Abstract
Neisseria meningitidis is a human commensal that can also cause life-threatening meningitis and septicemia. Despite growing evidence for RNA-based regulation in meningococci, their transcriptome structure and output of regulatory small RNAs (sRNAs) are incompletely understood. Using dRNA-seq, we have mapped at single-nucleotide resolution the primary transcriptome of N. meningitidis strain 8013. Annotation of 1625 transcriptional start sites defines transcription units for most protein-coding genes but also reveals a paucity of classical σ70-type promoters, suggesting the existence of activators that compensate for the lack of −35 consensus sequences in N. meningitidis. The transcriptome maps also reveal 65 candidate sRNAs, a third of which were validated by northern blot analysis. Immunoprecipitation with the RNA chaperone Hfq drafts an unexpectedly large post-transcriptional regulatory network in this organism, comprising 23 sRNAs and hundreds of potential mRNA targets. Based on this data, using a newly developed gfp reporter system we validate an Hfq-dependent mRNA repression of the putative colonization factor PrpB by the two trans-acting sRNAs RcoF1/2. Our genome-wide RNA compendium will allow for a better understanding of meningococcal transcriptome organization and riboregulation with implications for colonization of the human nasopharynx.
INTRODUCTION
The human microbiota is normally beneficial or neutral, but some members of these populations can cause disease and are responsible for more infections today than the so-called classical pathogens. Infections by opportunistic pathogens are not limited to immuno-compromised patients, but also include severe diseases in immune-competent individuals as those caused by meningococci and pneumococci (1,2). For example, Neisseria meningitidis colonizes the nasopharynx of ∼30% of the healthy population (3) and can cause life-threatening invasive meningococcal disease (IMD), manifesting as acute bacterial meningitis and/or sepsis (2). Additionally, N. meningitidis has been linked to large epidemics of acute meningitis in developing countries of sub-Saharan Africa (4).
Understanding how N. meningitidis causes disease has been notoriously difficult due to a lack of classical virulence factors (5). Notwithstanding recent evidence linking certain mobile genetic elements and CRISPR-Cas loci to invasiveness, pathogenicity or carriage (6–9), ‘classical’ protein-coding virulence genes that clearly separate hyperinvasive from carriage lineages remain absent in N. meningitidis (10–12). Therefore, alternative explanations for differences observed epidemiologically in meningococcal pathogenicity are needed.
One promising avenue to explaining these differences in pathogenicity is the study of regulatory differences in intergenic regions (IGR), affecting either gene regulation in a polar manner and/or coding for regulatory small RNAs (sRNAs) (13,14). Regarding post-transcriptional control in cis, RNA thermosensors have been identified in the 5΄ untranslated regions (UTRs) of several mRNAs that encode proteins essential for meningococcal resistance against immune killing (15). Cis-acting antisense RNAs are known to control pili expression in meningococci (16,17). Moreover, pioneering tiling microarray based analyzes of the meningococcal transcriptome in human blood or after in vitro exposure to different stresses predicted hundreds of intergenic sRNAs and antisense RNAs (18,19). Several intergenic sRNAs have been characterized experimentally in the N. meningitidis stress responses to oxygen tension (20), iron starvation (21–23) and perturbed envelope homeostasis (24). What is more, the RNA chaperone Hfq, whose sRNA–mRNA matchmaking function underlies post-transcriptional regulation in many other disease-related Gram-negative bacteria (25), is an important virulence-associated factor in N. meningitidis: Hfq-deficient strains are attenuated in ex vivo and in vivo disease models (26) and display altered expression of numerous proteins involved in general metabolism, stress responses and virulence (27). Neisserial Hfq binds sRNAs and mRNAs when expressed in Salmonella (28), further arguing for a conserved role for this protein in facilitating RNA-based regulation. Despite these advances, a fine-grained annotation of the global RNA output and its potential for post-transcriptional modulation, which has enabled a systems understanding of RNA networks in other important bacterial pathogens (29), is lacking in N. meningitidis.
Here, we report the primary transcriptome and sRNA repertoire of the hyperinvasive clinical isolate N. meningitidis strain 8013. To this end, we combined two approaches: differential RNA-seq (dRNA-seq; (30,31)) for genome-wide annotation of transcriptional start sites (TSSs) and 5΄ ends of processed transcripts to define the major expression units, promoter elements and sigma factor binding sites of this organism; and RNA co-immunoprecipitation (coIP) with Hfq and cDNA sequencing (RIP-seq; (32)) to draft the global network of Hfq-mediated post-transcriptional regulation. To demonstrate the utility of these post-genomic datasets, we have integrated them to predict Hfq-mediated regulatory interactions between newly annotated transcripts. Moreover, we introduce a chromosomal gfp reporter system for post-transcriptional regulation in N. meningitidis and use it to validate native interactions of two paralogous Hfq-associated sRNAs with a target mRNA that encodes a putative colonization factor of the human nasopharynx. Altogether, this combination of high-throughput approaches provides a valuable resource for the exploration of transcriptional and post-transcriptional control in N. meningitidis and its relationship to pathogenesis.
MATERIALS AND METHODS
Bacterial strains, plasmids and oligonucleotides
A complete list of bacterial strains used in this study is provided in Supplementary Table S1. A complete list of all plasmids, as well as information on their construction, is provided in Supplementary Table S2 and Supplementary Materials and Methods. A complete list of DNA oligonucleotides used as hybridization probes or for cloning is provided in Supplementary Table S3.
Bacterial growth and construction of mutant strains
Neisseria meningitidis cells were routinely grown at 37°C in 5% CO2 with 95% humidity on either blood agar plates, GC agar plates or in GCBL++ medium in 50 ml tubes with vigorous shaking. When appropriate, 100 μg/ml ampicillin, 50 μg/ml kanamycin or 20 μg/ml chloramphenicol (final concentration) was added to the medium. Details about the generation of Neisseria mutant strains are listed in Supplementary Materials and Methods.
Sample collection for dRNA-Seq analysis and RNA extraction
Samples for dRNA-Seq were collected from bacterial cultures grown to optical densities (OD)600 of 0.5 or 1.5, corresponding to the mid logarithmic and late logarithmic/early stationary growth phase, respectively. Samples were fixated by the addition of STOP Mix [95% (vol/vol) EtOH and 5% (vol/vol) phenol], frozen in liquid nitrogen and subsequently stored at −80°C until RNA preparation. For RNA extraction frozen bacterial cultures were thawed on ice, centrifuged and cell pellets were resuspended in a lysis solution consisting of 800 μl of 0.5 mg/ml lysozyme in Tris-EDTA (TE) buffer (pH 8.0) and 80 μl 10% sodium dodecyl sulphate (SDS). Bacterial cells were lysed by placing the samples for 1–2 min at 65°C in a water bath. Total RNA was extracted from the lysates using the hot phenol method (33).
RNA immunoprecipitation with Hfq (RIP-seq)
Neisseria expressing either 3× FLAG-tagged- or wild-type (WT) Hfq protein were grown in the presence of kanamycin until an OD600 of 0.5. For each strain cells equivalent to an OD600 of 50 were collected and subjected to Hfq coIP and control coIP as described in (32).
Northern blot analysis and RNA stability assay
For northern blot analysis, 5 μg of DNase I-treated total RNA was separated on an 8% polyacrylamide gel containing 8.3 M urea. RNA was transferred onto Hybond-XL membranes, and membranes were hybridized overnight at 42°C with γ32P-ATP end-labeled oligodeoxyribonucleotide probes (Supplementary Table S3). Signals were visualized on a Phosphorimager (Typhoon FLA 7000, GE Healthcare) and quantified with the AIDA software (Raytest). Details about rifampicin stability assays are listed in Supplementary Materials and Methods.
SDS-PAGE and western blot analysis
For protein analysis, samples corresponding to an OD600 of 0.5 from N. meningitidis cells grown to mid logarithmic phase were collected by centrifugation at 16 100 × g at 4°C for 2 min and dissolved in 50 μl Laemmli protein loading buffer. After incubation for 5 min at 95°C, 0.1 OD600 equivalents of samples were separated by 12% (vol/vol) SDS-polyacrylamide gelelectrophoresis (PAGE). For western blot analysis, protein samples corresponding to an OD600 of 0.01 or 0.001 were separated by 12% (vol/vol) SDS-PAGE and transferred to a Polyvinyliden-Fluorid (PVDF) membrane by semi-dry blotting as described in (34).
EMSA andin vitro structure probing
DNA templates that contain a T7 promoter sequence for in vitro transcription were generated by polymerase chain reaction. Oligos and DNA templates used to generate the individual T7 templates are listed in Supplementary Materials and Methods and Table S3. Details about in vitro T7 transcription, structure probing and footprinting assays, as well as gel-shift experiments, are listed in Supplementary Materials and Methods.
Construction and sequencing of cDNA libraries for dRNA-seq and Hfq RIP-seq
cDNA libraries of dRNA-seq samples were constructed by Vertis Biotechnology AG, Munich, Germany; for details, see the Supplementary Materials and Methods. cDNA libraries of Hfq RIP-seq samples were constructed according to instructions of the NEBnext Multiplex sRNA library Prep Set for Illumina according to the manufacter's instruction but omitting the terminator exonuclease (TEX) treatment. cDNA libraries were pooled and sequenced using either an Illumina HiSeq2000 machine (dRNA-seq) or a MiSeq machine (Hfq-RIPseq) in the single-read mode. The raw, de-multiplexed reads as well as the normalized coverage files of all cDNA libraries have been deposited in the National Center for Biotechnology Information's Gene expression omnibus (GEO) (35) and are accessible via the GEO accession number GSE85252. Statistics on cDNA library sequencing are provided in Supplementary Table S4. For a detailed description of the dRNA-seq and Hfq RIP-seq read mappings and their normalization see Supplementary Materials and Methods.
Transcription start site annotation and promoter detection
Transcription start sites were predicted based on the normalized wiggle files using TSSpredator (http://it.inf.uni-tuebingen.de/TSSpredator) (36) applying the ‘more strict’ parameter presets. To detect potential promoter motifs, sequence regions corresponding to 50 nt upstream of the TSS positions and the TSS position itself were extracted from the TSSpredator master table and analyzed with MEME version 4.8.1 (37). UTRs were defined as previously described (38) using the output of TSSpredator. Briefly, transcriptional units were defined based on NCBI CDS annotations, the TSS annotations described above and Rho-independent terminators predicted by RNIE (39). If a primary TSS and/or Rho-independent terminator was not predicted for a given CDS, an ad hoc 100 base UTR was added for the purposes of analyzing our RIP-seq data below.
Analysis of Hfq RIP-seq data
Read counts were summed for all genomic features contained in the NCBI annotation and additionally described above (UTRs, non-coding RNAs), and imported into R for further analysis. Enrichment analysis was conducted using the edgeR package (40). Normalization factors were calculated using the trimmed means of M values (TMM) method between flag-tagged Hfq RIP-seq libraries and non-flag-tagged mock pull-down control libraries, using only those features with greater than five reads in the control library. For enrichment analysis, all features containing less than five reads in the flag-tagged library were excluded. Features with a resulting Benjamini–Hochberg corrected P-value (Q-value) < 0.1 and log2 fold-change (log2 f.c.) ≥1 were considered significantly enriched.
RESULTS
The primary transcriptome of Neisseria meningitidis 8013
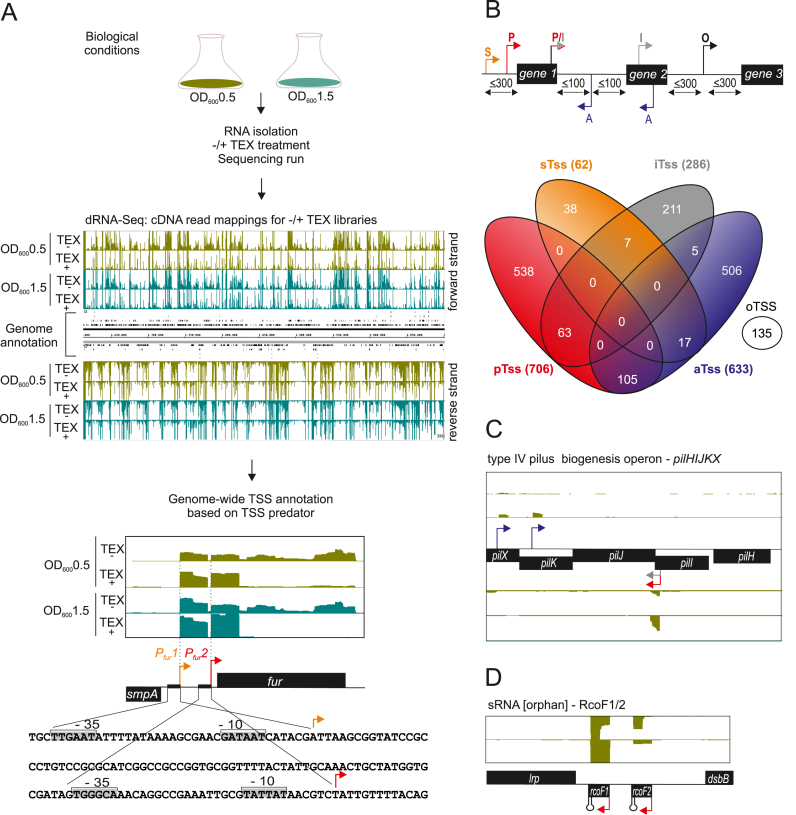
To define the primary transcriptome of N. meningitidis, we subjected serogroup C clinical isolate strain 8013 (41) to a dRNA-seq analysis with samples taken at two different representative phases of growth, i.e. the mid logarithmic and the early stationary phase (Figure 1A). Transcriptome profiling by dRNA-seq (31) differentiates between primary (5΄-PPP) and processed (5΄-P or 5΄OH) transcripts based on a characteristic enrichment pattern after RNA treatment with TEX, enabling precise TSS annotation. Sequencing reads from TEX− and TEX+ libraries were analyzed by TSSpredator (36) and detected TSS were assigned to five different classes: primary TSS (pTSS; main transcription start of a gene or operon), secondary TSS (sTSS; alternative start with lower expression), internal TSS (iTSS; start within a gene), antisense TSS (aTSS; transcript start antisense to a gene ±100 nt) and orphan TSS (oTSS; not associated with a gene) (Figure 1B). Mapping of two TSS upstream of the fur gene, encoding a global iron-responsive regulator, matched previously annotated promoters (42) and gives an illustration of the accuracy of this approach (Figure 1A).
Figure 1.
Differential RNA-seq and growth-phase based annotation of transcriptional start sites (TSSs). (A) (top) Schematic workflow for generating the biological, library and sequencing samples from strain N. meningitidis 8013 at two different growth phases (OD600 0.5 and 1.5) that were subjected to dRNA-seq analysis for genome-wide TSS annotation based on TSSpredator. (bottom) dRNA‐seq data from two growth phases OD600 0.5 and 1.5 were mapped to the fur gene. Red and orange arrows indicate a primary (pTSS) and secondary (sTSS) transcriptional start site and confirm initiation of RNA transcription as mapped with primer extension experiments in (42). Nucleotide sequence of the smpA- fur intergenic region (IGR) with promoter DNA elements are boxed and marked −10 and −35 at Pfur1 and Pfur2. (B) (Top) TSS classifications based on expression strength and genomic context: primary (P, red), secondary (S, orange), internal (I, gray), antisense (A, blue) or orphan (O, black). Venn diagrams were generated by VENNY (http://bioinfogp.cnb.csic.es/tools/venny/index.html). (C) Comparative annotation of primary TTSs (pTSSs) of the pilXKJIH operon. Detection of a pTSS (red arrow) for pilJ within the operon at two growth phases OD600 0.5 and 1.5 indicates a different genetic organization of their transcriptional units. Gray and blue arrows indicate an iTSS and an antisense TSS (aTSS), respectively. (D) Detection of an orphan TSS (oTSS; not associated with annotation) derived from dRNA-seq reads mapped to the intergenic lrp-dsbB region reveals two non-coding RNAs RcoF1 and RcoF2.
We identified a total of 1625 TSS at both growth stages (Supplementary Dataset S1). At a global level, a comparison of the dRNA-seq data shows that the majority of TSS are enriched and detected in both the logarithmic and early stationary phase samples (Supplementary Dataset S2). However, there are also differences in expression from TSS at different growth phases, probably due to promoters whose activities vary by growth phase dependent metabolite levels or transcription factors (see all loci listed in Supplementary Dataset S2). For example, there is one TSS of the rpoH gene that is only detected at early stationary phase (Supplementary Figure S1B). This gene encodes one of the two functional alternative sigma factors in N. meningitidis, and its expression has been shown to be upregulated in N. meningitis upon heat shock (43). This suggests that in Neisseria, which lack a general stress response sigma factor such as σS, the rpoH gene might be involved in stress responses during stationary phase. Figure 1B further shows a Venn diagram indicating the overlap between the five different TSS classes in N. meningitidis 8013. For example, 63 of the 706 pTSS (∼9%) are also classified as internal and 105 (∼15%) are also classified as antisense (Figure 1B and Supplementary Dataset S1). Since iTSS could reflect misannotated open reading frames (ORFs) (31) in the N. meninigitidis genome, we double-checked all 63 transcripts in question for alternative ribosome binding sites (RBSs) and start codons downstream of the respective iTSS but found no evidence for potential ORF misannotations.
N. meningitidis 8013 has 1918 annotated ORFs of which 1011 genes are predicted to be organized in 379 operons according to the DOOR 2.0 database of prokaryotic operons (44). As expected the majority of the 706 pTSS map upstream of ORFs, with 382 pTSS obtained for monocistronic genes and 240 pTSS obtained for genes in predicted operons (Supplementary Dataset S2). Among the 240 operon-associated pTSS, 187 precede the first gene in an operon, likely accounting for the expression of roughly a quarter (487) of all genes. The longest operon detected here consists of ten ORFs (NMV_0195-NMV_0204).
The remaining 53 operon-associated pTSS are derived from operon-internal ORFs and suggest alternative operon structures to those computationally predicted (Supplementary Dataset S2). For instance, the gene encoding the cell division protease FtsH, annotated as the second gene of the ftsJ-ftsH operon, has its own pTSS indicating its transcription as an independent unit (Supplementary Figure S1A and Supplementary Dataset S2). Conversely, our results link apparently isolated genes into transcriptional units. For example, of genes encoding sigma factors, rpoD is likely co-transcribed with the dnaG-secA operon as described for Escherichia coli (45) while rpoE appears to be transcribed from a polycistronic mRNA that spans six (NMV_2355-NMV2360) rather than four genes as previously postulated for N. meningitidis MC58 (Supplementary Dataset S2) (24).
There are 103 protein-coding genes that have been associated with the pathogenicity of N. meningitidis (46–48). We detect a pTSS for 40 of them (Supplementary Dataset S2) including the pil genes that encode the organism's type IV pilus. The Neisseria type IV pili are long filamentous surface structures that promote biofilm formation and adhesion to host cells, with an important role in meningococcal colonization and IMD (49,50). A pTSS was obtained for four out of five pil operons, namely operons pilT2-holB-pilZ-hp, pilG-pilD-coaE-hp, pilHIJKX and pilMNOPQ. The last two of these operons possess an additional internal pTSS (as depicted for pilHIJKX in Figure 1C), suggesting a more complex operon expression in which some proteins are translated from two rather than one poly-cistronic mRNA (Figure 1C), perhaps to ensure the correct protein stoichiometry in the type IV pilus complex. Furthermore, pTSS were detected for five monocistronic type IV pilus genes (pilE, pilT, pilF, pilC1, pilC2) that encode the structural subunit pilin and proteins involved in pilus assembly and adhesion, respectively. The polysaccharide capsule of serogroup C constitutes another important surface structure for meningococcal survival in the human host (46). It is encoded by the cps locus which contains three operon-like regions for capsule biosynthesis/transport and lipopolysaccharide (LPS) biosynthesis, respectively (51). We annotated a total of four pTSS in this locus, for the first gene of region A, comprising the capsule biosynthesis operon (cssA,-B,-C-csc-cssE-ctrG) and of regions D and D’, containing the LPS synthesis genes (rfbB1A1C1 and rfbB2A2C2) (52) and one for the ctrF gene, the second gene in region B comprising the capsule translocation genes (ctrE, -F) (53). We could not detect pTSS for any of the capsule transport genes in region C (ctrABCD) (Supplementary Dataset S2).
Finally, we also detected 135 oTSS in IGR. Since these TSS lack a clear association with flanking genes, they may represent previously unknown sRNA genes (Figure 1D) or ORFs of small peptides.
Transcriptome features of the N. meningitidis strain 8013
N. meningitidis not only has a small genome, about half the size (∼2.2 Mb) of that of well-characterized enteric pathogens such as Salmonella, it also seems to lack many classical mechanisms of primary gene regulation. There are only three functional sigma factors (σ70/rpoD; σH/rpoH and σE/rpoE), four two-component systems (NtrY/NtrX, MisR/MisS, NarQ/NarP and PilS/PilR homologs) (54) and <30 putative transcription factors (55,56) which contrasts with seven sigma factors, 62 two-component system proteins and over 300 predicted transcription factors in E. coli K-12 (57,58).
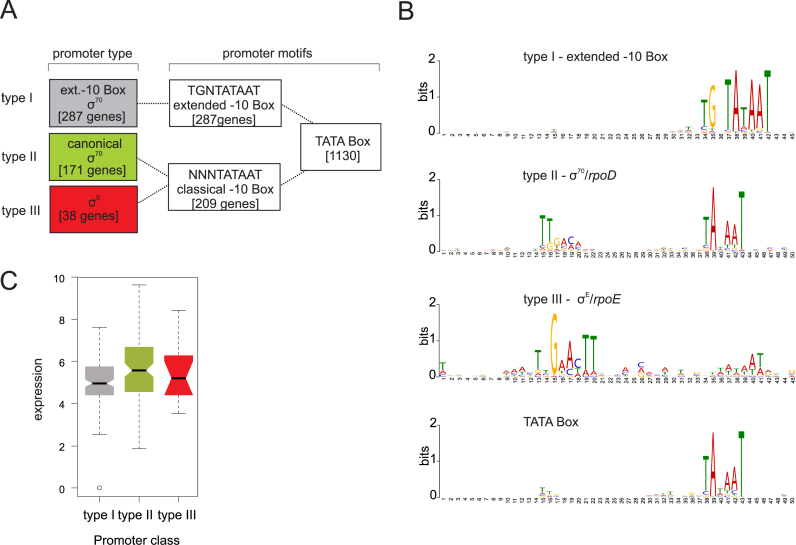
To better understand how transcription is initiated in N. meningitidis, we searched for putative promoter motifs in the upstream regions (−50 to +1) of all detected 1625 TSS. Predicting consensus sequences with the MEME toolkit (37) detected a −10 box upstream of the majority of the TSS (1130 sequences; ∼70%) (Figure 2A and B, Supplementary Dataset S3). In contrast, a conserved −35 box was absent in most N. meningitidis promoters, suggesting that their activity may require accessory activator proteins (59). Reanalyzing those 1130 sequences with a −10 box only, we found a −10 TG extension sequence in 25% of them, a key element of promoters with an extended −10 box that improves the efficiency of σ70 (type I) transcription in the absence of a −35 sequence (60). Interestingly, computational analyses further indicate that these extended −10 boxes are not preceded by any known AT-rich periodic sequence that is thought to contribute to highly curved DNA in compensation for the absence of a conserved −35 binding motif (61).
Figure 2.
Promoter motifs detected for TSS of N. meningitidis 8013 and expression strength. (A) Schematic overview of promoter types derived from a motif search based on 1625 TSS upstream sequences of N. meningitidis 8013. The number of motifs for each type of promoter detected for the two sigma factors σD and σE in the upstream sequences of the TSS are indicated. (B) Promoter motifs for each type within a fixed size of either 45 or 50 nt, that was applied for the search using the MEME toolkit are shown. (C) Box-and-whiskers plot depicting differences in the expression (log (basemean)) between genes with the three different promoter types. The line within each box gives the median and the upper and lower margins the upper and the lower quartile, respectively. The whiskers denote the highest and the lowest values, respectively, and the open circles outliers.
For 17% of the sequences (171 genes) with a canonical −10 box we could also identify a classical −35 box recognized by σ70 (type II) and an additional 38 with a canonical −35 box for σE (type III) dependent recognition (Figure 2A and B, Supplementary Dataset S3). In contrast to a recent dRNA-seq study in Neisseria gonorrhoeae (62) no promoter motif could be identified for the third sigma factor σH. Correlating the sigma factor dependency with expression level and functional class showed that type II promoters primarily drive highly expressed house-keeping genes (for example, those encoding ribosomal RNAs, tRNAs and ribosomal proteins). Genes with intermediate-to-low expression levels tend to possess a type I promoter (Figure 2C). This is reminiscent of results in E. coli where a perfect −35 box at optimal spacing is stronger than an extended −10 box promoter (63,64). Among the genes with an annotated function according to the COG database, genes involved in translation (COG J) constitute the largest group within the type I promoter genes (8%), whereas genes for DNA replication and repair (e.g. dinB, ihfA or rdgC as well as several transposase genes) (COG L) form the largest group within the type III promoter genes (16%). In contrast to type I and type III promoter genes, type II promoter genes were not enriched in a particular gene function (Supplementary Table S5). Interestingly, in addition to a type II promoter, some type IV pili genes (such as pilE) also possess a type III promoter, suggesting differential expression of the type IV pilus driven by different alternative sigma factors (Figure 2C and Supplementary Dataset S2).
σE promoters constitute a small minority (Figure 2A and B) and are associated with poorly expressed genes, which may reflect the absence of envelope stress under the growth conditions used here. This notwithstanding, our prediction of 38 new σE promoters (Supplementary Figure S2A and Supplementary Dataset S3) in addition to those 12 previously known (24,65) quadruples the size of the σE regulon in N. meningitidis, bringing it closer to the 89 σE controlled transcription units in E. coli. Newly discovered σE promoters in N. meningitidis are in front of dinB (polymerase IV), trxA (thioredoxin), ihfA (integration host factor) and secA (preprotein translocase SecA subunit), all of which function to combat environmental challenges such as oxidative stress, heat shock and misfolding of membrane proteins, in other words, classical functions of a σE regulon (66). Interestingly, seven genes (aceE, rpmB, trmE, pilE, trxA, php and secA) with a σE promoter possess additional type I or II promoters, and secA, the first gene in the operon that encodes sigma factor σ70, even possesses all three kinds of promoters (Supplementary Dataset S3).
UTRs of mRNAs are hot spots for post-transcriptional regulation, for instance serving as targets for trans-regulating sRNAs and RNA-binding proteins (67). Our dRNA-seq atlas of promoters now permits us to define 5΄ UTRs globally. In agreement with previous findings in N. gonorrhoeae (62) we determined a median 5΄ UTR length of 55 nt (minimal length 0 nt, maximal length 294 nt) in N. meningitidis 8013 based on 768 primary and sTSS (Supplementary Figure S2B and Supplementary Dataset S4). Twenty-four genes had 5΄ UTRs shorter than 10 nt; after manual inspection for potential alternative downstream start codons, 12 genes remain as leaderless mRNA genes (Supplementary Dataset S4). Regarding 3΄ UTRs, we could predict Rho-independent terminators for 706 coding genes of N. meningitidis 8013 (39) and determined the 3΄ UTR length for those genes with a median length of 63 nt (minimal length 8 nt, maximal length 492 nt) (Supplementary Figure S2C and Supplementary Dataset S4).
Antisense transcription has been found to be pervasive in many bacterial species (19,68), and a recent study using tiling arrays indicated over 260 antisense transcripts in N. meningitidis strain MC58 (19). In line with these findings, our dRNA-seq data suggest that 39% of the TSS initiated in the antisense direction (573 genes out of 1913 genes of N. meningitidis 8013 possess at least one aTSS). Among those, 236 have a pTSS identified by dRNA-seq (Supplementary Figure S2D and Supplementary Dataset S4). The majority of aTSS were active in both growth phases. However, we also found an example of an early log-phase specific asRNA associated with a putative zinc-type alcohol dehydrogenase (adh) whose mRNA is detected only at stationary growth phase. This suggests that this asRNA may repress the expression of the adh gene during early log-phase (Supplementary Figure S2E and Supplementary Dataset S4).
Different classes of small RNAs in N. meningitidis strain 8013
Beyond mRNAs, our dRNA-seq approach predicted a large number of sRNAs, including members of the emerging class of sRNAs derived from 3΄ mRNA regions (69). To predict sRNAs, we analyzed oTSS in IGR, requiring a distance of ≥300 nt relative to existing gene annotations. This analysis not only confirmed the expression of 20 previously described sRNAs but also predicted 45 new sRNA candidates (Table 1 and Supplementary Dataset S5) which were classified according to their genomic location: intergenic, 5΄ UTR, cis-encoded antisense and 3΄ UTR-derived sRNAs (Figure 3A and Supplementary Dataset S5).
Table 1. Non-coding RNAs of different Neisseria meningitidis strains.
| RNA classes* | 8013 | MC58 | WUE2594 | Z2491 |
|---|---|---|---|---|
| Structural RNAs | 6S RNA | 6S RNA | 6S RNA | 6S RNA |
| RNaseP | RNaseP | RNaseP | RNaseP | |
| tmRNA | tmRNA | tmRNA | tmRNA | |
| SRP | SRP | SRP | SRP | |
| Riboswitches | preQ riboswitch | preQ riboswitch | preQ riboswitch | preQ riboswitch |
| Tpp riboswitch | Tpp riboswitch | Tpp riboswitch | Tpp riboswitch | |
| sRNAs known (n°10) | AniSb | AniSb | AniSb | AniSb |
| NrrFc | NrrFc | NrrFc | NrrFc | |
| Bns1d | Bns1d | Bns1d | Bns1d | |
| Bns2d | Bns2d | Bns2d | Bns2d | |
| σEsRNAe | σEsRNAe | σEsRNAe | σEsRNAe | |
| G4-assoc. sRNAf | G4-assoc. sRNAf | - | - | |
| 0863-0864_Fg | 0863-0864_Fg | 0863-0864_Fg | 0863-0864_Fg | |
| 1400-1401_Fg | 1400-1401_Fg | 1400-1401_Fg | 1400-1401_Fg | |
| tracrRNAh | - | tracrRNAh – | tracrRNAh | |
| crRNAs (n°25) | - | crRNAs (n°22) | crRNAs (n°16) | |
| sRNAs this study (n°45) | NMnc0001 | NMnc0001 | NMnc0001 | NMnc0001 |
| NMnc0002 | NMnc0002 | NMnc0002 | NMnc0002 | |
| NMnc0003 | NMnc0003 | NMnc0003 | NMnc0003 | |
| NMnc0004 | - | - | - | |
| NMnc0005 | - | - | - | |
| NMnc0006 | - | - | - | |
| NMnc0007 | NMnc0007 | NMnc0007 | NMnc0007 | |
| NMnc0008 | NMnc0008 | NMnc0008 | NMnc0008 | |
| NMnc0009 | NMnc0009 | NMnc0009 | NMnc0009 | |
| NMnc0010 | NMnc0010 | NMnc0010 | NMnc0010 | |
| NMnc0011 | NMnc0011 | NMnc0011 | NMnc0011 | |
| NMnc0012 | NMnc0012 | NMnc0012 | NMnc0012 | |
| NMnc0013 | NMnc0013 | NMnc0013 | NMnc0013 | |
| NMnc0014 | - | - | - | |
| NMnc0015 | NMnc0015 | NMnc0015 | NMnc015 | |
| NMnc0016 | NMnc0016 | NMnc0016 | NMnc016 | |
| RcoF1 | RcoF1 | RcoF1 | RcoF1 | |
| RcoF2 | RcoF2 | RcoF2 | RcoF2 | |
| NMnc0019 | NMnc0019 | NMnc0019 | NMnc0019 | |
| NMnc0020 | NMnc0020 | NMnc0020 | NMnc0020 | |
| NMnc0021 | NMnc0021 | NMnc0021 | NMnc0021 | |
| NMnc0022 | NMnc0022 | NMnc0022 | NMnc0022 | |
| NMnc0023 | NMnc0023 | NMnc0023 | NMnc0023 | |
| NMnc0024 | - | - | - | |
| NMnc0025 | NMnc0025 | NMnc0025 | NMnc0025 | |
| NMnc0026 | NMnc0026 | NMnc0026 | NMnc0026 | |
| NMnc0027 | NMnc0027 | NMnc0027 | NMnc0027 | |
| NMnc0028 | NMnc0028 | NMnc0028 | NMnc0028 | |
| NMnc0029 | NMnc0029 | NMnc0029 | NMnc0029 | |
| NMnc0030 | - | - | - | |
| NMnc0031 | NMnc0031 | - | - | |
| NMnc0032 | NMnc0032 | NMnc0032 | NMnc0032 | |
| NMnc0033 | NMnc0033 | NMnc0033 | NMnc0033 | |
| NMnc0034 | NMnc0034 | NMnc0034 | - | |
| NMnc0035 | NMnc0035 | NMnc0035 | NMnc0035 | |
| NMnc0036 | NMnc0036 | NMnc0036 | NMnc0036 | |
| NMnc0037 | NMnc0037 | NMnc0037 | NMnc0037 | |
| NMnc0038 | NMnc0038 | NMnc0038 | NMnc0038 | |
| NMnc0039 | NMnc0039 | NMnc0039 | NMnc0039 | |
| NMnc0040 | - | NMnc0040 | NMnc0040 | |
| NMnc0041 | NMnc0041 | NMnc0041 | NMnc0041 | |
| NMnc0042 | NMnc0042 | - | - | |
| NMnc0043 | NMnc0043 | NMnc0043 | NMnc0043 | |
| NMnc0044 | NMnc0044 | NMnc0044 | NMnc0044 | |
| NMnc0045 | NMnc0045 | NMnc0045 | NMnc0045 | |
| Genome Reference | (41) | (123) | (124) | (125) |
*Detailed information concerning annotations of RNA classes is listed in Dataset S5.
aNumber of ORFs according to NCBI annotations (November 2016).
b(20)
c(22)
d(19)
e(24)
f(126)
g(18)
h(7)
n°-number, - sRNAs absent in N. meningitidis strains WUE2594, MC58, Z2491 based on conservation analyzes by basic local alignmentsearch tool (BLAST) searches regarding identity values below 40%.
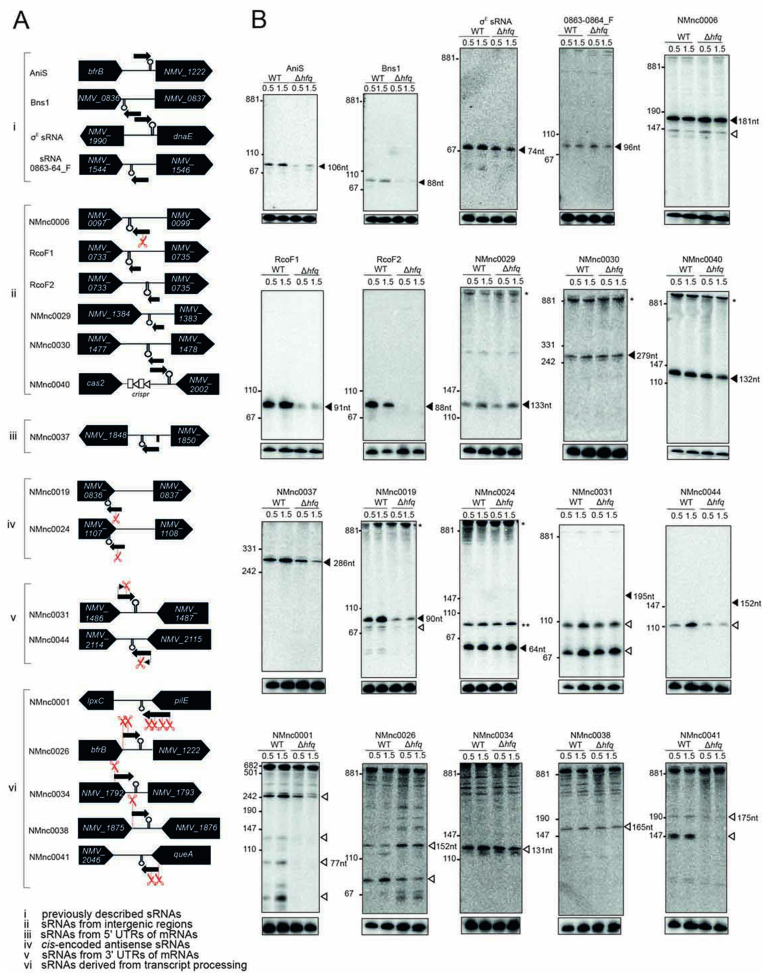
Figure 3.
Expression analysis of candidate sRNAs in N. meningitidis 8013. Total RNA was extracted at mid logarithmic (OD600 = 0.5) and late logarithmic/early-stationary (OD600 = 1.5) growth phases from wild-type (WT) and Δhfq N. meningitidis strains and analyzed by northern blot using labeled DNA probes complementary to the sRNAs (see Supplementary Table S3). (A) The genomic locations and relative orientations of (i) previously described sRNAs, (ii) sRNAs from IGR, (iii) sRNAs from 5΄ untranslated regions (UTRs) of mRNAs, (iv) cis-encoded antisense sRNAs, (v) sRNAs from 3΄ UTRs of mRNAs and (vi) sRNAs derived from transcript processing are shown. The designation in (vi) is the same as in (v) except that the promoters are shared between the sRNA and mRNA. Here, the mRNAs undergo ribonucleolytic cleavage to yield the sRNAs. Genes and sRNAs are shown in black. Arrows and scissors indicate TSS and processing sites, respectively. Black rectangle indicates the promoter of NMV_1848 and NMnc0037, respectively. (B) Gel images of blots from previously described sRNAs (i) and different classes of candidate sRNAs (ii–vi). The blots were probed for the housekeeping 5S rRNA as loading control. Filled triangles indicate bands for sRNAs derived from TSS and open triangles indicate bands derived from processing. Asteriks indicate signals of unclear origin, may represent longer read-through transcripts at the locus, result from cross hybridization of the probe with other abundant transcripts, or stem from undigested DNA trapped in the slot.
For independent validation of sRNA expression, we performed northern blot analysis of RNA samples from the same growth phases used in the dRNA-seq analysis. In addition to the 8013 WT strain, we included a Δhfq strain to determine the Hfq-dependence of sRNA expression (Figure 3B and Supplementary Dataset S5). As a positive control, we successfully detected the four previously described sRNAs AniS (20), Bns1 (19), σE sRNA (24) and sRNA 0863–0864_F (18) (Figure 3A and B). Of the 45 new sRNAs, twenty-two are canonical intergenic sRNAs characterized by their own promoters and Rho-independent terminators. Five of them possess a type I promoter, another five a type II promoter (Supplementary Dataset S5), and no promoter motif could be identified for the remaining 12 sRNA candidates. Using northern blots, we confirmed the expression of six sRNAs from IGR (Figure 3B).
The 5΄ UTR derived class of sRNAs are characterized by proximity to the translational start site of an ORF and (often) the presence of rho-independent terminator sequences at their 3΄ ends. Representative examples are NMnc0013 and NMnc0037. Both sRNAs were screened with the Rfam database (70), enabling us to identify NMnc0037 as a member of the glycine riboswitch family, which forms an on-switch when bound to glycine (71). Strikingly, the expression level of NMnc0037 was decreased in the Δhfq strain, suggesting that NMnc0037 may additionally act as a trans-acting sRNA (Figure 3B and Supplementary Dataset S5).
We also confirmed the expression of two of four tested cis-encoded antisense RNAs expressed from the opposite strand in the 3΄ CDS of genes NMV_1107 and NMV_0836 (Figure 3B and Supplementary Dataset S5). The former (NMV_1107) codes for a conserved protein with high homology to abortive infection bacteriophage resistance proteins (IPR011664) that protect bacteria against phages (72). NMV_0836 encodes a OsmC/Ohr family protein (IPR003718) that, in E. coli, is involved in oxidative stress regulation (73). These findings suggest that antisense transcription might add another layer of complexity in the regulation of stress responses and phage infections in N. meningitidis.
3΄ UTR-derived sRNAs are characterized by sharing a rho-independent terminator with the gene they are located by (69) (Supplementary Dataset S5). Of 17 predicted such sRNAs, we confirmed the expression of seven that were further sub-divided into sRNAs that possess their own promoters (NMnc0031/0044; enriched in the TEX+ samples; Figure 3B) or result from processing of mRNAs (NMnc0026, NMnc0034, NMnc0038, NMnc0041, NMnc0001; no enrichment by TEX treatment, but significant cDNA reads in the TEX− samples; Figure 3B). The majority of these 3΄ UTR-derived sRNAs accumulate as intermediate processing products. The expression levels of NMnc0044, NMnc0041 and NMnc0001 decreased in the absence of Hfq, suggesting that Hfq is required for their maturation and/or stability (Figure 3B, see also Figure 6 below). In addition, the processing of NMnc0026 also seems to be affected by absence of Hfq. In summary, we discovered 45 new meningococcal sRNAs belonging to different classes and showed that several of them accumulate in an Hfq-dependent manner. Of 37 sRNAs transcribed from their own TSS, only type I and II but no type III promoter motifs could be found for 26 sRNAs (Supplementary Dataset S5), indicating that most of these sRNAs are expressed from σ70 dependent promoters.
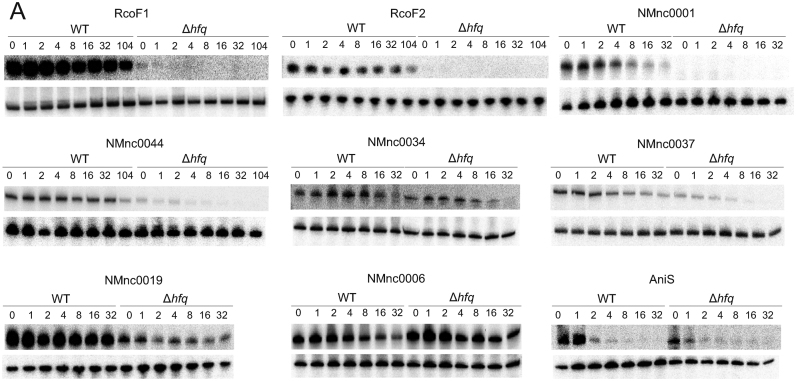
Figure 6.
Stability of Hfq binding sRNAs. (A) sRNA half-lives for RcoF1, RcoF2, NMnc0001, NMnc0044, NMnc0034, NMnc0037, NMnc0019, NMnc006 and AniS. At the top of each panel, northern blots of total RNA extracted at the indicated time points (minutes) after addition of rifampicin (250 μg/ ml) to terminate transcription in a N. meningitidis 8013 WT and an isogenic Δhfq strain is shown. Prior to the addition of rifampicin, strains were grown to OD600 of 0.5. 5S rRNA served as loading control.
N. meningitidis 8013 strain specific sRNAs
Given the high genetic diversity of N. meningitidis with currently over 12 000 multi locus sequence types deposited in the PubMLST database (http://pubmlst.org/neisseria/info/) (74), we asked whether our results are representative for the entire species and whether there might be differences in strains from other clonal complexes with impact on meningococcal virulence or epidemiology. Conservation of the 45 newly identified sRNAs was analyzed by basic local alignment search tool (BLAST) searches against three other prototypical N. meningitidis strains, MC58 (serogroup B), WUE2594 (serogroup A) and Z2491 (serogroup A); the three serogroups responsible for the majority of disease cases worldwide. This analysis revealed that 35 of the 45 newly identified sRNAs are conserved in all four strains including 8013 (40% sequence similarity cut-off), whereas 10 sRNAs are strain-specific (Table 1 and Supplementary Dataset S5).
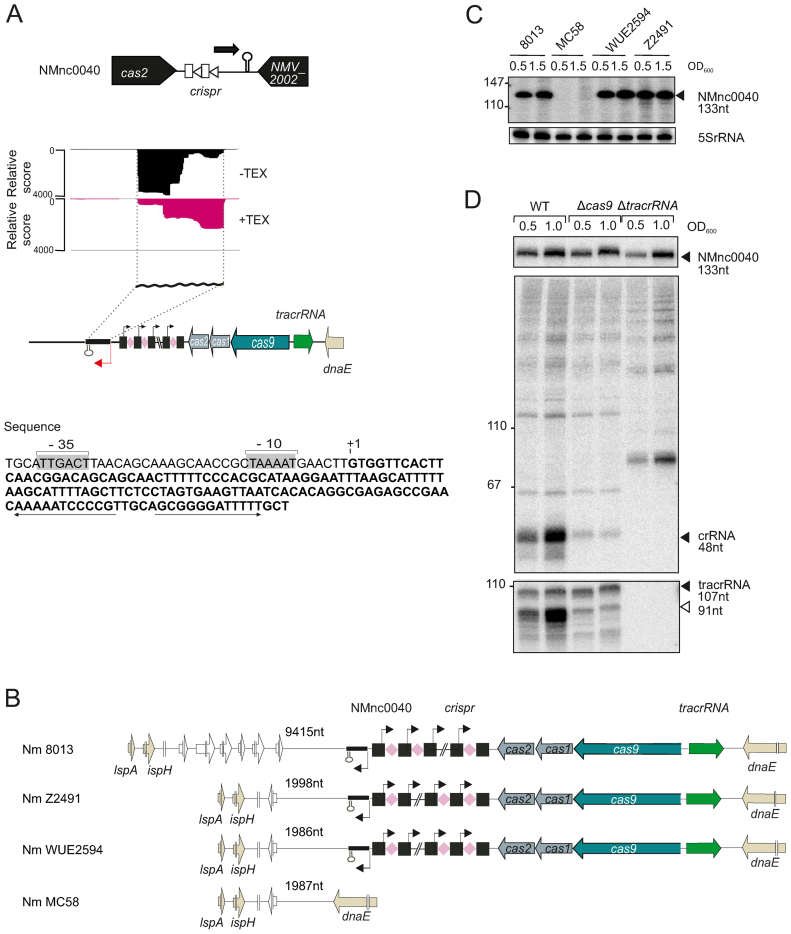
One sRNA candidate, NMnc0040, is located next to the CRIPSR-Cas locus in strain 8013 (Figure 4A). It is expressed from a σ70-like promoter in the opposite direction from the CRISPR array (Figure 4A). Based on a sequence comparison of 71 meningococcal strains with a completely sequenced and assembled genome available in the NCBI database, all 17 strains that possess a CRISPR/Cas locus also contained the neighboring NMnc0040. In turn, none of the 54 meningococcal genomes without a CRISPR/Cas locus have a NMnc0040 gene (here or elsewhere in the genome), suggesting that this RNA is indeed associated with the CRISPR/Cas locus (see Supplementary Table S7). Northern blot analysis, restricted to four strains representing epidemiologically important serogroups, confirmed that NMnc0040 appears to be present only in strains that harbor an intact type II CRISPR-Cas locus (Figure 4B and C) (7), suggesting that this sRNA may be functionally linked to the CRISPR-Cas system. To investigate a possible link between NMnc0040 and CRISPR-Cas, we examined its expression in strains lacking Cas9 and tracrRNA. However, NMnc0040 levels did not change in Δcas9 and ΔtracrRNA strains (7) as compared to the WT strain (Figure 4D), suggesting that NMnc0040 may regulate genes encoded elsewhere in the N. meningitidis genome or provide an alternative CRISPR-Cas function.
Figure 4.
A strain-specific sRNA, NMnc0040 is linked to the CRISPR-Cas locus. (A) (top) Genomic location of NMnc0040 sRNA. dRNA–seq reads of −/+ TEX libraries mapped to NMnc0040 sRNA. (bottom) Nucleotide sequence of NMnc0040 is indicated in bold letters. The −10 and −35 sites of the NMnc0040 promoter are boxed in light gray. ‘+1’ marks the TSS and the transcribed NMnc0040 sequence is given in bold letters. The rho-independent terminator is indicated by arrows. (B) Conservation analysis of NMnc0040 in four prototypical N. meningitidis strains. NMnc0040 is absent in MC58. In the WEU2594, Z2491 and 8013 strains NMnc0040 is linked to CRISPR-Cas locus and located upstream of the CRISPR array. The flanking gene at the 3΄ end is variable. Distances to flanking genes are indicated in nucleotides. (C) Total RNA from the four N. meningitidis strains 8013, MC58, WUE2594 and Z2491 belonging to three different serogroups was isolated at mid logarithmic (OD600 = 0.5) and late logarithmic/early stationary growth phase (OD600 = 1.5) and analyzed by northern blot using labeled DNA probes complementary to NMnc0040 (see Supplementary Table S3). The blots were probed for the housekeeping 5S rRNA as loading control. (D) Northern blot analysis of NMnc0040 sRNA, tracrRNA and crRNAs in 8013 WT, Δcas9 and ΔtracrRNA strains. Total RNA (5 μg per lane) isolated from WT, ΔtracrRNA and Δcas9 at mid logarithmic (OD600 = 0.5) and late logarithmic/early stationary growth phase (OD600 = 1.5) was analyzed on a northern blot using labeled DNA probes complementary to NMnc0040, crRNA and tracrRNA in N. meningitidis 8013.
Another interesting sRNA candidate is NMnc0031 (Supplementary Figure S3A), located on island of horizontal gene transfer E (IHT-E) (75) which was recently linked to hyperinvasive meningococcal lineages (6). Genome comparison revealed that IHT-E might derive from an integrated prophage that is absent in strains WUE2594 and Z2491, suggesting that NMnc0031 has a phage-related function (Supplementary Figure S3B). Northern blots confirmed its strain specificity and revealed that it exists in two different size forms (Supplementary Figure S3C).
The sRNA candidate NMnc0001 struck us as peculiar, being a processed product from a transcript encoding the major pilin (PilE) that forms the pilus fiber of type IV pili, a highly variable structure on the bacterial surface (Supplementary Figure S4A). Interestingly, we found expression of NMnc0001 in only three of four strains, even though our sequence analysis suggests NMnc0001 to be conserved among all four (Supplementary Figure S4B and C). In all strains where NMnc0001 is expressed it is located within the 3΄ UTR of the class I pilE genes. In contrast, in WUE2594 carrying a class II pilE gene with a different genetic context compared to class I pilE genes, NMnc0001 is located within a pilS cassette (Supplementary Figure S4A, center). The pilS cassettes serve as a reservoir for antigenic variation of class I pilE genes and are not transcribed, which consequently explains the lack of NMnc0001 expression. We speculate that NMnc0001, only expressed together with class I pili, may provide a trans-acting link between type IV pili variation and the regulation of other genes not directly related to type IV pilus function.
sRNAs and mRNAs that interact with Hfq in vivo
The neisserial RNA chaperone Hfq has been previously shown to globally regulate gene expression (26,27) and its ectopic expression in Salmonella provided evidence for its conserved function as global sRNA-binding protein (28). Understanding how Hfq controls Neisseria gene expression at the post-transcriptional level requires the identification of its repertoire of sRNA and mRNA ligands.
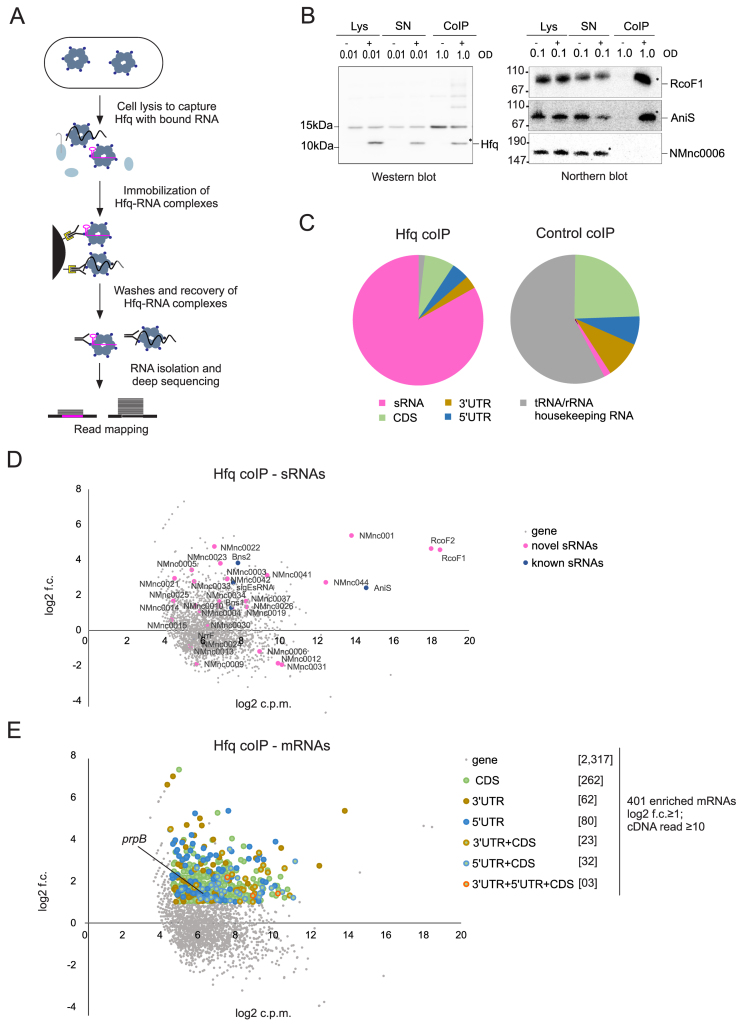
To identify RNA ligands of the native Hfq protein from N. meningitidis we performed RNA co-immunoprecipitation (coIP) and deep-sequenced RNA from a strain expressing a chromosomally encoded Hfq-3× FLAG protein (Figure 5A). CoIP was performed on extracts prepared from bacteria grown to mid logarithmic phase. Western blotting confirmed the successful immunoprecipitation of the tagged protein while northern blot verified co-precipitation of the sRNA RcoF1 (for ‘RNA regulating colonization factor’), indicating the integrity of the co-purified RNA (Figure 5B). The majority of sequences recovered with Hfq were derived from sRNAs (Figure 5C and Supplementary Dataset S6). Table 2 lists the top 10 Hfq-binding candidates with ≥50 reads from different classes of RNAs (sRNAs, CDSs, UTRs, rRNAs, tRNAs). RNA-seq libraries of Hfq coIP and control coIP data obtained from two biological replicates were compared with each other in terms of enrichment (log2 fold-change; log2 f.c.) and abundance (log2 counts per million reads; log2 c.p.m.). This revealed 19 of the newly identified sRNAs and four previously identified (Bns1, Bns2, AniS and σE sRNA) to be associated with Hfq (Figure 5D). Importantly, the three most abundant sRNAs are RcoF1, RcoF2 and AniS; these were enriched by 5- to 25-fold as compared to the control reaction.
Figure 5.
The repertoire of Hfq regulated sRNAs and mRNAs in N. meningitidis 8013. (A) Schematic workflow for generating co-immunoprecipitation (coIP) samples of WT and 3× FLAG tagged hfq strains grown in rich media to mid log phases (OD600 = 0.5) in two independent experiments. (B) Quality of RNA and protein samples was analyzed by western blots and northern blots. Lysate (Lys), supernatant (SN) and coIP samples were obtained from pull-down experiments with mouse anti-FLAG antibody performed in the presence (+) or absence (−) of the 3× FLAG tagged Hfq protein. OD600 equivalents of protein and RNA samples loaded on the gel are shown. Western blot with rabbit anti-FLAG antibody (left panels) and northern blot with probe against RcoF1 and AniS sRNAs as well as NMnc0006 sRNA as negative control (right panel) confirmed the success of Hfq pull-downs. The band indicated by a star within the coIP (+) lanes correspond to the sizes of purified Hfq proteins and co-purified RcoF1 sRNA. Size markers are given on the left (in kDa). (C) Pie chart for Hfq coIP and control coIP showing the relative proportions of all Hfq-associated sequences that unequivocally mapped to different classes of RNA sequences. (D) Scatter-plot of RIP-seq results. Axes represent log fold-change between the control coIP and Hfq coIP (y-axis) and abundance in log counts per million (x-axis) of cDNA reads obtained. New sRNAs and previously described sRNAs are depicted by pink and blue dots, respectively. (E) Scatter-plot analysis of RIP-seq results depicting 401 mRNAs and their associated segments (CDS, 5΄ UTR and 3΄ UTR) that were enriched (log2 f.c. ≥1; cDNA read ≥10) in the Hfq coIP, for example the 5΄ UTR of prpB gene. Axes are identical to above.
Table 2. Top 10 Hfq-binding candidates for each class of RNA.
| aclass | aannotated gene | reads | log2 f.c. | COG_ID | Class_ID | Process |
|---|---|---|---|---|---|---|
| sRNA | RcoF1 | 209162 | 4.6 | |||
| RcoF2 | 151733 | 4.6 | ||||
| AniS | 12697 | 2.4 | ||||
| NMnc0001 | 8456 | 5.3 | ||||
| NMnc0044 | 2959 | 2.7 | ||||
| NMnc0037 | 140 | 1.6 | ||||
| NMnc0026 | 134 | 1.3 | ||||
| Bns2 | 133 | 3.8 | ||||
| σEsRNA | 100 | 2.7 | ||||
| NMnc0003 | 83 | 2.9 | ||||
| CDS | NMV_0360 (rplU) | 1246 | 2.9 | |||
| NMV_0522 | 911 | 1.2 | COG0457 | R | Poorly characterized | |
| NMV_1824 (rplS) | 815 | 2.6 | ||||
| NMV_0019 (pilE) | 814 | 1.3 | ||||
| NMV_1510 (pilK) | 795 | 1.6 | ||||
| NMV_0215 (rne) | 672 | 2.2 | ||||
| NMV_2184 (app) | 658 | 1.2 | ||||
| NMV_2281 | 645 | 1.4 | COG1741 | S | Poorly characterized | |
| NMV_0045 (pilC2) | 567 | 1.6 | ||||
| NMV_1823 (trmD) | 562 | 1.8 | ||||
| 5΄ UTR | NMV_1222 | 12700 | 2.4 | COG1396 | K | Information storage and processing |
| NMV_0360 (rplU) | 645 | 3.9 | ||||
| NMV_2281 | 514 | 1.8 | COG1741 | S | Poorly characterized | |
| NMV_0522 | 395 | 3.0 | COG0457 | R | Poorly characterized | |
| NMV_1106 (nrdA) | 377 | 1.9 | ||||
| NMV_1646 (hfq) | 345 | 3.2 | ||||
| NMV_0309 | 343 | 2.0 | COG1611 | R | Poorly characterized | |
| NMV_1440 (sucA) | 309 | 3.0 | ||||
| NMV_1803 (rpsO) | 305 | 1.4 | ||||
| NMV_1593 | 246 | 3.6 | COG1469 | S | Poorly characterized | |
| 3΄ UTR | NMV_0019 (pilE) | 8503 | 5.3 | |||
| NMV_2115 | 2964 | 2.7 | COG0483 | G | Metabolism | |
| NMV_1651 | 769 | 3.5 | ||||
| NMV_2048 (queA) | 295 | 3.0 | ||||
| NMV_0526 | 192 | 1.9 | COG2018 | R | Poorly characterized | |
| NMV_1509 (pilX) | 126 | 1.6 | ||||
| NMV_0208 | 98 | 2.0 | COG1192 | D | Cell cycle control, cell division, Inorganic ion transport, metabolism | |
| NMV_0057 (comE1) | 97 | 2.6 | ||||
| NMV_2146 | 74 | 3.1 | COG4300 | P | ||
| NMV_1220 (bfrB) | 71 | 1.4 |
aTop-10 Hfq-binding candidates with their read-counts and log2 f.c. are listed for each class of RNA.
To further investigate the predicted interaction with Hfq, we determined changes in sRNA half-life in rifampicin stability assays for nine sRNAs (Figure 6), with AniS sRNA serving as a control for Hfq-dependent transcript stabilization (20). A decrease in stability was observed for four sRNAs (RcoF1, RcoF2, NMnc0044, NMnc0034). For example, the half-life of NMnc0044 declined from >40 min in the WT strain to ∼2 min in the Δhfq strain (Supplementary Figure S5B). Overall, we found a significant decrease in sRNA stability for all Hfq-bound sRNAs tested in the Δhfq strain (Wilcoxon rank sum test, p = 0.014; Supplementary Figure S5A).
In addition to sRNAs, we identified 401 Hfq-bound mRNAs (Figure 5E and Supplementary Dataset S6). Of the 47 proteins known to be expressed in an Hfq-dependent manner (as determined by protein levels on two-dimensional gels; (26,27)), we found a corresponding enrichment of seven mRNAs (acnB, glyA, godB, ibpB, prpB, argG and NMV_1914) in our Hfq RIP-seq data.
Our comprehensive TSS map offered the possibility of analyzing whether our newly annotated 5΄ and 3΄ UTRs were targeted by Hfq and to categorize Hfq-mRNA binding sites. The majority of the cDNAs of Hfq-bound mRNAs originated from the CDS (70%, 320 CDS) and the rest from UTRs (30%, 80 5΄ and 62 3΄ UTRs). These Hfq-bound mRNAs represent good candidates for being post-transcriptionally regulated by sRNAs, especially those where Hfq binds in the 5΄ UTR or early CDS where most sRNAs regulate mRNA translation and decay (67,76).
Identification of a colonization factor as target of two new sRNAs
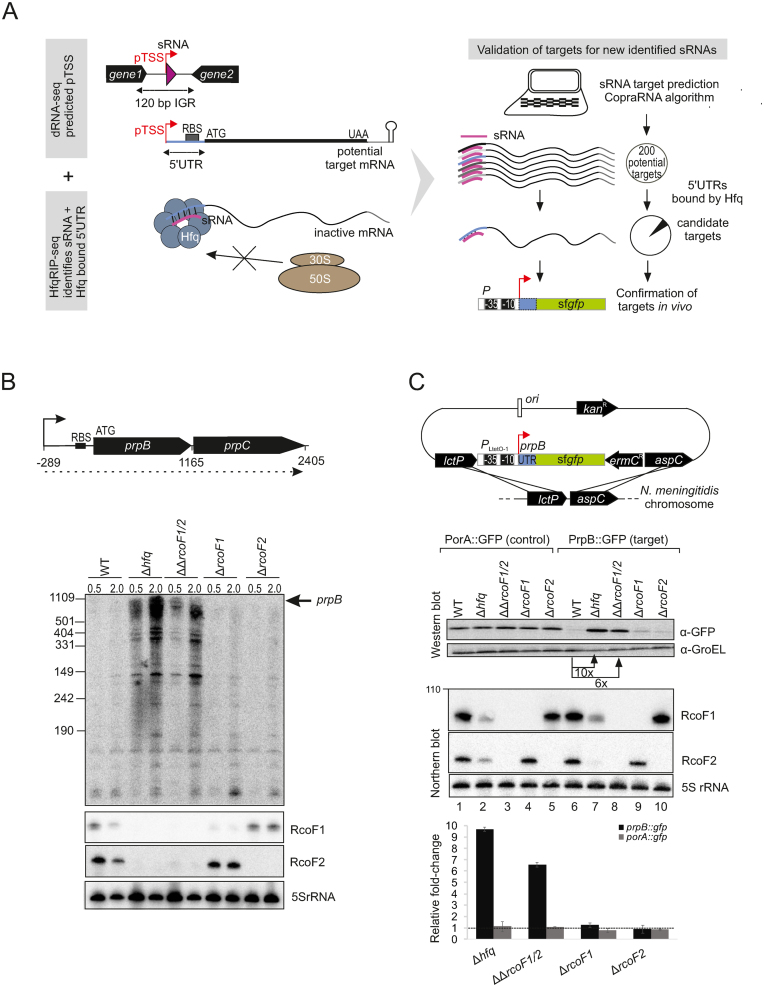
In order to predict mRNA regulated by sRNAs, we applied the CopraRNA algorithm (77) to the ten sRNAs with strongest enrichment in the Hfq RIP-seq data (Table 2). The predicted targets were further filtered for enrichment according to the Hfq RIP-seq data to reduce false-positive predictions (Figure 7A and Supplementary Table S6). For example, of 200 targets predicted for the highly abundant sRNAs RcoF1 and RcoF2, only 10 were clearly enriched with Hfq. Of the 10 mRNA targets, four were predicted to be bound by both of these paralogous sRNAs. One of the mRNAs predicted to interact with both RcoF1 and RcoF2 was prpB, encoding a methyl-citrate lyase, as part of the dicistronic prpB-prpC mRNA. Northern blot analysis revealed that both the single prpB and the dicistronic prpB-prpC mRNAs were upregulated upon deletion of hfq (23) or a double deletion of rcoF1 and rcoF2. This finding suggested a genetic link between RcoF1/2, Hfq and prpB expression (Figure 7B and Supplementary Figure S7).
Figure 7.
In vivo validation of prpB regulation by the two sRNAs RcoF1 and RcoF2. (A) (Left) Schematic representation for combining data gained from dRNA-seq and Hfq RIP-seq analysis to uncover and validate targets for newly identified sRNAs. dRNA-seq enriches for primary transcriptional start sides (pTSS) as indicated by red arrows and therewith uncovers sRNAs (pink triangles) located in IGR and other transcriptome features like 5΄ UTRs of mRNAs colored in blue. Hfq RIP-seq allows for the identification of sRNAs and 5΄ UTRs as targets of Hfq. 5΄ UTRs are target sites for Hfq-dependent sRNA regulation that leads to inhibition of ribosome binding, resulting in reduced translation. (Right) Schematic workflow for target prediction by the copra algorithm and validation via a superfolder gfp (sfgfp) reporter gene fusion, exemplified by the two newly identified sRNAs RcoF1 and RcoF2 and their target prpB. Hfq-bound 5΄ UTRs of mRNAs identified by Hfq-RIPseq (colored in blue) serve to further screen potential mRNA targets predicted for new sRNAs identified by dRNA-seq (colored in pink). (B) (top) The 1165-nt-long prpB transcript is part of the dicistronic mRNA prpB-prpC (dotted line) encoding a methylcitrate lyase and methylcitrate synthase, respectively. The TSS is indicated by arrows and the ribosome binding site (RBS) by a black bar. Numbers indicate the distance to the TSS. (bottom) The expression of prpB was analyzed during two growth phases in WT, Δhfq, ΔΔrcof1/2 double deletion mutants as well as Δrcof1 and Δrcof2 single deletion mutants at indicated optical densities (OD600) using a 32P-UTP labeled riboprobe. RcoF1 and RcoF2 were detected with JVO13282 and JVO13283. 5S rRNA served as loading control. (C) (top) Schematic illustration of the integrational vector pGCC2 for construction of translational superfolder gfp (sfgfp) fusions that were inserted into the lctP and aspC locus of N. meningitidis 8013 (127). The prpB 5΄ UTR and the first 15 amino acids of the prpB coding region (gray box) were fused to sfgfp reporter gene (green box) that is transcribed from the constitutive PLtetO-1 promoter as indicated. Individual elements are not drawn to scale. A sfgfp reporter fusion expressing the 5΄ UTR and the first 15 amino acids of the porA gene served as control. (bottom) N. meningitidis 8013 WT, Δhfq, ΔΔrcof1/2 double deletion mutants as well as Δrcof1 and Δrcof2 single deletion mutants expressing either the target-gfp or the control-gfp fusions were grown to mid logarithmic phase and RNA and protein samples were analyzed by northern blot and western blot. RcoF1 was detected with JVO-13282 and RcoF2 was detected with JVO-13283. 5S rRNA served as loading control. Whole cell protein fractions (OD600 = 0.01 for western blot) were detected with mouse anti-Gfp antiserum. GroEL served as control. Relative fold expression changes of prpB::gfp and porA::gfp (control) fusions in Δhfq, of both sRNAs RcoF1 and RcoF2 or of each sRNA alone determined by western blot analysis for Gfp in comparison with the respective WT backgrounds are represented in the bar plot. Error bars indicate the SDs among three biological replicates.
Validation of sRNA–mRNA interactions in vivo in Salmonella and E. coli has much benefitted from the availability of a plasmid-based green fluorescent protein (GFP) reporter system (34,78). To study the putative sRNA-mediated regulation of the prpB mRNA in vivo, we adapted this approach by integrating superfolder GFP (sfgfp) (78) in the chromosome of N. meningitidis (Figure 7A). To specifically measure post-transcriptional control, we fused a constitutive PLtetO-1 promoter to the 5΄ UTR and the first 15 codons of prpB, followed by the sfgfp reading frame. This construct was delivered on the pGCC2 vector for homologous recombination into the meningococcal lctP/aspC locus together with an erythromycin resistance gene (Figure 7C). A similar fusion of the porA gene was constructed as a control.
Western blot analysis of the PrpB::GFP fusion protein in N. meningitidis showed ∼6- and 10-fold upregulation of PrpB-GFP expression in strains deleted for either the RcoF1/2 sRNAs or hfq (Figure 7C, lanes 7 and 8). In contrast, PrpB::GFP levels in the two single deletion strains of RcoF1 or RcoF2 matched WT levels, indicating that these two sRNAs act redundantly to repress PrpB expression (Figure 7C, lanes 9 and 10). Importantly, the porA::sfgfp control fusion was not affected by either sRNAs or by Hfq (Figure 7C, lanes 2–5), ruling out non-specific effects on GFP expression itself. Together, these in vivo results provide strong evidence that these two sRNAs target the 5΄ UTR of prpB to post-transcriptionally repress this gene in an Hfq-dependent manner.
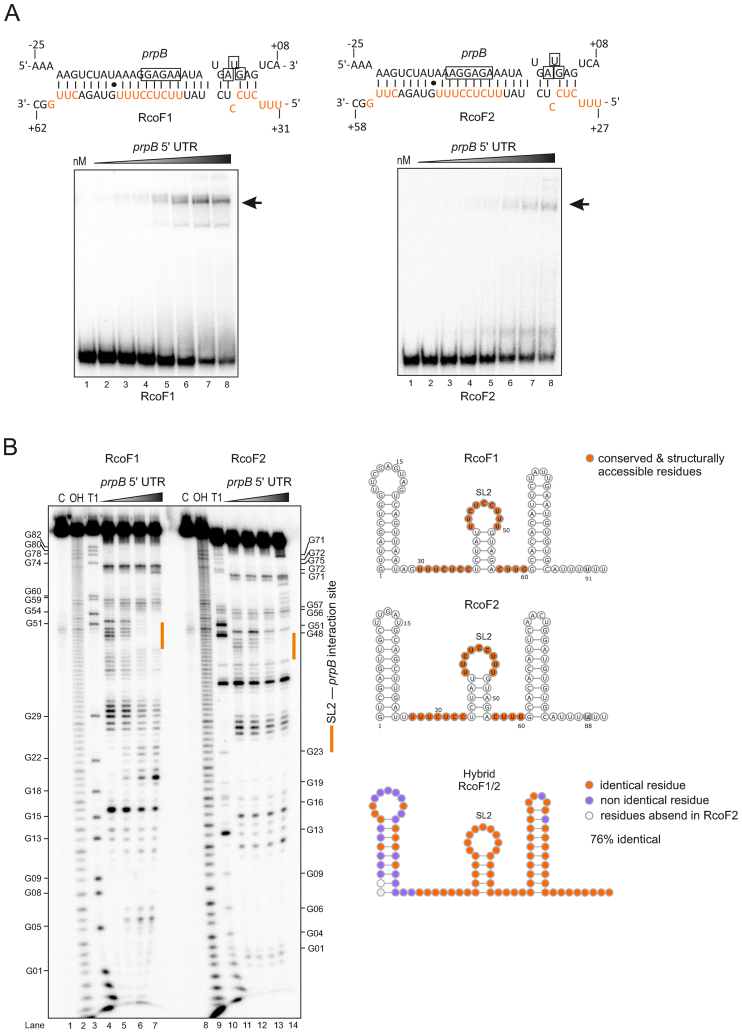
To directly monitor the interaction between sRNA and target, we performed in vitro gel-shift assays with radiolabeled RcoF1 or RcoF2 and increasing amounts of prpB mRNA leader. Each of the sRNAs shifted in a concentration-dependent manner, suggesting the formation of an sRNA–mRNA duplex (Figure 8A). Footprinting assays with radiolabeled sRNAs in the absence or presence of the prpB leader further validated a direct interaction and mapped the interaction sites at single-nucleotide resolution (Figure 8B). That is, probing of the RcoF1 and RcoF2 resulted in a clear footprint in its second stem loop region (Figure 8B). Combined with in silico analysis using the RNAhybrid algorithm (79), we predict that RcoF1 and RcoF2 are each able to sequester the Shine-Dalgarno sequence and start codon of the prpB mRNA, resulting in inhibition of translation initiation. Taken together, our combination of a global profiling approach (RIP-seq of Hfq), chromosomal sRNA deletion, the development of a new reporter system and in vitro RNA structure probing identifies the first example of sRNA-mediated control of a colonization-associated mRNA in N. meningitidis.
Figure 8.
In vitro gel-shift and structure probing assay show a direct interaction between the two sRNAs RcoF1/2 and the prpB leader. (A) (top) Predicted base-pairing interactions of RcoF1 and RcoF2 with their respective target mRNA prpB using RNA hybrid (79). The Shine-Dalgarno sequences and start codons of the mRNAs are boxed. Conserved and structurally accessible residues are shown in red. The proposed strength of interaction for each RNA duplex is −36.6 kcal/mol. (bottom) Gels-shift experiment with in vitro synthesized sRNAs (RcoF1/2) and prpB mRNA leader (−289 to +42 relative to the annotated start codon). About 0.04 pmol of each 32P-labeled sRNA was incubated with increasing concentrations of unlabeled prpB mRNA leader (lane 1–8: 0, 8, 16, 62.5, 125, 250, 500, 1000 nM). Arrows indicate the RNA-RNA duplex (B) (Left) In-line probing of 0.04 pmol of each 32P-labeled sRNA in the absence (lane 4, lane 11) or presence (lane 5–7, lane 12–14) of prpB mRNA leader. Untreated RNA (lane C), partially alkali (lane OH) or RNase T1 (lane T1) digested RcoF1 and RcoF2 served as ladders. (Right) Predicted secondary structures of sRNAs RcoF1 and RcoF2 using RNAfold. For both sRNAs nucleotides located within three single stranded regions (marked in orange) were predicted to base-pair with the 5΄ UTR of a dicistronic prpB-prpC mRNA. Identical residues (marked in orange) and non identical residues (marked in purple) of both sRNAs are represented by the hybrid sRNA structure. The designation SL2 represents loop 2 of the second Stem-loop structure.
DISCUSSION
Whole transcriptome RNA sequencing analyses have provided complete maps of transcriptional landscapes with coordinates for exact boundaries of transcripts and operons, as well as their relative abundances. However, conventional RNA-seq transcriptome maps provide little information as to whether transcripts are primary or processing products, and which of them are subject to posttranscriptional control. In this regard, the primary transcriptome and Hfq binding data presented here provide an unprecedented knowledge base for the human commensal pathogen N. meningitidis. Mapping a total of 1625 TSS by dRNA-seq, we identify 706 pTSS of annotated genes, many co-transcribed in polycistronic mRNAs, as well as novel TSS for non-coding RNAs and antisense transcripts (Figure 1B). We identified three different types of promoter sequences specific for σ70 (RpoD) and σE (RpoE) which are two out of three functionally active sigma factors in N. meningitidis. Combined with the results from a previous dRNA-seq analysis of N. gonorrhoeae (62), the absence of a canonical –35 box and abundance of extended −10 box promoters appears to be a common feature across the Neisseria species.
New types of neisseria sRNAs
A key finding from our analysis of the TSS mapping is the detection of 65 sRNAs, including 45 novel candidate sRNAs to this study. Inspection of their genomic location revealed four distinct classes of non-coding RNAs: intergenic, 5΄ UTR-derived, 3΄ UTR-derived and cis-encoded antisense RNAs. Intriguingly, more than one third of these sRNAs are generated from the 3΄ ends of mRNAs, with examples from both of the two general biogenesis pathways of such sRNAs, i.e. expression from ORF-internal promoters or processing of the parental mRNA (69). Following the recent discovery of many new members of this emerging class of sRNAs in E. coli, Salmonella and Streptomyces species (32,80–83) our results with N. menigitidis now reveal mRNA genes to be an important source of sRNAs in β-proteobacteria, too. Of note, sRNAs from mRNA 3΄ regions often provide missing links in important physiological circuits. For example, the characterization of MicL sRNA explained a previously observed σE-dependent repression of E. coli's major outer membrane lipoprotein (80) while CpxQ sRNA was found to be the previously unknown non-coding arm of the Cpx response to envelope stress (81). In this regard, it is tempting to speculate that some of the 3΄ derived sRNAs will help answer open questions in Neisseria biology.
One such promising candidate is NMnc0001, which we have shown to be an Hfq-dependent 3΄ processing fragment from the phase-variable pilE mRNA encoding the major component of the type IV pilus (Supplementary Figure S4A and B) (17). The type IV pili of Neisseria are long filamentous surface structures involved in DNA transformation, twitching motility and adherence to epithelial cells and therefore are key fitness determinants for residency within humans (84). Thus, type IV pili expression may respond to environmental changes and stress signals. In N. meningitidis, a number of proteins like CrgA and NafA participate in pilin regulation upon bacterial contact with host cells (85,86). Furthermore, pilE expression has been shown to be Hfq-dependent in a strain-specific manner: in strain H44/76, loss of Hfq leads to a reduction in PilE levels (27), while in MC58 absence of Hfq results in PilE upregulation (26). Because expression levels of NMnc0001 are synchronized with pilE expression levels, it could provide a regulatory arm to pilus expression, enabling crosstalk with physically unlinked genes in response to environmental changes.
Of the 45 new sRNAs that we have identified, 10 sRNAs were strain specific, in line with patterns seen across the bacterial phylogeny (87). One non-conserved sRNA, NMnc0040, is expressed from the CRISPR-Cas9 locus of N. menigitidis which we previously showed to function as a barrier to natural transformation (Figure 4) (7). Interestingly, a study in Francisella novicida showed that Cas9, without the help of another protein from the Francisella CRISPR-Cas type II locus, represses the synthesis of a bacterial lipoprotein and thereby enables the infecting bacteria to evade the Toll-like receptor 2-based innate immune response of its host (88). To do so, Francisella Cas9 requires both tracrRNA and another Cas9-dependent CRISPR-associated sRNA, scaRNA. Further studies will be required to understand whether NMnc0040 constitutes a functional analog of scaRNA in N. meningitidis, facilitating Cas9 mediated regulation of endogenous genes. Another strain-specific sRNA, NMnc0031, is located on the genomic island IHT-E of likely prophage origin (Supplementary Figure S3 A and B). We recently demonstrated that IHT-E is associated with hyperinvasive meningococcal strains (6). Therefore, knowledge of the functional elements of this island promise a better understanding of host-pathogen interactions of N. meningitidis. Taken together, the analysis of the TSS mapping presented here identified a gene architecture that reflects a structural complexity of the neisserial transcriptome similar to other lineages, such as γ-proteobacteria and ε-proteobacteria (36,89,90) and identified different classes of sRNAs with regulatory potential.
An unexpectedly large Hfq-RNA interactome
Comprehensive analysis of our RIP-seq data identified 23 sRNAs and 401 mRNAs as candidate targets of Hfq, constituting the first direct evidence for the existence of a global post-transcriptional network in N. meningitidis. Many of these transcripts are involved in cell energetics and metabolism, amino acid biosynthesis, oxidative stress responses and pathogenesis (Supplementary Dataset S5). Importantly, the Hfq-bound mRNAs include many transcripts (e.g. acnB, glyA, sodB, lbpB, prpB, argG and NMV_1914) whose protein expression differ between WT and Δhfq strains of N. meningitidis (26,27). Moreover, the evidence for Hfq binding to sRNAs and UTRs of mRNAs presented here bolsters previous predictions that Hfq is involved in both sRNA-dependent regulation at mRNA 5΄ regions and 3΄-end dependent processes in Neisseria (19). Most importantly, we provide a framework for elucidating specific sRNA functions in the meningococcal Hfq network. Comparing the 5΄ UTRs of Hfq-bound mRNAs with targets predicted for our top Hfq binding sRNAs using the CopraRNA algorithm, we identified 26 high-confidence mRNA targets for nine neisserial sRNAs (Supplementary Table S6 and Supplementary Dataset S5). Of the 26 different mRNA targets, six are predicted to be targeted by multiple sRNAs, suggesting overlapping modes of Hfq-mediated sRNA regulation in meningococci. A quite striking example is NMV_1044, encoding a putative MarC-family transporter, which may be regulated by six different sRNAs (RcoF1, RcoF2, NMnc0044, NMnc0037, Bns2, AniS). This suggests that N. meningitidis possesses post-transcriptional hubs similar to E. coli or Salmonella, in which the csgD, rpoS and ompD mRNAs are each controlled by between four and six different sRNAs (91–94).
Translational repression of prpB mRNA by two paralogous sRNAs and Hfq
Hfq-dependent sRNAs employ a range of mechanisms to negatively or positively regulate translation and stability of mRNAs (95). The cumulative results of extensive work in the γ-proteobacteria E. coli, Salmonella and Vibrio over the past decade (95–98) have suggested translational repression of mRNAs by sRNA-mediated sequestration of the RBS of the target mRNA to be the most common mechanism. Our study provides the first molecular characterization of Hfq-mediated translational repression in Neisseria, suggesting that this mechanism also operates in β-proteobacteria. Through genetic and in vitro RNA analyzes we predict that the paralogous RcoF1 and RcoF2 sRNAs each form an almost perfect RNA duplex of ∼24 bp in length with the 5΄ end of the prpB mRNA (Figure 8A). Not only do these duplexes sequester both the RBS and start codon of this target, a calculated change in free energy of −36.6 kcal/mol suggests that RcoF1/2 fully prevent association of prpB mRNA with 30S ribosomes (for background, see (99,100)). Our newly developed translational reporter system offers the opportunity to study the molecular determinants for translational control by RcoF1/2 in vivo.
The stabilization of the prpB mRNA in N. meningitidis mutants strains lacking either Hfq or the two sRNAs likely results from ribosome-mediated protection against a nuclease that turns over this messenger. This nuclease is currently unknown, but RNase E, which degrades the bulk of mRNA (101) and functions in Hfq-mediated regulations in other Gram-negative bacteria (102–104), is a likely candidate. In E. coli, sRNAs are known to guide RNase E to destroy mRNA targets in complex with Hfq (76,105). N. meningitidis encodes an RNase E homolog (NMV_0215) which has been shown to be essential by in vitro transposon mutagenesis (41). However, nothing is known so far about what role RNase E may play in RNA turnover in Neisseria.
Regarding their molecular architecture, RcoF1/2 deviate from most characterized Hfq-dependent sRNAs in that their mRNA base pairing regions are not fully unstructured but form a short stem-loop (nucleotides 37–55). The presentation of the anti-Shine Dalgarno sequence by a pyrimidine-rich loop is reminiscent of how CyaR sRNA recognizes the ompX mRNA in E. coli and Salmonella (106–108). Moreover, target recognition via pyrimidine-rich loops is a hallmark of regulatory sRNAs in Staphylococcus aureus which generally work without Hfq (109). Similar to these latter sRNAs, RcoF1/2 likely undergo a structural rearrangement as they recognize the prpB mRNA, as best visible in the structure probing experiment with RcoF1/2 (Figure 8B). Why these sRNAs are present in tandem is currently unclear. Similar sRNAs pairs exist in other organisms (110,111) and it has been suggested that this arrangement provides a fail-safe mechanism for stress responses (93).
Furthermore, given the significant difference in the expression of prpB between the Δhfq and ΔΔrcoF1/2 mutant strains (Figure 7C) our data may indicate the existence of another sRNA that represses prpB. Such candidate could be the σE sRNA (24) that was also predicted to interact with the prpB mRNA (Supplementary Table S6). Of note, E. coli and Salmonella possess numerous mRNAs, e.g. rpoS or csgD, that are regulated by up to six different sRNAs (93,112).
By regulating the putative colonization factor PrpB, RcoF1/2 may constitute the first trans-acting sRNAs with a direct host-associated function in N. meningitidis. The prpB gene is part of the methylcitrate cycle gene cluster located on a large genomic island in N. meningitidis, absent from closely related non-pathogenic Neisseria species such as N. lactamica. The methylcitrate cycle gene cluster allows meningococci to convert propionic acid to pyruvate, supporting growth and limiting the toxicity of propionic acid. The ability to utilize propionic acids is hypothesized to confer an advantage in colonizing the adult nasopharynx, which is rich in propionic acid generating bacteria (113). Of note, the ability of N. meningitidis to effectively colonize adults plays an important role in its transmission and disease epidemiology (114). Together, our data provide evidence for an important role of these two sRNAs in this species’ ecological niche in the host.
Outlook
Large-scale datasets are of increasing importance in biology, providing the raw material for integrative analyzes that can guide functional and molecular studies (29). Here, we have presented a transcriptomic compendium for N. meningitidis, including both transcription start sites and transcript interactions with Hfq. As this manuscript was in preparation, a study was published using transposon insertion sequencing to identify genes required for host cell colonization by a N. meningitidis serogroup A strain in an in vitro system (115). In further support of a central role of sRNAs in meningococcal infection biology (18), this study found 33 IGR containing sRNA candidates required for colonization of host cells. Integrating these and further studies should provide a solid foundation for the ongoing investigation of N. meningitidis infection biology, and particularly the role of post-transcriptional regulation in it.
One of the major challenges of studying N. meningitidis pathogenicity is the lack of defined virulence factors separating invasive and non-invasive isolates. This suggests a multifactorial pathogenicity, likely involving regulatory changes in the bacterium as well as variable host factors (116,117). New technologies, such as dual RNA-seq for simultaneous transcriptomics of pathogen and host (118,119), are beginning to provide direct insight into these interactions. Our study lays the groundwork for such future approaches, providing the transcriptional annotations needed to interpret the results of dual RNA-seq. One particularly exciting possibility is screening diverse N. meningitidis isolates with dual RNA-seq to define ‘molecular phenotypes’ (120)for these differing genotypes during association with host cells.
Finally, although we discovered a large Hfq network comprising more than 20 sRNAs and over 400 potential target mRNAs, it is clear that not all N. meningitidis sRNAs are bound by Hfq. Recent work using gradient profiling by sequencing (Grad-seq) has identified a new global RNA-binding protein in Salmonella, ProQ (121). An orthologous protein exists in meningococci (122), and may play a similar role. Regardless, it seems likely that additional RNA chaperones await discovery in Neisseria. Using Grad-seq to characterize the RNA–protein interactome of N. meningitidis, in combination with the RIP-seq technique presented here, is likely to uncover new biology, moving us toward a more complete understanding of post-transcriptional regulation in this important human pathogen.
ACCESSION NUMBER
RNA-Seq data are available at the NCBI GEO database, accession number GSE85252.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Erik Holmqvist for fruitful discussions and critical comments on the manuscript. We thank Biju Joseph Ampattu for help with the tracrRNA knockout plasmid and Gaurav Dugar for advice on experiments for deletion of sRNAs in Neisseria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [875/7-1, 875/7-2 to J.V.]; Alexander von Humboldt Foundation Research Fellowship (in part). The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Henriques-Normark B., Normark S.. Commensal pathogens, with a focus on Streptococcus pneumoniae, and interactions with the human host. Exp. Cell Res. 2010; 316:1408–1414. [DOI] [PubMed] [Google Scholar]
- 2. Stephens D.S., Greenwood B., Brandtzaeg P.. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007; 369:2196–2210. [DOI] [PubMed] [Google Scholar]
- 3. Caugant D.A., Maiden M.C.J.. Meningococcal carriage and disease-population biology and evolution. Vaccine. 2009; 27:B64–B70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greenwood B. Manson lecture. Meningococcal meningitis in africa. Trans. R. Soc. Trop. Med. Hyg. 1999; 93:341–353. [DOI] [PubMed] [Google Scholar]
- 5. Fredricks D.N., Relman D.a.. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin. Microbiol. Rev. 1996; 9:18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Joseph B., Schwarz R.F., Linke B., Blom J., Becker A., Claus H., Goesmann A., Frosch M., Müller T., Vogel U. et al. . Virulence evolution of the human pathogen Neisseria meningitidis by recombination in the core and accessory genome. PLoS One. 2011; 6:e18441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y., Heidrich N., Ampattu B.J., Gunderson C.W., Seifert H.S., Schoen C., Vogel J., Sontheimer E.J.. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol. Cell. 2013; 50:488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bille E., Zahar J.-R., Perrin A., Morelle S., Kriz P., Jolley K.a, Maiden M.C.J., Dervin C., Nassif X., Tinsley C.R.. A chromosomally integrated bacteriophage in invasive meningococci. J. Exp. Med. 2005; 201:1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bille E., Ure R., Gray S.J., Kaczmarski E.B., McCarthy N.D., Nassif X., Maiden M.C.J., Tinsley C.R.. Association of a bacteriophage with meningococcal disease in young adults. PLoS One. 2008; 3:e3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schoen C., Blom J., Claus H., Schramm-Glück A., Brandt P., Müller T., Goesmann A., Joseph B., Konietzny S., Kurzai O. et al. . Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3473–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schoen C., Tettelin H., Parkhill J., Frosch M.. Genome flexibility in Neisseria meningitidis. Vaccine. 2009; 27(Suppl. 2):B103–B111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Snyder L.A.S., Saunders N.J.. The majority of genes in the pathogenic Neisseria species are present in non-pathogenic Neisseria lactamica, including those designated as ‘virulence genes’. BMC Genomics. 2006; 7:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caldelari I., Chao Y., Romby P., Vogel J.. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb. Perspect. Med. 2013; 3:a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gripenland J., Netterling S., Loh E., Tiensuu T., Toledo-Arana A., Johansson J.. RNAs: regulators of bacterial virulence. Nat. Rev. Microbiol. 2010; 8:857–866. [DOI] [PubMed] [Google Scholar]
- 15. Loh E., Kugelberg E., Tracy A., Zhang Q., Gollan B., Ewles H., Chalmers R., Pelicic V., Tang C.M.. Temperature triggers immune evasion by Neisseria meningitidis. Nature. 2013; 502:237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cahoon L.a, Seifert H.S.. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009; 325:764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan F.Y.Y., Wörmann M.E., Loh E., Tang C.M., Exley R.M.. Characterization of a novel antisense RNA in the major pilin locus of Neisseria meningitidis influencing antigenic variation. J. Bacteriol. 2015; 197:1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fagnocchi L., Bottini S., Golfieri G., Fantappiè L., Ferlicca F., Antunes A., Guadagnuolo S., Del Tordello E., Siena E., Serruto D. et al. . Global transcriptome analysis reveals small RNAs affecting Neisseria meningitidis bacteremia. PLoS One. 2015; 10:e0126325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Del Tordello E., Bottini S., Muzzi A., Serruto D.. Analysis of the regulated transcriptome of Neisseria meningitidis in human blood using a tiling array. J. Bacteriol. 2012; 194:6217–6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fantappiè L., Oriente F., Muzzi A., Serruto D., Scarlato V., Delany I.. A novel Hfq-dependent sRNA that is under FNR control and is synthesized in oxygen limitation in Neisseria meningitidis. Mol. Microbiol. 2011; 80:507–523. [DOI] [PubMed] [Google Scholar]
- 21. Metruccio M.M.E., Pigozzi E., Roncarati D., Berlanda Scorza F., Norais N., Hill S.a, Scarlato V., Delany I.. A novel phase variation mechanism in the meningococcus driven by a ligand-responsive repressor and differential spacing of distal promoter elements. PLoS Pathog. 2009; 5:e1000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mellin J.R., Goswami S., Grogan S., Tjaden B., Genco C.A.. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 2007; 189:3686–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mellin J.R., McClure R., Lopez D., Green O., Reinhard B., Genco C.. Role of Hfq in iron-dependent and -independent gene regulation in Neisseria meningitidis. Microbiology. 2010; 156:2316–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huis in ’t Veld R.A.G., Willemsen A.M., van Kampen A.H.C., Bradley E.J., Baas F., Pannekoek Y., van der Ende A.. Deep sequencing whole transcriptome exploration of the σE regulon in Neisseria meningitidis. PLoS One. 2011; 6:e29002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chao Y., Vogel J.. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010; 13:24–33. [DOI] [PubMed] [Google Scholar]
- 26. Fantappiè L., Metruccio M.M.E., Seib K.L., Oriente F., Cartocci E., Ferlicca F., Giuliani M.M., Scarlato V., Delany I.. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect. Immun. 2009; 77:1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pannekoek Y., Huis in ’t Veld R., Hopman C.T.P., Langerak A.a.J., Speijer D., van der Ende A.. Molecular characterization and identification of proteins regulated by Hfq in Neisseria meningitidis. FEMS Microbiol. Lett. 2009; 294:216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sittka A., Sharma C.M., Rolle K., Vogel J.. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 2009; 6:266–275. [DOI] [PubMed] [Google Scholar]
- 29. Barquist L., Vogel J.. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu. Rev. Genet. 2015; 49:367–394. [DOI] [PubMed] [Google Scholar]
- 30. Sharma C.M., Vogel J.. Differential RNA-seq: the approach behind and the biological insight gained. Curr. Opin. Microbiol. 2014; 19C:97–105. [DOI] [PubMed] [Google Scholar]
- 31. Sharma C.M., Hoffmann S., Darfeuille F., Reignier J., Findeiss S., Sittka A., Chabas S., Reiche K., Hackermüller J., Reinhardt R. et al. . The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010; 464:250–255. [DOI] [PubMed] [Google Scholar]
- 32. Chao Y., Papenfort K., Reinhardt R., Sharma C.M., Vogel J.. An atlas of Hfq-bound transcripts reveals 3΄ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012; 31:4005–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blomberg P., Wagner, E.G.H. and Nordström, K. Control of replication of plasmid Rl: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990; 9:2331–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Urban J.H., Vogel J.. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007; 35:1018–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edgar R., Domrachev M., Lash A.E.. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dugar G., Herbig A., Förstner K.U., Heidrich N., Reinhardt R., Nieselt K., Sharma C.M.. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 2013; 9:e1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bailey T.L., Boden M., Buske F.a., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Holmqvist E., Wright P.R., Li L., Bischler T., Barquist L., Reinhardt R., Backofen R., Vogel J.. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016; 35:991–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gardner P.P., Barquist L., Bateman A., Nawrocki E.P., Weinberg Z.. RNIE: genome-wide prediction of bacterial intrinsic terminators. Nucleic Acids Res. 2011; 39:5845–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rusniok C., Vallenet D., Floquet S., Ewles H., Mouzé-Soulama C., Brown D., Lajus A., Buchrieser C., Médigue C., Glaser P. et al. . NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol. 2009; 10:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Delany I., Ieva R., Alaimo C., Rappuoli R., Scarlato V.. The iron-responsive regulator Fur is transcriptionally autoregulated and not essential in Neisseria meningitidis. J. Bacteriol. 2003; 185:6032–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guckenberger M., Kurz S., Aepinus C., Theiss S., Haller S., Leimbach T., Panzner U., Weber J., Paul H., Unkmeir A. et al. . Analysis of the heat shock response of Neisseria meningitidis with cDNA- and oligonucleotide-based DNA microarrays. J. Bacteriol. 2002; 184:2546–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mao X., Ma Q., Zhou C., Chen X., Zhang H., Yang J., Mao F., Lai W., Xu Y.. DOOR 2.0: presenting operons and their functions through dynamic and integrated views. Nucleic Acids Res. 2014; 42:D654–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burton Z.F., Gross C.A., Watanabe K.K., Burgess R.R.. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983; 32:335–349. [DOI] [PubMed] [Google Scholar]
- 46. Gordon D.L. Meningococcal disease and complement. Med. J. Aust. 1989; 150:286. [PubMed] [Google Scholar]
- 47. Echenique-Rivera H., Muzzi A., Del Tordello E., Seib K.L., Francois P., Rappuoli R., Pizza M., Serruto D.. Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathog. 2011; 7:e1002027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frosch M., Maiden M.C.J.. Frosch M, Maiden MCJ. Structure and genetics of the meningococcal capsule. Handbook of Meningococcal Disease. 2006; Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 145–162. [Google Scholar]
- 49. Coureuil M., Bourdoulous S., Marullo S., Nassif X.. Invasive meningococcal disease: a disease of the endothelial cells. Trends Mol. Med. 2014; 20:571–578. [DOI] [PubMed] [Google Scholar]
- 50. Trivedi K., Tang C.M., Exley R.M.. Mechanisms of meningococcal colonisation. Trends Microbiol. 2011; 19:456–463. [DOI] [PubMed] [Google Scholar]
- 51. Hammerschmidt S., Birkholz C., Zähringer U., Robertson B.D., Van Putten J., Ebeling O., Frosch M.. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol. Microbiol. 1994; 11:885–896. [DOI] [PubMed] [Google Scholar]
- 52. Harrison O.B., Claus H., Jiang Y., Bennett J.S., Bratcher H.B., Jolley K.A., Corton C., Care R., Poolman J.T., Zollinger W.D. et al. . Description and nomenclature of Neisseria meningitidis capsule locus. Emerg. Infect. Dis. 2013; 19:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tzeng Y., Datta A.K., Strole C.A., Michael A., Carlson R.W., Stephens D.S., Lobritz M.A.. Translocation and surface expression of lipidated serogroup b capsular polysaccharide in Neisseria meningitidis translocation and surface expression of lipidated serogroup B capsular polysaccharide in Neisseria meningitidis. Infect. Immun. 2005; 73:1491–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tzeng Y.-L., Zhou X., Bao S., Zhao S., Noble C., Stephens D.S.. Autoregulation of the MisR/MisS two-component signal transduction system in Neisseria meningitidis. J. Bacteriol. 2006; 188:5055–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Galperin M.Y. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005; 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Davidsen T., Tønjum T.. Meningococcal genome dynamics. Nat. Rev. Microbiol. 2006; 4:11–22. [DOI] [PubMed] [Google Scholar]
- 57. Ortet P., Whitworth D.E., Santaella C., Achouak W., Barakat M.. P2CS: updates of the prokaryotic two-component systems database. Nucleic Acids Res. 2015; 43:D536–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pérez-Rueda E., Collado-Vides J.. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000; 28:1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hook-Barnard I.G., Hinton D.M.. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene Regul. Syst. Bio. 2007; 1:275–293. [PMC free article] [PubMed] [Google Scholar]
- 60. Keilty S., Rosenberg M.. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem. 1987; 262:6389–6395. [PubMed] [Google Scholar]
- 61. Petersen L., Larsen T.S., Ussery D.W., On S.L.W., Krogh A.. Rpo D promoters in Campylobacter jejuni exhibit a strong periodic signal instead of a - 35 box. J. Mol. Biol. 2003; 326:1361–1372. [DOI] [PubMed] [Google Scholar]
- 62. Remmele C.W., Xian Y., Albrecht M., Faulstich M., Fraunholz M., Heinrichs E., Dittrich M.T., Müller T., Reinhardt R., Rudel T.. Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nucleic Acids Res. 2014; 42:10579–10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kumar A., Malloch R.A., Fujita N., Smillie D.A., Ishihama A., Hayward R.S.. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an ‘extended minus 10’ promoter. J. Mol. Biol. 1993; 232:406–418. [DOI] [PubMed] [Google Scholar]
- 64. Ponnambalam S., Chan B., Busby S.. Functional analysis of different sequence elements in the Escherichia coli galactose operon P 2 promoter. Mol. Microbiol. 1988; 2:165–172. [DOI] [PubMed] [Google Scholar]
- 65. Rhodius V.A., Suh W.C., Nonaka G., West J., Gross C.A.. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 2005; 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Feklístov A., Sharon B.D., Darst S.A., Gross C.A.. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014; 68:357–376. [DOI] [PubMed] [Google Scholar]
- 67. Fröhlich K.S., Haneke K., Papenfort K., Vogel J.. The target spectrum of SdsR small RNA in Salmonella. Nucleic Acids Res. 2016; 44:10406–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wade J.T., Grainger D.C.. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 2014; 12:647–653. [DOI] [PubMed] [Google Scholar]
- 69. Miyakoshi M., Chao Y., Vogel J.. Regulatory small RNAs from the 3΄ regions of bacterial mRNAs. Curr. Opin. Microbiol. 2015; 24:132–139. [DOI] [PubMed] [Google Scholar]
- 70. Nawrocki E.P., Burge S.W., Bateman A., Daub J., Eberhardt R.Y., Eddy S.R., Floden E.W., Gardner P.P., Jones T.A., Tate J. et al. . Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015; 43:D130–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.M., Ruzzo W.L., Breaker R.R.. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004; 306:275–279. [DOI] [PubMed] [Google Scholar]
- 72. Labrie S.J., Samson J.E., Moineau S.. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010; 8:317–327. [DOI] [PubMed] [Google Scholar]
- 73. Gordia S., Gutierrez C.. Growth-phase-dependent expression of the osmotically inducible gene osmC of Escherichia coli K-12. Mol. Microbiol. 1996; 19:729–736. [DOI] [PubMed] [Google Scholar]
- 74. Jolley K.A., Maiden M.C.J.. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010; 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hotopp J.C.D., Grifantini R., Kumar N., Tzeng Y.L., Fouts D., Frigimelica E., Draghi M., Giuliani M.M., Rappuoli R., Stephens D.S. et al. . Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology. 2006; 152:3733–3749. [DOI] [PubMed] [Google Scholar]
- 76. Massé E., Escorcia F.E., Gottesman S.. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003; 17:2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wright P.R., Georg J., Mann M., Sorescu D.A., Richter A.S., Lott S., Kleinkauf R., Hess W.R., Backofen R.. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014; 42:W119–W123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Corcoran C.P., Podkaminski D., Papenfort K., Urban J.H., Hinton J.C.D., Vogel J.. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 2012; 84:428–445. [DOI] [PubMed] [Google Scholar]
- 79. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R.. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Guo M.S., Updegrove T.B., Gogol E.B., Shabalina S.a, Gross C.a, Storz G.. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014; 28:1620–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chao Y., Vogel J.. A 3΄ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell. 2016; 61:352–363. [DOI] [PubMed] [Google Scholar]
- 82. Miyakoshi M., Chao Y., Vogel J.. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015; 34:1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kim H.M., Shin J.H., Cho Y.B., Roe J.H.. Inverse regulation of Fe-And Ni-containing SOD genes by a fur family regulator Nur through small RNA processed from 3΄UTR of the sodF mRNA. Nucleic Acids Res. 2014; 42:2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Obergfell K.P., Seifert H.S.. Mobile DNA in the pathogenic Neisseria. Microbiol. Spectr. 2015; 3:1–18. [DOI] [PubMed] [Google Scholar]
- 85. Kuwae A., Sjölinder H., Eriksson J., Eriksson S., Chen Y., Jonsson A.-B.. NafA negatively controls Neisseria meningitidis piliation. PLoS One. 2011; 6:e21749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Deghmane A.E., Giorgini D., Larribe M., Alonso J.M., Taha M.K.. Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol. Microbiol. 2002; 43:1555–1564. [DOI] [PubMed] [Google Scholar]
- 87. Lindgreen S., Umu S.U., Lai A.S.-W., Eldai H., Liu W., McGimpsey S., Wheeler N.E., Biggs P.J., Thomson N.R., Barquist L. et al. . Robust identification of noncoding RNA from transcriptomes requires phylogenetically-informed sampling. PLoS Comput. Biol. 2014; 10:e1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sampson T.R., Saroj S.D., Llewellyn A.C., Tzeng Y.-L., Weiss D.S.. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013; 497:254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Thomason M.K., Bischler T., Eisenbart S.K., Förstner K.U., Zhang A., Herbig A., Nieselt K., Sharma C.M., Storz G.. Global transcriptional start site mapping using dRNA-seq reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 2014; 197:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Papenfort K., Förstner K.U., Cong J.-P., Sharma C.M., Bassler B.L.. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E766–E775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Fröhlich K.S., Papenfort K., Berger A.a, Vogel J.. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 2012; 40:3623–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Serra D.O., Mika F., Richter A.M., Hengge R.. The green tea polyphenol EGCG inhibits E. coli biofilm formation by impairing amyloid curli fibre assembly and downregulating the biofilm regulator CsgD via the σE‐dependent sRNA RybB. Mol. Microbiol. 2016; 101:136–151. [DOI] [PubMed] [Google Scholar]
- 93. Holmqvist E., Reimegård J., Sterk M., Grantcharova N., Römling U., Wagner E.G.H.. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010; 29:1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Repoila F., Majdalani N., Gottesman S.. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 2003; 48:855–861. [DOI] [PubMed] [Google Scholar]
- 95. Wagner E.G.H., Romby P.. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015; 90:133–208. [DOI] [PubMed] [Google Scholar]
- 96. Feng L., Rutherford S.T., Papenfort K., Bagert J.D., Van Kessel J.C., Tirrell D.A., Wingreen N.S., Bassler B.L.. A Qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. 2015; 160:228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Melamed S., Peer A., Faigenbaum-Romm R., Gatt Y.E., Reiss N., Bar A., Altuvia Y., Argaman L., Margalit H.. Global mapping of small RNA-target interactions in bacteria. Mol. Cell. 2016; 63:884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jagodnik J., Brosse A., Le Lam T.N., Chiaruttini C., Guillier M.. Mechanistic study of base-pairing small regulatory RNAs in bacteria. Methods. 2016; doi:10.1016/j.ymeth.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 99. Papenfort K., Bouvier M., Mika F., Sharma C.M., Vogel J.. Evidence for an autonomous 5΄ target recognition domain in an Hfq-associated small RNA. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:20435–20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bouvier M., Sharma C.M., Mika F., Nierhaus K.H., Vogel J.. Small RNA binding to 5΄ mRNA coding region inhibits translational initiation. Mol. Cell. 2008; 32:827–837. [DOI] [PubMed] [Google Scholar]
- 101. Bandyra K.J., Luisi B.F.. Licensing and due process in the turnover of bacterial RNA. RNA Biol. 2013; 10:627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007; 10:134–139. [DOI] [PubMed] [Google Scholar]
- 103. Lalaouna D., Simoneau-Roy M., Lafontaine D., Massé E.. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim. Biophys. Acta. 2013; 1829:742–747. [DOI] [PubMed] [Google Scholar]
- 104. Chao Y., Li L., Girodat D., Förstner K.U., Said N., Corcoran C., Śmiga M., Papenfort K., Reinhardt R., Wieden H. et al. . In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell. 2017; 65:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Morita T., Maki K., Aiba H.. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005; 19:2176–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. De Lay N., Gottesman S.. The crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 2009; 191:461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Johansen J., Eriksen M., Kallipolitis B., Valentin-Hansen P.. Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP–CRP- and σE-dependent CyaR–ompX regulatory case. J. Mol. Biol. 2008; 383:1–9. [DOI] [PubMed] [Google Scholar]
- 108. Papenfort K., Pfeiffer V., Lucchini S., Sonawane A., Hinton J.C.D., Vogel J.. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 2008; 68:890–906. [DOI] [PubMed] [Google Scholar]
- 109. Bronesky D., Wu Z., Marzi S., Walter P., Geissmann T., Moreau K., Vandenesch F., Caldelari I., Romby P.. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu. Rev. Microbiol. 2016; 70:299–316. [DOI] [PubMed] [Google Scholar]
- 110. Wilderman P.J., Sowa N.A., FitzGerald D.J., FitzGerald P.C., Gottesman S., Ochsner U.A., Vasil M.L.. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9792–9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Guillier M., Gottesman S.. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006; 59:231–247. [DOI] [PubMed] [Google Scholar]
- 112. Storz G., Vogel J., Wassarman K.M.. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011; 43:880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Catenazzi M.C.E., Jones H., Wallace I., Clifton J., Chong J.P.J., Jackson M.A., Macdonald S., Edwards J., Moir J.W.B.. A large genomic island allows Neisseria meningitidis to utilize propionic acid, with implications for colonization of the human nasopharynx. Mol. Microbiol. 2014; 93:346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Christensen H., May M., Bowen L., Hickman M., Trotter C.L.. Meningococcal carriage by age: a systematic review and meta-analysis. Lancet Infect. Dis. 2010; 10:853–861. [DOI] [PubMed] [Google Scholar]
- 115. Capel E., Zomer A.L., Nussbaumer T., Bole C., Izac B., Frapy E., Meyer J., Bouzinba-Ségard H., Bille E., Jamet A. et al. . Comprehensive identification of Meningococcal genes and small noncoding RNAs required for host cell colonization. Mbio. 2016; 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pirofski L., Casadevall A.. Q and A: what is a pathogen? A question that begs the point. BMC Biol. 2012; 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Méthot P.-O., Alizon S.. What is a pathogen? toward a process view of host-parasite interactions. Virulence. 2014; 5:775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Westermann A.J., Barquist L., Vogel J.. Resolving host–pathogen interactions by dual RNA-seq. PLOS Pathog. 2017; 13:e1006033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Westermann A.J., Förstner K.U., Amman F., Barquist L., Chao Y., Schulte L.N., Müller L., Reinhardt R., Stadler P.F., Vogel J.. Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions. Nature. 2016; 529:496–501. [DOI] [PubMed] [Google Scholar]
- 120. Barquist L., Westermann A.J., Vogel J.. Molecular phenotyping of infection-associated small non-coding RNAs. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2016; 371, doi:10.1098/rstb.2016.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Smirnov A., Förstner K.U., Holmqvist E., Otto A., Günster R., Becher D., Reinhardt R., Vogel J.. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chaulk S., Lu J., Tan K., Arthur D.C., Edwards R.a., Frost L.S., Joachimiak A., Glover J.N.M.. N. meningitidis 1681 is a member of the FinO family of RNA chaperones. RNA Biol. 2010; 7:812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Tettelin H., Saunders N.J., Heidelberg J., Jeffries a.C., Nelson K.E., Eisen J.a, Ketchum K.a, Hood D.W., Peden J.F., Dodson R.J. et al. . Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000; 287:1809–1815. [DOI] [PubMed] [Google Scholar]
- 124. Schoen C., Weber-Lehmann J., Blom J., Joseph B., Goesmann A., Strittmatter A., Frosch M.. Whole-genome sequence of the transformable Neisseria meningitidis serogroup a strain WUE2594. J. Bacteriol. 2011; 193:2064–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Parkhill J., Achtman M., James K.D., Bentley S.D., Churcher C., Klee S.R., Morelli G., Basham D., Brown D., Chillingworth T. et al. . Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000; 404:502–506. [DOI] [PubMed] [Google Scholar]
- 126. Cahoon L.a, Seifert H.S.. Transcription of a cis-acting, noncoding, small RNA Is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog. 2013; 9:e1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Tobiason D.M., Seifert H.S.. Genomic content of Neisseria species. J. Bacteriol. 2010; 192:2160–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.