coding genes showed significant co-expression.
One possible explanation for the co-expression between circadian lncRNAs and nearby circadian protein coding genes is the existence of cis-regulation between them. Recent studies have shown that certain lncRNAs are originated from the enhancer loci and associated with enhancer function to regulate nearby protein-coding genes (51,52). To explore this possibility, we searched for enhancer regions among all 8705 lncRNAs clusters in mouse liver. We found that 1972 (23%) of them overlapped with enhancers. Among the 604 circadian lncRNA clusters, 281 of them overlapped with enhancers. There is a significant enrichment of enhancers in circadian lncRNA loci (Fisher's exact test, P-value < 2.2 × 10−16) (Figure 2D). Furthermore, we found that BMAL1 binding sites (Fisher's exact test, P-value < 2.2 × 10−16) or REV-ERBα binding sites (Fisher's exact test, P-value < 2.2 × 10−16) are also over-represented in these circadian lncRNA loci with enhancer marks (Figure 2D). Moreover, we found that among the 53 BMAL1-regulated circadian lncRNA clusters, a significantly higher proportion (57%) of them overlapped with enhancers, given the background of 23% of all lncRNA clusters overlapping with enhancers (Chi-square test, P-value = 1.2 × 10−8). Similarly, a significantly higher proportion (47%) of REV-ERBα regulated circadian lncRNA clusters overlapped with enhancers (Chi-square test, P-value < 2.2 × 10−16). This indicated that both BMAL1-regulated lncRNAs and REV-ERBα-regulated lncRNAs were over-represented at enhancer regions. This is consistent with the previous knowledge that active enhancer regions are often bound by TFs, polymerase, chromatin conformation factors and other protein complex to remodel chromatin state and facilitate enhancer function. With the significant co-occurrence of circadian lncRNAs, active enhancers and two key circadian TF binding sites, it raised the possibility that a significant proportion of circadian lncRNA loci may serve as enhancers for circadian regulation of gene expression.
In recent years, the concept of super-enhancer has been proposed and widely studied. Super-enhancers are believed to play particularly important functions in gene regulation, characterized by elevated binding signals of TFs, mediator complex and histone marks (33,53). Here we defined super-enhancers in mouse liver as those enhancers with distinctly higher intensities of H3K27ac marks than the rest of enhancers, based on the method in the study of Whyte et al. (33) (Supplementary Figure S1D). This resulted in 650 liver super-enhancers, which account for 1.8% of all enhancers in mouse liver. Among the 281 circadian lncRNA clusters within enhancer regions, 50 of them were located in super-enhancer regions. Interestingly, we found that circadian lncRNAs are even more likely associated with super-enhancers than regular enhancers (Chi-squared test, P-value = 0.026). On the other hand, the enhancer regions associated with lncRNAs tended to have much higher intensities of H3K27ac marks than the enhancers not associated with lncRNAs (KS test, P-value < 2.2 × 10−16) (Figure 2E). The enhancers associated with circadian lncRNAs have even higher levels of H3K27ac marks than those associated with lncRNAs (KS test, P-value = 0.0017). Taken together, there is a strong association between circadian lncRNAs and super-enhancers.
Subcellular localization of circadian lncRNAs
To investigate the subcellular localization of circadian lncRNAs, we analyzed a published dataset of circadian nascent-seq data in mouse liver (54) that captured the expression of nascent RNAs in the nucleus. The regular liver RNA-seq of dataset 2 was utilized as a measurement of RNAs in cytoplasm. The relative nucleus to cytoplasm ratio for a given transcript was estimated from the ratio between nascent-seq and regular RNA-seq data. We found that lncRNAs in general tend to have higher ratios than protein coding genes and thus are more likely to be localized in nucleus than in cytoplasm (Figure 3A, KS test P-value < 2.2 × 10−16). For example, two known nucleus-enriched lncRNAs, Fendrr (55) and Neat1 (56) have high ratios of 3.1 and 8.8 respectively. In comparison, two protein coding genes, Actb and Crot, have low ratios of 0.1 and 0.03 respectively. A circadian lncRNA transcript identified in our study, lnc_00179 that was then termed as lnc-Crot, has a ratio of 0.8 that is higher than those of protein coding genes Crot and Actb (Figure 3A). This nucleus enrichment of lnc-Crot was validated by qPCR of RNAs extracted separately from the nucleus and cytoplasm of mouse liver, showing around 20-fold increase in nucleus to cytoplasm ratio over those of Crot and Actb (Figure 3B). Furthermore, we conducted double RNA FISH in the hippocampal region of adult mouse brain labeling lnc-Crot and Crot RNA simultaneously. With DAPI staining labeling the whole nucleus, we observed that although both lnc-Crot and Crot can be found in the nucleus and cytoplasm, lnc-Crot was much more enriched in nucleus than Crot (Figure 3C). Circadian nascent-seq data also showed that 41% of circadian lncRNAs were already oscillating (cosine fitting P-value < 0.05) in the nucleus and 34% of them oscillated in phases within 4 h to the phases in RNA-seq data (Figure 3D). In comparison, only 23% of circadian protein coding genes oscillated with phase differences <4 h between nascent-seq and RNA-seq (Supplementary Figure S2A). The closer phase relationship between nuclear and cytoplasmic fractions of circadian lncRNAs may be explained by the shorter half-lives of lncRNAs than protein-coding genes, consistent with a previous kinetic model of RNA accumulation that shorter half-lives will lead to smaller phase delay between pre-mRNAs and mature mRNAs (49).
Previously a study discovered that eRNAs, a class of short non-coding RNAs that were transcribed at enhancers, also exhibited circadian oscillation detected by global run-on sequencing (GRO-seq) in mouse liver (22). In our study, we found that eRNAs were even more nucleus-localized than lncRNAs as shown in Figure 3A (KS test P-value < 2.2 × 10−16). Interestingly, a significant proportion (26%) of our circadian lncRNAs overlapped with circadian eRNAs and showed close circadian phases (Supplementary Figure S2B). The circadian eRNAs defined in the study of Fanget al. (22) also exhibited higher ratios of nucleus over cytoplasm expression than our circadian lncRNAs which in turn showed higher ratios than circadian protein coding genes (Supplementary Figure S2C). In summary, circadian lncRNAs transcribed from the same regions as circadian eRNAs showed similar circadian expression but were less nucleus localized than their eRNA counterpart.
Circadian lncRNAs in non-hepatic mouse tissues
It is known that lncRNA expression is more tissue-specific than protein coding genes (13,57). So we next investigated the circadian expression of liver circadian lncRNAs in other non-hepatic tissues. We examined circadian time-series RNA-seq data of 12 tissues other than liver sampled every 6 h for 48 h in the study of Zhang et al. (21). We quantified the expression of all liver lncRNAs identified in our study at each time point and analyzed their circadian expression in SCN, kidney, heart, lung, brown adipose, white adipose, muscle, adrenal gland, aorta, brain stem, cerebellum and hypothalamus. We found that, among the 604 circadian lncRNA clusters in liver, 299 lncRNA clusters show circadian expression in at least one other tissue (Supplementary Table S4). We provided a list of 13 circadian lncRNAs oscillating in at least six mouse tissues (Table 1). Eleven of them oscillated at highly consistent circadian phases across tissues (circular range test, P-value < 0.01). Among them, six circadian lncRNAs again overlapped with enhancers in liver. In addition, we found that three circadian lncRNAs are bidirectional to circadian protein coding genes including Wee1, Gab1 and Zfp653. They oscillated at similar circadian phases with their bidirectional genes. Three circadian lncRNAs were antisense to protein coding genes including Zbtb20, Klf13 and Ecsit. The antisense lncRNAs of Zbtb20 and Klf13 also showed similar phases with Zbtb20 and Klf13 respectively.
Table 1. Circadian lncRNAs oscillating in at least six mouse tissues.
| Chr | Start | End | ID | Cluster | Liver | Kidney | Heart | Brain stem | Adrenal gland | Muscle | Brown fat | White fat | Lung | Aorta | Cerebellum | Hypothalamus | SCN | Neighboring genes | Note |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr7 | 117263651 | 117265632 | lnc_07344 | cluster_7325 | 12.3 | 11.8 | 14.2 | 12.5 | 12.7 | 16.2 | 15.5 | 12.8 | 11.3 | 14.0 | 14.5 | NA | NA | Wee1 | bi; B |
| chr4 | 144553297 | 144555395 | lnc_10282 | cluster_5569 | 20.3 | 19.7 | NA | 15.8 | 19.3 | NA | 21.2 | 21.5 | 19.5 | 20.3 | 20.8 | NA | NA | Vps13d | in |
| chr7 | 120383763 | 120386730 | lnc_12450 | cluster_7071 | 22.3 | 22.0 | 23.5 | NA | 20.7 | 21.7 | 22.8 | 22.7 | 22.0 | 23.8 | NA | NA | NA | Bmal1 | sen; B,R,E |
| chr16 | 42884482 | 42912183 | lnc_02172 | cluster_3171 | 16.2 | 15.3 | 18.8 | NA | NA | NA | 19.0 | NA | 17.2 | 17.5 | 18.5 | 17.8 | NA | Zbtb20 | an; E |
| chr7 | 71042148 | 71044758 | lnc_10849 | cluster_6986 | 14.3 | 11.7 | 14.7 | NA | 11.0 | NA | 15.8 | NA | 10.7 | 14.2 | NA | 14.3 | 12.3 | Klf13 | an; B,R,E |
| chr9 | 21875445 | 21890575 | lnc_02176 | cluster_7921 | 5.3 | 5.3 | 2.5 | NA | 7.0 | NA | NA | 9.7 | 7.3 | 8.2 | NA | NA | NA | Zfp653,Ecsit | bi, an; B |
| chr4 | 144556433 | 144557622 | lnc_06077 | cluster_5570 | 20.3 | 19.8 | NA | NA | 19.2 | 20.5 | 18.8 | NA | 20.0 | 19.8 | NA | NA | NA | Vps13d | in; |
| chr17 | 15533165 | 15570545 | lnc_01930 | cluster_3254 | 12.0 | 14.0 | 15.3 | NA | NA | NA | NA | NA | 14.7 | 16.0 | 17.7 | NA | NA | Dll1 | in; B,R,E |
| chr4 | 126698544 | 126714258 | lnc_02286 | cluster_5522 | 12.8 | NA | 15.8 | 15.8 | 15.2 | NA | NA | NA | NA | NA | 18.7 | 18.0 | 16.8 | Sfpq | sen; E |
| chr13 | 35069616 | 35086013 | lnc_02666 | cluster_2087 | 9.2 | NA | 6.3 | 11.3 | 7.3 | NA | NA | NA | 5.5 | NA | NA | 10.8 | NA | Eci2 | sen; B,R,E |
| chr4 | 144550296 | 144553133 | lnc_06076 | cluster_5568 | 20.0 | 19.7 | NA | NA | 20.5 | NA | 22.7 | NA | 19.3 | 20.8 | NA | NA | NA | Vps13d | in; B,R,E |
| chr8 | 83404345 | 83405404 | lnc_07494 | cluster_7521 | 13.2 | 12.8 | 17.3 | NA | 14.7 | NA | 20.3 | NA | 12.5 | NA | NA | NA | NA | Gab1 | bi |
| chr5 | 5782334 | 5782661 | lnc_12024 | cluster_6159 | 14.5 | 15.3 | NA | NA | 18.2 | 21.0 | 18.5 | 19.8 | NA | NA | NA | NA | NA | Steap1 | in |
bi(bidirectional; in(intergenic; an(antisense); sen(sense overlapping); B(BMAL 1 binding); R(REV ERBα binding); E(Enhancer); NA(non-circadian).
To examine whether circadian lncRNAs also tend to be associated with enhancers in tissues other than liver, we analyzed a recently published data of enhancers and circadian RNA-seq data in mouse pancreas where samples were taken every 4 h for 48 h (58). Using the same pipeline of lncRNA assembly from RNA-seq in liver, we identified 7853 lncRNA clusters expressed in pancreas. Among them, 1914 lncRNA clusters showed circadian expression. After obtaining the active enhancers marked by H3K27ac in pancreas, we found that around 832 circadian lncRNA clusters overlapped with enhancer regions, again resulting in a significant enrichment of enhancers in circadian lncRNAs (Fisher's exact test, P-value = 0.007) (Figure 3E). Therefore, the significant association between circadian lncRNAs and enhancers also existed in tissues other than liver.
Comparative analysis of circadian lncRNAs
We next investigated the conservation of these circadian lncRNAs in species other than mouse. To this aim, we analyzed circadian RNA-seq data of rat liver (GEO accession GSE77572) as well as a RNA-seq data of WT rat liver (59). Rat liver lncRNAs were de novo assembled by a similar method to the one that we used for mouse liver. We obtained 6713 lncRNA clusters in rat liver and 790 lncRNA clusters with circadian expression (cosine fitting P-value < 0.01) (Supplementary Table S5). As lncRNAs are known for their poor sequence conservation (60,57), here we searched for lncRNA expression in orthologous intergenic regions (OIRs) defined as the intergenic regions flanked by orthologous neighboring genes between mouse and rat (40). Among 10771 OIRs between mouse and rat, 1975 and 1682 of them harbored lncRNAs in mouse and rat respectively. A significant number of OIRs, i.e. 608 of them, expressed lncRNAs in both mouse and rat livers (Fisher's exact test, P-value < 2.2 × 10−16, Figure 3F). However, only 36% of these 608 OIRs containing lncRNAs can be aligned between mouse and rat by BLAST (E-value < 1 × 10−10) and the aligned sequence identities of lncRNAs were significantly lower than those of protein coding genes (Supplementary Figure S4A). This indicates that the intergenic regions that express lncRNAs in mouse also tend to express lncRNAs in rat despite the poor sequence conservation of lncRNA transcripts between the two species. Combining mouse and rat circadian RNA-seq data, we found that 236 and 352 OIRs harbored circadian lncRNAs in mouse and rat respectively. Eighteen of them expressed circadian lncRNAs in both mouse and rat livers. Interestingly, six of these OIRs even showed lncRNA expression in the corresponding orthologous regions in human when we examined a published human liver RNA-seq data (Table 2). Among these 18 OIRs, 13 of them expressed circadian lncRNAs that overlapped enhancers in mouse liver. Almost all these enhancer regions had long stretches of elevated H3K27ac and H3K4me1 signals (Table 2). For example, the lncRNA flanked between Elovl5 and Gclc showed circadian expression peaking at CT0 in mouse liver while it showed circadian expression peaking at CT4 in the orthologous region in rat liver. This orthologous region in human also harbor a lncRNA expressed in human liver. In both mouse and rat livers, the circadian lncRNAs in these orthologous regions tended to follow similar circadian phases with their neighboring circadian protein-coding genes. Taken together, lncRNA transcription tends to be conserved despite of low sequence conservation of lncRNA transcripts and circadian transcription of a subset of enhancer-associated regions is conserved between mouse and rat.
Table 2. Orthologous intergenic regions containing circadian lncRNAs in both mouse and rat livers.
| Flanking genes | Mouse lncRNA phase (CT) | Rat lncRNA phase (CT) | Enhancer in mouse | Have lncRNAs in human |
|---|---|---|---|---|
| Bcl6_Lpp | 2.0 | 5.2 | Yes | Yes |
| Card10_Cdc42ep1 | 3.8 | 11.8 | ||
| Cebpa_Slc7a10 | 1.0 | 7.7 | Yes | |
| Col4a3bp_Hmgcr | 13.2 | 17.0 | ||
| Dnajb11_Ahsg | 6.0 | 2.2 | Yes | |
| Fam129a_Edem3 | 0.2 | 3.0 | ||
| Gclc_Elovl5 | 1.0 | 3.5 | Yes | Yes |
| Hes1_Cpn2 | 20.3 | 20.5 | Yes | Yes |
| Irf2bp2_Tomm20 | 1.8 | 13.8 | Yes | |
| Klf16_Fam108a | 8.3 | 20.7 | Yes | |
| Lrpprc_Ppm1b | 23.2 | 7.0 | Yes | |
| Ptpn9_Sin3a | 12.3 | 2.2 | Yes | |
| Scaf8_Tiam2 | 4.3 | 12.5 | Yes | |
| Serp2_Lacc1 | 4.2 | 7.7 | Yes | |
| Spag9_Tob1 | 22.0 | 4.0 | Yes | Yes |
| Tet3_Dguok | 3.2 | 8.3 | ||
| Tnks_Ppp1r3b | 19.0 | 20.0 | Yes | Yes |
| Tsku_Gucy2d | 18.7 | 20.0 | Yes |
Interactome of a circadian lncRNA, lnc-Crot, marked super-enhancer
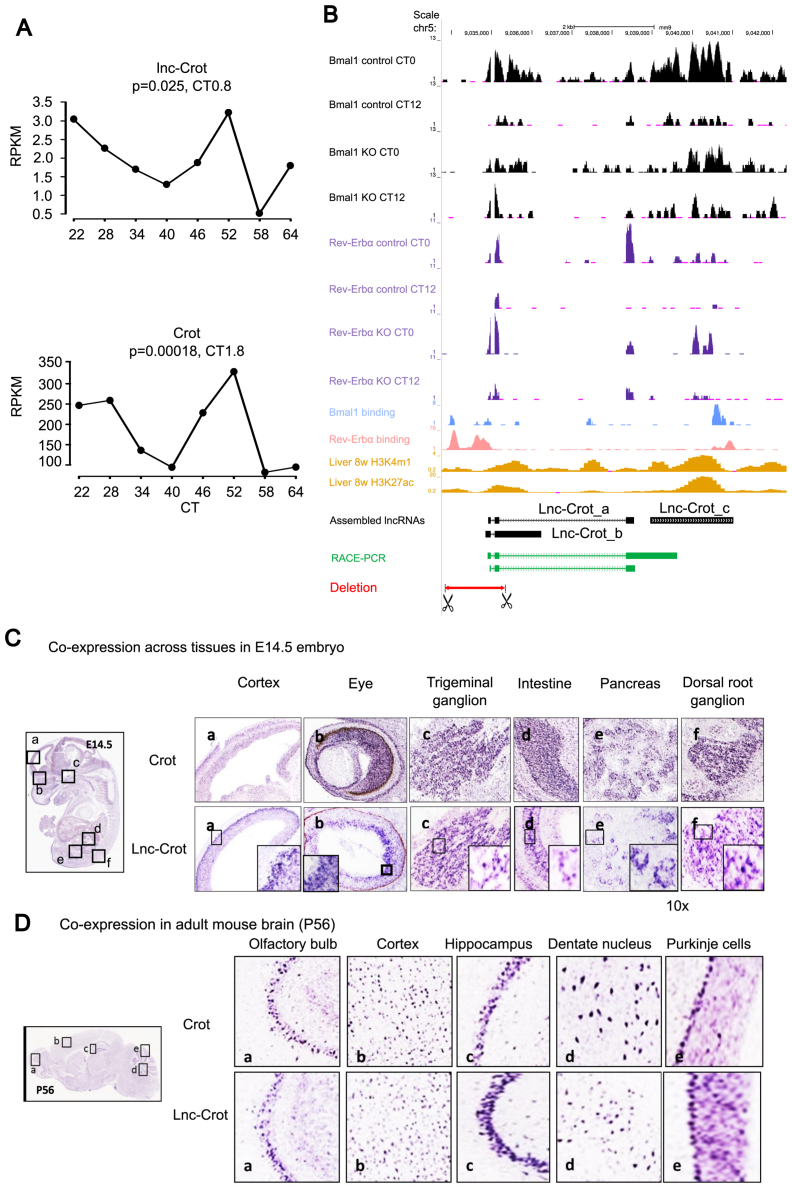
Among the circadian lncRNAs identified in this study, one particular lncRNA is located within a cluster of circadian protein-coding genes on the genome, including Crot, Dmtf1, Abcb4, Abcb1a, Rundc3b, Slc25a40 and Dbf4. This circadian lncRNA was named lnc-Crot because it is associated with a super-enhancer (Figure 4B) upstream of its nearest protein-coding gene, Crot and its circadian expression is shown in Figure 1D (lncRNA ID: Lnc_00179). Lnc-Crot showed strong positive correlation of circadian expression with its two nearest neighboring protein-coding genes, Crot (r = 0.85) and Dmtf1 (r = 0.79). The expression levels of lnc-Crot and Crot were oscillating with close circadian phases at CT23.5 and CT1.8 respectively in liver (Figure 4A) as well as in kidney (Supplementary Figure S3A). And the oscillation of lnc-Crot was already established in the nucleus with a similar circadian phase based on nascent-seq data (Supplementary Figure S3B). The enhancers defined from mouse liver GRO-seq (22) and 5΄ CAGE technology (61) were consistent with H3K27ac marks of enhancer at this lnc-Crot region (Supplementary Figure S3C). Using RACE assay, we obtained the full-length sequence of lnc-Crot (Figure 4B). Both BMAL1 and REV-ERBα binding sites were found on the promoter and gene body of lnc-Crot (Figure 4B) but not on the promoters of nearby circadian genes including Crot, Tmem243 and Dmtf1 (Supplementary Figure S3D). Circadian expression of lnc-Crot was disrupted when BMAL1 or REV-ERBα was knocked out, resulting in its downregulation at CT0 in BMAL1 knockout mice and upregulation at CT12 in REV-ERBα knockout mice (Figure 4B, Supplementary Figure S3E). This demonstrated that lnc-Crot was directly regulated by BMAL1 and REV-ERBα.
Figure 4.
Enhancer associated circadian lncRNA lnc-Crot and its co-expression with its neighboring gene Crot. (A) Circadian expression of lnc-Crot and its neighboring protein coding gene Crot in liver RNA-seq dataset 2. Significance of circadian oscillation and phase were calculated by cosine fitting method. (B) Lnc-Crot locus overlaps with enhancers and is bound by multiple transcription factors (TFs) as displayed by UCSC Genome Browser (mouse genome mm9) (http://genome.ucsc.edu). The tracks in black are RNA-seq of BMAL1 KO and control in liver and tracks in purple are RNA-seq of REV-ERBα KO and control in liver. BMAL1 binding (26,20), REV-ERBα binding (27,50) and histone marks were from ChIP-seq data in liver. Assembled gene models of three isoforms of lnc-Crot locus (lnc-Crot_a, lnc-Crot_b and lnc-Crot_c) were shown. Green track is the full-length gene model of lnc-Crot obtained by rapid amplification of cDNA ends (RACE)-PCR experiment. Red track is the deleted region of lnc-Crot by CRISPR-Cas9. (C) RNA-ISH of lnc-Crot and Crot across tissues in E14.5 mouse embryo (D) RNA-ISH of lnc-Crot and Crot across tissues in P56 adult mouse brain.
By qPCR, we showed that Crot and lnc-Crot were co-expressed (r = 0.8) across adult mouse tissues with elevated expression in liver and kidney (Supplementary Figure S3F). In E14.5 mouse embryo, RNA ISH showed that lnc-Crot and Crot have similar regional expression patterns with expression in pancreas, intestine, dorsal root ganglion, trigeminal ganglion, eye and cortex (Figure 4C). In adult mouse brain at postnatal 56 days, lnc-Crot and Crot also have similar expression in regions including olfactory bulb, cortex, hippocampus, dentate nucleus and Purkinje cells in cerebellum (Figure 4D).
We found that lnc-Crot OIRs flanked by Crot and Dmtf1 on the rat and human genomes also harbored actively transcribed lncRNA clusters in rat and human livers (Supplementary Figure S4B). In rat liver, the direction of the lncRNA transcription has been opposite to lnc-Crot in mouse liver. However, it still showed circadian expression with a moderate P-value = 0.1 according to circadian rat liver RNA-seq data. When we assayed its expression in rat livers by qPCR, it showed a significant circadian oscillation (P-value = 0.008) with a phase at CT5.7 (Supplementary Figure S3G), which is delayed for around 5 h from the phase of lnc-Crot in mouse liver. This is consistent with a general phase delay of 4–5 h of circadian genes in rat compared to mouse (25).
In order to examine the long-range interactions with the lnc-Crot locus, we conducted a 4C-seq experiment in mouse livers collected at CT6 (n = 3) and CT18 (n = 3) using the lnc-Crot region as the bait, where lnc-Crot expressed at relatively high levels at CT6 and low levels at CT18. 4C signals from three biological replicates at each circadian time were highly reproducible judging from their cross-sample correlations between 0.89 and 0.95. About 44% of 4C signals fell within chromosome 5 where lnc-Crot is situated and the signal profile around lnc-Crot region was displayed in Supplementary Figure S5A. To take into account the background interactions that decrease with the distance to the bait, we performed an analysis based on running-window smoothing and Loess regression (38) to select the significant interacting regions on chromosome 5. For each interacting region, we calculated a z-score to measure the difference between the observed 4C signals of this region and the expected values of background (62) (Materials and Methods section). To identify the specific regions or genes that may have different interaction strengths between CT6 and CT18, we performed Student's t-test on the z-scores of each interacting region across samples from the two circadian time points. Among all 2610 interacting regions, only 25 regions associated with nine genes showed significant difference in interaction strengths between CT6 and CT18 (Student'st-test, P-value < 0.05). It indicates that there are no overall significant changes in interactions between circadian time in our experiment (Figure 5A) and the long-range interactions are largely stable in 24 h similar to our previous finding (35). As such, we pooled the CT6 and CT18 samples together and selected the interacting regions present in at least two samples as the overall interacting regions. This resulted in 519 interacting regions associated with 120 genes.
We found that there were 36 genes with circadian expression among the lnc-Crot interacting genes. These include Dmtf1, Abcb4, Abcb1a, Rundc3b and Slc25a40 around the bait region on the genome (shown in Figure 5C). The proportion of circadian oscillating genes among the lnc-Crot interacting genes (30%) is higher than the proportion of circadian oscillating genes across the whole genome (21%, Chi-square test, P = 0.027). The circadian phases of 36 interacting circadian oscillating genes are enriched in the light phase between CT0 and CT8 (single-sided Fisher's exact test P = 0.034) compared with the overall phase distribution of circadian oscillating genes in mouse liver, where circadian oscillating genes were defined by a set of liver circadian microarray data with 1 h interval in 48 h (63) (Figure 5B). Therefore, circadian oscillating genes interacting with lnc-Crot tend to have circadian phases close to lnc-Crot. Taken together, we found that the super-enhancer-associated lnc-Crot tended to interact with the genes showing concerted circadian expression.
Furthermore, we examined a previously published high-throughput Hi-C data (64) for the long-range interactions of promoters of more than 20000 protein coding genes in mouse fetal liver cells (FLC) and embryonic stem cells (ESC). Consistent with our 4C result in liver, Crot and Tmem243 were also found interacting with lnc-Crot region in FLC and ESC while Abcb4 was found interacting with lnc-Crot in ESC shown by WashU EpiGenome Browser (65) (Figure 5C). The genomic region spanning 5Mb around lnc-Crot is one of highly interacted domains on chromosome 5 shared between FLC and ESC (Supplementary Figure S5B). Therefore, the interactome of lnc-Crot appears largely invariant across different cell types and developmental stages.
Functional enrichment analysis by DAVID (66) showed that the lnc-Crot interacting genes are enriched in membrane transport and ATP-related functions (Supplementary Table S6). Especially, the gene cluster close to the bait region consists of Abcb4, Abcb1b and Abcb1a and Slc25a40, all of which have known functions associated with membrane transporter activity. Interestingly, we observed that 36 lnc-Crot interacted genes showed differential expression in mouse liver after fasting in a previously published RNA-seq study (67), including the genes involved in metabolism (Crot, Plb1, Psph), membrane transport (Abcb4, Abcb1a, 9330182L06Rik, Slc46a3, Dpp6) and cell cycle (Dbf4, Ccng2, Cdk14). We validated the upregulation of lnc-Crot and Crot in mouse liver under fasting by qPCR comparing mice fasted for 24 h with mice under normal feeding condition (Supplementary Figure S6A). From several publicly available microarray data in GEO database, we further found that Crot expression was induced by ketogenic diet (GEO accession: GSE7699, Supplementary Figure S6B) and upregulated in circadian high-fat diet study (GEO accession: GSE52333, Supplementary Figure S6C) at CT16. This is consistent with the presence of PPARa and HNF4a binding sites on lnc-Crot locus from published ChIP-seq data (Supplementary Figure S3C), where PPARa and HNF4a were known as important TFs involved in core circadian feedback loops and regulating nutrient metabolism in rhythmic manners (68–70). Taken together, this indicates that the lnc-Crot locus is a super-enhancer at the center of an interactome integrating both circadian and metabolic regulation in mouse liver.
RNA-independent enhancer function of lnc-Crot locus
In order to investigate the function of lnc-Crot marked enhancer, we deleted a DNA region of lnc-Crot, where various TFs and enhancer histone marks are located (Supplementary Figure S3C), in Hepa1-6 cells by CRISPR-Cas9 technology. As the enhancer region was bound with TFs, we knocked down one of these factors, REV-ERBα, using siRNAs in both deleted cells and controlled cells and conducted RNA-seq. Among the 120 4C interacted genes, we found that the repression of 14 genes by REV-ERBα, shown by increased expression after REV-ERBα knockdown by siRNA in control cells, was lost after the genomic deletion (Figure 5D). This result suggests that these 14 genes were repressed by REV-ERBα through long-range interactions mediated by the enhancer and the deletion of this enhancer interrupted the regulation of REV-ERBα upon them. Among these genes, Crot, Mll3, 4933407H18Rik and Stard13 showed circadian expression with phases around CT4. This is consistent with our previous knowledge that circadian genes peaked around CT0 tend to be regulated by REV-ERBα (25). To examine whether the enhancer function of the lnc-Crot locus is dependent upon lnc-Crot RNA or its transcription activity, we over-expressed lnc-Crot in cis by inserting a constitutively expressed promoter upstream of the TSS of lnc-Crot (Supplementary Figure S3C) in Hepa1-6 cells by CRISPR-Cas9 and then the resulted transcriptome was sequenced by RNA-seq. Although the expression level of lnc-Crot was increased by more than 700-fold, the 14 genes regulated by REV-ERBα through lnc-Crot enhancer did not show obvious changes of expression after lnc-Crot in cis over-expression (Figure 5E). The result of Crot is validated by qPCR (Supplementary Figure S7A). This suggested that the enhancer function of lnc-Crot locus is not affected by lnc-Crot expression. Then we conducted in cis over-expression of another enhancer associated circadian lncRNA situated close to gene Rere, termed lnc-Rere (Supplementary Figure S7B). By qPCR, we again did not observe obvious expression changes of its neighboring genes Rere and Errif1 either (Figure 5F). Taken together, this suggested that the genomic loci marked by circadian lncRNAs such as lnc-Crot have enhancer function. But this enhancer function does not depend on the RNA or the transcription of the lncRNA transcripts. Our result points to a model that the lnc-Crot locus and nearby genes first form a scaffold upon which histone modifications and TF bindings then take place to confer tissue- and time-dependent expression (Figure 5G). The circadian lncRNAs that we examined may be a result of circadian transcription at the enhancer regions while the lncRNAs themselves do not influence the circadian gene regulation.
DISCUSSION
A major difference between our study and Zhang et al.'s study (21) is that we have comprehensively assembled lncRNAs de novo from RNA-seq data such that novel lncRNAs can be found while Zhang et al. only used existing gene models of around 1000 lncRNAs. For example, lnc-Crot has not been included in Zhang et al.'s study. It has been previously noted that a type of lncRNAs associated with enhancers shows tissue-specific expression in early embryos (71). Our study has extended this notion to a temporally regulated process, circadian rhythm. Fang et al. identified a number of circadian eRNA using GRO-seq (22). These eRNAs were bi-directionally or uni-directionally transcribed from both enhancer and promoter regions. They are usually short unstable transcripts and turn over rapidly thus less detected in cytoplasm. In our study, the circadian lncRNAs were identified by regular RNA-seq and appeared less nucleus-localized than eRNAs. Many of our circadian lncRNAs contained poly(A) tails and multiple exons. The circadian lncRNAs transcribed at enhancer regions identified in our study have been previously referred as elncRNAs (72). The elncRNAs and eRNAs may arise from the same origin when active transcription by Pol II polymerase takes place near the enhancer and promoter regions.
In this study, we found that the long-range interactions between lnc-Crot locus and nearby genes are tissue- and time-invariant. This is consistent with our previous 4C-seq result of circadian interactome for an enhancer upstream of REV-ERBα in mouse liver (35) but different from the finding of Aguilar-Arnal et al. (73). In that study, the interactome of Dbp locus was found to be varying at different time of day. The discrepancy may arise as we studied different genomic loci and we conducted our 4C-seq in mouse liver while Aguilar-Arnal et al. used 4C-chip in synchronized mouse embryo fibroblast (MEF) cells. The interactome of lnc-Crot locus is likely part of a topological-associated domain (TAD) revealed by Hi-C experiment (74). The disruption of TADs can rewire long-range regulatory architecture leading to functional changes in gene expression (75). Evidences suggest that TADs are large chromosome regions (∼Mbps) that are largely invariant between tissues or cell types. However, lnc-Crot expression is higher in liver and kidney than other tissues and the active enhancer marks and HNF4A binding sites at lnc-Crot locus were only found in these two tissues. Hi-C data suggest that the TAD involving lnc-Crot locus is even present in ESCs where a functional circadian clock is absent. Therefore, our result seems to suggest that the invariant interactions between lnc-Crot and nearby genes are necessary but not sufficient to establish tissue- and time-dependent expression of interacted loci. Histone modifications and TF bindings upon the stable scaffold are required to confer tissue- or time-dependent expression of interacted genes. LncRNAs have been previously found to be associated with enhancer functions in both RNA-dependent (51) and RNA-independent manners (76). Our study suggests that many rhythmically transcribed lncRNA transcripts like lnc-Crot fall into the latter category and can serve as the hallmarks of the enhancers that modulate long-range circadian gene regulation.
The interactome of lnc-Crot includes several protein-coding genes important for liver metabolism. Crot, carnitine O-octanoyl transferase located in peroxisome, plays an important role in carnitine shuttling during fatty acid β-oxidation. Abcb4 is liver-specific and involved in transporting phosphorlipid for bile synthesis, which is also under circadian control (77). Our study shows that lnc-Crot locus is a super-enhancer integrating both circadian and metabolic regulation in mouse liver. Circadian gene regulation through lnc-Crot interactome is likely involved in the temporal partition of fatty acid β-oxidation into the inactive (light) phase while fatty acid synthesis predominantly takes place in the active (dark) phase of mouse (78).
The circadian expression of Crot is highly conserved in vertebrates including in mouse peaking at CT0, in rat at CT5 and even in zebrafish at CT12 (79). In rat liver, we have observed circadian transcriptional activities in the intergenic region orthologous to mouse lnc-Crot locus even though the lncRNA itself is not conserved. In human liver, a lncRNA is also transcribed in the orthologous region although whether it shows circadian expression is unclear. In zebrafish, we did not find orthologous region of lnc-Crot. Therefore, the circadian enhancer regulating Crot is probably mammal-specific in spite of conserved circadian expression of Crot in vertebrates. It has been proposed that lncRNAs undergo rapid turnover in rodents giving rise to the tissue- and lineage-specific expression of lncRNAs (15). Our study suggests that circadian lncRNAs such as lnc-Crot evolve from the circadian transcriptional activities associated with the enhancers and therefore has shed new lights on the evolutionary origins of lncRNAs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Anna Marques (University of Lausanne, Switzerland) for providing the lncRNA models assembled from Robertson et al.'s RNA-seq data, Yi Liu (Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, China) for providing the RNA samples of liver-specific BMAL1-KO mice, Ying Xu (Soochow University, China) for providing REV-ERBα KO mice, Christina Thaller and Gregor Eichele (Max-Planck Institute for Biophysical Chemistry, Germany) for the helps on ISH. We are grateful to the experimental support of the Uli Schwarz public laboratory platform and Omics Core in PICB.
Author contributions: Z.F. and J.Y. conceived, designed and interpreted the experiments. M.Z., P.D.J., P.L., Y.Z., W.G. and Z.F. performed experiments. Z.F. and Y.X. performed the bioinformatics analysis. Z.F., J.Y., P.D.J., H.W. and M.Z. wrote and edited the manuscript. Z.Z. and J.Y. contributed reagents/materials/analysis tools.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chinese Academy of Sciences Strategic Priority Research Program [XDB02060006 to J.Y.]; National Natural Science Foundation of China [31571209 to J.Y., 31370762 to Z.Z.]; Natural Science Foundation of Shanghai [16ZR1448800 to H.W.]; National Basic Research Program of China [2013CB966802 to Z.Z.]; Chinese Academy of Sciences (to J.Y.); German Max-Planck Society (to J.Y.). Funding for open access charge: National Natural Science Foundation of China [31571209].
Conflict of interest statement. None declared.
REFERENCES
- 1. Thresher R.J., Vitaterna M.H., Miyamoto Y., Kazantsev a, Hsu D.S., Petit C., Selby C.P., Dawut L., Smithies O., Takahashi J.S. et al. . Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998; 282:1490–1494. [DOI] [PubMed] [Google Scholar]
- 2. Preitner N., Damiola F., Zakany J., Duboule D., Albrecht U., Schibler U.. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator University of Geneva University of Geneva. Cell. 2002; 110:251–260. [DOI] [PubMed] [Google Scholar]
- 3. Relógio A., Westermark P.O., Wallach T., Schellenberg K., Kramer A., Herzel H.. Tuning the mammalian circadian clock: robust synergy of two loops. PLoS Comput. Biol. 2011; 7:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pett J.P., Korenčič A., Wesener F., Kramer A., Herzel H.. Feedback loops of the mammalian circadian clock constitute repressilator. PLOS Comput. Biol. 2016; 12:e1005266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Panda S., Antoch M.P., Miller B.H., Su A.I., Schook A.B., Straume M., Schultz P.G., Kay S.A., Takahashi J.S., Hogenesch J.B.. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002; 109:307–320. [DOI] [PubMed] [Google Scholar]
- 6. Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A. et al. . Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ueda H.R., Hayashi S., Chen W., Sano M., Machida M., Shigeyoshi Y., Iino M., Hashimoto S.. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005; 37:187–192. [DOI] [PubMed] [Google Scholar]
- 8. Storch K.-F., Lipan O., Leykin I., Viswanathan N., Davis F.C., Wong W.H., Weitz C.J.. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002; 417:78–83. [DOI] [PubMed] [Google Scholar]
- 9. Na Y.J., Sung J.H., Lee S.C., Lee Y.J., Choi Y.J., Park W.Y., Shin H.S., Kim J.H.. Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp. Mol. Med. 2009; 41:638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang H., Fan Z., Zhao M., Li J., Lu M., Liu W., Ying H., Liu M., Yan J.. Oscillating primary transcripts harbor miRNAs with circadian functions. Sci. Rep. 2016; 6:21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hangauer M.J., Vaughn I.W., McManus M.T.. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013; 9:e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P. et al. . Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009; 458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-vega B., Regev A., Rinn J.L.. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011; 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P.. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011; 147:1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kutter C., Marques A.. Rapid turnover of long noncoding RNAs and the evolution of gene expression. Plos Genet. 2012; 8:e1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mukherjee N., Calviello L., Hirsekorn A., de Pretis S., Pelizzola M., Ohler U.. Integrative classification of human coding and non-coding genes based on RNA metabolism profiles. Nat. Struct. Mol. Biol. 2016; 24:86–96. [DOI] [PubMed] [Google Scholar]
- 17. Rinn J.L., Chang H.Y.. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012; 81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ulitsky I., Bartel D.P.. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013; 154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xue Z., Ye Q., Anson S.R., Yang J., Xiao G., Kowbel D., Glass N.L., Crosthwaite S.K., Liu Y.. Transcriptional interference by antisense RNA is required for circadian clock function. Nature. 2014; 514:650–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koike N., Yoo S.-H., Huang H.-C., Kumar V., Lee C., Kim T.-K., Takahashi J.S.. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012; 338:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B.. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang B., Everett L.J., Jager J., Briggs E., Armour S.M., Feng D., Roy A., Gerhart-Hines Z., Sun Z., Lazar M.A.. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014; 159:1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li W., Notani D., Rosenfeld M.G.. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016; 17:207–223. [DOI] [PubMed] [Google Scholar]
- 24. Robertson G., Schein J., Chiu R., Corbett R., Field M., Jackman S.D., Mungall K., Lee S., Okada H.M., Qian J.Q. et al. . De novo assembly and analysis of RNA-seq data. Nat. Methods. 2010; 7:909–912. [DOI] [PubMed] [Google Scholar]
- 25. Yan J., Wang H., Liu Y., Shao C.. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput. Biol. 2008; 4:e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rey G., Cesbron F., Rougemont J., Reinke H., Brunner M., Naef F.. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011; 9:e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cho H., Zhao X., Hatori M., Yu R.T., Barish G.D., Lam M.T., Chong L.-W., Ditacchio L., Atkins A.R., Glass C.K. et al. . Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012; 485:123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feng D., Liu T., Sun Z., Bugge A., Mullican S.E., Alenghat T., Liu X.S., Lazar M.A.. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011; 331:1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V et al. . A map of the cis-regulatory sequences in the mouse genome. Nature. 2012; 488:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Forrest A.R.R., Kawaji H., Rehli M., Kenneth Baillie J., de Hoon M.J.L., Haberle V., Lassmann T., Kulakovskiy I. V., Lizio M., Itoh M. et al. . A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rakhshandehroo M., Sanderson L.M., Matilainen M., Stienstra R., Carlberg C., De Groot P.J., Müller M., Kersten S.. Comprehensive analysis of PPARα-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007; 2007:26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt D., Wilson M.D., Ballester B., Schwalie P.C., Brown G.D., Marshall A., Kutter C., Watt S., Martinez-Jimenez C.P., Mackay S. et al. . Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010; 328:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Whyte W.a, Orlando D.a, Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.a. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao Z., Tavoosidana G., Sjölinder M., Göndör A., Mariano P., Wang S., Kanduri C., Lezcano M., Singh Sandhu K., Singh U. et al. . Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006; 38:1341–1347. [DOI] [PubMed] [Google Scholar]
- 35. Xu Y., Guo W., Li P., Zhang Y., Zhao M., Fan Z., Zhao Z., Yan J.. Long-range chromosome interactions mediated by cohesin shape circadian gene expression. PLoS Genet. 2016; 12:e1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tolhuis B., Blom M., Kerkhoven R.M., Pagie L., Teunissen H., Nieuwland M., Simonis M., de Laat W., van Lohuizen M., van Steensel B.. Interactions among polycomb domains are guided by chromosome architecture. PLoS Genet. 2011; 7:e1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. R Core Team R: a language and environment for statistical computing. R Foundationfor Statistical Computing. 2012; Viena: http://www.R-project.org/. [Google Scholar]
- 40. Volders P.-J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P.. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015; 43:D174–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smedley D., Haider S., Durinck S., Pandini L., Provero P., Allen J., Arnaiz O., Awedh M., Baldock R.. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015; 43:W589–W598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q. et al. . NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016; 44:D203–D208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. The ENCODE Project Consortium Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.a, Doyle F., Epstein C.B., Frietze S., Harrow J. et al. . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eichele G., Diez-Roux G.. High-throughput analysis of gene expression on tissue sections by in situ hybridization. Methods. 2011; 53:417–423. [DOI] [PubMed] [Google Scholar]
- 46. Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., Li J.. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013; 13:659–662. [DOI] [PubMed] [Google Scholar]
- 47. Ran F.A., Hsu P.D.P.P.D., Wright J., Agarwala V., Scott D.a, Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heigwer F., Kerr G., Boutros M.. E-CRISP: fast CRISPR target site identification. Nat. Methods. 2014; 11:122–123. [DOI] [PubMed] [Google Scholar]
- 49. Le Martelot G., Canella D., Symul L., Migliavacca E., Gilardi F., Liechti R., Martin O., Harshman K., Delorenzi M., Desvergne B. et al. . Genome-wide RNA Polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012; 10:e1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bugge A., Feng D., Everett L.J., Briggs E.R., Mullican S.E., Wang F., Jager J., Lazar M.A.. Rev-erb a and Rev-erb b coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012; 26:657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lai F., Orom U.a, Cesaroni M., Beringer M., Taatjes D.J., Blobel G.a, Shiekhattar R.. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature. 2013; 494:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W., Merkurjev D., Ohgi K.A., Meng D., Zhang J. et al. . lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013; 500:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pott S., Lieb J.D.. What are super-enhancers?. Nat. Genet. 2014; 47:8–12. [DOI] [PubMed] [Google Scholar]
- 54. Menet J.S., Rodriguez J., Abruzzi K.C., Rosbash M.. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012; 1:e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grote P., Wittler L., Hendrix D., Koch F., Währisch S., Beisaw A., Macura K., Bläss G., Kellis M., Werber M. et al. . The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013; 24:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A.. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007; 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. . The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Perelis M., Marcheva B., Moynihan Ramsey K., Schipma M.J., Hutchison A.L., Taguchi A., Peek C.B., Hong H., Huang W., Omura C. et al. . Pancreatic cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science. 2015; 350:aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu Y., Fuscoe J.C., Zhao C., Guo C., Jia M., Qing T., Bannon D.I., Lancashire L., Bao W., Du T. et al. . A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 2014; 5:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marques A.C., Ponting C.P.. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009; 10:R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. . An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ghavi-Helm Y., Klein F.a., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E.M.. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014; 512:96–100. [DOI] [PubMed] [Google Scholar]
- 63. Hughes M.E., DiTacchio L., Hayes K.R., Vollmers C., Pulivarthy S., Baggs J.E., Panda S., Hogenesch J.B.. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009; 5:e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schoenfelder S., Furlan-magaril M., Mifsud B., Tavares-cadete F., Sugar R., Javierre B., Nagano T., Katsman Y., Sakthidevi M., Wingett S.W. et al. . The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015; 25:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou X., Lowdon R.F., Li D., Lawson H.a, Madden P.aF., Costello J.F., Wang T.. Exploring long-range genome interactions using the WashU Epigenome browser. Nat. Methods. 2013; 10:375–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cavalieri D., De Filippo C.. Bioinformatic methods for integrating whole-genome expression results into cellular networks. Drug Discov. Today. 2005; 10:727–734. [DOI] [PubMed] [Google Scholar]
- 67. Chu Y., Gómez Rosso L., Huang P., Wang Z., Xu Y., Yao X., Bao M., Yan J., Song H., Wang G.. Liver Med23 ablation improves glucose and lipid metabolism through modulating FOXO1 activity. Cell Res. 2014; 24:1250–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schmutz I., Ripperger J.A., Baeriswyl-Aebischer S., Albrecht U.. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010; 24:345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang X., Downes M., Yu R.T., Bookout A.L., He W., Straume M., Mangelsdorf D.J., Evans R.M.. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006; 126:801–810. [DOI] [PubMed] [Google Scholar]
- 70. Reddy A.B., Maywood E.S., Karp N.A., King V.M., Inoue Y., Gonzalez F.J., Lilley K.S., Kyriacou C.P., Hastings M.H.. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology. 2007; 45:1478–1488. [DOI] [PubMed] [Google Scholar]
- 71. Wu H., Nord A.S., Akiyama J.a., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.a., Visel A.. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS Genet. 2014; 10:e1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vance K.W., Ponting C.P.. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014; 30:348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aguilar-Arnal L., Hakim O., Patel V.R., Baldi P., Hager G.L., Sassone-Corsi P.. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat. Struct. Mol. Biol. 2013; 20:1206–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. et al. . Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015; 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yoo E.J., Cooke N.E., Liebhaber S.A.. An RNA-independent linkage of noncoding transcription to long-range enhancer function. Mol. Cell. Biol. 2012; 32:2020–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Han S.S., Zhang R., Jain R., Shi H., Zhang L., Zhou G., Sangwung P., Tugal D., Atkins G.B., Prosdocimo D.a. et al. . Circadian control of bile acid synthesis by a KLF15-Fgf15 axis. Nat. Commun. 2015; 6:7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bass J., Takahashi J.S.. Circadian integration of metabolism and energetics. Science. 2010; 330:1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li Y., Li G., Wang H., Du J., Yan J.. Analysis of a gene regulatory cascade mediating circadian rhythm in Zebrafish. PLoS Comput. Biol. 2013; 9:e1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.