Abstract
HBO1, a member of the MYST family of histone acetyltransferases (HATs), is required for global acetylation of histone H3K14 and embryonic development. It functions as a catalytic subunit in multisubunit complexes comprising a BRPF1/2/3 or JADE1/2/3 scaffold protein, and two accessory proteins. BRPF2 has been shown to be important for the HAT activity of HBO1 toward H3K14. Here we demonstrated that BRPF2 can regulate the HAT activity of HBO1 toward free H3 and H4, and nucleosomal H3. Particularly, a short N-terminal region of BRPF2 is sufficient for binding to HBO1 and can potentiate its activity toward H3K14. The crystal structure of the HBO1 MYST domain in complex with this segment of BRPF2 together with the biochemical and cell biological data revealed the key residues responsible for the HBO1–BRPF2 interaction. Our structural and functional data together indicate that the N-terminal region of BRPF2 plays an important role in the binding of HBO1 and a minor role in the binding of nucleosomes, which provide new mechanistic insights into the regulation of the HAT activity of HBO1 by BRPF2.
INTRODUCTION
In eukaryotic cells, covalent modifications at specific sites of histones, such as acetylation, methylation, phosphorylation, ubiquitination and sumoylation can influence chromatin structure and dynamics, and hence regulate diverse cellular processes including transcription, DNA replication and repair, and cell cycle progression (1,2). Among the many histone modifications, lysine acetylation has been extensively studied and is well known to regulate chromatin accessibility (3,4). Lysine acetylation is deposited on chromatin by histone acetyltransferases (HATs) and removed by histone deacetylases (HDACs). Based on their sequence and structure similarities, HATs are grouped into four major families, namely Gcn5/PCAF, MYST, p300/CBP and Rtt109 (3). The MYST family HATs, including KAT5/Tip60/Esa1, KAT6a/MOZ/MYST3/Sas3, KAT6b/MORF/MYST4, KAT7/HBO1/MYST2, KAT8/MOF/MYST1/Sas2 are conserved in both sequence and structure from yeast to human in their catalytic HAT domains (also called MYST domains) with each containing a characteristic acetyl-CoA (AcCoA) binding motif and a zinc finger. However, these enzymes have different substrate specificities and function in distinct biological processes (5,6).
HBO1 of the MYST family was initially identified as a binding partner of the origin recognition complex (ORC) and was found to contain a conserved MYST domain and has the HAT activity toward histones H3 and H4 (Supplementary Figure S1A) (7–14). In addition, HBO1 can exert activity on some non-histone proteins including ORC2, MCM2 and CDC6 (15). Owing to its broad-spectrum acetyltransferase activities, HBO1 acts as a multifunctional protein in both transcription activation and DNA replication. Deletion of HBO1 in embryonic cells causes profound reduction of global H3K14 acetylation and embryonic lethality at E10.5 due to decreased expression of genes that regulate embryonic patterning (12). HBO1 can regulate DNA replication in two ways: it promotes the assembly of pre-replicative complex (Pre-RC) by acetylating H4K5/8/12 and interacting with Pre-RC components (10,15–18); and it also promotes the loading of CDC45 to activate DNA replication in S phase by acetylating H3K14 (14). However, HBO1 overexpression leads to excessive replication activation and cell proliferation, which could be a marker of tumorigenesis particularly among testicular germ cell tumors, breast adenocarcinomas and ovarian serous carcinomas (8).
HATs usually interact with scaffold proteins and other accessory partners to form HAT complexes and exert functions (3,19,20). HBO1 forms HAT complexes with scaffold proteins JADE1/2/3 or BRPF1/2/3, and accessory proteins ING4/5 and Eaf6 (6,9–11,14,21–23). The scaffold proteins bridge HBO1 and the accessory proteins, and the interplay and proper assembly modulate the substrate specificities and activities of the HAT complexes. In addition, HBO1 can bind to different scaffold proteins and this differential association also helps to determine the acetylation sites of histones or the substrate specificities of the HAT complexes. The HBO1–JADE1/2/3 complexes can acetylate both H3 and H4 on free histones but are more specific toward H4 (H4K5/8/12) in the context of chromatin (9–11,21,23). The HBO1–BRPF1/2/3 complexes can acetylate free H3 and H4 but prefer to acetylate H3 on chromatin with slightly varied specificities: the HBO1–BRPF1 complex can specifically acetylate H3K14/23 (11), and the HBO1–BRPF2 and HBO1–BRPF3 complexes have a high specificity toward H3K14 (13,14,22).
BRPF2 (also called BRD1 or BRL) is a scaffold protein of 1189 residues consisting of a PZP, bromo and PWWP domain, which are typical recognition modules involved in binding chromatin and/or specific histone modifications (Supplementary Figure S1A) (13,24). BRPF2 deficient mice exhibits profoundly decreased level of global acetylation of H3K14, similar to HBO1 deletion mice and remarkably impaired fetal liver erythropoiesis (13). BRPF2 and HBO1 largely co-localize in the genome and share a significant portion of their target genes involved in transcriptional regulation. The N-terminal region (residues 1–198) of BRPF2 is mapped to be responsible for its interaction with the MYST domain of HBO1 (13). These data indicate that the interaction between HBO1 and BRPF2 is important for the proper function of HBO1, particularly in the global acetylation of H3K14. However, the molecular mechanisms for how BRPF2 interacts with HBO1 and how the interaction regulates the function of HBO1 are unclear.
Here we demonstrate that a short N-terminal region of BRPF2 (residues 31–80) is sufficient to interact directly with HBO1 and additionally can enhance the HAT activity of HBO1 toward H3K14 in vitro. We also report the crystal structure of the MYST domain of HBO1 in complex with this BRPF2 segment, which together with biochemical and cell biological data reveals the key residues involved in the HBO1–BRPF2 interaction. Together, our structural and functional data indicate that the N-terminal region of BRPF2 plays an important role in the binding of HBO1 and a minor role in the binding of nucleosomes. These findings provide new mechanistic insights into the regulation of the HAT activity of HBO1 by BRPF2 and possibly other scaffold proteins.
MATERIALS AND METHODS
Cloning, expression and purification
cDNA fragments corresponding to the MYST domain of HBO1 (residues 336–611) and various BRPF2 fragments were all amplified with polymerase chain reaction from the cDNA library of human HEK 293T cells. The HBO1 and BRPF2 mutants were generated using the QuikChange® Site-Directed Mutagenesis kit (Strategene). All constructs were confirmed by DNA sequencing.
For in vitro HAT activity assay and structural study of HBO1, cDNA fragment corresponding to the MYST domain of HBO1 was cloned into the pET-22b plasmid (Novagen) with an N-terminal His6 tag. The plasmid was expressed in Escherichia coli BL21 (DE3) Codon-Plus strain (Novagen) and the transformed cells were grown at 37°C in Luria-Bertani medium until OD600 reached 0.8 and then induced with 0.2 mM Isopropyl β-D-1-Thiogalactopyranoside at 16°C for 24 h. The cells were harvested by centrifugation, resuspended in a lysis buffer [20 mM Tris–HCl (pH 8.0), 300 mM NaCl, 10% (w/v) glycerol and 1 mM phenylmethylsulfonyl fluoride] and then lysed by sonication. The precipitate was removed by centrifugation and the HBO1 protein was purified by affinity chromatography using a Ni-NTA column (QIAGEN) with the lysis buffer supplemented with 20 and 300 mM imidazole serving as the washing and elution buffer, respectively. The HBO1 protein was further purified by gel filtration using a Superdex 200 10/300 column (GE Healthcare) pre-equilibrated with the storage buffer containing 20 mM Tris–HCl (pH 8.0), 300 mM NaCl and 2 mM dithiothreitol.
For structural study of the HBO1–BRPF2 complex, cDNA fragment corresponding to the MYST domain of HBO1 was cloned into the pET-28a plasmid (Novagen) without tag and cDNA fragment corresponding to the N-terminal region of BRPF2 (residues 31–80) was cloned into the pET-22b plasmid with an N-terminal His6 tag. The two plasmids were co-expressed in E. coli BL21 (DE3) Codon-Plus strain as described above. The HBO1–BRPF2 complex was first purified with affinity chromatography using a Ni-NTA column. After removal of the N-terminal His6 tag of BRPF2 by Tobacco etch virus protease (TEV), the HBO1–BRPF2 complex was further purified with affinity chromatography using a Ni-NTA column and gel filtration chromatography using a Superdex 200 16/60 column (GE Healthcare).
For Glutathione S-transferase (GST) pull-down assays, cDNA fragment corresponding to the MYST domain of HBO1 was cloned into the pET-22b plasmid with an N-terminal His6 tag or the pGEX 4T-1 plasmid (GE Healthcare) with an N-terminal GST tag; and cDNA fragment corresponding to BRPF2 (residues 31–80) was cloned into the pGEX 4T-1 plasmid with an N-terminal GST tag or cloned into the pET-28-Sumo plasmid (Novagen) with an N-terminal His6-Sumo tag. The HBO1 and BRPF2 proteins were expressed in E. coli BL21 (DE3) Codon-Plus strain as described above and purified with affinity chromatography using a Ni-NTA column or glutathione-coated sepharose beads (GE Healthcare).
In vitro HAT activity assays
In vitro HAT activity assays of HBO1 were carried out at 30°C. The standard reaction solution (30 μl) consisted of 0.05 μM HBO1, 0.1 mM AcCoA and 20 μg/ml recombinant full-length H3 or H4 (NEB) or 0.05 μM nucleosomal core particles (NCPs), supplemented with the reaction buffer containing 50 mM Tris–HCl (pH 8.0), 300 mM NaCl and 10% (v/v) glycerol. The NCPs were reconstituted with 146-mer DNA and histone octamer using salt gradient deposition and the quality of the NCPs was assessed with various methods as described previously (25). For HAT activity assays of the HBO1–BRPF2 complex, the reaction solution was supplied with 0.05 μM wild-type (WT) or mutant Sumo–BRPF2. The reaction was stopped at 60 min by adding: sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. The reaction mixture was resolved by SDS-PAGE and the acetylated H3K14 was analyzed by western blotting with an antibody highly specific to acetylated H3K14 (Abcam).
In vitro GST pull-down assays
To validate the functional role of the key residues of BRPF2 involved in the interactions with HBO1, WT or mutant GST–BRPF2 (30 μg) was immobilized on glutathione-coated sepharose beads and then incubated with untagged HBO1 (100 μg) in a buffer containing 20 mM Tris–HCl (pH 8.0), 300 mM NaCl and 10% (w/v) glycerol at 4°C for 2 h. The beads were washed four times with the buffer and then analyzed with SDS-PAGE followed by Coomassie blue staining.
To analyze the interactions of HBO1 with different BRPF2 fragments, GST–HBO1 (30 μg) was loaded on glutathione-coated sepharose beads and incubated with WT or mutant Sumo–BRPF2 (100 μg) at 4°C for 2 h, and the beads were then analyzed as described above.
To analyze the potential binding of BRPF2 with H3, GST–BRPF2 (30 μg) was first loaded on glutathione-coated sepharose beads. After mixing with Bovine serum albumin (100 μg) (Sigma) to prevent nonspecific binding, histone mixture from calf thymus (100 μg) (Sigma) was added and incubated at 4°C for 2 h. The beads were washed four times with a buffer containing 20 mM Tris–HCl (pH 8.0), 1 mM PMSF, 2 mM DTT, 1 mM ethylenediaminetetraacetic acid, 1% NP-40 and 0.5 M NaCl, and then analyzed with SDS-PAGE followed by Coomassie blue staining.
Crystallization, data collection and structure determination
Prior to crystallization, the HBO1–BRPF2 complex was mixed with AcCoA (Sigma) at a molar ratio of 1:3. Crystallization was performed using the sitting drop vapor diffusion method at 16°C by mixing 1 μl protein solution (10 mg/ml) with 1 μl reservoir solution containing 2% Tacsimate (pH 8.0), 0.1 M Tris–HCl (pH 8.5) and 12% (w/v) polyethylene glycol 3350. The crystals were cryoprotected with the reservoir solution supplemented with 20% (v/v) glycerol and then flash frozen in liquid nitrogen. The diffraction data were collected at −175°C at beamline 19U1 of National Facility for Protein Science in Shanghai, China and were processed, integrated and scaled together with HKL3000 (26). The structure of the HBO1–BRPF2 complex was solved using the molecular replacement (MR) method as implemented in Phenix (27) using the structure of the MYST domain of human MOF (PDB code: 3QAH) (28) as the search model. Residues 39–62 of BRPF2 were clearly defined in the initial MR-phased electron density map and the model was manually built with COOT (29). Structure refinement was performed using Phenix (27) and Refmac5 (30). Structural analyzes were carried out using programs in CCP4 (31). The structure figures were generated using PYMOL (http://www.pymol.org). Statistics of the diffraction data, structure refinement and quality of the structure model are summarized in Table 1.
Table 1. Summary of diffraction data and structure refinement statistics.
| Diffraction data | |
| Wavelength (Å) | 0.9786 |
| Space group | C2 |
| Cell parameters | |
| a, b, c (Å) | 126.4, 39.3, 87.7 |
| α, β, γ (°) | 90.0, 122.1, 90.0 |
| Resolution (Å) | 50.0–2.40 (2.49–2.40)a |
| Observed reflections | 52 870 |
| Unique reflections (I/σ(I) > 0) | 14 322 |
| Average redundancy | 3.7 (3.7) |
| Average I/σ(I) | 18.0 (3.6) |
| Completeness (%) | 97.9 (97.9) |
| R merge (%)b | 7.8 (52.2) |
| Refinement and structure model | |
| Reflections (Fo ≥ 0σ(Fo)) | |
| Working set | 12 912 |
| Test set | 680 |
| R-factor/Free R-factorc | 0.233/0.260 |
| Average B factor (Å2) | |
| All atoms | 49.0 |
| Protein | 49.4 |
| Ligand/ion | 40.2/41.1 |
| Water | 38.5 |
| RMS deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.2 |
| Ramachandran plot (%) | |
| Most favored regions | 95.8 |
| Allowed regions | 3.9 |
aNumbers in parentheses refer to the highest resolution shell.
b R merge = ΣhklΣi|Ii(hkl)i - <I(hkl)> |/ΣhklΣiIi(hkl).
c R-factor = Σhkl| |Fo | - |Fc | |/Σhkl|Fo |.
Immunofluorescence microscopy analyzes
Immunofluorescence microscopy analyzes were performed to analyze the co-localization of HBO1 and WT or mutant BRPF2. Full-length BRPF2–GFP was cloned into the pEGFP-N3 vector (Clontech) and full-length Myc-HBO1 into the pcDNA3.0 vector (Invitrogen). HEK 293T cells were cultured on coverslips in Dulbecco's modified eagle's medium (Hyclone) supplemented with 10% fetal bovine serum (Biochrom), then transiently transfected with the plasmids using lipofectamin 2000 (Invitrogen). A total of 24 h after transfection, the cells were fixed with 4% paraformaldehyde at room temperature for 15 min. Then, the cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min and incubated sequentially with rabbit anti-Myc antibody and the secondary antibody. After incubating 4΄,6-diamidino-2-phenylindole (DAPI) (Sigma) at room temperature for 5 min, the coverslips were mounted on glass slides and the confocal images were acquired utilizing a Leica TCS SP8 confocal microscope with a 63× oil immersion lens.
In vivo immunoprecipitation and HAT activity assays
cDNAs encoding full-length Flag-BRPF2 and full-length His-HBO1 were cloned into the pCDH vector (SBI). HEK 293T cells were transfected with WT or mutant full-length Flag-BRPF2 and full-length His-HBO1. Cells were harvested 48 h post-transfection, and nuclear extracts were prepared using the nuclear and cytoplasmic protein extraction kit (Beyotime). Immunoprecipitation (IP) was performed based on the method described previously (32) with modifications. Specifically, the nuclear extracts were incubated with anti-Flag M2 magnetic beads (Sigma) at 4°C for 8 h. After washing three times with the buffer, the bound protein complexes were eluted with a 3× Flag peptide (Sigma) and then subjected to HAT assays using NCPs as the substrate. In HAT assays, the 30 μl reaction mixture consisted of 20 μl eluted protein complex, 0.1 mM AcCoA and 0.05 μM NCPs. The reaction was carried out at 30°C for 60 min, and the reaction mixture was analyzed with SDS-PAGE, Coomassie blue staining and western blotting with antibodies specific to His, Flag or acetylated H3K14.
RESULTS
A short N-terminal region (residues 31–80) of BRPF2 is sufficient to bind to HBO1 and can potentiate the HAT activity of HBO1
It was previously demonstrated that the scaffold proteins BRPF1/2/3 can interact with HBO1 and the accessory proteins ING4/5 and Eaf6 to form the HAT complexes, and the BRPF proteins contain two conserved domains homologous to those of the EPC proteins which serve as docking sites for HBO1 and the accessory proteins, respectively (11,13,14,22). The N-terminal region (1–198 residues) of BRPF2 containing the conserved domain I was identified to interact directly with the HBO1 MYST domain (13). Similarly, the N-terminal region (residues 1–127) of BRPF3 was shown to be sufficient for HBO1 binding (22). In addition, a short region N-terminal to the conserved domain I of both BRPF and JADE proteins was shown to be responsible for histone tail selection on chromatin for acetylation (11).
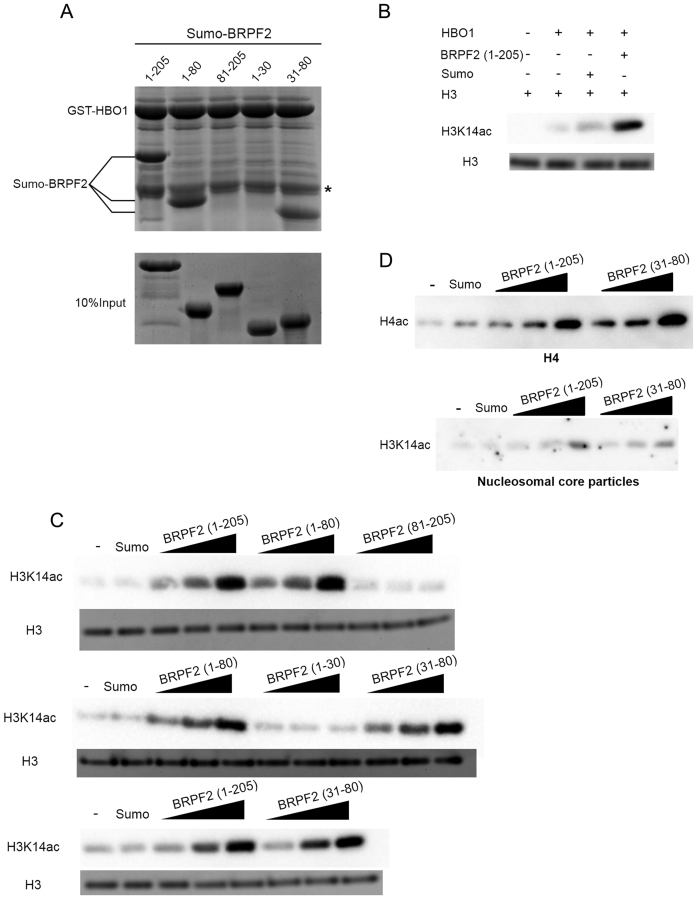
To confirm the interaction between BRPF2 and HBO1 and dissect the exact region of BRPF2 directly interacting with HBO1, we performed in vitro GST pull-down assays with GST-fused HBO1 MYST domain (residues 336–611) and Sumo-fused N-terminal fragments of BRPF2 (Figure 1A). As expected, BRPF2 (1–205) can bind tightly to the HBO1 MYST domain. Intriguingly, BRPF2 (1–80) and BRPF2 (31–80) can also bind to HBO1 comparable to BRPF2 (1–205), whereas BRPF2 (81–205) and BRPF2 (1–30) cannot, indicating that the N-terminal region of BRPF2 encompassing residues 31–80 is sufficient to bind to the HBO1 MYST domain.
Figure 1.
BRPF2 can potentiate the HAT activity of HBO1 towards H3K14 in vitro. (A) In vitro GST pull-down assay of the GST-tagged HBO1 MYST domain with different Sumo-tagged BRPF2 fragments. Equal loading of BRPF2 is shown. The band corresponding to the GST tag band is indicated by an asterisk. (B) Potentiation of the HAT activity of the HBO1 MYST domain by the N-terminal region of BRPF2 (1–205). In vitro HAT activity assay was performed on free full-length H3. The acetylation levels of H3 were analyzed with an antibody highly specific to acetylated H3K14 using half of the reaction mixture. Equal loading of H3 is shown with Coomassie blue staining of the other half of the reaction mixture. (C) In vitro HAT activity assay of the HBO1 MYST domain towards H3K14 in the presence of different Sumo–BRPF2 fragments. The molar ratio of BRPF2 and HBO1 was set at 0.5:1, 1:1 and 2:1, respectively. Equal loading of H3 is shown as in (B). (D) In vitro HAT activity assay of the HBO1 MYST domain toward free full-length H4 and nucleosomal H3 in the presence of Sumo–BRPF2 (1–205) and Sumo–BRPF2 (31–80).
To explore whether the interaction of BRPF2 with HBO1 has any effect on the function of HBO1 independent of the accessory proteins, we performed in vitro HAT activity assays of the HBO1 MYST domain towards H3K14 by itself and in the presence of different BRPF2 fragments. As shown in Figure 1B, compared with the HBO1 MYST domain alone, the acetylation level of H3K14 on free histone H3 is enhanced in the presence of BRPF2 (1–205). In addition, both BRPF2 (1–80) and BRPF2 (31–80) retain the potentiating effect on the HAT activity of the HBO1 MYST domain comparable to BRPF2 (1–205), but BRPF2 (81–205) and BRPF2 (1–30) have no effect (Figure 1C). Moreover, we found that BRPF2 (31–80) can also enhance the HAT activity of the HBO1 MYST domain towards free histone H4 and nucleosomal H3 at levels comparable to BRPF2 (1–205) (Figure 1D). It is noteworthy that the acetylation activities of the complexes containing the BRPF2 fragments towards NCPs are still inefficient (Figure 1D). Nevertheless, these results indicate that the N-terminal region (residues 31–80) of BRPF2 is responsible for binding the HBO1 MYST domain and this binding can potentiate the HAT activity of HBO1.
Structure of the HBO1–BRPF2 complex
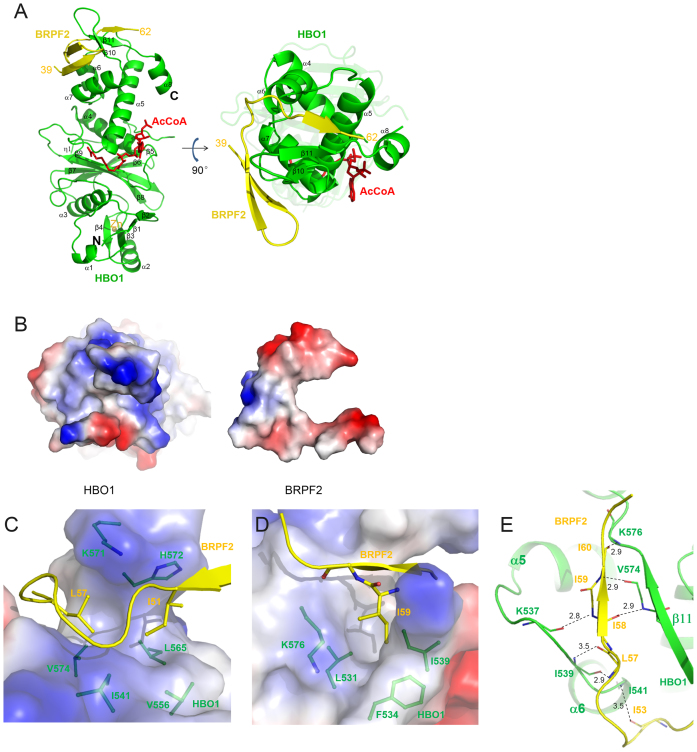
To investigate the molecular basis for how BRPF2 interacts with HBO1 and potentiates its HAT activity, we carried out structural studies of the HBO1 MYST domain (residues 336–611) alone and in complex with BRPF2 (31–80). HBO1 alone was unstable at high concentration and crystallization did not yield any crystals. Meanwhile, coexpression of the HBO1 MYST domain and BRPF2 (31–80) yielded a stable complex (HBO1–BRPF2) with a 1:1 molar ratio, which was purified with high purity and homogeneity (Supplementary Figure S1B and C). The HBO1–BRPF2 complex was crystallized in the presence of AcCoA and the crystal structure of the HBO1–BRPF2 complex was solved at 2.4 Å resolution by the MR method (Table 1). There is one HBO1–BRPF2 complex bound with an AcCoA in the asymmetric unit (Figure 2A). The MYST domain of HBO1 is well defined except for two short C-terminal regions (residues 586–591 and 607–611) (Supplementary Figure S2A and B). However, only the central portion of the BRPF2 fragment (residues 39–62) is visible; both the N-terminal and C-terminal regions are disordered. In addition, we also obtained crystals of the HBO1–BRPF2 (1–80) complex at the same crystallization condition as for the HBO1–BRPF2 (31–80) complex, and solved the structure of the HBO1–BRPF2 (1–80) complex at 2.5 Å resolution, which is however almost identical to that of the HBO1–BRPF2 (31–80) complex and did not reveal any additional residues at the N-terminus (data not shown).
Figure 2.
Structure of the HBO1–BRPF2 complex. (A) A ribbon representation of the overall structure of the HBO1–BRPF2 complex. The HBO1 MYST domain and the BRPF2 (residues 39–62) fragment are colored in green and yellow, respectively. The bound AcCoA and Zn2+ are shown with a red stick model and a gray sphere, respectively. (B) Electrostatic surface representations of HBO1 and BRPF2 showing good complementarity in both geometrical and electrostatic properties. (C and D) Hydrophobic interactions between HBO1 and BRPF2. HBO1 is shown with an electrostatic surface and BRPF2 is shown with a ribbon diagram. The residues involved in the hydrophobic interactions are shown with side chains. (E) Hydrophilic interactions between HBO1 and BRPF2. Both HBO1 and BRPF2 are shown with ribbon diagrams. The hydrophilic interactions are indicated with dotted lines and distances.
The MYST domain of HBO1 consists of 8 α-helices and 11 β-strands (Figure 2A and Supplementary Figure S2C). The central region, consisting of four β-strands (β6–β9) followed by an α-helix (α4), shows structural homology with the catalytic domains of other HATs, such as MOF, MOZ, Gcn5, p300 and Rtt109, albeit the N- and C-terminal structural elements flanking the central region are highly divergent compared with HATs from other families (3,33). Except for the region that interacts with BRPF2, the MYST domain of HBO1 superposes very well with these of other MYST family members. For example, the MYST domains of HBO1 and MOF (34) have a root-mean-square deviation of 1.07 Å over the aligned 245 Cα atoms (Supplementary Figure S3A). The characteristic motif A with the Arg/Gln-X-X-Gly-X-Gly/Ala sequence (35) is located in the β9–α4 loop and participates in the recognition and binding of AcCoA. Residues Cys368, Cys371, His384 and Cys388 form a classic zinc finger to coordinate a Zn2+ ion. Cofactor AcCoA is bound at the active site with the same conformation as that in MOF, and the catalytic residue Glu508 and the key residues Arg483, Gly485 and Gly487 of motif A of HBO1 assume the same positions and orientations as the equivalent residues of MOF (Supplementary Figure S3A).
Residues 39–62 of BRPF2 fold into a β-hairpin and an extra β-strand, which interact with the C-terminal region of the HBO1 MYST domain (Figure 2A). Interestingly, the interacting mode between HBO1 and BRPF2 is completely different from the MOF–MSL1 complex, in which MSL1 forms a long α-helix and interacts with the N-terminal part of the MOF MYST domain (36) (Supplementary Figure S3B). These results indicate the diversity in the interacting modes of HATs with their scaffold proteins.
Interactions between HBO1 and BRPF2
As shown in Figure 2A, BRPF2 wraps around the C-terminal region of HBO1. At the interaction interface, HBO1 and BRPF2 show good complementarity in both geometrical and electrostatic properties (Figure 2B). The HBO1–BRPF2 complex is stabilized by extensive hydrophobic and several hydrogen-bonding interactions and the interactions bury about 975.9 Å2 surface areas. Specifically, Ile51 and Leu57 of BRPF2 are embedded in a hydrophobic cleft formed by Ile541, Val556, Leu565, Val574, Lys571 and His572 of HBO1 (Figure 2C); and Ile59 of BRPF2 is inserted into a hydrophobic pocket formed by Leu531, Phe534, Ile539 and Lys576 of HBO1 (Figure 2D). Additionally, residues 53–60 of BRPF2 form several hydrogen bonds with Lys537, Ile539, Ile541, Val574 and Lys576 of HBO1 via their main chains (Figure 2E).
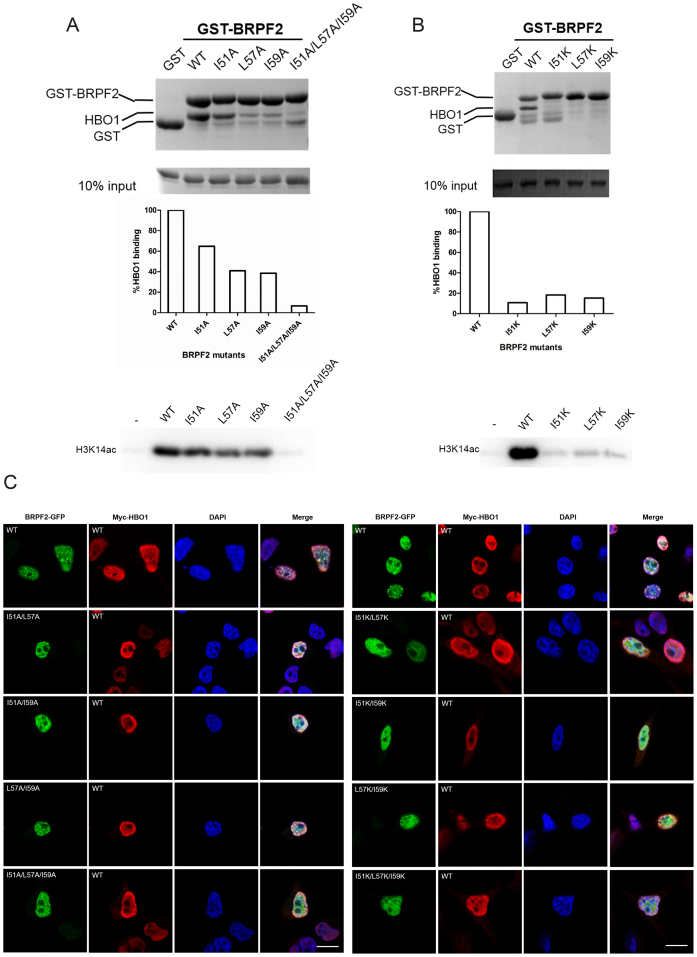
Sequence alignment shows that the three key residues (Ile51, Leu57 and Ile59) of BRPF2 involved in the hydrophobic interactions with HBO1 are highly conserved in other species as well as in BRPF1 and BRPF3 (Supplementary Figure S4A). To analyze their functional roles, we first performed mutagenesis and in vitro GST pull-down assays. The results show that single mutation of these residues to alanine significantly compromised the binding of BRPF2 with HBO1 and impaired its ability to potentiate the HAT activity of HBO1 on H3K14 (Figure 3A). Triple mutation of these residues to alanine almost abolished both the binding and potentiating abilities of BRPF2 (Figure 3A). Consistently, drastic mutation of these residues to lysine had more severe effects (Figure 3B).
Figure 3.
Validation of the functional roles of key residues of BRPF2. (A and B) In vitro functional analyzes of BRPF2 (31–80) mutants. BRPF2 (31–80) mutants with mutations of Ile51, Leu57 and/or Ile59 to (A) alanine or (B) lysine were analyzed. In vitro GST pull-down assays of wild-type (WT) and mutant GST–BRPF2 (31–80) with the HBO1 MYST domain were performed. Equal loading of HBO1 is shown. The amount of HBO1 that was pulled down was semi-quantitated. In vitro HAT activity assays of HBO1 toward H3K14 in the presence of WT or mutant Sumo–BRPF2 (31–80) were performed, and the acetylated H3K14 was detected with a specific antibody. (C) Localization of HBO1 and BRPF2 in transfected HEK 293T cells. Cells were co-transfected with full-length Myc-HBO1 and full-length WT or mutant BRPF2–GFP harboring mutations to alanine (left panel) or lysine (right panel). Immunofluorescence with anti-Myc antibody was performed to detect HBO1. Localization of HBO1 and BRPF2 was examined with confocal fluorescence microscopy. Scale bar represents 10 μm.
We then performed in vivo cell biological assays to examine the co-localization patterns of HBO1 and BRPF2. HEK 293T cells were co-transfected with the full-length HBO1 and BRPF2 in WT or mutants carrying single mutation I51A, L57A and I59A, double mutation I51A/L57A, I51A/I59A and L57A/I59A, or triple mutation I51A/L57A/I59A. As shown in Figure 3C, WT BRPF2 was co-localized with WT HBO1 with a relatively concentrated distribution pattern, in agreement with the previous chromatin immunoprecipitation (CHIP) assay results showing that both HBO1 and BRPF1/2 localized near the transcription starting sites of active genes in nucleus (11). In the cells co-transfected with BRPF2 mutants carrying single mutations, the co-localization pattern did not show notable changes (data not shown). However, in the cells co-transfected with BRPF2 mutants carrying double or triple mutations, HBO1 showed diffuse distribution in nucleoplasm and reduced co-localization with BRPF2, which could be attributed to the disruption of the HBO1–BRPF2 interaction by the mutations. Analyzes of BRPF2 mutants harboring drastic mutations to lysine showed similar results (Figure 3C). Together, the biochemical and cell biological data demonstrate that the identified key hydrophobic residues of BRPF2 are important for the HBO1–BRPF2 interaction and the biological function of the HBO1–BRPF2 complex.
The N-terminal region of BRPF2 has interaction with histones in vitro
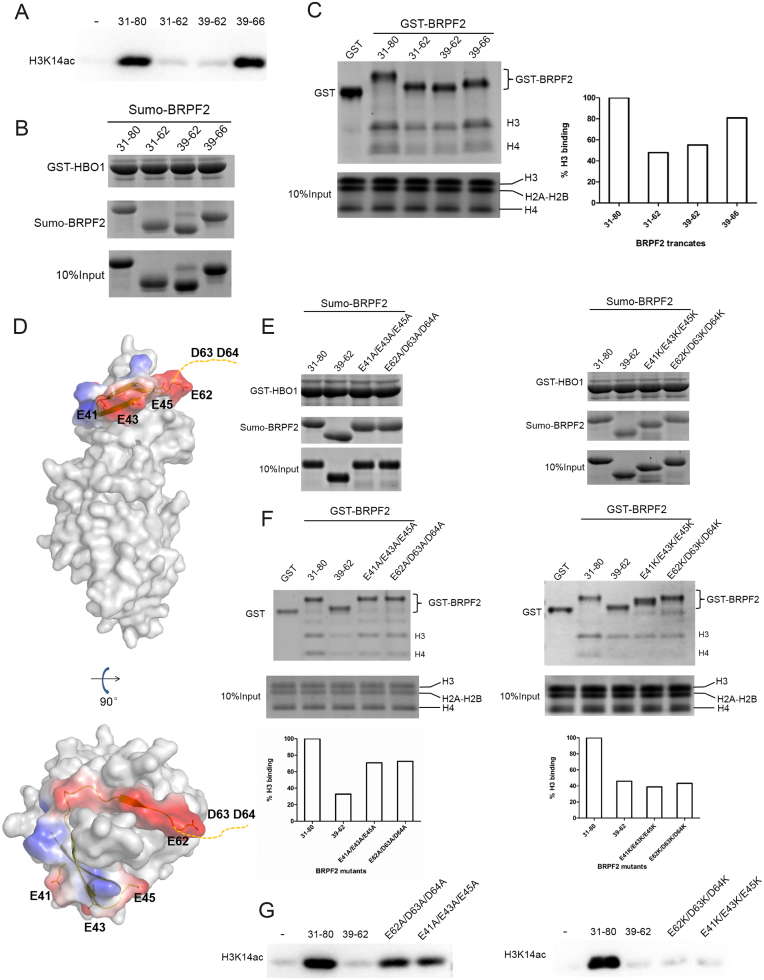
Our biochemical data show that BRPF2 (31–80) can potentiate the HAT activity of the HBO1 MYST domain toward free H3 and H4 as well as nucleosomal H3 (Figure 1B–D). Thus, we tried to co-crystallize the HBO1–BRPF2 complex with an H3 peptide (residues 1–21); unfortunately, the yielded structure is basically identical to that of the HBO1–BRPF2 complex: no histone peptide is bound in the complex. Intriguingly, although the structural data indicate that residues 39–62 of BRPF2 play an essential role in HBO1 binding, the biochemical data show that BRPF2 (39–62) was insufficient to potentiate the HAT activity of HBO1 (Figure 4A). Meanwhile, the BRPF2 fragment encompassing residues 39–66 could potentiate the HAT activity of HBO1 to an extent comparable to that of BRPF2 (31–80), but the BRPF2 fragment encompassing residues 31–62 could not (Figure 4A). On the other hand, in vitro GST pull-down assays showed that BRPF2 (31–62) and BRPF2 (39–62) could bind to HBO1 at similar levels as BRPF2 (31–80) and BRPF2 (39–66) (Figure 4B). These results prompted us to examine whether the N-terminal region of BRPF2 is also involved in histone binding. As the binding abilities of the HBO1–BRPF2 complexes with histones are not significantly affected (data not shown), which is reminiscent of the results that the binding partners of Rtt109 or Esa1 can potentiate the HAT activity but have only minor effects on the Km for histone peptide (37–40), thus, we analyzed the binding abilities of different BRPF2 fragments with free histones. BRPF2 (31–80) and BRPF2 (39–66) had the abilities to bind free H3 and H4; however, the binding of BRPF2 (31–62) and BRPF2 (39–62) to histones was weakened (Figure 4C). These data together indicate that the N-terminal region of BRPF2 (residues 31–80) can bind to both HBO1 and histones in vitro.
Figure 4.
Examination of potential binding of the N-terminal region of BRPF2 with histones in vitro. (A) In vitro HAT activity assays of HBO1 toward H3K14 in the presence of different Sumo–BRPF2 fragments. (B) In vitro GST pull-down assays of GST–HBO1 with different Sumo–BRPF2 fragments. Equal loading of Sumo–BRPF2 is shown. (C) In vitro GST pull-down assays of different GST–BRPF2 fragments with free H3 and H4 from histone mixture. The amount of histone H3 that was pulled down was semi-quantitated. (D) Two negatively charged surface patches in the N-terminal region of BRPF2. HBO1 is shown with gray surface. BRPF2 is shown with electrostatic surface and the conserved acidic residues are shown with side chains. Asp63 and Asp64 are invisible in the HBO1–BRPF2 structure and thus are indicated with a dash line. (E) In vitro GST pull-down assays of GST–HBO1 with WT or mutant Sumo–BRPF2 (31–80). Equal loading of Sumo–BRPF2 is shown. (F) In vitro GST pull-down assays of WT or mutant GST–BRPF2 (31–80) with free H3 and H4 from histone mixture. The amount of histone H3 that was pulled down was semi-quantitated. (G) In vitro HAT activity assays of HBO1 toward H3K14 in the presence of WT or mutant Sumo–BRPF2 (31–80).
It is common that histone-interacting proteins bind to histones with negatively charged regions (41–43). In the BRPF2 (39–66) fragment, there are two negatively charged segments consisting of acidic residues E41/E43/E45 and E62/D63/D64, respectively. Residues E41/E43/E45 do not participate in HBO1 binding in the HBO1–BRPF2 structure and instead form a negatively charged surface patch, which is solvent accessible and hence could be involved in histone binding (Figure 4D). Residues D63/D64 are disordered in the HBO1–BRPF2 structure and therefore are also not involved in HBO1 binding, but could be involved in histone binding as well. Additionally, these acidic residues are strictly conserved across different species (Supplementary Figure S4A). To examine whether these two regions are involved in histone binding and thereby potentiate the HAT activity of HBO1, we prepared four BRPF2 (31–80) mutants, namely E62A/D63A/D64A, E41A/E43A/E45A, E62K/D63K/D64K and E41K/E43K/E45K, in which the acidic residues were substituted with alanine or lysine. Although the mutants could bind to HBO1 at similar levels as WT BRPF2 (31–80) (Figure 4E), they exhibited compromised binding ability to free histones (Figure 4F) and had reduced potentiating effect on the HBO1 activity toward H3K14 (Figure 4G). As expected, the E62K/D63K/D64K and E41K/E43K/E45K mutants had more severe effects than the E62A/D63A/D64A and E41A/E43A/E45A mutants. These data together suggest that BRPF2 (31–80) might bind to histones in vitro via the two negatively charged surface patches.
The N-terminal region of BRPF2 plays an important role in the HBO1 binding in vivo
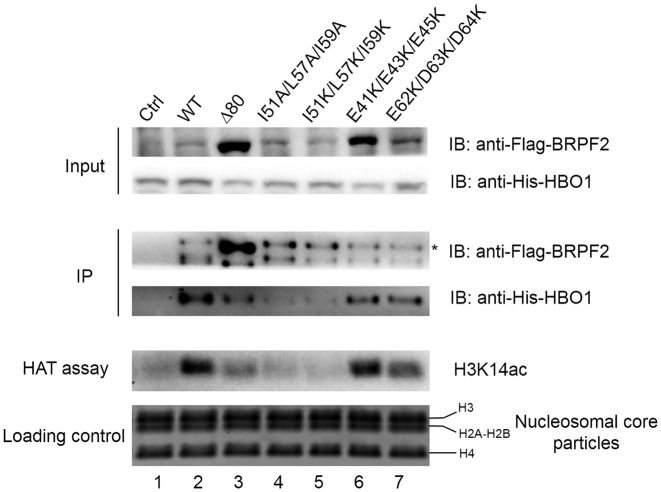
To investigate the functional role of the N-terminal region of BRPF2 in vivo, full-length His-HBO1 and full-length WT or mutant Flag-BRPF2 were co-expressed in HEK 293T cells and immunoprecipitated (IPed) with an anti-Flag antibody. The IPed HBO1–BRPF2 complex was then subjected to HAT activity assays using NCPs as the substrate. As shown in Figure 5, full-length HBO1 was readily co-purified with full-length WT BRPF2 and the IPed HBO1–BRPF2 complex displayed specific HAT activity toward H3K14 of the NCPs. In contrast, the binding of HBO1 with the BRPF2Δ80 mutant or the I51A/L57A/I59A and I51K/L57K/I59K BRPF2 mutants was substantially disrupted, yielding a significantly reduced amount of co-IPed HBO1 and thus exhibiting a substantially weakened HAT activity. These results are in agreement with the in vitro assay results using the N-terminal region of BRPF2 and the MYST domain of HBO1 and the results of the co-localization experiments using full-length BRPF2 and full-length HBO1 (Figures 1 and 3). However, the in vivo assay results show that the E41K/E43K/E45K and E62K/D63K/D64K mutations had only very minor effect on the HBO1 binding and the IPed complexes exhibited only slightly decreased HAT activity (Figure 5), which are in discrepancy with the in vitro assay results showing that the two mutants had moderate effects on the HBO1 binding and the HAT activity (Figure 4).
Figure 5.
Examination of the functional role of the N-terminal region of BRPF2 in vivo. HEK 293T cells were co-transfected with full-length His-HBO1 and WT or mutant Flag-BRPF2, and cells transfected with His-HBO1 alone served as the negative control. Immunoprecipitation (IP) experiments were performed with nuclear extracts of the transfected cells using an anti-Flag antibody. The IPed BRPF2–HBO1 complexes were then subjected to HAT activity assays using NCPs as the substrate. The nuclear extracts, the IPed complexes and the reaction mixture of the HAT activity assays were analyzed with sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting using antibodies specific to the Flag tag, His tag, and acetylated H3K14. The band corresponding to the full-length Flag-BRPF2 protein was denoted by an asterisk and the band below might represent the degraded Flag-BRPF2 protein. Equal loading of NCPs in the HAT activity assays were shown with SDS-PAGE and Coomassi blue staining.
As BRPF2 also contains a PZP domain, bromo domain and PWWP domain, which are typically involved in the binding of histones or chromatin and/or specific histone modifications (Supplementary Figure S1), it is very likely that in addition to the N-terminal region, these domains might also be involved and possibly play more important roles in the binding of chromatin. Indeed, the previous in vitro and in vivo data showed that the PZP, bromo and PWWP domains of the BRPF proteins are essential for their binding to chromatin or histones and for acetylation of chromatin (11,44–48). Based on these results, the discrepancy between our in vitro and in vivo data on the E41K/E43K/E45K and E62K/D63K/D64K mutations could be easily explained.
The in vitro HAT assays were carried out using the N-terminal fragment of BRPF2 (residues 31–80) and the MYST domain of HBO1 with free histones as the substrate. The histone substrate has interactions only with the HBO1 MYST domain and the two negatively charged patches of BRPF2 (31–80). Thus, mutations of the acidic residues of BRPF2 (31–80) could moderately impair its binding ability with histones and thus compromise the HAT activity of HBO1 toward H3K14. On the other hand, the in vivo HAT assays were performed using full-length HBO1 and full-length BRPF2 with the NCPs as the substrate. The full-length BRPF2 has interactions with the NCPs via the N-terminal region and the PZP, bromo and PWWP domains, the latters of which play more important roles in the binding of nucleosomes. Thus, mutations of the acidic residues in the N-terminal region of BRPF2 had only minor effects on the binding of BRPF2 with the NCPs and the HAT activity of HBO1 toward H3K14. As the in vivo HAT assays were performed in a native context and the results are more relevant to the biological function of BRPF2, the in vitro and in vivo data together indicate that the N-terminal region of BRPF2 plays an important role in the binding of HBO1 but a very minor role in the binding of nucleosomes.
DISCUSSION
Multisubunit HAT complexes are responsible for acetylating specific lysine residues on histones to promote transcription activation and DNA replication. HAT complexes usually exhibit higher activities and specificities toward histones than HAT enzymes alone (3,6,40). The HAT enzymes are the catalytic subunits, the accessory proteins contain different histone recognition modules such as PZP domain, chromo domain, bromo domain and PWWP domain that can recognize and bind chromatin on specific histone marks, and the scaffold proteins provide the docking sites for the HAT enzymes and the accessory proteins; and the interplay among these subunits can modulate the activities and substrate specificities of the HAT complexes. The crystal structure of the HBO1–BRPF2 complex reveals the molecular basis for the HBO1–BRPF2 interaction. Our in vitro data indicate that the N-terminal region of BRPF2 can bind to both the HBO1 MYST domain and free histones, and can potentiate the HAT activity of HBO1. In addition, our in vivo data indicate that the N-terminal region of BRPF2 plays an important role in the binding of HBO1 but a minor role in the binding of nucleosomes. These results not only provide new mechanistic insight into regulation of the HAT activity of HBO1 by BRPF2 but also have important implications in understanding the molecular mechanism underlying the regulation of other HAT complexes.
It is well established that scaffold proteins bridge HATs and their accessory proteins to modulate the specificity activities of the HAT complexes (3,40). Intriguingly, it was previously shown that the N-terminal region containing the first 20 residues of Epl1 Enhancer of Polycomb A (EPcA) is important for chromatin binding and the HAT activity of yeast NuA4 complex (49,50), and the corresponding region of Epl1 can bind to the N-terminal tail of H2A in nucleosomes (11,51). Very recently, it was demonstrated that deletion of the corresponding region of JADE1 changed the specificity of the MOZ–JADE1 and HBO1–JADE1 complexes from H4 to H3 (11). Removal of the corresponding region of BRPF1 also led to loss of the HAT activity on chromatin for both HBO1 and MOZ, and additionally, although the mutant HBO1 and MOZ complexes did not seem to significantly change histone tail specificity on chromatin, they showed a significant loss of histone tail specificity in the nucleosomal HAT activity assays (11). These results led to the proposal that this short N-terminal region of the scaffold proteins may bind directly to histones and dictates which histone tail on chromatin is acetylated by the HAT complexes (6,11).
Sequence alignment shows that this short N-terminal region of EPcA corresponds to residues 42–65 of BRPF2 (Supplementary Figure S4B), which is the region involved in the interaction with HBO1 revealed by this work. Our structural and functional data provide new insight into how the N-terminal region of BRPF2 regulates the HAT activity of HBO1. We demonstrate that in addition to binding to HBO1, the N-terminal region of BRPF2 can also interact with histones in vitro. Particularly, two negatively charged patches of BRPF2 consisting of E41/E43/E45 and E62/D63/D64 might be involved in the interaction with H3. This is consistent with the notion that histone-interacting proteins usually bind to histones with negatively charged regions (41–43). In the HBO1–BRPF2 structure, the distance between the potential histone-binding site of BRPF2 and the substrate-binding site of HBO1 is about 30–40 Å (Supplementary Figure S5). Thus, we speculate that the region of H3 around residues 30–50 might be involved in the interaction with BRPF2. The interaction of BRPF2 with this part of H3 may stabilize the conformation of the H3 tail for proper positioning of H3K14 at the active site of HBO1 for acetylation, resulting in enhanced activity of HBO1. This is reminiscent of the observation that yeast NuA4 complex could acetylate full-length H4 much faster than H4 peptides, leading to the suggestion that residues 21–52 of H4 might be involved in interaction with the NuA4 complex and thus enhances the HAT activity of Esa1 (39).
On the other hand, our in vivo functional data show that the N-terminal region plays a minor role in the binding of nucleosomes. This is consistent with the previous in vitro and in vivo functional data showing that the PZP, bromo and PWWP domains of BRPF proteins are essential for their binding to histones or nucleosomes and for acetylation of chromatin (11,44–48). Collectively, these data lead us to propose that the N-terminal region of BRPF2 can interact with HBO1 to stabilize its conformation, and additionally, it is also involved in the interaction with the N-terminal tails of histones although BRPF2 binds to nucleosomes mainly via the PZP, bromo and PWWP domains (Supplementary Figure S6). The concurrent binding of BRPF2 with both HBO1 and histones can properly position the N-terminal tails of histones at the active site of HBO1 for acetylation and thus enhance the HAT activity of HBO1. It is also possible that the binding of the N-terminal region of BRPF2 can stabilize HBO1 in a more physiological conformation and hence enhances its interaction with histone substrate, leading to the potentiation of the HAT activity of HBO1.
It has been demonstrated that HBO1 can form HAT complexes with scaffold proteins BRPF1/2/3 or JADE1/2/3, and HBO1 can bind to different scaffold proteins and this differential association can determine the substrate specificities of the HAT complexes (8–14,22,23). Sequence alignment shows that BRPF1/2/3 are highly conserved in the N-terminal region responsible for HBO1 binding and particularly at the sites that are involved in the hydrophobic interactions (Supplementary Figure S4A), suggesting that BRPF1 and BRPF3 might employ a similar mechanism to interact with and regulate the activity of HBO1 as BRPF2. This suggestion is supported by the functional data showing that although the HBO1–BRPF1/2/3 complexes can acetylate free histones, they all have high specificities towards H3K14 (11,13,14,22). On the other hand, it was reported that JADE1 binds to the HBO1 MYST domain via a conserved region called domain I (corresponding to residues 90–180 of JADE1 or residues 114–195 of BRPF2) (9), implying that JADE1 and its homologs JADE2/3 would bind to HBO1 in a manner different from that of BRPF1/2/3. The differed interacting modes may account for the differentiated substrate specificities of the BRPF- and JADE-containing HBO1 complexes.
ACCESSION NUMBER
The crystal structure of the HBO1–BRPF2 complex has been deposited with the Protein Data Bank under accession code 5GK9.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff members at BL19U1 of National Facility for Protein Science in Shanghai, China for technical support in diffraction data collection. We thank Dr. Jing Huang of our institute for providing the NCPs.
Authorship contributions: Y.T. performed the functional studies and structural analysis and drafted the manuscript. C.Z. participated in the IP experiments and data analyzes, and drafted the manuscript. J.Z. performed the cloning, protein purification and crystallization. S.X. carried out the structure determination. J.D. conceived the study, participated in the experimental design and data analyzes and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31521061]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010302]; Ministry of Science and Technology of China [2013CB910404]. Funding for open access charge: National Natural Science Foundation of China [31521061].
Conflict of interest statement. None declared.
REFERENCES
- 1. Suganuma T., Workman J.L.. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 2011; 80:473–499. [DOI] [PubMed] [Google Scholar]
- 2. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marmorstein R., Zhou M.M.. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 2014; 6:a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tessarz P., Kouzarides T.. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014; 15:703–708. [DOI] [PubMed] [Google Scholar]
- 5. Sapountzi V., Cote J.. MYST-family histone acetyltransferases: beyond chromatin. Cell Mol. Life Sci. 2011; 68:1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lalonde M.E., Cheng X., Cote J.. Histone target selection within chromatin: an exemplary case of teamwork. Gene Dev. 2014; 28:1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iizuka M., Stillman B.. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 1999; 274:23027–23034. [DOI] [PubMed] [Google Scholar]
- 8. Iizuka M., Takahashi Y., Mizzen C.A., Cook R.G., Fujita M., Allis C.D., Frierson H.F. Jr, Fukusato T., Smith M.M.. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene. 2009; 436:108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Avvakumov N., Lalonde M.E., Saksouk N., Paquet E., Glass K.C., Landry A.J., Doyon Y., Cayrou C., Robitaille G.A., Richard D.E. et al. . Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol. Cell. Biol. 2012; 32:689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doyon Y., Cayrou C., Ullah M., Landry A.J., Cote V., Selleck W., Lane W.S., Tan S., Yang X.J., Cote J.. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006; 21:51–64. [DOI] [PubMed] [Google Scholar]
- 11. Lalonde M.E., Avvakumov N., Glass K.C., Joncas F.H., Saksouk N., Holliday M., Paquet E., Yan K., Tong Q., Klein B.J. et al. . Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Gene Dev. 2013; 27:2009–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kueh A.J., Dixon M.P., Voss A.K., Thomas T.. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol. Cell. Biol. 2011; 31:845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mishima Y., Miyagi S., Saraya A., Negishi M., Endoh M., Endo T.A., Toyoda T., Shinga J., Katsumoto T., Chiba T. et al. . The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood. 2011; 118:2443–2453. [DOI] [PubMed] [Google Scholar]
- 14. Feng Y., Vlassis A., Roques C., Lalonde M.E., Gonzalez-Aguilera C., Lambert J.P., Lee S.B., Zhao X., Alabert C., Johansen J.V. et al. . BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 2016; 35:176–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iizuka M., Matsui T., Takisawa H., Smith M.M.. Regulation of replication licensing by acetyltransferase Hbo1. Mol. Cell. Biol. 2006; 26:1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miotto B., Struhl K.. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Gene Dev. 2008; 22:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miotto B., Struhl K.. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by geminin. Mol. Cell. 2010; 37:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burke T.W., Cook J.G., Asano M., Nevins J.R.. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 2001; 276:15397–15408. [DOI] [PubMed] [Google Scholar]
- 19. McCullough C.E., Marmorstein R.. Molecular basis for histone acetyltransferase regulation by binding partners, associated domains, and autoacetylation. ACS Chem. Biol. 2016; 11:632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klein B.J., Lalonde M.E., Cote J., Yang X.J., Kutateladze T.G.. Crosstalk between epigenetic readers regulates the MOZ/MORF HAT complexes. Epigenetics. 2014; 9:186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saksouk N., Avvakumov N., Champagne K.S., Hung T., Doyon Y., Cayrou C., Paquet E., Ullah M., Landry A.J., Cote V. et al. . HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell. 2009; 33:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yan K., You L., Degerny C., Ghorbani M., Liu X., Chen L., Li L., Miao D., Yang X.J.. The chromatin regulator BRPF3 preferentially activates the HBO1 acetyltransferase but is dispensable for mouse development and survival. J. Biol. Chem. 2016; 291:2647–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Foy R.L., Song I.Y., Chitalia V.C., Cohen H.T., Saksouk N., Cayrou C., Vaziri C., Cote J., Panchenko M.V.. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J. Biol. Chem. 2008; 283:28817–28826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCullagh P., Chaplin T., Meerabux J., Grenzelias D., Lillington D., Poulsom R., Gregorini A., Saha V., Young B.D.. The cloning, mapping and expression of a novel gene, BRL, related to the AF10 leukaemia gene. Oncogene. 1999; 18:7442–7452. [DOI] [PubMed] [Google Scholar]
- 25. Dyer P.N., Edayathumangalam R.S., White C.L., Bao Y., Chakravarthy S., Muthurajan U.M., Luger K.. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004; 375:23–44. [DOI] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 27. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun B., Guo S., Tang Q., Li C., Zeng R., Xiong Z., Zhong C., Ding J.. Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274. Cell Res. 2011; 21:1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 30. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 31. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. . Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doyon Y., Selleck W., Lane W.S., Tan S., Cote J.. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004; 24:1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marmorstein R., Trievel R.C.. Histone modifying enzymes: structures, mechanisms, and specificities. Biochim. Biophys. Acta. 2009; 1789:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu H., Moshkina N., Min J., Zeng H., Joshua J., Zhou M.M., Plotnikov A.N.. Structural basis for substrate specificity and catalysis of human histone acetyltransferase 1. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:8925–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wolf E., Vassilev A., Makino Y., Sali A., Nakatani Y., Burley S.K.. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell. 1998; 94:439–449. [DOI] [PubMed] [Google Scholar]
- 36. Kadlec J., Hallacli E., Lipp M., Holz H., Sanchez-Weatherby J., Cusack S., Akhtar A.. Structural basis for MOF and MSL3 recruitment into the dosage compensation complex by MSL1. Nat. Struct. Mol. Biol. 2011; 18:142–149. [DOI] [PubMed] [Google Scholar]
- 37. Berndsen C.E., Tsubota T., Lindner S.E., Lee S., Holton J.M., Kaufman P.D., Keck J.L., Denu J.M.. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat. Struct. Mol. Biol. 2008; 15:948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berndsen C.E., Albaugh B.N., Tan S., Denu J.M.. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry. 2007; 46:623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berndsen C.E., Selleck W., McBryant S.J., Hansen J.C., Tan S., Denu J.M.. Nucleosome recognition by the piccolo NuA4 histone acetyltransferase complex. Biochemistry. 2007; 46:2091–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Berndsen C.E., Denu J.M.. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr. Opin. Struct. Biol. 2008; 18:682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen S., Yang Z., Wilkinson A.W., Deshpande A.J., Sidoli S., Krajewski K., Strahl B.D., Garcia B.A., Armstrong S.A., Patel D.J. et al. . The PZP domain of AF10 senses unmodified H3K27 to regulate DOT1L-mediated methylation of H3K79. Mol. Cell. 2015; 60:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Y., Zhang L., Liu T., Chai C., Fang Q., Wu H., Agudelo Garcia P.A., Han Z., Zong S., Yu Y. et al. . Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex. Genes Dev. 2014; 28:1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dreveny I., Deeves S.E., Fulton J., Yue B., Messmer M., Bhattacharya A., Collins H.M., Heery D.M.. The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification. Nucleic Acids Res. 2014; 42:822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qin S., Jin L., Zhang J., Liu L., Ji P., Wu M., Wu J., Shi Y.. Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF). J. Biol. Chem. 2011; 286:36944–36955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu L., Qin S., Zhang J., Ji P., Shi Y., Wu J.. Solution structure of an atypical PHD finger in BRPF2 and its interaction with DNA. J. Struct. Biol. 2012; 180:165–173. [DOI] [PubMed] [Google Scholar]
- 46. Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Muller S., Pawson T. et al. . Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012; 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ullah M., Pelletier N., Xiao L., Zhao S.P., Wang K., Degerny C., Tahmasebi S., Cayrou C., Doyon Y., Goh S.L. et al. . Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol. Cell. Biol. 2008; 28:6828–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vezzoli A., Bonadies N., Allen M.D., Freund S.M., Santiveri C.M., Kvinlaug B.T., Huntly B.J., Gottgens B., Bycroft M.. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 2010; 17:617–619. [DOI] [PubMed] [Google Scholar]
- 49. Selleck W., Fortin I., Sermwittayawong D., Cote J., Tan S.. The saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the enhancer of polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 2005; 25:5535–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chittuluru J.R., Chaban Y., Monnet-Saksouk J., Carrozza M.J., Sapountzi V., Selleck W., Huang J., Utley R.T., Cramet M., Allard S. et al. . Structure and nucleosome interaction of the yeast NuA4 and piccolo-NuA4 histone acetyltransferase complexes. Nat. Struct. Mol. Biol. 2011; 18:1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang J., Tan S.. Piccolo NuA4-catalyzed acetylation of nucleosomal histones: critical roles of an esa1 tudor/chromo barrel loop and an epl1 enhancer of polycomb A (EPcA) basic region. Mol. Cell. Biol. 2013; 33:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.