Abstract
FASTK family proteins have been identified as regulators of mitochondrial RNA homeostasis linked to mitochondrial diseases, but much remains unknown about these proteins. We show that CRISPR-mediated disruption of FASTKD1 increases ND3 mRNA level, while disruption of FASTKD4 reduces the level of ND3 and of other mature mRNAs including ND5 and CYB, and causes accumulation of ND5–CYB precursor RNA. Disrupting both FASTKD1 and FASTKD4 in the same cell results in decreased ND3 mRNA similar to the effect of depleting FASTKD4 alone, indicating that FASTKD4 loss is epistatic. Interestingly, very low levels of FASTKD4 are sufficient to prevent ND3 loss and ND5–CYB precursor accumulation, suggesting that FASTKD4 may act catalytically. Furthermore, structural modeling predicts that each RAP domain of FASTK proteins contains a nuclease fold with a conserved aspartate residue at the putative active site. Accordingly, mutation of this residue in FASTKD4 abolishes its function. Experiments with FASTK chimeras indicate that the RAP domain is essential for the function of the FASTK proteins, while the region upstream determines RNA targeting and protein localization. In conclusion, this paper identifies new aspects of FASTK protein biology and suggests that the RAP domain function depends on an intrinsic nucleolytic activity.
INTRODUCTION
The human mitochondrial genome is 16.6 kb in length, encodes 2 rRNAs, 13 open reading frames and 22 tRNAs and is packaged into discrete structures known as nucleoids (1–2). It is transcribed into two long polycistronic transcripts which are processed into rRNAs, tRNAs and mRNAs. Although import of RNA has been reported, locally transcribed RNA accounts for the vast majority of RNA within mitochondria (3). The sequences encoding the two rRNAs and most of the mRNAs are closely flanked by tRNAs interspersed throughout the genome. To explain how the mature RNA sequences are generated, the ‘tRNA punctuation’ model (4) was proposed, according to which excision of the tRNAs by RNase P (5) and RNase Z (6) respectively, generates the mature rRNA, and most of the mature mRNA transcripts. However, a number of mRNAs, such as ATP8/6, CO1, CO3, CYB, ND5, and ND6, do not have flanking tRNAs at both ends and so their processing cannot be explained by this model. Little is known about the mechanisms by which the 5΄ and 3΄-ends of these latter mRNAs are generated. Of particular note are the two adjacent genes, ND5 and CYB, which lack an intervening tRNA and for which an unprocessed ND5–CYB precursor is often observed. Processing of ND5–CYB is not dependent upon RNase P (7), but has been reported to require the PPR motif-containing protein, PTCD2 (8). In addition, it was recently reported that expression of the ND6 mRNA, which is the only mRNA encoded on the light strand, and which has no tRNA at the 3΄-end, requires the presence of the two related proteins, FASTK and FASTKD2 (9–10).
FASTK and FASTKD2 are part of a family of proteins with six members in humans: FASTK, and FASTKD1 to FASTKD5. All are known to be mitochondrial RNA binding proteins (11–12), and display three poorly conserved domains: FAST_1, FAST_2 and RAP. FASTKD2 mutations have been linked to severe encephalomyopathy in human patients (13). Interestingly, modeling of the RAP domain of FASTKD2 suggested a similarity to a putative endonuclease-like protein from Neisseria gonorrhoeae (11,14). On the basis of BLAST searches, there appear to be similar proteins in other branches of the metazoan tree, such as Drosophila, but their roles in non-mammalian species have not been explored. Additionally, in chloroplasts, there are multiple proteins which contain FAST-like domains characterized as Octotricopeptide repeats (OPR) and/or RAP domains, which are known to interact with each other and participate in trans-splicing reactions, ribosomal 5΄-end processing, mRNA stabilization, and possibly translational activation (15–20).
FASTK and FASTKD2 are two FASTK proteins shown to affect the processing of mitochondrial RNA, specifically ND6 mRNA (9–10). In 2014, Wolf and Mootha published the results of a MitoString screen that identified FASTKD4 (TBRG4) as a protein that interacts with all mitochondrial RNAs and modulates the half-lives of multiple transcripts (21). More recently, Antonicka et al. reported that FASTKD5 upregulates several mitochondrial RNAs and leads to accumulation of precursor RNAs that lack tRNA at both ends (22). One of the characteristics of some members of the FASTK family is their submitochondrial localization within distinct RNA granules (MRGs) (9–10,13,22). In addition to FASTK proteins, the MRGs contain RNase P (9), DDX28 (23), GRSF1 (24–25) as well as other RNA binding proteins (9,26), and are likely to represent the sites in mitochondria where RNA processing and assembly of the small and large ribosomal subunits occurs (26). PTCD2, apparently required for the processing of the tRNA-less ND5–CYB junction, also localizes in the MRGs (manuscript in preparation), suggesting that the RNA processing in these MRGs is not restricted to the removal of tRNAs by RNase P. Finally, these complexes may have many similarities to the MIOREX complexes recently described in yeast and which correspond to ribosome-containing assemblies (27).
Here, we report the first characterization of the effect of FASTKD1 knock-out on mitochondrial RNA content and novel aspects of FASTKD4 mitochondrial regulation. We generated several chimeras between different FASTK family members to explore the function of their domains and provide the first test of the hypothesis that the RAP domain of FASTKD4 possesses nucleolytic activity.
MATERIALS AND METHODS
Cell culture and transfection
Cells were cultured in DMEM, 10% fetal bovine serum with 100 μg/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-Glutamine, supplied by GE Healthcare. Media for Rho0 cells was additionally supplemented with 110 μg/ml pyruvate and 50 μg/ml uridine (Sigma). Transfections were performed using calcium phosphate or lipofectamine (Invitrogen).
Immunofluorescence and microscopy
Immunofluorescence and microscopy were performed as described by Jourdain (9). Briefly, cells were fixed in 4% paraformaldehyde and immunostaining with antibodies to HA (Covance), FLAG (Sigma), FASTKD2 (Proteintech), or FASTKD4 (Santa Cruz Biotechnology) was performed in PBS containing 0.1% Triton X-100 and 3% (w/v) BSA (Sigma-Aldrich). Imaging was performed using a Zeiss LSM700 confocal microscope or Zeiss Axiophot microscope. Antibodies are listed in Supplementary Table S1.
RNA extraction and northern blotting
RNA extraction and northern blot analyses were performed as described by Jourdain (9). Briefly, total RNA was extracted with Tri-Reagent (Sigma-Aldrich) and 5–15 μg RNA were separated on a denaturing formaldehyde agarose gel and transferred via electrophoresis to a Nylon membrane (GE Healthcare). Strand-specific 32P -UTP labeled riboprobes (Supplementary Table S1) were transcribed using T7 polymerase (Bio-Rad), and hybridization was performed at 60°C in 50% formamide, 7% SDS, 0.2M NaCl, 80 mM sodium phosphate (pH 7.4), and 100 mg/ml salmon sperm DNA. Imaging was done with a phosphorimager (BioRad).
Cloning and viral production
For protein expression, cDNAs were cloned into pWPT (Addgene) in frame with a C-terminal HA-TEV-6HIS tag or 3× FLAG tag. Chimeric proteins were cloned using the Gibson Assembly system (28). Lentiviruses for protein expression were produced in HEK293T cells by co-transfecting the constructs of interest cloned into the pWPT vector with the viral plasmids psPAX2 and pMD2G (Addgene). After 2 days, the supernatant was collected, filtered through a 0.45 μm pore size filter, and used to infect 143B or HEK293T cells.
Reconstruction of full length FASTKD1
The existing clone of FastKD1 obtained from the group of Maria Simarro 12 harbored a 69 bp deletion resulting in loss of the 23 amino acid sequence AGEAHDPLVEALVTEAWRRLERF. The full length cDNA was reconstructed by oligonucleotide directed mutagenensis as follows: two PCR products were generated from the existing clone using the primer pairs KMU193-KMU201 (upstream fragment) and KMU194-KMU202 (downstream fragment), and the two products were gel purified and fused using the two external PCR primers KMU193 (carrying a 5΄ MluI site) and KMU194 (carrying a 3΄ PstI site). The resulting full length cDNA was digested with MluI and PstI and cloned into the same sites of the vector pWPT previously engineered to encode a downstream TEV protease cleavage site, followed by FLAG and 6xHis tags. The correct sequence of the resulting construct was verified by DNA sequencing.
CRISPR/Cas9 gene disruptions
CRISPR constructs were made by subcloning 20 nt guide RNAs into the pX330 CRISPR/Cas9 plasmid (Addgene). The ends of the selected guide sequences and the PAM motif formed a BstNI cut site. Guide sequences are listed in Supplementary Table S1. HEK293T or 143B cells were then transfected with the CRISPR construct using lipofectamine, and single cells were plated using a Beckman Coulter MoFlo Astrios cell sorter. After expanding cell populations, an approximately 200 bp region containing the predicted cut site was amplified and digested with BstNI to assay for a deletion in this region. This ∼200 bp region was then sequenced to confirm disruption of the gene. FASTKD4 gene disruption candidates were further confirmed by protein immunoblotting. For the double gene disruptions, FASTKD1-KO cells were transfected with the FASTKD4 CRISPR and screened for loss of FASTKD4 by protein immunoblot (Supplementary Figure S1).
Transcriptional blockage experiments
Transcription of mt-RNA was blocked as in Jourdain et al. (9). Briefly to block transcription, cells were treated with 1μg/ml ethidium bromide for the indicated time before RNA extraction. The resulting RNA was then blotted and probed as described above.
35S labeling experiments
Cells in culture were washed two times with PBS. They were then incubated for 30 min in media consisting of: DMEM lacking cysteine, methionine, and glutamine; supplemented with 10% FBS, Glutamax (Gibco) 1×, and 110 mg/l sodium pyruvate. Emetine was then added to a concentration of 1 μM, and after 5 min 200 μCi of 35S labeled cysteine and methionine (Easy Tag protein labeling mix, Perkin Elmer) was added. One hour later, cells were thrice washed with PBS, collected and resuspended. Equal amounts of protein were loaded and run on a 12–20% gradient gel. The gel was then stained with Coomassie blue, destained, dried and imaged using a phosphorimager.
Structural modeling and sequence alignment of RAP domains
Blast searching using yeast FASTK or FASTKD4 proteins allowed the identification of human, mouse, bovine, fish, fly and worm protein sequences in the FASTK family. All sequences were aligned using webserver and the alignment was manually edited to optimize loop positioning based on secondary structure prediction. FASTK residues 405–539 and FASTKD4 residues 493–621 corresponding to their RAP domain were submitted to the webserver Phyre to obtain a structural prediction (29). Confidence scores are 97.60% and 97.73% for FASTK and FASTKD4 sequences respectively. These predicted models were then manually aligned on the closest atomic model (homing endonuclease, pdb code 3r3p) and on the related Vsr endonuclease atomic structure (pdb code 1vsr).
RESULTS
FASTKD1 and FASTKD4 are mitochondrial proteins, with FASTKD1 enriched in MRGs
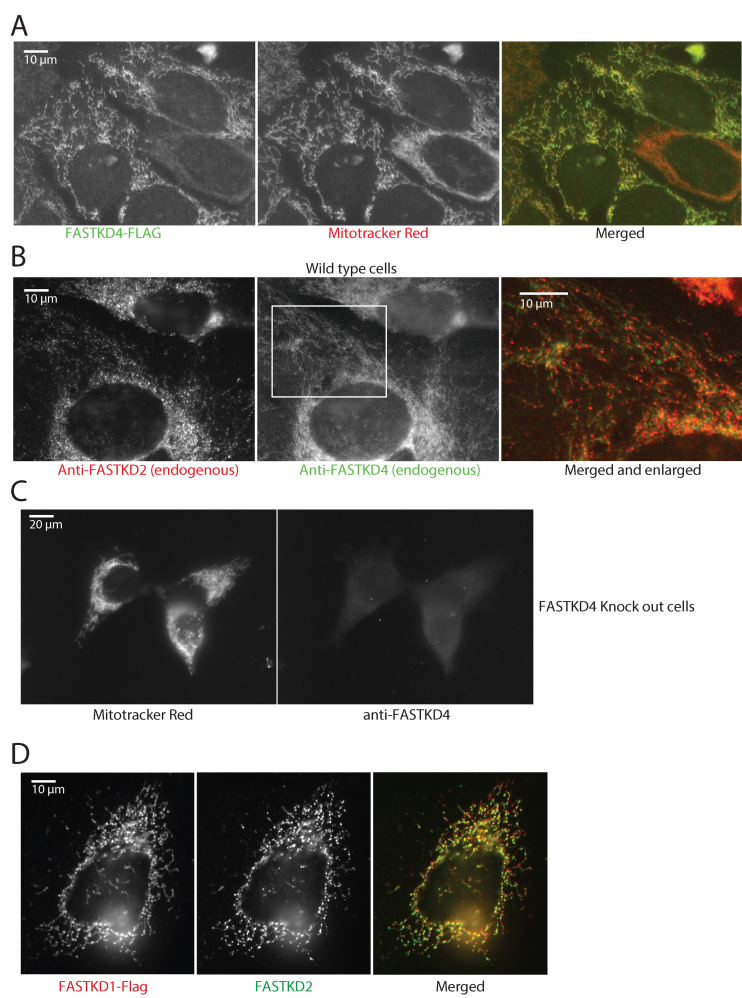
In previous studies, Wolf & Mootha have shown that FASTKD4 is found in the matrix of mitochondria, and is loosely associated with the inner mitochondrial membrane since it could be solubilized by alkali treatment of the mitochondrial membranes (21). Localization studies have reported that FASTK, FASTKD2 and FASTKD5 are enriched in MRGs (9,22,24). To investigate more precisely the subcellular localization of FASTKD1 and FASTKD4 and assess proper expression of our tagged constructs, we have performed immunofluorescence studies using antibodies against endogenous FASTKD4 or against the HA or FLAG tagged FASTKD1 or FASTKD4. Here, we confirm that FASTKD4 is a mitochondrial protein diffusely distributed throughout the mitochondrial matrix and displaying no preferential co-localization with MRGs, which we identified using an anti-FASTKD2 antibody (Figure 1A–C). This lack of foci was present even when observing the endogenous FASTKD4, ruling out the idea that FASTKD4 foci were simply obscured by overexpression of the protein. This is thus the first report of a FASTK family member, expressed at endogenous levels, which is not concentrated in MRGs. Previously, FASTKD1΄s localization was observed using a construct based on a predicted sequence which differs from the observed sequence by the presence of 23 amino acids near the N-terminus (9,12). We therefore corrected this sequence error and found that with the correct sequence near the N-terminus, FASTKD1 was preferentially localized to MRGs (Figure 1D).
Figure 1.
Submitochondrial localization of FASTKD1 and FASTKD4. (A) FASTKD4 has a mitochondrial distribution, but is not concentrated in foci. 143B cells expressing Flag-tagged FASTKD4 were immunostained for FASTKD4 using an anti-Flag antibody (left) and with Mitotracker Red to reveal mitochondria (center). Right: merged image. Flag staining is in green, and Mitotracker Red is in red. (B) FASTKD4 shows a diffuse mitochondrial staining whereas FASTKD2 is present in discrete foci. Immunostaining of endogenous FASTKD2 (left) and endogenous FASTKD4 (center) in 143B cells using anti-FASTKD2 and FASTKD4 antibodies respectively. In the right panel, the images are merged (FASTKD4 in green and FASTKD2 in red) and enlarged (the enlarged area is indicated with a white rectangle in the center panel). (C) Specificity of the anti-FASTKD4 antibody. FASTKD4-KO cells were labeled with Mitotracker Red (left) and anti-FASTKD4 antibody (right). No specific FASTKD4 signal is visible in the FASTKD4-KO cells. (D) FASTKD1 localizes to MRGs. 143B cells were transfected with pCi FASTKD1-FLAG. Cells were then immunostained with antibodies against endogenous FASTKD2 to label MRGs (middle panels) or FLAG to label FASTKD1 (left panels). The merge (right panel) shows that FASTKD1 is in MRGs.
Disruption of FASTKD1 and FASTKD4 genes differentially affect mitochondrial gene expression
Based on our previous studies showing that FASTK is required for expression of the ND6 mRNA (9), we hypothesized that other FASTK proteins may also regulate expression of specific mitochondrial RNAs. To test this, we used the CRISPR/Cas9 system to disrupt the FASTKD1 or FASTKD4 genes in both HEK293T and 143B cells. We obtained clones containing frameshift mutations (hereafter FASTKD1-KO or FASTKD4-KO) in both genes at the predicted cut sites (Supplementary Figure S1). To control for clonal or off target effects in the FASTKD1-KO or FASTKD4-KO cells, we exogenously expressed FASTKD1 or FASTKD4-FLAG, and compared the resulting RNA phenotypes by Northern blotting (Figure 2).
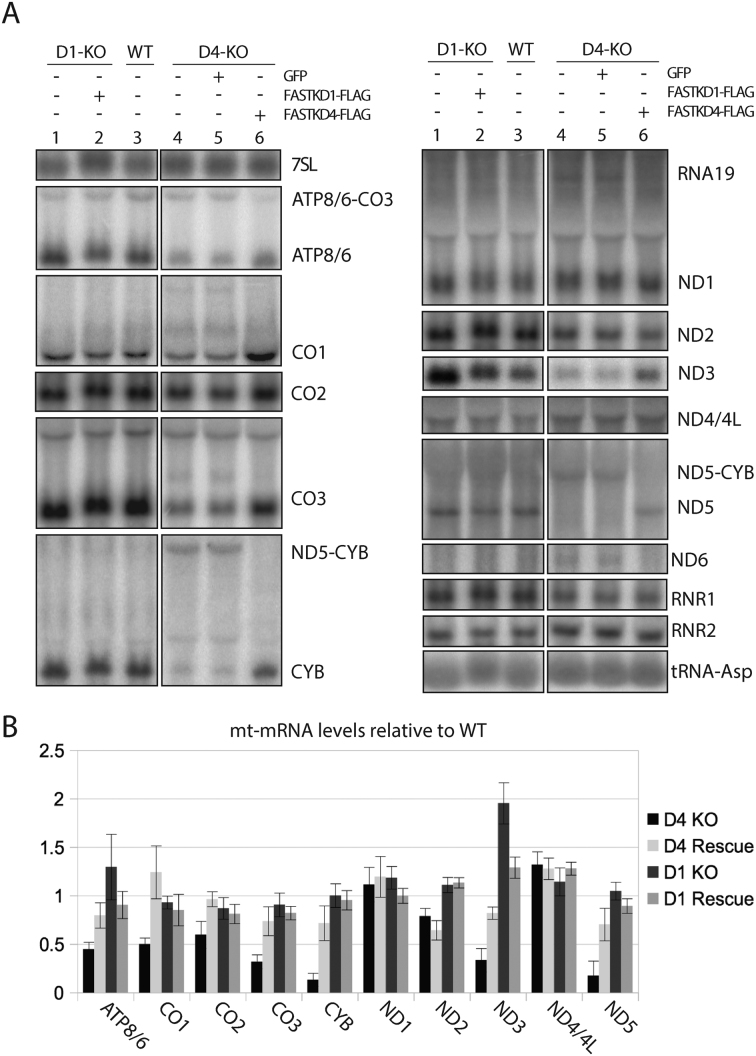
Figure 2.
FASTKD1 and FASTKD4 affect mitochondrial mRNA levels. (A) Northern blot analysis of mitochondrial RNAs. Gene disruption of FASTKD4 (D4-KO) reduces levels of multiple mitochondrial mRNAs and increases levels of multiple precursors (lane 4) relative to wild type cells (lane 3). In contrast, gene disruption of FASTKD1 (D1-KO, lane 1) leads to the over accumulation of ND3, and a slight over accumulation of ATP8/6 and ND1. Expression of Flag-tagged FASTKD4 (lane 6) or FASTKD1 (lane 2) in gene disrupted cells reversed this phenotype and restored mRNA levels, whereas expression of GFP did not (lane 5). All lanes are cropped from the same gel. (B) Quantification of RNAs levels in FASTKD1 and FASTD4 KO and rescue cells. RNA signal in the indicated cells is shown as a percent of the RNA signal in the wild-type cell line. Error bars indicate one standard deviation. 3 replicates were quantified for each condition.
As seen in Figure 2A, in FASTKD4-KO cells we observed statistically significant decreased levels of ATP8/6, CO1, CO2, CO3, CYB, ND3 and ND5 relative to the rescued and wild type cells. Interestingly, all these genes have atypical junctions except for CO2 and ND3, which are flanked at both ends by tRNAs. The results are mostly consistent with the previous observations of Wolf and Mootha (21). Interestingly, that study reported conflicting results for ND2 depending on the measurement method used. Our northern blot data agrees with the MitoString data reported by these authors (21) and does not conform to their contrasting qPCR results. Importantly, the analysis of RNA by Northern blot allowed us to detect a substantial accumulation of ND5–CYB precursor RNA, which was accompanied by a decrease in the mature forms of ND5 and CYB (Figure 2A). This key result was not detected using MitoString or qPCR in the Wolf and Mootha paper. Restoration of FASTKD4 expression in FASTD4-KO cells efficiently decreased accumulation of the ND5–CYB precursor and increased expression of mature forms of ATP8/6, CO1, CO2, CO3, ND3, ND5 and CYB (Figure 2A and B). We did not detect any other significant effect on other precursors that could be attributed to the presence or absence of FASTKD4. These results suggest that FASTKD4 could be a general factor involved in the processing or stabilization of mRNAs which lack flanking tRNAs at both ends.
Interestingly, when we analyzed the RNA from clones of the FASTKD1-KO cells, we saw a strong statistically significant increase in the level of ND3 mRNA compared to WT cells (Figure 2A), which was reversed by expression of FASTKD1-FLAG (Figure 2A). This is thus the second report about the loss of a FASTK protein leading to an increase in the steady state level of a processed mt-mRNA. Interestingly this also implies that FASTKD1 is not involved in the maturation of the mt-mRNAs, as previously concluded for FASTK and FASTKD5. It rather points towards a putative role of FASTKD1 in RNA stability-related processes.
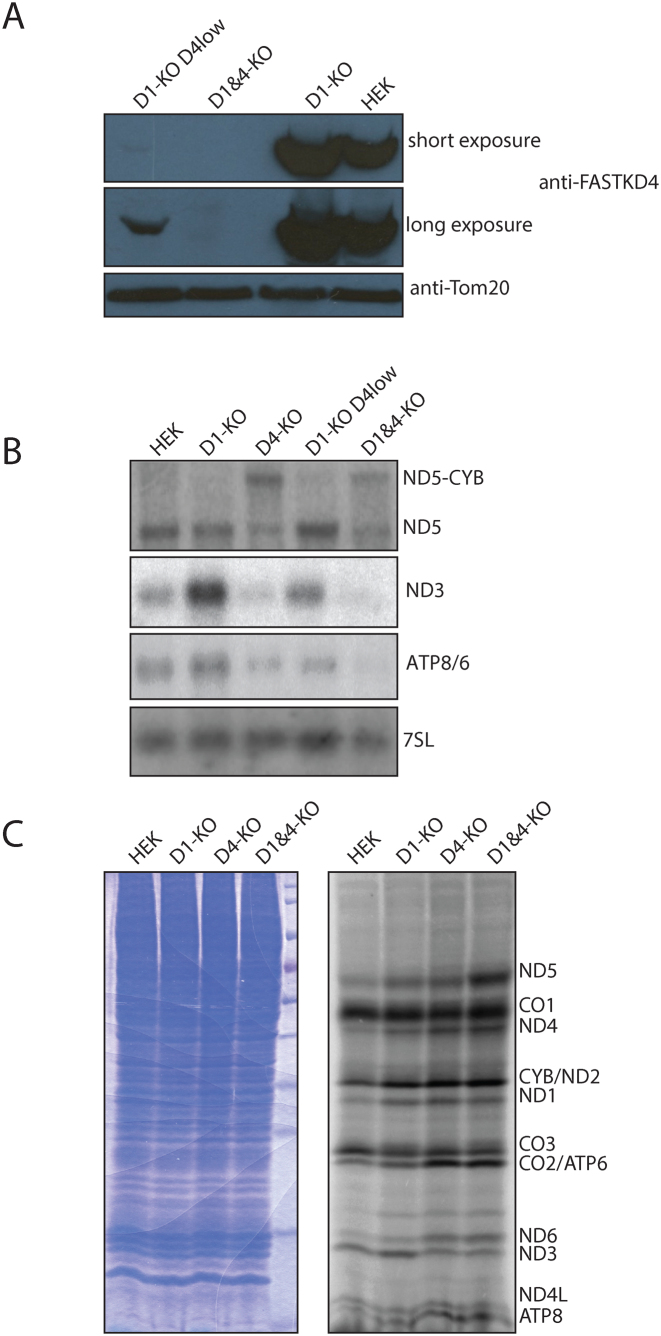
As described above, the ND3 mRNAs was affected differentially by FASTKD1 or FASTKD4 removal. To test whether loss of the FASTKD1 protein could rescue loss of FASTKD4, we used the CRISPR/Cas9 system to disrupt the FASTKD4 gene in FASTKD1-KO cells. We obtained a clone lacking FASTKD1 and FASTKD4 (hereafter D1&D4-KO) and assessed the functional consequences on mitochondrial RNA expression (Figure 3A). The D1&D4-KO cells showed a pattern of RNA expression that was nearly indistinguishable from the FASTKD4-KO cell line with accumulation of ND5–CYB precursor and strong reductions in mature ND3, ND5, and ATP8/6 mRNAs (Figure 3B). We conclude that FASTKD4 loss is epistatic to FASTKD1 loss.
Figure 3.
FASTKD1-FASTKD4 double gene disruption. (A) Western blot with anti-FASTKD4 (top panels) or anti-TOM20 (bottom panel) antibodies on purified mitochondria. Loading left to right: Incomplete knockout of FASTKD4 in a FASTKD1-KO cell line (D1-KO D4low), FASTKD1 and FASTKD4 gene disrupted cells (D1&D4-KO), the progenitor FASTKD1 gene disrupted cells (D1-KO), and wild type HEK293T cells (HEK). (B) Northern blot using antisense ND3, ATP8/6, ND5, and 7SL probes. Equal amounts of total RNA were loaded. Labels and colonies are the same as in Figure 3A, with the addition of RNA from a FASTKD4-KO clone (D4-KO). Note that loss of FASTKD4 is epistatic to the loss of FASTKD1, and small amounts of FASTKD4 are sufficient to prevent the RNA phenotype that results from FASTKD4 loss. (C) 35S labeled mitochondrial translation products. Cytoplasmic translation was blocked with Emetine, and mitochondrial protein was labeled with radioactive cysteine and methionine. Equal amounts of total cellular protein, as assayed by Bradford, were loaded. Left image: Coomassie staining as a loading control. Right image: 35S radiograph. Lanes left to right: HEK293T (HEK), FASTKD1 knockout cells (D1-KO), FASTKD4 knockout cells (D4-KO) and double FASTKD1 and FASTKD4 knockout cells (D1&40KO). Putative identities for the bands are shown on the basis of size. Note that translation is significantly altered, but that these alterations do not correlate with the changes in mRNA abundance, with the exception of ND3.
Mitochondrial translation in FASTKD1 and FASTKD4 KO cells
We wanted to know what effect these changes to mitochondrial RNA had on mitochondrial protein synthesis, and whether the observed loss of mRNA would also lead to a decrease in the abundance of the respective protein. We thus performed in vivo 35S cysteine and methionine labeling of mitochondrial encoded proteins. As expected from the northern blot analysis, the ND3 protein synthesis levels were decreased in the FASTKD4-KO and FASTKD1D4-KO cell lines (Figure 3C). On the other hand, the observed reductions in mature levels of CYB, ND5, ATP6/8, CO1, and CO3 mRNA did not appear to result in noticeably reduced synthesis of their translated product. Indeed in the case of ND5, the protein level was slightly increased in both the FASTKD1-KO and FASTKD4-KO cell lines, and even further increased in the double gene disruption FASTKD1D4-KO cell line. Likewise expression of the ND6 protein appears to have been increased in the cells in which FASTKD4 was disrupted despite no statistically significant alteration in ND6 RNA levels. In contrast to the results reported by Wolf and Mootha, who observed decreased steady state levels of CO2, we did not observe a decrease in the level of de-novo synthesis of CO2. Synthesis of CO2 is hard to distinguish from the synthesis of ATP6 by 35S labeling due to their similar sizes, however it should be noted that both CO2 and ATP6 mRNA levels are reduced in FASTKD4-KO cells. Since the intensity of the 35S band corresponding to the size of ATP 6 and CO2 did not decrease, it is not possible that synthesis rates of both CO2 and ATP6 were decreased in FASTKD4-KO cells, and thus it cannot be generalized that the reduced mRNA levels lead to reduced protein synthesis rate. On the basis of these results, we must consider the possibility that FASTK proteins may modulate translation of RNA and not simply the amount of RNA, or that the protein synthesis rate changes are indirect effects of a compensatory mechanism. Despite these differences in mitochondrial protein synthesis, we did not observe any defect in mitochondrial respiration on the basis of oxygen consumption measurements.
Specific function of FASTK family protein domains
While generating D1&D4-KO cells, a clone was identified in which the FASTKD4 protein signal was still detectable by western blot, but at dramatically reduced levels relative to wild type cells (Figure 3A, clone named D1-KO D4low). In these cells there was no obvious reduction in mature ND5 levels, yet a small amount of ND5–CYB precursor accumulated (Figure 3B). This result shows that the presence of FASTKD4, orders of magnitude below the normal protein level, is sufficient to allow normal processing of ND5–CYB precursor and prevents a loss of mature forms of ND5 and CYB mRNAs. The large effect due to a small amount of protein may suggest an enzymatic model over a stoichiometric model, although we do not know the molar amounts of FASTKD4 protein relative to the RNA to which it binds. In support of this hypothesis is the structural modeling described below.
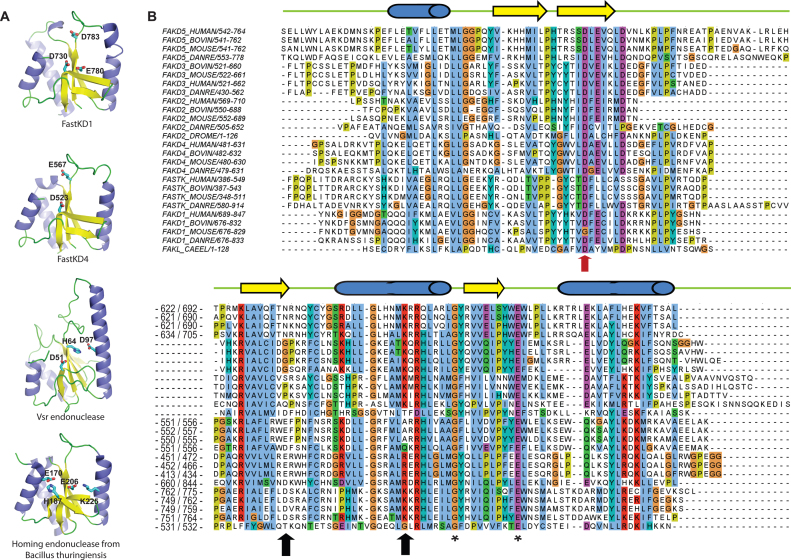
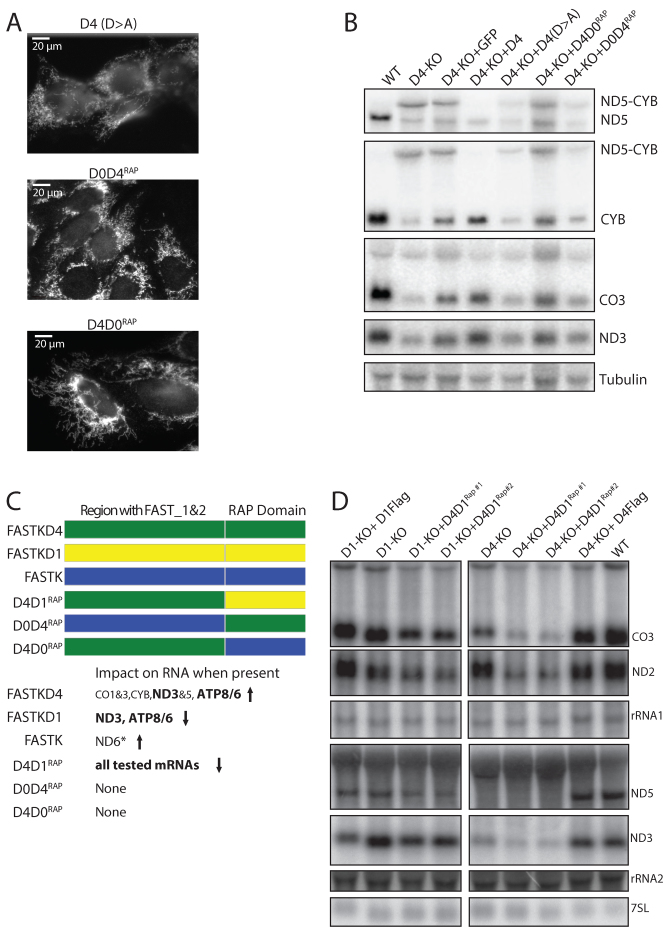
Structural modeling suggests that the RAP domain present in the FASTK protein family can adopt a PD-(D/E)-XK nuclease superfamily fold (14) (Figure 4A). Using an updated database we found that the RAP domains of FASTKD1 and FASTKD4 are best modeled on the structure of the bacterial VSR endonuclease, a verified endonuclease rather than on a putative endonuclease as previously proposed by Castello et al. (11). Interestingly, the VSR endonuclease was also identified as a close match for the RAP domain of related chloroplast proteins (20). Previous studies showed that VSR enzymatic activity can be abolished by a single D to A mutation in its active site (30). We note that the homologous Aspartate (D531 in FASTKD4) appears to be highly conserved in each RAP domain of the FASTK proteins (Figure 4B), including in a distant homologue in C. elegans. Therefore if the RAP domain functions similarly to the VSR endonuclease, mutation of this residue should render the protein non-functional. To perform an initial test of this nuclease hypothesis, we expressed FASTKD4 with a D531A mutation in FASTKD4-KO cells. This mutant protein still localized to mitochondria (Figure 5A) and was recognized by the anti-FASTKD4 antibody indicating that the mutant protein was expressed and likely correctly folded. However, expression of the mutant protein failed to rescue the level of mature ND3, CO3, CYB or ND5 mRNA and did not abolish the accumulation of the ND5–CYB precursor RNA or alter the ratio of mature ND5 to ND5–CYB precursor (Figure 5B). We have also previously mutated this residue in FASTKD2 and observed that the protein still formed foci (unpublished data), suggesting this D>A mutation does not interfere with correct folding. Thus, the residue D531 of the RAP domain is essential for the function of FASTKD4, possibly due to its role in the nucleolytic activity of the RAP domain.
Figure 4.
The RAP domain of FASTK family members shows structural similarities with PD-(D/E)XK nucleases. (A) Structural modeling of the human FASTKD1 and FASTKD4 proteins, compared to known PD-(D/E)-XK nucleases. Models and structures are shown as a schematic colored by secondary structure elements (α-helices blue, β-strands yellow, and loops green). Key residues suspected to be involved in the enzymatic activity are shown as sticks and colored by atom type (carbon cyan, oxygen red, nitrogen blue). (B) Alignment of protein sequences of FASTK proteins from metazoans (mammals, fish, flies, nematodes). Numbers in the alignment indicate the starting and last residues shown in the alignment as well as residues removed because of poor conservation. Predicted secondary structures are indicated on the top of the alignment. Three arrows point to the three residues responsible for the endonuclease activity in the PD-(D/E)-XK phosphodiesterase superfamily (14,29). The red colored arrow indicates the Aspartate mutated in subsequent experiments (note: FAKL_CAEL is the C. elegans protein B0564.7).
Figure 5.
Structure-function of FASTK family members. (A) Anti-Flag immunofluorescence showing expression of the mutant (D>A) and chimeric proteins (D0D4RAP and D4D0RAP) in the cells of Figure 5B had the expected mitochondrial localization. Note the punctuate staining visible in D0D4RAP expressing cells. (B) Chimeric FASTK/FASTKD4 proteins (D0D4RAP and D4D0RAP), and FASTKD4 with a D531→A531 mutation (D4 D>A) do not rescue FASTKD4 loss (D4-KO) while expression of wild type FASTKD4 does (D4). Total cellular RNA from WT cells and KO cells expressing GFP were included as controls. Northern blot with 32P labeled RNA probes antisense to ND5, CYB, CO3, ND3 and Tubulin were used. All lanes were cropped from the same gel. (C) Schematic showing the different chimeras that were generated. To the right is a summary of the effects on RNAs when the protein is expressed relative to when it is not expressed. An arrow pointing up indicates RNA levels are higher in the presence of the protein, while an arrow pointing down implies the opposite. * For data supporting the effect on ND6, see Jourdain et al. (9) (D) Expression of a chimera of FASTKD1 and FASTKD4 (D4D1RAP#1 and D4D1RAP#2) partially rescues the ND3 phenotype of FASTKD1 loss. Northern blot with 32P labeled antisense CO3, ND2, ND3, ND5, rRNA1, rRNA2 and 7SL RNA probes. Equal amounts of total RNA from the indicated cells were loaded for analysis. This reduction in ND3 levels was accompanied by reduction in other mRNAs showing the effect is not specific to ND3.
Partial analysis of the structure–function of FASTK proteins
We have shown previously that deletion of the RAP domain in FASTK impaired ND6 mRNA expression and localization to MRGs (9). To further investigate the role of the RAP domain, we decided to swap the RAP domain between FASTK proteins to test their importance for the overall function of the proteins. Therefore, we constructed chimeras of FASTK (D0) and FASTKD4 (which we termed D0D4RAP and D4D0RAP), as well as FASTKD1 and FASTKD4 (D4D1RAP) (Figure 5C).
We found that expression of neither the D0D4RAP nor D4D0RAP chimeras could prevent accumulation of the ND5–CYB precursor RNA or rescue the level of mature CO3, CYB, ND3 or ND5 mRNAs in FASTKD4-KO cells (Figure 5B). Similarly, neither D0D4RAP nor D4D0RAP could rescue the loss of ND6 mRNA in FASTK-KO cells (Supplementary Figure S2A). Interestingly, D0D4RAP, which did not prevent ND5–CYB precursor RNA accumulation, did localize in the MRGs, as expected for the wild type protein FASTK, while the other chimera, D4D0RAP, did not (Figure 5A). This indicates that the N-terminal region is extremely important for localization of FASTK proteins to foci, and is further supported by our observation that FASTKD1 with the correct N-terminal region also localizes to foci.
While we cannot be sure that the entire chimeras are all correctly folded, we observed that the D4D0RAP chimeras were recognized by the anti-FASTKD4 antibody when assayed by immunofluorescence suggesting that at least the FASTKD4 N-terminal domain of these chimeras is properly folded. Furthermore the localization of D0D4RAP to MRGs suggests that there was at least proper folding of the domains required for MRG localization.
FASTKD1 and FASTKD4 both affect the ND3 mRNA, and we found that D4D1RAP expression in FASTKD1-KO cells led to a significant reduction of ND3 level. Interestingly, this effect was not limited to ND3 but extended to all mitochondrial mRNAs, whereas rRNA and tRNA appeared unaffected (Figure 5D). These results suggest that this particular chimera is acting on additional mt-RNAs. FASTKD4 has been previously reported to interact with all the mt-mRNAs (21) and we suggest that the N-terminal domain of FASTKD4 in this chimera retargets the FASTKD1 RAP domain to additional RNAs. The RAP domain of FASTKD1 would be involved in the downregulation of mitochondrial RNAs, although with poor substrate specificity. Together these results reinforce the idea that the RAP domain is the functional domain whereas the region upstream of the RAP domain is responsible for subcellular localization and target specificity. These experiments highlight two novel functional aspects of the FASTK protein family: (i) the N-terminal region of the FASTK family members, which is expected to fold as a PPR-like domain formed by repetition of pairs of helices, apparently plays a role in targeting each FASTK protein to appropriate RNA(s); (ii) the RAP domain is likely to carry an enzymatic activity possibly with a defined specificity for each FASTK family member as the RAP domains are not interchangeable.
DISCUSSION
The goal of this study was to investigate the function of two members of the FASTK family, FASTKD1 and FASTKD4. We report that both proteins affect mitochondrial gene expression through regulation of specific RNA transcript levels.
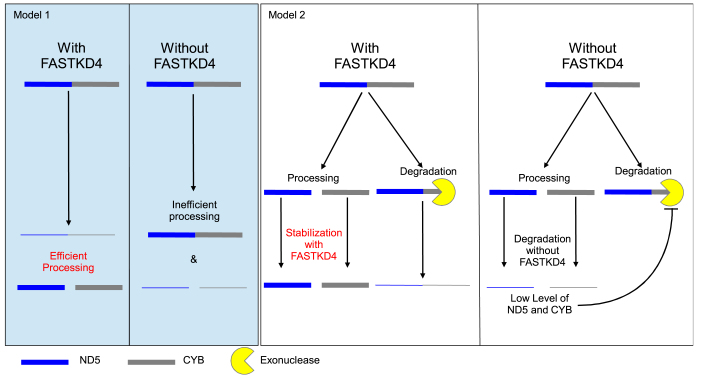
FASTKD4 is required to promote expression of the ND5, CYB, CO1, CO3 and ATP8/6 genes, which are not flanked by tRNAs on both ends, and of CO2 and ND3 genes, which are flanked by tRNAs on their 5΄ and 3΄ ends. In the absence of FASTKD4, the amount of these mRNAs is significantly decreased. One of the most striking phenotypes we observed in the FASTKD4-KO cells is an accumulation of the ND5–CYB precursor, which accompanied the drop in mature ND5 and CYB mRNAs. Two non-mutually exclusive hypotheses can be proposed to explain these results (see models 1 and 2, Figure 6).
Figure 6.
Possible (non-mutually exclusive) models to explain the action of FASTKD4. Model 1: FASTKD4 promotes processing of ND5–CYB precursor. In the absence of FASTKD4, the precursor remains mostly uncleaved, thus ND5 and CYB are made in low amounts. Model 2: FASTKD4 stabilizes multiple mature RNAs including ND5 and CYB. In the absence of FASTKD4, ND5, CYB (as well as ND3, CO1-3 and ATP8/6) are degraded much faster. In response to low levels of ND5 and/or CYB, degradation of the ND5–CYB precursor degradation is inhibited to favor production of ND5 and CYB.
In the first model (i) FASTKD4 is required for efficient processing of mRNAs with atypical junctions, including ND5–CYB precursor RNA (model 1, see Figure 6) while in the second model (ii) FASTKD4 is required to stabilize mature ATP8/6, CO1-3, CYB, ND3 and ND5 (model 2, see Figure 6).
In support of the processing model (Model 1), we found that all heavy strand mRNAs with atypical junctions, such as ND5 and CYB, were affected by the loss of FASTKD4, and interestingly, similar results have recently been reported for FASTKD5 (22). The authors of that study concluded similarly that loss of FASTKD5 causes a defect in the processing of atypical junctions. This model is further supported by previous experiments showing that in Chlamydomonas reinhardtii, the RAP domain-containing protein Raa3, together with the FAST domain-containing protein Raa1, plays a role in RNA trans-splicing (17). The processing of ND5–CYB into transcripts containing only ND5 or CYB would either require an internal cleavage (endonuclease activity) or alternatively both a 5΄-to-3΄ and 3΄-to-5΄ exonuclease activity to degrade ND5–CYB from either end. However, there is no known mitochondrial protein with 5΄ to 3΄ exoribonuclease activity, making the latter hypothesis unlikely. Instead, the apparent lack of cleavage of ND5–CYB, when FASTKD4 is mutated at a residue that is essential for endonuclease activity in a structurally similar protein, suggests that FASTKD4 may be carrying out this endonuclease activity. However, model 1 does not explain why in addition to mRNAs with atypical junctions, the level of the ND3 mRNA, which is flanked by tRNAs on both ends, is also significantly decreased or why ATP8/6 and CO3 precursor RNAs do not accumulate in FASTKD4-KO cells. Lastly it does not explain the combination of a high level of ND5–CYB precursor despite a high decay rate (Supplementary Figure S2C). One alternative hypothesis corresponding to Model 2 is that the phenotype displayed by FASTKD4-KO cells is due to increased degradation of the affected mature mRNAs. We observed that in these cells, the ND5–CYB precursor RNA accumulates to a level similar to that of mature ND5 mRNA in wild type cells (Figure 2B), despite being much less stable (Supplementary Figure S2C). If ND5–CYB was degraded at a faster rate than mature ND5, it would need to be produced at a higher rate than mature ND5 to accumulate to similar levels. From our observations, it appears that ND5–CYB and mature ND5 accumulate to similar steady state levels, despite a significantly higher decay rate for ND5–CYB. Therefore, we conjecture that production of ND5–CYB must exceed production of ND5. It is unlikely that the transcription of ND5–CYB is specifically increased, given the polycistronic nature of mitochondrial transcription and the lack of any global mt-mRNA upregulation. Instead, it is likely that normally the precursor is produced in excess of the amount needed for processing into mature ND5 or CYB, and it is possible that it is the rate of mature ND5 production, by processing of ND5–CYB precursor, that is modulated.
There are two possible fates for the ND5–CYB precursor: processing or degradation. If we assume that the processing of the ND5–CYB precursor transcript occurs normally in FASTKD4-KO cells, then its accumulation may be explained by decreased degradation (Model 2). Such a mechanism would serve as a feedback process to sense the amount of mature ND5 and CYB mRNAs. If their levels decrease, the degradation of the precursor is reduced to favor their production. Interestingly in the results of Antonicka et al. (22) the only instances of elevated precursor to mature mRNA ratios occurred when the mature mRNA was severely depleted, and thus model 2 may explain these results. Finally, Models 1 and 2 may be complementary and it is possible that FASTKD4 plays a role in both processing of precursor RNAs and stabilization of mature mRNAs.
Interestingly, unlike FASTKD4, which stabilizes ND3 mRNA, we found that FASTKD1 displays the opposite effect leading to ND3 degradation. The activity of FASTKD4 is likely epistatic to the activity of FASTKD1, since cells lacking both FASTKD1 and FASTKD4 display a phenotype similar to the phenotype of FASTKD4-KO cells. It is intriguing to note that these two proteins display a similar domain organization but ultimately produce opposite effects, at least on one particular mRNA.
To understand the mechanism by which these proteins act and produce different effects, we decided to construct chimeras of FASTK proteins. All FASTK family proteins are mitochondrial RNA binding proteins (11–12) which contain three poorly characterized domains: FAST_1, FAST_2 and RAP. To try and determine the contributions of these domains to the activity, we generated new combinations of these domains and observed the effect. We swapped the RAP domains between FASTKD1 and FASTKD4 to generate the D4D1RAP chimera and between FASTK and FASTKD4 to generate both the D0D4RAP and D4D0RAP chimeras. It was previously shown that the absence of FASTK in FASTK-KO cells leads to loss of ND6 mRNA, which can be rescued upon re-expression of the FASTK protein (9). Here, we show that neither the D0D4RAP nor the D4D0RAP chimeras were able to rescue ND6 mRNA levels in FASTK-KO cells. Similarly, the D4D0RAP and D0D4RAP chimeras were unable to rescue the ND5 and CYB mRNA phenotype in FASTKD4-KO cells. Collectively, these results indicate that the RAP domains of FASTK and FASTKD4 are not functionally equivalent. Interestingly, the RAP domain of FASTKD1 seems to carry out its function independently of its N-terminal region. Indeed, in FASTKD1-KO cells, which display elevated ND3 mRNA levels, expression of the D4D1RAP chimera leads to reduced ND3 mRNA levels suggesting that the RAP domain is a key determinant for the function of the chimeric protein. Interestingly, the chimera leads to decreased levels of not only ND3 mRNA, but also of all mitochondrial mRNAs tested. The results of Wolf and Mootha (21) suggest that FASTKD4 binds promiscuously to RNA, which may result in the D1 RAP domain of D4D1RAP being able to downregulate a much wider range of mt-RNAs. This result parallels the results found in plant chloroplasts, where it was also seen that a change to the OPR regions upstream of a RAP domain resulted in NCC1 and NCC2 recognizing and degrading different RNA targets (20).
We found that the RAP domains of FASTK family proteins display structural similarity with the PD-(D/E)-XK nuclease family (Figure 4A). This resemblance between RAP domains and these nucleases is apparently highly conserved, as RAP domains in plant chloroplast proteins also resemble PD-(D/E)-XK nucleases, specifically including the VSR endonucleases (20).
Proteins in the PD-(D/E)-XK family exhibit diverse functions, including endonuclease activity, exonuclease activity and other nucleic acid related activities such as attachment of eukaryotic 5΄ mRNA caps (14,31). Such an activity can explain many effects on mt-mRNA, just as PD-(D/E)-XK nucleases have a great variety of effects on nucleic acids. In brief, a nuclease may cleave at different positions on the transcript, with different bases required for cleavage. A nuclease activity could remove tags that would stabilize a mRNA or remove tags that mark it for degradation. Such a cleavage event may be linked to preparing an end for addition of a tag (31), or it may directly cleave a transcript triggering its decay.
Alignments, structural predictions and mutational analysis suggest that the RAP domains of FASTK proteins have an activity that is conferred by a PD-(D/E)-XK nuclease fold (Figure 4A). The active site of the PD-(D/E)-XK nuclease always contains a specific Aspartate residue which is critical for the enzymatic activity (30). We showed that FASTKD4 harboring a mutation of this conserved residue failed to rescue the FASTKD4-KO cells while the WT protein does complement the KO cells under the same experimental conditions (Figure 5B).
An enzymatic activity was also suggested by our observation that FASTKD4 can fulfill its function at very low, presumably sub-stoichiometric levels (Figure 3A and B). Taken together, these results are fully consistent with the conclusion that the RAP domain of FASTKD4 contains a functional PD-(D/E)-XK nuclease fold and that the function of this protein relies on a catalytic activity of the RAP domain.
Interestingly, mitochondrial translation seems relatively unaffected despite a large decrease in multiple mRNA levels in FASTKD4-KO cells and a large increase in ND3 mRNA level in FASTKD1-KO cells. It was previously reported by Larsson et al. (32) that there is a large excess of mitochondrial mRNA relative to the levels needed to maintain protein synthesis. In their study, these authors knocked out SLIRP and the result was a substantial reduction in mRNA levels without a corresponding decrease in protein translation rates. The reduction of mt-mRNA was apparently compensated by increased engagement of mRNAs with the 55S ribosomal subunits in addition to elevated levels of mt-ribosomes (32). A similar compensatory mechanism may be at work in our FASTKD4-KO cells, with the notable exception of the ND3 mRNA. Interestingly ND5 and CYB expression did not decrease despite a very severe reduction in their RNA levels. In this case, we must speculate that unprocessed ND5–CYB is able to be translated as other polycistronic mRNAs such as ATP8/6 or ND4/ND4L. Additional changes to de-novo translation may be occurring as part of a wider compensatory response to the altered mRNA levels in these KO cells due to a currently undetermined mechanism.
It is worth noting that Eberhard et al. previously reported that a FAST-like and RAP domain containing protein apparently modulated the translation of specific chloroplast mRNAs, although the mechanism of this modulation is unknown (19).
In conclusion, our results indicate that FASTKD1 and FASTKD4 regulate mitochondrial RNA expression, and in particular that they have opposite effects on the levels of ND3. Furthermore, a major determinant in the activity of these proteins lies in the C-terminal RAP domain. Strong reductions in the level of FASTKD4 can apparently compensate for the loss of FASTKD1 with regards to the steady state levels of ND3 RNA, although other mt-mRNAs are affected. However, a complete loss of FASTKD4 is epistatic over the loss of FASTKD1, and results in the loss of ND3. Similar to the results for FASTKD5, we show that FASTKD4 loss leads to elevated ND5–CYB precursor, and reductions in the mature forms of mRNAs with atypical junctions as well as ND3. On the basis of analyses using chimeric FASTK proteins, as well as structural modeling and mutagenesis of the RAP domain, we conclude that the N-terminal regions of the FASTK family proteins are responsible for RNA targeting, while the C-terminal RAP domain is essential for its activity and may contain a functional PD-(D/E)-XK nuclease fold.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all current and recently departed members of the JCM lab for their technical support, comments and assistance. We would additionally like to thank the lab of Maria Simarro for helpful correspondence. ST acknowledges the support of the Ligue genevoise contre le Cancer and the Worldwide Cancer Research.
Footnotes
Present address: Alexis A. Jourdain, Broad Institute of MIT & Harvard, 415 Main Street, Cambridge, MA 02142, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [310030B_160257/1 to J.-C.M.; 31003A_140924 and 31003A_124909 to S.T.]; IGE3; State of Geneva. Funding for open access charge: Swiss National Science Foundation [310030B_160257/1 to J.-C.M.; 31003A_140924 and 31003A_124909 to S.T.]
Conflict of interest statement. None declared.
REFERENCES
- 1. Spelbrink J.N. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. 2010; 62:19–32. [DOI] [PubMed] [Google Scholar]
- 2. Kukat C., Wurm C.A., Spåhr H., Falkenberg M., Larsson N.-G., Jakobs S.. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu. Rev. Biochem. 2011; 80:1033–1053. [DOI] [PubMed] [Google Scholar]
- 4. Ojala D., Montoya J., Attardi G.. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981; 290:470–474. [DOI] [PubMed] [Google Scholar]
- 5. Holzmann J., Frank P., Löffler E., Bennett K.L., Gerner C., Rossmanith W.. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008; 135:462–474. [DOI] [PubMed] [Google Scholar]
- 6. Rossmanith W. Localization of human RNase Z isoforms: dual nuclear/mitochondrial targeting of the ELAC2 gene product by alternative translation initiation. PLoS ONE. 2011; 6:e19152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanchez M.I.G.L., Mercer T.R., Davies S.M.K., Shearwood A.-M.J., Nygård K.K.A., Richman T.R., Mattick J.S., Rackham O., Filipovska A.. RNA processing in human mitochondria. Cell Cycle. 2011; 10:2904–2916. [DOI] [PubMed] [Google Scholar]
- 8. Xu F., Ackerley C., Maj M.C., Addis J.B.L., Levandovskiy V., Lee J., Mackay N., Cameron J.M., Robinson B.H.. Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem. J. 2008; 416:15–26. [DOI] [PubMed] [Google Scholar]
- 9. Jourdain A.A., Koppen M., Rodley C.D., Maundrell K., Gueguen N., Reynier P., Guaras A.M., Enriquez J.A., Anderson P., Simarro M. et al. . A mitochondria-specific isoform of FASTK is present in mitochondrial RNA granules and regulates gene expression and function. Cell Rep. 2015; 10:1110–1121. [DOI] [PubMed] [Google Scholar]
- 10. Popow J., Alleaume A.-M., Curk T., Schwarzl T., Sauer S., Hentze M.W.. FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA. 2015; 21:1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al. . Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 12. Simarro M., Gimenez-Cassina A., Kedersha N., Lazaro J.-B., Adelmant G.O., Marto J.A., Rhee K., Tisdale S., Danial N., Benarafa C. et al. . Fast kinase domain-containing protein 3 is a mitochondrial protein essential for cellular respiration. Biochem. Biophys. Res. Commun. 2010; 401:440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghezzi D., Saada A., D’Adamo P., Fernandez-Vizarra E., Gasparini P., Tiranti V., Elpeleg O., Zeviani M.. FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 2008; 83:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steczkiewicz K., Muszewska A., Knizewski L., Rychlewski L., Ginalski K.. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012; 40:7016–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kleinknecht L., Wang F., Stübe R., Philippar K., Nickelsen J., Bohne A.V.. RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell. 2014; 26:777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang F., Johnson X., Cavaiuolo M., Bohne A.V., Nickelsen J., Vallon O.. Two Chlamydomonas OPR proteins stabilize chloroplast mRNAs encoding small subunits of photosystem II and cytochrome b6f. Plant J. 2015; 82:861–873. [DOI] [PubMed] [Google Scholar]
- 17. Perron K., Goldschmidt-Clermont M., Rochaix J.-D.. A multiprotein complex involved in chloroplast group II intron splicing. RNA. 2004; 10:704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee I., Hong W.. RAP–a putative RNA-binding domain. Trends Biochem. 2004; 29:567–570. [DOI] [PubMed] [Google Scholar]
- 19. Eberhard S., Loiselay C., Drapier D., Bujaldon S., Girard-Bascou J., Kuras R., Choquet Y., Wollman F.-A.. Dual functions of the nucleus-encoded factor TDA1 in trapping and translation activation of atpA transcripts in Chlamydomonas reinhardtii chloroplasts. Plant J. Cell Mol. Biol. 2011; 67:1055–1066. [DOI] [PubMed] [Google Scholar]
- 20. Boulouis A., Drapier D., Razafimanantsoa H., Wostrikoff K., Tourasse N.J., Pascal K., Girard-Bascou J., Vallon O., Wollman F.A., Choquet Y.. Spontaneous dominant mutations in chlamydomonas highlight ongoing evolution by gene diversification. Plant Cell. 2015; 27:984–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wolf A.R., Mootha V.K.. Functional genomic analysis of human mitochondrial RNA processing. Cell Rep. 2014; 7:918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Antonicka H., Shoubridge E.A.. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015; 10:920–932. [DOI] [PubMed] [Google Scholar]
- 23. Tu Y.-T., Barrientos A.. The human mitochondrial DEAD-Box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep. 2015; 10:854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jourdain A.A., Koppen M., Wydro M., Rodley C.D., Lightowlers R.N., Chrzanowska-Lightowlers Z.M., Martinou J.-C.. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013; 17:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Antonicka H., Sasarman F., Nishimura T., Paupe V., Shoubridge E.A.. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013; 17:386–398. [DOI] [PubMed] [Google Scholar]
- 26. Silva D.D., Tu Y.-T., Amunts A., Fontanesi F., Barrientos A.. Mitochondrial ribosome assembly in health and disease. Cell Cycle. 2015; 14:2226–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kehrein K., Schilling R., Möller-Hergt B.V., Wurm C.A., Jakobs S., Lamkemeyer T., Langer T., Ott M.. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 2015; 10:843–853. [DOI] [PubMed] [Google Scholar]
- 28. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 29. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsutakawa S.E., Muto T., Kawate T., Jingami H., Kunishima N., Ariyoshi M., Kohda D., Nakagawa M., Morikawa K.. Crystallographic and functional studies of very short patch repair endonuclease. Mol. Cell. 1999; 3:621–628. [DOI] [PubMed] [Google Scholar]
- 31. Dias A., Bouvier D., Crépin T., McCarthy A.A., Hart D.J., Baudin F., Cusack S., Ruigrok R.W.. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009; 458:914–918. [DOI] [PubMed] [Google Scholar]
- 32. Lagouge M., Mourier A., Lee H.J., Spåhr H., Wai T., Kukat C., Silva Ramos E., Motori E., Busch J.D., Siira S. et al. . SLIRP regulates the rate of mitochondrial protein synthesis and protects LRPPRC from degradation. PLoS Genet. 2015; 11:e1005423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.