Abstract
Replication of the linear chromosomes of soil bacteria Streptomyces proceeds from an internal origin towards the telomeres, followed by patching of the resulting terminal single-strand overhangs by DNA synthesis using terminal proteins as the primer, which remains covalently bound to the 5΄ ends of the DNA. In most Streptomyces chromosomes, the end patching requires the single-strand overhangs, terminal protein Tpg, and terminal associated protein Tap. The telomere overhangs contain several palindromic sequences capable of forming stable hairpins. Previous in vitro deoxynucleotidylation studies indicated that Tap adds the Palindrome I sequence to Tpg, which is extended by a polymerase to fill the gap. In this study, the stringency of Palindrome I sequence was examined by an in vitro deoxynucleotidylation system and in vivo replication. Several nt in Palindrome I were identified to be critical for priming. While the first 3 G on the template were required for deoxynucleotidylation in vitro, deletions of them could be suppressed by the presence of dGTP. In vivo, deletions of these G were also tolerated, and the telomere sequence was restored in the linear plasmid DNA. Our results indicated that the truncated telomeres were repaired by extension synthesis by Tap on the foldback Palindrome I sequence.
INTRODUCTION
Soil bacteria of the genus Streptomyces possess linear chromosomes that are capped by terminal proteins (TP) covalently bound to the 5΄ ends of the DNA (reviewed in 1). The telomere sequences of typical Streptomyces chromosomes are highly conserved for approximately 170 bp, which contain several tightly-packed palindromic sequences with potential to form very stable secondary structures (2,3). On these chromosomes, the TPs are also highly conserved in sequence and size (185 aa), encoded by a terminally located tpg gene. Tpg contains a DNA-binding thumb domain of HIV reverse transcriptase (4,5) and a functional nuclear localization signal with unclear biological role, if any (6).
On most Streptomyces chromosomes, tpg is part of an operon with an equally conserved tap (telomere-associated protein) gene. Tap contains a DNA binding helix-turn-helix domain, and interacts with both Tpg and the telomere sequences (7). Both Tpg and Tap are required for synthesis of the linear chromosomes.
TP-capped linear plasmids are also present in many Streptomyces species. For them the TPs and telomeres sequences are more diversified (8). In some extreme cases, they differ from the chromosomal counterparts in sequences and sizes, such as those in the SCP1 plasmid of Streptomyces coelicolor A3 (2,9).
Replication of both the liner chromosomes and linear plasmids in Streptomyces proceeds in two steps: (i) bidirectional replication from an internal origin that leaves single-strand overhangs at the telomeres; (ii) TP-primed DNA synthesis that patches the single-strand gaps at the telomeres (‘end patching’) (10). Initiation of replication of the Streptomyces chromosomes at oriC is similar to those in other bacteria. In contrast, the end patching mechanism involving the single-strand telomere overhangs, Tpg and Tap is unique among bacteria.
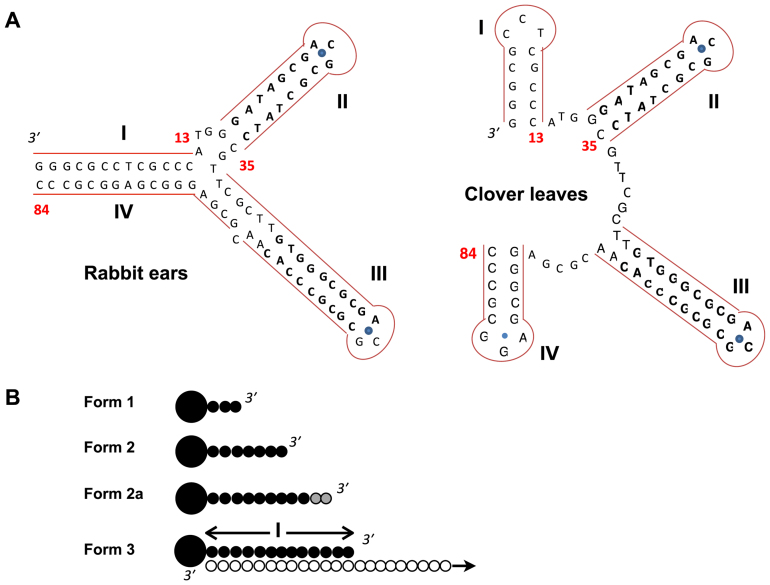
The single-strand telomere overhangs of most Streptomyces chromosomes (including the model species Streptomyces coelicolor and Streptomyces lividans) contain seven palindromes in the first 180 nt (3). The first 13 nt of the telomere is Palindrome I, which is conserved among most Streptomyces chromosomes. As part of the telomere overhangs, it may assume two alternative secondary structures of approximately equal thermodynamic stability (Figure 1A). In the Rabbit Ears structures, Palindrome I and part of Palindrome IV form a duplex. In the Clover Leaves structure, Palindromes I and IV each form a hairpin loop. Between these two palindromes, Palindromes II and III contain the binding sites for Tap (7).
Figure 1.
Structures of the telomere overhangs and the deoxynucleotidylation products. (A) Two most stable secondary structures of the 3΄ telomere overhang of the S. coelicolor chromosome. The secondary structures of the 330-nt 3΄ telomeric overhang of the S. coelicolor chromosome (TO330) were predicted using mfold. The two most stable structures differ only in the first 84 nt (TO84) spanning the first four palindromes (denoted by Roman numerals) as shown here. In the Rabbit Ears structure (ΔG = −140.1 kJ/mol), Palindrome I and part of Palindrome IV form a 13-bp duplex. In the Clover Leaves structure (ΔG = −140.9 kJ/mol), Palindromes I and IV form two separate hairpins. The Arabic numbers denote the positions of nucleotides from the 3΄ end. The G:A sheared pairing in the GNA loops is indicated by a blue dot. (B) Different forms of products of oligonucleotide elongation. Form 1. Tpg (large filled circle) linked to the first 3 nt (small filled circle). Form 2. Tpg linked to the first 7 nt. Form 2a. Tpg linked to the first 9 to 11 nt, the exact length of which was not determined (indicated by the shading). Form 3. Tpg linked to the first 13 nt (Palindrome I) and complexed with the template strand (open circles), the length of which varied in different experiments.
The hairpin formed by these palindromes in the overhangs consists of 3 nt with the sequence of GNA, in which the two purines probably form ‘sheared paring’, resulting in a highly compact conformation with a single unpaired nt in the loops (11). Such compact structure may account for the resistance of these hairpin structures to attack by single strand nucleases.
In other TP-capped linear replicons, such as the well-studied adenoviruses and Φ29 phage, TP-primed DNA synthesis is catalyzed by a B-family DNA polymerase. Streptomyces, however, do not possess any B-family DNA polymerase, and DNA polymerase I is not required for replication of the linear chromosomes (12). Instead, one of the translesion-synthesis DNA polymerases, DinB1 or DinB2, was genetically identified to be essential for end patching (13).
More recently, an in vitro deoxynucleotidylation system was established using purified, Tap, Tpg and DinB1, and single-strand telomere template (14). Unexpectedly, DinB1 was not required in the in vitro deoxynucleotidylation of Tpg. Instead, Tap was shown to act as a DNA polymerase that added the first 13 nt to Tpg. This was surprising, because Tap does not possess any DNA polymerase motif.
In vitro the first 13 nt are necessary and sufficient as the template (albeit at a low efficiency) for deoxynucleotidylation of Tpg by Tap. Longer single-strand overhangs that include the Tap binding site(s) are better templates for deoxynucleotidylation.
In vitro Tap adds only the first 13 nt to Tpg. Extension of synthesis beyond this oligonucleotide cannot be carried out by DinB1 added in vitro. Instead Escherichia coli Klenow fragment can perform the extension (14). It is possible that another factor (such as a helicase) was necessary for DinB1 to carry out the extension. Thus, end patching involves a unique two-stage priming process. In the first stage, Tpg and Tap act as a protein primer and a special DNA primase, respectively, to synthesize the first 13 nt. In the second stage, the 13 nt acts as a DNA primer for continuous DNA synthesis to fill the single-strand gaps.
In this study, we extended the in vitro deoxynucleotidylation studies to identify crucial sequences in Palindrome I by specific mutations. In the stem of the hairpin, several nt were identified that were important for priming and/or extension. The length of the stem appeared to be critical as well. Some changes in the 3-nt loop were tolerated, and some were not, presumably depending on the stability of the resulting loops.
The most interesting results concerned the first three G at the 3΄ end. Any base substitutions in them blocked priming, but deletions of up to all three G could be tolerated if dGTP was present in the deoxynucleotidylation system. Further investigation revealed that the missing G at the 3΄ ends were restored by the DNA polymerase activity of Tap acting on the foldback structure of the truncated template. Such repair of the terminal truncated templates was also demonstrated in vivo. These results indicated an additional role of Tap in maintaining the structural integrity of the Streptomyces telomeres.
MATERIALS AND METHODS
Prediction of secondary structure of single-strand DNA
The secondary structures of the telomere overhangs were predicted using the mfold server (http://unafold.rna.albany.edu/?q=mfold) based on a temperature of 30°C and an ionic strength of 50 mM NaCl.
In vitro deoxynucleotidylation
Single-stranded template DNA of various lengths was synthesized by Scientific Biotech (Taiwan) and Life Technologies (Japan). They bore the designation ‘TO’ (telomeric overhang) followed by a number indicating the length (in nt). For example, TO13 and TO35 designated the first 13 and 35 nt of the telomeric overhang, respectively.
Purification of Tap and Tpg and in vitro deoxynucleotidylation followed Yang et al. (14) with minor modifications. Typical priming reaction mixtures (in 20 μl) contained template DNA (in various concentrations), 60 nM Tap, 600 nM Tpg, a [α-32P]-labeled dNTP (0.165 μM), 1.0% glycerol, 50 mM NaCl and 50 mM imidazole in K buffer (10 mM Tris–HCl, pH 7.5, 7 mM MgCl2, 0.1 mM dithiothreitol). For extension synthesis, non-radioactive dNTPs prepared in deionized water were added to the reaction mixture to give a final concentration of 1 μM. The reaction mixture was typically incubated at 30°C for 60 min for TO13 and 10 min for TO35 and longer templates, and stopped by filtration through a G-25 spin columns pre-equilibrated with 0.1% SDS (for subsequent electrophoresis) or de-ionized water (for further manipulations). Reaction products were analyzed on 12% or 15% SDS-PAGE. After electrophoresis, the incorporated radioactivity was determined by autoradiography without an enhancer screen followed by analysis using ImageJ software of the National Institute of Health (https://imagej.nih.gov/ij/index.html).
Construction of linear plasmids with mutant Palindrome I
Escherichia coli-Streptomyces shuttle plasmid pLUS980 (15) and its derivatives were used for generating linear plasmids in transformed Streptomyces. pLUS980 contained a linear plasmid sequence consisting of a pair of telomeres of the S. lividans chromosome flanking a thiostrepton resistance gene (tsr), the autonomously replicating sequence (ARS) of linear plasmid pSLA2 (16), and the tap-tpg operon. Mutant telomere DNA was synthesized by polymerase chain reaction (PCR), and used to replace the wild-type telomeres on pLUS980 by restriction digestion and ligation (The oligonucleotides used in the PCR reactions are listed in Supplementary Data, Table S1). The resulting pLUS980 variant DNA was cut in the E. coli sequence with AseI, and used to transform S. lividans MR04 (17). Linear plasmid DNA in the thiostrepton-resistant transformants was detected by gel electrophoresis and restriction digestion.
Identification of linear plasmid and determination of terminal sequences
Transformants obtained from pLUS980 derivatives were initially screened for the presence of linear plasmids by agarose gel electrophoresis. DNA of approximately 13 kb was purified and digested by SpeI, which was expected to cut the linear plasmid DNA into two fragments of 5 and 8 kb (4).
To determine the telomere sequences of the linear plasmid, the plasmid DNA was purified from the agarose gel and treated with DNase-free RNase (Takara, Japan), followed by proteinase E digestion and phenol–chloroform extraction. Terminal deoxynucleotide transferase (Takara, Japan) was used to add protruding poly(A) to the 3΄ ends of the DNA. Two-step PCR was employed to amplify the telomeric DNA; two sets of primers were designed. The first set consisted of M13+polyT (GTAAAACGACGGCCAGTA + T15) and a sequence at 1.4 kb from the end (TCATCACTGACGAATCGA). The second set consisted of M13 and a sequence at 550 bp from the end (TGTGTTCAGTGACGCGAT). The PCR products were cloned into TA vector and subjected to sequencing according to Huang et al. (3).
RESULTS AND DISCUSSION
G1, G2, G3, C6 and T8 are required for the addition of the first 3 C to Tpg
In the previous in vitro deoxynucleotidylation study, Tap can add to Tpg the 13-nt Palindrome I sequence, which remains duplexed with the template (designated ‘Form 3’; Figure 1B) (14). The molecular weight of Form 3 varied with the lengths of the templates. In addition to Form 3, two intermediates were produced: Form 1 (apparent molecular weight 26 kD) corresponding to Tpg attached with 1 to 3 C, and Form 2 (apparent molecular weight 28 kD) corresponding to Tpg attached with the first 7 nt of Palindrome I (Figure 1B).
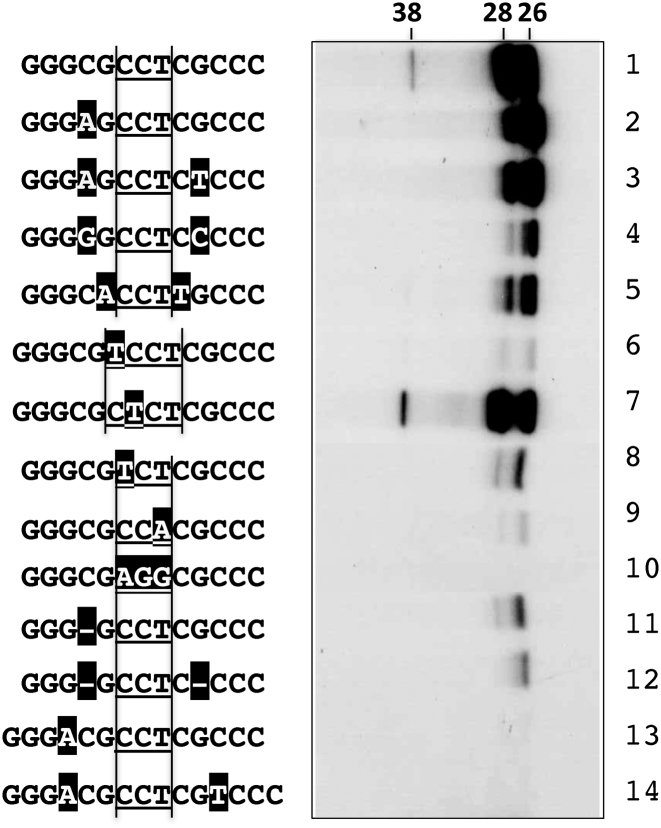
To examine the role of each nt in Palindrome I in the production of Form 1, a series of single-base transitional substitutions was performed on TO13 from the 3΄ end inward (Figure 2, left panel). Herein, all the template DNA sequences are presented and numbered in the 3΄ to 5΄ direction for better reference. Thus, the first variant contained a G1A substitution, the second one a G2A substitution, and so on. These substitution variants were tested for their ability to support production of Form 1 by measuring the incorporation of [α-32P]-dCMP to Tpg in the absence of the other dNTP.
Figure 2.
Involvement of the TO13 sequence in initial priming. In vitro deoxynucleotidylation was carried out in the presence of [α-32P]-dCTP and absence of the other dNTP. The template was TO13 and its variants. Left panel. Single transitional substitution variants of TO13. From the 3΄ end, one nt at a time was changed consecutively. The substitutions are shown shaded in black. The 3-nt loops are underlined and bracketed. The deoxynucleotidylated product Form 1, Tpg-(dCMP)3, exhibited a molecular weight of 26 kD. Right panel. Incremental restoration of the wild-type TO13 sequence from the surrogate template ST13. The ST13 sequence is shaded in black.
The absence of other Palindromes in the template simplified the interpretation of the results at the expense of lower efficiency due to the lack of Tap binding sites. TO13 supported the initial deoxynucleotidylation (lane 14), when the reaction time was increased from 10 to 60 min and the concentration of the template DNA was increased by 10 fold. Substitution in G1 to G3 (lanes 1–3), C6 (lane 6) and T8 (lane 8) eliminated Form 1 production, whereas the other substitutions did not. In fact, the G10A (lane 10) and C12T (lane 12) substitutions strongly increased deoxynucleotidylation.
In parallel, we constructed a surrogate template sequence ST13, AAATATTCTATTT, which represented TO13 with a transitional substitution in every nt (Figure 2, right panel, lane 1). Starting with ST13, variants were constructed in which the TO13 sequence was restored incrementally one nt at a time from the 3΄ end (lanes 2–14, right panel). These variants were tested for their ability to support the production of Form 1. As expected, the production of Form 1 was not detected (lanes 1–3) until the first three nt (GGG) were restored (lane 4). When the first 6 nt were restored, deoxynucleotidylation became stronger (lane 7). Unexpectedly, further restoration of the seventh nt diminished deoxynucleotidylation (lane 8). Beyond that, deoxynucleotidylation increased with the number of restored nt until it reached a maximum of 11 nt (lane 12), and then it decreased again after that. The maximum activity achieved with 11 restored nt mirrored the maximum activity observed in the C12T substitution (left panel lane 12).
These results showed the critical role of the first three G in the production of Form 1. The block of Form 1 production caused by the C6 or T8 substitution in the hairpin loop was unexpected, and suggested that the downstream CCT loop in the Clover Leaves structure played a role in the initial priming.
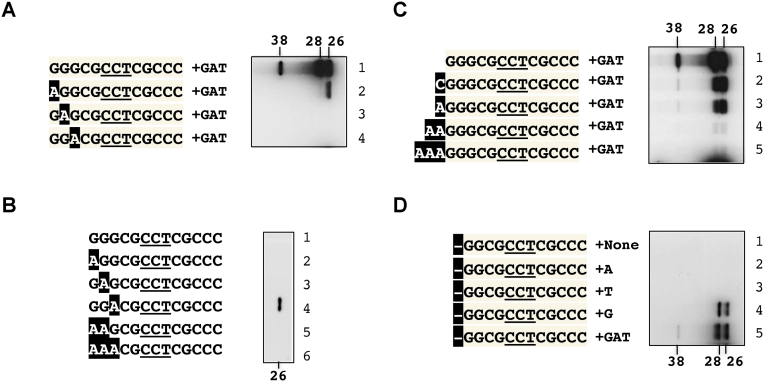
C4, G5, and G10 are important for elongation of Form 1
To test elongation of Form 1, non-radioactive dGTP, dTTP and dATP were added to the reactions (Figure 3). With TO13, Form 1 was extended to a length of ∼10 nt with an apparent molecular weight of 29 kD (left panel, lane 11). This slightly extended Form 2 was designated ‘Form 2a’ (Figure 1B). Without the supplements, no elongation was observed beyond Form 1 (lane 12). Supplemented with only dGTP only, a labeled product of approximately 28 kD (Form 2) was observed (lane 13).
Figure 3.
Involvement of the Palindrome I sequence in primer extension. The in vitro deoxynucleotidylation reactions were carried out as described in Figure 2 except for the addition of non-radioactive dGTP, dTTP, and dATP (+GAT). The templates were TO13 and selected substitution variants (left panel) and incremental restoration variants (right panel) in Figure 2. The products of TO13 in the presence of all three non-radioactive dNTP (lane 11), no non-radioactive dNTP (+none; lane 12), and only dGTP (+G, lane 13) provided the size markers. For reference, the same marker lanes are repeated in both panels, which were all electrophoresed in the same polyacrylamide gel.
Among the ten TO13 variants containing a substitution in position 4 to 13 (left panel, lanes 1–10), the C6T (lane 3) and T8C (lane 5) substitutions completely blocked deoxynucleotidylation. The C4T (lane 1) and G5A (lane 2) substitutions produced Form 1, but diminished elongation. The G10A substitution (lane 7) produced a relatively large quantity of Form 1, but elongation stopped at Form 2 (28 kD). The other substitutions, C7T (lane 4), C9T (lane 6), C11T (lane 8), C12T (lane 9), and C13T (lane 10) extended as far as TO13 did (lane 11).
The results with the incremental restoration variants (right panel) were in good agreement with the single substitution variants (left panel), except for the variant with eight restored nt (lane 6), which extended Form 1 to Form 2 (instead of Form 2a).
The reason why the substitutions in C4, G5 or G10 diminished elongation was not clear. Neither was it clear why the G10A substitution resulted in a strong efficiency of elongation that stopped at the seventh nt.
The two loop-closing base pairs in Palindrome I are involved in primer elongation
Interestingly, the central 7-nt CGCCTCG (the loop sequence underlined) loop segment in Palindrome I is identical to that in the single-strand promoter of phage N4 (18,19). NMR studies indicated that this sequence forms a 3-nt hairpin loop structure, which is as stable as its complementary sequence 5΄GCGGAGC3΄ (20). In the latter, the loop is closed by a G:A sheared base pairing, leaving a single unpaired G, whereas, in the former, all three nt in the loop are unpaired, but employ extensive base-stacking rearrangements to stabilize the hairpin structure with minor backbone distortion.
The role of CGCCTCG in priming was examined in in vitro deoxynucleotidylation using TO35, which contained Palindromes I and II, as the template. The inclusion of Palindrome II, which containing a Tap-binding site, in the template increased the efficiency of deoxynucleotidylation. With all four dNTP provided, Forms 1 and 2 were produced in relatively large quantities accompanied by a small quantity of Form 3 (of approximately 38 kD), which represented Tpg-CCCGCTTCGCGGG duplexed with the template (Figure 4, lane 1). The two relatively strong pausing sites at C3 and G7 were reported previously (14).
Figure 4.
Involvement of the two closing base pairs and loop of Palindrome I in deoxynucleotidylation. The effect of substitutions in the two closing bp and the loop of the Palindrome I hairpin on initial priming and extension were examined. TO35 was used as the template. The reaction time was 10 min with three non-radioactive dNTP added. Only the Palindrome I sequences were shown. The size markers (lane I) were provided by the products of TO35, 26 (Form 1), 28 (Form 2) and 38 kD (Form 3).
The C4A substitution in TO35 reduced the quantities of Forms 2 and 3 (lane 2). The C4A/G10T double substitution also yielded the same result (lane 3). Further reduction was seen in the C4G/G10C (lane 4) and G5A/C9T (lane 5) double (and complementary) substitutions. These results indicated that C4 and G5 were important for elongation beyond C3.
Deletion of C4 alone (lane 11) or accompanied by the complementary deletion of G10 (lane 12) strongly reduced all three products. Insertion of an A upstream of C4 alone (lane 13) or with a complementary insertion of T downstream of G10 (lane 14) eliminated them. These results indicated the importance of the sequence as well as the length of the stems in the initial priming and subsequent elongation.
The CCT loop is also important for elongation of the primer
To test the role of the CCT loop, a T was inserted into the loop to form a TCCT (lane 6) or a CTCT (lane 7) tetranucleotide loop. Interestingly, the TCCT loop almost eliminated the priming reaction completely, whereas the CTCT loop increased Form 3 production. The C6T (lane 8) or T8A substitution (lane 9) in the loop strongly reduced the initial priming and elongation. The substitution of the CCT loop sequence with its complementary sequence AGG (lane 10) eliminated priming.
In N4 phage, the CCT loop and the neighboring two-bp stems (CG:CG) are also crucial, mutations in which result in complete loss or impairment of the promoter function (18,19). The reason was attributed to the extensive interaction among C6 and T8 with the stem bases (20). In our study, the only variation (CTCT; lane 7) that did not reduce elongation contained an insertion of T but preserved C6 and T8.
The first three G are absolutely required for priming
Protein-priming DNA synthesis shares a sliding-back mechanism or its variation. The termini of these replicons possess repeated sequences of a single nt or short oligonucleotides. Deoxynucleotidylation initiates at an nt near the 3΄ end of the template that incorporates one unit of the repeats (one nt or oligonucleotide) to the TP. The TP-nt initiation product then slides (for single nt repeat) or jumps (for oligonucleotide repeat) to the first repeat to recover the complete sequence by extension synthesis.
In Φ29 and the related phage GA-1, initiation of DNA replication takes place at the 3΄ second nt of the template (3΄AAA and 3΄TTT, respectively) (21,22). The Φ29-related phage Nf and the Streptococcus pneumoniae phage Cp-1 initiate replication at the 3΄ third nucleotide of the template (3΄TTT) (23,24). E. coli phage PRD1 initiates at the 3΄ fourth nucleotide (3΄CCCC) (25). For adenovirus DNA, which has a duplication of GTA (3΄GTAGTA), initiation occurs at the 3΄ fourth to sixth template positions to form a TP-CAT initiation product, which jumps back to pair with the terminal GTA (26).
The telomere overhangs of typical Streptomyces chromosomes begin with a G triplet, which may also accommodate a sliding back mechanism. We investigated this possibility. Firstly, each of the three terminal G in TO35 was substituted by an A, and the resulting variants were tested for incorporation of [α-32P]-dCMP in the presence of non-radioactive dGTP, dATP and dTTP (Figure 5A). The results showed that the G1A variant produced labeled Form 1 (lane 2), but not the G2A or G3A variant (lanes 3–4). These results were consistent with the working of a sliding back mechanism, in which dCMP was incorporated opposite the 3΄ second or third G on the template.
Figure 5.
The critical role of the terminal sequences. The roles of the first three G are tested. (A) Effect of G1A (lane 2), G2A (lane 3), and G3A (lane 4) substitutions on TO35 on priming. [α-32P]-dCTP was used to label the products in the presence of non-radioactive dNTP. (B) Effect of G1A (lane 2), G2A (lane 3), G3A (lane 4), G1A/G2A and G1A/G2A/G3A substitutions on TO35 on priming. [α-32P]-dTTP was used to label the products in the absence of non-radioactive dNTP, and MgCl2 was replaced by MnCl2 in 1 mM. (C) Effect of addition of extra nt at the 3΄ end of TO35 on priming. [α-32P]-dCTP was used to label the products in the presence of non-radioactive dNTP. (D) Deletion of G1 (indicated by the hyphen) on TO35 on priming. [α-32P]-dCTP was used to label the products in the presence of non-radioactive dNTP as indicated. The reaction time was 30 min.
To distinguish whether the 3΄ second or third G is the initiation site, each of them was substituted by A and tested for incorporation of radioactive T in the reaction (Figure 5B). In these reactions, Mg+2 was replaced by Mn+2 as was used in the case of Φ29 (22). The results showed that only the variant containing the G3A substitution was incorporated with radioactive T (lane 4). Therefore, we concluded that initiation of deoxynucleotidylation takes place opposite the third G. The same results were obtained using a longer template TO52 (data not shown).
When the first two (lane 5) or three (lane 6) G were replaced by A, there was no incorporation of T, indicating that AA and AAA could not provide a sliding-back mechanism. The inability of AAA to provide initiation indicated that the one or both of the first two nt was also involved in initiation. Moreover, replacement of the first three G with three C also failed to incorporate [α-32P]-dGMP (data not shown).
These results indicated that the initiation of Tpg deoxynucleotidylation takes place at the third nt of the template and then slide back to the fist nt to recover the complete sequence by extension synthesis, similar to that in phage Nf and Cp-1.
A third question was whether the G triplet must reside at the 3΄ end of the template. This is an important issue, since copies of the Palindrome I sequence are found internally on Streptomyces chromosomes (e.g. seven in S. coelicolor). May these Palindrome sequences serve as the template for Tpg deoxynucleotidylation fortuitously? To address this question, three TO35 variants were created, in which one to three A were added to the 3΄ end. These variants were tested for deoxynucleotidylation in the presence of [α-32P]-dCTP and three non-radioactive dNTP. The results (Figure 5C, lanes 3–5) showed that deoxynucleotidylation was reduced ∼10 folds with each added A. Addition of a C (lane 2) also had a similar effect.
dGTP suppresses the deletion of terminal G of the telomere
We also constructed variant TO35-1 by removing the first 3΄ G fromTO35, and tested it for in vitro deoxynucleotidylation (Figure 5D). As expected it did not produce [α-32P]-dCMP labeled Form 1 in the absence of non-radioactive dNTP (lane 1). Interestingly, when the other dNTPs were added to the reaction, labeled Forms 1–3 were produced (lane 5). Adding non-radioactive dATP alone (lane 2) or dTTP alone (lane 3) yielded no labeled products, but adding non-radioactive dGTP yielded Forms 1 and 2 (lane 4).
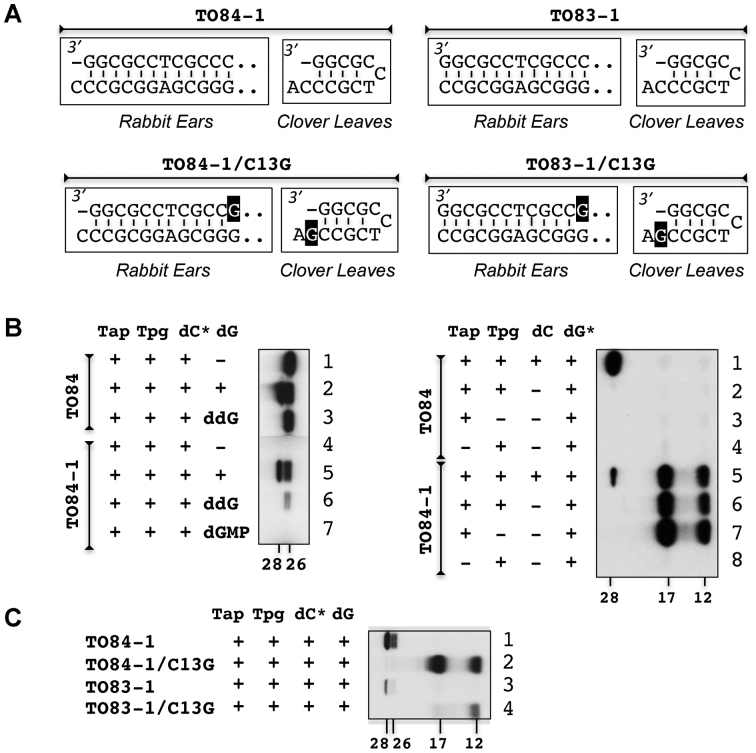
The ability of added dGTP to suppress the 3΄ G deletion and allow the production of Forms 1 and 2 was surprising. To investigate further, we created another pair of longer templates, TO84 and TO84-1. In TO84-1, the 3΄ G of TO84 was deleted (Figure 6A, top left panel). These two templates were tested for production of [α-32P]-dCMP-labeled Forms 1 and 2 (B, left panel). Palindromes I-IV, included in these templates, allowed formation of Rabbit Ears structure as well as Clover Leave structure (Figure 1A).
Figure 6.
Restoration of the missing G in the telomere overhang. (A) The four telomere templates used. Only their Rabbit Ears and Clover Leaves structures are shown (in boxes). TO84-1 is TO84 with G1 deletion. TO83-1 is TO84-1 with C84 deletion. TO84-1/C13G and TO83-1/C13G each further contained a C13G substitution (shaded in black). (B) Left panel. Effect of the G1 deletion in TO84-1 on priming. [α-32P]-dCTP (dC*) was used to label the products in the absence of non-radioactive dNTP (lanes 1, 4), non-radioactive dGTP (dG; lanes 2, 5), non-radioactive dideoxy GTP (ddG; lanes 3, 6), or non-radioactive dGMP (lane 7). Right panel. Incorporation of dGTP in TO84-1. [α-32P]-dGTP (dG*) was used to label the product in the presence or absence of non-radioactive dCTP (dC). In some reactions, Tpg (lanes 3, 7) or Tap (lanes 4, 8) was omitted. (C) Effect of the C13G substitution on extension synthesis repair. Incorporation of [α-32P]-dCTP was examined in the deoxynucleotidylation reaction in the presence of non-radioactive dGTP using the four template variants.
TO84, which spanned Palindromes I to IV, produced [α-32P]-dCMP-labeled Form 1 in the absence of dGTP (lane 1), and both Forms 1 and 2 in the presence of dGTP (lane 2) as expected. Substitution of dGTP with the chain terminator ddGTP blocked the production of Form 2, but not Form 1 (lane 3), also as expected. In comparison, TO84-1 did not produce Form 1 or 2 in the absence of dGTP (lane 4), but produced both in the presence of dGTP (lane 5), again indicating suppression of the 3΄ G deletion by the supplemented dGTP. Replacement of dGTP by ddGTP (lane 6), blocked the formation of Form 2 but not Form 1, indicating that the addition of one G was sufficient for suppression.
These results suggested a telomere repair mechanism in which the missing 3΄ G was restored by Tap-catalyzed extension synthesis using the template provided by the Clover Leaves and/or Rabbit Ears structure (Figure 6 A, top left panel). This model predicted that dGMP was added to the 3΄ end of the truncated template during the deoxynucleotidylation reaction. To test this hypothesis, [α-32P]-dGTP was used to replace [α-32P]-dCTP in the reaction (B, right panel).
In the presence of non-radioactive dCTP, both TO84 and TO84-1 produced 32P-labeled Form 2 (lanes 1, 5). TO84-1 produced less of it, but, interestingly, it produced two additional labeled products with apparent molecular weights of 17 and 12 kD (lane 5). Omission of dCTP in the reaction blocked the production of 32P-labeled Form 2 by either template (lanes 2, 6), but not the 17- and 12-kD products by TO84-1 (lane 6), indicating that the incorporation of dGTP did not require the presence of dCTP. Removal of Tpg from the reaction also did not block the 17- and 12-kD production (lane 7), indicating that these products were not attached to Tpg. Removal of Tap blocked the production of the 17- and 12-kD products (lane 8), indicating its role in the production.
These results indicated that the dGMP-labeled 17- and 12-kD products were not the result of deoxynucleotidylation of Tpg. Most likely they represented the restored template with a dGMP added at the 3΄ end. TO84-1 in either the Clover Leaves or Rabbit Ears structures (Figure 6A, top left panel) provided a 5΄ C as the template for the dGMP incorporation. It was possible that the 17- and 12-kD products represented the extension products in these two structures.
Repair of the truncated telomere occurs at the Clover Leaves structure
To investigate the origins of the 17- and 12-kD products, TO84-1/C13G, which was a variant of TO84-1 with a C13G substitution, was created (Figure 6A, lower left panel). Extension synthesis on TO84-1C13G was possible for both Rabbit Ears and Clover Leaves structures. However, in the Clover Leaves structure, extension synthesis would add a C, resulting in a defective telomere sequence. In the Rabbit Ears structure, extension synthesis would add a G, restoring the wild type sequence.
In the in vitro reaction in the presence of [α-32P]-dCTP and non-radioactive dGTP, TO84-1/C13G produced neither Form 1 nor Form 2, but the 17- and a 12-kD products instead (C, lanes 1, 2). The failure to produce Form 1 or 2 indicating that the missing 3΄ G was not restored on the template. One of the dCTP-labeled products would be the extension product of the Clover Leaves structure, but the identity of the other was unclear. It was not clear whether extension synthesis had occurred on the Rabbit Ears structure. If it had, the product could not be detected because the added G was not radioactive, and it was not a functional template probably due to the C13G mutation.
To investigate further, we removed C84 from the 5΄ end of TO84-1 to yield TO83-1. Lacking both 5΄ C84 and 3΄ G ends, the I-IV duplex formed in the Rabbit Ears structure of TO83-1 would be blunt-ended, and thus no extension synthesis was possible (A, right top). In contrast, extension synthesis remained possible in the Clover Leaves structure, which possesses a 5΄ C template. In the reaction containing [α-32P]-dCTP and non-radioactive dGTP, TO83-1 produced radioactive Forms 1 and 2 (C, lane 3), but in less quantity than TO84-1 (lane 1). The results supported the suppression of the G deletion by extension synthesis based on the Clover Leaves structure albeit with a low efficiency.
A variant of TO83-1, which contained a C13G substitution, was created and designated TO83-1/1C13G (A, lower right panel). Like TO83-1, extension synthesis on the Rabbit Ears structure was impossible, and, in the Clover Leaves structure, extension was possible, but the added nt (C) would result in a defective telomere template. When TO83-1/C13G was used as the template in the same deoxynucleotidylation reaction, no deoxynucleotidylation was detected, as expected. [α-32P]-dCTP labeled 12-kD product was detected, but not 17-kD product (C, lane 4). These results indicated that the 12-kD product was generated by extension synthesis on the Clover Leaves structure.
These results indicated that the Rabbit Ears structure was not involved in telomere restoration. It is possible that this conformation is not favorable for existence in the reaction conditions, or it is not a suitable substrate for repair. In either case, it is unlikely to be the 17-kD product, because no interconversion between the 12- and 17-kD products was observed by extensive heating or other denaturation treatments (data not shown). The identity of the 17-kD product remained unclear.
Deletion of up to 4 terminal nt may be restored
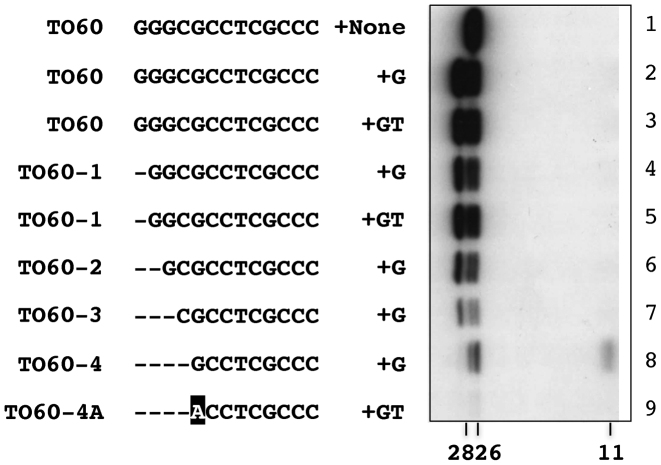
To find out the possible extent of deletions that might be repaired in vitro, a series of derivatives of TO60, which contained Palindromes I-III (for higher efficiency of deoxynucleotidylation), with a deletion of one to four terminal nt was created, and tested for incorporation of [α-32P]-dCTP in the presence of added dGTP (+G) or dGTP plus dTTP (+GT; Figure 7). Deletion of one G had a slight effect on the production of Forms 1 and 2 (lanes 4, 5). Deletions of 2 and 3 G further reduced production of Forms 1 and 2 (lanes 6, 7). When four nt (GGGC) were deleted (TO60-4), Form 1 was produced, but not Form 2 (lane 8). A further G5A substitution in TO60-4 (TO60-4A) blocked deoxynucleotidylation essentially completely (lane 9).
Figure 7.
The efficiency of restoration of missing terminal nt by Tap. The reactions were carried out as described in Figure 2 in the presence of no added non-radioactive dNTP (+none), non-radioactive dGTP (+G), or dGTP plus dTTP (+GT). The templates used were TO60 and its variants. TO60-1, TO60-2, TO60-3 and TO60-4 lacked 1, 2, 3 and 4 nt from the 3΄ end, respectively. TO60-4A lacked four nt from the 3΄ end and contained a G5A substitution. The 11 kD [α-32P]-dCTP product corresponds to the repaired TO60-4 (the repaired products of the other variants were not labeled and therefore undetectable).
For TO60-4, [α-32P]-dCTP was apparently used for restoration of the missing C4 as dictated by the fold-back template. The relatively low concentration of the [α-32P]-dCTP presumably account for the faint signal of the 11-kD repair product. For the other truncated templates, the repair products were expected to incorporate only non-radioactive G and not radioactive C, and therefore not detected. These results supported that the extension synthesis was based on the Palindrome I sequence.
Truncated telomeres may be repaired in vivo
To test whether some of the effects of various mutations in the telomere observed in vitro might also be observed in vivo, we employed an E coli-Streptomyces shuttle plasmid, pLUS980 (27), which harbored a linear plasmid sequence consisting of a pair of telomeres flanking the tsr (thiostrepton resistance) gene, the ARS of linear plasmid pSLA2, and the tap-tpg operon. When pLUS980 is cut by a restriction enzyme (e.g., AseI) in the E. coli sequence outside of the linear plasmid sequence, and introduced into Streptomyces, a Tpg-capped linear plasmid is found in the thiostrepton-resistant transformants (27).
Variants of Palindrome I were created. They were introduced into both telomeres on pLUS980 to avoid possible repair through intramolecular gene conversion. The resulting plasmid DNA was linearized with AseI, and used to transform S. lividans MR04 (17), a mutant in which the terminal sequences including the tap-tpg operon are deleted and the chromosome is circularized. The lack of chromosomal telomeres in MR04 prevented recombination between the telomere sequences on the plasmid and those on the chromosome. Thiostrepton-resistant transformants were selected and checked for the presence of linear plasmids.
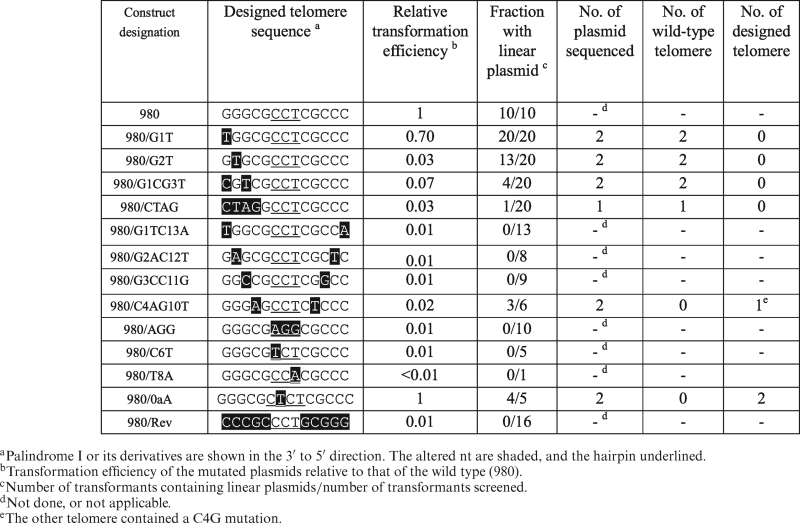
The variants in which the telomeres suffered substitutions of 1– 4 bp from the 3΄ ends generated linear plasmid albeit with decreasing efficiencies (980/G1T, 980/G1TG2T, 980/G1CG3T, 980/CTAG; Table 1). Since these telomere sequences were flanked by E. coli sequences, terminal nt substitutions were equivalent to terminal deletions. Thus, these results were consistent with the in vitro repair of terminal deletions of up to 4 bp (Figure 7). Of the linear plasmids produced by these variants, seven telomere sequences in all four cases were isolated and determined. The wild type Palindrome I sequences were restored in all of them.
Table 1. In vivo repair of Palindrome I sequences on linear plasmids.
|
Pairwise comparison provided useful information. While variant 980/G1T was repaired at a relatively high efficiency, 980/G1TC13A, with an additional C13A complementary substitution was not. For 980/G1TC13A, although restoration of the missing G1 was impossible on the Clover Leaves structure (with A13 being the incorrect template), it was possible on the Rabbit Ears structure. This confirmed the conclusion from the in vitro studies that the Rabbit Ears structure was not involved in telomere restoration.
Similarly, the two variants with G2A-C12T and G3C-C11G complementary substitutions did not produce any linear plasmids. The variant C4A-G10T, on the other hand, produced linear plasmids with the unaltered sequences. This was consistent with the in vitro observation that the fourth nt in the template could tolerate substitutions without affecting priming (Figures 2 and 4).
The in vivo results of variants in the loop sequence were also consistent with the in vitro results. Of the four variants tested, only 980/0aA, in which a T was inserted between C6 and C7 (CTCT loop), produced linear plasmids. The wild type sequence was not restored in two telomere sequences on these plasmids examined. The other three variant plasmids did not produce linear plasmids. In vitro, only the variant with the CTCT loop provided a strong template activity (Figure 4).
The extension repair is likely catalyzed by Tap
Based on these in vitro and in vivo results, we propose the following model of extension repair (Figure 8). The fold-back hairpin of Palindrome I of the 3΄ overhang serves as the template for the extension repair (A). The Tap–Tpg complex is attracted through binding to neighboring Palindromes II and III. Tap fills in the missing 3΄ nt and stops at the boundary of Palindrome I (B). Palindrome I is then opened up and Tap catalyzes deoxynucleotidylation of Tpg (C). The synthesis ends also at the boundary of Palindrome I, and Tap leaves.
Figure 8.
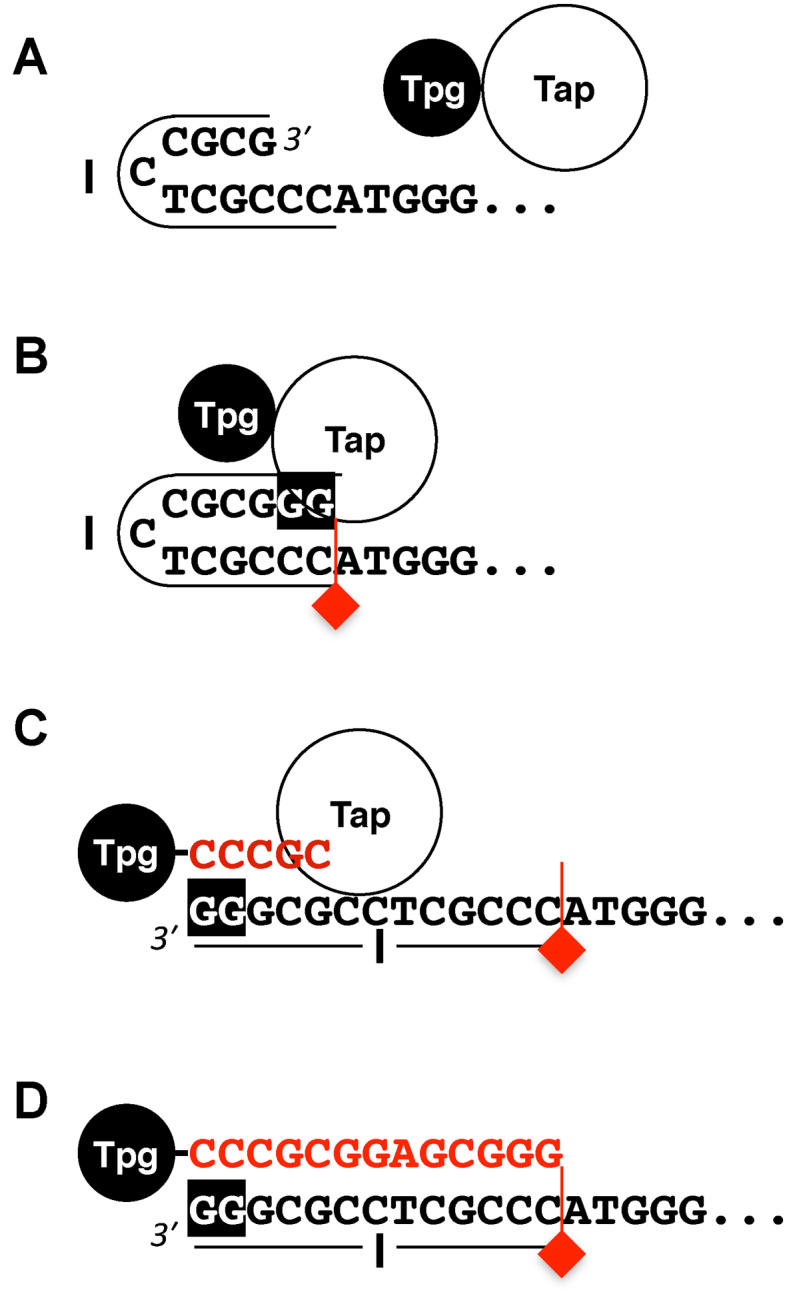
Model of telomere repair and priming by Tap. (A) The fold-back hairpin of Palindrome I (outlined) of the 3΄ overhang in the Clover Leaves structure serve as the template for repair, which is shown here with two missing G at the 3΄ end. The Tpg-Tap complex arrives through binding to neighboring Palindromes II and III (not shown). (B) Tap fills in the two missing G (shaded in black), and stops at the boundary of Palindrome I (indicated by the red inverted flag). (C) The hairpin is opened up and Tap deoxynucleotidylates Tpg using Palindrome I (underlined) as the template. The newly synthesized nt are shown in red. (D) The synthesis also stops at the boundary of Palindrome I, and Tap leaves.
We propose that the repair in vivo is catalyzed by Tap instead of a different DNA polymerase, because the other polymerase would likely carry out the extension synthesis beyond Palindrome I, resulting in a defective substrate for End Patching. On the other hand, The Tap-catalyzed Tpg-primed synthesis terminates after the completion of Palindrome I, and it is reasonable to assume it also stops at the same junction when it performs the repair synthesis. The signal that stops Tap during Tpg-primed extension would likely stops Tap during repair synthesis.
The Tap-catalyzed repair is important for telomere protection during replication
Previously, using the same experimental procedure, Qin and Cohen (28) observed repair of a truncated telomere with a 2- or 4-bp deletion on a linear plasmid. Because the other telomere on the plasmid was intact, these authors attributed the restoration to gene conversion between the truncated and intact telomeres. In view of our results, the telomere repair they observed could be mediated by the extension synthesis mechanism instead.
In each round of replication of linear plasmids and chromosomes in Streptomyces, the approaching replication forks displace the telomere DNA, and expose the 3΄ ends as single-stranded overhangs, which are potential targets for exonuclease attack. The Tap-catalyzed extension repair of the truncated telomeres may play an important role in maintaining the integrity of these linear replicons.
Generation of linear plasmids from linearized artificially constructed plasmid fragments (such as pLUS980), on the other hand, requires the precise trimming of extrinsic (E. coli) sequences flanking the telomere sequences. In the transformed mycelia, the linearized fragments harboring the linear plasmid sequence undergo rounds of bidirectional replication that progressively shorten the daughter DNA. Based on our in vitro observations, a functional telomere may be established by end patching only when the flanking E. coli sequence (of hundreds of bp) is completely removed and the end of the template 3΄ strands suffers no more than 4 nt of deletion. With no deletion, no repair is required. With 1- to 4-nt deletions, the truncated telomeres may be repaired by Tap-mediated extension synthesis. This suggested that, unless an unknown mechanism for precise trimming exists, most linear plasmids recovered from the transformants were repair products.
Moreover, the probability that a 3΄ telomere overhang is shortened to within 0 to 4 nt deletion would be extremely low. This notion was supported by the following observation. CY9804, a variant of pLUS980 containing two consecutive copies of the 330-bp telomere sequences at both termini, was constructed. MR04 transformed by AseI-linearized CY9804 DNA produced linear plasmids. There were four possible combinations for the presence of the telomere copies on the plasmids: (i) two copies at both ends; (ii) one copy at both ends; (iii) two at the left and one at the right and (iv) one at the left and two at the right. Interestingly, the four combinations were observed at approximately equal frequencies (data not shown). Given the probability of a functional telomere being established at an embedded telomere sequence being p, the probability of the telomere being established at the proximal copy instead of the distal copy would be (1 − p) • p. For these two probabilities to be approximately equal, i.e., p ≈ p • (1 − p), p must be exceedingly small.
No similar telomere repair mechanism has been reported in the TP-priming replication of the whole TP-capped replicons such as adenoviruses and Φ29. This is probably due to basic biochemical differences between the two systems. For Streptomyces linear replicons, replication is initiated at an internal origin, which, upon reaching the telomeres, produces 3΄ single-strand overhangs, and the TP-priming reaction takes place on these single-strand overhangs. In the likes of adenoviruses and Φ29, replication is initiated by TP-primed synthesis at the telomeres in a double stranded state. During the process, the parental TP-capped 5΄ strand is displaced. The presence of TP protects the overhangs from exonucleolytic attacks. This may account for the reported lack of a telomere repair mechanism for these TP-capped replicons.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chung Yuan Christian University and a research grant [MOST 103-2311-B-033-001] from the Ministry of Science and Technology, Taiwan, Republic of China. Funding for open access charge: Chung Yuan Christian University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kirby R., Chen C.W.. Dyson P. Streptomyces: Molecular Biology and Biotechnology. 2011; Norfolk: Caister Academic Press; 5–26. [Google Scholar]
- 2. Lin Y.-S., Kieser H.M., Hopwood D.A., Chen C.W.. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 1993; 10:923–933. [DOI] [PubMed] [Google Scholar]
- 3. Huang C.-H., Lin Y.-S., Yang Y.-L., Huang S.-w., Chen C.W.. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol. 1998; 28:905–926. [DOI] [PubMed] [Google Scholar]
- 4. Yang C.-C., Huang C.-H., Li C.-Y., Tsay Y.-G., Lee S.-C., Chen C.W.. The terminal proteins of linear Streptomyces chromosomes and plasmids: A novel class of replication priming proteins. Mol. Microbiol. 2002; 43:297–305. [PubMed] [Google Scholar]
- 5. Bao K., Cohen S.N.. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 2001; 15:1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai H.-H., Huang C.-H., Lin A.M., Chen C.W.. Terminal proteins of Streptomyces chromosome can target DNA into eukaryotic nuclei. Nucleic Acids Res. 2008; 36:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bao K., Cohen S.N.. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev. 2003; 17:774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C.W. Meinhardt F, Klassen R. Microbial Linear Plasmids. 2007; Berlin, Heidelberg: Springer-Verlag; 33–61. [Google Scholar]
- 9. Huang C.H., Tsai H.H., Tsay Y.G., Chien Y.N., Wang S.L., Cheng M.Y., Ke C.H., Chen C.W.. The telomere system of the Streptomyces linear plasmid SCP1 represents a novel class. Mol. Microbiol. 2007; 63:1710–1718. [DOI] [PubMed] [Google Scholar]
- 10. Chang P.C., Cohen S.N.. Bidirectional replication from an internal origin in a linear Streptomyces plasmid. Science. 1994; 265:952–954. [DOI] [PubMed] [Google Scholar]
- 11. Chou S.-H., Zhu L., Reid B.R.. Sheared purine-purine pairing in biology. J. Mol. Biol. 1997; 267:1055–1067. [DOI] [PubMed] [Google Scholar]
- 12. Huang T.W., Chen C.W.. DNA polymerase I is not required for replication of linear chromosomes in Streptomyces. J. Bacteriol. 2008; 190:755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsai H.H., Shu H.W., Yang C.C., Chen C.W.. Translesion-synthesis DNA polymerases participate in replication of the telomeres in Streptomyces. Nucleic Acids Res. 2012; 40:1118–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang C.C., Tseng S.M., Chen C.W.. Telomere-associated proteins add deoxynucleotides to terminal proteins during replication of the telomeres of linear chromosomes and plasmids in Streptomyces. Nucleic Acids Res. 2015; 43:6373–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang C.C., Sun W.C., Wang W.Y., Huang C.H., Lu F.S., Tseng S.M., Chen C.W.. Mutational analysis of the terminal protein Tpg of Streptomyces chromosomes: identification of the deoxynucleotidylation site. PLoS One. 2013; 8:e56322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qin Z., Cohen S.N.. Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol. Microbiol. 1998; 28:893–904. [DOI] [PubMed] [Google Scholar]
- 17. Redenbach M., Flett F., Piendl W., Glocker I., Rauland U., Wafzig O., Leblond P., Cullum J.. The Streptomyces lividans 66 chromosome contains a 1 Mb deletogenic region flanked by two amplifiable regions. Mol. Gen. Genet. 1993; 241:255–262. [DOI] [PubMed] [Google Scholar]
- 18. Dai X., Kloster M., Rothman-Denes L.B.. Sequence-dependent extrusion of a small DNA hairpin at the N4 virion RNA polymerase promoters. J. Mol. Biol. 1998; 283:43–58. [DOI] [PubMed] [Google Scholar]
- 19. Dai X., Greizerstein M.B., Nadas-Chinni K., Rothman-Denes L.B.. Supercoil-induced extrusion of a regulatory DNA hairpin. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:2174–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chou S.H., Tseng Y.Y., Chu B.Y.. Stable formation of a pyrimidine-rich loop hairpin in a cruciform promoter. J. Mol. Biol. 1999; 292:309–320. [DOI] [PubMed] [Google Scholar]
- 21. Illana B., Blanco L., Salas M.. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1. Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J. Mol. Biol. 1996; 264:453–464. [DOI] [PubMed] [Google Scholar]
- 22. Mendez J., Blanco L., Esteban J.A., Bernad A., Salas M.. Initiation of φ29 DNA replication occurs at the second 3΄ nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:9579–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin A.C., Blanco L., Garcia P., Salas M., Mendez J.. In vitro protein-primed initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3΄ nucleotide of the linear template: a stepwise sliding-back mechanism. J. Mol. Biol. 1996; 260:369–377. [DOI] [PubMed] [Google Scholar]
- 24. Longás E., Villar L., Lazaro J.M., de Vega M., Salas M.. Phage phi29 and Nf terminal protein-priming domain specifies the internal template nucleotide to initiate DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:18290–18295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caldentey J., Blanco L., Bamford D.H., Salas M.. In vitro replication of bacteriophage PRD1 DNA. Characterization of the protein-primed initiation site. Nucleic Acids Res. 1993; 21:3725–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. King A.J., van der Vliet P.C.. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 1994; 13:5786–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang C.H., Chen C.Y., Tsai H.H., Chen C., Lin Y.S., Chen C.W.. Linear plasmid SLP2 of Streptomyces lividans is a composite replicon. Mol. Microbiol. 2003; 47:1563–1576. [DOI] [PubMed] [Google Scholar]
- 28. Qin Z., Cohen S.N.. Survival mechanisms for Streptomyces linear replicons after telomere damage. Mol. Microbiol. 2002; 45:785–794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.