Abstract
RNA aptamers that bind HIV-1 reverse transcriptase (RT) inhibit HIV-1 replication, but little is known about potential aptamer-specific viral resistance. During replication, RT interacts with diverse nucleic acids. Thus, the genetic threshold for eliciting resistance may be high for aptamers that make numerous contacts with RT. To evaluate the impact of RT–aptamer binding specificity on replication, we engineered proviral plasmids encoding diverse RTs within the backbone of HIV-1 strain NL4-3. Viruses inhibited by pseudoknot aptamers were rendered insensitive by a naturally occurring R277K variant, providing the first demonstration of aptamer-specific resistance in cell culture. Naturally occurring, pseudoknot-insensitive viruses were rendered sensitive by the inverse K277R mutation, establishing RT as the genetic locus for aptamer-mediated HIV-1 inhibition. Non-pseudoknot RNA aptamers exhibited broad-spectrum inhibition. Inhibition was observed only when virus was produced in aptamer-expressing cells, indicating that encapsidation is required. HIV-1 suppression magnitude correlated with the number of encapsidated aptamer transcripts per virion, with saturation occurring around 1:1 stoichiometry with packaged RT. Encapsidation specificity suggests that aptamers may encounter dimerized GagPol in the cytosol during viral assembly. This study provides new insights into HIV-1's capacity to escape aptamer-mediated inhibition, the potential utility of broad-spectrum aptamers to overcome resistance, and molecular interactions that occur during viral assembly.
INTRODUCTION
Aptamers are structured nucleic acids that bind specific molecular targets with high affinity. Aptamers that target type 1 human immunodeficiency virus (HIV-1) reverse transcriptase (RT) bind RT at low nanomolar to picomolar concentrations and inhibit RT enzymatic activities by competing with primer-template substrates for access to the RT active site (1,2). When expressed in cultured human cells, they are encapsidated into nascent virions and inhibit the subsequent infection of target cells (3–6). Together, these qualities make aptamers attractive tools for dissecting viral pathogenesis, and several groups have explored their potential use in genetic therapies. However, the molecular and cellular interactions between aptamer and virus that govern antiviral specificity and aptamer encapsidation into the viral particle have not been clearly defined, and their propensity for eliciting aptamer-specific viral resistance is largely unexplored.
RT must interact with diverse nucleic acids with variable composition (RNA/RNA, RNA/DNA, DNA/RNA and DNA/DNA) and structures during HIV-1 replication. Because anti-HIV RT aptamers bind within the active site of the enzyme, any viral mutation that reduces RT binding to an inhibitory aptamer could potentially also reduce viral fitness by interfering with RT binding to viral genome (1,7). In support of this idea, RT mutations N255D and N265D, which confer resistance to DNA aptamer RT1t49 in enzymatic assays, were shown to induce processivity defects and to impair replication when introduced into viruses (7). On the other hand, aptamers from various structural families exhibit differential sensitivities to natural and engineered RT amino acid sequence variation in biochemical assays (8–13). Some of these RTs are derived from naturally occurring, viable viruses and are unlikely to represent false positives for resistance, although this biochemical insensitivity has not yet been validated in a biological context.
To date, four RNA aptamer structural families have been identified as potent inhibitors of HIV-1 RT: family 1 pseudoknot motif (F1Pk), family 2 pseudoknot motif (F2Pk), 6/5 asymmetric loop motif (6/5AL) and UCAA bulge motif (UCAA) (1,2,9,10). Aptamers from each of these structural families inhibit RT from the HIV-1 subtype B strain used as the original selection target (BH10), as well as RT from close relatives (HXB2 and NL4-3) (1,2,9–11). F1Pk aptamers fail to inhibit RT from most other subtypes when tested against a panel of phylogenetically diverse RTs in biochemical assays, and a single point mutation (R277K) was sufficient to abolish inhibition by F1Pk aptamers (8). In contrast, single-stranded DNA aptamers RT1t49 and R1T strongly inhibited every RT in the RT panel (12,13) by binding RT in a manner that mimics its binding to the natural substrate (14). The 6/5AL and UCAA aptamer structural families emerged from a selection performed to identify aptamers with increased RT affinity (9,10), but their abilities to inhibit diverse RT subtypes have not yet been described. In principle, broad-spectrum aptamers could enhance combination therapies and curtail the emergence of new viral escape mutants, provided that they exhibit similar broad-spectrum suppression of viral replication in cell culture.
When anti-RT RNA aptamers are expressed intracellularly in virus producer cells, they are encapsidated into nascent virions and inhibit replication by those viruses when they infect new target cell (3–5). A diverse array of molecular interactions drive encapsidation of RNA and other molecules into viral particles. During HIV replication, encapsidation of the RNA genome is mediated by the nucleocapsid (NC) domain of Gag (15). Encapsidation of tRNALys3, the primer for HIV-1 reverse transcription, is required for replication and occurs in part through interaction of tRNALys3 with the LysRS protein, which is co-packaged with Pol (16,17). Likewise, viral restriction factor APOBEC3G is also encapsidated into virions via interactions with NC, where it reduces the net fidelity of genome replication by hypermutating the nascent proviral DNA minus strand (18,19). HIV also encapsidates several other cellular RNAs and proteins. Most encapsidated cellular RNAs are packaged in proportion to their expression levels within the cell, suggesting a non-specific, passive mechanism for incorporation (20). However, a few cellular RNAs, such as 7SL, are encapsidated into viral particles to a greater extent than suggested by their expression levels in cells (20,21). While the mechanisms driving high efficiency encapsidation for some of those RNAs are not well understood, their preferential enrichment within virions suggests active encapsidation. Notably, while it is generally assumed that anti-RT aptamer encapsdiation is mediated through aptamer–RT interactions within the producer cell, this has not been directly demonstrated in cell culture experiments.
Here, we present evidence that stoichiometric encapsidation of inhibitory anti-HIV RT aptamers in proportion to the number of viral RTs is responsible for inhibition of HIV-1 replication. Quantification of aptamer transcripts relative to viral genomes established that inhibition correlates with the number of aptamer molecules incorporated, with maximal inhibition plateauing at around 1:1 stoichiometry with RT. Non-specific mechanisms may drive encapsidation of non-inhibitory aptamers, as higher levels of aptamer expression are required to encapsidate non-inhibitory transcripts to levels equal to those of the inhibitory aptamers, suggesting a non-specific encapsidation mechanism. Importantly, even when encapsidated to the level of inhibitory aptamers, these aptamers fail to inhibit viral replication. These observations have important implications for the dimerization state and accessibility of the GagPol polyprotein in the cytoplasm of producer cells during assembly. Additionally, we demonstrate that aptamer expression in target cells does not protect against subsequent infection, and therefore that prior encapsidation is required for aptamer-mediated inhibition of HIV replication. These data provide independent evidence in clear support of current models of HIV-1 capsid disassembly, in which the capsid does not immediately dissociate upon entry into the target cell cytoplasm (22). Finally, we evaluated potential broad-spectrum inhibitory activity for the four known aptamer structural families and demonstrate that specific structural families inhibit recombinant viruses that express a phylogenetically diverse panel of RT subtypes. Together, the data presented here expand our knowledge of aptamer–RT interactions, as well as the mechanism of HIV assembly, encapsidation and disassembly.
MATERIALS AND METHODS
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA). Restriction enzymes and T4 DNA ligase used for cloning purposes were purchased from New England Biolabs (Ipswitch, MA, USA).
Plasmids
Aptamers from the four RNA structural families were 70.05 (F1Pk), 70.08 (F2PK), 88.1 (6/5AL) and 80.103 (UCAA) (1,9,10). Sequences of aptamers and the arbitrary control can be found in Supplementary Table S1. Double-stranded DNA for each RNA was cloned into a pcDNA3.1-based plasmid to drive aptamer expression from the human cytomegalovirus (CMV) immediate early promoter, as previously described (4,23).The HIV-1NL3-4-derived plasmid (pNL4-3-CMV-EGFP) modified for single-cycle infectivity assays was kindly provided by Vineet KewalRamani (National Cancer Institute, Fredrick, MD, USA). This proviral vector lacks the genes encoding vif, vpr, vpu, nef and env, and carries enhanced green fluorescent protein (EGFP) in place of nef, expressed from a CMV immediate early promoter (24). Phylogenetically diverse RT have been previously described (8) (Figure 1 and Table 1) and were cloned in place of the NL4-3 RT by introducing silent sites for XmaI and NheI flanking the RT sequence region (Supplementary Figures S1 and 2). The vesicular stomatitis virus (VSV) glycoprotein-expressing plasmid, pMD-G, used for viral pseudotyping, was obtained from Invitrogen (Carlsbad, CA, USA). All plasmids were verified by sequencing.
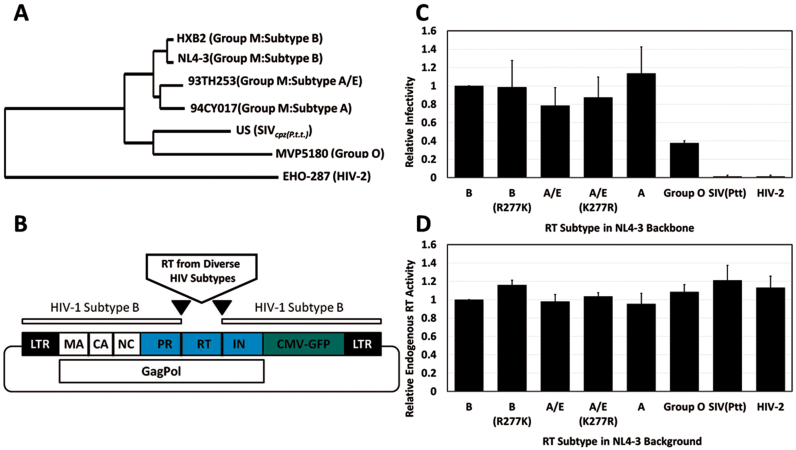
Figure 1.
Proviruses expressing diverse RT subtypes in the NL4-3 background are viable. (A) Phylogenetically diverse RT subtypes used in this study. (B) Schematic of cloning strategy using NL4-3 as the backbone for expression of phylogenetically diverse RT subtypes. (C) Infectivity of recombinant virus expressing phylogenetically diverse RT sequences was determined using flow cytometry and is shown as % EGFP-positive cells normalized to p24 for each sample. Values for the Subtype B proviral construct were set to 1. Values are shown as the mean ± SD for three experiments. (D) Endogenous RT activity was assessed for purified viral particles and normalized to p24. Values obtained for the Subtype B proviral construct were set to 1. Plasmid pNL4-3-CMV-GFPDeltaRT was used to establish assay background. Values are shown as the mean ± SD for three experiments.
Table 1. Strains of HIV used in the study.
| HIV strain | Group | % Identity | Accession number |
|---|---|---|---|
| HXB2 | HIV-1 (M:B) | 98.9 | K03455 |
| HXB2 (R277K) | HIV-1 (M:B) | 98.9 | K03455 |
| 93TH253 | HIV-1 (M:A/E) | 90.5 | U51189 |
| 93TH253 (K277R) | HIV-1 (M:A/E) | 90.5 | U51189 |
| 94CY017 | HIV-1 (M:A) | 90.4 | AF286237 |
| MVP5180 | HIV-1 (M:O) | 78.2 | L20571.1 |
| SIV-US | SIVcpz(P.t.t.) | 72.7 | AF103818 |
| EHO | HIV-2 (B) | 60.5 | U27200 |
Percent identity is based on the RT amino acid sequence.
Cell lines and transfections
The human embryonic kidney cell line, 293FT (Invitrogen, Carlsbad, CA, USA) was maintained in standard culture media containing Dulbecco's Minimum Essential Medium (Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma, St Louis, MO, USA), 2 mM L-glutamine (Gibco, Life Technologies, Grand Island, NY, USA), 1 mM non-essential amino acids (Gibco, Life Technologies, Grand Island, NY, USA) and 1 mM sodium pyruvate (Gibco, Life Technologies, Grand Island, NY, USA). CEM-T4 (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: CEM CD4+ Cells from Dr J.P. Jacobs) (25) and H9 (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: H9 from Dr Robert Gallo) (26–28) T cell lines were maintained in RPMI medium (Sigma, St Louis, MO, USA) supplemented with 10% FBS (Sigma, St Louis, MO, USA) and 2 mM L-glutamine (Gibco, Life Technologies, Grand Island, NY, USA). All cell lines were maintained at 37°C in 5% carbon dioxide. Transfections were performed with the indicated amounts of DNA on cells plated the previous day using polyethylenimine (PEI) at 3 μl per μg of DNA, as previously described (4,23).
Single-cycle infectivity and endogenous RT activity assays
For experiments evaluating encapsidated aptamer, 293FT cells were co-transfected with a mixture of recombinant pNL4-3-CMV-GFP (150 ng), pMD-G (50 ng) and aptamer-expressing plasmids (indicated dosages). Medium was changed ∼12 h post-transfection. Supernatant containing VSV-pseudotyped virus was harvested 48 h post-medium change. Infectivity was determined by adding viral supernatant to fresh 293FT cells such that arbitrary control infectivity levels were ∼10%. Infected cells were collected 24–48 h post-infection and fixed with 2% paraformaldehyde for analysis by flow cytometry. Infectivity was normalized to p24 ELISA values and to the arbitrary RNA control. Endogenous RT activity assays (29) were performed with cell-free virus produced in the presence or absence of aptamer using the EnzCheck Reverse Transcriptase Kit (Molecular Probes, Eugene, OR, USA) per manufacturer's directions. A virus lacking the RT sequence, pNL4-3-CMV-GFPDeltaRT, was used as a control to establish assay background. Briefly, virus-containing supernatant was syringe filtered and pelleted through a 20% sucrose cushion by centrifugation at 30 000 RPM for 2 h at 4°C. Pellets were resuspended in assay buffer containing 0.1% Triton X-100 and 1 mM dNTPs. Reactions were incubated at 37°C for 18 h. Picogreen solution was prepared and was added to each reaction to detect reverse transcription products from the viral genome template. Fluorescence was read on a Perkin Elmer AlphaLISA instrument (excitation 480 nm, emission 520 nm). Values shown represent endogenous RT activity after subtracting pNL4-3-CMV-GFPDeltaRT values and normalizing to the Arbitrary control value.
To evaluate the effect of aptamer expression in target cells, viruses were produced by co-transfecting 293FT cells in 10 cm dishes with proviral plasmid expressing subtype B RT (10 μg) and pMD-G (1 μg) complexed with PEI (3 μl/μg DNA) in the absence of aptamer expressing plasmid. Virus was harvested 48 h post media change. Separately, aptamer-expressing plasmids were transfected into fresh target 293FT cells at varying doses (1000–3000 ng). At varying times post-transfection (6, 24 or 48 h), these cells were challenged with virus produced in the absence of aptamer. EGFP-positive cells were counted and evaluated 24–48 h post-infection as above.
Cellular and viral RNA isolation and real time quantitative PCR
For isolation of total cellular RNA (4,23), transfected 293FT cells were collected by scraping into 1× phosphate buffered saline (PBS) and washed once with 1× PBS, followed by RNA isolation using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) per the manufacturer's instructions. RNA was precipitated twice and washed with 70% ethanol to remove any contaminants resulting from TRIzol RNA isolation procedures. Isolated RNA was subjected to DNase treatment using Turbo DNase (Ambion, Life Technologies, Grand Island, NY, USA) and quantified on a NanoDrop Spectrophotometer (Thermo-Fisher Scientific, Waltham, MA, USA). For isolation of viral RNA, 1.5 ml of cell-free virus was pelleted by centrifugation at 30 000 × g for 2 h at 4°C using an Eppendorf Centrifuge. Supernatant was discarded and viral RNA was isolated from the viral pellet using the TRIzol-LS reagent (Invitrogen, Carlsbad, CA, USA) per the manufacturer's instructions. Glycogen (2 μg) was added to each sample. Viral RNA was precipitated twice, washed with 70% ethanol and subjected to Turbo DNase treatment and NanoDrop quantification.
Total cellular and viral RNA were used to synthesize cDNA using random hexamer primers with the ImProm II Reverse Transcription System (Promega, Madison, WI, USA) per the manufacturer's instructions. The cDNA synthesis was performed using 500 ng RNA. ‘No RT’ and ‘non-template’ (NTC) controls were included. Prior to performing Real Time Quantitative PCR, cellular and viral cDNA, including ‘no RT’ and ‘NTC’ controls, were validated by endpoint PCR (40 cycles) using primers specific for aptamer constant regions, 18S rRNA and HIV-1 gag (4). Each primer set was previously validated for amplification efficiency (4). Viral cDNA was free of 18S rRNA signal, indicating no cellular contamination and ‘no RT’ and ‘NTC’ controls were free of contaminating signal. RT-qPCR was performed on each set of cDNA using iTaq Universal SYBR Green Master Mix (BioRad, Hercules, CA, USA) and primers specific to aptamer constant regions, 18S rRNA (cellular reference gene), or HIV-1 gag (viral genome reference gene) according to the manufacturer's instructions (4). Each sample was assayed in triplicate to determine technical variability. Relative quantities were determined using the relative quantity (2−ΔΔCT) method and normalized to the specified endogenous control and to the negative control (set to 1) as previously described (4). Samples were assayed on the ABI 7500 (Applied Biosystems, Foster City, CA, USA) and analyzed using ABI 7500 Software Version 2.3. Experiments were repeated three times, and two representative experiments are shown to demonstrate experimental reproducibility (Figure 4 and Supplementary Figure S5).
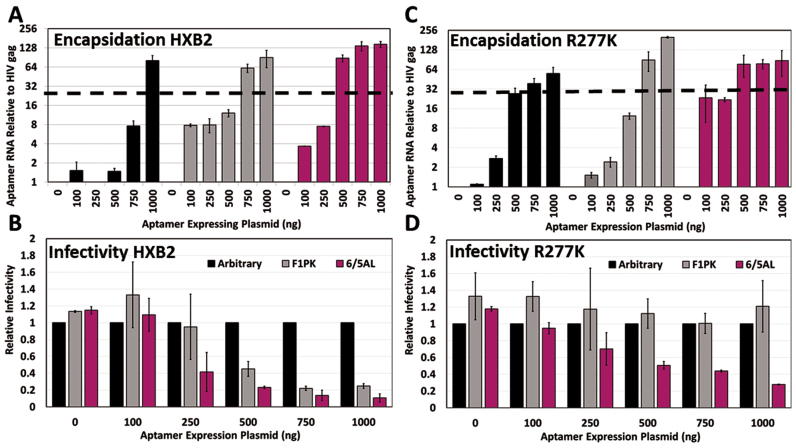
Figure 4.
Differential encapsidation levels for inhibitory and non-inhibitory aptamers at low doses. (A and C) Viral RNA was harvested from cell-free virus produced by co-transfection of aptamer expressing plasmids (0–1000 ng), DNA filler plasmid (1–1000 ng), proviruses expressing either HXB2 (A) or the HXB2 point mutant R277k (C) (150 ng) and pMD-G (50 ng). cDNA was synthesized from 500 ng of viral RNA and subjected to qRT-PCR using primers to amplify aptamers and HIV-1 gag. ‘No RT’ controls were included for each sample, and each sample was assayed in triplicate to determine technical variability. Samples were normalized to HIV-1 gag and the 0 ng control, and averaged. Values shown are the means of three technical replicates ±SD. A representative experiment of three total experiments is shown. An additional experiment can be found in the Supplement (Supplementary Figures S5B and C) to demonstrate experimental reproducibility. (B and D) VSV-pseudotyped virus generated as described in (A) and (C) was used to assess viral infectivity in 293FT cells by flow cytometry as described in Figure 2. Values are the mean % EGFP measured ± SD for three independent experiments.
Aptamer Kd determination
The F1Pk and 6/5AL aptamers and arbitrary control were 5΄-end labeled with 32P-ATP and polynucleotide kinase, then added to solutions containing various concentrations of subtype B RT or the R277K point mutant (0, 0.1, 0.3, 1, 3, 10, 100 or 300 nM final concentrations) in binding buffer (140 mM KCl, 1 mM MgCl2, 50 mM Tris–HCl, pH 7.5). Aptamer–RT binding reactions were incubated on ice for 15 min. Reactions were then loaded onto pre-wet nitrocellulose membranes. An unfiltered sample with 0 mM RT was used to define 100% signal. Samples were filtered through the nitrocellulose membranes under vacuum to capture aptamer–RT complexes. Membranes were transferred to scintillation vials with 4 ml of scintillation liquid for determination of radioactivity via scintillation counter. Percent bound to protein was calculated by subtracting the signal from the filtered ‘No protein’ sample from all values and then dividing by the signal from the unfiltered control representing ‘total input.’ Values at each RT concentration were fit into a Hill slope one-site Kd binding graph using Prism software version 6.2. Aptamer concentrations required for half-maximal inhibition (IC50 values) were calculated as described (11) by fitting the data to a standard two-state sigmoidal dose response curve: Y = 1/(1 + 10[x − log(IC50)]), where Y is the normalized fraction full-length product at a given aptamer concentration (x). Binding assays were performed in triplicate.
RESULTS
Proviruses that express phylogenetically diverse RT subtypes in the NL4-3 backbone are viable
To evaluate the effects of RT–aptamer binding interactions on viral replication, we utilized a single-cycle infectivity system to test aptamer-mediated effects against a panel of proviruses that express phylogenetically diverse RTs within a constant NL4-3 backbone (Figure 1 and Table 1) (4,23). Because only the RT portion is varied, this approach avoids potential complications from variations in other parts of the viral genome that could confound analysis of aptamer–RT interactions. Co-transfection of pNL4-3-CMV-EGFP proviral plasmid with pMD-G, which encodes glycoprotein from VSV, yields pseudotyped reporter viruses that are capable of a single round of infection and that can be scored by transduction of virally encoded EGFP. Single-cycle infectivity assays allow for evaluation of design parameters more rapidly than approaches that require generation of stable cell lines and multiple cycles of viral replication, and they accurately predict aptamer performance against replication-competent virus (4). Thus, they are suitable surrogates for multiple-cycle assays.
The RT-coding segment of plasmid pNL4-3-CMV-EGFP was replaced with silent cloning sites that were then used to insert eight naturally-occurring or site-specifically-mutated RTs (Figure 1; Supplementary Figures S1 and 2). Viability of the recombinant, VSV-pseudotyped viruses was determined by infecting 293FT cells and quantifying infectivity by flow cytometry to detect EGFP-positive cells. Infectivity data were normalized to p24 values determined by ELISA. All proviral plasmids produced viable virus, with the exception of those expressing RT sequences from SIVcpz or HIV-2 (Figure 1C). Endogenous RT activity was similar across the entire panel, even for proviruses expressing RT sequences from SIVcpz and HIV-2 (Figure 1D), ruling out intrinsic enzymatic activity as the cause for differential replication. Instead, these two RTs appear to be incompatible with one or more steps in NL4-3 replication, as has been observed previously for fully-infectious RT-SHIV recombinants expressing the HIV-2-like SIVsm RT and adjoining genes within an HIV-1 backbone (30). The remaining six recombinants were used in assessing potential inhibition by RNA aptamers.
Broad-spectrum aptamers inhibit phylogenetically diverse RT subtypes
RNA aptamers with different secondary structures are expected to make distinct molecular contacts with RT and therefore, to differ in their susceptibility to RT amino acid sequence variation. Here, we refer to aptamer insensitivity to this variation as ‘resistance’ and describe viruses that evade aptamer-mediated inhibition as ‘resistant.’ To determine whether RNA aptamers from each of the four structural families were capable of inhibiting the proviral panel, VSV-pseudotyped virus was generated in cells that expressed aptamers from each structural family (F1Pk, F2PK, 6/5AL and UCAA) or a non-inhibitory control (‘Arbitrary’). Aptamer or control plasmids were co-transfected in 293FT cells with recombinant proviral plasmids and pMD-G, and infectivity was evaluated by counting EGFP-positive cells upon infecting 293FT (Figure 2A and B), CEM-T4 (Figure 2B) or H9 (Figure 2B) cells with cell-free pseudotyped virus. In addition, cell-free virus harvested from 293FT cells was used to determine endogenous RT activity, using a provirus with the RT sequence deleted as a control.
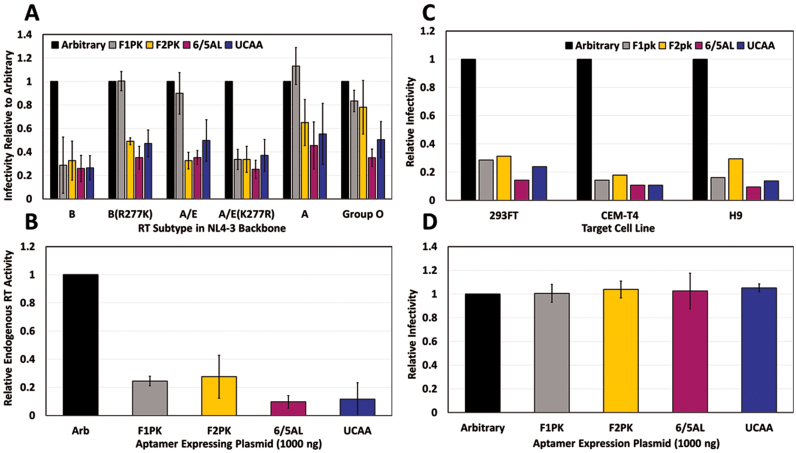
Figure 2.
Aptamer inhibition of diverse RT subtypes upon encapsidation within the viral particle. (A) Viral fractions generated in aptamer-expressing cells were used to infect fresh 293FT cells. Infectivity was determined using flow cytometry and is shown as % EGFP positive cells normalized to p24 values for each sample and the arbitrary control. Values are the mean ± SD for three experiments. (B) Endogenous RT activity was assessed as in Figure 1 using Subtype B virus produced as in (A). Values shown represent triplicate technical measurements (±SD) of a single experiment. (C) Subtype B virus produced in the presence of aptamer from (A) was used to infect 293FT, CEM-T4 and H9 cell lines. Infectivity was determined as in (A). Values shown represent a single experiment. (D) Virus produced in the absence of aptamer were used to infect aptamer-expressing 293FT cells in triplicate. Infectivity was measured as described in (A). Values are shown as the mean ± SD for four experiments. Additional data in Supplementary Figure S4.
As expected, all four aptamers inhibited endogenous RT activity (Figure 2C) and infection of all three cell types (Figure 2A and B) by viruses carrying RT from HIV-1 strain HXB2 (subtype B), which is 98.9% identical at the amino acid level to the BH10 RT used in the initial aptamer selections (1,2). In contrast, inhibition of viruses carrying other RTs varied according to the particular RT–aptamer combination used (Figure 2A). An R277K mutation in the Subtype B RT completely abolished viral inhibition by the F1Pk aptamer (Figure 2A and Supplementary Figure S3A), consistent with previous observations that the amino acid identity at position 277 in RT governs enzymatic inhibition by F1Pk aptamers (8). The same aptamer failed to inhibit virus carrying RT from two naturally-occurring K277 strains (Subtype A and an A/E recombinant), but inhibition was restored by mutation to R277 (Subtype A/E K277R). In contrast, the related F2Pk pseudoknot aptamer, shown to be insensitive to position 277 in enzymatic inhibition assays (8), overcame the resistance and inhibited viruses carrying RT from subtypes B, A and A/E. This same F2Pk aptamer was less effective against virus carrying RT from Group O, again mirroring prior enzymatic inhibition patterns (8). Finally, the most potent broad-spectrum inhibition was observed for the 6/5AL and UCAA aptamers, which inhibited all six recombinant viruses (Figure 2A).
These observations support three important conclusions. First, this is the first demonstration of viable aptamer resistance in cell culture, and it was achieved without apparent loss of fitness, as infectivity levels were similar for the natural HIV-1 variants (the subtype A and A/E strain) and for point mutations (subtype B R277K mutant) (Figure 1A). This is in contrast to previous observations of a strong replication defect for N255D/N265D single and double mutations (7). Second, because a single point mutation in RT determines whether a given virus is inhibited by, or resistant to, F1Pk aptamers, these data establish that RT is the genetic locus responsible for aptamer-mediated inhibition and resistance, at least for pseudoknot aptamers (Figure 2A and Supplementary Figure S3A). Finally, the observation that recombinant viruses carrying diverse RTs are inhibited by the 6/5AL and UCAA aptamers (and to a lesser extent by the F2Pk aptamer) (Figure 2A) establishes that broad-spectrum aptamers can overcome some resistance mutations, likely by making additional or different contacts with RT. By extension, diverse aptamer families may be susceptible to different—and potentially non-overlapping—spectra of resistance mutations.
Encapsidation is required for aptamer-mediated inhibition
Reverse transcription occurs in the cytoplasm of infected cells, but in the experiments above, the aptamers were expressed in virus-producing cells. Expressing the aptamers in target cells prior to infection could, in principle, also block infection, provided that aptamer transcripts can penetrate the formidable barrier of the capsid lattice. To determine whether prior encapsidation into the viral particle was required for aptamer-mediated inhibition, VSV-pseudotyped reporter virus encoding subtype B RT was generated in non-aptamer-expressing cells and used to infect target 293FT cells that had been previously transfected with varying amounts of aptamer-expressing plasmid (ranging from 1000 to 3000 ng) at various times prior to infection (6, 24 or 48 h). No aptamer-mediated inhibition of viral replication in target cells was observed relative to arbitrary control RNA at any of the dosages or transfection times using the aptamer-expressing plasmids from any of the four structural families (Figure 2D and Supplementary Figure S4). Similar results were obtained for a construct that directs transcription of the minimal core of aptamer 70.15 (F1Pk family; 70.15MC B0F0) (Supplementary Figure S4B) that had previously been reported to inhibit incoming viral infection, albeit in a different experimental system (31). These results establish that prior encapsidation is required for aptamer-mediated inhibition of replication in our system.
Stoichiometric encapsidation of inhibitory aptamers
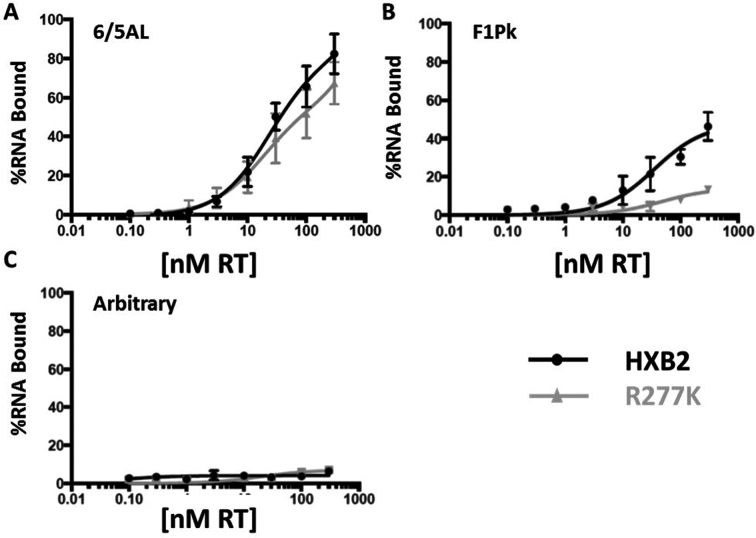
To determine whether differential molecular recognition within aptamer–RT complexes drives relative encapsidation, we measured the binding affinities for F1Pk and 6/5AL aptamers to subtype B RT and to the R277K point mutant. For the 6/5AL aptamer, binding was similar for purified subtype B RT and the R277K point mutant (Figure 3A). The F1Pk aptamer bound slightly less well than the 6/5AL aptamer to the Subtype B RT, but it barely interacted at all with the R277K mutant (Figure 3B), consistent with its inability to inhibit the mutant (8). Arbitrary control RNA did not bind either RT (Figure 3C). We next generated VSV-pseudotyped virus in the presence of F1Pk or 6/5AL aptamer or control RNA and measured infectivity and aptamer RNA levels in cellular and viral fractions as a function of co-transfected expression plasmid dose. As expected, cellular accumulation increased with plasmid dose (Supplementary Figure S5A). For subtype B RT, both the F1Pk and 6/5AL aptamers are encapsidated even at the lowest doses of input plasmid, and both the encapsidation levels (Figure 4A and Supplementary Figure S5B) and the potency of viral inhibition increased over this range (Figure 4B). For the non-inhibitory Arbitrary control RNA, comparable levels were encapsidated at the highest doses of input expression plasmid, but no viral inhibition was observed even at those high doses, suggesting non-specific incorporation (Figure 4B, D and Supplementary Figure S3B). In contrast, for the R277K mutant RT, the 6/5AL aptamer is encapsidated more efficiently than non-inhibitory transcripts at the lowest doses of input plasmid (Figure 4C and Supplementary Figure S5C) and only this aptamer inhibited infection by the R277K virus (Figure 4D). The F1Pk aptamer and arbitrary RNA control were encapsidated less efficiently than the inhibitory 6/5AL aptamer at the lowest plasmid doses, and neither one inhibited viral replication at any dose. All three RNAs accumulated in viruses at high doses of input plasmid (Figure 4A, C and Supplementary Figure S5B and C), indicating that a non-specific, non-productive packaging mechanism exists. In separate experiments, aptamers accumulated to high levels in cells (Supplementary Figure S3C). Similar non-specific packaging was observed at high input plasmid doses for additional aptamer–RT combinations (Supplementary Figure S3B).
Figure 3.
Aptamer–RT binding. The 6/5AL aptamer (A), F1Pk (B) and arbitrary aptamer (C) were 5΄-end labeled with 32P-ATP and added to aptamer–RT binding reactions containing various concentrations of subtype B RT or the R277K point mutant in binding buffer. Aptamer–RT binding reactions were incubated on ice for 15 minutes and reactions were then loaded onto pre-wet nitrocellulose membranes. Samples were allowed to flow through the nitrocellulose membranes via vacuum and aptamer–RT complexes were retained on the membrane. Radioactivity was determined via scintillation counter. The 0 nM RT reaction was used as a control and the amount of radioactivity present in the unfiltered sample was set to 100%. The protein-dependent signal was determined by subtracting the value obtained for the 0 nM control from the value obtained for the sample. Values at each RT concentration were fit into a one-site binding graph using Prism software version 6.2. For the 6/5AL aptamer, Kd values under the conditions of these assays were 81 nM for subtype B RT and 57 nM for the R277K point mutant.
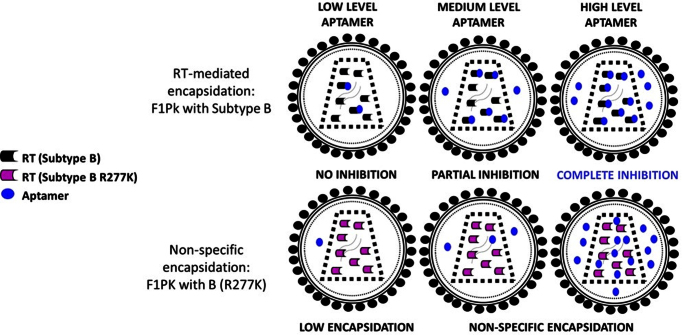
These data suggest that there are at least two mechanisms by which HIV-1 can package these aptamers (Figure 5). Inhibitory anti-RT aptamers bind their target RTs with high affinity and are recruited into viral particles more efficiently at low doses. This specific recruitment likely involves cytosolic interaction with the RT portion of GagPol during assembly. Non-inhibitory transcripts are incorporated non-specifically in proportion to their expression levels within producer cells. Notably, for all three inhibitory aptamer–RT combinations, viral suppression saturated once the level of recruited RNA surpassed 25 copies per viral genome (normalized to HIV-1 gag RNA), or about 50 copies per virion. This value is approximately equal to the number of RT dimers found in mature virions (31) and again supports preferential recruitment of inhibitory aptamers through high-affinity RNA–RT interactions during viral assembly. For the more potent 6/5AL aptamer, this threshold was reached at 500ng input plasmid for both the Subtype B and R277K mutant viruses; for the F1Pk aptamer, the threshold was reached at 750 ng input plasmid for the Subtype B virus (Figure 4). For both aptamers, little additional viral suppression was observed as the input plasmid doses was increased above that threshold.
Figure 5.
Model of the two proposed encapsidation mechanisms for anti-RT aptamers: at low aptamer doses, preferential encapsidation is mediated by specific aptamer–RT interactions that can result in RT inhibition. At high aptamer doses, encapsidation results from non-specific incorporation of both inhibitory and non-inhibitory aptamer transcripts into the budding viral particles.
DISCUSSION
Anti-RT aptamers are potent inhibitors of HIV-1 RT in biochemical assays and in cell culture. Their impact on replication provides a unique tool for dissecting molecular and cellular events in the viral life cycle and offers a promising genetic means of inhibiting HIV-1 infection. This study used single-cycle infectivity assays with proviral plasmids that express RTs from phylogenetically diverse HIV-1 subtypes in a constant NL4-3 background, thereby avoiding potential complications from variations in other regions of the viral genome and focusing attention on the RT–aptamer interaction.
Building upon previous findings that anti-RT aptamers accumulate in HIV-1 particles (4,5), we have established that aptamer encapsidation during viral assembly is required for inhibition of HIV-1 replication. This requirement parallels observations for other macromolecular inhibitors of HIV-1. For example, encapsidated APOBEC3G (A3G) interferes with accurate replication by hypermutating the nascent proviral DNA minus strand (32), but A3G does not alter replication when expressed in target cells (32). Similarly, RNA interference by short hairpin RNA against the viral genome is effective against HIV-1 when expressed in producer cells but not in target cells (33). Apparently neither A3G (∼43 kDa) nor RISC (∼140–500 kDa) can gain access to the viral genome while it is in its ssDNA or RNA form within the capsid. Similarly, neither the full-length aptamer transcripts (∼96 kDa) nor shorter forms (∼24 kDa) that can be generated by self-cleavage of embedded ribozymes can access RT prior to completion of reverse transcription.
Intact capsids clearly offer a formidable barrier to macromolecular inhibitors that target molecules located within the viral interior. The requirement for aptamer encapsidation is consistent with two current models of HIV-1 capsid uncoating, in which reverse transcription begins prior to the initiation of uncoating (22,34). In the first model (slow, partial cytoplasmic uncoating), the capsid remains assembled upon entry into the cell, followed by partial cytoplasmic dissociation, with a portion of capsid protein remaining associated with the reverse transcription complex at the time of nuclear entry. In the second model (uncoating at the nuclear membrane), the capsid lattice remains fully assembled until it reaches the nuclear pore complex, thereby assuring safe, non-immune activating transport of the genome to the nucleus. Specifically, either the aptamers are unable to penetrate the intact capsid to inhibit reverse transcription, or reverse transcription happens too quickly for sufficient aptamer RNA to accumulate in the capsid core within the target cell. Notably, recent experimental evidence supports the ability of dNTPs or small molecule drugs to diffuse in and out of the dynamic pores of the capsid core (35). However, the large size of our aptamer transcript is expected to preclude such diffusion and account for our observation that prior encapsidation is required for aptamer-mediated inhibition of replication.
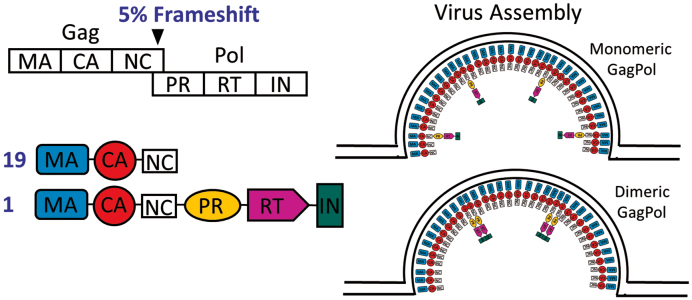
Our results suggest potential mechanisms of aptamer encapsidation via interactions with GagPol. We propose that anti-RT aptamers are recruited into virions by binding to dimeric GagPol polyprotein, either in the cytoplasm prior to GagPol recruitment to the membrane, or on the exposed cytosolic surface of GagPol at the plasma membrane (Figure 6). These interactions can provide a vehicle for the aptamer to enter the virus and subsequently gain access to the proteolytically processed RT, where it outcompetes the viral genome for access to the RT active site. The preferential recruitment of inhibitory aptamers indicates that their encapsidation is driven by interaction with RT prior to completion of assembly. The mature p66/p51 RT is formed by proteolytic cleavage of a p66/p66-like homodimer (36,37), but at the time when RT is exposed to the cytosol, it exists as a domain within the GagPol polyprotein. It is unlikely that an aptamer selected to bind mature p66/p51 heterodimer would also bind monomeric p66 within GagPol, as both RT subunits show close contact in a crystal structure of an F1Pk aptamer in complex with RT (38) and both subunits are protected by several different aptamers in mass spec footprinting (14). Although wild-type p66 cannot be maintained stably in solution in monomeric form, the p66/p66 homodimer is enzymatically active (39) and its enzymatic inhibition by aptamers is similar to that observed for mature RT (Supplementary Figure S6). The HIV-1 protease (PR) and integrase are also encoded within Pol and are also dimers in their mature forms (40,41), although the sequence of dimerization events that precede proteolytic processing from GagPol is poorly understood. While the active site of PR lies at the dimer interface (36,40), the enzyme is dormant until triggered by an as-yet-unknown signal after the virus buds from the plasma membrane. Thus, while our data support the possibility that the RT portion of GagPol dimerizes while exposed to the cytosol, it must do so without prematurely activating PR.
Figure 6.
Model of proposed dimerization of GagPol in the cytoplasm during viral assembly.
Finally, this work demonstrates for the first time that HIV-1 can exhibit aptamer-specific resistance as defined above and that broad-spectrum aptamers overcome such resistance. Because resistance was conferred by exchanging only the RT-encoding segment within a constant NL4-3 background, this work also established RT as the genetic locus for aptamer-mediated HIV-1 inhibition. F1Pk pseudoknot aptamers were the most abundant structural motif in early selections (1,2,10), and they have been the subject of numerous in vitro and cell-based studies (3–6,8,11). However, HIV-1 strains and mutants carrying K277 readily escaped inhibition with no apparent replication defect, indicating a low genetic barrier to resistance for that class of aptamers. In contrast, the 6/5AL, UCAA and F2Pk aptamers were insensitive to this mutation, and they exhibited broader-spectrum viral inhibition that reflected their relative abilities to inhibit diverse purified RT. For aptamers that target evolving pathogens such as HIV, specificity provides an opportunity for resistance. It is unknown whether HIV-1 can escape resistance by broad-spectrum aptamers without compromising fitness, or how many mutations would be required to achieve such resistance, although it seems likely that their genetic barriers to resistance may be high. Furthermore, non-overlapping sets of mutations may be associated with aptamers that make different molecular contacts, such that de novo resistance could potentially be averted by co-expressing aptamers from different structural classes. The footprint of the F1Pk pseudoknot aptamer T1.1 is relatively small compared to broad-spectrum ssDNA aptamers RT1t49(-5) and R1T, both of which have footprints that closely mimics that of primer-template substrates (14). The interfaces for broad-spectrum aptamers 6/5AL and UCAA are unknown and will be the subject of further studies.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the following individuals for their technical and conceptual contributions to this work: JinGeol Kim, Jay Kissel, Tarun Sharma, Angela Whatley and Drew Sawyer.
Footnotes
Present addresses:
Donald Burke and Margaret Lange, Department of Molecular Microbiology & Immunology, University of Missouri, Columbia, MO 65211, USA.
Mackenzie K. Callaway, Department of Biomedical Engineering, University of Minnesota, Twin Cities, MN 55455, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [NIH NIAID R01AI074389, NIH NIAID F32-AI085627]. Funding for open access charge: NIH [R01AI074389].
Conflict of interest statement. None declared.
REFERENCES
- 1. Burke D.H., Scates L., Andrews K., Gold L.. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J. Mol. Biol. 1996; 264:650–666. [DOI] [PubMed] [Google Scholar]
- 2. Tuerk C., MacDougal S., Gold L.. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:6988–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chaloin L., Lehmann M.J., Sczakiel G., Restle T.. Endogenous expression of a high-affinity pseudoknot RNA aptamer suppresses replication of HIV-1. Nucleic Acids Res. 2002; 30:4001–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lange M.J., Sharma T.K., Whatley A.S., Landon L.A., Tempesta M.A., Johnson M.C., Burke D.H.. Robust suppression of HIV replication by intracellularly expressed reverse transcriptase aptamers is independent of ribozyme processing. Mol. Ther. 2012; 20:2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Joshi P., Prasad V.R.. Potent inhibition of human immunodeficiency virus type 1 replication by template analog reverse transcriptase inhibitors derived by SELEX (systematic evolution of ligands by exponential enrichment). J. Virol. 2002; 76:6545–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Joshi P.J., North T.W., Prasad V.R.. Aptamers directed to HIV-1 reverse transcriptase display greater efficacy over small hairpin RNAs targeted to viral RNA in blocking HIV-1 replication. Mol. Ther. 2005; 11:677–686. [DOI] [PubMed] [Google Scholar]
- 7. Fisher T.S., Joshi P., Prasad V.R.. Mutations that confer resistance to template-analog inhibitors of human immunodeficiency virus (HIV) type 1 reverse transcriptase lead to severe defects in HIV replication. J. Virol. 2002; 76:4068–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Held D.M., Kissel J.D., Thacker S.J., Michalowski D., Saran D., Ji J., Hardy R.W., Rossi J.J., Burke D.H.. Cross-clade inhibition of recombinant human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus SIVcpz reverse transcriptases by RNA pseudoknot aptamers. J. Virol. 2007; 81:5375–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whatley A.S., Ditzler M.A., Lange M.J., Biondi E., Sawyer A.W., Chang J.L., Franken J.D., Burke D.H.. Potent inhibition of HIV-1 reverse transcriptase and replication by nonpseudoknot, ‘UCAA-motif’ RNA aptamers. Mol. Ther. Nucleic Acids. 2013; 2:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ditzler M.A., Lange M.J., Bose D., Bottoms C.A., Virkler K.F., Sawyer A.W., Whatley A.S., Spollen W., Givan S.A., Burke D.H.. High-throughput sequence analysis reveals structural diversity and improved potency among RNA inhibitors of HIV reverse transcriptase. Nucleic Acids Res. 2013; 41:1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Held D.M., Kissel J.D., Saran D., Michalowski D., Burke D.H.. Differential susceptibility of HIV-1 reverse transcriptase to inhibition by RNA aptamers in enzymatic reactions monitoring specific steps during genome replication. J. Biol. Chem. 2006; 281:25712–25722. [DOI] [PubMed] [Google Scholar]
- 12. Kissel J.D., Held D.M., Hardy R.W., Burke D.H.. Single-stranded DNA aptamer RT1t49 inhibits RT polymerase and RNase H functions of HIV type 1, HIV type 2, and SIVCPZ RTs. AIDS Res. Hum. Retroviruses. 2007; 23:699–708. [DOI] [PubMed] [Google Scholar]
- 13. Michalowski D., Chitima-Matsiga R., Held D.M., Burke D.H.. Novel bimodular DNA aptamers with guanosine quadruplexes inhibit phylogenetically diverse HIV-1 reverse transcriptases. Nucleic Acids Res. 2008; 36:7124–7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ditzler M.A., Bose D., Shkriabai N., Marchand B., Sarafianos S.G., Kvaratskhelia M., Burke D.H.. Broad-spectrum aptamer inhibitors of HIV reverse transcriptase closely mimic natural substrates. Nucleic Acids Res. 2011; 39:8237–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Comas-Garcia M., Davis S.R., Rein A.. On the selective packaging of genomic RNA by HIV-1. Viruses. 2016; 8:E246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kleiman L., Jones C.P., Musier-Forsyth K.. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010; 584:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saadatmand J., Guo F., Cen S., Niu M., Kleiman L.. Interactions of reverse transcriptase sequences in Pol with Gag and LysRS in the HIV-1 tRNALys3 packaging/annealing complex. Virology. 2008; 380:109–117. [DOI] [PubMed] [Google Scholar]
- 18. Schafer A., Bogerd H.P., Cullen B.R.. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004; 328:163–168. [DOI] [PubMed] [Google Scholar]
- 19. Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H.. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002; 418:646–650. [DOI] [PubMed] [Google Scholar]
- 20. Rulli S.J. Jr, Hibbert C.S., Mirro J., Pederson T., Biswal S., Rein A.. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007; 81:6623–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keene S.E., Telesnitsky A.. cis-Acting determinants of 7SL RNA packaging by HIV-1. J. Virol. 2012; 86:7934–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Campbell E.M., Hope T.J.. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 2015; 13:471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lange M.J., Burke D.H.. Screening inhibitory potential of anti-HIV RT RNA aptamers. Methods Mol. Biol. 2014; 1103:11–29. [DOI] [PubMed] [Google Scholar]
- 24. Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D.. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997; 15:871–875. [DOI] [PubMed] [Google Scholar]
- 25. Foley G.E., Lazarus H., Farber S., Uzman B.G., Boone B.A., McCarthy R.E.. Continuous culture of human lymphoblasts from peripheral blood of a child with acute leukemia. Cancer. 1965; 18:522–529. [DOI] [PubMed] [Google Scholar]
- 26. Popovic M., Read-Connole E., Gallo R.C.. T4 positive human neoplastic cell lines susceptible to and permissive for HTLV-III. Lancet. 1984; 2:1472–1473. [DOI] [PubMed] [Google Scholar]
- 27. Popovic M., Sarngadharan M.G., Read E., Gallo R.C.. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984; 224:497–500. [DOI] [PubMed] [Google Scholar]
- 28. Mann D.L., O’Brien S.J., Gilbert D.A., Reid Y., Popovic M., Read-Connole E., Gallo R.C., Gazdar A.F.. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res. Hum. Retroviruses. 1989; 5:253–255. [DOI] [PubMed] [Google Scholar]
- 29. Zhang H., Dornadula G., Pomerantz R.J.. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage for viral infection of nondividing cells. J. Virol. 1996; 70:2809–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ambrose Z., Boltz V., Palmer S., Coffin J.M., Hughes S.H., Kewalramani V.N.. In vitro characterization of a simian immunodeficiency virus-human immunodeficiency virus (HIV) chimera expressing HIV type 1 reverse transcriptase to study antiviral resistance in pigtail macaques. J. Virol. 2004; 78:13553–13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Briggs J.A., Simon M.N., Gross I., Krausslich H.G., Fuller S.D., Vogt V.M., Johnson M.C.. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004; 11:672–675. [DOI] [PubMed] [Google Scholar]
- 32. Zhang H., Yang B., Pomerantz R.J., Zhang C., Arunachalam S.C., Gao L.. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003; 424:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Westerhout E.M., ter Brake O., Berkhout B.. The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference. Retrovirology. 2006; 3:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Da Silva Santos C., Tartour K., Cimarelli A.. A novel entry/uncoating assay reveals the presence of at least two species of viral capsids during synchronized HIV-1 infection. PLoS Pathog. 2016; 12:e1005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jacques D.A., McEwan W.A., Hilditch L., Price A.J., Towers G.J., James L.C.. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature. 2016; 536:349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sluis-Cremer N., Arion D., Abram M.E., Parniak M.A.. Proteolytic processing of an HIV-1 pol polyprotein precursor: insights into the mechanism of reverse transcriptase p66/p51 heterodimer formation. Int. J. Biochem. Cell Biol. 2004; 36:1836–1847. [DOI] [PubMed] [Google Scholar]
- 37. Sharaf N.G., Poliner E., Slack R.L., Christen M.T., Byeon I.J., Parniak M.A., Gronenborn A.M., Ishima R.. The p66 immature precursor of HIV-1 reverse transcriptase. Proteins. 2014; 82:2343–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jaeger J., Restle T., Steitz T.A.. The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J. 1998; 17:4535–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Restle T., Muller B., Goody R.S.. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J. Biol. Chem. 1990; 265:8986–8988. [PubMed] [Google Scholar]
- 40. Wlodawer A., Miller M., Jaskolski M., Sathyanarayana B.K., Baldwin E., Weber I.T., Selk L.M., Clawson L., Schneider J., Kent S.B.. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989; 245:616–621. [DOI] [PubMed] [Google Scholar]
- 41. Petit C., Schwartz O., Mammano F.. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 1999; 73:5079–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.