Abstract
Hypomorphic mutations in DNA-methyltransferase DNMT3B cause majority of the rare disorder Immunodeficiency, Centromere instability and Facial anomalies syndrome cases (ICF1). By unspecified mechanisms, mutant-DNMT3B interferes with lymphoid-specific pathways resulting in immune response defects. Interestingly, recent findings report that DNMT3B shapes intragenic CpG-methylation of highly-transcribed genes. However, how the DNMT3B-dependent epigenetic network modulates transcription and whether ICF1-specific mutations impair this process remains unknown. We performed a transcriptomic and epigenomic study in patient-derived B-cell lines to investigate the genome-scale effects of DNMT3B dysfunction. We highlighted that altered intragenic CpG-methylation impairs multiple aspects of transcriptional regulation, like alternative TSS usage, antisense transcription and exon splicing. These defects preferentially associate with changes of intragenic H3K4me3 and at lesser extent of H3K27me3 and H3K36me3. In addition, we highlighted a novel DNMT3B activity in modulating the self-regulatory circuit of sense-antisense pairs and the exon skipping during alternative splicing, through interacting with RNA molecules. Strikingly, altered transcription affects disease relevant genes, as for instance the memory-B cell marker CD27 and PTPRC genes, providing us with biological insights into the ICF1-syndrome pathogenesis. Our genome-scale approach sheds light on the mechanisms still poorly understood of the intragenic function of DNMT3B and DNA methylation in gene expression regulation.
INTRODUCTION
DNA methylation plays an important role in epigenetic signaling, having an impact on gene regulation, chromatin structure, development and disease. Generally, most mammalian genomes are largely methylated except at active or ‘poised’ promoters, enhancers and CpG islands, where it has a repressive effect. Nevertheless, gene body DNA methylation has been associated with high expression levels (1).
DNA methylation is established and maintained by the combined function of three active DNA methyltransferases DNMT3A, DNMT3B and DNMT1 (2). Although it has been largely studied, much remains unknown regarding how genomic DNA methylation patterns are determined in human cells, and which are the mechanisms that guide recruitment and activity of DNMTs in vivo (1). Mouse models suggest that although exhibiting overlapping functions, DNMT3A and DNMT3B have unique expression patterns and genomic targets during development (3–5). In line with this view, latest genome-wide studies showed that gene body DNA methylation patterns of highly expressed genes is dependent on DNMT3B activity (6–8). DNA methylation in transcribed regions might potentially silence alternative promoters, antisense transcripts, transcription factor binding sites, retrotransposon elements (LINEs, SINEs, LTRs and other retroviruses) and other functional elements to ensure the efficiency of transcription (9–11).
In addition, it is increasingly clear that DNA methylation plays a role in the processing of mRNAs during transcription modulating the elongation or splicing (12–14). Accordingly, DNA methylation level has been recently found associated with inclusion rate of alternative exons (15,16). Modulation of alternative splicing might represent a specific and evolutionary conserved activity of de novo DNA methyltransferases, as suggested by knockdown studies of DNA methyltransferase 3 (dnmt3) in honeybee (17). A kinetic model, in which epigenetic modifications affect the rate of transcriptional elongation, and/or a recruitment model, in which adaptor proteins bind to epigenetic modifications recruiting splicing factors, have been proposed (18). However, how exactly chromatin factors influence the DNMT3B activity and methylation profiles at transcribed regions remains to be elucidated.
In humans, hypomorphic DNMT3B mutations are sufficient to cause majority of the rare autosomal recessive disorder Immunodeficiency, Centromere instability and Facial anomalies (ICF) syndrome (MIM 242860) cases, reported as ICF type1 (19–21). Patients are characterized by DNA hypomethylation and decondensation of specific heterochromatic and euchromatic regions, and show alterations in tissue-specific gene silencing (22,23). ICF1-specific DNA methylation defects give rise to severe chromosomal rearrangements only in lymphocytes, probably acting in the onset of immunological phenotype. Defective steps of B-cell terminal differentiation might contribute to the agammaglobulinemia in ICF syndrome, given that ICF peripheral blood only contain naive B cells, while memory and gut plasma cells are absent (24). Most ICF1 patients carry missense mutations in or near the catalytic domain of DNMT3B (21). Nonsense mutations always occur as compound heterozygous, highlighting that the DNMT3B protein is essential for life, according to mouse models (3,5,25). Mutations severely perturb the DNA methylation profile at satellite 2 and 3 of juxtacentromeric heterochromatin and at telomeric/subtelomeric repeats, where it associates with chromosomal instability and abnormal shortening of telomeres, respectively (26–28). Treatment of human lymphocytes with the DNA methylation inhibitor 5-azacytidine (5-AzaC) induces ICF-like pericentromeric chromosomal abnormalities and missegregation of chromosomes 1, 16 and 9, suggesting that it is the DNA hypomethylation to trigger these anomalies (29,30).
Defects in lymphoid-specific pathways directly associated with the impaired DNMT3B activity have not been identified yet. The main reason is that alterations of DNA methylation pattern were previously pursued through candidate gene approaches based on promoter region analysis, which mostly failed to clarify the molecular pathogenesis of ICF syndrome (22,31,32). In this view, we carried out an integrated genome-wide study by combining maps of DNMT3B binding sites, differentially methylated CpGs, H3K36me3, H3K4me3 and H3K27me3 histone marks and differentially expressed genes (DE-genes) in ICF1 patients derived B lymphoblast cell lines (B-LCLs). Examining the genome-wide effect of DNMT3B dysfunction in this disease-relevant model, we identified its target genes and gained a wider understanding of the protein function in modulating the intragenic regulation of transcription.
All together, our observations demonstrate that DNMT3B controls the proper mRNA transcription through the regulation of transcript-isoform TSS usage, cryptic-TSS expression, sense–antisense transcription and alternative exon splicing, by affecting the intragenic epigenetic signature and/or by directly interacting with pre-mRNA molecules. Since the in-depth epigenomic and transcriptomic defects in ICF1 syndrome have not been previously described, we provided novel insights into the molecular mechanisms associated to ICF1-specific DNMT3B mutations.
MATERIALS AND METHODS
Cell culture
Epstein Barr Virus immortalized lymphoblast cell lines (B-LCL) were derived from peripheral blood mononuclear cells (PBMCs) of two unrelated ICF patients with missense mutations in DNMT3B, ICF1p1 (female, compound heterozygous A603T and intron 22 G to A mutation resulting in insertion of three amino acids (STP) in DNMT3B, Coriell Cell Repository) and ICF1p2 (male, compound heterozygous V699G and R54X mutation in DNMT3B, provided by Dr R.S. Hansen). Control EBV-immortalized B-LCLs were from unaffected unrelated individuals (Ctrl1 and Ctrl4) and phenotypically normal parents of ICF1p1 patient (Ctrl2 and Ctrl3, female and male respectively, Coriell Cell Repository). The B-LCLs from the ICF patients’ parents had a similar passage history as the patients’ B-LCLs. We confirmed the genome-wide results in additional B-LCLs (kindly provided by Dr Francastel, INSERM) deriving from ICF1 patients (ICF1pT and ICF1pY, with homozygous mutations D817G/D817G and T775I/T775I, respectively).
For drug treatment, 1 × 106 control cells were plated and treated with 1 μM 5-AzaC (Sigma) for 24 h. Then, the medium containing 5-AzaC was removed and replaced with RPMI + FBS 10% and cells were harvested at different time points (48, 72 and 120 h) for further analyses.
For siRNA knockdown, endogenous DNMT3B was knocked down using the pre-designed siRNA Smart Pool (Dharmacon), while siGLO RNAi control was used as negative control (Dharmacon). The siRNA transfection was performed with Nucleofector transfection (Amaxa, Nucleofector, Kit V) according to the manufacturer's instructions. SiRNA treated cells were processed at 24, 48, 72 and 120 h post-transfection.
DNA extraction was performed using Wizard Genomic DNA (Promega). Total RNA was extracted using the TRIzol reagent (Life Technologies) and purified from residual DNA contaminant using the TURBO DNA-freeTM Kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. RNA quality was checked on the Agilent 2100 Bioanalyzer and quantity was measured using the Qubit instrument (Life Technologies). Whole cell lysates were extracted with ice-cold lysis buffer (100 mM Tris–HCl, pH 8, 1% NP-40, 0.2% SDS, 140 mM NaCl, 20 mM EDTA pH 8) supplemented with protease inhibitor cocktail (Roche Applied Sciences).
Reduced representation bisulfite sequencing (RRBS) and data processing
RRBS libraries were prepared and sequenced according to Illumina's instructions. High quality reads were aligned to the reference genome using the Bismark aligner (33) and methylation call was performed with methylation extractor script. We quantified the methylation level of 1 291 564 CpGs spanning a number of different functional regions in the genome and belonging to different CpG contexts, and with sequencing depth of at least 10 reads in each condition (the median depth was >23 reads among these CpGs in each condition). A detailed description of tools used for data processing is reported in Supplementary Experimental Procedures. Gene-specific DNA methylation level was evaluated by MethylMiner Methylated DNA Enrichment Kit (Invitrogen) and bisulfite sequencing using EpiTech Bisulfite Kit (Qiagen) according to the manufacturer's instructions. Gene-specific RT-qPCR were performed and data were expressed and normalized as log2(methylated/unmethylated), representing the enrichment of the target region in the methylated compared with the unmethylated fraction. Primer sequences are reported in Supplementary Table S1.
RNA sequencing and data processing
RNA isolation and library construction were performed according to Illumina's instructions. After mapping the reads to the reference genome using TopHat2 (34), differential gene expression between two different cell conditions was calculated using DESeq2 implemented in R (35). The relative isoform abundance estimation was performed using BitSeq (36) and the measurement of exon usage was carried out using DEXSeq (37), according to the script published in (38). Gene ontology analysis was performed using DAVID Bioinformatics Resource (39), while enrichment of specific pathways was analyzed using Ingenuity pathway analysis (IPA; http://www.ingenuity.com). A detailed description of tools used for data processing is reported in Supplementary Experimental Procedures.
Quantitative real time PCR
Total RNA from B-LCLs was reverse-transcribed using iScript cDNA Synthesis kit (Bio-Rad San Diego, CA, USA). Quantitative real-time PCR (qPCR) was performed using SsoAdvanced™ universal SYBR® Green supermix (Bio-Rad) on Bio-Rad iCycler according to the manufacturer's protocols. The ΔΔCt method was used to determine relative quantitative levels. GAPDH or ACTB genes were used to normalize the data, considering their unaltered expression in ICF1 samples (Supplementary Figure S1). Primer sequences for gene expression analysis are shown in Supplementary Table S1. All qPCR assays were performed according to the MIQE guidelines (40), and relative informations are provided in the supporting material Supplementary Table S2.
ChIP sequencing and data processing
Chromatin immunoprecipitation was performed as previously described (41). Suitable amount of chromatin was incubated with 5 μg of the indicated antibodies against H3K27me3, H3K4me3, H3K36me3, DNMT3A, CTCF (Abcam) and anti-DNMT3B (Diagenode). Immunoprecipitated complexes were recovered with protein A sepharose (Pharmacia), washed with low and high salt buffers, reverse-crosslinked, and purified. Immunoprecipitated samples were used for qPCR and results were expressed as site occupancy (normalization relative to a negative region amplified in the same sample) to exclude technical variations between samples. We first normalize to the input amount and then, the obtained %input is used to calculate the enrichment over %input of a negative region (GAPDH promoter, MB exon2 or ACTB promoter; Supplementary Figure S1). Mean of results deriving from independent ChIP replicates were reported. Primers sequences are reported in Supplementary Table S1. Immunoprecipitated samples for H3K4me3, H3K27me3, H3K36me3 and DNMT3B were used for preparing libraries and sequencing. 50bp reads were first analyzed for quality filtering and aligned to the reference genome. All the reported results were obtained by normalizing for the total library size. A detailed description of tools used for mapping, peak calling and identification of differentially enriched peaks is reported in Supplementary Experimental Procedures.
RNA immunoprecipitation (RIP) sequencing and data processing
Native and cross-linked RNA immunoprecipitation experiments were performed using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) and the Magna Nuclear RIP (Cross-Linked) RNA-Binding Protein Immunoprecipitation Kit (Millipore), respectively, according to the manufacturer's instructions. Antibodies for RIP assays were anti-DNMT3B (Diagenode), anti-DNMT3A (Abcam) and anti-hnRNP-LL (Aviva). Immunoprecipitated fractions were retro-transcribed and cDNAs were used for qPCR with gene-specific primers and/or for preparing libraries. Sequences (50 bp reads) were analyzed for quality filtering and aligned to the reference genome using TopHat2 (34). FPKMs were calculated using Cufflinks (42) on Galaxy (43) after selecting default parameters. A detailed description of tools used for data analysis is reported in Supplementary Experimental Procedures.
Flow cytometry analysis
Determination of cell surface expression of CD45RABC and CD45RO molecules was carried out by cytofluorimetric analysis using the FACS ARIA cell-sorting system and DIVA software (BD Biosciences). Direct immunofluorescence was performed using PerCP and FITC mouse anti-human CD45RABC and CD45RO antibodies respectively, along with the appropriate mouse IgG isotype controls (BioLegend). Staining, washing and analysis were performed following the manufacturer's recommendations.
Co-immunoprecipitation
Co-immunoprecipitation experiments were performed using Nuclear Complex Co-IP kit (Active Motif) and following the manufacturer's instructions. The antibodies used were the following: DNMT3B (Diagenode), and hnRNP-LL (Aviva). Input samples were also used as nuclear extract in protein expression assay.
Western blot
Whole cell and nuclear protein lysates were quantified by Bradford protein assay (Bio-Rad). Twenty microgram of total protein lysates were separated by SDS polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes (Millipore), blocked with 2% BSA in TBS buffer with 0.05% Tween20 and incubated with primary antibodies for 1 h at RT, followed by incubation with an appropriate HRP-conjugated secondary antibody for 1 h at RT. Densitometric analysis was performed using Typhoon Scan and ImageQuant software.
Statistical analysis
Gene expression, gene specific ChIP, RIP and methylation analysis were presented as means ± standard deviations (SD) from at least three independent experiments. Statistical analyses were performed using Student's t-test (two-tails). P-values were adjusted with BH method and we generally considered the following values as statistically significant: *P-adj <0.05; **P-adj <0.005; ***P-adj <0.0005. The significance of the overlapping between two gene lists was calculated using the hypergeometric test available in R. Statistics relative to DNA methylation comparison between samples was analyzed using Kolmogorov–Smirnov test and values were corrected with Bonferroni method. CpG methylation clustering diagram between samples and principle component analysis (PCA) of mean methylation levels were performed in R using the Methylkit package (44).
RESULTS
Altered CpG methylation affects intragenic regions in mutant-DNMT3B human cells
Previous candidate gene approaches were unsuccessful in identifying ICF1-specific DNA hypomethylation at CpG island promoters of differentially expressed genes (DE-genes), suggesting either that only few of them are DNMT3B direct targets or that differences in DNA methylation level are outside promoters (22,31). Methylome studies carried out in ICF1 B-LCLs suggested that DNA methylation defects might be more extensive than what supposed until now (45,46). However, these studies did not explore which DNA methylation defects are dependent on mutant-DNMT3B, how they functionally impact on gene expression regulation, and whether this effect occurs influencing other epigenetic marks.
In this light, we combined data from Whole Genome Bisulfite Sequencing (WGBS) (45), Reduced Representation Bisulfite Sequencing [RRBS; (47)], RNA-Seq and ChIP-Seq to map DNMT3B binding sites and histone marks (H3K4me3, H3K36me3 and H3K27me3) in B-LCLs derived from peripheral blood of two unrelated ICF patients with DNMT3B missense mutations, ICF1 patient 1 (ICF1p1) and ICF1 patient 2 (ICF1p2), compared to control B-LCLs. These cells represent a suitable model for ICF studies because of the central role of B cells in abnormal immunoglobulin production in ICF cases and the scarcity of primary B cells from patients, of which only few reach adulthood. Despite cell culturing, B-LCLs maintain highly significant ICF-specific differences in mRNA levels of immunoglobulin genes [(31,32) and the present study] and high frequencies of karyotype anomalies described in mitogen-stimulated ICF lymphocytes (27).
We identified 79 521 and 174 030 hypo-methylated CpGs in ICF1p1 and ICF1p2 respectively (Figure 1A and Supplementary Figure S2A–C). When we compared DNA methylation values from RRBS and the whole-genome bisulfite sequencing (WGBS), previously reported for one ICF1 sample (45), we found that they were significantly concordant (Supplementary Figure S2D). Moreover, because DNA methylation profile may be heavily influenced by immortalization and culture condition, we validated our results by comparing the RRBS results with DNA methylation assessment by Infinium HumanMethylation 450K in PBMCs of ICF1 patients (Dr. Francastel, personal communication; Supplementary Figure S2E).
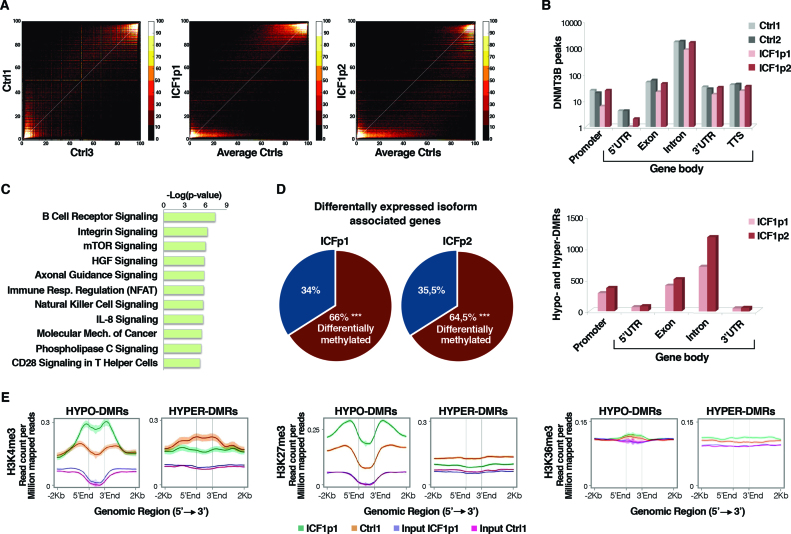
Figure 1.
Genome-wide analysis of CpG methylation, gene expression, DNMT3B and histone marks (H3K4me3, H3K27me3 and H3K36me3) profiles in ICF1-specific DNMT3B mutant cells. (A) Scatter plots and density color codes for DNA methylation data (RRBS) of all autosomes. Pairwise comparisons of methylation percentage between controls (Ctrls) and between ICF1p1 or ICF1p2 and average Ctrls are shown; (B) Distribution of DNMT3B peaks (upper panel) and hypo and hyper-DMRs (Δ ≥ |25%|; lower panel) along the genic features (promoter, 5΄UTR, exon, intron, 3΄UTR and TTS) calculated using Homer tool (85); (C) Enriched gene pathways (IPA) among the differential expressed isoform associated genes (DE-isoforms) (P-values corrected by BH method were considered); (D) Pie chart showing the percentage of DE-isoforms associated genes affected by DNA methylation changes (P-values <10−16 were calculated using one-tail hypergeometric test); (E) Density plots of H3K4me3, H3K27me3 and H3K36me3 (read count per million mapped reads) at the hypo- and hyper-DMRs (−2 kb upstream, +2 kb downstream).
Besides the expected hypomethylation, ICF1 samples also showed gain of methylation with 75 573 and 35 872 hypermethylated CpGs (ICFp1 and ICFp2 respectively; Supplementary Figure S2A). In contrast to observations in ICF1 iPSCs (48) and similar to a recent report in cancer (49), our results did not show substantial levels of DNA methylation at non-CpG sites in ICF1 or control B-LCLs (data not shown). We identified 2381 in ICF1p1 (1833 hypo- and 548 hyper-DMRs) and 3520 DMRs in ICF1p2 (3053 hypo- and 467 hyper-DMRs), which affected all the chromosomes (Supplementary Figure S3A). Of note, 74% of hypomethylated DMRs in ICF1p1 were shared with ICF1p2, while 13% of hypermethylated DMRs were shared between the two ICF1 samples. As previously reported, pericentromeric heterochromatin (PCH) and repetitive sequences (satellite, LINE, SINE, LTRs, etc.) were considerably hypomethylated in ICF1 samples (Supplementary Figure S3B and data not shown).
To assess the effect of inactivating mutations on the genome-wide binding pattern of DNMT3B protein, we performed ChIP-Seq using an antibody against the endogenous protein. We observed that the ability of mutant-DNMT3B to bind DNA was not impaired in terms of total number of peaks both at genic and intergenic regions (Supplementary Figure S3C), but rather in terms of their extension depending on the DNMT3B variant. Indeed, DNMT3B peaks larger than 100kb were increased in ICF1p1 sample, compared to ICF1p2 sample and two controls (data not shown). Given that the extensive loss of DNA methylation at pericentromeric heterochromatin represents a hallmark of ICF molecular phenotype (Supplementary Figure S3B), we examined the DNMT3B binding at these regions, finding that it was mostly unaltered in ICF1 samples compared to controls, as we observed at genes (Supplementary Figure S3D and E).
Because human DNMT3B protein has recently been proposed as responsible for intragenic DNA methylation, we examined the distribution of DNMT3B binding sites along the genic features. Remarkably, both wild-type and mutant-DNMT3B preferentially targeted gene bodies particularly at exons and introns rather than at promoters (Figure 1B, upper panel). Accordingly, most hypo and hyper-DMRs matched with CpG-rich regions at gene-bodies (Figure 1B, lower panel), indicating that most DNA methylation defects occurred at intragenic regions. CpG methylation level at mutant-DNMT3B genomic targets clearly decreased compared to controls, as demonstrated in both RRBS and WGBS, indicating that ICF1-specific mutations prevalently cause hypomethylation (Supplementary Figure S3F). Interestingly, the ICF1p1 sample exhibited a higher level of DNA hypermethylation (Supplementary Figure S2A) associated with a lower number of DNMT3B peaks at gene body regions (Figure 1B, upper panel) compared to ICF1p2 sample. This suggests that DNMT3A might be responsible for the aberrant DNA hypermethylation.
Taken together, these results indicate that ICF1-specific mutations in DNMT3B severely affect gene body CpG methylation.
Intragenic CpG methylation defects associate with altered expression of transcript isoforms
The Gene Ontology analysis of DMR-associated genes and DNMT3B target genes interestingly showed significant enrichment of the alternatively splicing functional category in both control and disease samples [∼45% of DNMT3B targets and DMR-genes, P-value: 3.53 × 10−35 and 2.56 × 10−5, respectively]. This observation together with the finding that DNMT3B-mediated CpG hypomethylation defects occur at intragenic regions suggested that DNMT3B might play a role in the regulation of alternative isoform expression.
To address this possibility and to investigate whether this process is perturbed by ICF1-specific DNMT3B mutations, we analyzed the RNA-Seq datasets using the BitSeq tool, which evaluates the exon–exon junction usage to measure the transcript isoform abundance (36; Supplementary Table S3).
Nearly 55% of differentially expressed isoform (DE-isoform) associated genes were shared by the two ICF1 samples when compared to controls. The newly identified genes belonged to molecular pathways relevant for ICF immunological phenotype (Figure 1C). The most significant pathway affected in both ICF1 samples was the B cell receptor signaling (36% of genes within the pathway showed differential isoform expression; Benjamini–Hochberg (BH) adjusted P-value: 1 × 10−3 and 1 × 10−7 for ICF1p1 and ICF1p2, respectively), presumably accounting for the agammaglobulinemia commonly reported in ICF patients (Supplementary Figure S4).
We found that roughly 65% of DE-isoform associated genes were affected by differential DNA methylation (TSS ± 2 kb) in both ICF1 samples (Figure 1D), supporting the hypothesis that ICF1-specific DNMT3B mutations might perturb the transcription from canonical TSSs by shifting it toward alternative isoform-specific TSSs. Moreover, it has been reported that histone modifications, in particular the H3K4me3, H3K36me3 and H3K27me3 marks, define the transcriptional status at intragenic isoform-specific TSS (50). Therefore, we wondered whether these histone marks had altered profiles in the context of DNA methylation changes in ICF condition. We carried out ChIP-Seq experiments and integrated the results with the RRBS, WGBS and RNA-Seq datasets.
First, we found that H3K4me3 level inversely changed at hypo- and hyper-DMRs, suggesting that CpG methylation defects associate with H3K4me3 changes throughout the genome (Figure 1E, left panel). Conversely, the repressive H3K27me3 mark level significantly increased only at hypo-DMRs indicating that a compensatory mechanism might occur to overcome specific events of loss of gene silencing (Figure 1E; middle panel). At the same time, H3K36me3 was not influenced by increase or decrease of DNA methylation at genome-wide level (Figure 1E; right panel) consistently with the other observations that H3K36me3 mark is not affected in the more severely demethylated cell line HCT116 DKO, which has lost 90% of DNA methylation (6,51).
We then decided to examine whether and how H3K4me3, H3K27me3 and H3K36me3 were involved in the TSS differential expression observed, dependently or not on DNA methylation changes. For this reason, we focused the analysis on differentially expressed TSS (DE-TSS) of annotated isoforms in ICF1 and control samples (Figure 2A and Supplementary Figure S5A; TSS ± 500 bp). We found CpG hypo- and/or hyper-methylation at a majority of them, indicating that ICF-specific DNMT3B mutations affect the isoform transcription by altering CpG methylation at their TSS. Next, we measured the histone marks level at the affected TSS finding that H3K4me3 mark showed the most significant alteration associated with DNA methylation changes, while H3K27me3 and H3K36me3 remained unaffected at most DE-TSS (Figure 2A). Similarly, no significant enrichment of these histone marks was observed at DE-TSS that were not differentially methylated.
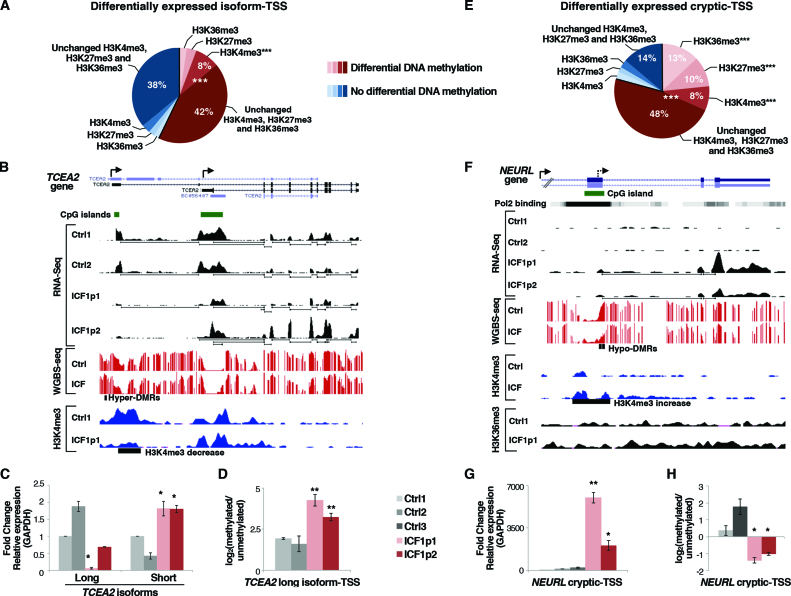
Figure 2.
DNMT3B dysfunction affects isoform-TSS usage and transcription from cryptic-TSS in ICF1 cells. (A) Pie chart showing the percentage of differentially expressed isoform-TSS (±500 bp) with at least five CpGs differentially methylated and/or H3K4me3, H3K27me3 and H3K36me3 marks changes, obtained by integrating the RNA-Seq, WGBS and ChIP-Seq datasets (P-values < 10−16 were calculated using one-tail hypergeometric test); (B) UCSC genome browser screenshot of TCEA2 gene showing differentially expressed long and short transcript isoforms, CpG methylation status and H3K4me3 enrichment at corresponding TSSs. The setting of vertical viewing range is the same for ICF1 and control samples in each experiment; (C) mRNA level (qPCR) of long and short isoforms at TCEA2 locus and, (D) CpG methylation level [log2 (methylated/unmethylated)] of TCEA2 long isoform-TSS in ICF1 samples compared to controls; (E) Pie chart showing the percentage of differentially expressed cryptic-TSS (CAGE-tag, Pol II binding sites, H3K4me3 peaks ± 500 bp) with at least five CpGs differentially methylated and/or histones marks changes (P-values < 10−16 were calculated using one-tail hypergeometric test); (F) UCSC genome browser screenshot representing a cryptic-TSS (Pol II binding site; UCSC Accession: wgEncodeEH001426) at NEURL gene associated with CpG hypomethylation, H3K4me3 increase and transcriptional upregulation, as shown by WGBS, RNA-Seq, H3K4me3 and H3K36me3 ChIP-Seq; (G) Expression level of NEURL cryptic-TSS and, (H) CpG methylation [log2 (methylated/unmethylated)] in ICF1 samples compared to controls. The enrichment of histone modification changes in differentially methylated category compared to the unchanged methylation category was evaluated using χ2-test. *P-adj <0.05; **P-adj <0.005; ***P-adj <0.0005.
Alteration of isoform transcription was found at TCEA2 gene, which resulted from the analysis of DE-isoform associated genes and further validated by qPCR. The long isoform transcribed from the most upstream TSS was abnormally silenced, while the transcription from the internal TSS of the short isoform was upregulated in both ICF1 samples (Figure 2B and C). Silencing of long isoform was linked to significant DNA methylation increase and concomitant reduction of H3K4me3 mark at TSS, while short isoform-TSS showed H3K4me3 increase but no CpG hypomethylation (Figure 2B and D and Supplementary Figure S5B). A gain of mutant-DNMT3B and DNMT3A binding was observed at the hypermethylated CpG island, supporting the idea that a methylating activity of both proteins is aberrantly recruited in ICF1 samples (Supplementary Figure S5C). To investigate whether DNA hypermethylation was directly responsible for the long isoform silencing we treated ICF1 cells with 5-AzaC and monitored the expression of the two TCEA2 isoforms upon DNA hypomethylation induction. We found reactivation of the long isoform, while the short isoform was simultaneously downregulated, confirming that the DNA methylation change alone is sufficient to disturb the relative level of isoform expression (Supplementary Figure S5D and E). Moreover, to examine whether mutant-DNMT3B was directly involved in long isoform downregulation, we reduced its expression using short interfering RNAs (siRNAs) in ICF1 cells. Indeed, repression of TCEA2 long isoform was attenuated upon DNMT3B knockdown (Supplementary Figure S5F and G), indicating that mutant DNMT3B protein was necessary for DNMT3A/3B complex to aberrantly methylate its TSS. Collectively, our findings support an essential role for DNMT3B activity in regulating the transcription of alternative transcript isoforms by differential TSS methylation, which correlates with H3K4me3 mark changes.
Activation of intragenic cryptic-TSS associates with altered CpG and histone methylation
Recent studies have proposed that cryptic or alternative promoters, marked by H3K4me3 and detected by the capped analysis of gene expression (CAGE) sequencing, may be characterized by promoter-like methylation in gene bodies, providing a mechanism to ensure proper transcriptional elongation (10,52–54). To deeper investigate the status of the aberrant transcription initiations in ICF1 samples, we predicted the position of intragenic cryptic-TSS by integrating several datasets from ENCODE Project. We overlapped CAGE tags, Polymerase II binding sites and H3K4me3-enriched regions filtering them for intragenic position and after excluding the annotated TSS.
We then measured the differential expression of the predicted intragenic cryptic-TSS between ICF1 samples and controls using DESeq tool (55) and subsequently searched for differentially methylated CpGs at these regions. We found that about 80% of DE cryptic-TSS were differentially methylated (Figure 2E and Supplementary Figure S6A; P-value < 10−16 in both comparisons).
Remarkably, H3K4me3, H3K27me3 and H3K36me3 marks were significantly affected only when associated with differential CpG methylation at the DE cryptic-TSS, suggesting that these histone marks tightly cooperate with DNA methylation to regulate the intragenic spurious transcription (Figure 2E).
Next, we investigated whether cryptic sites mapped to known repetitive elements, and we found the enrichment of two specific classes, LTRs and DNA repeats, at differentially methylated cryptic-TSS (Supplementary Figure S6B). Moreover, we examined the transcription directionality at cryptic-TSS calculating the directionality score that measures the transcriptional bias to either plus or minus strand of each cryptic-TSS. We found that the large majority of them were transcribed in the sense direction (Supplementary Figure S6C).
Among the compromised genes with a cryptic-TSS, we report the case of NEURL gene, where CpG island hypomethylation at the exon 5, corresponding to an intragenic Polymerase II binding site, resulted in the activation of a cryptic H3K4me3-enriched TSS in ICF1 samples (Figure 2F–H and Supplementary Figure S6D). The activation of this spurious intragenic TSS was linked to the expression of an aberrant short isoform including the last three exons of NEURL gene (Figure 2G). Either upon induction of CpG hypomethylation in control cells using 5-AzaC treatment or DNMT3B gene knockdown, we observed increased expression of the cryptic-TSS at NEURL gene, indicating that this altered regulatory mechanism is directly controlled by DNMT3B-mediated DNA methylation level (Figure S6E–G). Collectively, these results provide evidence that mutant-DNMT3B affects the proper regulation of gene expression also by inducing intragenic changes of CpG methylation and histone marks level at cryptic-TSSs.
Natural antisense transcription is impaired in mutant-DNMT3B cells and affects sense transcription by altering the epigenetic-based self-regulatory circuit
Intragenic DNA methylation and chromatin context have been involved in the regulation of the sense-antisense gene pair transcription (56). Therefore, we hypothesized that DNMT3B mutations might affect this epigenetic-based regulatory mechanism and that it might involve specific histone mark alteration. As a first step, we examined the RNA-Seq dataset and found 153 altered natural antisense transcripts (NATs). Then, by analyzing the CpG methylation and the H3K4me3, H3K27me3 and H3K36me3 marks status at TSS (±500 bp) of NATs, we highlighted that 68% showed significant CpG methylation changes, and altered histone marks were detected at a significant fraction of them (Figure 3A and Supplementary Figure S7A). Interestingly, 66 out of 153 were associated with altered expression of their sense gene and significant changes of CpG methylation, H3K4me3, H3K27me3 and H3K36me3 were observed at the TSS of these genes (Figure 3B). These results would suggest that DNMT3B activity is essential for the sense-antisense pairs transcriptional regulation by ensuring a proper epigenetic context.
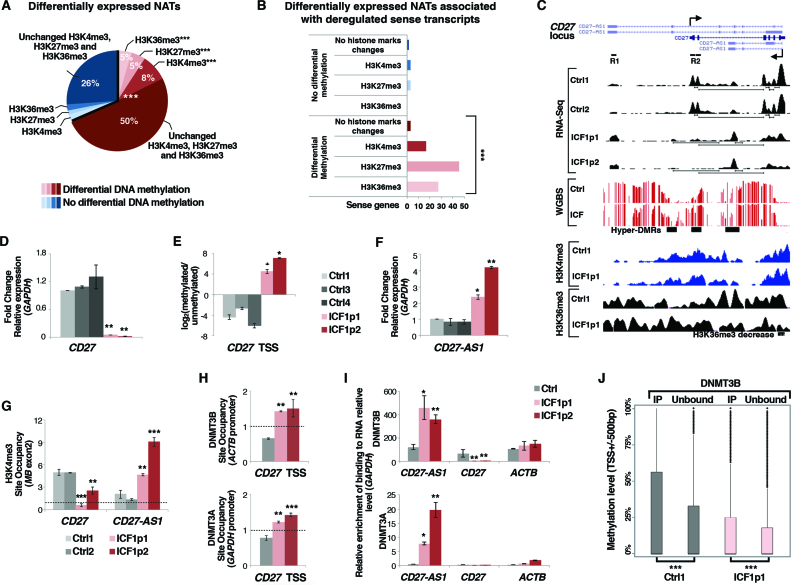
Figure 3.
DNMT3B dysfunction affects sense-antisense pair genes transcription in ICF1 cells. (A) Pie chart showing the percentage of differentially expressed natural antisense transcripts (±500 bp) with at least 5 CpGs differentially methylated and/or H3K4me3, H3K27me3 and H3K36me3 marks changes, obtained by integrating the RNA-Seq, WGBS and ChIP-Seq datasets (P-values << 10−16 were calculated using one-tail hypergeometric test); (B) Differentially expressed sense genes associated with deregulated NATs are significantly enriched for DNA methylation and H3K4me3, H3K27me3 and H3K36me3 changes (P-values << 10−16 were calculated using one-tail hypergeometric test). The enrichment of histone modification changes in differentially methylated category compared to the unchanged methylation category was evaluated using χ2-test; (C) UCSC genome browser screenshots of CD27 gene showing the aberrant silencing of the sense transcript and the concomitant overexpression of the antisense transcript CD27-AS1 (RNA-Seq), the CpG methylation status (WGBS), and H3K4me3 and H3K36me3 enrichments (ChIP-Seq); (D) mRNA, and (E) CpG methylation level [log2 (methylated/unmethylated)] of CD27 sense gene; (F) mRNA level of CD27-AS1 antisense in ICF1 samples compared to controls (gene specific primers were used to detect exclusively the antisense transcript; Supplementary Table S1); (G) H3K4me3 enrichment (ChIP-qPCR) at CD27 and CD27-AS1 TSS; (H) DNMT3B and DNMT3A enrichment to CD27 TSS sense region (ChIP-qPCR); (I) DNMT3B and DNMT3A interaction with CD27-AS1 antisense transcript and with CD27 sense and ACTB transcripts as negative controls (RIP-qPCR); (J) Boxplot showing the methylation level at TSS of sense genes, which expression is reduced compared to the antisense partner (antisense/sense ratio ≥ 1.5 FPKM) and which interacts with DNMT3B at transcript level comparing the immunoprecipitated (DNMT3B-interacting transcripts) and unbound RNA fractions in each cell line. *P-adj <0.05; **P-adj <0.005; ***P-adj <0.0005.
In an attempt to identify functionally relevant pairs of deregulated sense-antisense transcripts, we focused our attention on the memory B-cell marker CD27 gene, known to be aberrantly silenced in peripheral blood of ICF1 patients and ICF1 cell lines. By performing gene-specific qPCR, and CpG methylation level analysis, we highlighted that CD27 gene repression in ICF1p1 and p2 samples was linked to TSS CpG hypermethylation (Figure 3C–E). Intriguingly, the overlapping antisense long non-coding transcript CD27-AS1 showed upregulation in ICF1 samples, as revealed by isoform-specific analysis of RNA-Seq dataset and qPCR (Figure 3C and F).
To identify the mechanism responsible for the inverse transcriptional correlation between sense and antisense expression at CD27 locus and investigate how this mechanism is impaired in ICF1 condition, we first analyzed H3K4me3 and H3K36me3 marks at TSS and gene body of sense and antisense transcripts by ChIP assay. Interestingly, in control cells we observed high H3K4me3 level at the sense TSS, while it was poorly enriched at CD27-AS1 TSS (Figure 3G). Conversely, an opposite H3K4me3 profile was observed in ICF1 samples (lower level at CD27 TSS, higher level at CD27-AS1 TSS) according with CD27 sense silencing and higher antisense transcription in disease cells (Figure 3G).
Consistently with the active CD27 gene expression in control samples, H3K36me3 mark was enriched downstream the CD27 TSS along the gene body (Figure 3C). Because accumulation of H3K36me3 has been considered a general mechanism for reversible gene silencing when it occurs at promoters (57), we analyzed the enrichment of this histone mark at CD27-AS1 TSS. Notably, we detected a high H3K36me3 level overlapping the antisense promoter in control cells, which is presumably responsible for suppressing the antisense transcription (Figure 3C and Supplementary Figure S7B). Accordingly, H3K36me3 level was significantly reduced in ICF1 samples compared to control samples at CD27-AS1 TSS, being correlated with CD27-AS1 antisense upregulation in disease cells (Figure 3C and Supplementary Figure S7B).
We next examined the interaction of DNMT3A and DNMT3B proteins at the aberrantly hypermethylated CD27 promoter region by ChIP assay. The active DNMT3A and the catalytically inactive DNMT3B both interacted with the CD27 sense promoter only in ICF1 samples, suggesting that these proteins were aberrantly recruited and responsible for the hypermethylation and consequent gene silencing (Figure 3H). We also examined DNMT3A/B binding at CD27-AS1 TSS, without detecting any enrichment (Supplementary Figure S7C).
To prove that DNA hypermethylation was able to silence the CD27 gene in ICF1 condition, we induced CpG hypomethylation by treating the ICF1p2 cells with 5-AzaC and examined the expression of the sense transcripts at different time points. We found that CD27 gene repression was released, thus confirming that DNA hypermethylation was sufficient to aberrantly silence CD27 transcription in ICF1 samples (Supplementary Figure S7D and E). Interestingly, we observed a concomitant reduction of the CD27-AS1 transcription in the treated ICF1p2 cells, indicating that a self-regulatory circuit takes place at CD27 locus, which implies a transcriptional perturbation between sense and antisense transcripts (Supplementary Figure S7D). Similar effects on the CD27/CD27-AS1 reciprocal transcription were obtained when we ectopically silenced DNMT3B in CD27-hypermethylated ICF1p2 cells, suggesting that DNMT3B plays a role in this regulatory network, either by methylating itself with its residual catalytic activity and/or by tethering DNMT3A at CD27 TSS (Supplementary Figure S7F). Remarkably, in 5-AzaC-treated control cells CD27-AS1 resulted overexpressed and its TSS was hypomethylated, suggesting the CD27-AS1 transcription can be also directly modulated by CpG methylation itself (Supplementary Figure S7G and H).
Antisense expression has been shown to regulate transcription initiation by affecting DNA methylation at sense gene CpG islands/promoter in several contexts (58–61). While the process of antisense transcription may itself regulate sense gene transcritpion, in other instances it has been shown that NATs can directly regulate promoter DNA methylation via recruitment of DNMTs (62). Therefore, to explore the involvement of this specific mechanism we performed native RNA immunoprecipitation (RIP) assay followed by qPCR using antibodies against DNMT3B and DNMT3A. Then, we evaluated the interaction of the antisense transcript CD27-AS1 with the two DNA methyltransferases. Remarkably, in ICF1 cells CD27-AS1 was selectively pulled-down by both the antibodies (after normalization to the transcript amount), indicating that CD27-AS1 forms a complex with DNMT3A and DNMT3B proteins (Figure 3I). Given that DNMT3A and DNMT3B were also both enriched at CD27 sense promoter (Figure 3H), the recruitment of DNMTs at this region, mediated by the antisense non-coding transcript, may eventually leads to the aberrant CpG hypermethylation. All together, our findings demonstrate that DNMT3B dysfunction affects the antisense-mediated gene regulatory networks through altering the epigenetic marks and the ncRNA-dependent guide of DNMT3B.
Next, we wondered whether this NAT-mediated DNMT3s tethering at sense promoters might be more generally involved in the self-regulatory circuit of sense-antisense gene pairs and whether DNMT3B mutations affect this mechanism at wider scale in disease context.
Thus, we first selected the overexpressed antisense transcripts compared to their sense partner in ICF samples and controls. Next, we identified the overexpressed antisense transcripts interacting with DNMT3B, which we obtained sequencing the immunoprecipitated fraction of the RIP assay (RIP-Seq; Supplementary Figure S8A–D). Interestingly, we found that the DNMT3B-bound fraction of overexpressed antisense transcripts significantly correlated with the highest CpG methylation level at TSS of the sense partner, compared to the unbound fraction both in control and disease cells (Figure 3J). This suggests that the interaction between DNMT3B and non-coding antisense RNAs to the sense promoter might be a general mechanism to recruit the methyltransferase activity.
DNMT3B protein is actively involved in modulating exon exclusion through its RNA-binding property
Intragenic DNA methylation has been proposed as mechanism able to modulate alternative splicing events (63). Therefore, we hypothesized that DNMT3B mutations could affect the alternative splicing process and examined changes in exon skipping and inclusion between ICF1 and control samples using DEXSeq tool (37). We identified 3821 and 2330 differentially skipped and included exons, respectively, in ICF1p1 and 6436 and 3753 differentially skipped and included, respectively, in ICF1p2, and 42%-46% of them were differentially methylated (Figure 4A and Supplementary Figure S9A and B). However, this subtype was not significantly enriched among the differentially included exons, when compared with the total number of exons affected by altered DNA methylation in ICF1. This suggests that most of these events are a secondary effect of DNMT3B mutations or are caused by a regulatory DNMT3B function independent on methyltransferase activity.
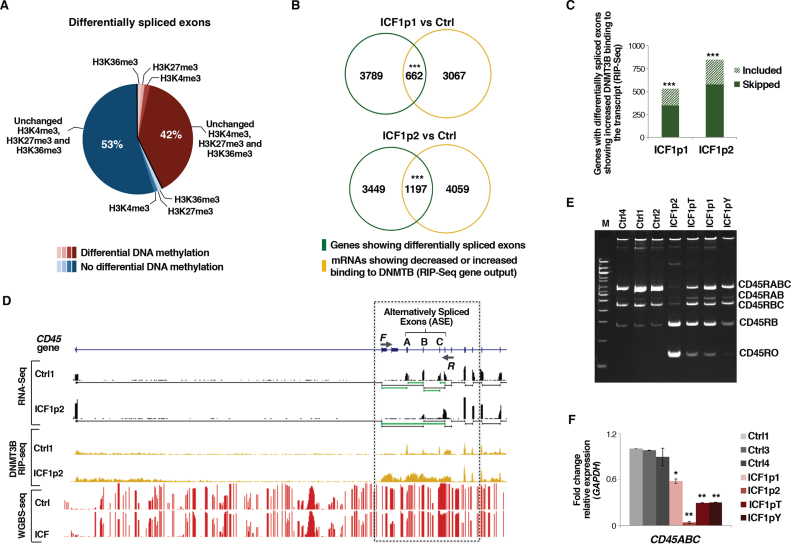
Figure 4.
DNMT3B dysfunction affects alternative exon splicing in ICF1 cells. (A) Pie chart showing the percentage of differentially spliced exonic parts with differentially methylated CpGs and/or H3K4me3, H3K27me3 and H3K36me3 marks changes (statistical analysis was performed using one-tail hypergeometric test, which shows no significant enrichment for these epigenetic modification at differentially included exons); (B) Venn diagram showing the overlap between genes with differentially spliced exons and genes with decreased or increased DNMT3B binding to their transcripts (RIP-Seq) (P-values << 10−16 were calculated using one-tail hypergeometric test); (C) Histogram showing the number of genes with differentially spliced exons and increased DNMT3B binding to the transcript (normalized to IgG fraction; RIP-Seq). Gain of DNMT3B binding significantly associates with skipping or inclusion of exons (P-values << 10−16 were calculated using one-tail hypergeometric test); (D) UCSC genome browser screenshots of PTPRC (CD45) gene showing exon 4–5–6 skipping (RNA-Seq) and CpG methylation status (WGBS); (E) Semi-quantitative PCR amplification of CD45 splicing isoforms in controls and ICF1 samples. Primers used are F and R as shown in D; (F) expression level (qPCR) of CD45RABC using isoform-specific oligonucleotides. *P-adj <0.05; **P-adj <0.005; ***P-adj <0.0005.
To discriminate between these two possibilities we analyzed RNA expression levels of all spliceosome components in control and ICF1 samples and found no significant alterations (Supplementary Table S3), implying that most of the changes in alternative splicing in ICF1 samples are a direct outcome of DNMT3B dysfunction.
Therefore, considering that DNMT3B is capable of interacting with components of the transcriptional machinery (64), we asked whether the RNA-binding ability of DNMT3B might play a role in the alternative exon inclusion process. First, we compared the list of genes showing differentially spliced exons with the list of DNMT3B-interacting genes (RIP-Seq dataset) and found a significant overlap (Figure 4B and Supplementary Figure S9C). Next, we selected genes with differentially spliced exons showing an increased binding of DNMT3B at their transcripts in ICF samples and measured the number of exon skipping or inclusion events. From this analysis, we found significant enrichment for either skipping or inclusion events, indicating that acquisition of DNMT3B binding to mRNAs similarly impacts on both categories of alternative splicing (Figure 4C). All together, our studies suggest that in human cells ICF1-specific mutations of DNMT3B may affect the alternative splicing of mRNAs through a novel function of the protein, implying its ability to interact with mRNA molecules.
Among the aberrantly spliced genes, we identified the Protein Tyrosine Phosphatase, Receptor type, C gene (PTPRC; also known as CD45), which is a trans-membrane protein tyrosine phosphatase essential for antigen receptor-mediated signaling in lymphocytes (65). Differences in exon skipping and glycosylation of the protein produce various isoforms of CD45 that are present in a cell-specific manner. CD45RO, the smallest isoform, distinguishes effector-memory T cells, whereas primary B cells are known to express predominantly the largest CD45 protein isoform (encoded by CD45RABC, including exons 4–5–6). In line with the in vivo findings, the cultured B cells and B-LCLs express almost uniquely the RABC and RBC isoforms (66).
By comparing control and ICF1p2 RNA-seq tracks we found that exons 4–5–6 were differentially spliced (Figure 4D) and this intriguingly correlated with an increased binding of DNMT3B protein to CD45 transcript in ICF1 sample (Figure 4D). To validate these observations, we performed isoform-specific PCR and found that ICF1 cells showed significantly higher CD45RO isoform expression than controls, while the CD45RABC isoform decreased, suggesting that DNMT3B activity is necessary to ensure a proper inclusion of the exons 4–5–6 (Figure 4E and F). The altered isoform transcription led to a change from CD45RABChigh/CD45ROlow population in control cells to CD45RABClow/CD45ROhigh population in ICF1 cells, which is more or less pronounced depending on DNMT3B mutations (Supplementary Figure S10A).
Given the fact that splicing occurs co-transcriptionally, epigenetic marks might affect the splicing decisions both by influencing the kinetics of transcriptional elongation (kinetic model), or through adaptor proteins that bind to epigenetic modifications and recruit splicing factors [recruitment model; (18)]. Both processes have been described for the splicing regulation mediated by DNA methylation (15,16,67). Therefore, we decided to examine thoroughly the involvement of these two mechanisms.
In line with WGBS results, we found that the alternative exons 4–5–6 were highly CpG methylated, either in control or ICF1 samples, and showed significant increase between the two conditions only in ICF1p2 cells (Figure 4D and Supplementary Figure S10B). Also, we analyzed the methylation status at single CpG level of exon 5, which has been reported alternatively spliced in naïve and activated CD4+ T cells according to its methylation level (15). We did not detect any changes at CpG methylation and consistently exon 5 did not show differential CTCF binding (Supplementary Figure S10C and D) (15). Similarly, we measured the enrichment of H3K36me3 marks at the spliced exons without finding any significant change (Supplementary Figure S10E).
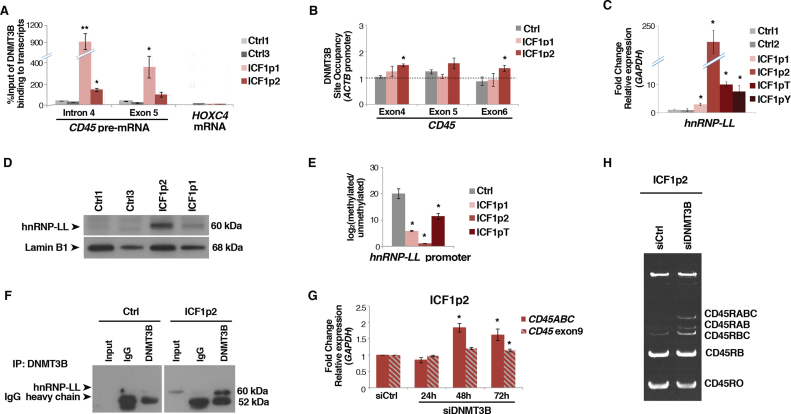
Because we did not detect epigenetic modifications affecting the elongation rate of CD45 gene, we explored in detail the mechanism suggested by our genome-wide studies (RIP-Seq), implying the direct binding of DNMT3B to the spliced exons of the transcript. Indeed, by carrying out native RNA immunoprecipitation with anti-DNMT3B we highlighted the interaction of the protein with CD45 pre-mRNA in ICF1 samples (Figure 5A), which was further confirmed by cross-linked RNA Immunoprecipitation (Supplementary Figure S10F). This interaction was absent when analyzing different genes, which represented negative controls (e.g. HOXC4, GAPDH, ACTB).
Figure 5.
DNMT3B modulates exon 4–5–6 skipping by physically interacting with hnRNP-LL and CD45 pre-mRNA. (A) Native RNA immunoprecipitation assay (RIP-qPCR) shows DNMT3B interaction with CD45 pre-mRNA in control and ICF1 samples. Binding to HOXC4 mRNA is reported as negative control; (B) DNMT3B enrichment at DNA (exons 4, 5 and 6) of CD45 gene by ChIP assay; (C and D) Expression level of hnRNP-LL in ICF1 samples and controls by qPCR and western blot, respectively; (E) CpG methylation level [log2 (methylated/unmethylated)] at hnRNP-LL promoter in ICF1 samples; (F) DNMT3B and hnRNP-LL physically interact in ICF1p2 sample as shown in co-immunoprecipitation experiments; (G) Expression level (qPCR) of CD45RABC and the constitutive exon9 using isoform specific oligonucleotides upon siDNMT3B and control siRNAs transfection; (H) Semi-quantitative PCR amplification of CD45 splicing isoforms in overexpressing-hnRNP-LL ICF1p2 sample upon siDNMT3B and control siRNAs transfection. *P-adj <0.05; **P-adj <0.005; ***P-adj <0.0005.
As suggested by the recruitment model, chromatin-binding proteins, which bind to specific epigenetic modifications, can gather RNA-binding proteins. These RNA-binding proteins are then transferred to the mRNA molecule as it is being transcribed and change its alternative splicing pattern. We thus, verified the hypothesis that DNMT3B may function according to this model. First, we analyzed the DNMT3B binding to DNA by ChIP assay and we found a significant increase of DNMT3B interaction to the CD45 exons 4–5–6 in ICF1 samples (Figure 5B). Interestingly, DNMT3A was enriched at these exons, indicating that high CpG methylation level at the exons was presumably ascribed to DNMT3A binding (Supplementary Figure S10G).
Then, we searched for molecules potentially participating to this aberrant exon skipping. CD45 gene alternative splicing is tightly controlled by the tissue-specific ribonucleoprotein heterogeneous nuclear RNA-binding protein L-Like (hnRNP-LL). Hn-RNP-LL expression is specifically induced in terminally differentiated lymphocytes, including effector T cells and plasma cells, where it mediates the transition from CD45RA or CD45RABC to CD45RO, respectively (68,69). Current models of combinatorial alternative splicing propose that hnRNP-LL cooperates with the heterogeneous ribonucleoprotein hnRNP-L on CD45 pre-mRNA, bridging exons 4 and 6 and looping out exon 5, thereby achieving full repression of the three variable exons (70).
Therefore, we first examined the expression of hnRNP-LL in ICF1 samples. Remarkably, hnRNP-LL was overexpressed in ICF1 cells compared to controls, and this correlated with CpG hypomethylation of its CpG island-associated promoter (Figure 5C–E and Supplementary Figure S11A). Conversely, the transcription of the protein interactor hnRNP-L was unaffected (data not shown). To confirm that overexpression of hnRNP-LL was able to modulate exons 4–5–6 skipping in an independent context, we treated control cells with 5-AzaC and verified that CpG hypomethylation led to the up-regulation of hnRNP-LL transcription (Supplementary Figure S11B and C) and resulted in the increased exons 4–5–6 exclusion (Supplementary Figure S11D).
Interestingly, we observed that the overexpressed hnRNP-LL interacted with CD45 pre-mRNA in ICF1 cells, by performing RNA immunoprecipitation using anti-hnRNP-LL antibody (Supplementary Figure S11E). Collectively, these results suggest that DNMT3B might be recruited to pre-mRNA by the increased expression of hnRNP-LL. To address this hypothesis, we performed a co-immunoprecipitation assay, showing that hnRNP-LL and DNMT3B proteins physically interacted in ICF1 cells, which suggests the formation of a protein complex perturbing the correct inclusion of exons 4–5–6 (Figure 5F). Absence of the interaction was observed in control cells where hnRNP-LL is not expressed, as expected (Figure 5F).
DNMT3B might exert an active role in promoting the aberrant exons 4–5–6 skipping in ICF1 cells by interacting with the unspliced exons or being a secondary effect of the increased hnRNP-LL activity. To discriminate between these two scenarios and demonstrate that the DNMT3B activity is functionally relevant to influence the exons 4–5–6 skipping, we knocked down DNMT3B in ICF1p2 cells, where hnRNP-LL shows the greatest overexpression, by transfecting DNMT3B siRNAs. The DNMT3B knockdown did not affect the hnRNP-LL protein level, as shown by western blot analysis (Supplementary Figure S11F). Interestingly, when we measured the inclusion level of exons 4–5–6, we found that DNMT3B knockdown partially reverted the exon skipping, increasing the abundance of the longest isoform CD45ABC (Figure 5G,H). This indicates that despite the presence of hnRNP-LL in ICF1p2 cells DNMT3B function directly influences this process. In order to verify if hnRNP-LL and DNMT3B are reciprocally regulated we knocked down the heteronucleoriboprotein in ICF1p2 cells. The western blot analysis showed an unaltered level of DNMT3B protein (Supplementary Figure S11G), indicating that there is not a regulation feedback between these two proteins.
Overall, these results demonstrate that ICF1-specific DNMT3B mutations lead to CpG hypomethylation and overexpression of hnRNP-LL, which is engaged to CD45 pre-mRNA in a protein complex through interacting with the mutant-DNMT3B. Here, DNMT3B activity specifically promotes aberrant exon skipping.
DISCUSSION
In this manuscript we tackled the important but still unanswered question of how hypomorphic mutations of DNMT3B affect the intragenic regulation of transcription in ICF1 condition. The integrative genome-wide study that we present shows that ICF1-specific mutations in DNMT3B catalytic domain slightly alter the ability of the protein to target DNA genomic regions, while the methyltransferase activity is rather impaired. The fact that mutant-DNMT3B is able to bind DNA is expected, considering that ICF1-specific mutations mainly affect the catalytic domain, while the DNA binding capability depends on the PWWP domain within the N-terminal region (71). By combining genomic profiles of DNMT3B binding and CpG methylation defects, we observed that the DNA hypomethylation is prevalently explained by the catalytically inactive mutant-DNMT3B, which may function as negative dominant. Conversely, given that mutations inhibit the DNMT3B catalytic activity, the gain of CpG methylation is presumably ascribed to DNMT3A, as suggested by the findings that DNMT3A and mutant-DNMT3B are both mistargeted at hypermethylated sites, e.g. TCEA2 long isoform TSS, CD27 sense TSS and CD45 exon 4-5-6. In support of this conclusion, it has recently been confirmed that DNMT3B may play an accessory role by inducing (recruiting and stimulating) DNMT3A methylation activity (8).
Our results demonstrate that most DNMT3B binding sites occur at intragenic positions of genes, where DNA methylation defects largely take place in ICF1 cells. This is a novel finding in the context of ICF1 syndrome studies and explains why previous attempts to identify profound differences in DNA methylation profile of deregulated genes failed using candidate gene approaches focused at CpG island-associated promoters (22,31,32). These results paved the way to the identification of unprecedented aspects related to DNMT3B deficient activity in ICF1 syndrome, such as the alteration of isoform transcription by modulating the epigenetic signature (DNA methylation and H3K4me3, H3K27me3 and H3K36me3) at intragenic TSS. Notably, we found deregulation of alternative isoforms at genes relevant for ICF1-specific phenotype.
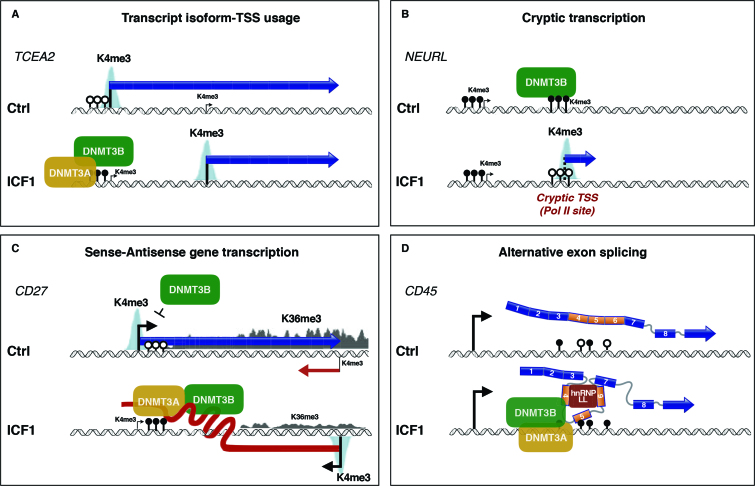
The large majority of affected genes were direct targets of DNMT3B, suggesting that proper binding and methylating activity of the protein is required for the appropriate relative abundance of transcript isoform. For instance, we found CpG-island hypermethylation overlapping the TCEA2 long isoform TSS dependent on the aberrant mutant-DNMT3B/3A methylating activity in ICF1 cells, which silences its transcription, thus promoting higher level of the short isoform transcript (Figure 6). This was associated with a significant change of H3K4me3 at the affected TSS, but not at H3K27me3 and H3K36me3 marks, indicating that alteration of CpG methylation is sufficient to impact on TSS selection and tightly cooperate with H3K4me3 levels. That CpG methylation changes alone are sufficient to determine the TSS usage is further confirmed upon induction of long-isoform TSS hypomethylation in 5-AzaC treated ICF1p2 cells, where we observed the rescue of the TCEA2 long isoform expression. Rescue of the long isoform level upon DNMT3B knockdown in ICF1 cells demonstrates that the binding activity of mutant DNMT3B might be important for DNMT3A recruitment to CpG island. Overall, together with the evidence that cryptic-TSS are illegitimately expressed in ICF1 samples this finding suggests that a proper intragenic DNMT3B-mediated CpG methylation contributes to achieve an efficient mRNA transcriptional initiation and elongation (Figure 6).
Figure 6.
Summarizing model showing the mechanisms affected by ICF1-specific DNMT3B dysfunction in the regulation of (A) alternate isoform-TSS usage, (B) cryptic transcription, (C) sense–antisense gene self-regulatory circuit and (D) alternative exon splicing.
Antisense transcription is increasingly being recognized as an important regulator of gene expression. The locus specificity of natural antisense RNAs (NATs) opposite sense genes indicates that they might be part of self-regulatory circuits that allow genes to regulate their own expression. Studies have shown that antisense expression can induce a threshold-dependent on-off switch on sense-gene expression (56).
Noncoding antisense loss or gain of expression in a sense-antisense gene pair has often been implicated in human disease (59,72). The antisense-mediated repression may involve transcriptional perturbation of the sense partner but may also include engagement of chromatin remodeling complexes, with local accumulation of histone modifications and DNA methylation. Alternatively, chromatin modulation based on the process of transcription per se might be involved (73). This model is supported by the association of Pol II with chromatin remodelers like Set2, which catalyzes the deposition of H3K36me3 in transcribed regions (74). H3K36me3 in turn may be recognized by chromatin factors correlating with transcriptional repression (75–77).
Despite the cases where the requirement of the RNA molecule is clearly necessary, it is presumable that several complementary mechanisms might act cooperatively to ensure the faithful regulation of overlapping gene pairs. In this light, a remarkable example is the memory B-cell marker CD27 gene, which transcription aberrantly switches from the sense to the overlapping antisense in ICF1 condition (Figure 6). In control cells, the high-level expression of sense CD27 gene associates with the spreading of a transcription-induced accumulation of H3K36me3. In turn, H3K36me3 enrichment over the CD27-AS1 TSS determines a repressive chromatin environment disfavoring the antisense transcription (Figure 6). We did not detect recruitment of DNMT3A/3B, which is known to bind H3K36me3, suggesting that different chromatin factors, like H3K4me3 demethylases or histone deacetylases, may be implicated.
Conversely, the epigenetic silencing of CD27-AS1 promoter (CpG methylation and H3K4me3 level) is altered in ICF1 cells, fostering its transcription. The overexpressed antisense transcript interacts with DNMT3B/3A proteins inducing their recruitment to CD27 sense TSS, which in turn acquires CpG hypermethylation and loses H3K4me3 mark. Strikingly, the epigenetic marks promote aberrant CD27 sense gene repression, which then correlates with a reduced H3K36me3 level at CD27-AS1, thereby sustaining the antisense transcription.
CD27 protein plays a key role during the lymphocyte differentiation to memory B-cells and CD27 gene expression is known to be severely affected in ICF condition. Indeed, complete lack of the CD19-CD27+ memory B-cell compartment in ICF patients in contrast to controls was detected from biochemical studies of ICF patients’ peripheral blood (24). Peripheral B-cell maturation blockage was thought to contribute to agammaglobulinemia and to immune response dysfunction, which are the main clinical signs of the disease. Strikingly, our findings provided a molecular explanation to CD27 aberrant silencing in ICF cells, contributing to decipher the molecular mechanisms underlying the disease pathogenesis.
There is a growing evidence for co-transcriptional regulation of splicing decisions and the current view is that CpG methylation may modulate the alternative splicing process, at least at a fraction of transcripts (63). Our findings point to a crucial role of DNMT3B in this process. According to our observations, DNMT3B might play a structural role acting as an adaptor protein, able to bind methylated DNA together with DNMT3A, and to recruit hnRNPs, which in turn tether splicing factors to the pre-mRNA. We proved that DNMT3B modulates the exon splicing through a mechanism implying the physical interaction of endogenous protein with mRNA molecules. The RNA binding capability of DNMTs was predicted based on several evidences, particularly involving non-coding RNAs (78,79). However, here we report the first example of interaction between DNMT3B and pre-mRNA molecules, which has the function to modulate the alternative exon splicing. At a genome-scale level we demonstrate that DNMT3B/mRNA interaction occurs in physiological conditions, suggesting a more general involvement of DNMT3B in mediating the alternative splicing. Further studies will be necessary to dissect the contribution of this regulatory mechanism to gene expression program in normal cells.
In disease context, this DNMT3B/mRNA binding alters the proper sequence of events leading to the inclusion of alternative exons 4–5–6 at CD45 gene. We deciphered the molecular mechanism underlying the illegitimate exon exclusion in ICF1 samples, finding that DNMT3B is recruited to these exons and physically interacts with hnRNP-LL, which is ectopically expressed. To exclude that exons 4–5–6 skipping in ICF1 cells was exclusively dependent on hnRNP-LL overexpression we induced the DNMT3B knockdown in highly expressing hnRNP-LL ICF1 cells, and we were able to revert the exons 4–5–6 skipping. This provides direct evidence that DNMT3B does have an active role in modulating exon splicing at CD45 gene. Here we describe a mechanism, which does not involve CTCF mediated regulation, which are instead observed at CD45 exon5 in naïve and activated CD4+ T cells (15,80), indicating that specific epigenetic-based regulation of alternative splicing occurs in different cell contexts.
Notably, one study in embryonic stem cells suggests that DNMT3B would be able to interact with proteins involved in various aspects of Pol II-mediated transcription, as for instance PAF1 complex components, and heterogeneous ribonucleoproteins (64). We report a specific interaction between DNMT3B and hnRNP-LL in somatic cells, which would confirm this observation.
Within the complex with hnRNP-LL, the interaction between DNMT3B and pre-mRNA is expected to be direct as suggested by the native RNA immunoprecipitation and it may occur through the catalytic domain, which has been reported able to bind RNA molecules in case of DNMT1 protein (78). Mechanistically, the regulation of alternative exon inclusion/exclusion at DNMT3B-interacting pre-mRNAs, might involve a still unknown role of the protein in stimulating the m6A or the m5C RNA methylation process. Indeed, the m6A RNA methylation has been reported as a modulator of mRNA splicing (81). Whatever the machinery involved, it is remarkable that it might represent a general DNMT3B-mediated mechanism of mRNA alternative exon splicing, as suggested by the RIP-Seq results in control cells. Strikingly, we identified a significant correlation between DNMT3B binding to mRNA transcripts and their alternative exon splicing. It will be interesting to assess the biological consequence of the altered transcriptome at global protein level.
Collectively, our results support the concept that gene body CpG methylation impacts on multiple regulatory mechanisms at intragenic level, including the repression of cryptic transcription from alternative promoters, selection of the transcribed isoform, antisense transcription and alternative exon splicing (9,10,82). Our findings contribute to decipher how the epigenetic modifications, which occur co-transcriptionally, interfere with a proper resultant mRNA transcript (in size and exon-intron content). Here, we provide evidence that DNMT3B plays a key role in regulating each of these events highlighting that dysfunction of DNA methylating activity or its aberrant ncRNA-mediated recruitment causes altered control of mRNA transcription and processing. These novel aspects concerning DNMT3B function deserve to be deeply studied, because they represent a conceptual proof that DNA methylation and mRNA processing functionally converge, pointing to a central role for DNMT3B protein.
Besides providing us with novel insights into DNMT3B-dependent transcriptional regulatory network, our integrative multi-omic approach contributes to clarify the disease-associated molecular defects. A terminal B-cell differentiation blockage at the transitional B-cell stage was reported in ICF1 patients (24). Consistently, we found that ICF samples exhibited transcriptional defects at genes involved in the B cell receptor-signaling pathway. Deregulation of this pathway typically causes the absence of circulating mature B cells and of all immunoglobulin isotypes, accompanied by the accumulation of pre‐B cells in the bone marrow (83). Of note, among the newly identified differentially expressed genes we found either receptor-type PTPs, (e.g. PTPRC, PTPRJ and PTPRB) and nonreceptor-type PTPs (e.g. PTPN13). These are known to positively or negatively regulate lymphocyte activation and development, thus providing novel candidate genes contributing to the immune response defects in ICF1 patients.
The most striking example was CD45, which main role in B and T lymphocytes promotes cells activation. Remarkably, CD45 activity is negatively modulated by dimerization (84). Larger CD45RABC isoforms exist predominantly as monomeric active phosphatase, while the smaller size of the CD45RO extracellular domain facilitates dimerization, rendering it less active and increasing the signal transduction threshold. In T or B cells this would contribute to cessation of the primary immune response (66). In this light, the aberrant exon skipping detected in ICF cells might perturb the CD45 protein activity by increasing the abundance of the CD45RO isoform.
In summary, we have performed an integrated epigenomic and transcriptomic analysis in ICF1-derived B-LCL. Compared to previous classical transcriptional studies, we identified a new level of DNMT3B-mediated transcriptional regulation, which highlighted defects in gene pathways and functions predicted and long sought, thus representing a step forward towards a more careful characterization of the ICF1 immune phenotype. Furthermore, our study highlights the power of integrative analyses to clarify the DNMT3B-mediated intricate regulatory epigenetic network modulating the proper tissue-specific transcriptome.
ACCESSION NUMBER
Data for the RNA-seq, ChIP-seq, RRBS and RIP-seq in ICF1 and Ctrl cell lines have been deposited in the GEO database with accession number GSE95747.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Tom Roberts, Maria Strazzullo, Giovanna Del Pozzo and Andrea Riccio for helpful discussion, careful reading and critical comments of the manuscript. We also thank Sarah Teichmann, Henk Stunnenberg for their valuable help in the initial phases of the study and Maria Frola, Marianna Mastroianni for their help in the validation and organization of the data. Holger Heyn, Juan Sandoval and Manel Esteller are acknowledged for providing us with the WGBS data. Luigi Leone and Vincenzo Mercadante (NGS Core Facility) are acknowledged for technical assistance during histone marks ChIP-Sequencing experiments. We thank the IGA Technology Service (Udine) for RRBS, RNA-Seq and ChIP-Seq experiments and Laura Pisapia and Pasquale Barba (IGB FACS Facility) for surface phenotypic cell analysis. We are grateful to Giulio Pavesi, Francesco Russo, Dario Righelli and Marta Milo for bioinformatic support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
‘Epigenomics Flagship Project (EPIGEN) Italian Ministry University and Research—CNR’ [M.R.M., C.A. and M.D.E.]; UE Initial Training Network Project ‘DISCHROM’ [238242 to M.R.M. and M.D.E]; Telethon [GGP15209 to M.R.M.]. Funding for open access charge: Telethon GGP15209.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jeltsch A., Jurkowska R.Z.. New concepts in DNA methylation. Trends Biochem. Sci. 2014; 39:310–318. [DOI] [PubMed] [Google Scholar]
- 2. Jurkowska R.Z., Jurkowski T.P., Jeltsch A.. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2011; 12:206–222. [DOI] [PubMed] [Google Scholar]
- 3. Okano M., Bell D.W., Haber D.A., Li E.. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999; 99:247–257. [DOI] [PubMed] [Google Scholar]
- 4. Chen T., Ueda Y., Dodge J.E., Wang Z., Li E.. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003; 23:5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ueda Y., Okano M., Williams C., Chen T., Georgopoulos K., Li E.. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006; 133:1183–1192. [DOI] [PubMed] [Google Scholar]
- 6. Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G.. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014; 26:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schubeler D.. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015; 520:243–247. [DOI] [PubMed] [Google Scholar]
- 8. Duymich C.E., Charlet J., Yang X., Jones P.A., Liang G.. DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells. Nat. Commun. 2016; 7:11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kulis M., Heath S., Bibikova M., Queiros A.C., Navarro A., Clot G., Martinez-Trillos A., Castellano G., Brun-Heath I., Pinyol M. et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012; 44:1236–1242. [DOI] [PubMed] [Google Scholar]
- 10. Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D., Johnson B.E., Hong C., Nielsen C., Zhao Y. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010; 466:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolff E.M., Byun H.M., Han H.F., Sharma S., Nichols P.W., Siegmund K.D., Yang A.S., Jones P.A., Liang G.. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010; 6:e1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anastasiadou C., Malousi A., Maglaveras N., Kouidou S.. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell Biol. 2011; 30:267–275. [DOI] [PubMed] [Google Scholar]
- 13. Chodavarapu R.K., Feng S., Bernatavichute Y.V., Chen P.Y., Stroud H., Yu Y., Hetzel J.A., Kuo F., Kim J., Cokus S.J. et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010; 466:388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gelfman S., Cohen N., Yearim A., Ast G.. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res. 2013; 23:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S.. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011; 479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yearim A., Gelfman S., Shayevitch R., Melcer S., Glaich O., Mallm J.P., Nissim-Rafinia M., Cohen A.H., Rippe K., Meshorer E. et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015; 10:1122–1134. [DOI] [PubMed] [Google Scholar]
- 17. Li-Byarlay H., Li Y., Stroud H., Feng S., Newman T.C., Kaneda M., Hou K.K., Worley K.C., Elsik C.G., Wickline S.A. et al. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:12750–12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iannone C., Valcarcel J.. Chromatin's thread to alternative splicing regulation. Chromosoma. 2013; 122:465–474. [DOI] [PubMed] [Google Scholar]
- 19. Hagleitner M.M., Lankester A., Maraschio P., Hulten M., Fryns J.P., Schuetz C., Gimelli G., Davies E.G., Gennery A., Belohradsky B.H. et al. Clinical spectrum of immunodeficiency, centromeric instability and facial dysmorphism (ICF syndrome). J. Med. Genet. 2008; 45:93–99. [DOI] [PubMed] [Google Scholar]
- 20. Matarazzo M.R., De Bonis M.L., Vacca M., Della Ragione F., D’Esposito M.. Lessons from two human chromatin diseases, ICF syndrome and Rett syndrome. Int. J. Biochem. Cell Biol. 2009; 41:117–126. [DOI] [PubMed] [Google Scholar]
- 21. Weemaes C.M., van Tol M.J., Wang J., van Ostaijen-ten Dam M.M., van Eggermond M.C., Thijssen P.E., Aytekin C., Brunetti-Pierri N., van der Burg M., Graham Davies E. et al. Heterogeneous clinical presentation in ICF syndrome: correlation with underlying gene defects. Eur. J. Hum. Genet. 2013; 21:1219–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin B., Tao Q., Peng J., Soo H.M., Wu W., Ying J., Fields C.R., Delmas A.L., Liu X., Qiu J. et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis, and immune function. Hum. Mol. Genet. 2007; 17:690–709. [DOI] [PubMed] [Google Scholar]
- 23. Matarazzo M.R., Boyle S., D’Esposito M., Bickmore W.A.. Chromosome territory reorganization in a human disease with altered DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:16546–16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blanco-Betancourt C.E., Moncla A., Milili M., Jiang Y.L., Viegas-Pequignot E.M., Roquelaure B., Thuret I., Schiff C.. Defective B-cell-negative selection and terminal differentiation in the ICF syndrome. Blood. 2004; 103:2683–2690. [DOI] [PubMed] [Google Scholar]
- 25. Velasco G., Hube F., Rollin J., Neuillet D., Philippe C., Bouzinba-Segard H., Galvani A., Viegas-Pequignot E., Francastel C.. Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:9281–9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeanpierre M., Turleau C., Aurias A., Prieur M., Ledeist F., Fischer A., Viegas-Pequignot E.. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum. Mol. Genet. 1993; 2:731–735. [DOI] [PubMed] [Google Scholar]
- 27. Gisselsson D., Shao C., Tuck-Muller C.M., Sogorovic S., Palsson E., Smeets D., Ehrlich M.. Interphase chromosomal abnormalities and mitotic missegregation of hypomethylated sequences in ICF syndrome cells. Chromosoma. 2005; 114:118–126. [DOI] [PubMed] [Google Scholar]
- 28. Yehezkel S., Segev Y., Viegas-Pequignot E., Skorecki K., Selig S.. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 2008; 17:2776–2789. [DOI] [PubMed] [Google Scholar]
- 29. Ji W., Hernandez R., Zhang X.Y., Qu G.Z., Frady A., Varela M., Ehrlich M.. DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat. Res. 1997; 379:33–41. [DOI] [PubMed] [Google Scholar]
- 30. Prada D., Gonzalez R., Sanchez L., Castro C., Fabian E., Herrera L.A.. Satellite 2 demethylation induced by 5-azacytidine is associated with missegregation of chromosomes 1 and 16 in human somatic cells. Mutat. Res. 2012; 729:100–105. [DOI] [PubMed] [Google Scholar]
- 31. Ehrlich M., Buchanan K.L., Tsien F., Jiang G., Sun B., Uicker W., Weemaes C.M., Smeets D., Sperling K., Belohradsky B.H. et al. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphogenesis genes. Hum. Mol. Genet. 2001; 10:2917–2931. [DOI] [PubMed] [Google Scholar]
- 32. Gatto S., Della Ragione F., Cimmino A., Strazzullo M., Fabbri M., Mutarelli M., Ferraro L., Weisz A., D’Esposito M., Matarazzo M.R.. Epigenetic alteration of microRNAs in DNMT3B-mutated patients of ICF syndrome. Epigenetics. 2010; 5:427–443. [DOI] [PubMed] [Google Scholar]
- 33. Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Glaus P., Honkela A., Rattray M.. Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics. 2012; 28:1721–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anders S., Reyes A., Huber W.. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012; 22:2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cho V., Mei Y., Sanny A., Chan S., Enders A., Bertram E.M., Tan A., Goodnow C.C., Andrews T.D.. The RNA-binding protein hnRNPLL induces a T cell alternative splicing program delineated by differential intron retention in polyadenylated RNA. Genome Biol. 2014; 15:R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. da Huang W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 40. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 41. Matarazzo M.R., Lembo F., Angrisano T., Ballestar E., Ferraro M., Pero R., De Bonis M.L., Bruni C.B., Esteller M., D’Esposito M. et al. In vivo analysis of DNA methylation patterns recognized by specific proteins: coupling CHIP and bisulfite analysis. Biotechniques. 2004; 37:666–673. [DOI] [PubMed] [Google Scholar]
- 42. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Eberhard C. et al. Tathe Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016; 44:W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E.. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012; 13:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heyn H., Vidal E., Sayols S., Sanchez-Mut J.V., Moran S., Medina I., Sandoval J., Simo-Riudalbas L., Szczesna K., Huertas D. et al. Whole-genome bisulfite DNA sequencing of a DNMT3B mutant patient. Epigenetics. 2012; 7:542–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simo-Riudalbas L., Diaz-Lagares A., Gatto S., Gagliardi M., Crujeiras A.B., Matarazzo M.R., Esteller M., Sandoval J.. Genome-wide DNA methylation analysis identifies novel hypomethylated non-pericentromeric genes with potential clinical implications in ICF syndrome. PLoS One. 2015; 10:e0132517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith Z.D., Gu H., Bock C., Gnirke A., Meissner A.. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009; 48:226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang K., Wu Z., Liu Z., Hu G., Yu J., Chang K.H., Kim K.P., Le T., Faull K.F., Rao N. et al. Selective demethylation and altered gene expression are associated with ICF syndrome in human-induced pluripotent stem cells and mesenchymal stem cells. Hum. Mol. Genet. 2014; 23:6448–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Berman B.P., Weisenberger D.J., Aman J.F., Hinoue T., Ramjan Z., Liu Y., Noushmehr H., Lange C.P., van Dijk C.M., Tollenaar R.A. et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 2012; 44:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hahn M.A., Wu X., Li A.X., Hahn T., Pfeifer G.P.. Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. PLoS One. 2011; 6:e18844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schubeler D.. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015; 520:243–247. [DOI] [PubMed] [Google Scholar]
- 52. Illingworth R.S., Gruenewald-Schneider U., Webb S., Kerr A.R., James K.D., Turner D.J., Smith C., Harrison D.J., Andrews R., Bird A.P.. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010; 6:e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deaton A.M., Webb S., Kerr A.R., Illingworth R.S., Guy J., Andrews R., Bird A.. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011; 21:1074–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kulis M., Queiros A.C., Beekman R., Martin-Subero J.I.. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim. Biophys. Acta. 2013; 1829:1161–1174. [DOI] [PubMed] [Google Scholar]
- 55. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pelechano V., Steinmetz L.M.. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013; 14:880–893. [DOI] [PubMed] [Google Scholar]
- 57. Wagner E.J., Carpenter P.B.. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012; 13:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tufarelli C., Stanley J.A., Garrick D., Sharpe J.A., Ayyub H., Wood W.G., Higgs D.R.. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 2003; 34:157–165. [DOI] [PubMed] [Google Scholar]
- 60. Lyle R., Watanabe D., te Vruchte D., Lerchner W., Smrzka O.W., Wutz A., Schageman J., Hahner L., Davies C., Barlow D.P.. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 2000; 25:19–21. [DOI] [PubMed] [Google Scholar]
- 61. Latos P.A., Pauler F.M., Koerner M.V., Senergin H.B., Hudson Q.J., Stocsits R.R., Allhoff W., Stricker S.H., Klement R.M., Warczok K.E. et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012; 338:1469–1472. [DOI] [PubMed] [Google Scholar]
- 62. Li Q., Su Z., Xu X., Liu G., Song X., Wang R., Sui X., Liu T., Chang X., Huang D.. AS1DHRS4, a head-to-head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14110–14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lev Maor G., Yearim A., Ast G.. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015; 31:274–280. [DOI] [PubMed] [Google Scholar]
- 64. Rigbolt K.T., Prokhorova T.A., Akimov V., Henningsen J., Johansen P.T., Kratchmarova I., Kassem M., Mann M., Olsen J.V., Blagoev B.. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 2011; 4:rs3. [DOI] [PubMed] [Google Scholar]
- 65. Rhee I., Veillette A.. Protein tyrosine phosphatases in lymphocyte activation and autoimmunity. Nat. Immunol. 2012; 13:439–447. [DOI] [PubMed] [Google Scholar]
- 66. Hermiston M.L., Xu Z., Weiss A.. CD45: a critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 2003; 21:107–137. [DOI] [PubMed] [Google Scholar]
- 67. Maunakea A.K., Chepelev I., Cui K., Zhao K.. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013; 23:1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chang X., Li B., Rao A.. RNA-binding protein hnRNPLL regulates mRNA splicing and stability during B-cell to plasma-cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E1888–E1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Oberdoerffer S., Moita L.F., Neems D., Freitas R.P., Hacohen N., Rao A.. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008; 321:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Preussner M., Schreiner S., Hung L.H., Porstner M., Jack H.M., Benes V., Ratsch G., Bindereif A.. HnRNP L and L-like cooperate in multiple-exon regulation of CD45 alternative splicing. Nucleic Acids Res. 2012; 40:5666–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Qiu C., Sawada K., Zhang X., Cheng X.. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 2002; 9:217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ladd P.D., Smith L.E., Rabaia N.A., Moore J.M., Georges S.A., Hansen R.S., Hagerman R.J., Tassone F., Tapscott S.J., Filippova G.N.. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 2007; 16:3174–3187. [DOI] [PubMed] [Google Scholar]
- 73. Loos F., Loda A., van Wijk L., Grootegoed J.A., Gribnau J.. Chromatin-mediated reversible silencing of sense-antisense gene pairs in embryonic stem cells is consolidated upon differentiation. Mol. Cell. Biol. 2015; 35:2436–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li J., Moazed D., Gygi S.P.. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 2002; 277:49383–49388. [DOI] [PubMed] [Google Scholar]
- 75. Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P. et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005; 123:581–592. [DOI] [PubMed] [Google Scholar]
- 76. Xie L., Pelz C., Wang W., Bashar A., Varlamova O., Shadle S., Impey S.. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011; 30:1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R.Z., Ragozin S., Jeltsch A.. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 2010; 285:26114–26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Di Ruscio A., Ebralidze A.K., Benoukraf T., Amabile G., Goff L.A., Terragni J., Figueroa M.E., De Figueiredo Pontes L.L., Alberich-Jorda M., Zhang P. et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013; 503:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schmitz K.M., Mayer C., Postepska A., Grummt I.. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010; 24:2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Marina R.J., Sturgill D., Bailly M.A., Thenoz M., Varma G., Prigge M.F., Nanan K.K., Shukla S., Haque N., Oberdoerffer S.. TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. EMBO J. 2016; 35:335–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fu Y., Dominissini D., Rechavi G., He C.. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014; 15:293–306. [DOI] [PubMed] [Google Scholar]
- 82. Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E. et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013; 23:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Durandy A., Kracker S., Fischer A.. Primary antibody deficiencies. Nat. Rev. Immunol. 2013; 13:519–533. [DOI] [PubMed] [Google Scholar]
- 84. Xu Z., Weiss A.. Negative regulation of CD45 by differential homodimerization of the alternatively spliced isoforms. Nat. Immunol. 2002; 3:764–771. [DOI] [PubMed] [Google Scholar]
- 85. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.