Abstract
Epigenetic mechanisms, including chromatin structure, chromatin dynamics and histone modifications play an important role for maintenance and differentiation of pluripotent embryonic stem cells. However, little is known about the molecular mechanisms of adult stem cell specification and differentiation. Here, we used intestinal stem cells (ISCs) as a model system to reveal the epigenetic changes coordinating gene expression programs during these processes. We found that two distinct epigenetic mechanisms participate in establishing the transcriptional program promoting ISC specification from embryonic progenitors. A large number of adult ISC signature genes are targets of repressive DNA methylation in embryonic intestinal epithelial progenitors. On the other hand, genes essential for embryonic development acquire H3K27me3 and are silenced during ISC specification. We also show that the repression of ISC signature genes as well as the activation of enterocyte specific genes is accompanied by a global loss of H2A.Z during ISCs differentiation. Our results reveal that, already during ISC specification, an extensive remodeling of chromatin both at promoters and distal regulatory elements organizes transcriptional landscapes operating in differentiated enterocytes, thus explaining similar chromatin modification patterns in the adult gut epithelium.
INTRODUCTION
Together with transcription factors, the structure and organization of chromatin play important roles in the establishment and maintenance of different transcriptional programs during development and differentiation. The fundamental element of chromatin, the nucleosome core, is a multi-subunit structure consisting of four histone types (1). Each histone has the potential to be differentially altered by a number of covalent modifications. Biochemical and genetic studies showed that post-translational modifications of core histones modulate gene expression. For example, the acetylation of lysine residues within histone tails increases the accessibility of the chromatin template to the transcriptional machinery (2). Consistently, genome-wide studies mapping the distribution of modified histones identified high levels of histone acetylation in promoter regions of active genes (3) and enhancers (4).
In contrast, tri-methylation of histone H3 at lysine 27 (H3K27me3) is required for the repression of genes instructing developmental patterning and differentiation, such as transcription factors, receptors and signaling molecules (5). Analogous to histone modifications, histone variants also regulate gene expression (6). For instance, the histone variant H2A.Z influences the accessibility of chromatin structure at promoters and enhancer elements and is required for embryonic stem cells (ESCs) maintenance and differentiation (7). The organization and composition of nucleosomes, in turn, affect DNA methylation, an essential epigenetic modification in eukaryotes (8). Genetic studies in mice revealed that the establishment and maintenance of DNA methylation patterns are crucial for early steps of embryonic development (8).
Recent genetic and chromatin profiling analyses provided new insights on the functions of chromatin modifications during the maintenance and differentiation of adult stem cells. Most of the studies were focused on the adult intestinal stem cells (ISCs), which are essential for continuous renewal of the gut epithelium (9). In contrast to ESCs, loss of the DNA methyltransferase Dnmt1 has modest effects on homeostasis of the adult intestinal stem cells (ISCs) (10). Accordingly, DNA methylation patterns are very similar between adult ISCs and terminally differentiated enterocytes (10–12). Loss of essential components of the Polycomb Repressive Complex 2, which positions the H3K27me3 mark, in the adult ISCs leads to cell cycle arrest and spontaneous differentiation towards the secretory lineage, yet has little effect on enterocyte specific differentiation program (13–15). Consistently, H3K27me3 patterns are very similar between adult ISCs and enterocytes (14). This raises the question of which epigenetic control(s) determine changes in gene expression programs during differentiation of the gut epithelium. Furthermore, how epigenetic mechanisms influence the establishment of the ISC identity during embryonic development is not known.
To address these questions, we established transcriptome and chromatin profiles for the embryonic intestinal epithelium at two different stages, prior to and after the specification towards stem cells (16), as well as for the adult ISCs and their most abundant differentiated progenies, enterocytes. Based on the distribution of H3K27me3, H3K4me3 and H3K27Ac, the histone variant H2A.Z, as well as DNA methylation, in these four cell populations we found that the embryonic intestinal epithelium is very different to the adult ISCs at the transcriptional and chromatin levels. Genes essential for embryonic development are the main targets of H3K27me3 mediated repression during the transition from embryonic progenitors to adult ISCs. In contrast, the majority of ISC signature and enterocyte specific genes lose DNA methylation during adult ISCs specification. We further show that the loss of H2A.Z accompanies activation of many enterocyte specific genes during ISCs differentiation. Finally, our data demonstrate that already during embryogenesis, major chromatin changes at TSS and distal regulatory elements prepare appropriate transcriptional landscapes for later activation of the enterocyte specific genes during ISC differentiation.
MATERIALS AND METHODS
Mice
Lgr5EGFP-Cre-ERT mice were obtained from Jackson laboratory. Bl6/N and CD1 mice were from Charles Rivers. Mouse colonies were maintained in a certified animal facility in accordance with European guidelines.
Isolation of embryonic intestinal epithelial cells using flow cytometry
Small intestines were dissected from mouse embryos at indicated stages, cut in pieces of 2 mm and incubated for 5–10 min with 0.15 mg/ml collagenase (Sigma) in PBS at 37°C with shaking at 800 rpm. Single cell suspensions were collected by centrifugation at 200g for 3 min, washed twice and resuspended in PBS supplemented with 2% fetal calf serum. Cells were stained with PerCP-eFluor® 710-conjugated anti-EpCAM 1:400 (eBioscience), PE-conjugated anti-CD45 at 1:400 (BD Biosciences) and anti-CD31 at 1:400 (BD Biosciences) antibody for 15 min at room temperature. Living cells were gated by DAPI dye exclusion. Embryonic intestinal epithelial cells were isolated as EpCAM+CD31−CD45−DAPI−. Fluorescence-activated cell sorting was performed using BD FACS Aria III SORP cell sorter (85 μM nozzle) and analysed using FlowJo software.
Isolation of adult enterocytes and ISCs using flow cytometry
Small intestines were dissected from 2 months old Lgr5EGFP-Cre-ERT males, cut in pieces of 2 mm2, placed into 50 ml conical tubes and washed three times for 10 min with 40 ml of PBS/5 mM EDTA on a rocking plate at room temperature. Villi were mechanically removed by gentle shaking three times and trituration using a 10 ml pipette. Villi material was collected by centrifugation at 200g for 3 min and washed twice with PBS. Crypts were separated from the intestine by vigorous shaking two times passing through a 70 μM filter (BD Falcon). Crypt material was collected by centrifugation at 200g for 3 min, washed twice with PBS and incubated for 5–10 min with 0.15 mg/ml collagenase (Sigma) and 0.1 mg/ml DNase I (Qiagen) at 37°C on a rocking plate. Single cell suspensions were washed twice and resuspended in PBS supplemented with 2% fetal calf serum. Cells were stained with PerCP-eFluor® 710-conjugated anti-EpCAM 1:400 (eBioscience), PE-conjugated anti-CD45 at 1:400 (BD Biosciences) and anti-CD31 at 1:400 (BD Biosciences) antibody for 30 min at room temperature. Living cells were gated by DAPI dye exclusion. Enterocytes were isolated as EpCAM+CD31−CD45−DAPI−. ISCs were isolated as Lgr5-EGFPhighCD31−CD45−DAPI−. Fluorescence-activated cell sorting was performed using BD FACS Aria III SORP cell sorter (85 μM nozzle) and analysed using FlowJo software.
RNA-sequencing
Fifty thousand EpCAM+ embryonic intestinal cells, Lgr5-EGFPhigh ISCs or enterocytes were isolated by FACS directly in 300 μl of RTL buffer (Qiagen) supplemented with 1% β-mercaptoethanol and stored at −80°C. For each replicate, RNA was isolated from pools of 20 embryos (E12.5) or one adult gut using RNeasy micro-kit (Qiagen). 50 ng of total RNA was used for cDNA synthesis using Ovation v2.0 kit (NuGEN) according to manufacturer's instructions. After cDNA fragmentation (Covaris), libraries were prepared using NEBNext v2.0 kit (NEB) according to manufacturer's instructions. Three independent RNA extractions, cDNA synthesis, library preparations and sequencing experiments were performed.
MBD-sequencing
Two hundred thousand EpCAM+ embryonic intestinal cells, Lgr5-EGFPhigh ISCs or enterocytes were isolated by FACS and stored at −80°C. For each replicate, DNA was isolated using phenol: chloroform extraction. 1 ug of genomic DNA was fragmented to 150–250 bp using Covaris. MBD pull-downs were performed using Methylminer Methylated DNA Enrichment kit (Invitrogen) according to manufacturer's instructions. Libraries were prepared using NEBNext v2.0 kit (NEB) according to manufacturer's instructions. Two independent DNA extractions, MBD pull-downs, library preparations and sequencing experiments were performed for each stage.
ChIP-sequencing
Single cell suspensions of embryonic or adult intestinal cells were fixed for 10 min with 1% PFA at RT, washed twice with PBS and stained with PerCP-eFluor® 710-conjugated anti-EpCAM 1:400 (eBioscience), PE-conjugated anti-CD45 at 1:400 (BD Biosciences) and anti-CD31 at 1:400 (BD Biosciences) antibody for 15–30 min at RT. For H3K27Ac ChIP, samples were dissected and all the time processed in the presence of 20 mM Na butyrate. Two hundred fifty thousand EpCAM+ embryonic intestinal cells, Lgr5-EGFPhigh ISCs or enterocytes were isolated by FACS and stored at −80°C. To obtain fragments ∼350–700 bp, chromatin was resuspended in 150 ul of sonication buffer (1% SDS, 50 mM Tris–Cl pH 8.0, 10 mM EDTA) and sonicated for 56 cycles 10 s ON/ 90 s OFF (Diagenode) at 4°C. Anti-H3K27Ac (Abcam 4729), H3K27me3 (Millipore #17-622), H2A.Z (Active Motif, #39113), H3K4me3 (Millipore #07-473) antibodies were used. 20 uL of Protein A/G sepharose beads (Sigma, P6486 and E3403) were pre-incubated for 3 h with 0.5 ug of antibody in RIPA buffer, then washed three times with RIPA buffer. The sonicated chromatin was diluted till final volume 750 ul and incubated with antibody-bead slurry at 4°C overnight. Beads were washed three times with 1 ml of RIPA buffer and once with TE. DNA was eludet and incubated with RNAse A for 30 min, followed by overnight incubation with PK. DNA was extracted using phenol: chloroform. Libraries were prepared using NuGEN Ovation Ultralow Library System V2 (NuGEN) according to manufacturer's instructions. Two independent ChIP pull-downs, library preparations and sequencing experiments were performed for each stage and each antibody. Whole genomic DNA obtained after the sonication for each stage were used as an input control.
RNA-sequencing data analysis
Reads were mapped to the mouse (mm9) genome using STAR (http://support.illumina.com/ sequencing/sequencing_software/igenome.html), allowing up to two mismatches and discarding all secondary alignments and reads mapping to more than 10 loci. Quality of the sequencing was assessed with FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc) and RSeQC (17). To quantify the reads per gene, HTSeq-count (18) was used in the default union mode. To normalize, transform and test the differential expression of genes between conditions we used the DESeq2 (19). For calling differential expression between conditions, the read counts were fitted to a negative binomial (NB) generalized linear model (GLM) and the default Wald test was used. Genes with RPKM≥2 were considered as expressed. Genes, expression of which would differ log2 ≥1 and FDR ≤0.01, were considered as differentially expressed. Hierarchical clustering was performed using Euclidian distance as a similarity metric (Gene Cluster 3.0 software, http://bonsai.hgc.jp/∼mdehoon/software/cluster/). Heatmap was obtained using Treeview (http://jtreeview.sourceforge.net/). The GO enrichment analysis was performed using DAVID (20). P-values were determined by a hypergeometric and ANOVA tests.
ChIP-sequencing data analysis
Reads were converted to FASTQ format and were mapped to the mouse genome (mm9) using Bowtie version 12.5, allowing up to two mismatches and retaining not exceeding a base quality of 70, only the best position. Duplicates were removed with Picard (http://http://broadinstitute.github.io/picard/). For the H2A.Z, H3K4me3 and H3K27Ac ChIPs, peaks were called using MACS version 2.1.0.20150420 (21) with a q-value cutoff of 1e−5 and the default M-fold of 5.50. Peaks were assigned to genes based on RefSeq gene annotation. The closest gene promoter was defined as a window of ±3 kb around the TSS. H2A.Z and H3K4me3 marks were called with the broad option of MACS activated. For the H3K27me3 ChIP, we used SICER version 1.1 with the same parameters used in (14). We used DiffBind (http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/ DiffBind.pdf) to call different peaks between conditions in the default edgeR (22) model. Peaks with width ≥450 bp, and FDR ≤0.01 were considered for further analyses. Sequencing tracks were visualized with UCSC genome browser (http://genome.ucsc.edu/). P-values were determined by a hypergeometric and ANOVA tests.
MBD-sequencing data analysis
After mapping reads and removing duplicates as with the ChIP-seq datasets, peaks were called using MACS version 1.4.2 with a bandwidth of 200 and fragment size of 180, coinciding with the average fragment length of the library. Data was downstream processed as described in the ChIP-seq section.
RNA in situ hybridization and histological techniques
Embryonic small intestines were dissected, fixed overnight in 4% formaldehyde in PBS at 4°C, dehydrated and embedded in paraffin. 10 μm sections were rehydrated, treated 15 min with 6% H2O2, 10 min with 20 μg/ml proteinase K, post-fixed with 4% formaldehyde for 15 min, treated with acetic anhydride, equilibrated with 2× SSC and hybridized overnight at 63°C with digoxygenin labeled RNA probes for Axin2, Kcne3 and Sfrp5. Sections were washed and incubated overnight with sheep anti-digoxygenin antibody 1:3000 (Roche). The signal was revealed using NBT/BCIP (Roche). Images were acquired with Leica DM2500 microscope. For antibody staining, small intestines were dissected, fixed for 20 min in 1% formaldehyde in PBS at 4°C, incubated overnight in 30% sucrose, embedded in OCT and kept at −80°C. Immunohistochemical analyses were performed on 10 μm cryosections using 1:1000 anti-EpCAM antibody (rat, eBiosciences), anti-H2A.Z 1:1000 (Active Motif, #39113) and H3K27Ac 1:1000 (Abcam 4729), followed by 1:300 Alexa566-conjugated secondary antibody (Invitrogen), Images were acquired with Leica AF7000 microscope.
RESULTS
Defining transcriptional programs during specification, maintenance and differentiation of the adult ISCs
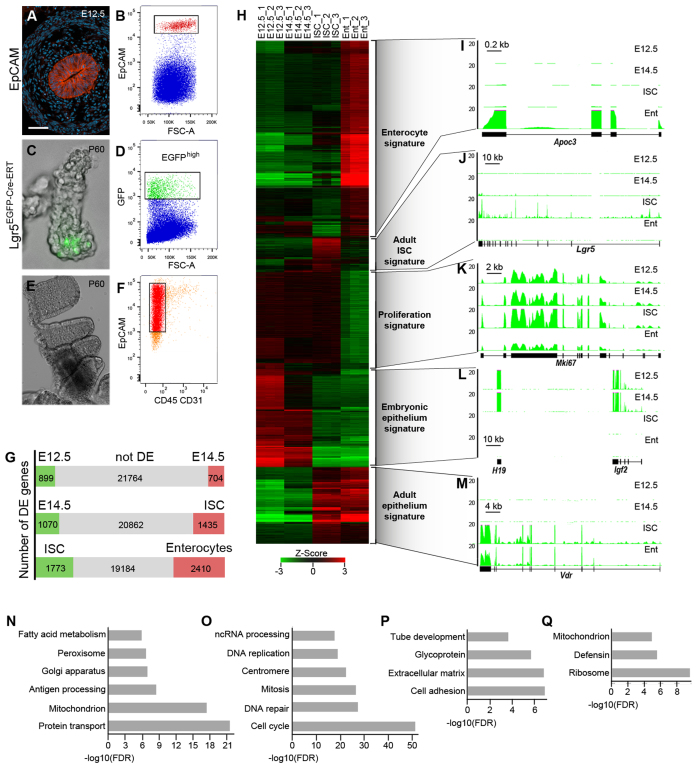
To follow the changes in the transcriptional programs accompanying the specification, maintenance and differentiation of the adult ISCs we sequenced whole transcriptomes of embryonic intestinal epithelium progenitors (at E12.5 and E14.5), adult ISCs and their differentiated progenies, the majority of which are absorptive enterocytes. EpCAM positive embryonic gut epithelium was isolated from dissected small intestines using fluorescence activated cell sorting (FACS) (Figure 1A and B and Supplementary Figure S1A). Adult ISCs were purified on the basis of GFP fluorescence from crypts of Lgr5GFP-Cre-ERT mice (Figure 1C and D and Supplementary Figure S1B; (23)), whereas enterocytes (EpCAM+ CD45− CD31−) were isolated from villi (Figure 1E and F and Supplementary Figure S1C). RNA-sequencing analyses revealed 1603 differentially expressed genes between E12.5 and E14.5 embryonic epithelial cells (RPKM ≥ 2, log2 ≥ 1, FDR ≤ 0.01) (Figure 1G and Supplementary Tables S1 and S2), suggesting very dynamic changes of the transcriptome during gut development. 2505 genes were differentially expressed in E14.5 embryonic epithelium versus adult ISCs (Figure 1G and Supplementary Tables S3 and S4), indicating that different transcriptional programs operate in these cell populations. Finally, over 4100 transcripts were significantly changed in enterocytes relative to the adult ISCs (Figure 1G and Supplementary Tables S5 and S6).
Figure 1.
Transcriptional programs during specification, maintenance and differentiation of the adult ISCs. (A) Immunostaining for EpCAM in E12.5 mouse small intestine. EpCAM (red) specifically labels epithelial cells. DAPI staining (blue) shows nuclei. (B) Fluorescence-Activated Cell Sorting (FACS) plot showing purification of EpCAM+ cells (red) from E12.5 mouse small intestine (n > 10). (C) Immunofluorescence for Lgr5-EGFP expressing adult ISCs (green). (D) FACS plot showing isolation of Lgr5-EGFPhigh adult ISCs (green) (n > 10). (E) Image of intestinal villi. (F) FACS plot showing isolation of EpCAM+CD31−CD45− enterocytes (red) (n > 10). Scale bar: 27 μm (A), 100 μm (C), 0.5 mm (E). (G) Pair-wise comparisons of gene expression between successive cell populations. The differentially expressed genes are shown in green or red. (H) Heat map showing clustering of differentially expressed genes for each cell population. (I–M) Representative examples of gene expression patterns for ‘enterocyte’ (I), ‘adult ISC’ (J), ‘proliferation’ (K), ‘embryonic epithelium’ (L) and ‘adult epithelium’ (M) signatures. The y-axis indicates the coverage normalized by library size (reads per million). (N–Q) Gene ontology analysis of ‘enterocyte’ (N), ‘proliferation’ (O), ‘embryonic epithelium’ (P) and ‘adult epithelium’ (Q) signatures.
Using hierarchical clustering of differentially expressed transcripts across all cell types, we identified five major groups of co-regulated genes (Figure 1H–M). Genes up-regulated or specifically expressed in enterocytes formed the largest group (2142 genes, 34%), here referred to as ‘enterocyte signature’ (Figure 1H and I). Consistent with the functions of absorptive enterocytes, Gene Ontology (GO) term analysis showed enrichment for protein transport, ion transport, fatty acid metabolism, mitochondria, Golgi apparatus and peroxisome (Figure 1N). The next group consisted of 1164 (19%) genes specifically down-regulated in the differentiated enterocytes compared to the embryonic epithelial cells and adult ISCs (Figure 1H). Many of these genes encoded cell cycle regulators representing a ‘proliferation signature’ (Figure 1K). Accordingly, GO analysis revealed enrichment for cell cycle progression, mitosis, centromere, DNA damage repair, ribosome biogenesis and ncRNA processing (Figure 1O). 1070 transcripts (17%) found in both E12.5 and E14.5 embryonic epithelium and down-regulated in the adult cell populations constituted the list of ‘embryonic epithelium signature’ (Figure 1H). This cluster included imprinted genes (Igf2-H19, Dlk1-Meg3-Rian and Peg3-Zim loci), factors regulating transcription (Dnmt3a, Dnmt3b, Sox11 and Tcf7l1), signalling molecules (Bmp2, Igf2, Ihh, Pdgfc and Shh) and multiple extracellular matrix proteins (Fn1, Fras1, Gjc1/Cx45 and Lama1) highlighting the importance of intercellular crosstalk during gut tube development (Figure 1L and P).
Notably, our analysis showed that genes down-regulated from E12.5 to E14.5 would either be even less expressed or stay at the same expression level during the adult stages (757 out of 900 genes, P < 10−20, hypergeometric test), indicating that the early embryonic transcriptional programs are not re-activated in adult cells. The genes whose expression increased in the adult small intestinal epithelium compared to the embryonic stages (628 genes, 10%) defined the ‘adult intestinal epithelium signature’ (Figure 1H and M). This cluster was the most highly enriched for antimicrobial response transcripts (Figure 1Q). Again, our data showed that genes up-regulated from E12.5 to E14.5 would either be further activated or stay at the same expression level in adults (524 out of 704 genes, P < 10−20, hypergeometric test), suggesting that an important switch establishing the adult transcriptional programs occurs already during embryogenesis, between E12.5 and E14.5.
The only exception was the group of ‘adult ISCs signature’ genes (395 genes, 6%), which were up-regulated in the adult ISCs but got silenced or strongly down-regulated in differentiated enterocytes (Figure 1H and J, and Supplementary Table S7). Based on transcriptome dynamics during embryogenesis we further subdivided the adult ISCs marker genes into two sub-clusters. The first group of genes (Lgr5, Axin2, Kcne3, Slc12a2 and Smoc2) was activated in the embryonic epithelium between E12.5 and E14.5 (Figure 1J). All these genes are known targets of Wnt signalling (24). The second group of genes (Olfm4, Kcnq1 and Lrig1) was up-regulated specifically in the adult ISCs. The expression of those genes is not directly controlled by Wnt signalling (24), implying at least two modes of transcriptional regulation for the ‘ISCs signature’ genes.
Multiple genetic studies have shown that an inhibition of Bmp pathway is important for the maintenance and proliferation of adult ISCs (25). Furthermore, Bmp signalling plays an important role during differentiation of the embryonic intestinal epithelium (26). Consistently, we detected higher expression of Bmp1 and Bmp2 as well as Bmpr1b and Bmpr2 receptors in the embryonic epithelium compared to the adult ISCs (Supplementary Tables S1–S3). Furthermore, in differentiated enterocytes Bmp1, Bmp2 and Bmp8a were upregulated compared to the adult ISCs (Supplementary Table S6), indicating that the expression of some Bmp ligands anti-correlates with adult ISC transcriptional program. In summary, we defined changes in transcriptional programs during specification, maintenance and differentiation of the adult ISCs and showed that the ‘ISC signature’ genes get activated during embryogenesis after E12.5.
Developmental regulators are the main targets of H3K27me3 mediated repression during ISC specification
The global patterns of histone modifications show little change during differentiation of the adult ISCs (14). We explored the potential importance of dynamic changes in histone marks during development and specification of the ISCs by mapping the repressive H3K27me3 as well as modifications associated with active transcription, H3K4me3 and H3K27Ac. We performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) using 2×105 FACS purified embryonic intestinal epithelial cells at two time points of development (E12.5 and E14.5), adult Lgr5+ ISCs and enterocytes. We identified over ten thousand regions positive either for H3K4me3 (peak width ≥ 450 bp, FDR ≤ 0.01) or for H3K27me3 (peak width ≥ 1000 bp, FDR ≤ 0.01) in each cell population. Over thirty thousand H3K27Ac positive sites (peak width ≥ 450 bp, FDR ≤ 0.01) were detected either in embryonic epithelium or adult ISCs, and only ∼19 000 sites were identified in differentiated enterocytes. Peaks called for each dataset were assigned to the genome features including promoters, 5΄ and 3΄ UTRs, exons, introns and distal elements, defined as regions positioned at least 3 kb from a gene body. Consistent with previous reports, 35–40% of H3K27me3 positive peaks were found at promoters, and a similar fraction at intergenic regions (Supplementary Figure S2A). Around 45% of H3K27Ac enriched sites were located within promoters, 22% within introns and 24% within intergenic regions (Supplementary Figure S2A). In contrast, around 86% of H3K4me3 positive peaks mapped to promoters while less than 8% were found in intergenic regions (Supplementary Figure S2A).
The majority of genes (80%) associated with H3K27me3 were transcriptionally silent at all stages examined (Supplementary Figure S2B). These include known Polycomb targets from the Hox, Dlx, Tbx, Six and Zic families (Supplementary Figure S2C-E). We therefore focused on the 6200 differentially expressed genes that we previously identified (Figure 1G and H). Among those, few genes were differentially marked by H3K27me3 between E12.5 and E14.5 (Figure 2A and D). Furthermore, we found moderate correlation between changes in H3K4me3 and H3K27Ac levels and gene expression between embryonic stages (Figure 2B–D). Specifically, hundreds of poised genes belonging to the ‘adult epithelium’ and ‘enterocyte’ clusters gained H3K4me3 at their promoters at E14.5 (Figure 2B, D and Supplementary Figure S2B). These genes were activated subsequently during ISC differentiation (Figure 2B and D), indicating that extensive changes of chromatin marks at TSS, establishing enterocyte specific transcriptional programs, take place already in embryonic epithelium.
Figure 2.
Dynamics of histone marks during specification and differentiation of ISCs. (A–C) Pair-wise comparisons showing changes in H3K27me3 (A), H3K4me3 (B) and H3K27Ac (C) levels at the TSS of differentially expressed genes between successive stages. (D) Heat maps showing distributions of H3K27me3 (blue), H3K4me3 (red) and H3K27Ac (magenta) at the TSS of differentially expressed genes for five transcriptional signatures defined by RNA expression. The heat maps were generated using ChIP/input fold enrichment ratios. (E and F) Representative examples of chromatin and gene expression profiles for ‘adult ISC’ (E) and ‘embryonic epithelium’ (F) signatures. The y-axis indicates the coverage normalized by library size (reads per million).
Strong changes in the distribution of H3K27me3 mark occurred during the transition from the embryonic progenitors to the adult ISCs (Figure 2A). In ISCs, we observed enrichment for H3K27me3 over the TSS of 616 differentially expressed genes. Two third (373 genes) of those had gained the mark in ISCs compared to E14.5 epithelium. Strikingly, one third of the genes belonging to the ‘embryonic epithelium’ cluster gained H3K27me3 over their promoters in ISCs (Figure 2D and F), suggesting a key role for this mark in silencing developmental regulators in adult cells. Many of these genes are essential for embryonic gut morphogenesis, with the most prominent being Prdm1 (27,28).
A quarter of genes (863) carrying H3K27me3 mark at their promoters was also positive for H3K4me3 in adult ISCs. The majority of genes with bivalent promoters was silent or expressed at very low levels in all intestinal cell types examined. In contrast to ESCs, promoters of the genes from the Hox, Dlx and Zic families were only H3K27me3 positive (Supplementary Figure S2C-E) both in embryonic and in adult intestinal cells. Interestingly, the majority of differentially expressed bivalent genes was from the ‘embryonic epithelium’ cluster (168 out of 328) (Figure 2F). In contrast, fewer genes (56 out of 328) belonged to the ‘enterocyte’ cluster. This suggests that in the adult ISCs (and in strong contrast with ESCs) bivalent H3K4me3/H3K27me3 promoters usually characterize genes that were expressed at an earlier developmental stage and were subsequently silenced in ISCs, rather than genes poised for activation upon differentiation. Of note, a subset of the genes classified as bivalent was expressed only in a subset of ISCs. Indeed, there are significant differences in gene expression profiles, amongst ISCs, along the anterior–posterior axis of the small intestine (16,29).
Half (317 out of 560 genes) of the differentially expressed genes positive for H3K27me3 in the E14.5 embryonic epithelium lost this mark during ISC specification (Figure 2A and D). Some of them (44 out of 317) belonged to the ISCs signature (Figure 2E). Notably, the pattern of H3K27me3 distribution was very diverse for the ISC signature genes. While Ascl2, Cd44, Notch1 and Smoc2 promoters were marked in embryos (Figure 2E), Lgr5 promoter was only marked in enterocytes (Supplementary Figure S2F), indicating that H3K27me3 mediated repression is gene and stage specific. Interestingly, the adult ISC signature genes showed very dynamic changes in H3K4me3 and H3K27Ac at their promoters during both specification and differentiation of the adult ISCs (Figure 2D–E and Supplementary Figure S2F), supporting the notion that chromatin changes could be involved in tightly coordinated transcription of the regulators of ISC specification and maintenance.
The other groups of genes that lost H3K27me3 mark during ISC specification belonged to the ‘adult intestine’ and ‘enterocyte’ classes, implying that some enterocyte specific genes lose this repressive mark prior to their activation, already in ISCs. Accordingly, we detected few genes regulated by H3K27me3 during differentiation of the ISCs towards the enterocyte lineage (Figure 2D). 218 genes lost H3K27me3 during ISCs differentiation towards enterocytes (Figure 2A), with the most prominent being the Cdkn2a/b gene cluster (Supplementary Figure S2G). On the other hand, 98 genes gained H3K27me3 in enterocytes (Figure 2A). Several ISC signature genes, including Lgr5, Msi1 and Slc12a2 were on that list (Supplementary Figure S2F). The majority of genes downregulated in enterocytes did not gain the H3K27me3 mark but lost H3K27 acetylation at their promoters (‘proliferation signature’, Figure 2C). In summary, strong changes in distribution of H3K27me3 mark take place during transition from embryonic progenitors to adult ISCs. A large proportion of genes essential for the development of the small intestine acquire H3K27me3 in adult ISCs, suggesting its role during ISCs specification.
‘ISC signature’ genes are the main targets of 5mC mediated repression in the embryonic epithelium
Only a small fraction of ISC signature genes showed differential methylation of H3K27 during the specification and differentiation of the intestinal stem cells. This prompted us to assess whether changes in DNA methylation, a distinct repressive mechanism, were associated with the regulation of ISC signature genes. Methylated regions of DNA from whole genome were isolated using Methyl Binding Domain pull-down followed by sequencing (MBD-seq). The size of DNA fragments was between 120 to 170 bp, which provides a resolution at nucleosome level. We have detected up to three hundred thousand 5mC positive regions in embryos (FDR≤0.01) and over two hundred thousand in adults, respectively. Since MBD-sequencing does not provide quantitative data and weakly methylated DNA regions (methylation ratio <0.2) are underrepresented in MDB pull-down experiments (30), we performed comparative analyses on a qualitative level, considering only the presence or absence of the peaks.
We observed few differences in DNA methylation profiles between two embryonic stages. 68 genes (7%) lost DNA methylation upon their activation from E12.5 to E14.5 (Figure 3A and Supplementary Figure S3A). These genes belonged to the ‘adult epithelium’ and ISC signature clusters. Only 15 genes showed gain in DNA methylation and were downregulated between E12.5 and E14.5 (Figure 3A). Consistent with previous studies (10,11) we observed very similar DNA methylation patterns between adult ISCs and enterocytes. Only 5 genes were differentially methylated in the adult cells (Figure 3A and Supplementary Figure S3B), indicating that the regulation of either stem cell or enterocyte specific genes does not rely on changes in DNA methylation status in the adult gut. Overall, changes in DNA methylation patterns were not only confined to few genes but also to one or two peaks per gene.
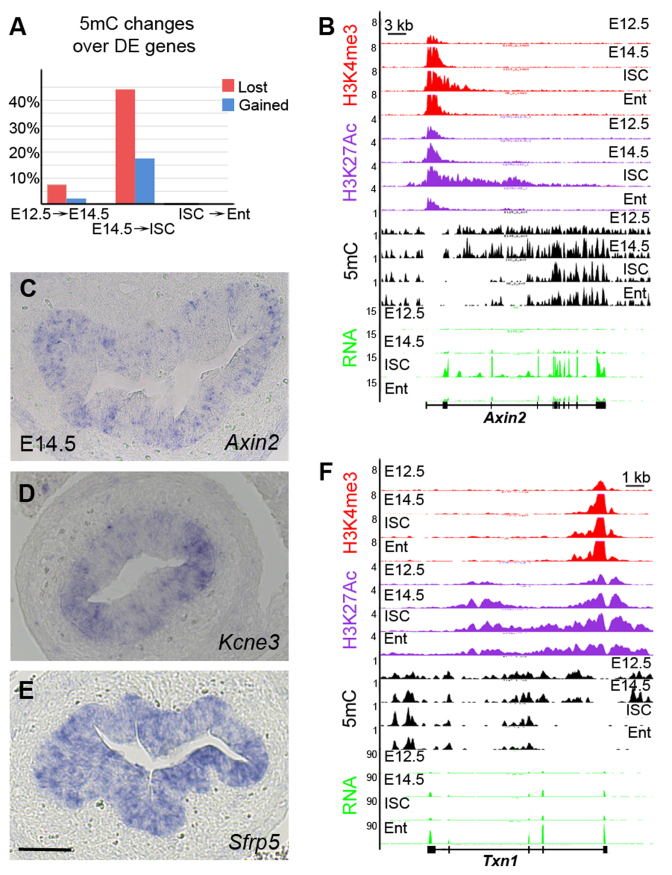
Figure 3.
Dramatic changes in DNA methylation during ISC specification. (A) Pair-wise comparisons showing changes in 5mC distribution over the differentially expressed genes between successive stages. Silent genes that lost 5mC mark upon activation are indicated in red, whereas genes that were downregulated and gained 5mC mark are indicated in blue. (B) Representative example of chromatin and gene expression profiles in four cell types for ISC marker Axin2. The y-axis indicates the coverage normalized by library size (reads per million). (C–E) Expression patterns of Axin2 (C), Kcne3 (D) and Sfrp5 (E) in the embryonic small intestine at E14.5. Only a subset of the embryonic intestinal epithelial cells expresses Axin2 or Kcne3, whereas Sfrp5 is expressed in all epithelial cells. Scale bar: 50 μM. (F) Representative example of chromatin and gene expression profiles in four cell types for enterocyte specific Txn1. The y-axis indicates the coverage normalized by library size (reads per million).
In striking contrast, 473 genes (44% of upregulated genes) lost DNA methylation during the transition from embryo at E14.5 to adult ISCs (Figure 3A). Importantly, differentially methylated regions were very large, spanning up to 20 kb or more in some cases (Figure 3B and Supplementary Figure S3C). Moreover, they were often located within the first intron of the genes. Loss of DNA methylation was accompanied by gain of H3K27Ac and H3K4me3 modifications over the same regions. The highest proportion of genes undergoing changes in DNA methylation patterns was found in the ISC signature cluster (114 out of 395 ISC signature genes, 29%). Between them were Olfm4, Axin2, Scl12a2, Kcne3 and Lrig1 (Figure 3B and Supplementary Figure S3C). During ISC differentiation, these genes were silenced without any changes in DNA methylation pattern but lost H3K27Ac at their TSS. Further comparative analysis of DNA methylation and H3K27Ac modification profiles revealed that the ISC signature genes carried both marks at E14.5, indicating that this group of genes were expressed only in a subset of embryonic epithelium. Indeed, RNA in situ hybridization on tissue sections showed that the ISC signature genes were activated in some embryonic epithelial cells (Figure 3C–E).
The ‘enterocyte’ (207 genes) and ‘adult epithelium’ clusters (152 genes) composed the rest of the genes losing DNA methylation in ISCs (Figure 3F), indicating that the chromatin landscapes are already prepared in ISCs for further transcriptional changes in differentiated progenies. A majority of genes that gained DNA methylation (66%, 166 genes) were embryonic genes silenced in adult ISCs (Supplementary Figure S3D). The number of embryonic genes silenced via DNA methylation was significantly lower compared to those marked by H3K27me3, underlying the importance of the H3K27me3 modification in this process. To conclude, strong changes in DNA methylation patterns underlie transcriptional changes during the switch from the embryonic progenitors to adult ISCs. These changes occurred most often in both promoter and intron regions of differentially expressed genes.
Enterocytes specific genes are associated with H2A.Z in stem cells
While acetylation of H3K27 strongly correlated with changes in gene expression, H3K27me3 and DNA methylation marks were more stable between ISCs and enterocytes. Therefore, we examined whether the distribution of H2A.Z (6) correlates with gene activity and could be implicated in the regulation of gene expression during both specification and differentiation of the stem cells in the gut epithelium. We detected over eleven thousand H2A.Z positive peaks in each cell type. 62% of H2A.Z positive regions we found at promoters, 8% within introns and 27% at distal elements (Supplementary Figure S4A). 70% of genes with H2A.Z promoter occupancy were transcriptionally active (Supplementary Figure S4B). Very little difference in distribution of H2A.Z was detected between two embryonic stages. Only 158 out of 4614 differentially expressed genes showed different occupancy of H2A.Z at their promoters (Figure 4A). During transition from embryonic to adult ISCs stage, 178 differentially expressed genes lost and 177 gained H2A.Z at their promoters (Figure 4A). These data indicate that the distribution of H2A.Z does not change significantly and does not correlate with strong transcriptional changes observed during ISC specification.
Figure 4.
H2A.Z levels decreases during ISC differentiation. (A) Pair-wise comparisons showing changes in H2A.Z levels at the TSS of differentially expressed genes between successive stages. (B and C) Immunostainings for H2A.Z (B, yellow) and H3K27Ac (C, yellow) in the adult mouse small intestine. H2A.Z levels are decreased in enterocytes (Ent) compared to the crypt cells. Red arrowheads point on the ISCs. In contrast, equal amounts of H3K27Ac are detected in both enterocytes and crypt cells. DAPI staining (blue) shows nuclei. Scale bar: 50 μM. (D) Representative example of chromatin and gene expression profiles in four cell types for Txn1. The y-axis indicates the coverage normalized by library size (reads per million).
During ISCs differentiation, we observed a great bias towards the loss of H2A.Z over promoter regions (896 lost versus 73 gained) (Figure 4A). Further histological analysis confirmed that H2A.Z levels were globally lower in the nuclei of enterocytes compared to ISCs (Figure 4B and C). Interestingly, 421 genes activated in enterocytes lost H2A.Z mark at their promoters, suggesting that the presence of H2A.Z negatively regulated their transcription in ISCs (Figures 3F and 4D). The majority of these genes (300 out of 421, 71%) were already H3K4me3 and H3K27Ac positive in ISCs and only a minor fraction (77 out of 421, 18%) gained those marks in enterocytes. The other group of genes that lost H2A.Z was Polycomb targets carrying H3K27me3 mark at their promoters in ISCs. Those genes (158 out of 616, 25%) were silent or expressed at low levels in both adult cell types but further lost H2A.Z and H3K4me3 upon differentiation. 51 out of 133 genes (38%, P < 10−4, hypergeometric test) downregulated in enterocytes and losing H2A.Z belonged to ISC signature genes (Supplementary Figure S4C), indicating that the expression of ISC signature genes is tightly regulated at multiple layers with involvement of histone modifications but also histone variants. Taken together, H2A.Z occupancy strongly correlates with the presence of H3K4me3 and H3K27Ac marks at gene promoters and correlates with active transcription. On the other hand, loss of H2A.Z seems to facilitate transcription of enterocyte specific genes during differentiation of the adult ISCs and suggests a repressive function for this mark in stem cells.
Dynamic chromatin landscapes at distal elements
The presence of H3K27Ac and H2A.Z in intergenic regions is indicative of putative distal enhancers (4,31). We observed very dynamic enrichment for H3K27Ac mark across embryonic and adult cell populations at distal sites (further than 3 kb from TSS) located near differentially expressed genes (Figure 5A). More than 30% of all regions were marked only in one cell type (Figure 5A). Consistent with the idea that H3K27Ac is present at active enhancers, the distribution of H3K27Ac at distal sites correlated with a higher transcriptional activity of nearby genes (Figure 5B, P < 10−4, ANOVA test). In contrast, genes linked only to H2A.Z positive regions did not show enhanced transcription (Figure 5B).
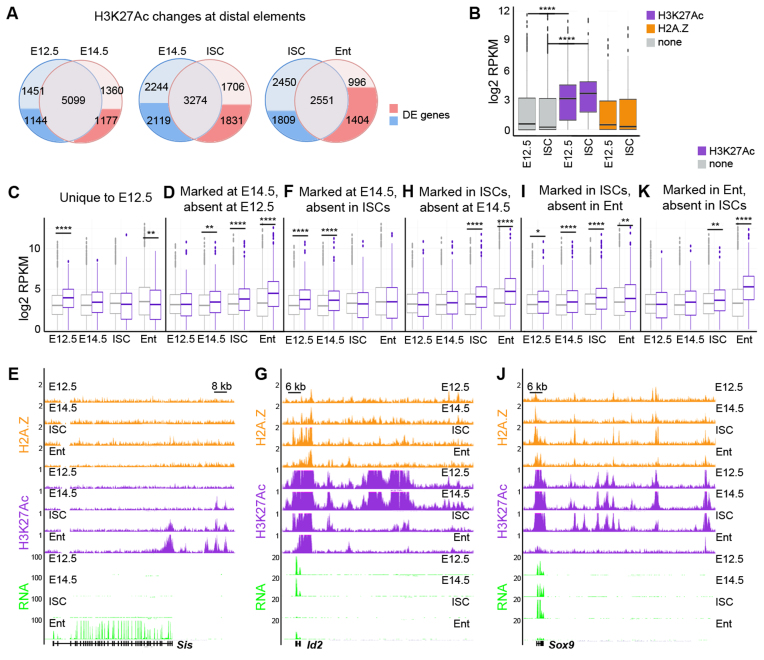
Figure 5.
Epigenetic changes at distal elements during specification and differentiation of the ISCs. (A) Pair-wise comparisons showing changes in H3K27Ac levels at distal elements between successive stages. The distal elements linked to differentially expressed genes are indicated in dark blue and red. (B) Box and whisker plots showing correlations of RNA expression levels (log2) with the presence of either H3K27Ac positive (magenta) or H2A.Z positive (orange) distal elements in all studied cell populations. Genes linked to H3K27Ac distal elements (magenta) are expressed at higher levels than non-linked genes (gray). The expression levels of genes linked to H2A.Z distal elements (orange) do not differ from non-linked genes (gray). ****P < 10−4, by ANOVA test. (C, D, F, H, I, K) Box and whisker plots showing correlations of RNA expression levels (log2) for genes linked to H3K27Ac positive distal elements (magenta) versus control genes (gray) at E12.5 (C), at E14.5 absent at E12.5 (D), at E14.5 absent in ISCs (F), in ISCs absent at E14.5 (H), in ISCs absent in Enterocytes (I) and in Enterocytes (K). Genes linked to H3K27Ac distal elements (magenta) are expressed at higher levels than not linked genes (gray). Furthermore, many distal elements are pre-marked by H3K27Ac prior to the expression of the linked genes, for example enterocyte specific genes at E14.5 (Ent in d) ****P < 10−10, **P < 10−4, *P < 10−2, by ANOVA test. (E, G, J) Representative examples of chromatin and gene expression profiles in four cell types for Sis (E), Id2 (G) and Sox9 (J) loci. The y-axis indicates the coverage normalized by library size (reads per million).
Comparative analysis linking H3K27Ac positive distal regions with differentially expressed genes showed that 34% of the regions unique for E12.5 were assigned to the genes specifically expressed at this developmental stage and downregulated in other cell types. Accordingly, genes immediately adjacent to H3K27Ac regions were expressed at significantly higher levels (P < 10−20, ANOVA test) at E12.5 compared to other stages and to the genes, which did not have H3K27Ac positive region in their proximity (Figure 5C). Interestingly, the same analysis showed that genes, which acquired H3K27Ac positive regions in their proximity at E14.5 (compared to E12.5), were expressed at higher levels at E14.5 but also in adult ISCs and enterocytes, when compared to the genes lacking H3K27Ac regions (Figure 5D, P < 10−4 for E14.5 and P < 10−20 for ISCs or enterocytes, ANOVA test). This suggests that some of the distal elements positive for H3K27Ac could be shared between E14.5 and adult stages. Indeed, we observed that 721 out of 1177 (61%) of distal elements, which gained H3K27Ac mark at E14.5, were linked to genes belonging to ‘enterocyte’ and ‘adult epithelium’ signatures (Figure 5E). Furthermore, some genes belonging to ISC signature, including Axin2, Ascl2, Cd44, Slc12a2, Olfm4 and Lrig1, showed enrichment of H3K27Ac on distal elements already at E14.5 (Supplementary Figure S5A-B). Half of H3K27Ac positive regions detected at E14.5 but not in adult ISCs were specific for embryonic epithelium (Figure 5A). 40% (846 out of 2119 elements) of embryo-specific peaks were linked to genes downregulated from E14.5 to adult ISCs (Figure 5F–G), including multiple imprinted genes as well as transcription factors critical for gut morphogenesis, such as Prdm1 and Id2 (16,27,28,32).
Consistent with the activation of numerous adult and stem cell specific genes, over 3500 H3K27Ac positive regions were detected in the adult ISCs compared to the embryonic stages (Figure 5A). Of those, 1831 were linked to differentially expressed genes. 17% of H3K27Ac regions were assigned to ‘adult epithelium’ signature genes and were shared between both adult cell types (Figure 5H). However, the largest fraction (817 out of 1831, 45%) of H3K27Ac positive distal elements was assigned to genes belonging to ‘enterocyte’ signature, indicating that many regulatory elements with potential function in enterocytes are already pre-marked in stem cells (Supplementary Figure S5C). Moreover, for genes linked to multiple H3K27Ac positive islands we observed cell type specific changes in regulatory element landscapes. For example, the enterocyte specific gene Sis was linked to two distal elements pre-marked already at E14.5 and in ISCs, and it acquired one more in enterocytes (Figure 5E). The TSS of the gene was H3K27Ac positive only in ISCs and enterocytes, indicating that the acquisition of H3K27Ac mark at distal elements preceded that at promoters. Accordingly, the expression of Sis was gradually increasing from E14.5 to ISCs (32 times) and to enterocytes (32 times) (Figure 5E).
12% of ISC specific H3K27Ac positive distal elements were assigned to ISC signature genes, including Olfm4, Lrig1, Prom1, Slc12a2, Smoc2, Edn1 and Axin2 (Supplementary Figure S5A-B). Consistent with the loss of expression for ISC signature genes during differentiation, H3K27Ac mark was lost over all these elements in enterocytes. The largest number (483 out of 1809, 27%) of H3K27Ac peaks observed in ISCs but not in enterocytes were assigned to ‘proliferation signature’ genes, such as Myc, Sox9 and Mki67 (Figure 5I, J and Supplementary Figure S5D). Interestingly, the Sox9 gene displayed different H3K27Ac-positive distal elements in its vicinity, when comparing embryos and adult ISCs (Figure 5J), suggesting the involvement of different regulatory programs controlling Sox9 expression in these cell types. Moreover, a comparative analysis of the intestinal and hair follicle stem cells (HFSCs) (33) revealed different H3K27Ac landscapes around Sox9. While H3K27Ac marked over ten distal elements in ISCs, only three H3K27Ac positive regions were identified in HFSCs (Supplementary Figure S5E). Two of those were overlapping with the intestinal epithelium enhancers. This suggests that the transcriptional regulation of the common stem cell marker Sox9 might be partly shared, yet it differs between intestinal and skin lineages.
In agreement with the activation of enterocyte specific genes during ISCs differentiation, 996 out of 1404 (71%) H3K27Ac positive regions were detected specifically in enterocytes and were linked to ‘enterocyte signature’ genes (Figure 5K). Thus, our data demonstrate that the specification and differentiation of the adult ISCs is accompanied by dynamic changes in H3K27Ac at distal regulatory elements. Importantly, analysis of H3K27Ac positive distal regions revealed that the enterocyte specific transcriptional landscape is pre-established already during embryogenesis at E14.5.
Recent studies shed light on the relationship between DNA methylation states and enhancer activity in cultured epithelial and cancer cells (34–36). We therefore analysed distribution of 5mC and 5-hydroxymethylcytosine (5hmC) (37) at H3K27Ac positive distal elements in intestinal epithelial cells. Here, we considered that at least one MBD/ MeDIP peak (150 bp) must overlap with H3K27Ac positive region. We found that 15–22% of H3K27Ac positive distal elements overlapped with 5mC peaks (Supplementary Figure S6A), which is in agreement with the known antagonistic relationship between H3K27Ac and DNA methylation (38). Over 75% of H3K27Ac/5mC distal elements were found in every cell type. We focused then on the cell type specific distal elements (∼300 elements). This analysis did not reveal either significant enrichment or loss for any gene signature linked to double positive H3K27Ac/5mC compared to H3K27Ac distal elements in any cell type examined. In contrast to 5mC, we found that 58% of H3K27Ac distal elements overlapped with 5hmC peaks in ISCs and differentiated enterocytes (Supplementary Figure S6B), indicating that 5hmC correlates well with the presence of H3K27Ac at distal elements and with the transcriptional activity of the linked genes. Moreover, 60–70% of H2K27Ac/5hmC double positive distal elements were cell type specific, suggesting that double positive H3K27Ac/5hmC elements might play an important role during maintenance and differentiation of ISCs.
H2A.Z and H3K27Ac mark distinct classes of distal elements
Next, we investigated whether changes in H2A.Z correlated with different enrichment of H3K27Ac at distal elements and with alterations in gene expression. Strong differences (over 50% of the peaks) in H2A.Z distribution at distal elements were found between embryonic and adult stages as well as between ISCs and differentiated enterocytes (Figure 6A). In contrast, only 1220 (10% of H2A.Z positive regions) regions were different between E12.5 and E14.5 (Figure 6A). Similar to H3K27Ac, most of the distal elements (665 out of 1713, 39%), which gained H2A.Z in ISCs, were assigned to genes belonging to ‘enterocyte signature’ (Supplementary Figure S5C). Furthermore, genes (547 out of 1920, 28%) that lost H2A.Z at distal elements and were downregulated in enterocytes belonged to the ‘proliferation’ signature (Supplementary Figure S5D). Consistent with the global loss of H2A.Z in enterocytes, only few regions linked to differentially expressed genes gained this histone variant upon activation.
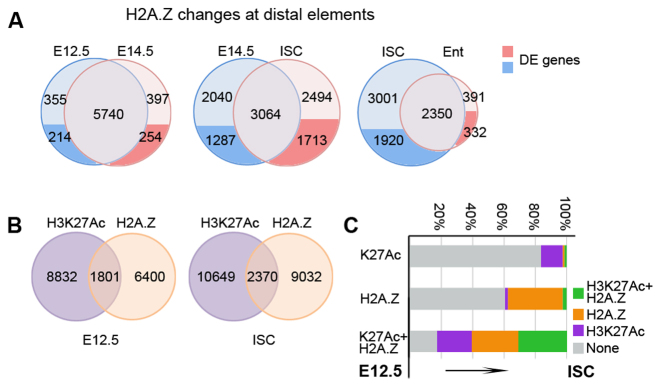
Figure 6.
H3K27Ac and H2A.Z mark distinct set of inter and intragenic elements. (A) Pair-wise comparisons showing changes in H2A.Z levels at distal elements between successive stages. The distal elements linked to differentially expressed genes are indicated in dark blue and red. (B) Overlap between stage specific H3K27Ac and H2A.Z positive distal elements. (C) Changes of chromatin marks at distal elements during transition from embryonic epithelium to adult ISCs. The distal elements covered by a certain chromatin mark at E12.5 are displayed on the left y-axis. The percentage of distal elements in ISCs is shown on x-axis. Most regions positive for either H3K27Ac or H2A.Z at E12.5 are negative for both marks in ISCs (grey). In contrast, most distal elements double positive at E12.5 stay either double positive (green) or become single H3K27Ac (magenta), or H2A.Z (orange) positive in ISCs.
The strong overlap between H2A.Z and H3K27Ac at TSS suggests that these chromatin marks could also coincide at distal regions. However, our analysis identified that only 22% of H2A.Z positive distal regions, including both inter and intragenic, showed an overlap with H3K27Ac peaks (Figure 6B). This was true for both differentially expressed as well as stably expressed genes in all cell types. For example, 57% of differentially expressed genes were linked to both H2A.Z and H3K27Ac positive regions, which were not overlapping, suggesting that the majority of H2A.Z and H3K27Ac distal elements could function independently for the regulation of gene expression. We further tested how H2A.Z, H3K27Ac and double positive regulatory regions evolve during transition from embryonic epithelium to the adult ISCs. The majority of regions labelled only by one mark, either H2A.Z (60%) or H3K27Ac (83%), in embryonic epithelium lost the mark in adult ISCs (Figure 6C). Furthermore, less than 2% of single labelled regions acquired a second mark in adult ISCs, indicating that the majority of double positive regulatory elements acquired both H2A.Z and H3K27Ac during the specification of the adult ISCs de novo. Interestingly, the double positive regions found in embryos remained either double (31%) or single H2A.Z (30%), or H3K27Ac (22%) positive in ISCs (Figure 6C), indicating that these regions are more stable between embryonic and adult stages. GO term analysis revealed that double positive distal elements were linked to genes encoding factors regulating transcription (P < 10−14 for E12.5 and P < 10−9 for ISCs). In summary, out data show that in contrast to TSS, H3K27Ac and H2A.Z mark different sets of inter- and intragenic distal regulatory elements.
DISCUSSION
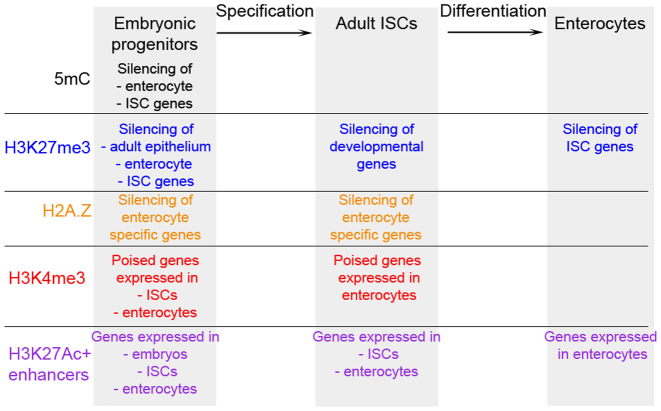
In this study, we report the first genome-wide characterization of the transcriptome and chromatin profiles of the embryonic intestinal epithelium prior to and during the specification of adult ISCs. Our results reveal highly dynamic changes at the molecular level during gut development. We define sets of signature genes associated with different stages of ISCs specification, maintenance and differentiation. Furthermore, we show that large-scale epigenetic changes in the distribution of modified histones, the histone variant H2A.Z and DNA methylation in genic and intergenic regions accompany the specification and differentiation of the adult intestinal stem cells (Figure 7). Our results suggest that DNA methylation greatly impacts specification of ISCs from embryonic progenitors by negatively regulating the expression of ISC signature genes. Furthermore, Polycomb mediated repression is also likely critical for the specification of ISCs during embryogenesis since a substantial number of developmental genes acquires H3K27me3 mark at their promoters in ISCs. Strikingly, we found that the repression of ISC signature genes as well as the activation of enterocyte specific genes was associated with decrease in H2A.Z levels during ISCs differentiation. Moreover, we observed strong changes in the distribution of histone modifications associated with transcriptional activity at putative regulatory regions during specification and differentiation of the ISCs. Unexpectedly, we detected little overlap between H3K27Ac and H2A.Z positive distal regulatory regions both in embryonic progenitors and adult ISCs.
Figure 7.
Epigenetic changes during the specification and differentiation of ISCs. In embryonic progenitors, ISC signature, adult epithelium and enterocyte specific genes are repressed by DNA methylation (5mC) or Polycomb complexes (H3K27me3). On the other hand, promoters of ISC and enterocyte specific genes are poised for activation and marked by H3K4me3. Moreover, distal elements linked to embryonic, ISC and enterocyte specific genes are H3K27Ac positive. Upon specification, ISC signature, adult epithelium and enterocyte specific genes lose 5mC and H3K27me3, whereas developmental genes get silenced and acquire H3K27me3. During differentiation of the adult ISCs, while enterocyte specific genes lose H2A.Z and get activated, ISC signature genes get silenced.
Epigenetic changes during the specification of the adult ISCs
Lgr5+ intestinal stem cells became a classical model system to study mechanisms of tissue maintenance and differentiation. Yet, the mechanisms of ISCs specification during embryogenesis are poorly understood. Our data show that distinct transcriptional programs operate in the embryonic intestinal epithelium compared to the adult ISCs. Both the gene expression and chromatin profiles revealed that ISC signature genes are activated after E12.5. Proportional to transcriptional activity we detected an increase in the levels of H3K27Ac at the TSS of ISC signature genes at E14.5. Simultaneously, we observed the presence of either methylated DNA or H3K27me3 mark at the promoters of these genes at both embryonic stages but not in the adult ISCs. This indicates that ISC signature genes are activated only in a subset of embryonic epithelial cells by E14.5, which we further confirmed by RNA in situ hybridization.
Our chromatin and transcriptome data demonstrate that in embryonic epithelium different ISC signature genes have different modes of repression, such as DNA methylation (Axin2, Scl12a2, Kcne3, Lrig1 and Olfm4), H3K27me3 modification (Ascl2, Cd44, Nfia and Smoc2) or none of them (Lgr5) implying that distinct mechanisms likely operate for their activation. Furthermore, some of the ISC signature genes were already expressed at E14.5 whereas the others were detected only in adult ISCs. This step-wise acquisition of the ISC transcriptional program suggests that their specification requires multiple mechanisms working together to establish ISC identity. The extensive changes in DNA methylation patterns, spanning up to 20 kb, over developmental and ISC signature genes make this epigenetic mark a candidate for mediating ISC specification during embryogenesis. Another major feature of ISC specification is the repression of a considerable number of developmental genes and gain of H3K27me3 over their promoters. Therefore, Polycomb mediated repression might play an essential role for the correct specification of adult ISCs.
Epigenetic changes during the maintenance and differentiation of adult ISCs
The changes in chromatin states during specification of ISCs are in strong contrast to the chromatin landscapes in adult cells. In the small intestine over 4000 genes (almost half of the whole active transcriptome) are differentially expressed between stem cells and enterocytes. However, the profiles of chromatin marks associated with gene repression are very similar between stem cells and differentiated enterocytes. Our results show that all enterocyte specific genes lose DNA methylation already during ISC specification. Furthermore, many enterocyte specific genes lost H3K27me3 mark already in ISCs. The most prominent exception was the Cdkn2a/b cluster, encoding inhibitors of cell cycle progression. Therefore, our data suggest that chromatin landscapes operating in differentiated enterocytes are established during the specification of embryonic progenitors towards the adult ISCs. In contrast, genes essential for differentiation along the secretory lineage, including Neurod1, Neurog3 and Atoh1, were H3K27me3 positive at all stages examined, indicating that Polycomb mediated repression prevents differentiation of the ISCs towards secretory cells (13).
A prominent loss of H2A.Z over promoters of enterocyte specific genes upon differentiation implies that this histone variant could play repressive functions in the ISCs, when DNA methylation and H3K27me3 patterns do not change much. Remarkably, the promoters of these enterocyte specific genes were both H3K4me3 and H3K27Ac positive in ISCs, indicating that they could be poised by H2A.Z for further activation. Moreover, the majority of these enterocyte specific genes was H2A.Z positive in embryonic epithelium, implying that H2A.Z could prevent precocious differentiation of the embryonic progenitors towards enterocytes during development. A recent study in C. elegans revealed that H2A.Z is required for the repression of DREAM (Dimerization Partner, RB-like, E2F and multi-vulval class B) complex target genes, which regulate G2/M-specific progression during the cell cycle (39). It is possible that a certain class of transcription factors in concert with H2A.Z coordinates repression of enterocyte specific genes in ISCs and in the embryonic epithelium. Consistent with the genome-wide decrease in the number of H2A.Z positive regions, we have observed a loss of H2A.Z at the transcriptional and protein levels specifically in enterocytes. Whether loss of H2A.Z is a general mark of differentiation remain unknown. However, it has been shown that H2A.Z levels are elevated in various cancers (40–42).
Stage-specific activation of putative distal regulatory elements
Our time course analysis of potential distal regulatory elements revealed very dynamic regulatory landscapes directing biological processes essential for the specification, maintenance and differentiation of the ISCs. Thousands of stage specific H3K27Ac and H2A.Z positive distal elements were identified at each time point indicating that chromatin marks at the regulatory elements are more transient than at the promoters. For instance, genes belonging to the proliferation signature were downregulated and lost H2A.Z at the associated distal elements in enterocytes. However, H2A.Z occupancy at the promoters of these genes was constant. The stage specific differences in the distribution of H3K27Ac and H2A.Z over distal regulatory regions reflected very well the transition from embryonic progenitors to ISCs, which takes place between E12.5 to E14.5. On the other hand, many distal regulatory elements linked to enterocyte specific genes acquired H3K27Ac and H2A.Z marks either at E14.5 or in ISCs, indicating that not only repressive but also activating chromatin marks pre-establish enterocyte specific transcriptional landscape already in ISCs.
The strong correlation between the presence of H3K27Ac positive distal elements and the activity of the nearest in silico associated genes was remarkable. This correlation was observed for all gene clusters at specific developmental or differentiation stage, reflecting very well underlying biological processes. For example, the distal elements marked by H3K27Ac in ISCs but not in embryonic epithelium were in close proximity and associated with the genes belonging to the ISC signature, adult epithelium and enterocyte clusters. Consistently, these genes were expressed in the adult tissues. In contrast, the distal elements marked by H3K27Ac in ISCs but not in enterocytes were connected to the ISC signature and proliferation clusters. Accordingly, genes positively regulating proliferation were transcribed in all cell types except of differentiated enterocytes. Our data thus indicate that H3K27Ac positive distal elements are good indicators of cell identity as well as its proliferation and differentiation status.
H3K27Ac and H2A.Z marked distinct and largely non-overlapping sets of distal elements. A majority of genes were linked to both H3K27Ac and H2A.Z positive elements. Many genes encoding transcription factors essential for intestinal development and homeostasis were linked to double (overlapping) H3K27Ac/H2A.Z positive regions. Accordingly, these elements were more stable between different stages. Interestingly, genes that were linked only to H2A.Z positive distal elements were either silent or expressed at low levels at all stages examined suggesting that, in the absence of H3K27Ac, H2A.Z could mark silent or poised regulatory elements. However, only a negligible fraction (2%) of H2A.Z positive regions acquired H3K27Ac during the transition from embryonic to ISC stage, leaving the question about their potential functions open.
Our analyses showed that most H3K27Ac positive (active) distal elements overlapped with 5hmC peaks in ISCs and differentiated enterocytes. Importantly, the majority of H2K27Ac/5hmC double positive distal elements was cell type specific suggesting that double positive H3K27Ac/5hmC elements might play an important role for tissue specific regulation of gene expression. In contrast, only a minor fraction of H3K27Ac positive distal elements overlapped with 5mC peaks in embryonic progenitors, adult ISCs and differentiated enterocytes. The majority of H3K27Ac/5mC double positive distal elements were not cell type specific. In agreement with our data, the comparative analyses of epigenetic and transcriptional changes accompanying differentiation of skin stem cells revealed few distal elements decorated by both H3K27Ac and 5mC marks (36). In skin stem cells depletion of Dnmt3b, which is responsible for positioning of 5mC at both gene bodies and enhancers, leads to decrease in transcriptional activity of many genes (36). However, it is not known whether depletion of 5mC at enhancers or gene bodies affects transcription of the target genes. Thus, functions of 5mC at distal elements remain poorly understood.
Of note, in the adult intestine both ISCs and enterocytes have a large number of stage specific H3K27Ac distal elements. In contrast, during differentiation of the skin stem cells over 10 000 H3K27Ac positive elements are lost and only 110 are gained specifically in keratinocytes (36), suggesting that there are very important differences between gene regulatory programs operating during differentiation of the adult intestinal and skin stem cells. In summary, our results suggest that profound changes in chromatin states are likely critical for controlling the specification of the ISCs during embryogenesis and their differentiation along the absorptive lineage in the adult gut.
ACCESSION NUMBERS
The datasets supporting the conclusions of this article are available in the NCBI Gene Expression Omnibus repository [NCBI GEO: GSE89684].
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Hartwig and I. Schaefer (Cytometry platform), M. Mendez-Lago, C.-T. Han, C. Werner and H. Lukas (Genomics platform), and S. Ritz and M. Hanulova (Microscopy platform). L. Fauk, K. Weiser and T. Dehn for assistance with mouse breeding. We thank T. Montavon for valuable comments on the manuscript.
Author contributions: J.K., B.M., C.K. and N.S. performed experiments and analysed the data. S.S. performed computational analysis of the data. N.S. designed the study and wrote the manuscript with inputs from J.K. and S.S.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Boehringer Ingelheim Foundation; EU Marie Curie CIG program [PCIG12-GA-2012-333859 FaME]; University of Mainz (to N.S.). Funding for open access charge: IMB core funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Allis C.D., Jenuwein T.. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016; 17:487–500. [DOI] [PubMed] [Google Scholar]
- 2. Lee D.Y., Hayes J.J., Pruss D., Wolffe A.P.. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993; 72:73–84. [DOI] [PubMed] [Google Scholar]
- 3. Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A.. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007; 130:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Piunti A., Shilatifard A.. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016; 352:aad9780. [DOI] [PubMed] [Google Scholar]
- 6. Weber C.M., Henikoff S.. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014; 28:672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu G., Cui K., Northrup D., Liu C., Wang C., Tang Q., Ge K., Levens D., Crane-Robinson C., Zhao K.. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013; 12:180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012; 13:484–492. [DOI] [PubMed] [Google Scholar]
- 9. Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013; 154:274–284. [DOI] [PubMed] [Google Scholar]
- 10. Sheaffer K.L., Kim R., Aoki R., Elliott E., Schug J., Burger L., Schübeler D., Kaestner K.H.. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014; 28:652–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaaij L.T., van de Wetering M., Fang F., Decato B., Molaro A., van de Werken H.J., van Es J.H., Schuijers J., de Wit E., de Laat W., Hannon G.J. et al. DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol. 2013; 14:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Camp J.G., Frank C.L., Lickwar C.R., Guturu H., Rube T., Wenger A.M., Chen J., Bejerano G., Crawford G.E., Rawls J.F.. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. 2014; 24:1504–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiacchiera F., Rossi A., Jammula S., Zanotti M., Pasini D.. PRC2 preserves intestinal progenitors and restricts secretory lineage commitment. EMBO J. 2016; 35:2301–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jadhav U., Nalapareddy K., Saxena M., O’Neill N.K., Pinello L., Yuan G.C., Orkin S.H., Shivdasani R.A.. Acquired tissue-specific promoter bivalency is a basis for PRC2 necessity in adult cells. Cell. 2016; 165:1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koppens M.A., Bounova G., Gargiulo G., Tanger E., Janssen H., Cornelissen-Steijger P., Blom M., Song J.Y., Wessels L.F., van Lohuizen M.. Deletion of polycomb repressive complex 2 from mouse intestine causes loss of stem cells. Gastroenterology. 2016; 151:684–697. [DOI] [PubMed] [Google Scholar]
- 16. Nigmatullina L., Norkin M., Dzama M.M., Messner B., Sayols S., Soshnikova N.. Id2 controls specification of Lgr5+ intestinal stem cell progenitors during gut development. EMBO J. 2017; doi:10.15252/embj.201694959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang L., Wang S., Li W.. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012; 28:2184–2185. [DOI] [PubMed] [Google Scholar]
- 18. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2012; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:R550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang D.W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A.. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007; 8:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007; 449:1003–1007. [DOI] [PubMed] [Google Scholar]
- 24. Clevers H., Loh K.M., Nusse R.. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014; 346:1248012. [DOI] [PubMed] [Google Scholar]
- 25. He X.C., Zhang J., Tong W.G., Tawfik O., Ross J., Scoville D.H., Tian Q., Zeng X., He X., Wiedemann L.M. et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 2004; 36:1117–1121. [DOI] [PubMed] [Google Scholar]
- 26. Haramis A.P., Begthel H., van den Born M., van Es J., Jonkheer S., Offerhaus G.J., Clevers H.. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004; 303:1684–1686. [DOI] [PubMed] [Google Scholar]
- 27. Harper J., Mould A., Andrews R.M., Bikoff E.K., Robertson E.J.. The transcriptional repressor Blimp1/Prdm1 regulates postnatal reprogramming of intestinal enterocytes. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10585–10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muncan V., Heijmans J., Krasinski S.D., Büller N.V., Wildenberg M.E., Meisner S., Radonjic M., Stapleton K.A., Lamers W.H., Biemond I. et al. Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat. Commun. 2011; 2:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X., Yamamoto Y., Wilson L.H., Zhang T., Howitt B.E., Farrow M.A., Kern F., Ning G., Hong Y., Khor C.C. et al. Cloning and variation of ground state intestinal stem cells. Nature. 2015; 522:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harris R.A., Wang T., Coarfa C., Nagarajan R.P., Hong C., Downey S.L., Johnson B.E., Fouse S.D., Delaney A., Zhao Y. et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010; 28:1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ku M., Jaffe J.D., Koche R.P., Rheinbay E., Endoh M., Koseki H., Carr S.A., Bernstein B.E.. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 2012; 13:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Russell R.G., Lasorella A., Dettin L.E., Iavarone A.. Id2 drives differentiation and suppresses tumor formation in the intestinal epithelium. Cancer Res. 2004; 64:7220–7225. [DOI] [PubMed] [Google Scholar]
- 33. Adam R.C., Yang H., Rockowitz S., Larsen S.B., Nikolova M., Oristian D.S., Polak L., Kadaja M., Asare A., Zheng D. et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015; 521:366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Charlet J., Duymich C.E., Lay F.D., Mundbjerg K., Dalsgaard Sørensen K., Liang G, Jones P.A.. Bivalent regions of cytosine methylation and H3K27 acetylation suggest an active role for DNA methylation at enhancers. Mol. Cell. 2016; 62:422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. King A.D., Huang K., Rubbi L., Liu S., Wang C.Y., Wang Y., Pellegrini M., Fan G.. Reversible regulation of promoter and enhancer histone landscape by DNA methylation in mouse embryonic stem cells. Cell Rep. 2016; 17:289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rinaldi L., Datta D., Serrat J., Morey L., Solanas G., Avgustinova A., Blanco E., Pons J.I., Matallanas D., Von Kriegsheim A. et al. Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell. 2016; 19:491–501. [DOI] [PubMed] [Google Scholar]
- 37. Kim R., Sheaffer K.L., Choi I., Won K.J., Kaestner K.H.. Epigenetic regulation of intestinal stem cells by Tet1-mediated DNA hydroxymethylation. Genes Dev. 2016; 30:2433–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bartke T., Vermeulen M., Xhemalce B., Robson S.C., Mann M., Kouzarides T.. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010; 143:470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Latorre I., Chesney M.A., Garrigues J.M., Stempor P., Appert A., Francesconi M., Strome S., Ahringer J.. The DREAM complex promotes gene body H2A.Z for target repression. Genes Dev. 2015; 29:495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hua S., Kallen C.B., Dhar R., Baquero M.T., Mason C.E., Russell B.A., Shah P.K., Liu J., Khramtsov A., Tretiakova M.S. et al. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol. Syst. Biol. 2008; 4:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vardabasso C., Gaspar-Maia A., Hasson D., Pünzeler S., Valle-Garcia D., Straub T., Keilhauer E.C., Strub T., Dong J., Panda T. et al. Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol. Cell. 2015; 59:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang H.D., Kim P.J., Eun J.W., Shen Q., Kim H.S., Shin W.C., Ahn Y.M., Park W.S., Lee J.Y., Nam S.W.. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget. 2016; 7:11412–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.