Abstract
tRNAs are synthesized as precursor RNAs that have to undergo processing steps to become functional. Yeast Trz1 is a key endoribonuclease involved in the 3΄ maturation of tRNAs in all domains of life. It is a member of the β-lactamase family of RNases, characterized by an HxHxDH sequence motif involved in coordination of catalytic Zn-ions. The RNase Z family consists of two subfamilies: the short (250–400 residues) and the long forms (about double in size). Short form RNase Z enzymes act as homodimers: one subunit embraces tRNA with a protruding arm, while the other provides the catalytic site. The long form is thought to contain two fused β−lactamase domains within a single polypeptide. Only structures of short form RNase Z enzymes are known. Here we present the 3.1 Å crystal structure of the long-form Trz1 from Saccharomyces cerevisiae. Trz1 is organized into two β-lactamase domains connected by a long linker. The N-terminal domain has lost its catalytic residues, but retains the long flexible arm that is important for tRNA binding, while it is the other way around in the C-terminal domain. Trz1 likely evolved from a duplication and fusion of the gene encoding the monomeric short form RNase Z.
INTRODUCTION
Transfer RNAs (tRNAs) play the key role in mRNA decoding during translation by the ribosome. They are usually synthesized as long precursors and subsequently processed by removing the 5΄ and 3΄ extensions (1). Maturation of tRNA further requires the addition of a CCA-motif to the 3΄ terminus (in cases where it is not encoded) and chemical modifications of nucleotides in various positions. The 5΄ end is trimmed by either the RNase P ribozyme or a protein only variant (1,2), while the 3΄ processing can involve either endo- or exonucleolytic cleavages (3–5). In eukaryotic cells, tRNAs are mainly processed endonucleolytically and the exonucleolytic pathway is minor. The order of 5΄ and 3΄ processing events is variable and is likely to be tRNA specific.
RNase Z is the enzyme responsible for the 3΄ endonucleolytic cleavage of tRNA precursors in all three domains of life, as shown in Methanococcus janaschii (6), Bacillus subtilis (7), Drosophila melanogaster (8), Arabidopsis thaliana (9), human mitochondria (10) and Schizosaccharomyces pombe (11). RNase Z enzymes belong to the family of Zn-dependent β-lactamases with a highly-conserved Zn-coordinating signature motif HxHxDH (in which x represents any hydrophobic amino acid). They are further classified according to their sequence length into two groups: the short form RNase ZS (also termed ELAC1 in humans, between 250 and 400 residues) present in bacteria, archaea and eukaryotes and the long form RNase ZL (also termed ELAC2 in humans, between 750 and 900 residues) found exclusively in eukaryotes (12,13). Some eukaryotes (D. melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae and S. pombe) only have the RNase ZL form while others (Homo sapiens, Mus musculus, Danio rerio, A. thaliana) have both the RNase ZS and RNase ZL forms, usually with differential sub-cellular localizations. Mutations in the human RNase ZL have been associated with an increased risk of prostate cancer (14,15) and cardiomyopathies in humans (16), showing the importance of this enzyme in general cell well-being.
The crystal structures of RNase ZS enzymes from B. subtilis, Thermotoga maritima, Escherichia coli and H. sapiens are known (17–20). They all adopt the β-lactamase fold and their HxHxDH motifs are involved in coordinating the enzymatically essential Zn ions. A flexible arm (also called the exosite) is inserted between the third and fourth β-strands of the α-helix flanked central β-core. This arm is composed of a compact globular domain extruded from the β−lactamase core by an extended two-stranded stalk. All the documented RNase ZS structures form similar homodimers with the two subunits in a head to head arrangement. The structure of B. subtilis RNase Z (BsuTrz) bound to tRNA was very informative on how the two subunits of the dimer play different functional roles (18). The tRNA substrate is wedged between the flexible arm of one subunit and the α7 helix of the other. The α7 helix thus helps guide the 3΄ end of the tRNA toward the active site of the subunit opposite that containing the clamping flexible arm.
Saccharomyces cerevisiae has been shown to have tRNA 3΄ endonucleolytic activity localized both in the nucleus and in the mitochondria, coded by the TRZ1 gene (17,21,22). The Trz1 protein is of the long form RNase ZL type and is composed of two domains connected by a long linker (about 60 residues). The C-terminal domain (CTD) shares significant sequence identity with the RNase ZS enzymes while the sequence of the N-terminal domain (NTD) diverges from that of the β-lactamase family. Despite this sequence divergence it was predicted that the NTD would also have a β−lactamase fold (23). While the N-terminal half of Trz1 has lost the HxHxDH, the PxKxRN (P-loop) and the AxDx motifs important for the cleavage reaction, it was predicted to contain the flexible arm, involved in tRNA binding. On the other hand, the CTD contains a full set of catalytic residues, but has lost the flexible arm insertion. In this manuscript we present the crystal structure of the full length Trz1 at a resolution of 3.1Å. Our data confirm the presence of two similar β-lactamase domains, but only the CTD contains a fully structured catalytic center. The structure suggests an evolutionary scenario whereby Trz1 evolved from a gene fusion event between two copies encoding the short form RNase Z.
MATERIALS AND METHODS
Protein expression and purification
The ORF encoding Trz1 was cloned into the pET-45 vector (Novagen) with the N-terminus in fusion with a 6xHis-tag and a linker containing an enterokinase cleavage site (underlined) (the linker coded for the following peptide MAHHHHHHVG TGSNDDDDKS PDPNWELVYT ARLQEF). Trz1 was expressed at 15°C o/n using the transformed E. coli BL21 Gold (DE3) strain and 2YT medium, supplemented with 100 μg/ml ampicillin. Cells were harvested by centrifugation, suspended in 20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 5 mM β-mercaptoethanol plus 10% glycerol and stored at −20°C. Cells were lysed by sonication, purified by Ni-NTA agarose column (Qiagen), followed by an ion-exchange column (Mono Q 5/50 GL, GE healthcare) in a buffer containing 20 mM Tris–HCl, pH 7.5, 10 mM β-mercaptoethanol and 10% glycerol, using a gradient between 50 mM (low salt) and 500 mM (high salt) NaCl. The final gel filtration (column Superdex 200 Hiload 16/60, GE healthcare) was carried out in a buffer containing 20 mM Tris–HCl, pH 7.5, 10 mM β-mercaptoethanol, 10% glycerol and 100 mM NaCl. The Se-Methionine labeled version of Trz1 was prepared using standard protocols and purified in the same way as the native protein (24).
Structure determination of Trz1
Crystallization screening (Qiagen PEGs kit) of Trz1 was performed at 18°C by the sitting drop vapor diffusion method, using a CARTESIAN pipetting robot. Crystals of seleno-methionine labeled Trz1 were obtained in 0.1 M Hepes, pH 7.5, 25% (w/v) PEG 8K, using a protein concentration of 5 mg/ml supplemented with 1 mM dGMP. The protein was mixed with reservoir solution at a 1:1 ratio. Crystals of approximate dimension 60 × 120 × 60 μm appeared within three days. Crystals were cryo-cooled using mother liquor supplemented with 20% glycerol as cryo-protectant. SAD data were collected on the Proxima1 beamline at the SOLEIL synchrotron radiation facility (Saint-Aubin, France) using a Pilatus detector. The data were processed by the XDS package (25). The crystals had the space group P41 and contained two copies of Trz1 per asymmetric unit (Table 1). The positions of the Se atoms were located using the SHELX C/D/E programs (26). The program Profess from the CCP4 suite (K. Cowtan) was successful in finding the non-crystallographic symmetry operator. The positions of 22 Se-sites were refined and the phasing was performed by the program Phaser SAD pipeline from CCP4 suite (27). After several cycles, 36 sites were finally identified and the resolution extended to 3.39 Å. The phases were further improved by map density modification using the program Parrot (K. Cowtan, CCP4 suite). Both copies in the asymmetric unit were used to complete the model. We used the program Buccaneer for automatic model building (28) and refined the model with the program Refmac5 to 3.1 Å resolution (29). The coordinates were deposited at the Protein Data Bank (PDB code: 5MTZ).
Table 1. Data collection and refinement statistics.
| Trz1-SeMet | |
|---|---|
| X-ray source | Proxima 1 |
| Wavelength (Å) | 0.9792 |
| Temperature (K) | 100 |
| Unit-cell parameters (Å, °) | a = 136.0, b = 136.0, c = 115.8, α = β = γ = 90.0 |
| Space group | P41 |
| Resolution limitsa (Å) | 49.6–3.1 (3.3–3.1) |
| Number of observationsa | 589 612 (91 190) |
| Number of unique reflectionsa | 76 016 (12 111) |
| R-measa (%) | 25.8 (180.0) |
| Completenessa (%) | 99.7 (98.5) |
| I/σa (I) | 8.13 (1.13) |
| CC(1/2) (%) | 99.1 (52.0) |
| Anomal Corra,b | 38 (19) |
| SignAnoa,b,c | 1.14 (0.96) |
| Number of non-hydrogen atoms (Protein/other) | 12 426/14 |
| R/Rfree (%) | 18.58/25.43 |
| R.M.S.D. Bonds (Å)/angles (°) | 0.01/1.61 |
| Average temperature factors (Å2) (Protein/other) | 83.29/85.53 |
aValues in parentheses refer to the highest resolution shell.
bValues for resolution limits: 49.61–3.5 (3.78–3.5).
cMean anomalous difference in units of its estimated standard deviation (|F(+)−F(−)|/Sigma). F(+), F(−) are structure factor estimates obtained from the merged intensity observations in each parity class.
RESULTS
Trz1 is composed of two β-lactamase domains connected by a long linker
No crystal structure was previously available for a RNase ZL. To probe the mechanistic differences between short and long form RNase Z enzymes we set out to obtain structural information on Trz1, the long form RNase Z from yeast. Recombinant Trz1 supplemented with a polyhistidine-tag was expressed in E. coli and purified according to standard protocols. Small crystals in the spacegroup P41 were obtained in 0.1 M HEPES buffer pH 7.5 and 25% PEG8K. The structure was solved at 3.1 Å resolution using a complete SAD data set collected on Se-methionine substituted crystals (Table 1), which diffracted better than the native protein crystals (data not shown) and which were therefore used for the refinement. The refinement yielded an R-factor of 18.6% (Rfree 25.4%) and a model with good stereochemistry. The crystals had two copies of Trz1 in the asymmetric unit with an r.m.s.d. of 0.71 Å (Supplementary Figure S1A). The current model includes regions 1–204, 246–623 and 636–820 in one copy and regions 1–204, 247–623 and 637–809 in the other. A phenylalanine was also visible in both copies at the N-terminus introduced by the fusion with the affinity tag which was not removed from the protein construct. There was clear residual electron density for four Zn2+ ions and two phosphate moieties. The protein regions with missing electronic density are likely explained by their mobility in the crystal.
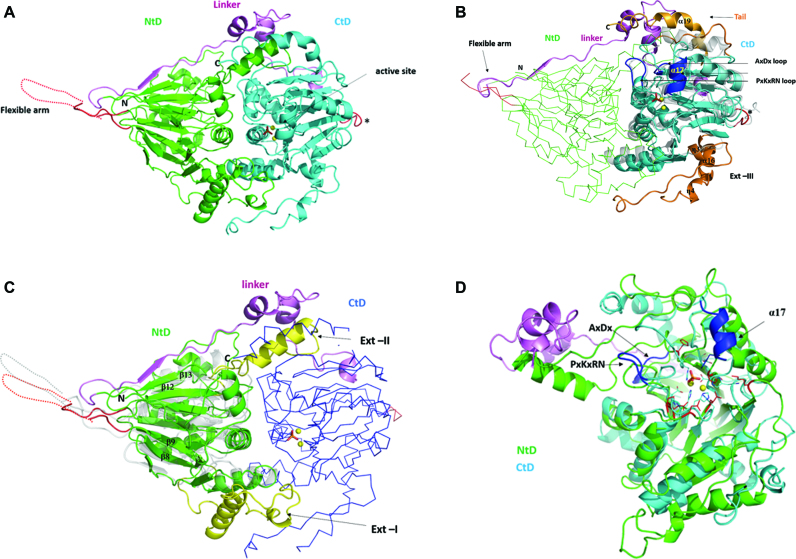
Trz1 is composed of two main domains connected by an extensive linker (Figure 1A). The NTD is contained between residues 1 and 404 and the CTD between residues 466 and 818. As predicted by sequence analysis, the CTD belongs to the β-lactamase family of Zn-dependent hydrolases. Its structure is very like that of the BsuTrz (PDB code: 2FK6, Z-factor 28.8, r.m.s.d. = 2.3 Å for 251 aligned Cα positions and 24% sequence identity) (Figure 1B) and the human ELAC1 (PDB code: 3ZWF, Z-factor 26.4, r.m.s.d. = 2.3 Å for 237 aligned Cα positions and 24% sequence identity). Despite weak sequence identity with the RNase Z family members, the NTD of Trz1 also possesses a β-lactamase fold (Figure 1C). The NTD consists of two opposing seven-strand β-sheets sandwiched by α-helices. The first four β-strands (β7, β8, β9, β10 and β14, β1, β2, β3) are antiparallel and the next three are parallel in each of the sheets. The CTD also consists of two opposing β-sheets sandwiched by α-helices, but only one of the β-sheets has seven strands (β28, β16, β17, β18 antiparallel; β19, β20, β21 parallel); the other is six-stranded (β22, β23, β24, antiparallel; β25, β26, β27 parallel) and lacks the first β-strand compared to BsuTrz. The linker between the two domains forms an elongated peptide stretch (residues 405–423) that extends the β-sheet of the NTD, followed by two short α-helices and a second irregular stretch that interacts with the CTD (Figure 1A).
Figure 1.
Structure of Trz1. (A) Cartoon presentation of the Trz1 structure. The N-terminal domain (NTD) and C-terminal domain (CTD) are shown in green and cyan, respectively, and the linker is shown in purple. Two zinc ions are shown as yellow spheres and the phosphate ion is shown as sticks (present in the active site). The region corresponding to the unresolved flexible arm protruding from the NTD is shown as a dashed line and the loop replacing the flexible arm in the CTD is in red (labeled with an asterisk). (B) The CTD of Trz1 superimposed on BsuTrz (transparent gray, PDB code: 1y44). The core of the CTD is shown in cyan and the extensions are in orange. The conserved tRNA recognition elements α17 and loop PxKxRN and the nearby AxDx loop in the CTD are in blue. The NTD of Trz1 is shown as a green ribbon and the linker region is shown in purple. (C) The NTD of Trz1 superimposed on BsuTrz (transparent gray). The β-lactamase core of the NTD is shown in green and the extensions are in yellow. The CTD of Trz1 is shown as blue ribbon. (D) Superimposition of the NTD (green) and CTD (cyan) of Trz1 (linker in purple).
The two domains of Trz1 both possess long extensions from the core, which are not present in RNase ZS sequences, here named Ext-I, Ext-II, Ext-III and Tail (Figure 1B and C). Ext-I of the NTD is situated between strands β8 and β9 (residues 136–191); it contains a helix (α4) and an irregular peptide stretch that mainly interact with the N-terminal core. Ext-II from the NTD is a long connection between strands β12 and β13 (residues 319–356) that contains a helix (α7, residues 330–342) inserted between the core of the CTD and the helical part of the linker peptide (Figure 1C). This linker therefore contributes considerably to the interaction surface between the two domains. Ext-III is in the CTD, between the β-strands β21 and β22 (residues 600–659). The tail of the CTD (residues 792–818) also has a long helix (α19, residues 795–809) that together with the linker helices and helix α7 (from the NTD), forms a helical bundle sub-domain (Figure 1B).
N- and C-terminal domain comparison
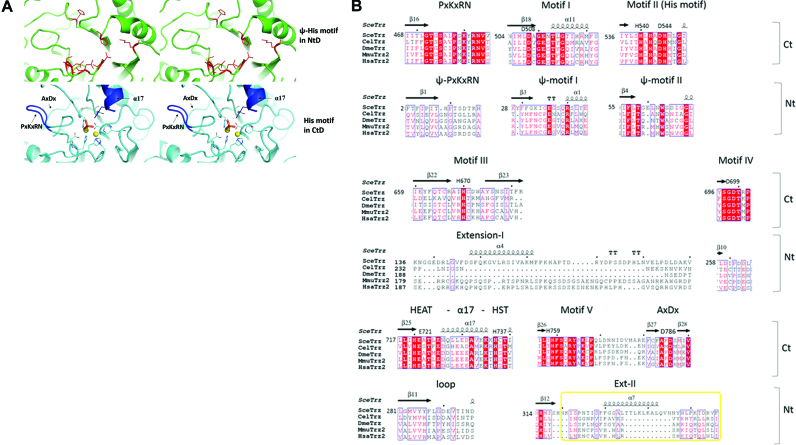
The two domains of Trz1 superimpose well (r.m.s.d. = 3.3 Å for 228 aligned Cα positions and 13% sequence identity, Z-factor = 15.1; Figure 1D) but they also have a few significant structural differences. First, the key catalytic Zn-binding residues are only present in the CTD and are replaced in the NTD by ‘pseudo Ψ-motifs’ (Figure 2A and B). Residual electron density at the active site situated in the CTD of Trz1 revealed the presence of two Zn ions separated by 3.6 Å that are bridged by a phosphate ion bound by His737 (Figure 3D). The two Zn ions are further coordinated by five histidines (His540, His542, His545, His670 and His759) and two aspartate residues (Asp544 and Asp699). The coordination sphere of the two Zn ions in the Trz1 CTD is perfectly superimposable on that of the Zn ions in the RNase ZS structures (Figure 3C). Thus, the Trz1 CTD is clearly the catalytically active domain while the Trz1 NTD has lost its catalytic residues during evolution. It is interesting to compare this to the situation in the BsuTrz holo-enzyme crystal structure where the active site of the A subunit contained two coordinated Zn ions while the active site in the B subunit lacked bound metal ions and was in an inactive conformation (15). The backbones of the ψ-motif II residues are superimposable in the two domains, but the central HxHxDH signature of motif II is lost in the NTD (Figure 2A and B). The same holds for motif I (Asp): the backbone is superimposable for both domains but Asp508 which helps to stabilize His545 in motif II is lost in the NTD. Motifs III (His), IV (Asp) and V (His) are absent in the NTD and the presence of the Ext-I and Ext-II extensions completely changes the conformation of the corresponding regions. The HEAT (Glu721) -α17- HST (His737) motif, which is essential for both binding and cleavage of the tRNA precursor, is present in the CTD but is replaced in the NTD by a short loop (residues 278–295). Also the PxKxRN and AxDx motifs, involved in pre-tRNA binding and cleavage, are only present in the CTD (Figure 2A). The AxDx motif is conserved in the eukaryotic RNase ZL and bacterial RNase ZS sequences. This loop is involved in the cleavage reaction and has the potential to form hydrogen bonds with the PxKxRN loop. It was shown that an aspartate to alanine mutation in the AxDx motif affected cell viability of S. pombe (30,31).
Figure 2.
Active site motifs. (A) Stereo views of the Ψ-histidine motif in NTD (upper panels) and Zn-binding His-motif in CTD (lower panels). (B) Structure based sequence alignment of the conserved motifs in C- and NTDs: PxKxRN loop, motif I to V and AxDx motif. RNase ZL sequences from Saccharomyces cerevisiae (Sce = Trz1), Caenorhabditis elegans (Cel), Drosophila melanogaster (Dme), Mus musculus (Mmu) and Homo sapiens (Hsa) are represented.
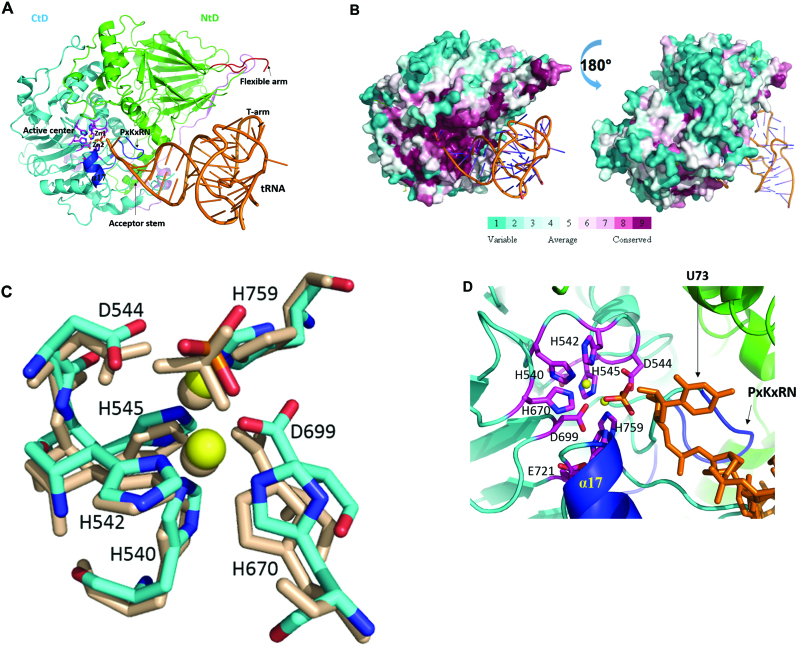
Figure 3.
Model of Trz1 in complex with tRNA. (A) This model was generated by superimposition of Trz1 onto the BsuTrz/tRNA complex (PDB code: 2FK6). The N- and C-terminal halves and linker of Trz1 are shown in green/cyan and purple, respectively. For clarity, BsuTrz was omitted in the figure and only the tRNA (orange) is shown. Arrows indicate the position of the flexible arm and acceptor stem. The active center residues of Trz1 are shown as pink sticks, and zinc ions as yellow spheres. The α17 helix and the ‘PxKxRN’ and AxDx loops are shown in blue. (B) Sequence conservation projected onto the surface of Trz1 as generated with default parameters by the consurf webserver (http://consurf.tau.ac.il). The complex is shown in the same orientation as for panel A. (C) Zoom on the superimposition of the active site residues of Trz1 and BsuTrz. Only Trz1 residues are labeled and BsuTrz residues are shown as gray sticks. (D) Zoom on the active site of the Trz1/tRNA complex. The tRNA is shown as orange sticks and the uracil 73 is labeled.
Another important functional difference between the NTD and CTD in the regions implicated in the binding of tRNA. In the crystal structure of the RNase ZS/tRNAThr complex (7,18), tRNA is wedged between the flexible arm of one subunit and the α7 helix of the other. In Trz1, the flexible arm between residues 203 and 251 is disordered in the NTD and replaced by a short loop in the CTD (Figure 1A) suggesting this half of Trz1 does not interact with a tRNA substrate (see ‘Discussion’ section). Superimposing the Trz1 NTD onto RNase ZS structures showed that the flexible arm emanates from the same position on the surface in both forms of RNase Z, i.e. between the third and fourth β-strand of the core domain (Figure 1C). A second region involved in tRNA binding provided by the alpha-helix between the HEAT and the HST motifs (α7 in the BsuTrzS crystal structure) is present in the CTD (α17, Figure 3A) but replaced by a short loop in the NTD. Thus the two domains of Trz1 clearly have distinct biochemical roles.
Trz1 mimics the RNase ZS dimer
Trz1 superimposes well onto the BsuTrz dimer (Supplementary Figure S2A). The NTD and CTD interact tightly and are in the same position as the two subunits of the RNase ZS dimers. The major interface between the two domains is contributed by three major α−helices, namely α1/α11, α2/α13 and α3/α14 from the NTD and CTD, respectively. Interaction between the domains is reinforced by the linker region and by the extensions emanating from the NTD (Supplementary Figure S2B and C). BsuTrz alone showed an asymmetry in the two subunits of the dimer: in one subunit, the protruding arm was disordered and in the other it was stabilized through crystal contacts (17). In the complex of BsuTrz with substrate tRNAThr this flexible arm reaches out from the core of the protein to embrace the T-arm of the tRNA substrate, while the acceptor end of the tRNA plunges into the active site of the other subunit (18). One of the subunits of the BsuTrz dimer is responsible for the main contacts with the tRNA substrate while the other subunit provides the catalytic site competent for tRNA cleavage. This structure shows that the dimeric short form RNase Z can process two tRNAs simultaneously.
In contrast, Trz1 consists of two domains that mimic the structure of the dimeric RNase ZS. The NTD lacks the Zn-binding residues but has a protruding arm that is likely involved in clamping of the tRNA substrate. The CTD has a properly formed active site responsible for the cleavage reaction. The tRNA substrates are likely bound and processed in a very similar way by the long and short RNase Z forms, except that RNase ZL can process only one tRNA at a time. Our structural data confirm that RNase ZL likely originated through gene duplication and divergent evolution of the two original subunits, resulting in the loss of the flexible arm and catalytic site from the NTD and CTD, respectively.
DISCUSSION
Eukaryotes encode at least one long form RNase Z (yeast, H. sapiens), but many organisms have both the long and short form of this tRNA 3΄ processing enzyme (e.g. H. sapiens, A. thaliana). The short RNase Z proteins are active as homodimers, whereas the long forms act as monomers. Our crystal structure of Trz1 strengthens the hypothesis that RNase ZL arose from a gene duplication-fusion event from an RNaseZS. Despite a considerable sequence divergence between the N- and C-terminal halves of Trz1, both possess a very similar β−lactamase structure. The crystal structure of Trz1 clearly explains why RNase ZL is monomeric while the short form is dimeric. The two β-lactamase domains of Trz1 can be superimposed on the two subunits of the short form RNase Z and clearly mimic the structure of the dimer (Figure 1B and C). The interactions between the NTD and CTD of Trz1 are reinforced by numerous contacts provided by the long extensions emanating from the N-terminal half and by the linker. To analyze the functional organization of the RNase ZL forms, we created a model of a Trz1/tRNA complex by superimposing the structure of Trz1 onto that of the BsuTrz/tRNAThr complex (PDB code: 2FK6; Figure 3A). The two Trz1 domains superimpose very well with the BsuTrz dimer and tRNAThr fits perfectly onto the Trz1 surface. A minor steric clash is generated between tRNAThr and residues 760 and 769 of Trz1, situated in a long loop that is probably subject to conformational changes upon substrate binding. Although the structure and association of the two Trz1 domains strongly mimic the dimer of BsuTrz, the functional roles of the two domains are clearly separated. First, the flexible arm present in the NTD of Trz1, although disordered in absence of substrate tRNA, is well positioned to play the same role in tRNA clamping as for BsuTrz. In the CTD, the flexible arm is replaced by a short loop instead that is clearly too short to embrace the T arm of the tRNA, but the α17 helix (equivalent to helix α7 in BsuTrz) is present. In our model the 3΄-end of the tRNAThr substrate fits precisely into the active centre of the CTD and establishes contacts with Trz1 that are identical to those observed for the BsuTrz/tRNAThr complex (Figure 3D). In addition, the acceptor stem of tRNAThr is bound by the α17 helix and the PxKxRN loop motif of the CTD. The active site region of the C-terminal Trz1 domain is positively charged and highly conserved (Figure 3B), as would be expected. We conclude that our model of the Trz1/tRNA complex obtained by simple superimposing the Trz1 and the BsuTrz/tRNAThr complex structures highlights the functional segregation between the NTD and CTD: the NTD primarily recognizes the tRNA substrate and the CTD is responsible for the cleavage reaction. The complete catalytic machinery used by BsuTrz is perfectly superimposable onto that of the CTD of Trz1. The histidines from motifs II, III and V, the aspartates from motif I, II, IV, a phosphate ion and a water molecule provide a pseudo-octahedral coordination of the two zinc ions, constituting the catalytic centre (Figure 3D). By analogy with what was proposed for BsuTrz, the negatively charged Asp544 (Asp67 in BsuTrz) likely acts as a general base and deprotonates a neighboring water (17,23). The nucleophilic hydroxyl group then attacks the cleavable phosphate linkage of the tRNA substrate that is polarized by the two zinc ions, resulting in the breakage of the phospho-diester bond between nucleotide 73 and 74. Then the 3΄ position of tRNA restores its hydroxy-group by protonation through a water molecule.
A number of mutations found in the human ELAC2 gene have been connected with various diseases. Trz1 and ELAC2 share 21% sequence identity and 49% similarity (the sequence alignment for some RNaseZL orthologs is represented in Supplementary Figure S2F). With the help of the structure of Trz1 we explored the locations of these mutations. ELAC2 has been tagged as a prostate cancer susceptibility gene (15). The related mutations are S217L, A541T and R781H, three residues that are not conserved in Trz1 (Supplementary Figure S2F). A fourth mutation is a guanine insertion after base 1641, causing premature termination of ELAC2 translation (14). None of the mis-sense mutations affect ELAC2 catalytic activity (14), suggesting that their link to cancer is more likely due to another problem such as a defect in its association with other partners. Ser217 is found in a region corresponding to Ext-I in Trz1 (residues 136–190) whose sequence is not conserved among species; Arg781 is in the tail of ELAC2 that corresponds to the disordered C-terminal region of Trz1. Ala541 aligns with Met535 in Trz1, a residue of the hydrophobic core of the CTD. The positions of these mutations in the 3D structure of Trz1 are consistent with the possibility that they affect protein–protein interactions.
ELAC2 mutations were also found to be associated with hypertrophic cardiomyopathy, which is caused by a mitochondrial RNA processing defect (16). The four related mutations are F154L, L423F, T520I and a STOP mutation at Arg211. Phe154 (Tyr102 in Trz1) is at the end of helix α3 at the interface between the two domains. Thr520 (Thr513 in Trz1) is in the conserved Motif I of the CTD. Introduction of the T520I mutation into TRZ1 caused a phenotype indicative of impaired respiratory chain function, a reduced growth rate on medium containing non-fermentable carbon sources; a higher petite frequency is also observed with this mutant, indicating quantitatively reduced mitochondria translation (16). Mutation of the equivalent in DmeTrzLΔ23 (Thr471) reduced the activity 10-fold (32). It is interesting to note that Thr520 is part of helix α1, which is also at the interface between the NTD and CTD. Leu423 is located in the beginning of the linker region between the strands β14 and β15, and is not conserved in Trz1. Some of the cardiomyopathy-linked mutations thus clearly affect catalysis, while others may have subtler structural effects or affect protein–protein interactions as proposed for the mutations associated with prostate cancer susceptibility.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of the Proxima1 beamlines at the SOLEIL synchrotron for assistance during the diffraction data collections.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chinese Scholarship Council Scholarship (to M.M.); Agence Nationale de la Recherche (subtilRNA2, ARNr-QC and Labex Dynamo) (to C.C.); French Infrastructure for Structural Biology (FRISBI ANR-10-INSB-0501). Funding for open access charge: Nationale de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hartmann R.K., Gossringer M., Spath B., Fischer S., Marchfelder A.. The making of tRNAs and more - RNase P and tRNase Z. Prog. Mol. Biol. Transl. Sci. 2009; 85:319–368. [DOI] [PubMed] [Google Scholar]
- 2. Rossmanith W. Of P and Z: mitochondrial tRNA processing enzymes. Biochim. Biophys. Acta. 2012; 1819:1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spath B., Canino G., Marchfelder A.. tRNase Z: the end is not in sight. Cell Mol. Life Sci. 2007; 64:2404–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hopper A.K., Pai D.A., Engelke D.R.. Cellular dynamics of tRNAs and their genes. FEBS Lett. 2010; 584:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Phizicky E.M., Hopper A.K.. tRNA biology charges to the front. Genes Dev. 2010; 24:1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schiffer S., Rosch S., Marchfelder A.. Assigning a function to a conserved group of proteins: the tRNA 3΄-processing enzymes. EMBO J. 2002; 21:2769–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pellegrini O., Nezzar J., Marchfelder A., Putzer H., Condon C.. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis. EMBO J. 2003; 22:4534–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dubrovsky E.B., Dubrovskaya V.A., Levinger L., Schiffer S., Marchfelder A.. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3΄ ends in vivo. Nucleic Acids Res. 2004; 32:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Canino G., Bocian E., Barbezier N., Echeverria M., Forner J., Binder S., Marchfelder A.. Arabidopsis encodes four tRNase Z enzymes. Plant Physiol. 2009; 150:1494–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rossmanith W. Localization of human RNase Z isoforms: dual nuclear/mitochondrial targeting of the ELAC2 gene product by alternative translation initiation. PLoS One. 2011; 6:e19152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gan X., Yang J., Li J., Yu H., Dai H., Liu J., Huang Y.. The fission yeast Schizosaccharomyces pombe has two distinct tRNase Z(L)s encoded by two different genes and differentially targeted to the nucleus and mitochondria. Biochem. J. 2011; 435:103–111. [DOI] [PubMed] [Google Scholar]
- 12. Vogel A., Schilling O., Spath B., Marchfelder A.. The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties. Biol. Chem. 2005; 386:1253–1264. [DOI] [PubMed] [Google Scholar]
- 13. Condon C., Putzer H.. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002; 30:5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takaku H., Minagawa A., Takagi M., Nashimoto M.. A candidate prostate cancer susceptibility gene encodes tRNA 3΄ processing endoribonuclease. Nucleic Acids Res. 2003; 31:2272–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tavtigian S.V., Simard J., Teng D.H., Abtin V., Baumgard M., Beck A., Camp N.J., Carillo A.R., Chen Y., Dayananth P. et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 2001; 27:172–180. [DOI] [PubMed] [Google Scholar]
- 16. Haack T.B., Kopajtich R., Freisinger P., Wieland T., Rorbach J., Nicholls T.J., Baruffini E., Walther A., Danhauser K., Zimmermann F.A. et al. ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2013; 93:211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li de la Sierra-Gallay I., Pellegrini O., Condon C.. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005; 433:657–661. [DOI] [PubMed] [Google Scholar]
- 18. Li de la Sierra-Gallay I., Mathy N., Pellegrini O., Condon C.. Structure of the ubiquitous 3΄ processing enzyme RNase Z bound to transfer RNA. Nat. Struct. Mol. Biol. 2006; 13:376–377. [DOI] [PubMed] [Google Scholar]
- 19. Ishii R., Minagawa A., Takaku H., Takagi M., Nashimoto M., Yokoyama S.. Crystal structure of the tRNA 3΄ processing endoribonuclease tRNase Z from Thermotoga maritima. J. Biol. Chem. 2005; 280:14138–14144. [DOI] [PubMed] [Google Scholar]
- 20. Kostelecky B., Pohl E., Vogel A., Schilling O., Meyer-Klaucke W.. The crystal structure of the zinc phosphodiesterase from Escherichia coli provides insight into function and cooperativity of tRNase Z-family proteins. J. Bacteriol. 2006; 188:1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y., Beck A., Davenport C., Shattuck D., Tavtigian S.V.. Characterization of TRZ1, a yeast homolog of the human candidate prostate cancer susceptibility gene ELAC2 encoding tRNase Z. BMC Mol. Biol. 2005; 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skowronek E., Grzechnik P., Spath B., Marchfelder A., Kufel J.. tRNA 3΄ processing in yeast involves tRNase Z, Rex1, and Rrp6. RNA. 2014; 20:115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Redko Y., Li de la Sierra-Gallay I., Condon C.. When all's zed and done: the structure and function of RNase Z in prokaryotes. Nat. Rev. Microbiol. 2007; 5:278–286. [DOI] [PubMed] [Google Scholar]
- 24. Doublie S. Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol. Biol. 2007; 363:91–108. [DOI] [PubMed] [Google Scholar]
- 25. Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sheldrick G.M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cowtan K. Completion of autobuilt protein models using a database of protein fragments. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 30. Zhao Z., Su W., Yuan S., Huang Y.. Functional conservation of tRNase ZL among Saccharomyces cerevisiae, Schizosaccharomyces pombe and humans. Biochem. J. 2009; 422:483–492. [DOI] [PubMed] [Google Scholar]
- 31. Wang Z., Zheng J., Zhang X., Peng J., Liu J., Huang Y.. Identification and sequence analysis of metazoan tRNA 3΄-end processing enzymes tRNase Zs. PLoS One. 2012; 7:e44264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zareen N., Hopkinson A., Levinger L.. Residues in two homology blocks on the amino side of the tRNase Z His domain contribute unexpectedly to pre-tRNA 3΄ end processing. RNA. 2006; 12:1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.