Abstract
N 6-methyladenosine (m6A) is the most abundant internal modification in eukaryotic messenger RNA (mRNA), and affects almost every stage of the mRNA life cycle. The YTH-domain proteins can specifically recognize m6A modification to control mRNA maturation, translation and decay. m6A can also alter RNA structures to affect RNA–protein interactions in cells. Here, we show that m6A increases the accessibility of its surrounding RNA sequence to bind heterogeneous nuclear ribonucleoprotein G (HNRNPG). Furthermore, HNRNPG binds m6A-methylated RNAs through its C-terminal low-complexity region, which self-assembles into large particles in vitro. The Arg-Gly-Gly repeats within the low-complexity region are required for binding to the RNA motif exposed by m6A methylation. We identified 13,191 m6A sites in the transcriptome that regulate RNA–HNRNPG interaction and thereby alter the expression and alternative splicing pattern of target mRNAs. Low-complexity regions are pervasive among mRNA binding proteins. Our results show that m6A-dependent RNA structural alterations can promote direct binding of m6A-modified RNAs to low-complexity regions in RNA binding proteins.
INTRODUCTION
N 6-methyladenosine (m6A) is the most abundant internal modification of messenger RNAs (mRNAs) and has known functions in RNA splicing, export, translation and decay (1). m6A is a reversible and dynamic modification that is introduced at a subset of RRACH motifs (R = A/G, H = A/C/U) through the activity of the m6A methyltransferase complex, composed of methyltransferase-like 3 (METTL3), METTL14 and Wilms tumor 1 associated protein (2,3), and reversed by two m6A demethylases, fat mass and obesity associated protein and AlkB family member 5 (ALKBH5) (4,5). Consistent with the dynamic character of m6A modifications, m6A sites span a wide range of modification fractions (6), and cellular conditions such as heat shock can lead to changes in m6A modification levels and patterns (7,8). Dynamic m6A modifications add another dimension to the regulation of the life cycle of cellular RNAs.
m6A modifications impact the fate of modified RNAs by influencing their interactions with m6A ‘reader’ proteins. Proteins containing YT521-B homology (YTH) domains directly bind the N6-methyl group of m6A, resulting in the regulation of mRNA splicing, translation, and decay (9–17). m6A modifications also influence biological RNA structures, as shown both in biophysical studies in vitro (18) and in transcriptome-wide mapping studies in vivo (19,20). m6A-induced changes in RNA structure can alter the accessibility of protein binding sites, thereby influencing protein binding. The m6A reader protein HNRNPC (heterogeneous nuclear ribonucleoprotein C) binds m6A-modified RNA through this ‘m6A-switch’ mechanism, in which the m6A-mediated destabilization of an RNA hairpin exposes a single-stranded HNRNPC binding motif (20). Given the impact of RNA structure on protein binding, m6A-switch-like changes in RNA structure likely have a widespread effect on the binding of many proteins to m6A-modified RNAs. In addition to HNRNPC, other m6A reader proteins lacking a YTH domain have been shown to regulate pri-miRNA processing and mRNA translation (21,22). These reader proteins might recognize m6A-modified RNAs by directly binding the m6A base or through m6A-induced changes in RNA structure.
mRNAs interact with proteins throughout their life cycle. These interactions constitute a crucial and widespread mechanism for the regulation of mRNA transcription, processing, translation and stability. Interactome studies have expanded the number of RNA binding proteins identified in the human genome to ∼1,500 (23,24). Many of the newly discovered RNA binding proteins do not possess canonical RNA binding domains. Furthermore, it has become increasingly clear that intrinsically disordered regions and low-complexity sequences have widespread roles in RNA binding (24). Low-complexity sequences are protein regions with an amino acid composition that is highly biased in such a way that they are unlikely to fold into a globular structure. Low-complexity sequences have been implicated in the formation of both normal and pathological ribonucleoprotein assemblies, including cellular granules as well as protein aggregates characteristic of neurodegenerative diseases (25–27). Moreover, RNA binding proteins with low-complexity regions frequently function as protein–protein interaction hubs and are therefore ideally positioned for the regulation of RNA–protein interactions and RNA metabolism (24). Among the m6A reader proteins identified so far, several have been shown to bind m6A-modified RNAs using globular YTH domains or RNA recognition motifs (12–15,20). Although some of these m6A reader proteins contain low-complexity sequences, these regions have not been implicated in the binding of m6A-modified RNAs. Given their increasingly recognized roles in RNA biology, low-complexity regions likely contribute to the recognition of m6A-modified RNAs by some m6A readers.
Here, we report that heterogeneous nuclear ribonucleoprotein G (HNRNPG) is a new m6A reader protein that uses a low-complexity region to recognize a motif exposed by m6A modification. We first identified HNRNPG as a protein that preferentially binds to the m6A-modified form of a hairpin from the long noncoding RNA (lncRNA) metastasis associated lung adenocarcinoma transcript 1 (MALAT1). HNRNPG binds a purine-rich region that can overlap with the m6A consensus sequence and is exposed upon m6A modification. Moreover, HNRNPG binding is mediated by a low-complexity region rather than a canonical RNA binding domain. Transcriptome-wide studies further identified 13,191 high-confidence m6A sites bound by HNRNPG, while HNRNPG knockdown and m6A methyltransferase knockdown led to correlated changes in mRNA splicing. Thus, HNRNPG uses its low-complexity region to bind purine-rich sequences exposed upon m6A modification of RNA, and thereby functions in the regulation of gene expression and alternative splicing.
MATERIALS AND METHODS
Mammalian cell culture, siRNA knockdown and cell fractionation
Human cervical adenocarcinoma cell line HeLa (CCL-2) and human embryonic kidney (HEK) cell line HEK293T (CRL-11268) were obtained from the American Type Culture Collection (ATCC) and cultured under standard conditions. Control siRNA (1027281, Qiagen), METTL3 siRNA (SI04317096, Qiagen), METTL14 siRNA (SI00459942, Qiagen) or HNRNPG siRNA (SI00700084 and SI00700077, Qiagen) was transfected into HEK293T cells at a concentration of 20–50 nM, using Lipofectamine RNAiMAX (13778100, Invitrogen) according to the manufacturer's instructions. Cells were collected 48 h after transfection, shock-frozen in liquid nitrogen and stored at −80°C for further studies.
Nuclear and cytoplasmic extracts were isolated using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (78833, Thermo Scientific) according to the manufacturer's instructions.
Western blotting
Western blots were performed using standard procedures. Briefly, 10–30 μg protein samples were separated on 4–12% polyacrylamide Bis-Tris gels (NP0336BOX, Invitrogen) and transferred to polyvinylidene fluoride membranes (IPVH00010, Millipore). The blots were probed with METTL3- (15073-1-AP, Proteintech), METTL14- (HPA038002, Sigma), HNRNPG- (sc-14581 and sc-48796, Santa Cruz Biotechnology) or GAPDH- (A00192-40, Genscript) specific primary antibody, followed by rabbit anti-goat IgG-HRP (sc-2768, Santa Cruz Biotechnology) or goat anti-rabbit IgG-HRP (ab97051, Abcam) secondary antibody, and then visualized by enhanced chemoluminescence (RPN2109, GE Healthcare).
Protein expression
For expression of the N-terminal RNA recognition motif (N-RRM, residues 1–83) and C-terminal RNA binding domain (C-RBD, residues 334–391) of human HNRNPG protein, sequences encoding these HNRNPG domains were amplified by polymerase chain reaction (PCR) from human HeLa cDNA libraries (637203, Clontech) and then subcloned into pGEX-6p-1 expression vectors using BamHI and XhoI restriction sites. Plasmid DNA was transformed into Escherichia coli BL21-CodonPlus(DE3)-RP or BL21-CodonPlus(DE3)-RIL cells (Agilent). The transformed bacteria were grown to saturation at 37°C, 200 rpm in Luria–Bertani Lennox medium with 100 μg/ml ampicillin, then diluted 1:100, grown in the same culture medium to an absorbance of ∼0.6 at 600 nm, and induced with 1 mM isopropyl β-D-1-thiogalactoside (IPTG). The bacteria were grown an additional 16–22 h at 18°C, 200 rpm, then harvested and sonicated at 4°C. GST-fusion proteins were isolated from the soluble lysate using glutathione-Sepharose beads and stored in 10 mM Tris-Cl (pH 7.4), 100 mM KCl, 2.5 mM MgCl2, 30% glycerol at −80°C.
RNA oligos
The following RNA oligos were synthesized by Q.D. and purified by high-performance liquid chromatography (HPLC) and/or denaturing gel electrophoresis, as previously described (28).
The RNA oligos used in Figures 1, 2C–D, and 3C and Supplementary Figure S2A:
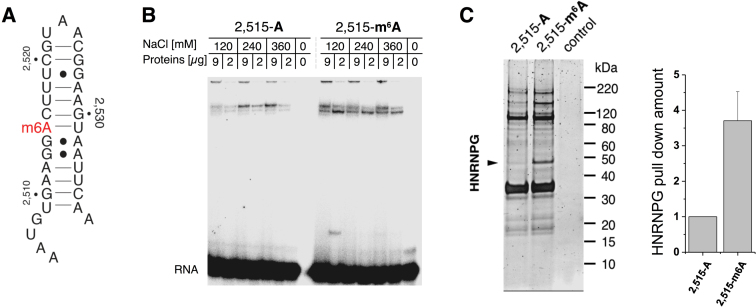
Figure 1.
HNRNPG preferentially binds an m6A-modified hairpin in MALAT1. (A) Secondary structure of the 34-nt hairpin derived from positions 2,505–2,538 of MALAT1, including the m6A site at position 2,515. The methylated form of the hairpin is termed 2,515-m6A and the unmethylated form is termed 2,515-A. (B) Gel shift showing binding of HeLa nuclear extract to the MALAT1 hairpin in both its unmethylated (2,515-A) and methylated (2,515-m6A) forms. (C) Left: denaturing gel of the proteins pulled down by the unmethylated and methylated MALAT1 hairpins. In the control, no RNA was used as bait. Right: quantification of relative HNRNPG pull-down with the unmethylated and methylated hairpins, normalized to pulled-down Histone H1.2 (HIST1H1C). Data shown as mean; error bar = standard deviation; n = 4 biological replicates.
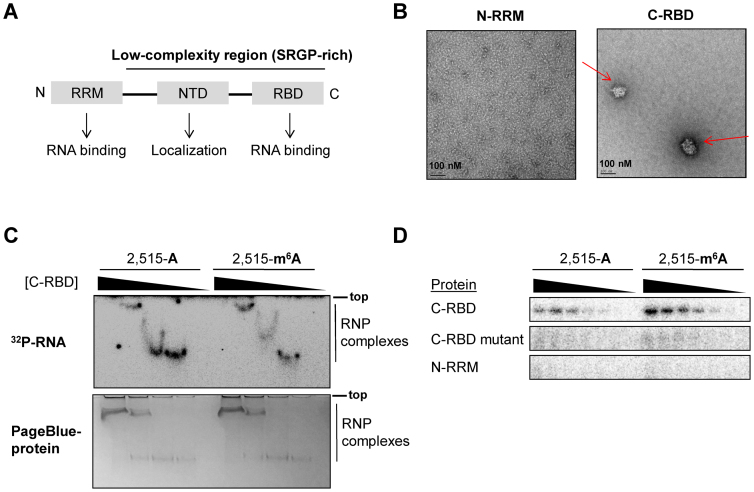
Figure 2.
HNRNPG uses a low-complexity region to bind the MALAT1 hairpin. (A) Domain structure of HNRNPG, including an N-terminal RNA recognition motif (RRM) and an SRGP-rich low-complexity region, which contains the nascent transcripts targeting domain (NTD) and a C-terminal RNA binding domain (RBD). (B) Electron microscopy images of the N-terminal RRM (N-RRM) and C-terminal RBD (C-RBD) of HNRNPG at 5 μM concentration. C-RBD aggregates are marked by arrows. (C) Gel shift showing the ribonucleoprotein (RNP) complexes that form upon binding of the C-RBD of HNRNPG (0–20 μM) to the unmethylated and methylated MALAT1 hairpins. The free RNA is not shown, as it has run much farther down the gel. Top: 32P-labeled RNA gel; bottom: same gel stained for protein. (D) Ultraviolet cross-linking of the HNRNPG C-RBD, C-RBD mutant and N-RRM (0–5 μM) to the unmethylated and methylated MALAT1 hairpins. In the C-RBD mutant, all three RGG repeats in the C-RBD were mutated to FGG repeats.
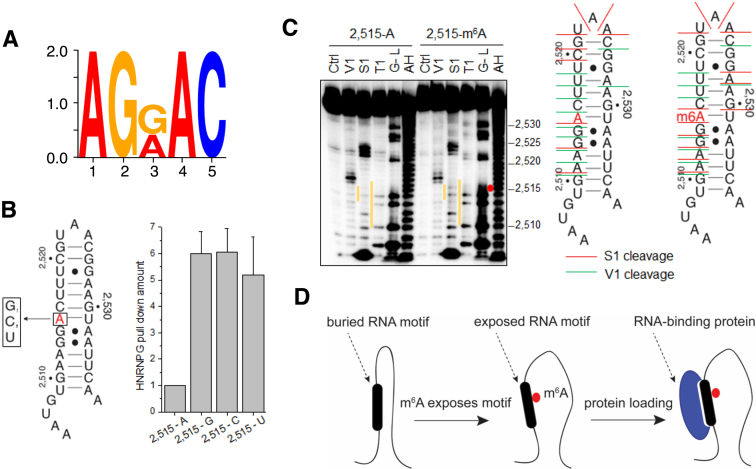
Figure 3.
m6A alters RNA structure to recruit HNRNPG. (A) Sequence logo of the most enriched motif within HNRNPG PAR-CLIP peaks. (B) Left: secondary structure of the MALAT1 hairpin, showing the A-2,515-to-G/C/U mutations that were introduced at the m6A site. Right: quantification of relative HNRNPG pull-down with the original (2,515-A) and mutated (2,515-G/C/U) MALAT1 hairpins, normalized to pulled-down HIST1H1C. Data shown as mean; error bar = standard deviation; n = 3 biological replicates. (C) Left: structural probing of the unmethylated and methylated MALAT1 hairpins. The orange lines indicate regions with marked differences between the unmethylated and methylated hairpins. The location of the m6A residue is indicated by a red dot. Ctrl, no nuclease added; V1; RNase V1 digestion; S1, S1 nuclease digestion; T1, RNase T1 digestion; G-L, G-ladder; AH, alkaline hydrolysis. Right: secondary structure of the unmethylated and methylated MALAT1 hairpins, marked at their S1 nuclease (red lines) and V1 nuclease (green lines) cleavage sites. (D) Model showing that m6A disrupts RNA structure, exposes a motif that includes the m6A site, and recruits an RNA binding protein.
2,515-A: 5΄- AAUGUGAAGGACUUUCGUAACGGAAGUAAUUCAA-Biotin;
2,515-m6A: 5΄- AAUGUGAAGGm6ACUUUCGUAACGGAAGUAAUUCAA-Biotin.
The RNA oligos used in Figure 3B:
2,515-A: 5΄- AAUGUGAAGGACUUUCGUAACGGAAGUAAUUCAA-Biotin;
2,515-G: 5΄- AAUGUGAAGGGCUUUCGUAACGGAAGUAAUUCAA-Biotin;
2,515-C: 5΄- AAUGUGAAGGCCUUUCGUAACGGAAGUAAUUCAA-Biotin;
2,515-U: 5΄- AAUGUGAAGGUCUUUCGUAACGGAAGUAAUUCAA-Biotin.
The RNA oligos used in Supplementary Figure S2B:
#1: 5΄-AAUGUGAUGGm6ACUUUCGUAACGGAAGUAAAUCAA-Biotin;
#2: 5΄-AAUGUGAAAGm6ACUUUCGUAACGGAAGUAGUUCAA-Biotin;
#3: 5΄-AAUGUGAAGAm6ACUUUCGUAACGGAAGUGAUUCAA-Biotin;
#4: 5΄-AAUGUGAAGGUCUUUCGUAACGGAAGm6AAAUUCAA-Biotin;
#5: 5΄-AAUGUGAAGGm6AGUUUCGUAACGGAACUAAUUCAA-Biotin.
The RNA oligos used in Supplementary Figure S2C were transcribed in vitro with T7 RNA polymerase and purified by denaturing gel electrophoresis:
2,518-U & 2,528-A:
5΄-GAAAAAUGUGAAGGACUUUCGUAACGGAAGUAAUUCAAGATCA;
2,518-A & 2,528-A:
5΄-GAAAAAUGUGAAGGACUAUCGUAACGGAAGUAAUUCAAGATCA;
2,518-U & 2,528-U:
5΄-GAAAAAUGUGAAGGACUUUCGUAACGGUAGUAAUUCAAGATCA;
2,518-A & 2,528-U:
5΄-GAAAAAUGUGAAGGACUAUCGUAACGGUAGUAAUUCAAGATCA.
RNA pull-down, gel shift and cross-linking
The in vitro pull-down assays were performed as described (7). The eluted protein samples were separated on 4–12% polyacrylamide Bis-Tris gels (NP0321BOX, Invitrogen) and stained with SYPRO Ruby Protein Gel Stain (S12000, Thermo Scientific) according to the manufacturer's instructions. Proteins in gel slices or in the entire pulled-down protein sample were digested with trypsin and identified using LC–MS/MS by the Donald Danforth Plant Science Center (Washington University, St Louis, MO, USA).
For the gel shift in Figure 1B, the gel-purified 5΄ 32P-labeled RNA oligos were refolded by heating at 90°C for 1 min, then at 30°C for 5 min. A total of 3 μl nuclear extract and 6 μl refolded RNA were incubated together at room temperature for 30 min, and then at 4°C for 2 h. Each sample was mixed with 1 μl 50% glycerol, separated on an 8% polyacrylamide, 44.5 mM Tris–borate (pH 8.3 at 25 °C, pH 8.9 at 5 °C), and 1 mM Na2EDTA (ethylenediaminetetraacetic acid) native gel at 5 °C, and visualized by phosphorimaging using the Personal Molecular Imager (Bio-Rad).
For the gel shift and ultraviolet cross-linking assays in Figure 2C and D, gel-purified 5΄ 32P-labeled RNA oligos were refolded by incubating at 90°C for 1 min, then at room temperature for 3 min. The refolded RNA was combined with purified recombinant N-RRM, C-RBD or C-RBD mutant protein either in 10 mM Tris-Cl (pH 7.4), 100 mM KCl, 2.5 mM MgCl2 (gel shift) or in 10 mM HEPES (pH 8.0), 50 mM KCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.05% Triton X-100, 5% glycerol, 1 mM dithiothreitol (DTT), 10 μg/ml salmon sperm DNA (cross-linking) and the RNA–protein mixture was incubated for 30 min at 30°C, 600 rpm. For the gel shift assay in Figure 2C, each sample was mixed with 1 μl 50% glycerol, separated on an 8% polyacrylamide, 44.5 mM Tris–borate (pH 8.3 at 25 °C, pH 8.9 at 5 °C) and 1 mM Na2EDTA native gel at 5 °C, and visualized by phosphorimaging using the Personal Molecular Imager (Bio-Rad). Ultraviolet cross-linking was conducted at 254 nm, 150 mJ/cm2. The cross-linked protein samples were separated on 4–12% polyacrylamide Bis-Tris gels, visualized by phosphorimaging, and stained in PageBlue Protein Staining Solution (24620, Thermo Scientific).
RNA structural probing by RNase V1/S1
The synthetic RNA oligos were 5΄ end-labeled with γ-32P-ATP by T4 polynucleotide kinase (70031, Affymetrix), gel purified and re-folded. Structural probing assays with RNase T1, nuclease S1, and RNase V1 were performed as previously described (6).
Electron microscopy
The purified GST-fusion C-RBD or N-RRM domain of the HNRNPG protein was diluted to 5 μM in 10 mM Tris-Cl (pH 7.4), 100 mM KCl, 2.5 mM MgCl2. The 5 μl sample solution was spotted on a 400 mesh carbon grid, washed with 1–2 drops of water, stained with 2 drops of 1% uranyl acetate, blotted and dried in air. Images were obtained at 68.6K× magnification on an FEI Tecnai G2 F30 Super Twin scanning transmission electron microscope.
PAR-CLIP and PAR-CLIP–MeRIP
Photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) was performed as previously reported (29) with the following modifications. HEK293T cells in 15-cm plates treated following normal PAR-CLIP procedures were lysed and digested with a combination of RNase I (AM2295, Ambion, 12 μl diluted 1:50 with H2O) and Turbo DNases (2 μl) for 3 min at 37°C, shaking at 1100 rpm. The lysate was then immediately cleared by spinning at 14 000 rpm, 4°C for 30 min and placed on ice for further use. HNRNPG binding sites were identified by PARalyzer v1.1 with default settings (30).
The PAR-CLIP–methylated RNA immunoprecipitation (MeRIP) experiment applied m6A-antibody immunoprecipitation (31) to the HNRNPG PAR-CLIP RNA samples (from HEK293T cells in eight 15-cm plates). The HNRNPG PAR-CLIP RNA sample was incubated with m6A-specific antibody (202003, SYSY), RNase inhibitor (80 units, Sigma-Aldrich), human placental RNase inhibitor (NEB) in 200 μl 1× IP buffer (50 mM Tris-Cl (pH 7.4), 750 mM NaCl and 0.5% (vol/vol) Igepal CA-630) at 4°C for 2 h under gentle shaking conditions. For each PAR-CLIP–MeRIP experiment, 20 μl Protein A beads (10002D, Thermo Scientific) were washed twice with 1 ml 1× IP buffer, blocked by a 2-h incubation with 100 μl 1× IP buffer supplemented with BSA (0.5 mg/ml), RNasin and human placental RNase inhibitor, and then washed twice with 100 μl 1× IP buffer. The pre-blocked Protein A beads were then combined with the prepared immuno-reaction mixture and incubated at 4°C for 2 h, followed by three washes with 100 μl 1× IP buffer. Finally, the RNA was eluted by 1-h incubation with 20 μl elution buffer (1× IP buffer and 6.7 mM m6A, Sigma-Aldrich) under gentle shaking conditions, and then purified by ethanol precipitation. The purified RNA sample (IP) as well as the input PAR-CLIP RNA sample (input control) were used for library construction.
Libraries for both PAR-CLIP and PAR-CLIP–MeRIP experiments were prepared using the TruSeq Small RNA Sample Preparation Kit (RS-200-0012, Illumina) according to the manufacturer's instructions, and then sequenced by Illumina HiSeq 2000 with single-end 50-bp reads. The control and IP samples from PAR-CLIP–MeRIP experiments were sequenced together in two lanes of one flow cell, and the reads from two lanes of each sample were combined for analysis. (The control and knockdown (KD) samples from METTL3/L14 KD experiments were sequenced and combined for analysis in the same manner.) The raw sequencing data was trimmed using the Trimmomatic computer program version 0.30 to remove adaptor sequences, and mapped to the human genome version hg19 by Bowtie 1.0.0 (32) without any gaps and allowing for at most two mismatches. Approximately 20 million reads were mapped to hg19 for each sample.
Detection and distribution analysis of m6A sites within HNRNPG binding sites
Detection of HNRNPG-bound m6A sites by PAR-CLIP–MeRIP involves comparing the read counts of the IP sample with those of the control (Ctrl) sample as follows: (i) we identified all AGRAC motifs within HNRNPG PAR-CLIP peaks; (ii) we performed transcriptome-wide scanning to compare read counts of each AGRAC motif in (1) from both Control and IP samples to calculate the fold change score, score = log2 (CountsIP/CountsControl). AGRAC motifs with log2 (CountsIP/CountsControl) larger than 0.5 were considered to be HNRNPG-bound m6A sites.
Detection of HNRNPG-bound m6A sites by PAR-CLIP involves comparing the read counts of the METTL3/L14 KD sample with that of the control (Ctrl) sample as follows: (i) we identified all AGRAC motifs within HNRNPG PAR-CLIP peaks; (ii) we performed transcriptome-wide scanning to compare read counts of each AGRAC motif in (1) from both Control and METTL3/L14 KD samples to calculate the fold change score, score = log2 (CountsKD/CountsControl). AGRAC motifs with log2 (CountsKD/CountsControl) <−0.5 were considered to be HNRNPG-bound m6A sites.
High-confidence m6A (HC m6A) sites within HNRNPG binding sites fulfill the following two requirements: (i) log2 (CountsIP/CountsControl) > 0.5; (ii) log2 (CountsKD/CountsControl) <−0.5. Pie charts illustrating the distribution of high-confidence HNRNPG-bound m6A sites within each segment were made using the following hierarchy: intron > ncRNA > 3΄UTR > 5΄UTR > CDS > intergenic.
RNA-sequencing
RNA-seq experiments were performed on two replicates of RNA samples from HNRNPG knockdown (KD1 and KD2) as well as control HEK293T cells 48 h after transfection. Total RNA was extracted using the RNeasy Plus kit (74104, Qiagen). Libraries were prepared using the TruSeq Stranded mRNA LT Sample Prep Kit (RS-122-9005DOC, Illumina). KD and control samples were sequenced together in four lanes in one flow cell. All samples were sequenced by Illumina HiSeq 2000 with paired-end 100-bp reads. The reads from the four lanes of each sample were combined for all analyses. The RNA-seq data were mapped using the splice-aware alignment algorithm TopHat version 1.1.4 (33) based on the following parameters: tophat –num-threads 8 –mate-inner-dist 200 –solexa-quals –min-isoform-fraction 0 –coverage-search-segment-mismatches 1. Gene expression level changes were analyzed using Cuffdiff (34). Approximately 140–200 million reads were mapped for each sample. Differential splicing was determined using DEXSeq (35) based on Cufflinks-predicted, non-overlapping exons.
Evolutionary conservation, graphics and statistical analysis
Phylogenetic conservation analysis was performed by comparing PhyloP scores at high-confidence m6A-containing AGRAC motifs to those at randomly selected AGRAC sequences. The PhyloP scores were accessed from the precompiled PhyloP scores (36) (ftp://hgdownload.soe.ucsc.edu/goldenPath/hg19/phyloP46way/) for primates and vertebrates. P-values were evaluated using the Mann–Whitney–Wilcoxon test. The random selection was done separately for primates and for vertebrates.
Sequence logos were generated using the WebLogo package. R statistical package was used for all statistical analysis (unless stated otherwise).
RT-PCR quantification
Total RNA was extracted from HEK293T cells and reverse transcribed using the SuperScript III First-Strand Synthesis System (18080-051, Life Technologies). In order to validate the splicing changes identified from our RNA-seq data, we performed RT-PCR measurements using Taq DNA Polymerase (Thermo Scientific) under the following conditions: 95°C for 3 min, 30 cycles of [95°C for 30 s, 55°C for 30 s, 72°C for 1 min], and then finally 72°C for 10 min. For the target alternatively spliced exon, we designed and used primers annealing to both neighboring constitutive exons. The PCR products were separated on a 10% polyacrylamide, 89 mM Tris–borate (pH 8.3 at 25 °C) and 2 mM Na2EDTA gel and stained with SYBR Gold Nucleic Acid Gel Stain (S11494, Thermo Scientific).
To validate the gene expression level changes identified from our RNA-seq data, we performed RT-qPCR measurements using Power SYBR Green PCR Master Mix (4367659, Thermo Scientific) under the following conditions: 50°C for 3 min, followed by 95°C for 10 min, 40 cycles of [95°C for 15 s, 60°C for 1 min], and then 40°C for 1 min, 95°C for 15 s, and finally 60°C for 30 s.
The primer sequences are listed below (Gene name: forward primer; reverse primer): C20orf72: ACAGCGGATGATTCTGGAAC; TTCCTGGGGTGAAAGTATGC; NASP: TGTGCATGTGGAAGAGGAAG; GAAGGTGTGCATGTGGAAGA; CTNNB1: GAAAATCCAGCGTGGACAAT; CAGGACTTGGGAGGTATCCA; PKIA (primers_1): CCTGGTTTCCCCAAAGAAGT; TGATTGGAAACCTTCTTGTCTTT; PKIA (primers_2): TGGTAGCAATGACTGATGTGG; ACTTGCAGAGGAAACCAGGA; RHBDF2 (primers_1): AGAGCCAGAGACCCAAGACA; CCCAAGACTCAGAGAGGCA; RHBDF2 (primers_2): GAGTACCCAGGAAGCTGCAC; TACAGATGCTCCGGTGTCAA; SCD (primers_1): TGTTCGTTGCCACTTTCTTG; GGGGGCTAATGTTCTTGTCA; SCD (primers_2): CTCCACTGCTGGACATGAGA; AATGAGTGAAGGGGCACAAC; SFXN2 (primers_1): GCCAGACTGGTCTCGAACTC; ACGGTCCCCTTTTTAGCACT; SFXN2 (primers_2): CCTGGGATTGGTCGAAAAG; AAATGCCACCAGTTACAGCC; ANKRD52 (primers_1): CTGTGCCGAGACTTTAAGGG; GCGAGTATCCGCTGTAATCC; ANKRD52 (primers_2): AGACGCTGGTGAATCTGGAC; GCTGTAAGCACCTCCACACA; MBNL1 (primers_1): AATATCTTCATCCACCCCCA; TTGGCTAGTTGCATTTGCTG; MBNL1 (primers_2): GCTGCATCTGTCTATGCCAA; CGAATTTCCAAGCTGCTTTC; VPRBP (primers_1): GCTGACAAAAGAGGCTGACC; GCTGAGGATGAGCAGTAGGG; VPRBP (primers_2): TGATAGAATATGGCCCAGCG; CCAATTGCAGGCAATAGAAA; ZBTB4 (primers_1): TTCCATGCCTTTGGATCTTC; ATTTGGGGGTCAAGATAGGG; ZBTB4 (primers_2): GCTCACTTCAGCCCCACTAC; AGACGAGGAAGAGGAGGAGG; SSU72 (primers_1): GCACTTCCCGACATACCTGT; GCACAATGACAGCAGCATCT; SSU72 (primers_2): AAATAAGAGAATCAAGCCCCG; TTCCACCACCTGGTCATACA; SPOPL (primers_1): GCTGGAGTCGTAACTCGGAA; CGCTCCTAAACTTCTTCCCC; SPOPL (primers_2): GGAGGTTTGTTCTGGTGCAT; GCCCTTAAGAAGCACACTGG; EDEM1 (primers_1): AGCCTCCTTTCTGCTCACAG; GGTGTTTTCAAAAGCAGGGA; EDEM1 (primers_2): ATGAGCATCTTCGGGAATTG; AACTCATGAGGTTTCGGCCT.
RESULTS
HNRNPG preferentially binds m6A-modified RNA
In order to identify nuclear m6A reader proteins, we conducted an RNA pull-down assay using methylated and unmethylated forms of a 34-nt hairpin from the lncRNA MALAT1 (Figure 1A). This hairpin contains a single m6A site corresponding to position 2,515 of MALAT1, which is 63% m6A-modified in HeLa cells (6). Following incubation with HeLa nuclear extract, the methylated and unmethylated forms of the MALAT1 hairpin pulled down different protein complexes, which were resolved on native gels (Figure 1B). Denaturing gel electrophoresis (Figure 1C) and mass spectrometry (Supplementary Figure S1) further revealed that the protein HNRNPG was enriched in the pull-down with the m6A-methylated hairpin. These results identify HNRNPG as an m6A reader protein that preferentially binds an m6A-modified hairpin from the lncRNA MALAT1.
HNRNPG is a ubiquitously expressed RNA binding protein encoded by the gene RBMX (RNA binding motif protein, X chromosome) (37). A dominant function of HNRNPG is the regulation of alternative splicing (38,39), but HNRNPG also functions in other cellular processes including DNA repair (40), sister chromatid cohesion (41), and transcriptional regulation (42). Notably, HNRNPG has been shown to either positively or negatively regulate the inclusion of several disease-related exons (38,39). In addition, HNRNPG plays an important role in neural development (43), and a frameshift mutation in the C-terminal region of HNRNPG causes an intellectual disability syndrome in humans (44).
HNRNPG binds the MALAT1 hairpin through a low-complexity region
HNRNPG is composed of an N-terminal globular RNA recognition motif of ∼90 amino acids, followed by ∼300 amino acids of low-complexity sequence, in which two-thirds of the amino acid residues are serine, arginine, glycine, and proline (Figure 2A). In addition to the N-terminal RNA recognition motif (N-RRM), which binds A/C-rich sequences in single-stranded RNA (37), the C-terminal 58 amino acids of the low-complexity sequence have been shown to bind an RNA hairpin with an A/G-rich motif (45). This C-terminal RNA binding domain (C-RBD) is highly conserved across species (45), and a frameshift mutation in this region causes an intellectual disability syndrome in humans (44). However, the RNA sequence and structural preferences for the HNRNPG C-RBD have yet to be determined.
Due to the extensive low-complexity region of HNRNPG, the full-length recombinant protein was difficult to express and purify by us and others (45). Instead, we purified each of the two known RNA binding domains of HNRNPG, the N-RRM and the C-RBD, fused to an N-terminal glutathione S-transferase (GST). Similar to other low-complexity sequences (25,46), the purified C-RBD self-assembled into higher-order structures, forming aggregates of ∼100 nm in size that were visible by electron microscopy (Figure 2B). We conducted gel shift assays to evaluate the binding of the N-RRM and C-RBD to the methylated and unmethylated MALAT1 hairpins. While the N-RRM did not shift the MALAT1 hairpin, the C-RBD shifted both hairpins with high cooperativity (Figure 2C). However, quantification was difficult to analyze due to the protein concentration dependent aggregation of the C-RBD. Therefore, we turned to ultraviolet cross-linking assays (Figure 2D). Similar to the gel shift assays, the N-RRM did not cross-link to the MALAT1 hairpin, whereas the C-RBD cross-linked to both hairpins and more strongly to the m6A-modified MALAT1 hairpin.
The C-RBD contains three Arg-Gly-Gly (RGG) repeats. Since RGG motifs frequently function in RNA binding (47), we evaluated the role of the RGG motif by introducing arginine-to-phenylalanine mutations in the three RGG repeats of the C-RBD (Figure 2D; Supplementary Figures S1C and 2A). Mutating one or two of the RGG motifs did not alter cross-linking efficiency. However, introducing mutations in all three RGG motifs abolished cross-linking to both the methylated and unmethylated MALAT1 hairpins, suggesting that the RGG repeats in the C-RBD mediate MALAT1 hairpin binding.
m6A alters RNA structure and increases the accessibility of an HNRNPG binding motif
To examine transcriptome-wide binding sites of HNRNPG, we performed PAR-CLIP against HNRNPG in HEK293T cells. HNRNPG PAR-CLIP of biological replicates yielded a total of 354,057 HNRNPG binding sites. AGRAC (R = A/G) was the most enriched motif among HNRNPG-bound RNAs, accounting for ∼30% (106,300) of HNRNPG binding sites identified by PAR-CLIP (Figure 3A). The AGRAC motif overlaps with the m6A consensus motif RRACH, indicating that a large fraction of HNRNPG binding sites could be m6A-modified.
The preferential binding of HNRNPG to m6A-modified RNA can be mediated by direct recognition of m6A, or through an effect of m6A on the accessibility of its RNA binding motif. Mutating the m6A site in the MALAT1 hairpin to G, C, or U led to increased pull-down of HNRNPG from nuclear extract (Figure 3B). This result indicates that the preferential binding of HNRNPG to the methylated hairpin does not depend on the presence of the N6-methyl group of the m6A residue.
We then investigated whether m6A modification influences the secondary structure of the MALAT1 hairpin using structural mapping. We used S1 nuclease, which specifically cuts single-stranded regions and RNase V1, which specifically cuts double-stranded, stacked regions of RNA (Figure 3C). Upon m6A modification, the MALAT1 hairpin became more single-stranded in the region surrounding the m6A site as shown by both increased S1 cuts and decreased V1 cuts in the region around the m6A site. Our structural mapping results are consistent with m6A disrupting the hairpin structure and increasing the accessibility of its surrounding nucleotides. The affected region in the MALAT1 hairpin included the AGGAC sequence, which matches the AGRAC motif found in the dominant HNRNPG binding sites identified by PAR-CLIP (Figure 3A).
To evaluate the role of the AGGAC sequence for HNRNPG binding to the MALAT1 hairpin, we switched individual base pairs involving the AGG[m6A]C, in order to disrupt the sequence while minimizing the effect on hairpin structure. Each of these pairwise mutations in the MALAT1 hairpin led to decreased HNRNPG pull-down from nuclear extract, with pairwise mutations at the m6A position having the smallest effect (Supplementary Figure S2B). This result suggests that the AGG[m6A]C sequence is important for HNRNPG binding, with the exception of the m6A within this motif, which influences protein binding mainly by disrupting RNA structure to expose the surrounding bases. In addition, mutations that disrupted the structure of the MALAT1 hairpin led to increased cross-linking of the purified C-RBD protein, while double mutations that restored the hairpin structure reduced cross-linking back to the wild-type hairpin level (Supplementary Figure S2C). These results, together with the structural mapping data, indicate that m6A modification of the MALAT1 hairpin promotes HNRNPG binding by increasing the accessibility of the AGGAC motif (Figure 3D).
Transcriptome-wide identification of m6A sites facilitating HNRNPG interactions
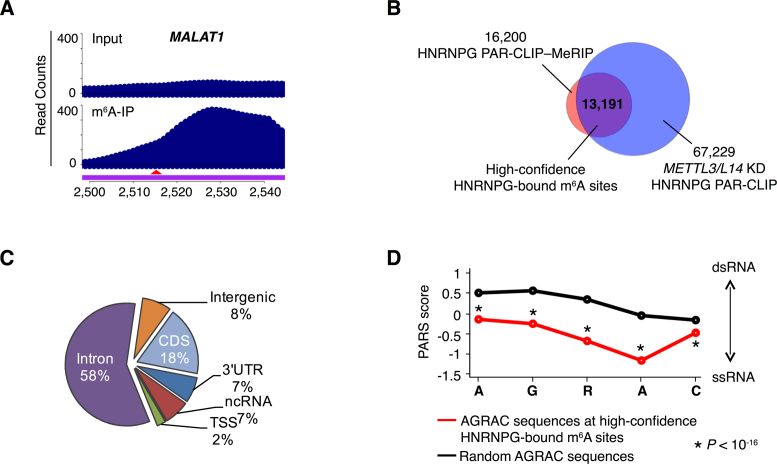
To identify m6A-modified HNRNPG binding sites, we conducted HNRNPG PAR-CLIP followed by MeRIP of biological replicates (20). In this HNRNPG PAR-CLIP–MeRIP experiment, HNRNPG-bound RNAs were isolated by PAR-CLIP (‘input’ sample) and then used as the input for MeRIP to enrich the m6A-containing HNRNPG-bound RNAs (‘IP’ sample). HNRNPG PAR-CLIP–MeRIP yielded a total of 16,200 m6A-modified HNRNPG binding sites (log2 (IP/input) > 0.5), including the m6A at position 2,515 of MALAT1 (Figure 4A). In addition, we conducted HNRNPG PAR-CLIP in cells depleted of either of the core m6A methyltransferase components METTL3 and METTL14 (Supplementary Figure S3A). Sites with decreased HNRNPG occupancy upon METTL3 or METTL14 knockdown were considered as m6A methyltransferase-dependent HNRNPG binding sites. We identified 67,229 AGRAC motifs that were located within HNRNPG PAR-CLIP peaks but showed decreased HNRNPG binding following either METTL3 or METTL14 knockdown. Among these sites, 37,750 showed decreased HNRNPG binding in both METTL3 and METTL14 knockdown cells (Supplementary Figure S3B). A total of 13,191 m6A methyltransferase-dependent AGRAC sites were also identified as m6A-modified HNRNPG binding sites by PAR-CLIP–MeRIP (Figure 4B and C; Supplementary Table S1). In addition, these AGRAC sites were highly conserved across species (Supplementary Figure S3C). We designate these 13,191 sites as high-confidence HNRNPG-bound m6A sites.
Figure 4.
HNRNPG binds m6A-modified RNAs transcriptome-wide. (A) PAR-CLIP–MeRIP input and IP (m6A-IP) read counts in a region of the MALAT1 transcript. The red arrowhead indicates the m6A site at position 2,515. (B) Identification of high-confidence HNRNPG-bound m6A sites (purple) as the overlap between m6A-modified HNRNPG binding sites, identified by HNRNPG PAR-CLIP–MeRIP (pink) and m6A methyltransferase-dependent HNRNPG-bound AGRAC sites, identified by HNRNPG PAR-CLIP in m6A methyltransferase (METTL3 and METTL14) knockdown HEK293T cells (blue). (C) Regional distribution of high-confidence HNRNPG-bound m6A sites. (D) Comparison of the structure of AGRAC sequences at high-confidence HNRNPG-bound m6A sites (red) versus random AGRAC sequences (black) in human polyadenylated RNAs, based on parallel analysis of RNA structure (PARS) data (48). The x-axis denotes nucleotide position; the y-axis shows the PARS score. Positive PARS scores indicate double-stranded conformation; negative scores indicate single-stranded conformation. P–value, Mann–Whitney U test.
Notably, using transcriptome-wide data from parallel analysis of RNA structure of human polyadenylated RNAs (48), we found that AGRAC sequences at high-confidence HNRNPG-bound m6A sites were less structured than random AGRAC sequences (Figure 4D), suggesting that the m6A-dependent structural change in the MALAT1 hairpin could be generalized to HNRNPG-bound AGRAC motifs transcriptome-wide.
m6A-dependent HNRNPG binding regulates gene expression and alternative splicing
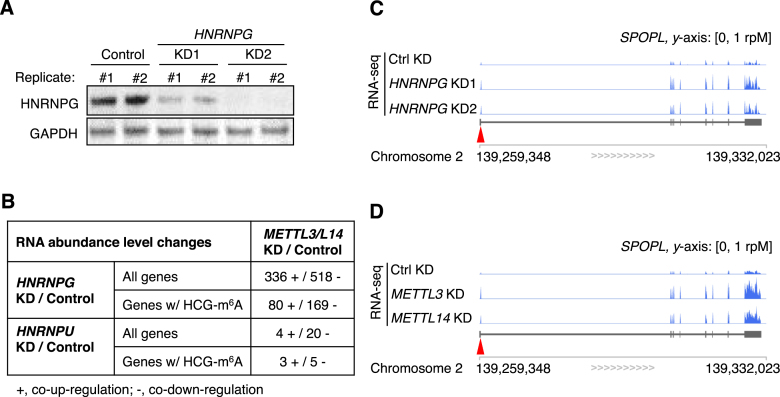
HNRNPG regulates the transcription and alternative splicing of multiple genes (38,39,42). To examine the effect of HNRNPG on gene expression and splicing transcriptome-wide, we conducted mRNA sequencing in HEK293T cells depleted of HNRNPG with two different siRNAs (Figure 5A). If HNRNPG binding to m6A-modified RNAs contributes to its function as a regulator of transcription and splicing, then perturbations to cellular m6A modifications should affect the expression and splicing of HNRNPG-regulated genes. To test this prediction, we compared our HNRNPG knockdown mRNA-seq data to our previous mRNA-seq data from cells with a global reduction in m6A due to knockdown of METTL3 or METTL14 (20). Using Cuffdiff2 (34), we found that hundreds of m6A-containing RNA transcripts were similarly up- or downregulated by both HNRNPG knockdown and m6A methyltransferase knockdown (Figure 5B–D). These results were validated by RT-qPCR (Supplementary Figure S4A). As a control, gene expression changes in cells depleted of the mRNA binding protein HNRNPU did not correlate with gene expression changes upon METTL3 or METTL14 knockdown (Figure 5B) (49), consistent with the observation that HNRNPU did not preferentially bind m6A-modified RNA in nuclear pull-down assays (Supplementary Figure S1A).
Figure 5.
m6A-dependent HNRNPG binding regulates mRNA abundance. (A) Western blot showing depletion of HNRNPG with two different siRNAs (KD1 and KD2) for mRNA-seq experiments. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (B) Number of genes with correlated changes in expression upon HNRNPG knockdown and m6A methyltransferase (METTL3 or METTL14) knockdown. HCG-m6A, high-confidence HNRNPG-bound m6A site. mRNA-seq data from HNRNPU knockdown HEK293T cells (Gene Expression Omnibus, GSE34995 (49)) were analyzed as a control. (C and D) mRNA-seq reads for SPOPL (speckle-type POZ protein-like) transcripts in control, HNRNPG knockdown (C) and m6A methyltransferase knockdown (D) cells. The arrowhead indicates the m6A site.
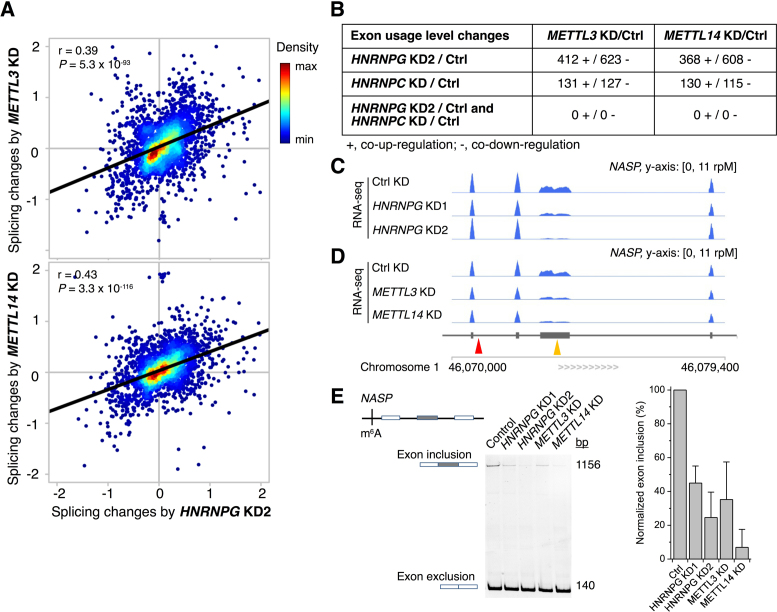
Next, we used DEXSeq (35) to examine changes in alternative splicing. Splicing changes resulting from HNRNPG knockdown were correlated with splicing changes resulting from m6A methyltransferase knockdown (Figure 6A and Supplementary Figure S5A–B). In particular, ∼1,000 exons in genes with high-confidence HNRNPG-bound m6A sites were similarly up- or downregulated by HNRNPG knockdown and m6A methyltransferase knockdown (Figure 6B). For example, both HNRNPG knockdown and m6A methyltransferase knockdown reduced the inclusion of an exon in the NASP transcript, which encodes nuclear autoantigenic sperm protein (Figure 6C and D). HNRNPG PAR-CLIP–MeRIP revealed an m6A-modified HNRNPG binding site in a nearby intron, suggesting that HNRNPG binding to this m6A-modified site might promote inclusion of the alternatively spliced exon. The HNRNPG- and m6A methyltransferase-dependent regulation of exon inclusion in NASP and two other transcripts was validated by RT-PCR (Figure 6E; Supplementary Figure S5C and D). These results suggest that HNRNPG binding to m6A-modified transcripts can regulate the alternative splicing of nearby exons.
Figure 6.
m6A-dependent HNRNPG binding regulates alternative splicing. (A) Splicing changes of annotated differentially expressed exons upon HNRNPG knockdown with siRNA KD2 (x-axis) and m6A methyltransferase (METTL3 or METTL14) knockdown (y-axis), by log ratio of normalized counts relative to control knockdown, log2 (KD/Control). Pearson's correlation coefficient r and P-values are shown for each panel. (B) Number of exons for which changes in exon usage are correlated upon HNRNPG, HNRNPC, and/or m6A methyltransferase (METTL3 or METTL14) knockdown. For HNRNPG knockdown, only exons in genes with high-confidence HNRNPG-bound m6A sites were counted. For HNRNPC knockdown, only exons in genes with high-confidence m6A-switches were counted (20). (C and D) mRNA-seq reads for NASP transcripts in control, HNRNPG knockdown (C) and m6A methyltransferase knockdown (D) cells. The yellow arrowhead indicates the alternatively spliced exon; the red arrowhead indicates the m6A site. (E) Reverse transcription and PCR (RT-PCR) validating differential exon usage in NASP. Data shown as mean; error bar = standard deviation; n = 3 biological replicates.
The m6A reader protein HNRNPC also binds m6A-modified RNAs through an m6A-switch mechanism. Previous results revealed thousands of RNA transcripts and hundreds of exons that are similarly up- or downregulated by HNRNPC knockdown and m6A methyltransferase knockdown (20). However, <10% of these transcripts and none of these exons were also co-regulated by HNRNPG and m6A methyltransferase knockdown (Supplementary Figure S4B and Figure 6B). Thus, while HNRNPC and HNRNPG both regulate gene expression and alternative splicing by binding to m6A-modified transcripts, the target genes regulated by m6A-dependent binding of HNRNPC are largely distinct from those regulated by m6A-dependent binding of HNRNPG. These results suggest that m6A-promoted HNRNPG and HNRNPC binding influence different subsets of gene transcripts.
DISCUSSION
In this study, we identified HNRNPG as a new m6A ‘reader’ protein that uses a low-complexity region to bind m6A-modified RNAs. Rather than directly recognizing the N6-methyl group, HNRNPG binds a purine-rich motif that becomes accessible upon m6A modification of an RNA hairpin. We identified 13,191 high-confidence HNRNPG-bound m6A sites transcriptome-wide and showed that HNRNPG and m6A modification cooperate to regulate gene expression and alternative splicing. These results demonstrate a role for the low-complexity domain of HNRNPG in the binding of m6A-modified RNAs, and suggest that the m6A-dependent binding of HNRNPG functions in the regulation of gene expression.
In the m6A-switch mechanism, m6A modulates protein binding by inducing an RNA structural change that alters the accessibility of a protein binding site. In the case of the m6A reader HNRNPC, m6A modification disrupts base-pairing and exposes a U-rich binding site for HNRNPC in the opposite strand (20,50). HNRNPG binding to m6A-modified RNA also depends on an m6A-induced structural change. However, in contrast to HNRNPC, HNRNPG binds to a purine-rich motif that includes the m6A site. Since the HNRNPG binding motif overlaps with the m6A site, HNRNPG binding could compete with the binding of direct m6A readers such as the YTH domain proteins. HNRNPG might also modulate the binding of other m6A readers that use RNA structural changes as a readout for m6A. Although we found no indication that HNRNPG and HNRNPC cooperate to regulate the alternative splicing of m6A-containing transcripts, positive or negative cooperativity between these and other m6A readers may play a role in the regulation of other cellular processes.
Unlike previously described m6A reader proteins, which all bind m6A-modified RNA through folded domains, HNRNPG binds to m6A-modified RNA through a low-complexity region that self-assembles into large particles in vitro (Figure 2B). Interactome studies have revealed that many RNA binding proteins lack canonical RNA binding domains, and suggest that intrinsically disordered and low-complexity regions play an important role in RNA binding (24). In fact, low-complexity sequences are prevalent in RNA binding proteins, and an estimated one-third of RNA binding proteins are highly disordered (23). Unstructured RGG repeats are particularly common in RNA binding proteins and have been shown to mediate structure- and sequence-specific RNA binding (47). In addition to their widespread role in RNA binding, low-complexity domains frequently function in protein–protein interactions (24), are necessary and sufficient for the formation of granule-like structures in vitro (25), and are common sites of disease mutations that disrupt the balance between the assembly and clearance of ribonucleoprotein granules in the cell (24,26). Several other m6A reader proteins contain low-complexity regions, but these regions have not been shown to directly bind m6A-modified RNAs. The low-complexity regions of the YTH proteins function in their recruitment to various cellular bodies (12,13,15). The RNA recognition motifs of the m6A reader HNRNPA2B1 show only a slight preference for m6A-modified RNAs in vitro (22), and it is yet to be determined whether this protein directly binds to m6A. HNRNPA2B1 also has a glycine-rich low-complexity domain with four RGG motifs, which might contribute to the preferential binding of HNRNPA2B1 to m6A-modified RNA. Moreover, HNRNPA2B1 may also use an m6A-switch mechanism to selectively bind m6A-modified RNAs. Thus, it is possible that HNRNPA2B1 and HNRNPG both use low-complexity regions and an m6A-switch mechanism to bind m6A-modified transcripts.
Our results reveal that HNRNPG regulates the alternative splicing of ∼1,000 exons in m6A-modified transcripts, but the mechanism for m6A-dependent splicing regulation by HNRNPG remains unclear. HNRNPG might recruit or compete with splicing factors for binding to m6A-modified transcripts, similar to the m6A reader YTHDC1 (15). In addition, both the N-terminal RRM and other parts of the low-complexity region of HNRNPG could contribute to the m6A-dependent regulation of splicing. RNA binding proteins frequently contain multiple repeats or combinations of RNA binding domains. This modular domain structure can contribute to RNA binding affinity or specificity, define the spacing between binding sites, organize RNA topology, or allow a single protein to simultaneously interact with multiple RNA molecules (51). Given that the regulation of alternative splicing by RNA binding proteins is highly position-dependent (52), the role of HNRNPG in splicing regulation is likely to be strongly dependent on the relationship between its different RNA binding domains.
The reversibility of m6A modifications allows cells to dynamically regulate modification levels and fine-tune the fate of RNA transcripts. Indeed, modification fractions vary widely among different m6A sites (6) and are regulated in response to external stimuli (7,8). The heterogeneity and dynamic nature of m6A modification make it a useful mechanism for control of RNA fate. In addition to its role in binding m6A-modified RNA, the low-complexity region of HNRNPG likely functions in granule formation. Cellular granules play an important role in regulating the localization of both RNA transcripts and RNA binding proteins (26), which can in turn influence RNA splicing, translation and turnover, all of which are processes regulated by m6A (9–17). The relationship between HNRNPG granule formation and binding to m6A-modified RNAs could be a key to its function in the m6A-dependent regulation of gene expression and alternative splicing.
Many cellular functions of m6A are mediated by m6A reader proteins. HNRNPG is the second m6A reader protein shown to use the m6A-switch mechanism (20), strongly suggesting that m6A-mediated changes in RNA structure might impact interactions with many other RNA binding proteins. At the same time, HNRNPG differs from previously discovered m6A reader proteins in that it uses a low-complexity region to bind to m6A-modified RNA. Moreover, HNRNPG influences a large number of alternative splicing events in an m6A-dependent manner. In the future, it will be important to investigate the mechanisms by which interactions between the low-complexity region of HNRNPG and m6A-modified RNA modulate alternative splicing in the cell.
ACCESSION NUMBER
All RNA sequencing data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE74085.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Pan laboratory for comments and discussions. We also thank Y.C. Leung, Y. Pigli, J. Yue, J. Liu, Y. Yue, K. Chen, M. Yu, and Y. Chen for technical assistance. The authors would like to thank the generous support of the University of Chicago Biological Sciences Division and the Frank Family Endowment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM113194 to T.P., K01HG006699 to Q.D., F30GM120917 to K.I.Z.]; NIH Medical Scientist Training Program Grant [T32GM007281]; University of Chicago Biological Sciences Division and Frank Family Endowment (K.I.Z.). Funding for open access charge: NIH [R01GM113194].
Conflict of interest statement. None declared.
REFERENCES
- 1. Liu N., Pan T.. N6-methyladenosine-encoded epitranscriptomics. Nat. Struct. Mol. Biol. 2016; 23:98–102. [DOI] [PubMed] [Google Scholar]
- 2. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ping X.-L., Sun B.-F., Wang L., Xiao W., Yang X., Wang W.-J., Adhikari S., Shi Y., Lv Y., Chen Y.-S. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S.-H. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.-G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu N., Parisien M., Dai Q., Zheng G., He C., Pan T.. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013; 19:1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 8. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3΄ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J.. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014; 10:927–929. [DOI] [PubMed] [Google Scholar]
- 10. Theler D., Dominguez C., Blatter M., Boudet J., Allain F.H.-T.. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014; 42:13911–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo S., Tong L.. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.-B.. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiao W., Adhikari S., Dahal U., Chen Y.-S., Hao Y.-J., Sun B.-F., Sun H.-Y., Li A., Ping X.-L., Lai W.-Y. et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell. 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- 16. Haussmann I.U., Bodi Z., Sanchez-Moran E., Mongan N.P., Archer N., Fray R.G., Soller M.. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016; 540:301–304. [DOI] [PubMed] [Google Scholar]
- 17. Lence T., Akhtar J., Bayer M., Schmid K., Spindler L., Ho C.H., Kreim N., Andrade-Navarro M.A., Poeck B., Helm M. et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature. 2016; 540:242–247. [DOI] [PubMed] [Google Scholar]
- 18. Roost C., Lynch S.R., Batista P.J., Qu K., Chang H.Y., Kool E.T.. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 2015; 137:2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spitale R.C., Flynn R.A., Zhang Q.C., Crisalli P., Lee B., Jung J.-W., Kuchelmeister H.Y., Batista P.J., Torre E.A., Kool E.T. et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015; 519:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T.. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015; 518:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.-B., Jaffrey S.R.. 5΄ UTR m6A promotes cap-independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F.. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. 2015; 162:1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neelamraju Y., Hashemikhabir S., Janga S.C.. The human RBPome: from genes and proteins to human disease. J. Proteomics. 2015; 127:61–70. [DOI] [PubMed] [Google Scholar]
- 24. Calabretta S., Richard S.. Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem. Sci. 2015; 40:662–672. [DOI] [PubMed] [Google Scholar]
- 25. Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012; 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramaswami M., Taylor J.P., Parker R.. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013; 154:727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012; 149:768–779. [DOI] [PubMed] [Google Scholar]
- 28. Dai Q., Fong R., Saikia M., Stephenson D., Yu Y., Pan T., Piccirilli J.A.. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic Acids Res. 2007; 35:6322–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A.-C., Munschauer M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Corcoran D.L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R.L., Keene J.D., Ohler U.. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011; 12:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G.. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013; 8:176–189. [DOI] [PubMed] [Google Scholar]
- 32. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anders S., Reyes A., Huber W.. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012; 22:2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heinrich B., Zhang Z., Raitskin O., Hiller M., Benderska N., Hartmann A.M., Bracco L., Elliott D., Ben-Ari S., Soreq H. et al. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J. Biol. Chem. 2009; 284:14303–14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hofmann Y., Wirth B.. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-β1. Hum. Mol. Genet. 2002; 11:2037–2049. [DOI] [PubMed] [Google Scholar]
- 39. Wang Y., Wang J., Gao L., Stamm S., Andreadis A.. An SRp75/hnRNPG complex interacting with hnRNPE2 regulates the 5΄ splice site of tau exon 10, whose misregulation causes frontotemporal dementia. Gene. 2011; 485:130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J.. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012; 14:318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Matsunaga S., Takata H., Morimoto A., Hayashihara K., Higashi T., Akatsuchi K., Mizusawa E., Yamakawa M., Ashida M., Matsunaga T.M. et al. RBMX: a regulator for maintenance and centromeric protection of sister chromatid cohesion. Cell Rep. 2012; 1:299–308. [DOI] [PubMed] [Google Scholar]
- 42. Zhao S., Korzan W.J., Chen C.-C., Fernald R.D.. Heterogeneous nuclear ribonucleoprotein A/B and G inhibits the transcription of gonadotropin-releasing-hormone 1. Mol. Cell. Neurosci. 2008; 37:69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsend-Ayush E., O'Sullivan L.A., Grützner F.S., Onnebo S.M.N., Lewis R.S., Delbridge M.L., Marshall Graves J.A., Ward A.C.. RBMX gene is essential for brain development in zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2005; 234:682–688. [DOI] [PubMed] [Google Scholar]
- 44. Shashi V., Xie P., Schoch K., Goldstein D. b., Howard T. d., Berry M. n., Schwartz C. e., Cronin K., Sliwa S., Allen A. et al. The RBMX gene as a candidate for the Shashi X-linked intellectual disability syndrome. Clin. Genet. 2015; 88:386–390. [DOI] [PubMed] [Google Scholar]
- 45. Kanhoush R., Beenders B., Perrin C., Moreau J., Bellini M., Penrad-Mobayed M.. Novel domains in the hnRNP G/RBMX protein with distinct roles in RNA binding and targeting nascent transcripts. Nucl. Austin Tex. 2010; 1:109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schwartz J.C., Wang X., Podell E.R., Cech T.R.. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013; 5:918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thandapani P., O’Connor T.R., Bailey T.L., Richard S.. Defining the RGG/RG motif. Mol. Cell. 2013; 50:613–623. [DOI] [PubMed] [Google Scholar]
- 48. Wan Y., Qu K., Zhang Q.C., Flynn R.A., Manor O., Ouyang Z., Zhang J., Spitale R.C., Snyder M.P., Segal E. et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014; 505:706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huelga S.C., Vu A.Q., Arnold J.D., Liang T.Y., Liu P.P., Yan B.Y., Donohue J.P., Shiue L., Hoon S., Brenner S. et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012; 1:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou K.I., Parisien M., Dai Q., Liu N., Diatchenko L., Sachleben J.R., Pan T.. N6-Methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J. Mol. Biol. 2016; 428:822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fu X.-D., Ares M. Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014; 15:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.