Abstract
Certain chemical modifications confer increased stability and low immunogenicity to in vitro transcribed mRNAs, thereby facilitating expression of therapeutically important proteins. Here, we demonstrate that N1-methyl-pseudouridine (N1mΨ) outperforms several other nucleoside modifications and their combinations in terms of translation capacity. Through extensive analysis of various modified transcripts in cell-free translation systems, we deconvolute the different components of the effect on protein expression independent of mRNA stability mechanisms. We show that in addition to turning off the immune/eIF2α phosphorylation-dependent inhibition of translation, the incorporated N1mΨ nucleotides dramatically alter the dynamics of the translation process by increasing ribosome pausing and density on the mRNA. Our results indicate that the increased ribosome loading of modified mRNAs renders them more permissive for initiation by favoring either ribosome recycling on the same mRNA or de novo ribosome recruitment.
INTRODUCTION
Expression of therapeutically important proteins by introducing in vitro transcribed mRNAs into mammalian cells is a highly promising and innovative drug development concept. Transfection with mRNA offers many advantages over DNA-based technologies. First, gene transfer using mRNA poses no risk of undesirable and potentially deleterious chromosomal integration. Second, mRNA transfer is more efficient than DNA transfection in both total amount taken up and the number of targeted cells. Third, mRNA directs protein expression almost immediately after reaching the cytoplasm.
Significantly, the incorporation of modified nucleotides into therapeutic mRNAs improves their performance in cell culture and in animals, which leads to a reduction of the applied dose and improved safety for patients (1–3). In addition, cell-based studies have shown that the presence of modified nucleotides in synthetic mRNAs reduces their immune stimulatory activity (1). While non-modified mRNAs activate the interferon inducers, Toll-like receptors and retinoic acid-inducible gene I protein, pseudouridine (Ψ) or 2-thiouridine-containing mRNAs fail to do so (4). In addition, modified nucleotides in mRNA reduce the activation of RNA-dependent protein kinase (PKR) (5,6). PKR is one of four kinases known to phosphorylate the α-subunit of translation initiation factor 2 (eIF2α) and repress translation (7,8). Other eIF2α kinases in mammals are PKR-like endoplasmic reticulum kinase (PERK), general control non-derepressible-2 (CCN2) and heme-regulated inhibitor. eIF2, which is composed of three subunits α, β and γ forms a ternary complex with guanosine 5΄-triphosphate (GTP) and methionyl initiator tRNA (Met–tRNAi). The role of the eIF2•GTP•Met–tRNAi complex is to deliver Met–tRNAi to the 40S ribosomal subunit. Following GTP hydrolysis, eIF2-GDP is released from the ribosome and is subsequently converted to eIF2-GTP with the aid of eIF2B. The affinity of phosphorylated eIF2 for eIF2B is dramatically enhanced, resulting in the sequestration of eIF2B, which impairs the regeneration of the ternary complex and attenuates translation. Since the level of eIF2 is higher than that of eIF2B, even low amounts of phosphorylated eIF2α are sufficient to block the activity of eIF2B (8). PKR is activated by double-stranded RNA, such as that formed during virus infection and requires dimerization and autophosphorylation of the protein. However, in vitro transcribed mRNAs can also activate PKR (5,6,9). This activation is due to stable secondary structure in the mRNA 5΄ untranslated region (UTR), such as the trans-activation response (TAR) region of human immunodeficiency virus 1 mRNA (9), but can also occur because of the generation of double-stranded RNA during in vitro transcription (10).
A significant proportion of synthetic mRNA transfected into cells is degraded in the endosomes, making it unavailable to the translation machinery (11). The restriction of protein expression from in vitro transcribed mRNA has also been linked to activation of the interferon-induced 2΄-5΄-oligoadenylate synthetases (OAS) (12). Activated OAS produce short 2΄-5΄-linked oligomers (termed 2–5A) using adenosine triphosphate (ATP) as a substrate. Binding of 2–5A to RNase L monomers induces their dimerization and activation (13). Remarkably, nucleoside modifications in RNA reduce activation of the OAS/RNase L system and cleavage of single-stranded RNAs (12).
Cell-based assays measure new protein expression many hours after transfection, making it difficult to study direct effects, the kinetics of protein synthesis and roles of different regulatory mechanisms. To circumvent these limitations, we recapitulated the stimulation of translation conferred by the modified nucleosides 5-methylcytidine (5 mC) and N1-methyl-pseudouridine (N1mΨ) and their combination (5 mC/N1mΨ) in cell-free extracts. We demonstrate that N1mΨ outperforms 5 mC and 5 mC/N1mΨ in translation. In cell-free extracts, phosphorylation of eIF2α is stimulated by the addition of standard, but not modified, in vitro transcribed mRNAs. Enhancement of eIF2α phosphorylation inhibits the translation of other mRNAs in trans. Unexpectedly and significantly, however, prevention of eIF2α phosphorylation by addition of recombinant GADD34 and K3L proteins, albeit reduced, did not completely negate the translational superiority of N1mΨ-containing Firefly luciferase (Luc) mRNA over standard unmodified mRNA, suggesting the existence of an additional mechanism. We report the impediment of ribosome movement at defined sites in the modified mRNA, resulting in the increase of the abundance and size of polysomes. We suggest that increased ribosome occupancy of the modified mRNA facilitates initiation or intra-polysomal ribosome recycling, ultimately enhancing the overall translation rate above the contribution of reduced immunogenicity.
MATERIALS AND METHODS
Cells and proteins
Human embryonic kidney (HEK293T) and HeLa S3 cells were maintained in Dulbecco's modified eagle's medium supplemented with 2 mM L-glutamine, 10% foetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin. Krebs-2 ascites carcinoma cells were propagated in mice (14). Mouse embryonic fibroblasts (MEF) wild-type (WT) and MEF that have a homozygous knockin mutation (Ser 51 to Ala) in the eIF2α gene (A/A) (15) were kindly provided by Dr Maria Hatzoglou. The mutant eIF2α is not a substrate for eIF2 kinases. MEF cell lines were cultured as described above. Recombinant proteins N-terminally truncated GADD34 (Δ1-240) (referred as GADD34) and K3L were expressed in bacteria and purified as reported (16,17). For the preparation of native eIF2 from rabbit reticulocyte lysate (RRL) see (18).
mRNA preparation
Conventional and modified polyadenylated Luc and enhanced green fluorescent protein (GFP) mRNAs were prepared by T7 polymerase in vitro transcription (New England Biolabs) and purified with spin columns (Life Technologies). All four nucleoside triphosphates in the reaction, natural and modified, were applied at a final concentration of 1.8 mM. The used nucleoside modifications were the following: 5 mC, N1mΨ, 5 mC and N1mΨ (5 mC/N1mΨ) or 5 mC and Ψ (5 mC/Ψ). The DNA template was generated by polymerase chain reaction amplification of codon-optimized sequences, which were obtained as custom-made plasmids (DNA2.0). To increase stability and template activity, all mRNAs were capped using the Vaccinia enzyme m7G capping system (New England Biolabs). For quality assurance, the mRNA preparations were analyzed by denaturing agarose gel electrophoresis and capillary RNA electrophoresis (Agilent). The purity of the mRNA was >80% for full-length transcripts (Supplementary Figure S1).
mRNA transfection
One day prior to transfection, HEK293T or MEF cells were seeded into 96-well plates at a density of 6 × 104 cells/well. mRNA (90 ng) was transfected into ∼90% confluent cells using TransIT-mRNA transfection kit as recommended by the manufacturer (Mirus). After culturing for 4.5 h, cells were lysed in 100 μl of Passive lysis buffer (Promega) with a single freeze-thaw cycle. The lysates were clarified by centrifugation. Aliquots (12 μl) of the 100-fold diluted samples were assayed for luc activity using the Luc assay system (Promega) and Lumat LB 9507 bioluminometer (Berthold Technologies).
In vitro translation assays
Translation-competent S10 extracts from Krebs and HeLa cells untreated or treated with micrococcal nuclease (RNase) were prepared as described previously (14,19,20). Translation of Luc or GFP mRNAs was carried out using standard techniques (14,21). To prevent eIF2α phosphorylation, extracts were supplemented with either GADD34 or the GADD34/K3L protein combination. Translation in RNase-treated RRL was carried out as recommended by the manufacturer (Promega). To increase m7G cap-dependency of the system, the final concentration of KCl in RRLs was increased by 40 mM (22). RNase-untreated RRL (Promega) was used as described (23) with slight modifications (22). Reaction mixtures (12.5 μl) were incubated at 30°C for the times indicated in the figure legends. Reactions were stopped by 30-fold dilution with 0.6 mM cycloheximide solution in phosphate buffered saline (PBS). Aliquots of the samples (3 μl) were withdrawn to measure luc activity. When 35S-methionine labeling of proteins was conducted, reactions were stopped with sodium dodecyl sulphate (SDS)-sample buffer. The measurement of 35S-methionine incorporation into trichloroacetic acid-insoluble material and analysis of the translation products by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (14% acrylamide) and autoradiography were done as described (19). To monitor the kinetics of luc and GFP synthesis in real time, the components of in vitro translation reactions were assembled in a standard white 96-well round bottom plate (Corning, NY, USA) covered with a self-adhesive fluorescence-compatible seal (BioRad, Hercules, CA, USA). Incubation was at 30°C inside the plate reader Cytation 3 (BioTek, Winooski, VT, USA). These assays made use of the HeLa 1-step coupled IVT kit (ThermoFisher Scientific, Waltham, MA, USA) or RNase-treated RRL (Promega). The reaction mixtures (20 μl) were supplemented with 5 μl (500 ng) of unmodified or modified GFP or Luc mRNAs. The fluorescence and chemiluminescence signals in the reactions with GFP and Luc mRNAs, respectively, increase over time and are considered proportional to the occurring synthesis of the full-size proteins. Kinetics of GFP production in each translation reaction was monitored for 90–180 min with the following settings: excitation −485 nm, emission −515 nm. The recording sensitivity was typically adjusted with a photomultiplier (PMT) gain of 80. The distance of the reading head was set to 1 mm above the plate and a reading speed to one per sample every 11–17 s. Each cell-free reaction with Luc mRNA (25 μl) was supplemented with 1-μl of 15 mg/ml D-Luciferin solution in PBS. Reactions were monitored for 90–180 min in luminescence mode. The recording sensitivity was typically adjusted with a PMT gain of 130. The distance of the reading head was set to 7 mm above the plate and a reading speed to one per sample every 11–17 s.
Analyses of formation of 80S initiation complexes and polysomes
Unmodified or modified Luc mRNAs were radiolabeled in their poly(A) tails using [α-32P]ATP and yeast poly(A) polymerase (24). For analysis of 80S initiation complex formation, the 32P-poly(A)-labeled mRNA (∼4•106 cpm, 200 ng) was incubated in a total reaction volume of 50 μl with untreated Krebs extract or RRL in the presence of all the translational components and cycloheximide (0.6 mM). After incubation at 30°C for 15 min, the reactions were stopped by 5-fold dilution with ice-cold polysome (P) buffer (15 mM Tris–HCl, pH 7.5, 15 mM Mg(OAc)2, 0.3 M NaCl and 0.2 mg/ml heparin) containing 0.6 mM cycloheximide. 80S ribosomal complexes were resolved by centrifugation (Beckman SW41 rotor, 37 000 rpm for 2 h at 4°C) through 7.5–45% sucrose gradients prepared with buffer P. Fractions (0.35 ml) were collected manually from the top of the gradients and radioactivity was measured by liquid scintillation counting. For polysome profiling experiments, cycloheximide was omitted from the reaction mixtures. Polysomes were formed at 30°C for 15 or 30 min. The samples were then diluted with buffer P containing cycloheximide and subjected to centrifugation through sucrose gradients as described above.
Western blotting
Proteins were resolved by SDS-10% PAGE, transferred onto a nitrocellulose membrane and detected using western lightning chemiluminescence kit (Perkin-Elmer). The primary antibodies were anti-eIF2α (pS52 phosphospecific, Invitrogen; 1:2500 dilution) and anti-eIF2α (total, Cell Signaling; 1:1000 dilution). Secondary HRP-conjugated anti-rabbit antibody (1:5000) was from GE Healthcare. The membrane was first probed with the phosphospecific eIF2α antibody and then after stripping with the total eIF2α antibody. Vertical slab gel isoelectric focusing separation of unphosphorylated and phosphorylated forms of eIF2α in RRL was done as described (25) using a mini-gel format. The running settings were the following: 30 min at 100 V, 18 h at 200 V, 1.5 h at 500 V. The focused proteins were transferred onto a Polyvinylidene Difluoride membrane and probed with the total eIF2α antibody as described above. For quantifications of signals, ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used.
Immunoprecipitation
The 5 mC/N1mΨ nucleoside modified Luc mRNA was translated in RNase treated RRL (100 μl) in the presence of [35S]Methionine for 120 min under standard conditions. The reaction was stopped by addition of 100 μl of 2% SDS in TNE (Tris-NaCl-ethylenediaminetetraacetic acid [EDTA]) buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA). Five minutes later, 10% Triton X-100 was added to the samples to a final concentration of 1%. Following further addition of 1 ml of 1% Triton X-100 in TNE buffer, samples were mixed with antibodies directed against either the N-terminal or C-terminal portion of luc protein (EPR17789 from Abcam [ab185923] and C-12 from Santa Cruz [sc-74548], respectively) that were immobilized on protein G-Sepharose (20 μl). After overnight incubation at 4°C and washing the beads with TNE buffer containing 1% Triton X-100 (1 ml, three times), bound proteins were dissolved in SDS-sample buffer, resolved by SDS-PAGE and detected by autoradiography.
Protein phosphorylation assays
Assays were conducted with HeLa S10 extracts as described previously (9,26). Reaction mixtures (12 μl) contained 50% (V/V) S10 extract, 2.5 mM spermidine, 1 mM Mg(OAc)2 and 20 μM (5 μCi) [γ-32P]ATP. Unmodified or modified Luc and GFP mRNAs (4 μg/ml) were added where indicated. After incubation at 30°C for 15 min, the phosphorylated proteins were analyzed by SDS-PAGE (9% acrylamide) and autoradiography.
Northern blot analysis
Untreated or RNase-treated Krebs extracts were incubated with Luc or N1mΨ–Luc mRNAs (4 μg/ml) at 30°C. At the indicated times, 12.5 μl aliquots of the reaction mixtures were withdrawn and the translation was stopped by the addition of SDS-proteinase K solution (20). Following incubation for 15 min at room temperature, total RNA was extracted with phenol–chloroform and precipitated with ethanol. RNA was separated on formaldehyde-1% agarose gels and transferred onto nylon membranes (Hybond-N, GE Healthcare). To confirm equal RNA loading, the blots were stained with Blot Stain Blue (Sigma) and the intensities of bands of 18S ribosomal RNA (rRNA) were measured using NIH Image J. software. RNA was then hybridized with ∼300 bp-long fragment of randomly primed 32P-labeled luc cDNA using ExpressHyb hybridization solution (Clontech Laboratories, Inc), as described by the manufacturer. The blots were exposed to X-ray films. Bands of Luc mRNA were quantified using a Typhoon PhosphorImager (GE Healthcare).
Statistical analysis
Data were analyzed by two-tailed unpaired Student'st-test.
RESULTS
Incorporation of N1mΨ into mRNA enhances protein expression in cells
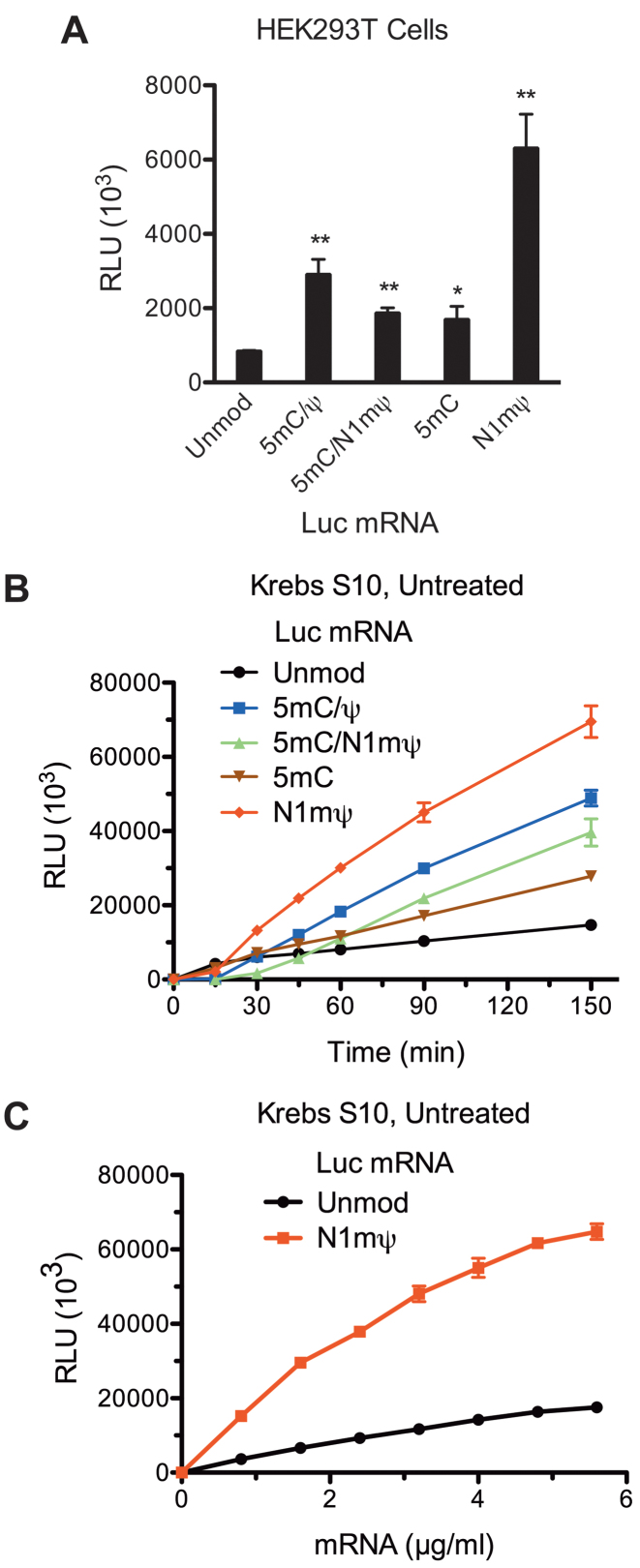
Global substitution of Ψ for uridine in in vitro transcribed mRNA is beneficial for protein expression (6,12). Furthermore, single N1mΨ or double 5 mC/N1mΨ-modified mRNAs were recently reported to outperform Ψ-containing mRNAs in mammalian cell lines and mice (27). To study the mechanism of enhanced capability of N1mΨ-modified mRNAs to express proteins, Luc mRNAs containing either none, one (5 mC or N1mΨ) or two (5 mC/N1mΨ) types of modified nucleosides were transfected in HEK293T cells and luc activity was monitored 4.5 h post-transfection. In addition, we tested the combination of 5 mC and Ψ nucleoside modifications (5 mC/Ψ), as this was reported to potentiate protein expression (28). All the modified mRNAs produced higher amounts of luc than the standard Luc mRNA (Figure 1A). The most dramatic stimulation of luc synthesis (7.4-fold) was elicited by the N1mΨ nucleoside modification. Combination of N1mΨ and 5 mC was not superior to N1mΨ alone (2.2-fold stimulation). The double 5 mC/Ψ modification of Luc mRNA produced a similar stimulatory effect (∼3-fold). Thus, optimal protein expression may not require extensive mRNA modification.
Figure 1.
Recapitulation of the translational enhancement by modified nucleosides in mRNA in Krebs extract. (A) Nucleoside modifications conferring enhanced translation to Luc mRNA in cells. Luc mRNAs, either not containing (Unmod) or containing the 5 mC/Ψ, 5 mC/N1mΨ (N1-methyl-pseudouridine), 5 mC and N1mΨ nucleoside modifications, were transfected into HEK293T cells. Cells were lysed 4.5 h after transfection and luc activity was measured in 1% aliquots of the lysates. (B) Time course analysis of luc synthesis in untreated Krebs extracts supplemented with unmodified or 5 mC/Ψ, 5 mC/N1mΨ, 5 mC and N1mΨ-incorporated Luc mRNAs (4 μg/ml). At the indicated time points after beginning of translation at 30°C, 1-μl aliquots of the reaction mixtures were assayed for luc activity. (C) Unmodified and N1mΨ-modified Luc mRNA dose response of translation in Krebs extract. Luc and N1mΨ–Luc mRNAs were translated in untreated Krebs extracts at the indicated concentrations. Following incubation at 30°C for 4 h, 1-μl aliquots of the translation mixtures were assayed for luc activity. Data are means from three assays ± SD (*P < 0.05, **P < 0.01). Relative luciferase units (RLU).
Stimulation of mRNA translation in vitro by nucleoside modifications
In cell-based assays, factors other than mRNA translation, e.g. cellular uptake, endosomal activity and delivery to the translation machinery, can determine the efficiency of protein expression. To analyze mRNA translation independent of these factors, we used a cell-free in vitro translation system derived from Krebs cells (14). To recapitulate the physiological conditions, we initially used Krebs extracts that were not micrococcal nuclease (RNase) treated. Luc mRNAs containing unmodified or modified (5 mC/Ψ, 5 mC/N1mΨ, 5 mC and N1mΨ) nucleosides were programmed into extracts and time course of luc synthesis was followed (Figure 1B). Consistent with the results in cells, the translation of N1mΨ–Luc mRNA was more sustainable than Luc mRNA, ultimately yielding more luc activity (∼4.7-fold more after 150 min). The enhancement of N1mΨ–Luc mRNA translation was independent of mRNA concentration (Figure 1C). Activities of other modified mRNAs, i.e. those with 5 mC/Ψ, 5 mC/N1mΨ and 5 mC nucleosides, were intermediate between those of Luc and N1mΨ–Luc mRNAs (Figure 1B). As with cells, combining the N1mΨ nucleoside modification with 5 mC significantly reduced its stimulatory effect. Thus, the translational enhancement imparted by modified nucleotides in mRNA in a cell-based system can be recapitulated in a Krebs extract.
Stability of Luc mRNA in Krebs extract is not altered by N1mΨ nucleoside modifications
In vitro transcribed unmodified mRNAs were previously shown to activate the antiviral OAS/RNase L system more potently than RNAs containing modified nucleosides (12). Accordingly, greater resistance of modified mRNA to cleavage by RNase L has been suggested to explain the enhanced protein production (12). We thus sought to determine whether the N1mΨ nucleoside modifications stabilize Luc mRNA in Krebs extracts. Northern blot analyzes of Luc and N1mΨ–Luc mRNA decay in untreated (Supplementary Figure S2A and B) or RNase-treated (Supplementary Figure S2C and D) Krebs extracts showed that both mRNAs are quite stable in these systems, as more than 50% remained intact after 150 min of incubation. We conclude that the OAS/RNase L RNA surveillance mechanism is inactive in the Krebs extract, making this system ideal for reliable comparison of translation efficiencies of unmodified and modified mRNAs.
mRNA modifications decrease the rate of polypeptide elongation
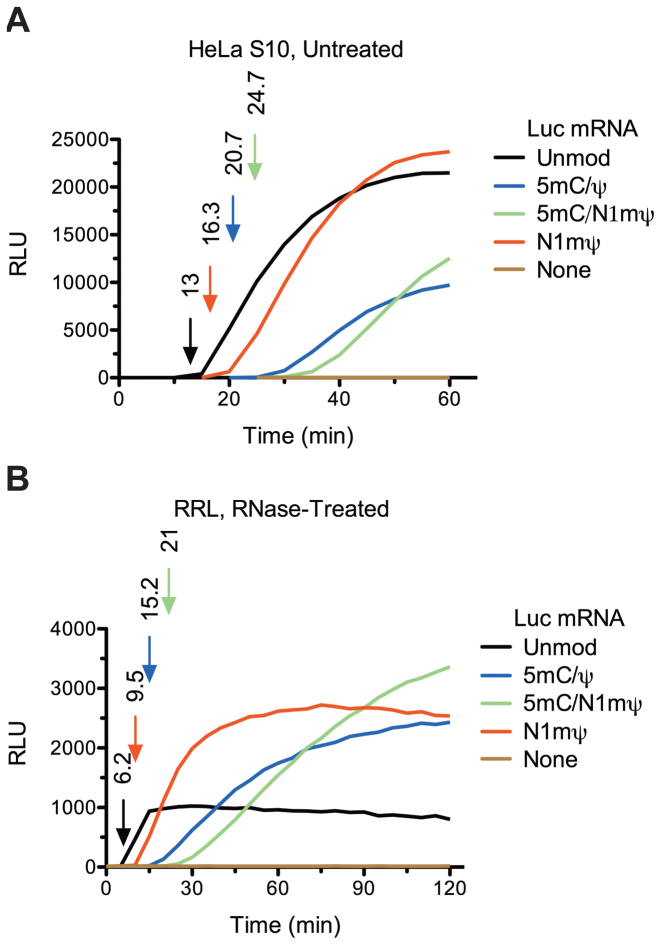
Curiously, the first appearance of luc activity occurred after a longer time lag in the extracts translating modified than unmodified Luc mRNA. This delay in luc synthesis was especially prominent for 5 mC/N1mΨ–Luc mRNA (Figure 1B). Previously, the first time point at which the luminescence signal becomes higher than the background level in Luc mRNA-programmed reactions has proved a reliable measure of translation velocity (29). To examine the lag between the start of translation and the appearance of luminescence signal in more detail, we used 3-min time increments to monitor luc activity. For the N1mΨ–Luc mRNA translation, luc activity was first detected after 15 min of incubation, as compared to the lag of ∼9-min in the control reaction (Supplementary Figure S3A). Treatment of Krebs extract with RNAse increased the lag of luc appearance by ∼1.3-fold for both Luc and N1mΨ–Luc mRNAs (Supplementary Figure S3A and B). To further increase the time resolution, we monitored protein synthesis in real time using HeLa and RRL in vitro translation systems. The smooth kinetic curves recorded in this assay allowed measurement of total translation time of mRNAs with high accuracy. The timing of the first appearance of luc activity was 13 min in HeLa extract and 6.2 min in RRL translating unmodified Luc mRNA (Figure 2A and B). This corresponds to translation velocity of ∼0.7 and 1.5 amino acids/s, in HeLa extract and RRL, respectively (given the length of Luc protein of 550 amino acids). Similar kinetics of unmodified mRNA translation was reported for other cell-free systems (29,30). However, in mammalian cells, a faster translation rate (5.5 amino acids/s) has been reported (31). In our extracts, nucleoside modifications in Luc mRNA delayed the appearance of luc activity in the following order: N1mΨ < 5 mC/Ψ < 5 mC/N1mΨ (Figure 2A and B). The extension of the single translation cycle by nucleoside modifications in mRNA is a likely consequence of the reduced elongation rate. For example, for N1mΨ–Luc mRNA, the rate of elongation would be expected to be ∼1.3- and ∼1.5-fold lower than on Luc mRNA in HeLa extract and RRL, respectively (Figure 2A and B). By the same estimate, incorporation of 5 mC/N1mΨ in Luc mRNA reduces elongation rate by 1.9- to 3.4-fold.
Figure 2.
Kinetics of luc synthesis in cell-free extracts translating unmodified or modified Luc mRNAs as determined by real-time measurements of Luc activity. Unmodified or 5 mC/Ψ, 5 mC/N1mΨ and N1mΨ-incorporated Luc mRNAs (20 μg/ml) were translated in untreated HeLa S10 extract (A) or RNase-treated RRL (B). The representative kinetic curves of luc synthesis and the background levels (None) are shown. The first time point at which the recorded signal is significantly above the background are taken as the durations of single translation cycle of mRNAs (indicated by arrows in matching colors). For details, see ‘Materials and Methods’ section.
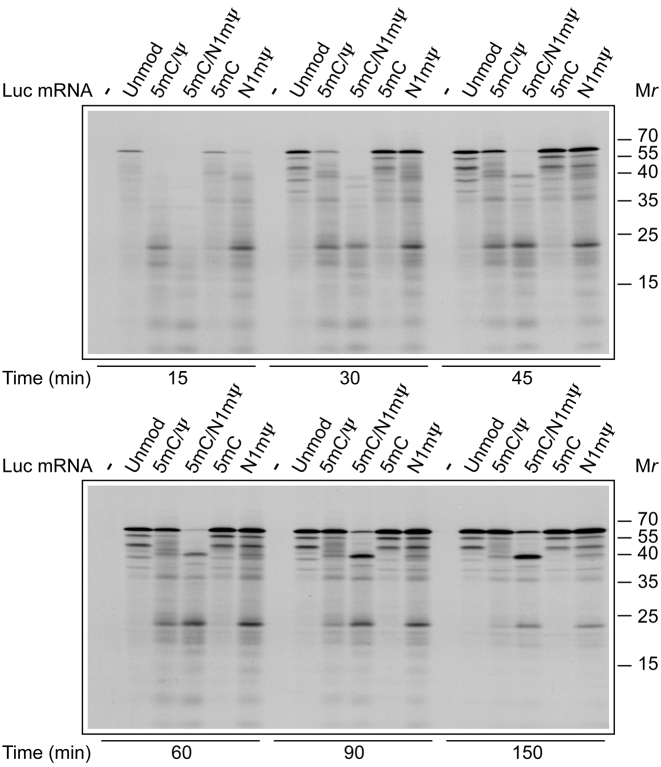
Steps other than elongation (e.g. initiation and termination of translation as well as protein folding) can potentially determine the first time point at which luc signal is detected in Luc mRNA programmed reactions. To investigate the impact of nucleoside modifications on the different steps of translation, we examined the kinetics of incorporation of 35S-methionine in RNase-treated Krebs extracts supplemented with unmodified or modified mRNAs. Translation time course analysis demonstrated that 5 mC/Ψ, 5 mC/N1mΨ and N1mΨ-containing Luc mRNA requires a longer time for synthesis of full-size luc protein as compared to the unmodified mRNA. After a short incubation of 15 min, the 62 kDa full-size luc protein while being prominent in the Luc and 5 mC-Luc mRNA programmed reactions, was barely detectable for the 5 mC/Ψ, 5 mC/N1mΨ and N1mΨ-containing Luc mRNAs (Figure 3). Likewise, the 5 mC/Ψ, 5 mC/N1mΨ and N1mΨ nucleoside modifications caused a delayed synthesis of the full-size luc in RRL (Supplementary Figure S4A). Thus, the delayed appearance of a luminescent signal in extracts programmed with modified Luc mRNAs (Figure 2) is not because of retarded folding, but rather because of slow synthesis of the luc protein.
Figure 3.
Time course of synthesis of polypeptides in Krebs extracts programmed with unmodified or modified Luc mRNAs. Unmodified or 5 mC/Ψ, 5 mC/N1mΨ, 5 mC and N1mΨ-incorporated Luc mRNAs (4 μg/ml) were translated in RNase-treated Krebs extracts in the presence of 35S-methionine. At the indicated time points, aliquots of the reaction mixtures were withdrawn and fixed with SDS-sample buffer. Translation products were analyzed by SDS-PAGE and autoradiography. Molecular mass markers are indicated on the right.
Interestingly, the translation of 5 mC/Ψ, 5 mC/N1mΨ and N1mΨ–Luc mRNAs yielded more nascent polypeptides or premature terminated products in both Krebs extract and RRL as compared to the unmodified mRNA (e.g. ∼20 kDa polypeptide (p20); Figure 3 and Supplementary Figure S4A). In addition, the 5 mC/N1mΨ–Luc mRNA produced a truncated protein of ∼40 kDa (p40). Although detectable for 5 mC/Ψ and N1mΨ Luc mRNAs as well, this product was much less abundant for the latter mRNAs. The formation of shortened luc polypeptides strongly suggests that ribosome movement is slowed down at the precise sites of modified mRNAs (32). However, there is a slight possibility that p20 and p40 are synthesized from alternative initiation sites in the 5 mC/N1mΨ–Luc mRNA. These polypeptides would be expected to differ from the full-size Luc protein with respect to their N-terminal amino acid sequence. Based on the results in Supplementary Figure S4B, it is evident that this scenario is highly unlikely. In this experiment, an N-terminal luc antibody efficiently immunoprecipitated p20, p40 and the full-length Luc protein formed in RRL. In contrast, and consistent with their C-terminal truncation, p20 and p40 failed to react with an antibody raised against the C-terminus of luc (Supplementary Figure S4B). Furthermore, inspecting the reading frame of Luc mRNA does not reveal potential translation start sites for p20 and p40 (i.e. the sites with the purine in position in position −3 and the G in position +4 relative to the A of AUG) (33). Interestingly, p20 differs from p40 with respect to its kinetics of accumulation in the 5 mC/N1mΨ–Luc mRNA-programmed reactions. While the amount of p40 as well as the full-size luc increased with time, the amount of p20 remained almost the same (Figure 3 and Supplementary Figure S4A). Thus, p20 seems to be a translational intermediate associated with stalled ribosomes rather than a terminal product of translation.
Importantly, while incorporation of N1mΨ in Luc mRNA delayed the appearance of luc in Krebs extract, the N1mΨ-modified and unmodified mRNAs directed 35S-methionine incorporation with similar initial kinetics (Supplementary Figure S3C). Furthermore, in RRL, neither N1mΨ nor other nucleoside modifications in Luc mRNA reduced the initial rate of 35S-methionine incorporation (Supplementary Figure S4C). We thus suspect that the negative impact of the decreased elongation rate on the translation of modified mRNAs is counterbalanced by increased initiation rate (see below).
If elongation were more limiting for the translation of the N1mΨ–Luc than Luc mRNA, than the translation of the former would be expected to be more sensitive to inhibition by suboptimal concentrations of elongation inhibitors (34). In support of this prediction, cycloheximide dose-response inhibition of luc synthesis directed by N1mΨ–Luc mRNA was stronger than that directed by Luc mRNA (∼3.2-fold, as judged by the values for the half maximal inhibitory concentration, IC50; Supplementary Figure S5A). In contrast, sequestering the cap-binding initiator factor eIF4E by adding increasing concentrations of the cap analog m7GpppG inhibited less the translation of the N1mΨ–Luc than that of Luc mRNA (Supplementary Figure S5B). The translational resistance of 5 mC/Ψ, 5 mC/N1mΨ and 5 mC-containing Luc mRNAs to inhibition by the cap analog was intermediate between those exhibited by Luc and N1mΨ–Luc mRNAs (Supplementary Figure S5C).
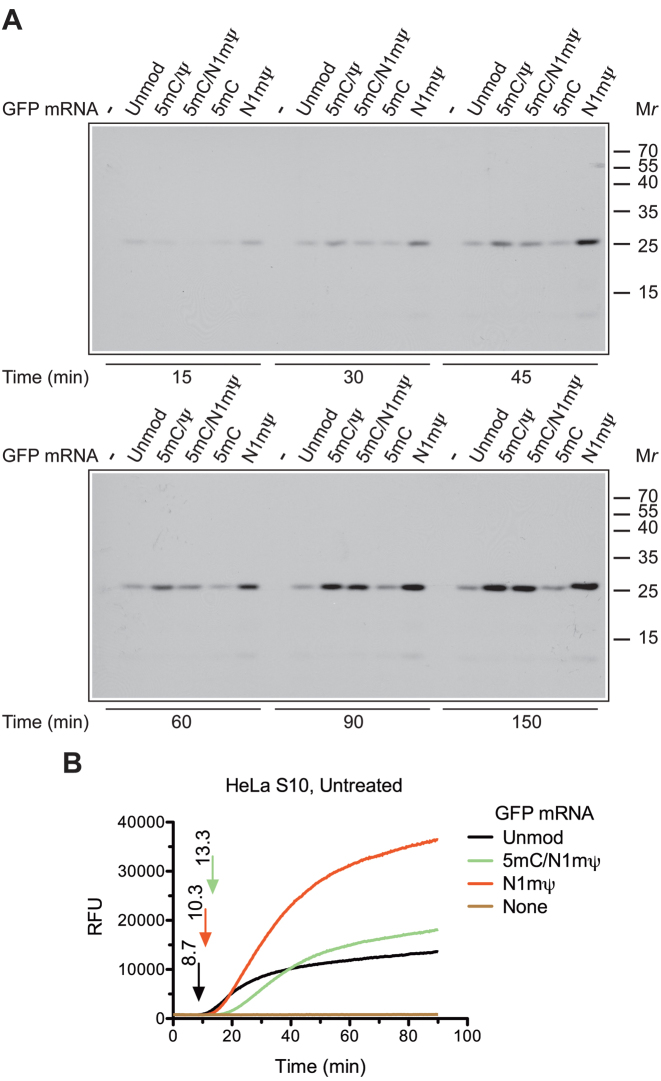
To generalize our conclusions, we also compared the translation of unmodified and modified GFP mRNAs. As for Luc mRNA, the translation of the N1mΨ–GFP mRNA in Krebs extract produced the highest amount of the full-size GFP protein as compared to GFP mRNAs that contain unmodified or other modified nucleosides (Figure 4A). However, we did not observe the synthesis of incomplete GFP polypeptides for any of the modified mRNAs. In addition, although a delay in the appearance of GFP fluorescence was observed for both N1mΨ and 5 mC/N1mΨ-containing GFP mRNA in a HeLa extract (with N1mΨ < 5 mC/N1mΨ), this delay was less pronounced than that for the Luc signal (compare Figure 4B with Figure 2A). This suggests that some features of GFP mRNA attenuate the effect of nucleoside modifications on the duration of the single translation cycle. These features could include mRNA length, RNA secondary structure or the sequence that determine the precise location of each nucleoside modifications.
Figure 4.
Time course of GFP synthesis in cell-free extracts translating unmodified or modified GFP mRNAs. (A) Unmodified or 5 mC/Ψ, 5 mC/N1mΨ, 5 mC and N1mΨ-incorporated GFP mRNAs (4 μg/ml) were translated in RNase-treated Krebs extracts in the presence of 35S-methionine. At the indicated time points, aliquots of the reaction mixtures were withdrawn for analysis by SDS-PAGE and autoradiography. The positions of molecular mass markers are indicated on the right. (B) Real-time kinetic analysis of active GFP synthesis in untreated HeLa S10 extract programmed with unmodified or 5 mC/N1mΨ and N1mΨ-incorporated GFP mRNAs (see the legend to Figure 2A for details). Relative fluorescence units (RFU).
Nucleoside modifications in mRNA promote translation by attenuating eIF2α phosphorylation
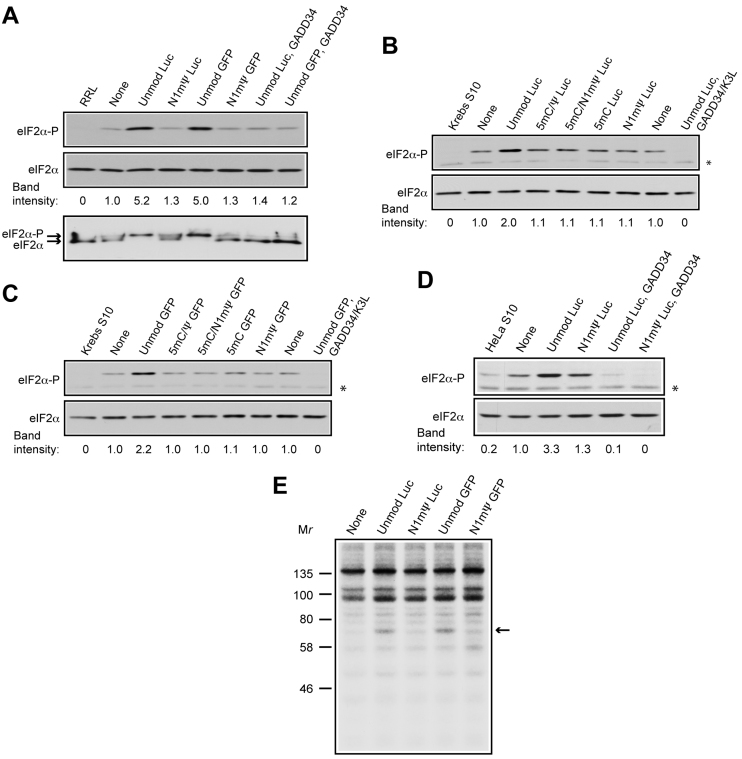
It is plausible that the translational superiority of modified mRNAs is determined by more efficient initiation or re-initiation of translation, which are generally the rate-limiting steps. One cause of translational enhancement could be the attenuation of eIF2α phosphorylation by nucleoside modifications in mRNA (6). We therefore asked how eIF2α phosphorylation status in our extracts is changed upon the addition of unmodified and modified mRNAs. Western blotting using an antibody against eIF2α phospho-Ser-51 revealed substantial phosphorylation of eIF2α following incubation of RNase-untreated RRL, Krebs or HeLa S10 extracts in the absence of mRNA (Figure 5A, top panel and Figure 5B–D—compare lanes none with RRL/Krebs S10/HeLa S10). This is most likely evoked by ATP and creatine phosphate as shown before (16). Strikingly, the addition of Luc or GFP mRNA to the extracts increased eIF2α phosphorylation over the baseline level in the control samples that lack mRNA (2–5.2-fold; Figure 5A, top panel and Figure 5B–D). Significantly, neither N1mΨ-containing nor other modified mRNAs elicited this effect. Consistent with published data (16), the recombinant protein GADD34, either alone or in combination with K3L, prevented eIF2α phosphorylation when added to the extracts. The human stress-inducible GADD34 protein promotes dephosphorylation of eIF2α by recruiting the phosphatase PP1 (35), while the vaccinia virus K3L protein inhibits the phosphorylation of eIF2α by acting as a pseudosubstrate (36). As RRL exhibited the greatest eIF2α phosphorylation response to addition of unmodified mRNA (∼5-fold), we chose this system to evaluate the overall degree of eIF2α phosphorylation using a combination of isoelectric focusing and western blotting (25). Incubation of RRL with unmodified Luc or GFP mRNA resulted in nearly complete phosphorylation of eIF2α (Figure 5A, bottom panel). In contrast, only partial conversion of unphosphorylated to phosphorylated form of eIF2α was observed in the water control or N1mΨ mRNA-supplemented samples. As expected, the inclusion of GADD34 in the reaction mixture inhibited eIF2α phosphorylation in the presence of unmodified Luc and GFP mRNAs.
Figure 5.
Unmodified, but not modified, mRNAs induce eIF2α phosphorylation and RNA-dependent protein kinase (PKR) activation in cell extracts. (A–D) Western blot analyzes of eIF2α phosphorylation in RNase untreated RRL (A, top panel), Krebs (B and C) or HeLa (D) S10 extracts not supplemented (none) or supplemented with unmodified or 5 mC/Ψ, 5 mC/N1mΨ, 5 mC and N1mΨ-incorporated Luc or GFP mRNAs (4 μg/ml), as indicated. GADD34 (6 μg/ml), either alone or in combination with K3L (16 μg/ml), was present where indicated. Extracts were either not incubated (RRL, Krebs S10 and HeLa S10) or incubated at 30°C for 30 min (A, top panel) or 60 min (B–D). The blots for Phospho-eIF2α (eIF2α-P) and total eIF2α (loading control) are shown. Phosphorylation (band intensity) of eIF2α normalized to the phosphorylation in the mRNA minus samples (none) is indicated below each panel. Asterisks indicate unspecific bands. (A, bottom panel) Phosphorylation states of eIF2α in RRL as analyzed by a combination of isoelectric focusing and western blotting. Arrows indicate the positions of phosphorylated and unphosphorylated forms of eIF2α. (E) Effects of unmodified and N1mΨ-incorporated mRNAs on protein phosphorylation in HeLa S10 extract. The assays were conducted in the absence (none) or presence of unmodified or N1mΨ-incorporated Luc and GFP mRNAs and [γ-32P]ATP as indicated. Molecular mass markers are indicated on the left. Arrow indicates the position of a phosphorylated 68-kDa protein.
To relate eIF2α phosphorylation to PKR activation, protein phosphorylation assays were carried in HeLa S10 extracts supplemented with unmodified or N1mΨ-incorporated mRNAs and [γ-32P]ATP. Addition of unmodified Luc or GFP mRNAs to the extracts resulted in the phosphorylation of ∼68 kDa protein (Figure 5E), which has been shown previously to correspond to PKR (9,26). In contrast, the N1mΨ-containing mRNAs induced no or very little phosphorylation of this protein. Thus, the diminished eIF2α phosphorylation in the presence of N1mΨ-containing mRNA could be explained by the failure of the mRNA to activate PKR. In agreement, PKR is feebly activated in cells by the Ψ-modified as compared to unmodified mRNA (6). As the phosphorylation of the 68-kDa protein did not occur in the absence of mRNA (Figure 5E, none), we conclude that the basal eIF2α phosphorylation is not mediated by PKR.
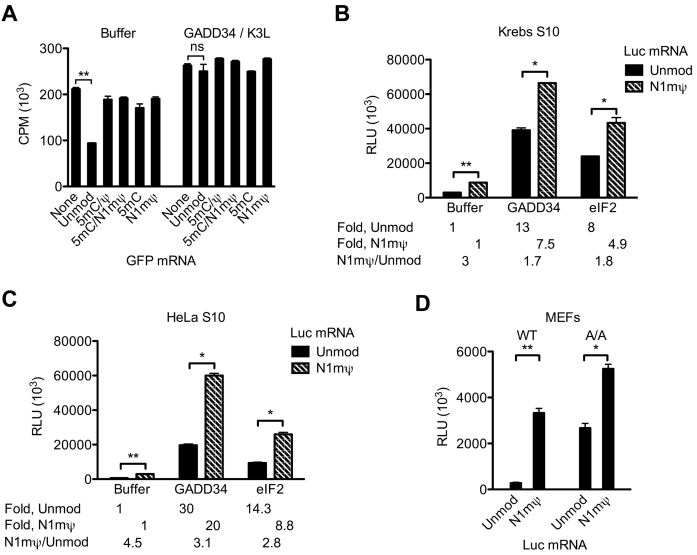
To determine whether the induction of eIF2α phosphorylation by unmodified mRNAs is sufficient to cause general translation inhibition, total cellular protein synthesis was monitored in RNase-untreated Krebs extract pre-incubated with unmodified or modified GFP mRNAs. Pre-incubation of the extract with GFP mRNA decreased 35S-methionine incorporation by 2.3-fold as compared to control (Figure 6A). In contrast, the effect of the 5 mC/Ψ, 5 mC/N1mΨ, 5 mC and N1mΨ nucleoside containing GFP mRNAs on endogenous protein synthesis was minimal (<1.2-fold inhibition). Importantly, trans-inhibition of 35S-methionine incorporation by GFP mRNA was averted when eIF2α phosphorylation was inhibited by the GADD34/K3L protein combination (Figure 6A).
Figure 6.
Unmodified, but not modified, mRNA induces general eIF2α phosphorylation-dependent translation repression. (A) Endogenous protein synthesis in the untreated Krebs extract as affected by unmodified or 5 mC/Ψ, 5 mC/N1mΨ, 5 mC and N1mΨ-incorporated GFP mRNAs. The extracts were pre-incubated at 30°C for 15 min with the indicated mRNAs (4 μg/ml) in the absence (Buffer) or presence of GADD34 (6 μg/ml) and K3L (16 μg/ml). 35S-methionine was then added to the samples, and the incubation was allowed to proceed for 60 min. 35S-methionine incorporation was measured in 1-μl aliquots of the samples. (B and C) GADD34 and eIF2 preferentially stimulate the in vitro translation of Luc as compared to N1mΨ–Luc mRNA. The untreated Krebs (B) or HeLa (C) S10 extracts were programed with unmodified or N1mΨ-incorporated Luc mRNA (4 μg/ml) in the absence or presence of GADD34 (6 μg/ml) or eIF2 (40 μg/ml). Following incubation at 30°C for 120 min, luc activity was measured in 1-μl aliquots of extracts. Fold stimulation of translation of Luc and N1mΨ–Luc mRNAs by GADD34 or eIF2 relative to Buffer controls and the ratios of translation of N1mΨ–Luc to Luc mRNA are indicated. (D) The 5 mC/Ψ nucleoside modification confers enhanced translation to Luc mRNA in wild-type (WT) and eIF2α-phosphorylation deficient (A/A) MEFs. Luc mRNAs, either not containing (Unmod) or containing the N1mΨ nucleoside modification, were transfected into WT or A/A MEFs. Cells were lysed 4.5 h after transfection. Luc activity was measured in 1% aliquots of the lysates. Data are means from three assays ± SD (*P < 0.05, **P < 0.001, ns—non-significant).
To determine whether the superior translation of the N1mΨ-containing Luc mRNA is exclusively due to the reduction of eIF2α phosphorylation, the translation of Luc and N1mΨ–Luc mRNAs was analyzed in Krebs and HeLa extracts in the presence of GADD34 (Figure 6B and C). Inhibition of basal and PKR-induced eIF2α phosphorylation in Krebs extract dramatically stimulated the translation of Luc mRNA (13-fold) (Figure 6B). By comparison, de-repression of N1mΨ–Luc mRNA translation by GADD34 was less pronounced (7.5-fold), consistent with inability of this mRNA to increase eIF2α phosphorylation above the basal level (Figure 5B). Similarly, inhibition of eIF2α phosphorylation in HeLa extract stimulated more the translation of Luc than N1mΨ–Luc mRNA (30- versus 20-fold) (Figure 6C). Thus, the Luc mRNA is more repressed by eIF2α phosphorylation than the N1mΨ–Luc mRNA. However, although significantly reduced, the translational difference between the N1mΨ–Luc and Luc mRNAs was still detectable in GADD34-supplemented extracts (1.7- to 3.1-fold; Figure 6B and C). In agreement with this, adding the initiation factor eIF2 reduced but did not negate the difference in the extent of Luc and N1mΨ–Luc mRNA translation. Therefore, the significantly reduced eIF2α phosphorylation only partially explains the superior translation of N1mΨ–Luc mRNA. To confirm the ability of the N1mΨ nucleoside modification to stimulate translation in eIF2α phosphorylation-independent manner in cells, Luc and N1mΨ–Luc mRNAs were transfected in MEFs homozygous for the Ser51 to Ala mutation in eIF2α (A/A). The mutant eIF2α cannot be phosphorylated (15). In WT MEFs, the Luc mRNA was 11 times less active than N1mΨ–Luc mRNA, which closely resembles the situation with HEK293T cells (Figure 6D). This relatively low expression of Luc mRNA was largely overcome in the A/A cells, as expected. However, the N1mΨ–Luc mRNA was expressed to a higher level than Luc mRNA even in the absence of eIF2α phosphorylation (∼2-fold).
Facilitation of polysome assembly by nucleoside modifications in mRNAs
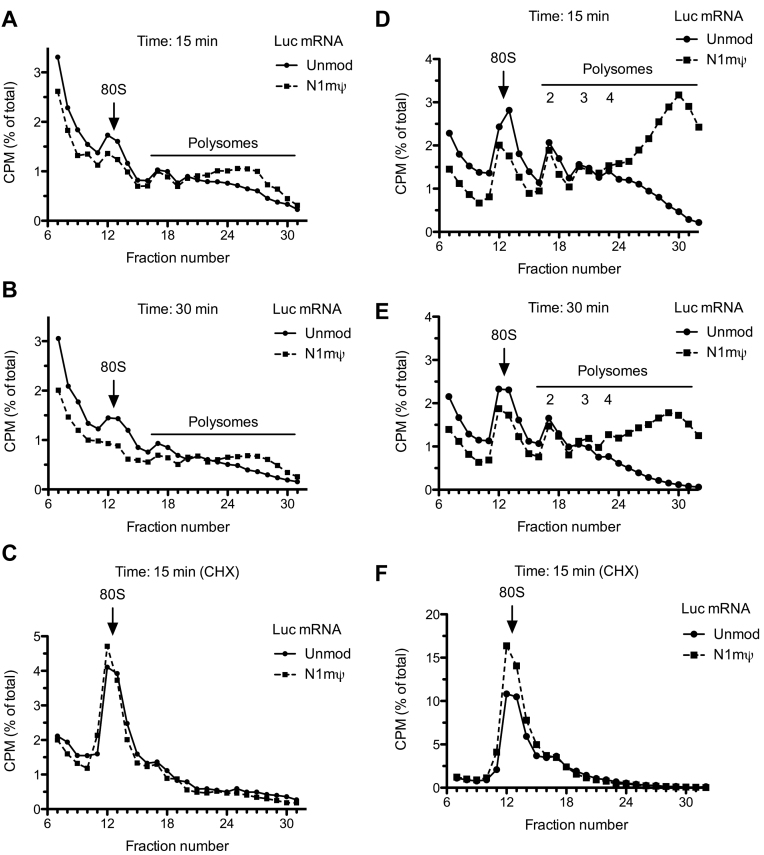
Increased initiation or decelerated elongation on modified mRNAs would be expected to increase polysome size and abundance. To test this, we analyzed polysome formation on 32P-labeled Luc and N1mΨ–Luc mRNAs in Krebs extracts. Both mRNAs were associated with polysomes after 15 or 30 min incubation (Figure 7A and B). However, the association of the N1mΨ–Luc mRNA with heavy polysomes was more pronounced than the Luc mRNA. In parallel, we characterized binding of the first ribosome to these mRNAs (80S initiation complex formation) using the elongation inhibitor cycloheximide. Interestingly, despite a higher proportion of N1mΨ–Luc mRNA that reached the state of translation after 15 min of incubation as compared to the Luc mRNA, these mRNAs formed 80S initiation complexes with similar efficiency (Figure 7C). Hence, it is plausible that binding of the first ribosome to unmodified mRNA in Krebs extract precedes eIF2α phosphorylation. In agreement, kinetic analysis has shown that the formation of 80S initiation complex on Luc mRNA in Krebs extract is already complete after 5 min of incubation, while eIF2α phosphorylation requires longer time to develop (10–15 min; Supplementary Figure S6A and B). To generalize these findings, Luc and N1mΨ–Luc mRNAs were subjected to polysome profiling after 15 and 30 min of incubation in untreated RRL. Strikingly, in this system, the polysome profile of N1mΨ–Luc mRNA differed from that of Luc mRNA much more than in the Krebs extract (Figure 7D and E). In particular, the N1mΨ nucleoside modification increased Luc mRNA association with heavy polysomes (arbitrarily defined as those containing more than four ribosomes) by more than 3.1- and 5.5-fold after 15 and 30 min of incubation, respectively. At variance with the Krebs extract, the proportion of N1mΨ–Luc mRNA engaged in 80S initiation complexes was ∼1.4 higher than Luc mRNA (Figure 7F). The 5 mC/Ψ–Luc and 5 mC/N1mΨ–Luc mRNAs were also more efficient than the Luc mRNA in 80S initiation complex formation in RRL (Supplementary Figure S7A and B). This may reflect the higher sensitivity of initiation in RRL to inhibition by eIF2α phosphorylation (7). Thus, the increase in polysome size on the N1mΨ–Luc mRNA in RRL is determined not only by the decreased elongation rate, but also by enhanced initiation during the pioneer round (and probably subsequent rounds) of translation. Inhibition of eIF2α phosphorylation preferentially de-represses the translation of the Luc mRNA as compared to N1mΨ–Luc (Figure 6B and C). In agreement with this observation, adding GADD34 to RRL preferentially increased the recruitment of ribosomes by the Luc mRNA (compare Supplementary Figures S8A and B). However, the increase of ribosome binding was insufficient to offset the difference between Luc and N1mΨ–Luc mRNAs with regard to their polysome distribution.
Figure 7.
Incorporation of N1mΨ in Luc mRNA increases the fraction of translated mRNA in cell-free extracts. Krebs extracts (A–C) or RRLs (D–F) untreated with RNase were incubated with 3΄ end labeled Luc or N1mΨ–Luc mRNAs. Polysomes were analyzed after 15 min (A and D) or 30 min (B and E) of incubation of the reaction mixtures at 30°C. 80S initiation complex formation in cycloheximide-supplemented Krebs extract (C) or RRL (F) is also shown. For details, see ‘Materials and Methods’ section. In the profiles of RRL, the discernible peaks of light polysomes (disomes, trisomes and qudrosomes) are indicated. In Krebs extract after 15 and 30 min of incubation, the formation of heavy polysomes (fractions 24–31) was more efficient on N1mΨ–Luc than Luc mRNA (A and B). In RRL, the engagement of N1mΨ–Luc mRNA in heavy polysomes, which contain more than four ribosomes (fractions 24–32), was >3.1-fold and >5.5-fold greater than Luc mRNA after 15 min (D) and 30 min (E) of incubation, respectively. In RRL, the N1mΨ–Luc mRNA was more efficient than Luc mRNA in 80S initiation complex formation (∼1.4-fold, fractions 11–15, panel F). Six top fractions of the gradients are omitted for greater clarity.
In conclusion, incorporation of N1mΨ nucleoside modification in both Luc and GFP mRNA enhances the initiation step of translation, in part by suppressing eIF2α phosphorylation. In addition, polysome formation and growth on the N1mΨ-containing Luc mRNA is enhanced due to the reduction of elongation rate.
DISCUSSION
Incorporation of Ψ or N1mΨ into cell-transfected mRNA enhances protein expression by increasing both translation and stability of the mRNA (6,12,27). Hence, the use of intact cells makes it hard to study the role of translation in the stimulation of protein synthesis. As shown here, both Krebs and HeLa cell-free translation extracts as well as RRL faithfully recapitulate the increase in protein expression from modified mRNAs. Moreover, Luc and N1mΨ–Luc mRNAs exhibit similar stabilities in Krebs extract, making it possible to uncouple the contribution from mRNA stabilization to overall protein synthesis. In all the in vitro translation systems, incorporation of N1mΨ in Luc and GFP mRNAs dramatically enhanced translation. In this and a related study (27), the major cause of this stimulation has been assigned to the reduced activation of PKR and phosphorylation of eIF2α.
Surprisingly, the rate of polypeptide chain elongation is decreased on modified mRNAs. Notwithstanding that the time needed for initiation on N1mΨ–Luc mRNA in Krebs extract and RRL is not longer than that on Luc mRNA (Figure 7C and F), the N1mΨ–Luc mRNA translation showed ∼1.5-fold longer delay in the first appearance of luc protein and activity (Figures 2 and 3; Supplementary Figures S3B and S4A). Furthermore, it appears that modified nucleosides preferentially affect translational dynamics at specific sites of the mRNA. This is evidenced by a higher abundance of truncated proteins from modified mRNA translation. In principle, site-specific ribosome clustering on mRNA should occur if the time of clearance of a ribosome occupied RNA fragment (∼30 nt) were greater than the time needed for initiation (32). In our case, the appearance of a prominent ∼20 kDa translational intermediate among the products of translation of 5 mC/Ψ, 5 mC/N1mΨ and N1mΨ–Luc mRNAs indicates the existence of a ribosome pausing site within the first 400 nt of the Luc open reading frame. That the velocity of ribosome movement is uneven along the mRNA is very well established and tRNA selection being suggested as a major rate-limiting step in ribosome progression (29,32,37–39). It is plausible that modified nucleosides in mRNA impinge on the stability of the codon-anticodon duplex and consequently, on the speed of ribosome decoding. Another possibility is that modified nucleosides stabilize mRNA secondary structures, akin to Ψ in tRNA and rRNA (40). Data from ribosome profiling and single molecule imaging experiments indicate that stable secondary structures in mRNA pose a hurdle for translating ribosomes (39,41,42). Finally, it cannot be excluded that some of the mRNA-binding proteins (43,44) whose affinity for RNA is altered by nucleoside modifications modulate the elongation rate.
Our data support the general tenet that initiation rather than elongation largely determines overall translation efficiency (8,45). If this were not the case, the decelerated elongation rate on modified mRNAs would have been deleterious for translation. The major mechanism that enhances the translation of modified mRNAs is the attenuation of eIF2α phosphorylation. While standard in vitro transcribed mRNAs increase eIF2α phosphorylation and impose general translation repression in Krebs extracts, mRNAs bearing nucleoside modifications fail to do so (Figures 5 and 6A). The eIF2α phosphorylation response to the unmodified mRNAs correlates with the phosphorylation of the 68-kDa protein previously identified as PKR (9,26). Consistently, in intact cells, Ψ-containing mRNA activates PKR and increases eIF2α phosphorylation to a lesser extent than standard mRNA (6). It is possible that the altered secondary structures in modified mRNAs cannot be recognized well by PKR (6,9).
The N1mΨ–Luc mRNA is associated with heavier polysomes than Luc mRNA (Figure 7). This is the expected consequence of a faster initiation rate and a slower ribosome movement along the N1mΨ–Luc mRNA. Interestingly, we did not detect enhancement of 80S initiation complex formation on the N1mΨ-incorporated Luc mRNA in Krebs extract although in RRL it was clearly observed. This could be explained by the delay of eIF2α phosphorylation in Krebs extracts relative to 80S initiation complex formation (Supplementary Figure S6A and B). Low phosphorylation of eIF2α in Krebs extract at early time points can also account for a relatively smaller difference between polysome formation on Luc and N1mΨ–Luc mRNAs as compared to the translation difference (which are measured after 15–30 and 60–150 min of incubation, respectively).
Significantly, because phosphorylation of eIF2α was similarly reduced by all nucleoside modifications in Luc and GFP mRNAs, but the N1mΨ-containing mRNAs were translated better than other modified mRNAs, the attenuation of eIF2α phosphorylation is not the only mechanism that leads to the enhancement of translation of the N1mΨ-modified mRNAs. Consistently, the reduction in eIF2α phosphorylation to a nearly undetectable level in Krebs and HeLa S10 extract by the addition of GADD34 did not completely offset the translational advantage of the N1mΨ–Luc mRNA over the Luc mRNA (Figure 6B and C). Moreover, the N1mΨ–Luc mRNA outperformed Luc mRNA in eIF2α phosphorylation-deficient MEF cells (Figure 6D). We conclude that the superb translation activity of N1mΨ–nucleoside modified mRNA is partially due to increased ribosome density resulting from the deceleration of elongation. Strikingly, in the steady state of translation, a significant fraction of heavy-loaded polysomes in wheat germ and HeLa cell extracts has been shown to acquire a double-raw ‘circular’ structure (46–48). These compact polysomes exhibit slow exchange with free ribosomes and mRNA suggesting that terminating ribosomes predominantly initiate translation on the same mRNA template. Furthermore, there is evidence to suggest that this circular translation mode does not involve scanning of the 5΄ UTR, which is dependent on eIF4F (46). Since eIF4F-cap interaction is generally the rate-limiting step in translation (49,50), one would expect that the permissiveness of the mRNA for ribosome recruitment should increase once ribosome packing reaches a critical level. Higher translational resistance of N1mΨ–Luc than Luc mRNA to inhibition by a cap analog is consistent with more efficient recycling of ribosomes from the 3΄- to 5΄-end of the N1mΨ–Luc mRNA in an eIF4F-independent manner (Supplementary Figure S5B). Obviously, overall translation would diminish if ribosome pausing on the mRNA were too frequent so that it affects ribosome processivity (i.e. the ability of initiating ribosome to traverse the entire open reading frame of mRNA). Based on the timing of appearance of luc protein and activity in Krebs extract and RRL the 5 mC/Ψ and 5 mC/N1mΨ nucleoside modifications in Luc mRNA reduce elongation rates stronger that the single N1mΨ or 5 mC modification (Figures 1B, 2A, B, 3 and Supplementary Figure S4A). Such an extent of ribosome movement retardation is probably too strong to ensure an optimal translation output. In addition, a very slow elongation rate could lead to the formation of premature terminated proteins, such as p40 in the 5 mC/N1mΨ–Luc mRNA-programmed reactions. It is noteworthy that the cap-independent mechanism of ribosome recruitment has recently been suggested for mRNAs that contain N6-methyladenosine (m6A) nucleoside modification in the 5΄ UTR (51). These mRNAs purportedly binds eIF3 that is a critical component of the 43S pre-initiation complex. We consider this mechanism irrelevant for the mRNAs used in this study. As shown, all the employed nucleoside modifications only relaxed, but did not eliminate, cap-dependence of Luc mRNA translation (Supplementary Figure S5B and C).
In summary, we provide strong biochemical evidence that the increased translation from nucleoside-modified mRNAs is due to the enhancement of the initiation step of translation and that the attenuation of eIF2α phosphorylation plays an important role in this process. We also discovered a role of modified nucleosides in mRNA in increasing polysome complexity and suggest that tight ribosome packing and cooperation on modified mRNA confers an advantage for initiation or ribosome recycling. Our results are of broad significance given the presence of modified nucleosides (e.g. 5 mC, Ψ, m6A, m1A, inosine and 2΄-O-methylated nucleosides) in almost all cellular mammalian mRNA (52–60). Although only reported for 18S rRNA (61), the natural N1mΨ nucleoside modification could be present in mRNA, as it might not be distinguished from Ψ in the N-cyclohexyl-N'-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate (CMC)-based high-throughput analysis (52). Significantly, for the m1A methylation, a dynamic response to stimuli and correlation with elevated translation has been recently documented (57,60). In light of our data, the role of natural chemical modifications of mRNA would be not only to suppress mRNA recognition by the innate immune system, but also to increase the robustness of translation of select mRNAs by promoting ribosome recycling. Finally, ribosome pausing has emerged as an important means to fine-tune the folding of the nascent polypeptide chains (62). Thus, the natural nucleoside modifications in mRNA can assist in proper folding of proteins by determining the location and potency of ribosome pausing sites on the mRNA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dan Dominissini for critical reading of the manuscript, Hiroaki Imataka for plasmids encoding proteins GADD34 and K3L, Maria Hatzoglou for WT and A/A MEFs, and Sandra Perreault for excellent technical assistance.
Footnotes
Present address: Tirtha Chakraborty, and Matthias John, CRISPR Therapeutics, Cambridge, MA 02139, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Moderna Therapeutics; Canadian Institute of Health Research (CIHR) [MOP-7214 to N.S.]. Funding for open source: CIHR [MOP-7214 to N.S.].
Conflict of interest statement. Y.M.C., T.C., V.P. and M.J. are employees of Moderna. N.S. is a paid consultant of Moderna. Y.V.S. has none to declare.
REFERENCES
- 1. Sahin U., Kariko K., Tureci O.. mRNA-based therapeutics–developing a new class of drugs. Nat. Rev. Drug Discov. 2014; 13:759–780. [DOI] [PubMed] [Google Scholar]
- 2. Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Spater D., Xu H., Tabebordbar M., Gorbatov R. et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013; 31:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011; 29:154–157. [DOI] [PubMed] [Google Scholar]
- 4. Kariko K., Buckstein M., Ni H., Weissman D.. Suppression of Rna recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005; 23:165–175. [DOI] [PubMed] [Google Scholar]
- 5. Nallagatla S.R., Toroney R., Bevilacqua P.C.. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr. Opin. Struct. Biol. 2011; 21:119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Kariko K.. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010; 38:5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ehrenfeld E., Hunt T.. Double-stranded poliovirus RNA inhibits initiation of protein synthesis by reticulocyte lysates. Proc. Natl. Acad. Sci. U.S.A. 1971; 68:1075–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sonenberg N., Hinnebusch A.G.. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009; 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edery I., Petryshyn R., Sonenberg N.. Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell. 1989; 56:303–312. [DOI] [PubMed] [Google Scholar]
- 10. Kariko K., Muramatsu H., Ludwig J., Weissman D.. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011; 39:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmid M., Jensen T.H.. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008; 33:501–510. [DOI] [PubMed] [Google Scholar]
- 12. Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Kariko K.. Nucleoside modifications in RNA limit activation of 2΄-5΄-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011; 39:9329–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chakrabarti A., Jha B.K., Silverman R.H.. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011; 31:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Svitkin Y.V., Sonenberg N.. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 2004; 257:155–170. [DOI] [PubMed] [Google Scholar]
- 15. Zeenko V.V., Wang C., Majumder M., Komar A.A., Snider M.D., Merrick W.C., Kaufman R.J., Hatzoglou M.. An efficient in vitro translation system from mammalian cells lacking the translational inhibition caused by eIF2 phosphorylation. RNA. 2008; 14:593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mikami S., Kobayashi T., Yokoyama S., Imataka H.. A hybridoma-based in vitro translation system that efficiently synthesizes glycoproteins. J. Biotechnol. 2006; 127:65–78. [DOI] [PubMed] [Google Scholar]
- 17. Mikami S., Kobayashi T., Machida K., Masutani M., Yokoyama S., Imataka H.. N-terminally truncated GADD34 proteins are convenient translation enhancers in a human cell-derived in vitro protein synthesis system. Biotechnol. Lett. 2010; 32:897–902. [DOI] [PubMed] [Google Scholar]
- 18. Pisarev A.V., Unbehaun A., Hellen C.U., Pestova T.V.. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007; 430:147–177. [DOI] [PubMed] [Google Scholar]
- 19. Svitkin Y.V., Sonenberg N.. A highly efficient and robust in vitro translation system for expression of picornavirus and hepatitis C virus RNA genomes. Methods Enzymol. 2007; 429:53–82. [DOI] [PubMed] [Google Scholar]
- 20. Svitkin Y.V., Pause A., Lopez-Lastra M., Perreault S., Sonenberg N.. Complete translation of the hepatitis C virus genome in vitro: membranes play a critical role in the maturation of all virus proteins except for NS3. J. Virol. 2005; 79:6868–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Svitkin Y.V., Herdy B., Costa-Mattioli M., Gingras A.C., Raught B., Sonenberg N.. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 2005; 25:10556–10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Svitkin Y.V., Yanagiya A., Karetnikov A.E., Alain T., Fabian M.R., Khoutorsky A., Perreault S., Topisirovic I., Sonenberg N.. Control of translation and miRNA-dependent repression by a novel poly(A) binding protein, hnRNP-Q. PLoS Biol. 2013; 11:e1001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rifo R.S., Ricci E.P., Decimo D., Moncorge O., Ohlmann T.. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007; 35:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Svitkin Y.V., Evdokimova V.M., Brasey A., Pestova T.V., Fantus D., Yanagiya A., Imataka H., Skabkin M.A., Ovchinnikov L.P., Merrick W.C. et al. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J. 2009; 28:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Savinova O., Jagus R.. Use of vertical slab isoelectric focusing and immunoblotting to evaluate steady-state phosphorylation of eIF2 alpha in cultured cells. Methods. 1997; 11:419–425. [DOI] [PubMed] [Google Scholar]
- 26. Petryshyn R., Chen J.J., London I.M.. Growth-related expression of a double-stranded RNA-dependent protein kinase in 3T3 cells. J. Biol. Chem. 1984; 259:14736–14742. [PubMed] [Google Scholar]
- 27. Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T.. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control Release. 2015; 217:337–344. [DOI] [PubMed] [Google Scholar]
- 28. Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010; 7:618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu C.H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M.S., Liu Y.. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 2015; 59:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vassilenko K.S., Alekhina O.M., Dmitriev S.E., Shatsky I.N., Spirin A.S.. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 2011; 39:5555–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wolin S.L., Walter P.. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988; 7:3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997; 16:2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mathews MB., Sonenberg N., Hershey J.W.B.. Mathews MB, Sonenberg N, Hershey JWB. Origins and principles of translational control. Translational Control in Biology and Medicine. 2007; NY: Cold Spring Harbor Laboratory Press; 1–40. [Google Scholar]
- 35. Novoa I., Zeng H., Harding H.P., Ron D.. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 2001; 153:1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carroll K., Elroy-Stein O., Moss B., Jagus R.. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J. Biol. Chem. 1993; 268:12837–12842. [PubMed] [Google Scholar]
- 37. Richter J.D., Coller J.. Pausing on polyribosomes: make way for elongation in translational control. Cell. 2015; 163:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee T.H., Blanchard S.C., Kim H.D., Puglisi J.D., Chu S.. The role of fluctuations in tRNA selection by the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tuller T., Waldman Y.Y., Kupiec M., Ruppin E.. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Charette M., Gray M.W.. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000; 49:341–351. [DOI] [PubMed] [Google Scholar]
- 41. Chen C., Zhang H., Broitman S.L., Reiche M., Farrell I., Cooperman B.S., Goldman Y.E.. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 2013; 20:582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pop C., Rouskin S., Ingolia N.T., Han L., Phizicky E.M., Weissman J.S., Koller D.. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014; doi:10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 44. Dethoff E.A., Chugh J., Mustoe A.M., Al-Hashimi H.M.. Functional complexity and regulation through RNA dynamics. Nature. 2012; 482:322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ingolia N.T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014; 15:205–213. [DOI] [PubMed] [Google Scholar]
- 46. Bonderoff J.M., Lloyd R.E.. Time-dependent increase in ribosome processivity. Nucleic Acids Res. 2010; 38:7054–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kopeina G.S., Afonina Z.A., Gromova K.V., Shirokov V.A., Vasiliev V.D., Spirin A.S.. Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA. Nucleic Acids Res. 2008; 36:2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Afonina Z.A., Myasnikov A.G., Shirokov V.A., Klaholz B.P., Spirin A.S.. Conformation transitions of eukaryotic polyribosomes during multi-round translation. Nucleic Acids Res. 2015; 43:618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Topisirovic I., Svitkin Y.V., Sonenberg N., Shatkin A.J.. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA. 2011; 2:277–298. [DOI] [PubMed] [Google Scholar]
- 50. Merrick W.C. eIF4F: a retrospective. J. Biol. Chem. 2015; 290:24091–24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R.. 5΄ UTR m(6)A promotes cap-independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V.. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014; 515:143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 54. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3΄ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T.. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012; 40:5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A., Fabris D., Agris P.F.. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011; 39:D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M.S., Dai Q., Di Segni A., Salmon-Divon M., Clark W.C. et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016; 530:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., Leon-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014; 159:148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li X., Zhu P., Ma S., Song J., Bai J., Sun F., Yi C.. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015; 11:592–597. [DOI] [PubMed] [Google Scholar]
- 60. Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C.. Transcriptome-wide mapping reveals reversible and dynamic N-methyladenosine methylome. Nat. Chem. Biol. 2016; 12:311–316. [DOI] [PubMed] [Google Scholar]
- 61. Brand R.C., Klootwijk J., Planta R.J., Maden B.E.. Biosynthesis of a hypermodified nucleotide in Saccharomyces carlsbergensis 17S and HeLa-cell 18S ribosomal ribonucleic acid. Biochem. J. 1978; 169:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Komar A.A. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009; 34:16–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.