Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that attenuate expression of their mRNA targets. Here, we developed a new method and an R package, to easily infer candidate miRNA–mRNA target interactions that could be functional during a given biological process. Using this method, we described, for the first time, a comprehensive integrated analysis of miRNAs and mRNAs during human normal plasma cell differentiation (PCD). Our results reveal 63 miRNAs with significant temporal changes in their expression during normal PCD. We derived a high-confidence network of 295 target relationships comprising 47 miRNAs and 141 targets. These relationships include new examples of miRNAs that appear to coordinately regulate multiple members of critical pathways associated with PCD. Consistent with this, we have experimentally validated a role for the miRNA-30b/c/d-mediated regulation of key PCD factors (IRF4, PRDM1, ELL2 and ARID3A). Furthermore, we found that 24 PCD stage-specific miRNAs are aberrantly overexpressed in multiple myeloma (MM) tumor plasma cells compared to their normal counterpart, suggesting that MM cells frequently acquired expression changes in miRNAs already undergoing dynamic expression modulation during normal PCD. Altogether, our analysis identifies candidate novel key miRNAs regulating networks of significance for normal PCD and malignant plasma cell biology.
INTRODUCTION
Plasma cells are highly specialized cells representing the end stage of B cell differentiation. They play an important role in humoral immunity by synthesizing and secreting antibodies protecting the host against infections (1). Activation of B cells leads to their differentiation into a transitional preplasmablast (prePB), a highly proliferating cell population (2). These preplasmablasts further differentiate into plasmablasts (PBs), which can develop into quiescent long-lived plasma cells after migrating to survival niches in the bone marrow (3,4). On the transcriptional level, the differentiation of B cells into plasma cells is associated with substantial and coordinated changes in the gene expression profile (4), which fall into two main categories: the loss of B cell-associated transcripts and the acquisition of plasma cell gene expression program. These changes are tightly guided by two sets of stage-specific transcription factors (TFs) that repress each other: i) B cell TFs (PAX5, BCL6 and BACH2) maintaining the B cell fate and ii) plasma cell TFs (IRF4, BLIMP1 and XBP1) that are required to extinguish the B cell genes and activate the antibody-secreting cell (ASC) program (4,5). Plasma cell differentiation (PCD) is initiated by the transcription factor IRF4, which activates PRDM1 (encoding BLIMP1) (6). BLIMP-1 coordinates PCD by inducing plasma cell-specific genes including XBP-1 and silencing the B cell gene-expression program in plasma cells (5,7). It induces the transcription of immunoglobulin genes, which is substantially increased from plasmablast to plasma cell stages (4). Furthermore, BLIMP1 regulates the expression switch from the membrane-bound form of the immunoglobulin to its secreted form by activating the transcription-elongation factor ELL2, which results in the secretion of large amounts of immunoglobulins (4,7).
To achieve this elevated antibody production, the endoplasmic reticulum (ER) of ASCs undergoes expansion in a process that requires continuous ER stress and activation of the unfolded protein response (UPR), resulting in adjustment of protein synthesis, enhancement of the ER folding capacity, increased degradation of misfolded proteins and enhanced ER biogenesis (8–10). The transcription factor XBP-1, a downstream of BLIMP1 activated by the UPR (11), plays a central role in regulating the UPR gene-expression program (12), and as a consequence, is essential for the secretion of immunoglobulins by plasma cells (12,13).
Although the role of the complex network of transcription factors involved in PCD has been investigated, mechanisms regulating key PCD transcription networks remain poorly known. MicroRNAs (miRNAs) are single-stranded non-coding RNAs of about 18–24 nucleotides that regulate gene expression by binding complementary sites in target messenger RNAs (mRNAs), typically resulting in the degradation of target mRNAs or the inhibition of protein translation (14). Recent studies have shown that miRNAs participate in various biological functions including differentiation and cell fate decision (15,16), immune system, tumorigenesis and cell death (17). Furthermore, there is an increasing recognition of the role of miRNAs in multiple myeloma, a plasma cell (PC) malignancy characterized by an accumulation of malignant PCs within the bone marrow (18–25). Research groups have started to address the role of miRNAs in PCD (26). However, little is known about miRNA expression during human PCD as well as about the full extent to which individual miRNAs regulate fundamental processes during PCD. A complete delineation of miRNA and their target expression during normal PCD is essential to understand the role of miRNAs in plasma cell malignancies.
We analyzed the expression profile of miRNAs and mRNAs during human PCD to infer miRNA–target relationships, as well as in multiple myeloma tumor plasma cells. We developed a method and an R package (miRTarget, https://github.com/kassambara/miRTarget), that uses miRNA and mRNA expression profiles across PCD cell subpopulations to infer candidate miRNA–target interactions that could be active and functional in PCD. We inferred miRNA–target relationships from sequence-based prediction, experimentally validated target interactions curated from the literature, published data from miRNA perturbation experiments and inverse expression relationships between miRNAs and their target mRNAs.
Our results reveal 63 miRNAs with significant temporal changes in their expression during normal PCD. We derived a high-confidence network of 295 target relationships comprising 47 miRNAs and 147 targets. These relationships include new examples of miRNAs that are likely to coordinately regulate multiple members of critical pathways associated with PCD. We have experimentally validated a role for the miRNA-mediated regulation of key PCD transcription factors. Furthermore, our work demonstrates that PCD stage-specific miRNAs are aberrantly overexpressed in multiple myeloma cells (MMCs) compared to the normal counterparts, suggesting that MMCs frequently acquired expression changes in miRNAs already undergoing dynamic expression modulation during normal PCD.
MATERIALS AND METHODS
Cell populations and miRNA/mRNA expression profiling
Preplasmablasts (prePBs), plasmablasts (PBs), and plasma cells (PCs) were generated using a 3-step in vitro model starting from peripheral blood memory B cells (MBCs) as reported (2,27). We performed three experiments starting from purified memory B cells of three different healthy donors. miRNA and mRNA expression profiling were obtained using Affymetrix microarrays as described in the Supplementary Data. To identify miRNAs associated with multiple myeloma (MM), we have analyzed publicly available miRNA expression data of purified normal bone marrow plasma cells (BMPCs) from healthy donors (n = 3), purified MMCs from newly diagnosed multiple myeloma patients (n = 62) and of human myeloma cell lines (HMCLs, n = 20) representative of the molecular heterogeneity of the patients (28) (ArrayExpress accession number: E-MTAB-1363) (25). We compared MMCs and HMCLs to the healthy counterpart BMPCs by using the limma R package (adjusted P-value ≤ 0.05).
Microarray data analysis
The workflow of data processing and analysis is outlined in Figure 1. All statistical analyses and data visualization were performed with the R/Bioconductor statistic software packages as described in the Supplementary Data. The miRNA expression data were normalized with Affymetrix miRNA QC tool using the default settings. Affymetrix U133P2 mRNA expression data were preprocessed using RMA algorithm (29). To decrease the number of uninformative features, we used a filtering step: features, for which the mean expression, in at least one subgroup, did not reach a signal intensity of 6 in a log2 scale, were removed (Supplementary Figure S2). Differential expression analysis of miRNA and mRNA data sets were performed using the limma R/Bioconductor package (30) (see Supplementary Data). We were interested in identifying expression changes during each plasma cell differentiation transition. Therefore, we performed the differential expression analysis between two consecutives cell subpopulations, as reported in previous studies (2,27,31).
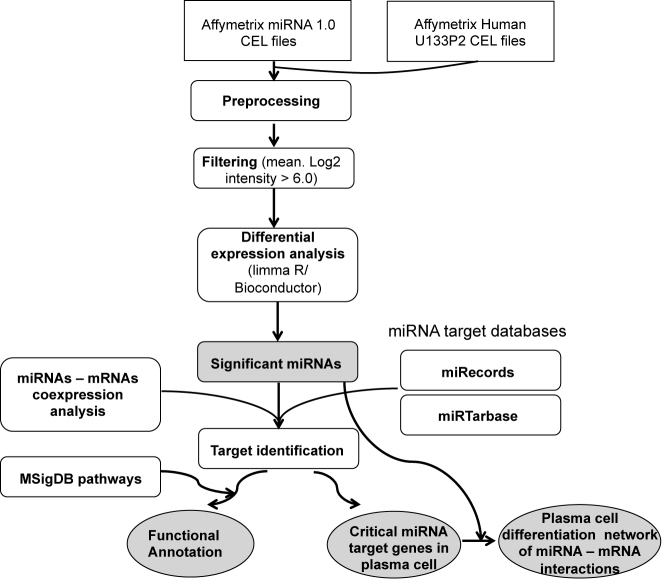
Figure 1.
Computational workflow of data processing and analysis. miRNA and mRNA array data were pre-processed as described in the Supplementary Data. A filtering step was included to select only actively expressed features. Differential expression analysis was performed using the ‘limma’ R package to identify significant differentially expressed miRNAs during plasma cell differentiation. Predicted and validated miRNA targets were collected from miRecords and miRTarbase databases. Co-expression analysis and identification of potential targets were performed directly on the paired miRNA–mRNA data. For a given miRNA, we kept only targeted genes that are differentially expressed during plasma cell differentiation and which expression is negatively correlated to the miRNA. Functional and pathway analysis of miRNA–targeted genes were performed using the clusterProfiler R package (35) and the MSigDB (34) gene set collections.
Inferring putative miRNA targets using the miRTarget R package
We developed the miRTarget R package to automatize and facilitate the procedure of inferring and validating miRNA–mRNA regulatory relationships. miRTarget includes a procedure to easily identify the predicted and validated mRNA targets given one or more miRNAs as input (https://github.com/kassambara/miRTarget). This is performed based on the union of two popular databases, miRTarBase (a database of experimentally validated miRNA targets, http://mirtarbase.mbc.nctu.edu.tw/, release 6.0) (32) and miRecords (http://c1.accurascience.com/miRecords/, version: 27 April 2013) (33). The miRecords database includes validated miRNA targets and also an integrated sequence-based miRNA target prediction resource from 11 popular miRNA target prediction programs (diana, microinspector, miranda, mirtarget2, mitarget, nbmirtar, pictar, pita, rna22, rnahybrid, targetscan). We retained only experimentally validated target genes (in either miRecords and/or miRTarbase databases) as well as those genes predicted by at least 5 of 11 available miRNA target prediction programs (in the miRecords database). The optimal number of target prediction programs can be adjusted in the miRTarget package and the rationale of choosing 5 programs is illustrated in the Supplementary Figure S1. Of the potential miRNA–target genes, only differentially expressed genes between studied cell subpopulations are kept for further analyses (limma R package, adjusted P-value for multiple testing correction ≤ 5%, fold change ≥ 2). To obtain a high-confidence list of candidate miRNA–target interactions, miRTarget package provides a convenient R function to perform correlation analysis between the expression values of each miRNA and the expression values of its deregulated target genes. Negative correlation between a given miRNA and a predicted target mRNA could be the first indicator of a potential direct interaction.
Canonical pathway enrichment analysis
Canonical pathway enrichment analysis were performed using the Molecular Signatures Database (MSigDB, v5) (34) gene set collections and the clusterProfiler R/Bioconductor package (35) as described in the Supplementary Data.
Quantitative RT-PCR for miRNA/mRNA expression validation
miRNA and mRNA expressions were measured by quantitative RT-PCR as described in the Supplementary Data. The miRNA expression was normalized to endogenous RNU6B levels using the ΔΔCt method. The mRNA expression was normalized to beta-2 microglobulin expression. All measurements were performed in triplicates following manufacturer's protocol. The primer references are in the Supplementary Table S1.
Cell culture
AMO1 and JJN3 were cultured in RPMI supplemented with 10% fetal bovine serum (FBS). XG7 were cultured in RPMI supplemented with 10% FBS and IL-6 (2 ng/ml). All cell lines were grown in 37°C humidified cell culture incubators with CO2 maintained at 5%.
miRNA transfection
Plasma cell myeloma derived cell lines AMO1 (with high expression of miR-30b/d/e) and XG7 (with low expression of miR-30b/d/e) were transfected with antisense oligonucleotides anti-miR-30/b/d/e and precursor miRNAs pre-miR-30b/d/e, respectively (see the Supplementary Data). The JJN3 cell line were transfected with antisense oligonucleotides anti-miR16 and anti-miR106b. The references of the used oligonucleotides sequences are in Supplementary Table S2. miRNA/mRNA levels were measured by quantitative RT-QPCR as mentioned above.
Data access
miRNA expression data from this study are available on GEO. mRNA expression data are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/, E-MTAB-1771, E-MEXP-2360 and E-MEXP-3034) (2,27).
RESULTS
miRNA expression profiling during human plasma cell differentiation
miRNA expression was profiled for four in-vitro human plasma cell differentiation subpopulations: memory B cells (MBCs), preplasmablasts (prePB), plasmablasts (PBs) and plasma cells (PCs). The outline of computational analysis steps is depicted in Figure 1. A multi-step approach was performed to identify significantly differentially expressed (SDE) miRNAs. First, the data were preprocessed as outlined in the Method section. After pre-processing, a total of 119 miRNAs was identified as transcriptionally active, which we defined as a mean intensity > log2 (64) in at least one cell subpopulation (Supplementary Figure S2).
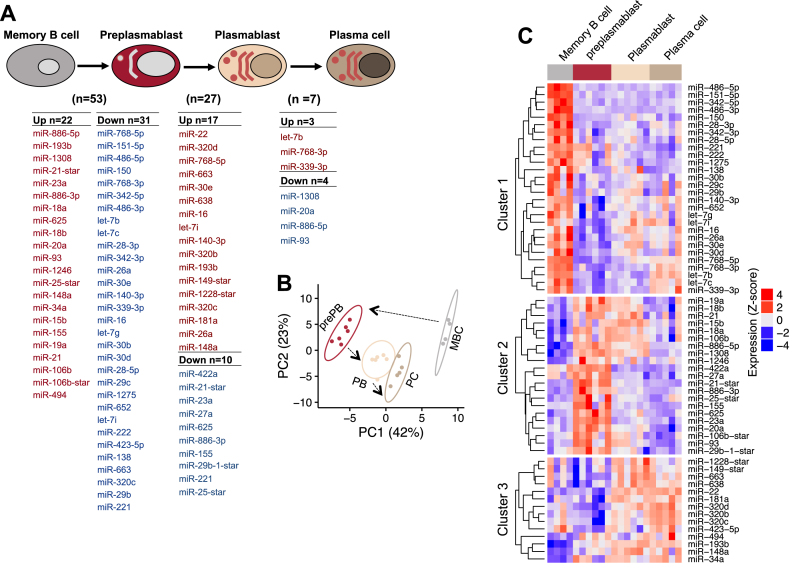
The modulation of miRNA expression patterns during plasma cell differentiation program were analyzed by generating several contrasts comparing two consecutive cell populations using the limma R package (adjusted P-value ≤ 0.05, fold change ≥ 2). The resulting significant miRNAs between the different plasma cell differentiation (PCD) transitions is shown in Figure 2, where major changes were scored between memory B cells and preplasmablasts. A total list of 63 unique miRNAs was identified as significantly differentially expressed in one or more steps during plasma cell differentiation (Figure 2 and Supplementary Table S3). Fifty three miRNAs were significantly differentially expressed during MBC to prePB transition (up: 22 miRNAs, down: 31 miRNAs); 27 miRNAs during prePB to PB transition (up: 17 miRNAs, down: 10 miRNAs); 7 miRNAs during PB to PC transition (up: 3 miRNAs, down: 4 miRNAs) (Figure 2A).
Figure 2.
Differentially expressed miRNAs during plasma cell differentiation. Differentially expressed miRNAs were identified using the limma package (adjusted P-value ≤ 0.05 and fold change ≥ 2). (A) List of differentially expressed miRNAs between two consecutive cell subpopulations. Upregulated miRNAs are represented by red color, while downregulated miRNAs are represented by blue color. miRNAs are sorted according to the P-value in ascending order. (B) Principal component analysis grouping plasma cell differentiation cell subpopulations according the expression profile of the significantly differentially expressed miRNAs. Labels represent cell subpopulations: MBC (memory B cell), prePB (preplasmablast), PB (plasmablast), PC (plasma cell). (C) Heat maps representing the expression profile of the differentially expressed miRNA. Expression values were log2 transformed and standardized before the analyses. Three miRNA clusters are detected by k-means.
In unsupervised principal component analysis (PCA) of the 63 miRNA expression levels, B cell to plasma cell subpopulations were segregated according to their developmental stage (Figure 2B). Further analyses, performed on the 63 miRNAs, identified three main clusters (Supplementary Figure S3) detected by the K-means algorithm. Hierarchical clustering based on the expression profiles of significantly regulated miRNAs is provided in Figure 2C showing the expression profile of the three miRNA clusters. Cluster 1 is expressed at greater level in MBCs compared to the other cell subtypes. Of notes, members of the miR-30 family (miR-30b/d/e) were among cluster 1 miRNAs, which expression decreased significantly during plasma cell differentiation. In agreement with this, it has been reported that miR-30 family members regulate PRDM1, a key plasma cell transcription factor (36). Cluster 2, mainly overexpressed in prePBs; and cluster 3, overexpressed in PBs and PCs. The analysis suggested a close relationship between PBs and PCs, with heat maps showing a clear demarcation between the prePBs and PBs/PCs.
Inferring miRNA targets by prediction and integrative analysis of miRNA and mRNA expression
We developed the miRTarget R package to facilitate the procedure of inferring and validating miRNA–mRNA regulatory relationships (https://github.com/kassambara/miRTarget, see Materials and Methods). It uses the union of two popular databases, miRTarBase (32), a database of experimentally validated miRNA targets, and miRecords (33).
The miRecords database includes validated miRNA targets and also an integrated sequence-based miRNA target prediction resource from 11 popular miRNA target prediction programs. We retained only experimentally-validated target genes and/or those genes predicted by at least 5 of 11 available miRNA target prediction programs. A total list of 11 972 unique microRNA–target interactions were identified for the 63 miRNAs, involving 4802 unique genes. In order to reduce false positive, we combined our miRNA and mRNA expression data sets. Of the 4802 potential miRNA–targeted genes, 1933 were differentially expressed between the four B cell to plasma cell populations (adjusted P-value ≤ 5%, fold change ≥ 2). To obtain a high-confidence list of candidate miRNA–target interactions, we performed Pearson's correlation analysis between the mean expression values of each miRNA and the mean expression values of its deregulated target genes using miRTarget R package developed by our group (https://github.com/kassambara/miRTarget). Negative correlation between a given miRNA and a predicted target mRNA could be the first indicator of a potential direct interaction. Finally, the combination of target prediction, differential expression (adjusted P-value ≤ 5%, fold change ≥ 2) and correlation filters (r < –0.6, Supplementary Figure S4) yielded 1596 negative miRNAs–mRNAs interactions involving 51 unique miRNAs and 948 unique mRNAs (Supplementary Table S4). At least 316 of the 1596 putative target interactions have experimental support in other cell types (comprising 272 unique genes and 35 miRNAs) according to miRecords and miRTarbase databases (Supplementary Table S4).
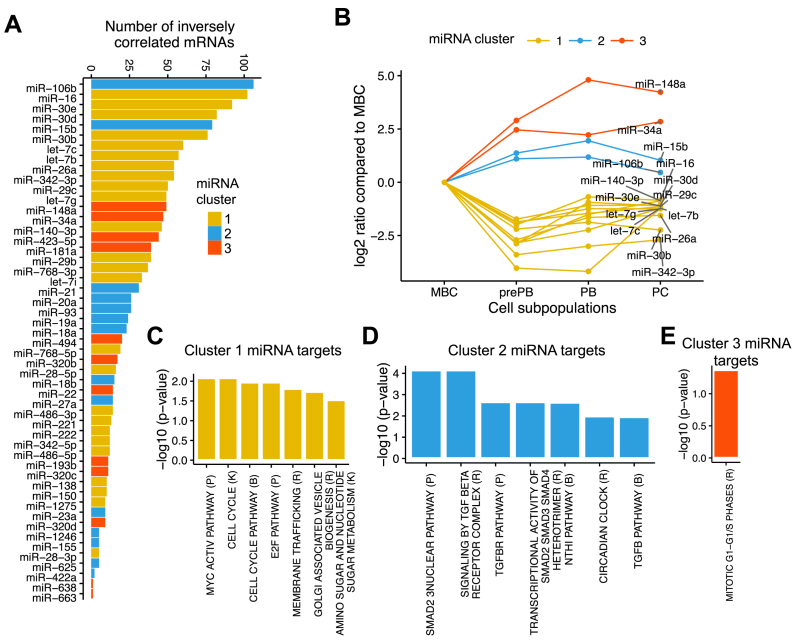
Next, we ranked the miRNAs based on the number of inversely correlated mRNA targets (Figure 3A). The first 15 miRNAs with most negatively correlated events includes miR-16, miR-30e, miR-30d, miR-30b, let-7c, let-7b, miR-26a, miR-342-3p, miR-29C, let-7g and miR-140-3p belonging to miRNA cluster 1; miR-106b and miR-15b in cluster 2; and miR-148a and miR-34a belonging to cluster 3 (Figure 3A). Figure 3B shows the mean expression of these top 15 associated miRNAs. Among the identified miRNAs highly associated to target gene expression, several have been related to plasma cell differentiation in literature, e.g. members of miR-30 family, miR-34a and miR-148a (26,36). Interestingly, our prediction reveals that these miRNAs target key genes coding for transcription factors controlling B cell or plasma cell identities. The miR-30 family overexpressed in MBCs, targets two well-known plasma cell transcription factors, IRF4 and PRDM1 mRNAs. The interaction between miR-30 family and PRDM1 has previously been studied and is functionally relevant in PCD context (36). The miR-148a and miR-34a, overexpressed in PCs, target BACH2 and FOXP1 B cell transcription factors as previously reported (26). The other miRNA–mRNA relationships represent unappreciated functional miRNA–target relationships with a potential role in plasma cell differentiation (Supplementary Table S4).
Figure 3.
Inferring miRNA targets and identification of the canonical pathways enriched in the targets. The combination of miRNA target prediction databases, differential expression and correlation filters have been used to identify miRNA targets. This analysis yielded 1596 negative miRNAs–mRNAs interactions involving 51 unique miRNAs and 948 unique mRNAs. The clusterProfiler R/Bioconductor package and MSigDB gene set collections are used for functional annotation. For each list, the top seven enriched pathways is shown (adjusted P-value ≤ 0.05). (A) Number of inversely correlated mRNAs per miRNAs. (B) Expression profiles of the top 15 miRNAs that show the highest association with gene expression of their target sets. The log2 ratio compares the expression from preplasmablast (prePB), plasmablast (PB), plasma cell (PC) to memory B cell (MBC). (C) Pathways enriched in genes targeted by miRNAs cluster 1. (D) Pathways enriched in genes targeted by miRNAs cluster 2. (E) Pathways enriched in miRNA cluster 3.
Functional analysis of dynamically regulated miRNAs
To better understand which and how biological functions are affected by the differentially expressed miRNAs during PCD, we performed functional annotations using the clusterProfiler R package (35) and MSigDB (34) canonical pathway gene set collections. We inferred the functions of deregulated miRNAs from their target genes. miRNAs that were more abundantly expressed in MBCs and down-regulated during PCD (cluster 1 miRNAs) are predicted to interact with genes coding for proteins involved cell cycle (CCND2, CCNE1/2, CDC25A, CDK6, CHEK1, E2F3, HDAC2 and RB1), MYC activation pathway (BCAT1, CAD, CCND2, CDC25A, E2F3, EIF4E, HMGA1, MTDH, POLR3D, SHMT1 and TAF12), E2F pathway (CCNE1/2, CDC25A, E2F3/6/7, MYBL2, RB1, RRM2 and TFDP1), membrane trafficking (AP2M1, AP4E1, ARCN1, CLTC, COPZ1, CPD, DNAJC6, IGF2R, MYO6, TPD52, TXNDC5, VPS25 and YIPF6), Golgi associated vesicle budding and biogenesis (AP4E1, CLTC, CPD, DNAJC6, IGF2R, TPD52, TXNDC5 and YIPF6), amino sugar and nucleotide sugar metabolism (GALE, GFPT1, GNPNAT1, NANP, PGM1, UGDH and UGP2) (Figure 3C and Supplementary Table S5). This observation suggests a potential link between the miRNA downregulation during the MBC to prePB transition and an advantage in cell proliferation, organelle biogenesis and metabolism. Conversely, the majority of miRNAs that were up-regulated at prePB stage (cluster 2) appeared to interact with genes assigned to the functional categories of TGF-β signaling pathway (Figure 3D and Supplementary Table S5). miR-106b, the miRNA with the highest number of inversely correlated target mRNAs, appeared to interact with transcripts from TGF-β signaling pathway. These genes included BAMBI, BMPR2, CCNT2, ITCH, TGFBR2, SKI and SMAD7. miRNAs from cluster 2, including miR-106b, are downregulated during prePB to PB transition leading, possibly, to the activation of TGF-β signaling pathway, which is known to be involved in cell differentiation (37). Finally, cluster 3 miRNAs, up-regulated in plasma cell subpopulations appeared to interact with transcripts from mitotic G1-G1/S phases (CCND2, CKS1B, DYRK1A, E2F3, MCM7, MYBL2, MYC, RRM2 and SKP1) (Figure 3E and Supplementary Table S5). These interactions could be responsible for the low proliferation state characterizing normal plasma cells.
Network of miRNAs regulating genes with critical role in plasma cell differentiation
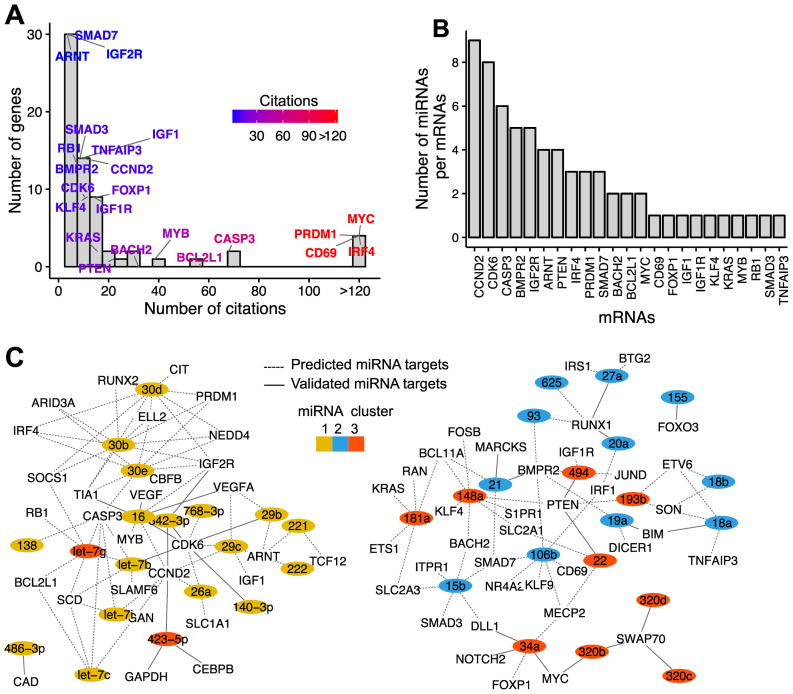
The temporal dynamic in the levels of miRNA expression at different PCD stages raised the hypothesis that miRNAs may control gene expression of key proteins regulating PCD. Therefore, we extracted the 1596 miRNA–mRNA pairs with strong negative correlation coefficient (r < –0.6), comprising 51 unique miRNAs and 948 unique mRNAs. We next investigated the extent of our current knowledge about the 948 miRNA–targeted genes in plasma cell differentiation by assessing PubMed abstracts and annotations using two key words (i) gene name + B cell differentiation and (ii) gene name + plasma cell differentiation. A citation index is computed for each gene as the average number of citations obtained using the two key words. Genes with a mean citation index ≥1 are kept as genes with functional relevance in PCD. This literature-based filter yielded 295 miRNA–mRNAs pairs corresponding to a network of 47 miRNAs and 141 target mRNAs (Supplementary Table S6). Figure 4 shows targets mRNAs with at least three citations, as well as, the number of miRNAs per target mRNAs. Our analysis reveals miRNAs interacting with critical genes, highly cited as involved in plasma cell differentiation, such as MYC, PRDM1, CD69, IRF4, CASP3, BCL2L1, MYB, BACH2, BIM1 and PTEN, with a citation index ranging from 20 to 300 (Figure 4A and Supplementary Table S6). Other miRNA targeted genes with a strong functional role in PCD included KRAS, FOXP1, IGF1R, KLF4, CDK6, CCND2, IGF1, TNFAIP3, SMAD3/7, BMPR2, RB1, IGF2R and ARNT (citation index: 3–19) (Figure 4A and Supplementary Table S6). We identified also several genes, less studied in normal plasma cell differentiation context, but with a potential importance in plasma cell biology, including PCAF, EED, NFAT5, HDAC2, DNMT3B, SKP1, CKS1B, PIK3R1 and RAB23 (citation: < 3) (Supplementary Table S6).
Figure 4.
Network of miRNAs regulating critical genes in plasma cell differentiation. Using literature-based filtering, we identified 141 miRNA targets with functional relevance in plasma cell differentiation. For better readability, we show only miRNA targeted mRNAs with at least 3 citations (39 miRNAs and 66 miRNA targets). (A) Distribution of the number of citations per gene. Some key genes with functional evidence in plasma cell differentiation are shown. Color intensity indicates the number of citations (blue: low citations, red: high citations). (B) Genes are ordered according to the number miRNAs. (C) Network of miRNA–target interactions. For better readability, Inferred network comprises miRNA targeted mRNAs with at least 3 citations (144 putative target interactions between 39 miRNAs and 66 target mRNAs). Edge line type represents the status of the interaction for a given miRNA–mRNA pair. Solid line types are miRNA - mRNA interactions with experimental support in the literature in other tissues. Dashed line types are predicted interactions. miRNAs are colored by cluster relationships (cluster 1:yellow, cluster 2: blue, cluster 3: red).
Many of these genes are potentially targeted by more than 2 miRNAs (Figure 4B and C): 8 miRNAs for CCND2 and CDK6; 6 miRNAs for BMPR2; 5 miRNAs for CASP3, 4 miRNAs for IGF2R, IRF1 and PTEN; and 3 miRNAs for BACH2, BCL11A, BIM, IRF4 and PRDM1 (Figure 4B and C). At least 57 out of the 295 putative target interactions have experimental support in the literature, and 3 interactions have functional relevance in plasma cell differentiation on the basis of earlier studies (Figure 4C and Supplementary Table S6). Interactions with strong functional evidence include pairs such as miR-148a:BACH2, miR-34a:FOXP1 (26) and the interaction between miR-30 family and PRDM1 (36).
The network also showed several possible and less studied target interactions with genes frequently studied in normal plasma cell differentiation, such as those with MYC (miR-320b:MYC, miR-34a:MYC), CD69 (miR-106b:CD69, miR-130b:CD69), IRF4 (miR-30 family:IRF4), CASP3 (miR-30e:CASP3, let-7 family:CASP3), BCL2L1 (let-7c/g:BCL2L1), BACH2 (miR-15:BACH2, miR-130b:BACH2, miR-135:BACH2) and BIM (miR-18a:BIM, miR-19a:BIM and miR-20a:BIM) (Figure 4C and Supplementary Table S6). Altogether, these data highlight new potential roles of miRNAs during PCD.
miR-106b, miR-15b and miR-21 modulation of TGF-β signaling, autophagy and EZH2 expression
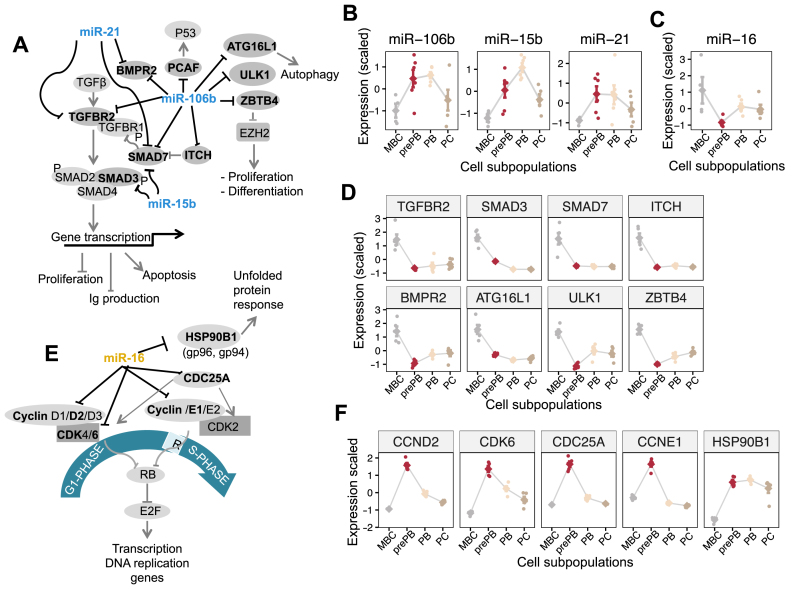
Several miRNAs that have been widely studied in other tissues were represented by many putative target interactions in the inferred PCD miRNA–mRNA network, including miR-106b, miR-16, members of miR-30 family and miR-15b (Figures 3A and 4C). We hypothesized that miRNA dysregulation would be of particularly importance in PCD when miRNAs coordinately regulate multiple components of a pathway or a biological process critical for plasma cell differentiation. miR-106b, miR-15b and miR-21, from miRNA cluster 2, were represented with several putative target relationships. At least five of these targets (TGFBR2, SMAD3, SMAD7, ITCH and BMPR2) encode known components of the transforming growth factor (TGF)-β signaling pathway (Figure 5A), which have been demonstrated to play an important role in regulating B cell activation and differentiation (37). In this pathway, TGF-β binds to the receptor TGFBR2, which recruits and phosphorylates TGFBR1, leading to activation of intracellular SMAD2 and SMAD3 by phosphorylation (P). Phosphorylated SMAD2 and SMAD3 subsequently bind to SMAD4 and translocate to the nucleus to initiate target gene expression in cooperation with co-factors and other transcription factors. Our analysis shows strong negative associations among miRNAs (miR-106b, miR-15b, miR-21) and TGF-β pathway genes during PCD (Figure 5B and D). Additionally, we found a strong negative interaction between miR-106b and several other genes encoding for protein involved in autophagy (ATG16L1 and ULK1) and ZBTB4, a transcription repressor regulating EZH2, which is know to be critical in B lymphocyte biology (38–40). Consistent with these findings, we experimentally showed that the inhibition of miR-106b, using antisense oligonucleotides, induced a significant upregulation of 7/7 tested predicted targets—PCAF, ATG16L2, ULK1, ZBTB4, TGFBR2, SMAD7 and BMPR2—in JJN3 myeloma cell line relative to experiments with control antisense oligonucleotides (P ≤ 0.01, one-tailed t-test, n = 3, mean ± S.E.M., Supplementary Figure S5).
Figure 5.
miRNA regulation of TGF-β pathway components, autophagy, ZBTB4/EZH2 axis and cell cycle. Several miRNAs were represented by many putative target interactions in the inferred plasma cell differentiation miRNA–mRNA network. miR-106b, miR-15b and miR-21, from miRNA cluster 2 (blue color), were represented with several putative target relationships encoding known components of the transforming growth factor (TGF)-β signaling pathway, autophagy and the ZBTB4/EZH2 axis. miR-16, from miRNA cluster 1 (yellow color) targets G1/S transition genes and HSP90B1. The figure shows the pathways as well as the expression profile of the different miRNAs and target mRNAs. Expression values are standardized before plotting. (A) Association between miR-106b, miR-15b, miR-21 and predicted target mRNAs: TGF-β pathway (TGFBR2, SMAD3, SMAD7, ITCH and BMPR2), autophagy (ATG16L1 and ULK1) and ZBTB4. (B) Expression profile of miR-106b, miR15b and miR-21 during plasma cell differentiation. (C) Expression profile of miR-16 during plasma cell differentiation. (D) Expression profile of miR-106b, miR15b and miR-21 target mRNAs. (E) Association between miR-16 and target mRNAs: G1/S transition genes (CCND2, CCNE1, CDK6 and CDC25A) and HSP90B1. (F) Expression profile of miR-16 target mRNAs.
miR-16 targets cooperatively regulate cell cycle progression
The miR-16, belonging to miRNA cluster 1, had multiple inferred target interactions in the PCD network, and four of these genes, CCND2, CCNE1, CDK6 and CDC25A, encode critical components of G1/S phase cell cycle progression (Figure 5E). In this pathway, CCND2 in association with CDK6, CCNE1 and CDK2 cooperate to phosphorylate RB. This phosphorylation prevents RB binding to E2F leading to the activation of E2F-mediated transcription and driving cells from G1 into S phase. CDC25A is a phosphatase that activates CDK2 by removing two phosphate groups. We observed a very strong inverse correlation between miR-16 expression (Figure 5C) and CCND2, CCNE1, CDK6 and CDC25A expression during PCD (Figure 5F). We found that the inhibition of miR-16, using antisense oligonucleotides, induced a significant upregulation of 4/4 tested predicted targets - CCND2, CCNE1, CDK6 and CDC25A—in JJN3 myeloma cell line relative to experiments with control antisense oligonucleotides (P ≤ 0.01, one-tailed t-test, n = 3, mean ± S.E.M., Supplementary Figure S5). Additionally, our analysis reveals potential interaction between miR-16 and HSP90B1 (also known as gp96 or gp94), a major endoplasmic reticulum chaperone mediating unfolded protein response (UPR), a process known to play a critical role in B cell development and plasma cell differentiation (41–43). Taken together, our data suggest a functional role of miR-16 as a possible master regulator of cell cycle during MBC to prePB transition, as well as, a potential regulator of UPR.
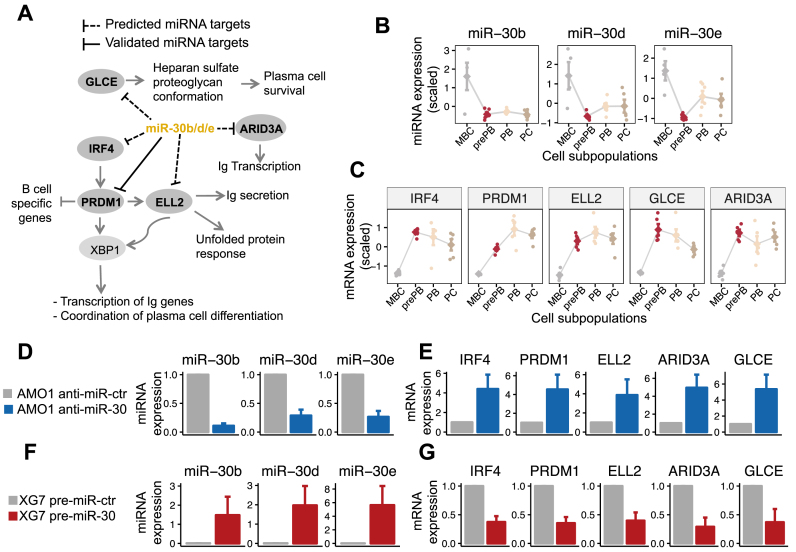
miR-30 family and coordination of plasma cell differentiation
Members of the miR-30 family, in miRNA cluster 1, had multiple inferred targets that have been described to play a critical role in coordinating of plasma cell differentiation, including IRF4, PRDM1, ELL2, ARID3A and GLCE (Figure 6A). In this pathway, IRF4 activates PRDM1, which encodes BLIMP-1, an essential transcription factor for plasma cell generation. BLIMP-1 silences B-cell specific genes, induces the transcription of immunoglobulin genes and activates the transcription elongation factor ELL2, which drives Ig secretory-specific mRNA production and the unfolded protein (7,44). ARID3A, also known as Bright in mouse, is a member of the ARID (A+T-rich interaction domain) family of proteins, which is required for both hematopoietic stem cell and B-cell lineage development (45,46). The glucuronyl C5-epimerase (GLCE) controls heparan sulfate conformation which is crucial for recruitment of factors that control plasma cell survival (47). We observed strong anti-correlation of miR-30 family expression (Figure 6B) and the inferred target genes (Figure 6C). One earlier study provided evidences for direct interaction between miR-30 family and PRDM1 (36). We experimentally tested the putative target interaction between members of miR-30 family and IRF4, PRDM1, ELL2, ARID3A and GLCE in two plasma cell myeloma derived cell lines (AMO1 and XG7). As illustrated in Supplementary Figure S6, the expression values of miR30 family members are variable in primary multiple myeloma (MM) samples from patients. To validate the identified targets of miR-30 family, we selected two cell lines, AMO1 and XG7, with respectively high and low miR30 expression. Therefore, we inhibited miR-30b/d/e in AMO1 and overexpressed them in XG7. Inhibition of miR-30b/d/e using antisense oligonucleotides (Figure 6D) resulted to a significant upregulation of IRF4, PRDM1, ELL2, ARID3A and GLCE in AMO1 cell line relative to experiments with control antisense oligonucleotides (P ≤ 0.0001, one-tailed t-test, n = 4, mean ± S.E.M., Figure 6e). Overexpression of miR-30b/d/e using precursor miRNAs (Figure 6F), caused at least 60% reduction of IRF4, PRDM1, ELL2, ARID3A and GLCE expression in XG7, relative to experiments with control precursor miRNA (Figure 6G). Taken together, these data strongly suggest that miR-30 family controls plasma cell differentiation by targeting several crucial genes encoding proteins required in plasma cell generation.
Figure 6.
miR-30 family in coordinating plasma cell differentiation and experimental validation. Four biological replicates are performed for experimental validation. The means and the standard error of the mean are displayed in the bar plots. (A) Association between miR-30 family members and predicted target genes with critical role in plasma cell differentiation, including IRF4, PRDM1, ELL2, ARID3A and GLCE. (B) Expression profile of miR-30 family members during plasma cell differentiation. (C) Expression profile of miR-30 target mRNAs. Expression values are standardized before plotting. (D) Inhibition of miR-30b/d/e in AMO1 cell line. Relative expression of miR-30b/d/e 48 h after transfection with antisense oligonucleotides anti-miR-30b/d/e and anti-miR-control in AMO1 cells. (E) Effects of miR-30b/d/e inhibition on the expression level of target mRNAs. (F) Overexpression of miR-30b/d/e in XG7 cell line. Expression of miR-30b/d/e 48 after transfection with precursor miRNAs pre-miR-30b/d/e and pre-miR-control in XG7 cells. (G) Effects of miR-30b/d/e overexpression on the expression level of target mRNAs.
PCD miRNAs associated with multiple myeloma
We have just described the potential crucial role of miRNAs in coordinating normal PCD. We hypothesized that the deregulation of miRNA regulatory networks can be involved in tumorigenesis. Not surprisingly, multiple pieces of evidence from integrating miRNA and mRNA expression profiles point out miRNAs playing a role in multiple myeloma pathophysiology (MM), a plasma cell malignancy characterized by a clonal accumulation of malignant plasma cells within the bone marrow (18–25). Consequently, we next cross-referenced the list of our 63 differentially expressed miRNAs, during normal plasma cell differentiation (PCD), with different lists of deregulated miRNAs during MM pathogenesis, obtained from previous studies (18–25). Notably, 24 of those 63 miRNAs were reported to be differentially expressed (in at least two different studies) between normal and malignant plasma cells and/or overexpressed in high-risk MM patients (Supplementary Table S7). Among these miRNAs, 11 were in miRNA cluster 1 expressed at a higher level in MBCs (let-7b/c/i, miR-16, miR-221, miR-222, miR-26a, miR-28-3p, miR-29b and miR-30b/d); 10 in cluster 2 highly expressed in prePBs (miR-106b, miR-155, miR-15b, miR-18a, miR-19a, miR-20a, miR-21, miR-23a, miR-27a and miR-625); and three in miRNA cluster 3 (miR-148a, miR-181a and miR-34a) (Supplementary Table S7). Furthermore, 10 of these miRNAs have been reported to be overexpressed in high-risk MM patients, including (let-7c/i, miR-106b, miR-148a, miR-16, miR-18a, miR-19a, miR-20a, miR-21 and miR-29b) (20). One miRNA (miR-155) was described to be associated with t (4,14) cytogenetic group; one (miR-21) with t (11,14) and 11 miRNAs with del13q, including miR-16 and miR-34a (21,22) (Supplementary Table S7). Additionally, using publicly available miRNA expression data (ArrayExpress accession number: E-MTAB-1363), we found that 31 out of the 63 PCD miRNAs were significantly differentially expressed in primary multiple myeloma tumor plasma cells and/or in myeloma cell lines compared to the normal bone marrow plasma cells (Supplementary Table S8). These miRNAs include members of miR-30 family, miR-16 and miR-106b. These results demonstrate that plasma cell differentiation stage-specific miRNAs deregulation could play a role in myelomagenesis and MM pathophysiology.
DISCUSSION
miRNAs are known to be involved in many key cellular processes and different diseases. In this study, we used a powerful bioinformatics pipelines to analyze the temporal dynamic of miRNAs expression as well as to infer probable active and functional miRNA–target interactions in human plasma cell differentiation (PCD). We identified 63 significantly differentially expressed miRNAs (Figure 2A) associated with three main dynamic profiles: early expression at MBCs step followed by repression (miRNAs cluster 1), delayed up-regulation at prePBs step followed by down-regulation (miRNAs cluster 2) and delayed up-regulation from PBs to PCs steps (miRNAs cluster 3) (Figure 2C).
It is well established that identification of miRNA targets is crucial for deciphering the exact role of individual miRNAs or groups of related miRNAs in a given cell. In recent years, several algorithms have been developed for predicting miRNA targets based on sequence complementarity of the miRNA and the mRNAs (48). However, there is no single algorithm that can efficiently predict all targets with a minimal number of false positives (48). A straightforward approach to refine target gene predictions could be correlation analysis between miRNAs and mRNA expression profiles in combination with sequence-based target prediction. For this, we developed an R package, named miRTarget, to easily build a highly confident list of miRNA–mRNA target interaction from integrating correlation analysis and target predictions with miRecords and miRtarbase.
Using this approach, we identified 1596 negative miRNAs–mRNAs target interactions involving 51 unique miRNAs and 948 unique mRNAs (Supplementary Table S4). Interestingly, at least 316 of the 1596 putative target interactions (comprising 272 unique genes and 35 miRNAs) have experimental support in the literature according to miRecords and miRTarbase databases (Supplementary Table S4). Pathway analysis revealed that these miRNA target genes are mainly involved in cell cycle, MYC activation pathway, membrane trafficking, golgi associated vesicle biogenesis and budding, and signaling by TGF-beta. Overall, our approach provides valuable information for identification of potent miRNA target genes as well as the elucidation of dynamic gene expression changes and the underlying related biological processes.
We next searched to extract, from the 1596 miRNA–mRNA target interactions, the network of miRNAs regulating critical genes associated with plasma cell differentiation, critical genes being defined as genes that have at least 1 citation index associated with PCD. We have inferred a PCD network of 295 miRNA – mRNA target interactions, comprising 47 miRNAs and 141 target mRNAs. These candidate interactions show strong evidence of regulatory activity in earlier studies in other tissues. Furthermore, several miRNA families that have been widely studied in the literature, were represented by many target interaction during PCD, and there is functional evidence for at least 57 interactions from earlier experimental studies in the literature.
We acknowledge that miRNA and mRNA expression negative associations can only be identified if the mRNA is degraded after being targeted. Therefore, our approach may miss some miRNA–target interactions that primarily influence mRNA translation efficiency. Our PCD network of candidate miRNA–target relationships also sacrifices sensitivity in favor of specificity by applying stringent fold change-, correlation- and literature-based filters, and it may also discard biologically relevant interactions. However, the released miRNAs and mRNAs expression data sets allows rapid exploration and visualization of any miRNA–mRNA association independent of any filters.
This study highlights at least four cases in which miRNA families are predicted to coordinately target and regulate multiple members of a PCD-related pathway. In the first case, our method predicts that the miR-30 family directly targets and regulates IRF4 and PRDM1, ELL2, ARID3A and GLCE, four genes encoding proteins critical for PCD. Although earlier studies have shown that members of the miR-30 family directly target PRDM1 (36), IRF4, ELL2, ARID3A and GLCE have not been reported as a functional target of the miR-30 family. Our analysis suggests that the inhibition of miR-30 family expression is a potent mechanism for setting up the gene expression program required to the differentiation of B cell to plasma cell.
In the second case, our analysis identified a strong negative correlation between miR-16 and four cell cycle genes (CCND2, CCNE1, CDK6 and CDC25A) as well as HSP90B1, involved in unfolded protein response. miR-16 is present at a higher level in MBCs and downregulated during MBCs to prePBs transition. Consistent with this downregulation, prePB stage is characterized by an active proliferation (2). Five studies experimentally support direct miR-16 target interaction with CCND2, CCNE1, CDK6 and CDC25A (49–52) in other contexts. Taken together, our data are consistent with earlier observations and support a functional role of miR-16 as a possible master regulator of cell cycle during MBC to prePB transition.
We also identified that three miRNAs (miR-106b, miR-15b and miR-21), expressed at prePB stage, contribute to inhibit five genes (TGFBR2, SMAD3, SMAD7, ITCH and BMPR2) encoding components of the TGF-β signaling pathways. Furthermore, several of these miRNA–mRNA interactions have been validated in the literature: members of miR-106 family:TGFBR2 (53), miR-106b:ITCH (54), miR-106b:SMAD7 (55,56), miR-15b:SMAD3 (57), miR-21:TGFBR2 (58,59), miR-21:BMPR2 (60) and miR-21:SMAD7 (61). TGF-β signaling pathway is an important regulator of B cell activation and differentiation (37). There is evidence that TGF-β inhibits B cell proliferation through the induction of the transcription factor ID3, induction of the cyclin-dependent kinase inhibitor p21 (which inhibits the progression of cell cycle through the G1 and S phase) repression of the expression of c-MYC and ATM (62,63). Early studies show also that TGF- β has inhibitory function on antibody production (64) and induces apoptosis by inhibiting NF-κB activation (65). Altogether, these data suggest that the overexpression of miR-106b, miR-15b and miR-21, at prePB stage, modulates the TGF-β pathway leading to cell proliferation and plasma cell differentiation. Additionally, our analysis reveals interactions between miR-106b and several other genes encoding for protein involved in autophagy (ATG16L1 and ULK1) and ZBTB4, a transcription repressor regulating EZH2, which is known to be critical in B lymphocyte biology (38–40). During lymphopoiesis, EZH2 is strongly expressed in proliferating cells, such as human germinal center B cells, and plasmablasts, suggesting an important role in cell cycle regulation and in lymphocyte division (38–40). Accordingly, lower H3K27me3 levels have been reported in resting B cells compared to activating B cells (66). These data highlight a role of miR-106b and EZH2 deregulation in association to cell cycle activation in prePBs.
We observed a very strong inverse correlation between miR-106b and these genes (Figure 5). Furthermore, early studies provide experimental support for the direct miR-106b target interaction with ATG16L1 (67), ULK1 (68) and ZBTB4 (69). As described in previous studies, autophagy plays a key role in PC for sustainable immunoglobulin production (70).
Furthermore, our work demonstrates that 24 PCD stage-specific miRNAs are aberrantly overexpressed in multiple myeloma cells (MMCs) compared to their normal counterpart and/or are associated with high risk myeloma, suggesting that MMCs frequently acquired expression changes in miRNAs already undergoing dynamic expression modulation during normal PCD. Interestingly, these miRNAs include miR-21, which is upregulated in myeloma plasma cells compared to the normal counterparts (18,22). miR-21 plays a key role in tumor progression and is significantly upregulated in several human cancers (71). Furthermore, it has been recently shown that the Epstein–Barr virus (EBV)–encoded EBNA2, which is needed for the transforming capacity of B cells in vitro, significantly increases miR-21 expression (72). Additionally, there is a strong evidence that in vivo antagonism of miR-21 exerts anti-multiple myeloma activity, providing the rationale for clinical development of miR-21 inhibitors in this still incurable disease (73).
In conclusions, we describe, for the first time, a comprehensive integrated analysis of miRNAs and mRNAs during plasma cell differentiation. Of particular note, our analysis identified a high-confidence network of 295 miRNA – mRNA target relationships comprising 47 miRNAs and 141 mRNA targets. These relationships include new examples of miRNAs (miR-30, miR-106b and miR-16) that appear to coordinately modulate multiple members of critical pathways associated with PCD, including IRF4/PRDM1 axis, TGF-β signaling pathway, autophagy, ZBTB4/EZH2 axis and cell cycle. In addition, our work demonstrates that PCD stage-specific miRNAs are aberrantly overexpressed in multiple myeloma cells (MMCs) compared to the normal counterparts. Altogether, our results demonstrate that miRNAs may be important in controlling PCD and malignant plasma cell biology. Finally, this rich data set should prove valuable for researchers exploring plasma cell biology.
ACCESSION NUMBERS
miRNA expression data from this study are available in GEO under accession number GSE86604. mRNA expression data are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/, E-MTAB-1771, E-MEXP-2360 and E-MEXP-3034).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Microarray Core Facility of IRMB, CHU Montpellier.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French INCA (Institut National du Cancer) Institute [2012-109/087437 and PLBIO15-256]; Languedoc Roussillon CRLR (R14026FF), Fondation de France [201400047510]; ITMO Cancer (MM&TT) and the France Génomique National infrastructure, funded as part of the « Investissements d’Avenir » program managed by the Agence Nationale pour la Recherche [ANR-10-INBS-09]. Funding for open access charge: INCA (Institut National Du Cancer), France.
Conflict of interest statement. None declared.
REFERENCES
- 1. Shapiro-Shelef M., Calame K.. Regulation of plasma-cell development. Nat. Rev. Immunol. 2005; 5:230–242. [DOI] [PubMed] [Google Scholar]
- 2. Jourdan M., Caraux A., Caron G., Robert N., Fiol G., Reme T., Bollore K., Vendrell J.P., Le Gallou S., Mourcin F. et al. Characterization of a transitional preplasmablast population in the process of human B cell to plasma cell differentiation. J. Immunol. 2011; 187:3931–3941. [DOI] [PubMed] [Google Scholar]
- 3. Jourdan M., Cren M., Robert N., Bollore K., Fest T., Duperray C., Guilloton F., Hose D., Tarte K., Klein B.. IL-6 supports the generation of human long-lived plasma cells in combination with either APRIL or stromal cell-soluble factors. Leukemia. 2014; 28:1647–1656. [DOI] [PubMed] [Google Scholar]
- 4. Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M.. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015; 15:160–171. [DOI] [PubMed] [Google Scholar]
- 5. Shaffer A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K. et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002; 17:51–62. [DOI] [PubMed] [Google Scholar]
- 6. Klein U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., Dalla-Favera R.. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 2006; 7:773–782. [DOI] [PubMed] [Google Scholar]
- 7. Minnich M., Tagoh H., Bonelt P., Axelsson E., Fischer M., Cebolla B., Tarakhovsky A., Nutt S.L., Jaritz M., Busslinger M.. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat. Immunol. 2016; 17:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gass J.N., Gunn K.E., Sriburi R., Brewer J.W.. Stressed-out B cells? Plasma-cell differentiation and the unfolded protein response. Trends Immunol. 2004; 25:17–24. [DOI] [PubMed] [Google Scholar]
- 9. Ron D., Walter P.. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell. Biol. 2007; 8:519–529. [DOI] [PubMed] [Google Scholar]
- 10. Goldfinger M., Shmuel M., Benhamron S., Tirosh B.. Protein synthesis in plasma cells is regulated by crosstalk between endoplasmic reticulum stress and mTOR signaling. Eur. J. Immunol. 2011; 41:491–502. [DOI] [PubMed] [Google Scholar]
- 11. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K.. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001; 107:881–891. [DOI] [PubMed] [Google Scholar]
- 12. Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A. et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004; 21:81–93. [DOI] [PubMed] [Google Scholar]
- 13. Reimold A.M., Iwakoshi N.N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E.M., Friend D., Grusby M.J., Alt F., Glimcher L.H.. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001; 412:300–307. [DOI] [PubMed] [Google Scholar]
- 14. Pasquinelli A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012; 13:271–282. [DOI] [PubMed] [Google Scholar]
- 15. Shivdasani R.A. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006; 108:3646–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ivey K.N., Srivastava D.. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010; 7:36–41. [DOI] [PubMed] [Google Scholar]
- 17. Schickel R., Boyerinas B., Park S.M., Peter M.E.. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008; 27:5959–5974. [DOI] [PubMed] [Google Scholar]
- 18. Pichiorri F., Suh S.S., Ladetto M., Kuehl M., Palumbo T., Drandi D., Taccioli C., Zanesi N., Alder H., Hagan J.P. et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:12885–12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lionetti M., Biasiolo M., Agnelli L., Todoerti K., Mosca L., Fabris S., Sales G., Deliliers G.L., Bicciato S., Lombardi L. et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009; 114:e20–e26. [DOI] [PubMed] [Google Scholar]
- 20. Zhou Y., Chen L., Barlogie B., Stephens O., Wu X., Williams D.R., Cartron M.A., van Rhee F., Nair B., Waheed S. et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:7904–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gutierrez N.C., Sarasquete M.E., Misiewicz-Krzeminska I., Delgado M., De Las Rivas J., Ticona F.V., Ferminan E., Martin-Jimenez P., Chillon C., Risueno A. et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia. 2010; 24:629–637. [DOI] [PubMed] [Google Scholar]
- 22. Chi J., Ballabio E., Chen X.H., Kusec R., Taylor S., Hay D., Tramonti D., Saunders N.J., Littlewood T., Pezzella F. et al. MicroRNA expression in multiple myeloma is associated with genetic subtype, isotype and survival. Biol. Direct. 2011; 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pichiorri F., De Luca L., Aqeilan R.I.. MicroRNAs: new players in multiple myeloma. Front. Genet. 2011; 2:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lionetti M., Musto P., Di Martino M.T., Fabris S., Agnelli L., Todoerti K., Tuana G., Mosca L., Gallo Cantafio M.E., Grieco V. et al. Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia. Clin. Cancer Res. 2013; 19:3130–3142. [DOI] [PubMed] [Google Scholar]
- 25. Seckinger A., Meissner T., Moreaux J., Benes V., Hillengass J., Castoldi M., Zimmermann J., Ho A.D., Jauch A., Goldschmidt H. et al. miRNAs in multiple myeloma - a survival relevant complex regulator of gene expression. Oncotarget. 2015; 6:39165–39183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsai D.Y., Hung K.H., Lin I.Y., Su S.T., Wu S.Y., Chung C.H., Wang T.C., Li W.H., Shih A.C., Lin K.I.. Uncovering microRNA regulatory hubs that modulate plasma cell differentiation. Sci. Rep. 2015; 5:17957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jourdan M., Caraux A., De Vos J., Fiol G., Larroque M., Cognot C., Bret C., Duperray C., Hose D., Klein B.. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood. 2009; 114:5173–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moreaux J., Klein B., Bataille R., Descamps G., Maiga S., Hose D., Goldschmidt H., Jauch A., Reme T., Jourdan M. et al. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica. 2011; 96:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P.. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003; 4:249–264. [DOI] [PubMed] [Google Scholar]
- 30. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kassambara A., Reme T., Jourdan M., Fest T., Hose D., Tarte K., Klein B.. GenomicScape: an easy-to-use web tool for gene expression data analysis. Application to investigate the molecular events in the differentiation of B cells into plasma cells. PLoS Comput. Biol. 2015; 11:e1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chou C.H., Chang N.W., Shrestha S., Hsu S.D., Lin Y.L., Lee W.H., Yang C.D., Hong H.C., Wei T.Y., Tu S.J. et al. miRTarBase 2016: updates to the experimentally validated miRNA–target interactions database. Nucleic Acids Res. 2016; 44:D239–D247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T.. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009; 37:D105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang J., Jima D.D., Jacobs C., Fischer R., Gottwein E., Huang G., Lugar P.L., Lagoo A.S., Rizzieri D.A., Friedman D.R. et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009; 113:4586–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li M.O., Wan Y.Y., Sanjabi S., Robertson A.K., Flavell R.A.. Transforming growth factor-beta regulation of immune responses. Annu. Rev. Immunol. 2006; 24:99–146. [DOI] [PubMed] [Google Scholar]
- 38. Good-Jacobson K.L. Regulation of germinal center, B-cell memory, and plasma cell formation by histone modifiers. Front. Immunol. 2014; 5:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Su I.H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., Tarakhovsky A.. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 2003; 4:124–131. [DOI] [PubMed] [Google Scholar]
- 40. Herviou L., Cavalli G., Cartron G., Klein B., Moreaux J.. EZH2 in normal hematopoiesis and hematological malignancies. Oncotarget. 2016; 7:2284–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu B., Li Z.. Endoplasmic reticulum HSP90b1 (gp96, grp94) optimizes B-cell function via chaperoning integrin and TLR but not immunoglobulin. Blood. 2008; 112:1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Staron M., Yang Y., Liu B., Li J., Shen Y., Zuniga-Pflucker J.C., Aguila H.L., Goldschneider I., Li Z.. gp96, an endoplasmic reticulum master chaperone for integrins and Toll-like receptors, selectively regulates early T and B lymphopoiesis. Blood. 2010; 115:2380–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosenbaum M., Andreani V., Kapoor T., Herp S., Flach H., Duchniewicz M., Grosschedl R.. MZB1 is a GRP94 cochaperone that enables proper immunoglobulin heavy chain biosynthesis upon ER stress. Genes Dev. 2014; 28:1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park K.S., Bayles I., Szlachta-McGinn A., Paul J., Boiko J., Santos P., Liu J., Wang Z., Borghesi L., Milcarek C.. Transcription elongation factor ELL2 drives Ig secretory-specific mRNA production and the unfolded protein response. J. Immunol. 2014; 193:4663–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nixon J.C., Ferrell S., Miner C., Oldham A.L., Hochgeschwender U., Webb C.F.. Transgenic mice expressing dominant-negative bright exhibit defects in B1 B cells. J. Immunol. 2008; 181:6913–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Webb C.F., Bryant J., Popowski M., Allred L., Kim D., Harriss J., Schmidt C., Miner C.A., Rose K., Cheng H.L. et al. The ARID family transcription factor bright is required for both hematopoietic stem cell and B lineage development. Mol. Cell. Biol. 2011; 31:1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reijmers R.M., Groen R.W., Kuil A., Weijer K., Kimberley F.C., Medema J.P., van Kuppevelt T.H., Li J.P., Spaargaren M., Pals S.T.. Disruption of heparan sulfate proteoglycan conformation perturbs B-cell maturation and APRIL-mediated plasma cell survival. Blood. 2011; 117:6162–6171. [DOI] [PubMed] [Google Scholar]
- 48. Muniategui A., Pey J., Planes F.J., Rubio A.. Joint analysis of miRNA and mRNA expression data. Brief. Bioinform. 2013; 14:263–278. [DOI] [PubMed] [Google Scholar]
- 49. Linsley P.S., Schelter J., Burchard J., Kibukawa M., Martin M.M., Bartz S.R., Johnson J.M., Cummins J.M., Raymond C.K., Dai H. et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 2007; 27:2240–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Q., Fu H., Sun F., Zhang H., Tie Y., Zhu J., Xing R., Sun Z., Zheng X.. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008; 36:5391–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang F., Fu X.D., Zhou Y., Zhang Y.. Down-regulation of the cyclin E1 oncogene expression by microRNA-16-1 induces cell cycle arrest in human cancer cells. BMB Rep. 2009; 42:725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zubillaga-Guerrero M.I., Alarcon-Romero Ldel C., Illades-Aguiar B., Flores-Alfaro E., Bermudez-Morales V.H., Deas J., Peralta-Zaragoza O.. MicroRNA miR-16-1 regulates CCNE1 (cyclin E1) gene expression in human cervical cancer cells. Int. J. Clin. Exp. Medicine. 2015; 8:15999–16006. [PMC free article] [PubMed] [Google Scholar]
- 53. Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luo Z.L., Luo H.J., Fang C., Cheng L., Huang Z., Dai R., Li K., Tian F.Z., Wang T., Tang L.J.. Negative correlation of ITCH E3 ubiquitin ligase and miRNA-106b dictates metastatic progression in pancreatic cancer. Oncotarget. 2016; 7:1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith A.L., Iwanaga R., Drasin D.J., Micalizzi D.S., Vartuli R.L., Tan A.C., Ford H.L.. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012; 31:5162–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu D., Shin H.S., Lee Y.S., Lee Y.C.. miR-106b modulates cancer stem cell characteristics through TGF-beta/Smad signaling in CD44-positive gastric cancer cells. Lab. Invest. 2014; 94:1370–1381. [DOI] [PubMed] [Google Scholar]
- 57. Tijsen A.J., van der Made I., van den Hoogenhof M.M., Wijnen W.J., van Deel E.D., de Groot N.E., Alekseev S., Fluiter K., Schroen B., Goumans M.J. et al. The microRNA-15 family inhibits the TGFbeta-pathway in the heart. Cardiovasc. Res. 2014; 104:61–71. [DOI] [PubMed] [Google Scholar]
- 58. Mishra S., Deng J.J., Gowda P.S., Rao M.K., Lin C.L., Chen C.L., Huang T., Sun L.Z.. Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (TGFBR2) expression in prostate cancer. Oncogene. 2014; 33:4097–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim Y.J., Hwang S.J., Bae Y.C., Jung J.S.. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009; 27:3093–3102. [DOI] [PubMed] [Google Scholar]
- 60. Qin W., Zhao B., Shi Y., Yao C., Jin L., Jin Y.. BMPRII is a direct target of miR-21. Acta Biochim. Biophys. Sinica. 2009; 41:618–623. [DOI] [PubMed] [Google Scholar]
- 61. Li Q., Zhang D., Wang Y., Sun P., Hou X., Larner J., Xiong W., Mi J.. MiR-21/Smad 7 signaling determines TGF-beta1-induced CAF formation. Sci. Rep. 2013; 3:2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lebman D.A., Edmiston J.S.. The role of TGF-beta in growth, differentiation, and maturation of B lymphocytes. Microbes Infect./Institut Pasteur. 1999; 1:1297–1304. [DOI] [PubMed] [Google Scholar]
- 63. Roes J., Choi B.K., Cazac B.B.. Redirection of B cell responsiveness by transforming growth factor beta receptor. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:7241–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kehrl J.H., Thevenin C., Rieckmann P., Fauci A.S.. Transforming growth factor-beta suppresses human B lymphocyte Ig production by inhibiting synthesis and the switch from the membrane form to the secreted form of Ig mRNA. J. Immunol. 1991; 146:4016–4023. [PubMed] [Google Scholar]
- 65. Arsura M., Wu M., Sonenshein G.E.. TGF beta 1 inhibits NF-kappa B/Rel activity inducing apoptosis of B cells: transcriptional activation of I kappa B alpha. Immunity. 1996; 5:31–40. [DOI] [PubMed] [Google Scholar]
- 66. Baxter J., Sauer S., Peters A., John R., Williams R., Caparros M.L., Arney K., Otte A., Jenuwein T., Merkenschlager M. et al. Histone hypomethylation is an indicator of epigenetic plasticity in quiescent lymphocytes. EMBO J. 2004; 23:4462–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhai Z., Wu F., Chuang A.Y., Kwon J.H.. miR-106b fine tunes ATG16L1 expression and autophagic activity in intestinal epithelial HCT116 cells. Inflammatory Bowel Dis. 2013; 19:2295–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wu H., Wang F., Hu S., Yin C., Li X., Zhao S., Wang J., Yan X.. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell. Signal. 2012; 24:2179–2186. [DOI] [PubMed] [Google Scholar]
- 69. Yang W.S., Chadalapaka G., Cho S.G., Lee S.O., Jin U.H., Jutooru I., Choi K., Leung Y.K., Ho S.M., Safe S. et al. The transcriptional repressor ZBTB4 regulates EZH2 through a MicroRNA-ZBTB4-specificity protein signaling axis. Neoplasia. 2014; 16:1059–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pengo N., Scolari M., Oliva L., Milan E., Mainoldi F., Raimondi A., Fagioli C., Merlini A., Mariani E., Pasqualetto E. et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat. Immunol. 2013; 14:298–305. [DOI] [PubMed] [Google Scholar]
- 71. Pan X., Wang Z.X., Wang R.. MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol. Ther. 2010; 10:1224–1232. [DOI] [PubMed] [Google Scholar]
- 72. Rosato P., Anastasiadou E., Garg N., Lenze D., Boccellato F., Vincenti S., Severa M., Coccia E.M., Bigi R., Cirone M. et al. Differential regulation of miR-21 and miR-146a by Epstein-Barr virus-encoded EBNA2. Leukemia. 2012; 26:2343–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Leone E., Morelli E., Di Martino M.T., Amodio N., Foresta U., Gulla A., Rossi M., Neri A., Giordano A., Munshi N.C. et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin. Cancer Res. 2013; 19:2096–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.