Abstract
Short interspersed nuclear elements (SINEs) are retrotransposons evolutionarily derived from endogenous RNA Polymerase III RNAs. Though SINE elements have undergone exaptation into gene regulatory elements, how transcribed SINE RNA impacts transcriptional and post-transcriptional regulation is largely unknown. This is partly due to a lack of information regarding which of the loci have transcriptional potential. Here, we present an approach (short interspersed nuclear element sequencing, SINE-seq), which selectively profiles RNA Polymerase III-derived SINE RNA, thereby identifying transcriptionally active SINE loci. Applying SINE-seq to monitor murine B2 SINE expression during a gammaherpesvirus infection revealed transcription from 28 270 SINE loci, with ∼50% of active SINE elements residing within annotated RNA Polymerase II loci. Furthermore, B2 RNA can form intermolecular RNA–RNA interactions with complementary mRNAs, leading to nuclear retention of the targeted mRNA via a mechanism involving p54nrb. These findings illuminate a pathway for the selective regulation of mRNA export during stress via retrotransposon activation.
INTRODUCTION
Approximately 10% of mammalian genomic sequence consists of repetitive elements referred to as short interspersed nuclear elements (SINEs) (1). SINEs are non-autonomous, noncoding retrotransposons transcribed by RNA Polymerase III (RNA Pol III). Alu elements are the predominant SINE family in the human genome, while the murine genome harbors two major and unrelated SINE families, B1 and B2.
Both human and murine SINEs are evolutionarily derived from endogenous RNA Pol III transcripts. Though Alu and B1 elements are both derived from 7SL RNA, the RNA component of the signal recognition particle, they evolved independently of each other following the primate-rodent split (2–4). Conversely, B2 elements are derived from transfer RNA (tRNA) (5). Human and murine SINE elements have independent evolutionary origins, but they exhibit a similarly biased genomic distribution in gene-rich regions and a preference for an antisense orientation within genes (6–8). The evolutionary convergence of both SINE location and orientation suggests a functional advantage, perhaps at the level of gene expression regulation.
Although traditionally viewed as parasitic elements, numerous functions related to gene regulation and pathogenesis have been discovered for both silent and transcriptionally active SINE loci. At the genomic sequence level, SINEs have evolved into a variety of regulatory elements including enhancers and transcription factor binding sites (9–16). SINEs located within genes can also impact the fate of the transcribed gene, even if they are not independently expressed. For example, when these ‘embedded’ SINEs are located within RNA Pol II-transcribed RNAs, they can serve as polyadenylation signals (17,18). Additionally, they can form intra- or intermolecular complementary base pair interactions with other embedded SINE sequences. Such interactions can promote nuclear retention of mRNA (19), as well as facilitate mRNA or long ncRNA degradation via the Staufen-mediated RNA decay (SMD) pathway (20–22).
In germ cells and during early embryonic development, SINE elements are transcriptionally active (23). In contrast, in healthy somatic cells, SINE elements are typically transcriptionally repressed. However, SINE ncRNA levels rapidly and robustly accumulate in response to a variety of stresses, including heat shock, noxious chemicals, and viral infection, and functions for these ncRNAs are beginning to be discovered (24–30). For example, Alu and B2 ncRNAs induced during heat shock can repress gene expression by interacting directly with RNA Pol II and preventing it from establishing contacts with the promoter during closed complex formation (31–33). Upon entering the cytoplasm, SINE ncRNAs activate components of the innate immune response linked to inflammation, and thus their constitutive expression can lead to pathogenic outcomes such as age-related macular degeneration (AMD) (34,35). SINE ncRNAs are robustly induced by multiple viruses, and we recently demonstrated that one outcome of this induction during murine gammaherpesvirus 68 (MHV68) infection is activation of NF-κB, a component of the innate immune response (30). Interestingly, in the case of MHV68 infection the virus hijacks the IKKβ kinase from the NF-κB pathway to stimulate viral transcription (36), and thus SINE induction benefits the viral lifecycle (30). Presumably, additional functions exist for SINE ncRNAs in cell signaling and/or individual or global gene regulation.
The mechanisms behind SINE transcriptional regulation are not fully understood, although both DNA CpG methylation and trimethylation of histone H3 (H3K9me3) have been implicated (37–41). However, it is unclear how many of the individual SINE loci retain transcriptional potential, or whether the strong positional and orientation bias of SINEs impacts SINE RNA transcription or function. These questions cannot be addressed without a strategy to globally monitor repetitive element expression that discriminates between RNA Pol III-derived SINE RNAs and SINEs that are embedded within RNA Pol II transcripts but not independently expressed as ncRNAs.
Here, we present an approach to selectively profile RNA Pol III-derived SINE RNA, and use it to characterize B2 SINE expression induced upon infection with the gammaherpesvirus MHV68. Our analysis revealed 28 270 transcriptionally active B2 loci, many of which reside within annotated RNA Pol II genes. Transcribed SINE RNAs could impact gene expression in a variety of ways, and we demonstrate that one such mechanism involves intermolecular RNA–RNA interactions between SINE RNA and complementary mRNAs, leading to nuclear retention of the targeted mRNA. Thus, transcriptional activation of retrotransposon elements can impact the trafficking of individual mRNAs. Collectively, our findings provide a platform for analyzing how activation of specific SINE loci contributes to cellular gene regulation during stress.
MATERIALS AND METHODS
Cells, viruses, and infections
NIH 3T3 and NIH 3T12 mouse embryo fibroblasts were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) with 10% fetal calf serum (FBS; Invitrogen, HyClone). The green fluorescent protein (GFP)-expressing WT MHV68 bacterial artificial chromosome (BAC) and the MHV68 ORF37 R443I (ΔHS) point mutant BAC that produces virus defective for mRNA degradation activity were previously described (42,43). MHV68 BAC-derived virus was produced by transfecting BAC DNA into NIH 3T3 cells using SuperFect (Qiagen). Virus was then amplified in NIH 3T12 cells and titered by plaque assay on NIH 3T3 cells. For infections, NIH 3T3 cells were incubated with virus at a multiplicity of infection of 5 for 1 h at 37°C. The media was then removed and cells were washed with 1X PBS two times and then replaced with growth media and grown for an additional 24 h prior to harvesting. Where indicated, the RNA Polymerase III inhibitor ML-60218 (EMD Millipore; 40 μM) was added to cells 4 h prior to infection and maintained throughout the course of the experiment.
Nucleic acid isolation, quantitation and A-to-I editing analysis
For analysis of gene expression by RT-qPCR, total RNA was isolated with TRIzol (Invitrogen) in accordance with the manufacturer's instructions. cDNA was synthesized from 1 μg of RNA with random hexamers (Integrated DNA Technologies) and SuperScript II reverse transcriptase (Invitrogen). qPCR was performed using the DyNAmo ColorFlash SYBR green qPCR kit (Thermo Scientific) with appropriate primers (Supplementary Table S1).
For small RNA northern blot analysis, total RNA was separated on 8% polyacrylamide–7 M urea gels and electrotransferred at 4°C to Amersham Hybond-N+ membranes in 0.5× TBE buffer for 16 h at 15 V. Membranes were probed using 32P-end labeled probes (Supplementary Table S1) overnight at 55°C. Blots were washed three times in 0.1X SSC for 10 min each before exposed to phosphoimager screens overnight. For mRNA northern blot analysis total RNA was resolved on 1.2% agarose-formaldehyde gels and transferred to Hybond-N+ membranes by capillary action.
For primer extension, 15 μg of total RNA was incubated with 6 pmol 32P-labeled primer (Supplementary Table S1) in 10 μl of Buffer A (250 mM KCl, 10 mM Tris, pH 7.5 and 1 mM EDTA) for 1 h at 55°C. 40 μl of Buffer B (10 mM Tris–HCl pH8.8, 5 mM MgCl2, 5 mM DTT, 1 mM dNTP) and 0.5 μl AMV reverse transcriptase (Promega) were added and incubated at 42°C for 1 h. Products were phenol-chloroform extracted, ethanol-precipitated and resuspended in RNA loading dye before being resolved on 8% polyacrylamide–7 M urea gels.
To assess A-to-I editing in the SGOL2 3΄UTR, total RNA was isolated from NIH3T3 cells infected with MHV68 for 24 h. Total RNA was reverse transcribed using an SGOL2 gene-specific primer located 3΄ of the antisense B2 SINE (Supplementary Table S1). PCR was performed using Kapa HiFi DNA Polymerase (KapaBiosystems) and primers flanking the antisense B2 SINE in the SGOL2 3΄UTR (Supplementary Table S1). The resulting PCR products were inserted into the pCR-Blunt II-TOPO vector using the Zero Blunt TOPO PCR cloning kit (Invitrogen). 30 colonies were sequenced, of which 27 contained the SGOL2 3΄UTR.
The polyadenylation site within the SGOL2 3΄UTR was mapped using the 3΄ RACE System for Rapid Amplification of cDNA Ends (Invitrogen). The resulting 3΄RACE PCR products were inserted into the pCR-Blunt II-TOPO vector using the Zero Blunt TOPO PCR cloning kit (Invitrogen). A total of 24 SGOL2-positive clones were sequenced and mapped back to the SGOL2 gene (NCBI reference sequence NM_199007.2) to determine the location of the 3΄ end.
To knockdown B2 SINEs in MHV68-infected NIH3T3 cells, 2΄-O-methyl-phosphothioate antisense oligonucleotides targeting B2 SINE RNA or GFP RNA (negative control) (Supplementary Table S1) were electroporated into NIH3T3 cells using the Neon transfection system (Invitrogen). The media was replaced 12 h post-electroporation and the cells were infected as described above.
SINE-seq
200 μg total RNA was primer extended using an oligonucleotide specific for B2 SINEs (Supplementary Table S1) as described above, with the exception that the primer was not radioactively labeled. Primer extension products were resolved on 8% polyacrylamide–7 M urea gels. For isolation of B2 products, the region of the gel corresponding to 130–180 bp was isolated and placed in a 10 ml polypropylene tube containing 5 ml of G50 buffer (20 mM Tris–HCl, 300 mM NaCl, 2 mM EDTA, 0.2% SDS), flash frozen in liquid N2, and allowed to incubate overnight at RT. The following day the G50 buffer containing the isolated B2 products was phenol-chloroform extracted and ethanol precipitated. The B2 extension products were further 3΄ poly dG-tailed using 20 U of terminal deoxynucleotidyl transferase (TDT, Fermentas) and 0.25 mM dGTP. After 3΄ poly dG-tailing, the cDNAs were used as templates for 10 cycles of PCR amplification using an oligoC and B2 extension oligo that contained barcoded sequencing adapters (Supplementary Table S1). B2 SINE-seq libraries were sequenced on a MiSeq instrument at the UC Berkeley QB3 Vincent J. Coates Genomics Sequencing Laboratory.
SINE-Seq data analysis
SINE-Seq was performed on two biological replicates. 175 bp single end MiSeq generated reads were first subject to a windowed adaptive quality and adapter trimming using trim galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The G-tail was used to find the beginning of any adapter sequences, whereupon the reads were trimmed. The reads were then aligned to the mouse mm10 reference genome using bwa (44). Non-unique alignments were discarded giving 8257630 and 7946798 uniquely mapped reads for each replicate. Reads mapping to known B2 SINEs (these were part of the UCSC repeatmasker track) were counted and normalized RPKM values were computed using the BamUtil tool (http://genome.sph.umich.edu/wiki/BamUtil). B2 SINEs with an RPKM of ≥5 were categorized as expressed, and those expressed in both replicates were used in subsequent analyses (Supplementary Table S2). Intersection analysis of expressed B2 SINEs with genomic features (Intron, 3΄-UTR, 5΄-UTR, promoter, and other intergenic regions) obtained from UCSC table browser was performed using bedtools (45). Promoters were defined as 4kb upstream of known TSS and other intergenic regions were defined as regions that were not in genic or promoter regions. Consensus finding software RSAT matrix-scan (http://rsat.ulb.ac.be/rsat/) and sequence logo program weblogo (46) were used to find consensus A and B boxes in B2 SINEs. Expressed B2 SINEs were analyzed using the GREAT tool, which provided gene ontology information for the genes proximal to expressed B2 SINEs (46–48). Raw data are available in the NCBI GEO database under accession number GSE85518 (49).
ChIP-qPCR
ChIP was performed as described previously, except using a Covaris focused sonicator for chromatin shearing (50). Cross-linked chromatin was immunoprecipitated using anti-POLR3A (clone ab96328; Abcam) and normal rabbit IgG (clone 2729; Cell Signaling). Following reversal of cross-links, DNA was purified using a PCR purification spin column (Fermentas) and resuspended in 50 μl of dH2O; 1 to 2 μl of DNA was used for quantitative PCR (qPCR) with the DyNAmo ColorFlash SYBR green qPCR kit (Thermo Scientific) with appropriate primers (Supplementary Table S1). Signals obtained by qPCR were normalized to the input DNA.
Cell fractionation, siRNA knockdowns, western blotting and immunoprecipitations
Subcellular fractionation was performed using the REAP method as published (51), with the minor modification of using one 10-cm plate for each fractionation condition.
p54nrb (NONO) and control siRNA smart pools (Dharmacon) were transfected using RNAiMAX (Life Technologies). To determine knockdown efficiency, cells were lysed in NET-2 buffer (50 mM Tris–HCl [pH 7.6], 150 mM NaCl, 3 mM MgCl2, 10% glycerol, 0.5% Nonidet P-40), and protein concentrations were determined by Bradford assay. Equivalent quantities of each sample were resolved by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and incubated with the indicated antibodies (anti-p54nrb ab70335, Abcam; anti-GAPDH ab8245, Abcam). Western blots assays were developed with HRP-conjugated Protein G (EMD Millipore) or HRP-conjugated secondary antibodies (Southern Biotech), and ECL reagents (Pierce).
For immunoprecipitations, mock- or MHV68-infected cells were cross-linked with 1% formaldehyde for 10 min at RT followed by quenching of the cross-linking with 0.25 M glycine for 5 min. Cross-linked cells were resuspended in RIPA buffer (25 mM Tris–HCl (pH 7.6), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) and lysed by sonication with a branson sonicator. 5 μg of anti-p54nrb (clone ab70335, Abcam), anti-mIgG antibody (Cell Signaling), or 50 μl protein G magnetic beads were added to the extracts and rotated overnight at 4°C, whereupon 50 μl of pre-washed protein G magnetic beads were added rotated for an additional 2 h. Antibody-bead complexes were then isolated on a magnetic stand and washed three times with high salt RIPA buffer containing 500 mM NaCl and then eluted in 1× SDS loading buffer. Samples were incubated at 55°C for 1 h prior to western blot analysis. Anti-dsRNA (J2; Scicons) immunoprecipiations were preformed as described above but cells were not formaldehyde cross-linked and immunoprecipitations were preformed on REAP prepared nuclear fractions that were adjusted to 150 mM NaCl, 3 mM MgCl2, 10% glycerol and 0.5% Nonidet P-40.
Oligoaffinity chromatography
For oligoaffinity chromatography ∼200 million cells were psoralen cross-linked as described previously (6). Briefly, cells were washed with phosphate-buffered saline (PBS) twice and then incubated with 5 μM psoralen (Sigma) in PBS for 10 min. A first round of psoralen cross-linking was carried out with 365 nm UV light (1.2 kJ/m2/min; UVP) for 5 min at room temperature, after which excess psoralen was removed by two sequential washes with PBS. A second round of psoralen cross-linking was then performed in the presence of 365 nm UV light (1.2 kJ/m2/min) for 15 min at room temperature. RNA was then isolated as described above, except using Trizol pre-warmed to 60°C. The RNA pellet was resuspended in hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris 7.0, 1 mM EDTA, 15% formamide and RNAse inhibitor). 40 pmol of each 5΄-TEG biotinylated SGOL2 antisense oligonucleotide (Supplementary Table S1) was added and the mixture was rotated end-over-end at 37°C for 6 h. 100 μl of streptavidin-magnetic C1 beads were blocked with 500 ng/ml yeast total RNA, 100 μg/ml glycogen and 1 mg/ml BSA for 1 h at room temperature. Blocked C1 beads were added and the reaction was mixed for another 2 h at 37°C. Complexes were captured by magnets (Invitrogen) and washed five times with wash buffer (2× SSC, 0.5% SDS, and RNAse inhibitor). After the final wash, beads were resuspened in elution buffer (50 mM Tris pH 7.0, 100 mM NaCl, 1 mM EDTA). Beads in elution buffer were heated to 70°C for 5 min before separating the elution buffer from the beads. Psoralen crosslinks were reversed by irradiating samples under a 254 nm lamp for 10 min.
RESULTS
A genome-wide SINE activation map reveals extensive transcription of SINE loci during infection
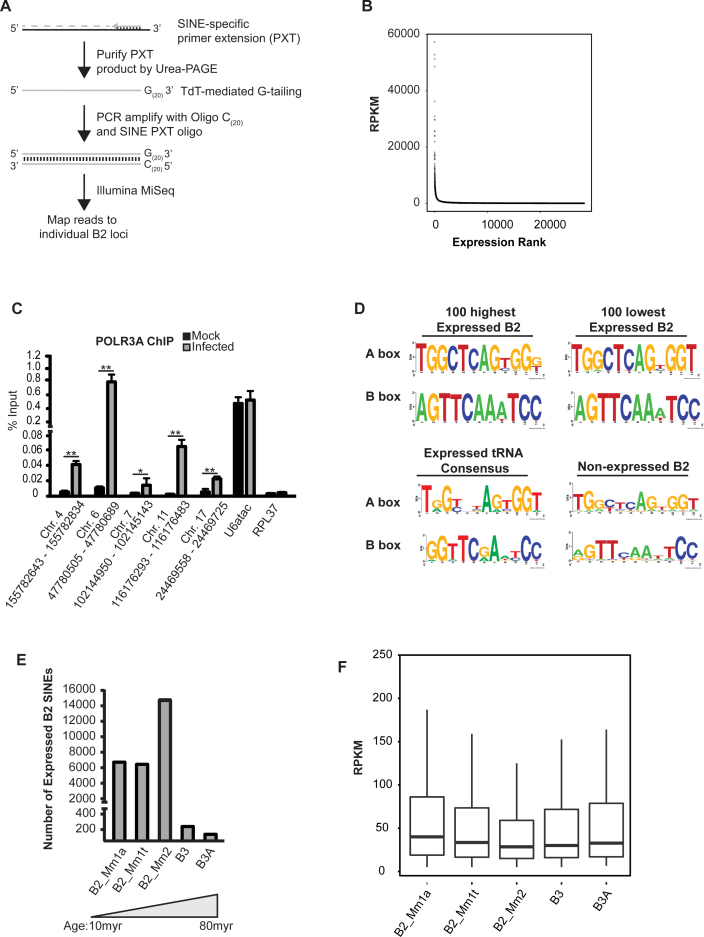
Many SINE sequences are embedded within mRNAs and other noncoding RNAs, and there is a high degree of sequence conservation between individual loci. For these reasons, conventional RNA-sequencing approaches cannot distinguish between sequence tags corresponding to bona fide SINE RNAs transcribed by RNA Pol III and SINE sequences that are not independently transcribed but instead embedded within other RNAs. We therefore generated SINE transcript-specific sequencing libraries, using an approach we termed SINE-seq, and identified RNA Pol III-transcribed SINE loci (Figure 1A). Selectivity for SINE RNAs was achieved first by reverse transcription (RT) of total RNA using an oligonucleotide complementary to a highly conserved region of B2 SINEs. These were then enriched through denaturing gel electrophoresis and purification of reverse transcription products that migrated between 130 and 180 nt, which corresponds to the size of RT products reaching the 5΄ transcription start-site of SINE RNAs. Although SINEs are repetitive, using 175 base pair long-reads, sufficient sequence divergence exists to identify individually active loci upon high-throughput sequencing of libraries generated from the size-selected cDNAs.
Figure 1.
SINE-seq profiling of MHV68-induced B2 SINE expression. (A) Schematic of SINE-seq method. (B) Dot plot describing dynamic range and extent of B2 SINE induction. (C) Chromatin from MHV68-infected NIH3T3 cells 24 hpi was subjected to ChIP using antibodies against RNA Pol III or control IgG. Recovered DNA was detected by qPCR. (D) MEME analysis of A and B box elements with the promoters of transcribed and nontranscribed SINE elements. Consensus A and B Box sequences for expressed tRNA genes from the mouse mm10 reference genome are shown for comparison. (E) Stratification of active B2 loci based on their genetic age. (F) Box plots of RPKM for individual SINE subfamilies. Statistical significance was determined by Student t test (*, P < 0.05; **, P < 0.01).
Using this technique, we examined the profile of B2 SINEs expressed 24 h post-MHV68 infection of NIH3T3 cells, as we have previously demonstrated their robust transcriptional induction under these conditions (30). We restricted our bioinformatics analysis of SINE-seq reads to those that map to a single SINE locus in the murine genome. Using these parameters, we identified 28 270 transcriptionally active B2 SINE loci in infected cells (Figure 1B). There was a >10 000 fold range in the expression of B2 RNA, with 808 of the transcriptionally active B2 loci exhibiting >1000 RPKM and the remainder being weakly expressed (Figure 1B). We were unable to identify specific features common to the robustly expressed set of SINE loci, such as proximity to RNA Pol II transcribed genes or B2 SINE subfamily. RNA Pol III occupancy of several of the loci identified as induced by SINE-seq were independently evaluated by chromatin immunoprecipitation (ChIP) analysis for POLR3A, the largest subunit of RNA Pol III (Figure 1C). We observed increased POLR3A occupancy at each of these SINE loci in infected cells, whereas no increased occupancy was observed at the Pol III-transcribed U6atac promoter, and no signal above background was detected at the control RNA Pol II-transcribed RPL37 promoter (Figure 1C). These data confirm that there is a specific induction of individual SINE loci upon infection, rather than a generalized increase in RNA Pol III occupancy.
The abundance of activated SINE loci coupled with the wide range in B2 RNA expression prompted us to explore whether transcriptional potential or expression level correlated with the strength of the A and B boxes of the internal RNA Pol III promoter. It has been suggested that the vast majority of B2 SINE loci have been genetically inactivated by promoter mutations (52), although there have been no prior systematic analyses of transcriptionally active SINE loci. MEME analysis on the highest 100 expressed, lowest 100 expressed, and 100 non-expressed B2 loci identified internal A and B box elements in all examined B2 loci (Figure 1D). However, only the expressed B2 loci contained highly conserved A and B box sequences, while those of non-expressed B2 loci were less well conserved. The presence of strong consensus sequences within both the highest expressed and weakest expressed SINEs indicate SINE RNA levels are not solely dictated by the RNA Pol III promoter sequence. Additionally, directed mutagenesis studies have demonstrated that the B-box is the most critical region for TFIIIC binding to tRNA genes, and that a highly conserved cytosine in the fifth position of the B box is critical for high affinity binding of TFIIIC (53). In both the highest and lowest expressed B2 loci a cytosine is present at the fifth position of the B box, while it is absent in most non-expressed B2 loci (Figure 1D). The lack of strong consensus sequences within the non-expressed B2 loci supports the hypothesis that large-scale mutational inactivation of SINE loci has occurred.
The B2 family of SINEs is grouped into several subfamilies of different genetic ages, with acquisition occurring between 10 and 80 million years ago. Assuming large-scale mutational inactivation has occurred over time, the older B2 SINE subfamilies should exhibit less transcriptional potential. Indeed, only ∼1% of transcriptionally active SINE loci identified in our dataset belong to the oldest B3 and B3A subfamilies (Figure 1E). Interestingly, however, despite the fact that few B3 and B3A subfamilies are transcriptionally active, the mean expression value for distinct SINE subfamilies is not statistically different (Figure 1F).
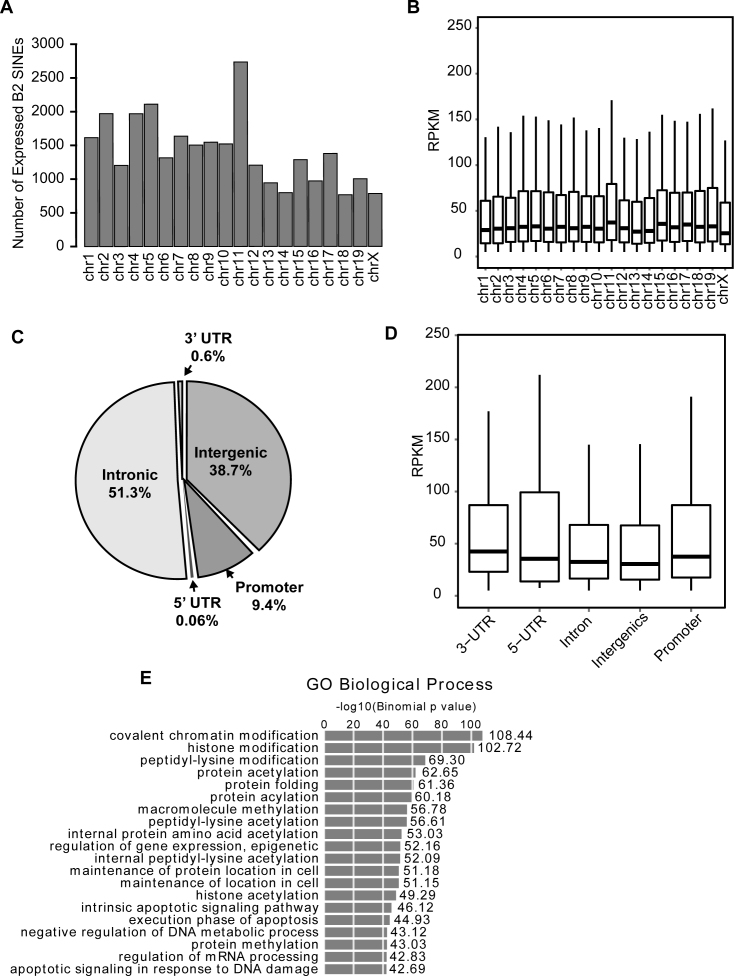
Active B2 loci were present on all chromosomes except the Y chromosome (Figure 2A). While the abundance of SINE loci varies on each chromosome, ∼7% of SINEs on each chromosome were transcriptionally induced, and SINE RNA expression levels did not correlate with chromosome of origin (Figure 2B). Examination of genomic features proximal to transcriptionally active B2 SINEs revealed that nearly 60% of active B2 loci reside within or near annotated RNA Pol II expressed loci (Figure 2C and D). Interestingly, many of these RNA Pol II loci are ontologically associated with the regulation of gene expression (Figure 2E). Furthermore, as many transcriptionally active SINEs map within promoters (defined as <4kb upstream of a transcription start site), introns and 5΄ and 3΄UTRs, under conditions of simultaneous transcription these active SINE loci have the potential to regulate their cognate RNA Pol II-transcribed genes.
Figure 2.
Genomic properties of transcriptionally activated SINE loci. (A) Histograms showing number of transcriptionally active SINEs per chromosome. (B) Box plots (whiskers extend +1.5 interquartile range (IQR)/5 RPKM) of RPKM for SINE expression on indicated chromosome. (C) Pie chart indicting the location of all transcriptionally active B2 SINEs identified by SINE-seq. (D) Box plots of RPKM for SINE expression from indicated genomic region. (E) Genomic coordinates of active B2 loci were analyzed using GREAT to retrieve gene ontology (GO) associations of SINE-proximal RNA Pol II loci.
B2 SINE RNA forms intermolecular base pairs with mRNA
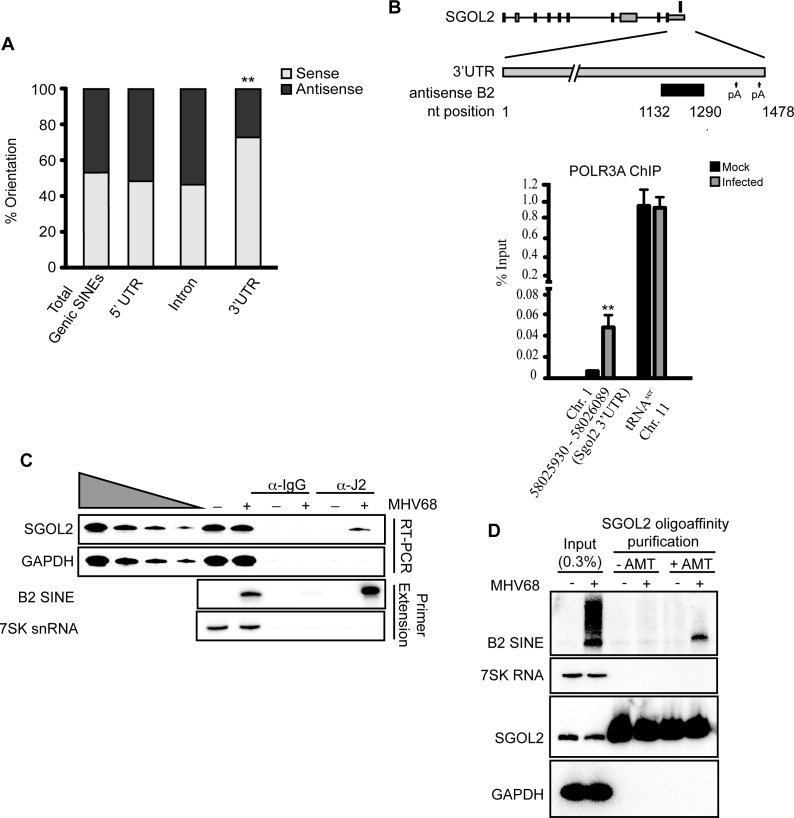
Approximately 50% of transcriptionally active SINE loci fall within genomic sequences corresponding to the noncoding regions of pre-mRNA, and could regulate processing or fate of their cognate mRNAs in a variety of ways (Figure 2C). We focused on SINEs derived from sequences corresponding to mRNA 3΄ UTRs, as this region is a frequent target for gene regulation. B2 RNAs present in the antisense orientation have the potential to form intermolecular base pairs with the mRNA on the opposite strand (54). Thus, they might be restricted to a subset of mRNAs that use SINE RNAs to regulate mRNA fate, as suggested by the fact that active B2 loci in 3΄UTRs are enriched in the sense orientation (p = 0.0164), while there is no preference for orientation for active B2 loci in introns and 5΄UTRs (Figure 3A).
Figure 3.
B2 SINE RNA forms intermolecular base pair interactions with SGOL2 mRNA. (A) Analysis of the orientation of active SINEs compared to RNA Pol II loci, displayed as percent sense versus antisense. (B) Chromatin from MHV68-infected NIH3T3 cells 24 hpi was subjected to ChIP using antibodies against RNA Pol III. Recovered DNA was detected by qPCR. A schematic (not drawn to scale) of the SGOL2 locus is shown above. The antisense B2 SINE is depicted as a black box, with nucleotide positions within the SGOL2 3΄ UTR indicated. Polyadenylation sites (pA) detected by 3΄ RACE at nucleotide positions 1345–1361 and 1463–1478 are noted by the arrows. The rectangle above the diagram represents the single transcriptionally active B2 SINE. (C) Extracts were prepared from Mock- or MHV68-infected cells at 12 hpi and immunoprecipitated with anti-J2 antibodies. Recovered RNA was subjected to RT-semi quantitative PCR (RT-PCR) analysis or primer extension. (D) Mock- or MHV68-infected cells were either untreated or cross-linked with the psoralen derivative AMT at 12 hpi and subjected to SGOL2 oligoaffinity chromatography. Recovered RNA was subjected to cross-link reversal and analyzed by northern blotting with the indicated probes. Statistical significance was determined by Student's t test (**P < 0.01).
We identified 81 transcriptionally active B2 loci present in the antisense orientation within sequences corresponding to mRNA 3΄ UTRs (Supplementary Table S2). Many of these loci contained multiple SINE elements, making it difficult to distinguish between potential intermolecular and intramolecular interactions. We therefore focused our analysis on the SGOL2 locus, which was identified by SINE-seq as having a single, highly expressed antisense B2 SINE located within its 3΄UTR. We confirmed by ChIP for POLR3A that the SGOL2-derived SINE was induced in infected cells, whereas RNA Pol III occupancy of the tRNAser promoter was unchanged (Figure 3B). Given the possibility that convergent transcription of the SINE and SGOL2 might prevent RNA Pol II from reaching the 3΄end of the SGOL2 gene, we performed 3΄-rapid amplification of cDNA ends (RACE) to compare the SGOL2 mRNA produced in mock and MHV68-infected NIH3T3 cells. We detected use of 2 polyadenylation sites within the SGOL2 gene, one located at nts 1345–1361 following the stop codon and a second at nts 1463–1478 following the stop codon (diagramed in Figure 3B). Among the 13 SGOL2-positive clones sequenced from mock-infected cells, 11 mapped to the first polyA site and 2 mapped to the second polyA site. Similarly, among the 11 SGOL2-positive clones sequenced from MHV68-infected cells, 8 mapped to the first polyA site and 3 mapped to the second polyA site. These data suggest that full-length SGOL2 mRNA is produced in both mock- and MHV68-infected NIH3T3 cells at a similar frequency.
To first test for the possibility that the SGOL2-derived SINE RNA bound the SGOL2 mRNA, we immunoprecipitated (IP) dsRNAs from extracts of mock- or MHV68-infected cells using the dsRNA-specific antibody J2 (55). Primer extension analysis revealed that both B2 SINE RNA and the SGOL2 mRNA were enriched in eluates of J2 IPs specifically in MHV68-infected cells (Figure 3C). This result indicates that a dsRNA structure within the SGOL2 transcript is formed specifically during MHV68 infection. Neither 7SK nor the GAPDH controls, which lack transcriptionally active B2 SINEs, were detectable in the J2 IPs.
We next used 4-aminomethyltrioxalen (AMT) crosslinking to detect direct B2 SINE RNA-SGOL2 mRNA interactions. AMT, a psoralen derivative, is a reversible crosslinker specific for double-stranded nucleic acids; upon irradiation at 365 nm, covalent adducts form between adjacent pyrimidine bases, while irradiation at 254 nm reverses the crosslinks (56–58). Thus, AMT crosslinking can be used to directly detect RNA–RNA interactions (58,59). Purification of the SGOL2 mRNA via oligonucleotide affinity chromatography of extracts cross-linked with AMT led to a selective co-purification of the B2 SINE RNA (Figure 3D). The interaction occurred specifically in MHV68-infected cells in which the B2 SINE within the SGOL2 locus is transcribed, and no co-purification of the highly abundant 7SK snRNA or the GAPDH mRNA controls occurred. These data demonstrate that infection-induced B2 RNAs engage in intermolecular RNA–RNA interactions with mRNA, as demonstrated for SGOL2.
SINE transcription is linked to selective control of mRNA export
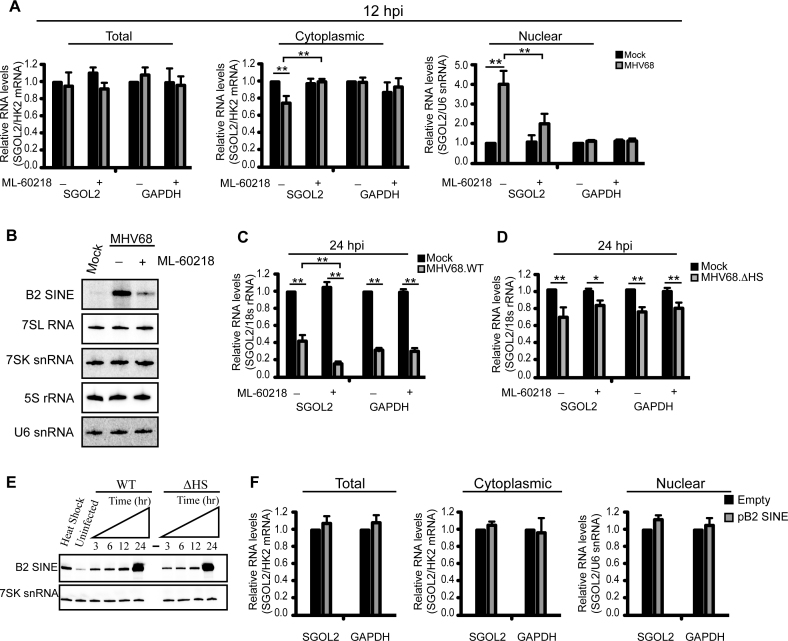
Intermolecular RNA–RNA interactions affect diverse processes in the gene expression cascade. Thus, we examined whether infection-induced B2 SINE RNA impacted the abundance or localization of SGOL2 or control GAPDH mRNAs. Both northern blotting and RT-qPCR analysis showed no change in SGOL2 or GAPDH mRNAs between mock or MHV68-infected NIH3T3 cells at 12 hpi (Figure 4A and Supplementary Figure S1A). However, subcellular fractionation revealed a specific increase in SGOL2 mRNA levels within the nucleus during MHV68 infection (Figure 4A and Supplementary Figure S1A). This phenotype was linked to B2 SINE RNA expression, as pretreatment of the cells with the RNA Pol III-specific inhibitor ML-60218 prior to infection to reduce the expression of B2 RNAs partially relieved the SGOL2 mRNA export block (Figure 4A and Supplementary Figure S1A) (60). Steady state levels of other RNA Pol III transcribed RNAs remained unchanged during the duration of ML-60218 treatment, presumably do the long half-life of these RNAs relative to the rapidly degraded B2 RNAs (Figure 4B) (31,61). We also assessed the role of B2 RNAs in preventing SGOL2 nuclear export by specifically depleting B2 RNAs using 2΄-O-methyl phosphorothioate antisense oligonucleotides (ASOs) (Supplementary Figure S1B). Similar to our observations with the ML-60218 treated cells, depletion of B2 RNA by ASO treatment rescued the SGOL2 mRNA export block in MHV68-infeced cells, whereas expression of control ASOs targeting GFP had no effect (Supplementary Figure S1C). These results indicate that B2 RNA, but not other RNA Pol III transcripts, impacts SGOL2 mRNA export during MHV68 infection.
Figure 4.
SGOL2 subcellular distribution is affected by B2 SINE expression. (A) RT-qPCR analyses of SGOL2 and GAPDH mRNA levels in cytoplasmic and nuclear fractions from mock- or MHV68-infected NIH3T3 cells. Cells were pretreated with either DMSO or 40 μM ML-60218 for 4 h prior to infection. (B) Northern blot analysis of RNA Pol III transcribed RNAs from cells described in (A). (C) NIH3T3 cells were pretreated with DMSO or ML-60218 (40 μM) 4 h prior to being infected with MHV68 at an MOI of 5. Total RNA was isolated at 24 hpi and the levels of the indicated RNAs were monitored by RT-qPCR. (D) NIH3T3 cells were treated as described in (C) and then infected with MHV68.ΔHS. Total RNA was isolated at 24 hpi and the levels of the indicated RNAs were monitored by RT-qPCR. (E) Total RNA was isolated from NIH3T3 cells infected with either WT MHV68 or MHV68.ΔHS at the indicated times and the levels of B2 SINE and 7SK snRNA were analyzed by primer extension. (F) RT-qPCR analysis of subcellular fractions from NIH3T3 cells transfected with the indicated plasmids. Error bars represent the mean with SEM of ≥3 independently performed experiments. Statistical significance was determined by Student's t test (*P < 0.05; **P < 0.01).
We next evaluated SGOL2 mRNA distribution within cells using a functional assay in which we measured its susceptibility to an MHV68 encoded mRNA-specific nuclease (mRNase), termed SOX. SOX expression is induced ∼12 hpi, and specifically targets cytoplasmic mRNA for degradation (62). Thus, the population of SGOL2 retained in the nucleus in a B2 SINE-dependent manner should escape degradation by the viral mRNase. Indeed, while both SGOL2 and GAPDH mRNAs were depleted in cells infected with MHV68 at 24 hpi, blocking B2 SINE transcription by ML-60218 treatment prior to infection selectively enhanced the depletion of SGOL2 RNA (Figure 4C). This result is consistent with an increased percentage of SGOL2 mRNA residing in the cytoplasm in the absence of B2 RNA. Importantly, ML-60218 treatment did not affect SGOL2 steady state RNA levels during infection with a viral mutant with impaired SOX mRNase activity (MHV68ΔHS), despite the fact that MHV68ΔHS induced B2 RNA expression to a similar extent (Figure 4D and E). Thus, B2 RNA retains a proportion of SGOL2 RNA in the nucleus, where it cannot be accessed by the cytoplasmic viral mRNase.
Finally, we considered whether the SINE RNA-mediated nuclear retention of the SGOL2 mRNA required the B2 RNA to be transcribed from the native SGOL2 locus, or whether it could be mediated by an exogenously expressed SINE RNA. We tested this possibility by overexpressing a plasmid-borne B2 SINE RNA, complementary to the embedded B2 SINE within the SGOL2 3΄UTR, in uninfected cells where the native SGOL2-derived SINE was silent. Though B2 SINE RNA was robustly expressed from the plasmid, we did not detect nuclear retention of the SGOL2 mRNA. This suggests that the B2 RNA-mediated nuclear retention, at least in the case of SGOL2, originates as a result of overlapping transcription within the 3΄UTR of specific transcripts (Figure 4F).
p54nrb regulates the nuclear distribution of SGOL2-B2 duplexes
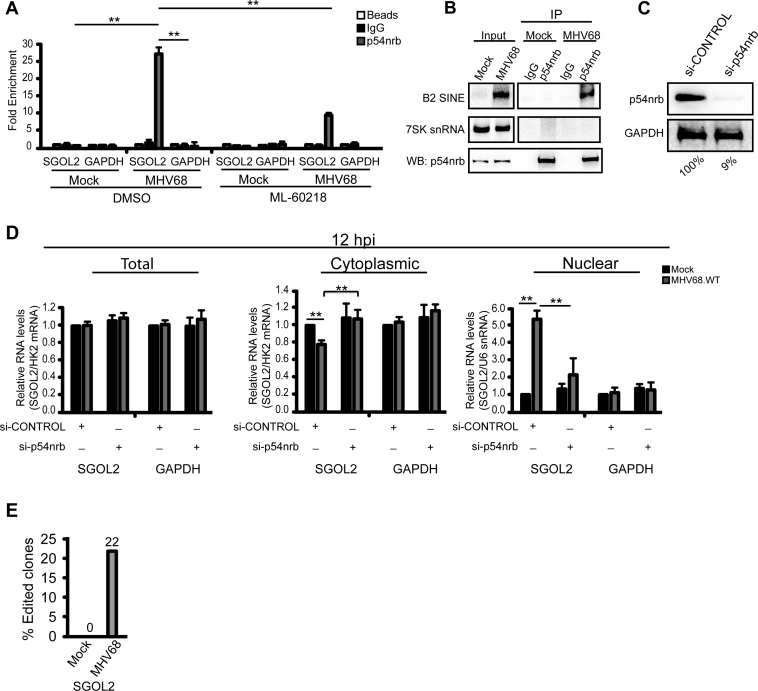
The SINE RNA-induced nuclear retention of mRNA is similar to the fate of mRNAs that contain inverted SINE sequences embedded within their transcript (19). Although not independently transcribed by RNA Pol III, because the SINE sequences are inverted and complementary, they can form long intramolecular dsRNA structures that promote nuclear retention. Nuclear retention of mRNAs containing inverted SINEs requires the nuclear protein p54nrb (63). We therefore tested whether p54nrb might similarly be required for nuclear retention of SGOL2 during MHV68 infection. First, we examined whether p54nrb was associated with SGOL2 mRNA (Figure 5A). To preclude post-lysis interactions, cells were formaldehyde cross-linked prior to p54nrb IP and RT-qPCR analysis. We detected a specific interaction between p54nrb and the SGOL2 mRNA in an infection-dependent manner (Figure 5A). No interaction was detected between p54nrb and the control GAPDH mRNA in mock or infected cells. Consistent with a role for B2 RNAs mediating this interaction, inhibiting B2 RNA transcription by treatment of MHV68-infected cells with ML-60218 reduced the association between p54nrb and SGOL2 (Figure 5A). Furthermore, IP of cross-linked p54nrb complexes also enriched B2 RNAs (Figure 5B).
Figure 5.
p54nrb regulates the nuclear distribution of SGOL2-B2 SINE RNA duplexed RNAs. (A) Cells were pretreated with DMSO or ML-60218 (40 μM) 4 h prior to mock or MHV68 infection. At 12 hpi cells were formaldehyde cross-linked and immunoprecipitated with IgG, α-p54nrb, or a no antibody control (beads). Recovered RNAs were detected by RT-qPCR. (B) Mock- or MHV68-infected cells were formaldehyde cross-linked 12 hpi and whole cell extracts were immunoprecipitated with IgG or α-p54nrb antibodies. Recovered RNAs were northern blotted for B2 RNA and 7SK snRNA. Western blot analysis confirmed the immunoprecipitation of p54nrb. (C) Western blot analysis of NIH3T3 cells transfected with si-CONTROL or si-p54nrb. GAPDH serves as a loading control for quantifying the efficiency of p54nrb depletion, indicated below. (D) NIH3T3 cells were transfected with si-CONTROL or si-p54nrb 36 hr before infection with MHV68. At 12 hpi, RNA was isolated from nuclear and cytoplasmic fractions and analyzed by RT-qPCR. (E) Bar graph showing number of RT-PCR clones demonstrating A-to-I editing. Error bars represent the mean with SEM of ≥3 independently performed experiments. Statistical significance was determined by Student's t test (*P < 0.05; **P < 0.01).
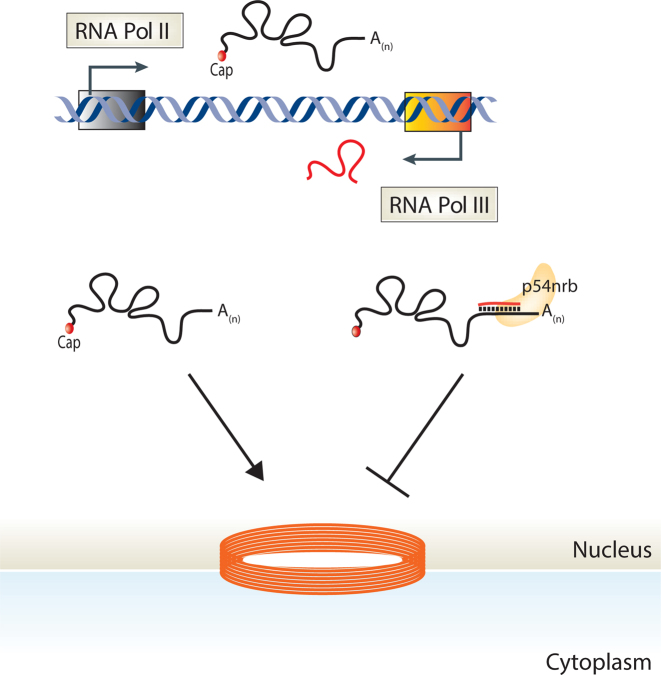
We next tested whether p54nrb, like B2 RNA, was required for SGOL2 nuclear retention. NIH3T3 fibroblasts were treated with either p54nrb or control siRNAs and subsequently mock or MHV68-infected for 12 h (Figure 5C). Knockdown of p54nrb prevented accumulation of SGOL2 in the nucleus (Figure 5D). p54nrb interacts with inosine containing RNAs, and dsRNA structures formed through inverted SINE sequences are also subjected to A-to-I editing (63–68). We thus examined whether the antisense B2 SINE embedded within the SGOL2 mRNA underwent A-to-I editing by cloning the 3΄UTR of SGOL2 from total RNA and sequencing individual clones. As expected, we detected A-to-I editing specifically in the B2 RNA binding region of the SGOL2 3΄ UTR, and specifically during MHV68 infection, in 22% of our clones (Figure 5E). These results demonstrate that both transcribed SINE RNA and embedded non-transcribed SINEs use a parallel mechanism for regulating mRNA export in a transcript-specific manner (Figure 6).
Figure 6.
Model depicting how SINE ncRNA-mRNA intermolecular interactions facilitate an mRNA export block. SINE loci are transcriptionally activated in response to many stresses. Antisense transcribed SINEs generate RNAs with the potential to base pair with complementary SINE sequences within mRNA. SINE ncRNA–mRNA intermolecular interactions recruit the protein p54nrb, leading to a block in mRNA export.
DISCUSSION
Our assembly of the first transcriptome-wide map of B2 SINE activation highlights the remarkable breadth of transcriptionally competent SINEs, and provides a straightforward means to probe SINE activation profiles under a variety of developmental and stress conditions. Historically, identifying active RNA Pol III-transcribed SINE loci relied on either ChIP-seq analyses of RNA Pol III or the use of extensive computational algorithms to analyze RNA-seq data (69–76). However, unlike SINE-seq, these techniques are limited by challenges associated with retrotransposon analyses. In particular, the extremely high copy number and sequence similarity of SINE elements within the murine genome, and the frequent location of SINE elements within introns or untranslated regions of RNA Pol II transcripts have likely resulted in a marked underestimation of transcriptionally active loci.
Our finding that many of the B2 loci transcribed in response to MHV68 infection cluster proximal to or within genes associated with the regulation of gene expression raise the intriguing question of whether distinct subsets of SINEs may be transcribed in response to different stimuli, and that SINE RNAs may therefore have evolved roles in regulating gene expression specific to each stimulus. This hypothesis is supported by a previous analysis of six human Alu loci, which revealed that viral infection induces a subset distinct from those induced by heat shock (77). Additionally, two recent RNA Pol III ChIP-seq studies in human K562 and HeLa cells each reported ∼1000 RNA Pol III-bound Alu loci, though only ∼2% of the loci overlapped, indicating that SINE transcriptional potential may also be cell type specific (37,78). Our characterization of how SINE RNAs can impact mRNA export through intermolecular RNA–RNA interactions, coupled with recent reports linking SINE RNAs to both gene expression control and immune signaling provide examples of distinct mechanisms of SINE RNA-based regulation (30,31,34,35,79,80).
Intermolecular RNA–RNA interactions are used by many noncoding RNAs as a mode of substrate recognition (81). The interaction between a B2 SINE RNA and its cognate SGOL2 mRNA demonstrates that transcribed SINEs also participate in this type of regulation. The B2 RNA-SGOL2 mRNA interaction promotes nuclear retention of the SGOL2 mRNA via a mechanism involving p54nrb. This mechanism is reminiscent of that which mediates nuclear retention of mRNA containing pairs of inverted SINE elements, though it is important to note that the mechanism we describe here is mediated via intermolecular rather than intramolecular RNA–RNA interactions (19,63). Interestingly, it was recently reported that expressed 4.5SH RNA, a repetitive element highly homologous to B1 SINE RNA, is similarly capable of mediating the nuclear retention of mRNAs containing embedded antisense B1 RNA elements (82). Whether 4.5SH-mediated nuclear retention requires the RNA-binding protein p54nrb is unknown. In addition to the role of p54nrb, nuclear retention of mRNA containing inverted SINE elements also requires the long nuclear-retained noncoding RNA NEAT1 (83). It was recently reported that the protein arginine methyltransferase CARM1 regulates nuclear retention of mRNAs containing inverted SINE elements via two distinct mechanisms: methylation of p54nrb, which reduces its affinity for mRNAs containing inverted SINEs, and as a transcriptional repressor for NEAT1 (84). It will be of interest to examine the role of CARM1 in nuclear retention of RNAs that engage in intermolecular interactions with expressed B1 or B2 RNAs.
How SGOL2 retention may affect viral infection or other cellular responses to stress is currently unclear, as its only characterized function is linked to chromosome segregation during gametogenesis (85–87). However, its regulation serves as a model for how SINE RNAs can impact the transport of specific mRNAs. Our data suggest the SINE-SGOL2 interaction likely occurs as a result of overlapping transcription within the SGOL2 3΄UTR, as nuclear retention of the SGOL2 mRNA did not occur when the B2 SINE was expressed ‘in trans’ from a plasmid. This intermolecular interaction may be favored in the context of overlapping transcription because of limiting SINE RNA abundance and/or increased efficiency of binding prior to completion of RNA folding or ribonucleoprotein assembly of each individual transcript. Alternatively, antisense transcription within an mRNA may result in transcriptional collisions between the two opposing polymerases resulting in the production of prematurely terminated mRNA, potentially resulting in a similar block in mRNA nuclear export. While this may occur at other loci, we disfavor this mechanism for the SGOL2 locus for several reasons. These include our detection of RNA synthesis 3΄ of the antisense B2 sequence, the similar SGOL2 3΄ polyA site usage in mock and infected cells, and the observation that SGOL2 nuclear retention is dependent on p54nrb. In addition to occurring at loci engaged in overlapping transcription, this type of regulation could presumably also occur in trans through interactions with embedded SINE elements in mRNAs transcribed from independent loci. In this scenario, akin to how a single microRNA can modulate the expression of multiple mRNAs, the transcriptional activation of a single SINE element has the potential to regulate multiple mRNAs. In this regard, additional experiments to map the specific target mRNA residues involved in B2 RNA binding through mutational analyses would be informative. Recently, several methods have been developed capable of globally mapping inter- and intra-molecular RNA–RNA interactions and the application of them to MHV68-infected cells should reveal the full breadth of SINE RNA–RNA interactions (88–90).
Independent of B2 RNA function, the act of B2 SINE transcription also provides the opportunity for regulation of cellular gene expression through alterations in the local chromatin environment. For instance, transcription of a B2 SINE facilitates the temporal activation of genes in the murine growth hormone (GH) locus by converting the local chromatin structure from a heterochromatic to a more permissive euchromatic state (49). Similarly, the recruitment of TFIIIC, a general transcription factor for RNA Pol III, to SINEs within the somatosensory cortex of mice exposed to novel enriched environmental conditions promoted the transcriptional activation of TFIIIC occupied SINE-proximal genes (91). It is therefore perhaps notable that active SINEs are located proximal to specific classes of genes, including those related to chromatin regulation.
SINE elements within genes are enriched for an antisense orientation. However, our analysis of the orientation of the transcriptionally active subset of B2 loci with respect to the host RNA Pol II gene revealed that within 5΄UTRs and introns, there is no preference for orientation. Interestingly, few of the transcriptionally active B2 loci reside within 5΄UTRs, perhaps because of the potential for transcriptional interference between the two polymerases. In contrast to 5΄UTRs and introns, active B2 loci within 3΄UTRs are overwhelmingly in the sense orientation. As shown for SGOL2, antisense B2 loci within 3΄UTRs can result in the formation of dsRNA structures that impede mRNA export, and the preference for sense orientation presumably restricts this regulation to specific loci. In the case of intronic antisense B2 transcription, intronic sequences are rapidly removed by the spliceosome leaving little time for the formation of dsRNA structures, thus minimizing orientation-specific effects. This would suggest an active mechanism is responsible for either the preferential inhibition of antisense B2 SINEs in 3΄UTRs, or conversely, the preferential induction of sense B2 SINEs in 3΄UTRs.
In unstressed somatic cells, SINE RNAs as well as the RNA Pol III occupancy of B2 SINE loci are virtually undetectable, suggesting a widespread redistribution of the RNA Pol III transcription machinery during stresses such as infection. What mechanism drives the selective induction of B2 SINE loci but not other RNA Pol III genes, as well as the basis for which of the SINE loci with intact promoters are transcribed remain key open questions. Stratification of the SINE-seq data with regard to B2 SINE subfamily age revealed a strong correlation between younger subfamilies and transcriptional activity, as only 1% of transcriptionally active B2 loci belong to the oldest B2 SINE subfamilies. This result fits well with the hypothesis that mutational inactivation has silenced a large fraction of SINE loci. However, the age of the subfamily is not associated with the expression level, as both the highest and lowest expressed B2 loci possess conserved A and B box promoter elements. Thus, other non-promoter sequences within or surrounding the SINEs may impact expression, perhaps in a posttranscriptional manner. For example, variations in SINE sequence would affect the secondary structure of SINE RNA, potentially impacting protein binding or accessibility to ribonucleases. Coupled with the fact that nearly 30 000 SINE loci become active during infection, this makes likely that SINE RNA expression has broad functional consequences.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank members of the Glaunsinger lab, Laurent Coscoy, and members of the Coscoy lab for insightful discussions and critical reading of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Damon Runyon Cancer Research Foundation fellowship [DRG 2121-12]; Vanderbilt University Medical Center Development Funds [1-04-500-9151 to J.K.]; NIH [CA136367 and CA160556]; Burroughs Wellcome Foundation Investigators in the Pathogenesis of Infectious Disease Award; W.M. Keck Foundation Distinguished Young Investigator Award (to B.G.). B.G is an investigator of the Howard Hughes Medical Institute. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1. Deininger P.L., Batzer M.A.. Mammalian retroelements. Genome Res. 2002; 12:1455–1465. [DOI] [PubMed] [Google Scholar]
- 2. Kriegs J.O., Churakov G., Jurka J., Brosius J., Schmitz J.. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007; 23:158–161. [DOI] [PubMed] [Google Scholar]
- 3. Weiner A.M. An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell. 1980; 22:209–218. [DOI] [PubMed] [Google Scholar]
- 4. Ullu E., Tschudi C.. Alu sequences are processed 7SL RNA genes. Nature. 1984; 312:171–172. [DOI] [PubMed] [Google Scholar]
- 5. Daniels G.R., Deininger P.L.. Repeat sequence families derived from mammalian tRNA genes. Nature. 1985; 317:819–822. [DOI] [PubMed] [Google Scholar]
- 6. Tsirigos A., Rigoutsos I.. Alu and b1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLoS Computat. Biol. 2009; 5:e1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C., Gentles A.J., Jurka J., Karlin S.. Genes, pseudogenes, and Alu sequence organization across human chromosomes 21 and 22. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:2930–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Korenberg J.R., Rykowski M.C.. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988; 53:391–400. [DOI] [PubMed] [Google Scholar]
- 9. Su M., Han D., Boyd-Kirkup J., Yu X., Han J.D.. Evolution of Alu elements toward enhancers. Cell Rep. 2014; 7:376–385. [DOI] [PubMed] [Google Scholar]
- 10. Bourque G., Leong B., Vega V.B., Chen X., Lee Y.L., Srinivasan K.G., Chew J.L., Ruan Y., Wei C.L., Ng H.H. et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008; 18:1752–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laperriere D., Wang T.T., White J.H., Mader S.. Widespread Alu repeat-driven expansion of consensus DR2 retinoic acid response elements during primate evolution. BMC Genomics. 2007; 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Polak P., Domany E.. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006; 7:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt D., Schwalie P.C., Wilson M.D., Ballester B., Goncalves A., Kutter C., Brown G.D., Marshall A., Flicek P., Odom D.T.. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012; 148:335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sundaram V., Cheng Y., Ma Z., Li D., Xing X., Edge P., Snyder M.P., Wang T.. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014; 24:1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roman A.C., Gonzalez-Rico F.J., Molto E., Hernando H., Neto A., Vicente-Garcia C., Ballestar E., Gomez-Skarmeta J.L., Vavrova-Anderson J., White R.J. et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011; 21:422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roman A.C., Benitez D.A., Carvajal-Gonzalez J.M., Fernandez-Salguero P.M.. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:1632–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roy-Engel A.M., El-Sawy M., Farooq L., Odom G.L., Perepelitsa-Belancio V., Bruch H., Oyeniran O.O., Deininger P.L.. Human retroelements may introduce intragenic polyadenylation signals. Cytogenet. Genome Res. 2005; 110:365–371. [DOI] [PubMed] [Google Scholar]
- 18. Chen C., Ara T., Gautheret D.. Using Alu elements as polyadenylation sites: a case of retroposon exaptation. Mol. Biol. Evol. 2009; 26:327–334. [DOI] [PubMed] [Google Scholar]
- 19. Chen L.L., DeCerbo J.N., Carmichael G.G.. Alu element-mediated gene silencing. EMBO J. 2008; 27:1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gong C., Maquat L.E.. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3΄ UTRs via Alu elements. Nature. 2011; 470:284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gong C., Tang Y., Maquat L.E.. mRNA-mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2013; 20:1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang J., Gong C., Maquat L.E.. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013; 27:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bachvarova R. Small B2 RNAs in mouse oocytes, embryos, and somatic tissues. Dev. Biol. 1988; 130:513–523. [DOI] [PubMed] [Google Scholar]
- 24. Fornace A.J. Jr, Mitchell J.B.. Induction of B2 RNA polymerase III transcription by heat shock: enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Res. 1986; 14:5793–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jang K.L., Latchman D.S.. HSV infection induces increased transcription of Alu repeated sequences by RNA polymerase III. FEBS Lett. 1989; 258:255–258. [DOI] [PubMed] [Google Scholar]
- 26. Liu W.M., Chu W.M., Choudary P.V., Schmid C.W.. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995; 23:1758–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panning B., Smiley J.R.. Activation of RNA polymerase III transcription of human Alu repetitive elements by adenovirus type 5: requirement for the E1b 58-kilodalton protein and the products of E4 open reading frames 3 and 6. Mol. Cell. Biol. 1993; 13:3231–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Panning B., Smiley J.R.. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology. 1994; 202:408–417. [DOI] [PubMed] [Google Scholar]
- 29. Williams W.P., Tamburic L., Astell C.R.. Increased levels of B1 and B2 SINE transcripts in mouse fibroblast cells due to minute virus of mice infection. Virology. 2004; 327:233–241. [DOI] [PubMed] [Google Scholar]
- 30. Karijolich J., Abernathy E., Glaunsinger B.A.. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-kappaB pathway. PLoS Pathogens. 2015; 11:e1005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Allen T.A., Von Kaenel S., Goodrich J.A., Kugel J.F.. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004; 11:816–821. [DOI] [PubMed] [Google Scholar]
- 32. Mariner P.D., Walters R.D., Espinoza C.A., Drullinger L.F., Wagner S.D., Kugel J.F., Goodrich J.A.. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008; 29:499–509. [DOI] [PubMed] [Google Scholar]
- 33. Yakovchuk P., Goodrich J.A., Kugel J.F.. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:5569–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A. et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011; 471:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tarallo V., Hirano Y., Gelfand B.D., Dridi S., Kerur N., Kim Y., Cho W.G., Kaneko H., Fowler B.J., Bogdanovich S. et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012; 149:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dong X., Feng H., Sun Q., Li H., Wu T.T., Sun R., Tibbetts S.A., Chen Z.J., Feng P.. Murine gamma-herpesvirus 68 hijacks MAVS and IKKbeta to initiate lytic replication. PLoS Pathogens. 2010; 6:e1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Varshney D., Vavrova-Anderson J., Oler A.J., Cowling V.H., Cairns B.R., White R.J.. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat. Commun. 2015; 6:6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu W.M., Maraia R.J., Rubin C.M., Schmid C.W.. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994; 22:1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kondo Y., Issa J.P.. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J. Biol. Chem. 2003; 278:27658–27662. [DOI] [PubMed] [Google Scholar]
- 40. Ichiyanagi K., Li Y., Watanabe T., Ichiyanagi T., Fukuda K., Kitayama J., Yamamoto Y., Kuramochi-Miyagawa S., Nakano T., Yabuta Y. et al. Locus- and domain-dependent control of DNA methylation at mouse B1 retrotransposons during male germ cell development. Genome Res. 2011; 21:2058–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Englander E.W., Wolffe A.P., Howard B.H.. Nucleosome interactions with a human Alu element. Transcriptional repression and effects of template methylation. J. Biol. Chem. 1993; 268:19565–19573. [PubMed] [Google Scholar]
- 42. Adler H., Messerle M., Wagner M., Koszinowski U.H.. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 2000; 74:6964–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Richner J.M., Clyde K., Pezda A.C., Cheng B.Y., Wang T., Kumar G.R., Covarrubias S., Coscoy L., Glaunsinger B.. Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PLoS Pathogens. 2011; 7:e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Turatsinze J.V., Thomas-Chollier M., Defrance M., van Helden J.. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 2008; 3:1578–1588. [DOI] [PubMed] [Google Scholar]
- 49. Lunyak V.V., Prefontaine G.G., Nunez E., Cramer T., Ju B.G., Ohgi K.A., Hutt K., Roy R., Garcia-Diaz A., Zhu X. et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007; 317:248–251. [DOI] [PubMed] [Google Scholar]
- 50. Listerman I., Sapra A.K., Neugebauer K.M.. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006; 13:815–822. [DOI] [PubMed] [Google Scholar]
- 51. Suzuki K., Bose P., Leong-Quong R.Y., Fujita D.J., Riabowol K.. REAP: A two minute cell fractionation method. BMC Res. Notes. 2010; 3:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ichiyanagi K. Epigenetic regulation of transcription and possible functions of mammalian short interspersed elements, SINEs. Genes Genet. Syst. 2013; 88:19–29. [DOI] [PubMed] [Google Scholar]
- 53. Baker R.E., Gabrielsen O., Hall B.D.. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J. Biol. Chem. 1986; 261:5275–5282. [PubMed] [Google Scholar]
- 54. Pelechano V., Steinmetz L.M.. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013; 14:880–893. [DOI] [PubMed] [Google Scholar]
- 55. Schonborn J., Oberstrass J., Breyel E., Tittgen J., Schumacher J., Lukacs N.. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991; 19:2993–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cimino G.D., Gamper H.B., Isaacs S.T., Hearst J.E.. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985; 54:1151–1193. [DOI] [PubMed] [Google Scholar]
- 57. Isaacs S.T., Shen C.K., Hearst J.E., Rapoport H.. Synthesis and characterization of new psoralen derivatives with superior photoreactivity with DNA and RNA. Biochemistry. 1977; 16:1058–1064. [DOI] [PubMed] [Google Scholar]
- 58. Rabin D., Crothers D.M.. Analysis of RNA secondary structure by photochemical reversal of psoralen crosslinks. Nucleic Acids Res. 1979; 7:689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Calvet J.P., Pederson T.. Heterogeneous nuclear RNA double-stranded regions probed in living HeLa cells by crosslinking with the psoralen derivative aminomethyltrioxsalen. Proc. Natl. Acad. Sci. U.S.A. 1979; 76:755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu L., Pan J., Thoroddsen V., Wysong D.R., Blackman R.K., Bulawa C.E., Gould A.E., Ocain T.D., Dick L.R., Errada P. et al. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot. Cell. 2003; 2:256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wagner S.D., Yakovchuk P., Gilman B., Ponicsan S.L., Drullinger L.F., Kugel J.F., Goodrich J.A.. RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J. 2013; 32:781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Covarrubias S., Richner J.M., Clyde K., Lee Y.J., Glaunsinger B.A.. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J. Virol. 2009; 83:9554–9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Z., Carmichael G.G.. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001; 106:465–475. [DOI] [PubMed] [Google Scholar]
- 64. Athanasiadis A., Rich A., Maas S.. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004; 2:e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bazak L., Levanon E.Y., Eisenberg E.. Genome-wide analysis of Alu editability. Nucleic Acids Res. 2014; 42:6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Carmi S., Borukhov I., Levanon E.Y.. Identification of widespread ultra-edited human RNAs. PLoS Genet. 2011; 7:e1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim D.D., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A.. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004; 14:1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Daniel C., Silberberg G., Behm M., Ohman M.. Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol. 2014; 15:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Conti A., Carnevali D., Bollati V., Fustinoni S., Pellegrini M., Dieci G.. Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-Seq data. Nucleic Acids Res. 2015; 43:817–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Canella D., Bernasconi D., Gilardi F., LeMartelot G., Migliavacca E., Praz V., Cousin P., Delorenzi M., Hernandez N., Cycli X.C.. A multiplicity of factors contributes to selective RNA polymerase III occupancy of a subset of RNA polymerase III genes in mouse liver. Genome Res. 2012; 22:666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Carriere L., Graziani S., Alibert O., Ghavi-Helm Y., Boussouar F., Humbertclaude H., Jounier S., Aude J.C., Keime C., Murvai J. et al. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res. 2012; 40:270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kutter C., Brown G.D., Goncalves A., Wilson M.D., Watt S., Brazma A., White R.J., Odom D.T.. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat. Genet. 2011; 43:948–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Oler A.J., Alla R.K., Roberts D.N., Wong A., Hollenhorst P.C., Chandler K.J., Cassiday P.A., Nelson C.A., Hagedorn C.H., Graves B.J. et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 2010; 17:620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Canella D., Praz V., Reina J.H., Cousin P., Hernandez N.. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010; 20:710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Roberts D.N., Stewart A.J., Huff J.T., Cairns B.R.. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:14695–14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Alla R.K., Cairns B.R.. RNA polymerase III transcriptomes in human embryonic stem cells and induced pluripotent stem cells, and relationships with pluripotency transcription factors. PLoS One. 2014; 9:e85648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li T.H., Schmid C.W.. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001; 276:135–141. [DOI] [PubMed] [Google Scholar]
- 78. Moqtaderi Z., Wang J., Raha D., White R.J., Snyder M., Weng Z., Struhl K.. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat. Struct. Mol. Biol. 2010; 17:635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chu W.M., Ballard R., Carpick B.W., Williams B.R., Schmid C.W.. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 1998; 18:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hu Q., Tanasa B., Trabucchi M., Li W., Zhang J., Ohgi K.A., Rose D.W., Glass C.K., Rosenfeld M.G.. DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nat. Struct. Mol. Biol. 2012; 19:1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cech T.R., Steitz J.A.. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014; 157:77–94. [DOI] [PubMed] [Google Scholar]
- 82. Ishida K., Miyauchi K., Kimura Y., Mito M., Okada S., Suzuki T., Nakagawa S.. Regulation of gene expression via retrotransposon insertions and the noncoding RNA 4.5S RNAH. Genes Cells. 2015; 20:887–901. [DOI] [PubMed] [Google Scholar]
- 83. Chen L.L., Carmichael G.G.. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009; 35:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hu S.B., Xiang J.F., Li X., Xu Y., Xue W., Huang M., Wong C.C., Sagum C.A., Bedford M.T., Yang L. et al. Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. Genes Dev. 2015; 29:630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rattani A., Wolna M., Ploquin M., Helmhart W., Morrone S., Mayer B., Godwin J., Xu W., Stemmann O., Pendas A. et al. Sgol2 provides a regulatory platform that coordinates essential cell cycle processes during meiosis I in oocytes. Elife. 2013; 2:e01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Llano E., Gomez R., Gutierrez-Caballero C., Herran Y., Sanchez-Martin M., Vazquez-Quinones L., Hernandez T., de Alava E., Cuadrado A., Barbero J.L. et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 2008; 22:2400–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lee J., Kitajima T.S., Tanno Y., Yoshida K., Morita T., Miyano T., Miyake M., Watanabe Y.. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat. Cell Biol. 2008; 10:42–52. [DOI] [PubMed] [Google Scholar]
- 88. Lu Z., Zhang Q.C., Lee B., Flynn R.A., Smith M.A., Robinson J.T., Davidovich C., Gooding A.R., Goodrich K.J., Mattick J.S. et al. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016; 165:1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sharma E., Sterne-Weiler T., O’Hanlon D., Blencowe B.J.. Global mapping of human RNA–RNA interactions. Mol. Cell. 2016; 62:618–626. [DOI] [PubMed] [Google Scholar]
- 90. Aw J.G., Shen Y., Wilm A., Sun M., Lim X.N., Boon K.L., Tapsin S., Chan Y.S., Tan C.P., Sim A.Y. et al. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol. Cell. 2016; 62:603–617. [DOI] [PubMed] [Google Scholar]
- 91. Crepaldi L., Policarpi C., Coatti A., Sherlock W.T., Jongbloets B.C., Down T.A., Riccio A.. Binding of TFIIIC to sine elements controls the relocation of activity-dependent neuronal genes to transcription factories. PLoS Genet. 2013; 9:e1003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.