Abstract
Variations in mutation rates across the genome have been demonstrated both in model organisms and in cancers. This phenomenon is largely driven by the damage specificity of diverse mutagens and the differences in DNA repair efficiency in given genomic contexts. Here, we demonstrate that the single-strand DNA-specific cytidine deaminase APOBEC3B (A3B) damages tRNA genes at a 1000-fold higher efficiency than other non-tRNA genomic regions in budding yeast. We found that A3B-induced lesions in tRNA genes were predominantly located on the non-transcribed strand, while no transcriptional strand bias was observed in protein coding genes. Furthermore, tRNA gene mutations were exacerbated in cells where RNaseH expression was completely abolished (Δrnh1Δrnh35). These data suggest a transcription-dependent mechanism for A3B-induced tRNA gene hypermutation.
Interestingly, in strains proficient in DNA repair, only 1% of the abasic sites formed upon excision of A3B-deaminated cytosines were not repaired leading to mutations in tRNA genes, while 18% of these lesions failed to be repaired in the remainder of the genome. A3B-induced mutagenesis in tRNA genes was found to be efficiently suppressed by the redundant activities of both base excision repair (BER) and the error-free DNA damage bypass pathway. On the other hand, deficiencies in BER did not have a profound effect on A3B-induced mutations in CAN1, the reporter for protein coding genes. We hypothesize that differences in the mechanisms underlying ssDNA formation at tRNA genes and other genomic loci are the key determinants of the choice of the repair pathways and consequently the efficiency of DNA damage repair in these regions.
Overall, our results indicate that tRNA genes are highly susceptible to ssDNA-specific DNA damaging agents. However, increased DNA repair efficacy in tRNA genes can prevent their hypermutation and maintain both genome and proteome homeostasis.
1. Introduction
Single-stranded DNA (ssDNA) accumulates lesions and mutations at a higher rate than intact double stranded DNA. This hypermutability of ssDNA is due to the inability of major repair pathways including base excision repair (BER) and nucleotide excision repair (NER) to repair lesions on ssDNA, as well as the enhanced susceptibility of nitrogenous bases exposed in ssDNA to damaging agents [1–6]. Recent studies on model organisms and analysis of mutation data in whole genome- or exome-sequenced cancers has highlighted the role of the APOBEC (apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like) family of cytidine deaminases as potent ssDNA-specific mutagens [5–8]. The APOBEC3 enzymes normally function to restrict the spread of retrotransposons and propagation of retroviruses by mutagenizing ssDNA formed during replication of a virus or a mobile element. However, upon localization to the nucleus, APOBEC3 can mutagenize the host genome (reviewed in [5–7]). Various studies, using model organisms ectopically expressing the APOBEC3 enzymes and other known DNA cytidine deaminases have demonstrated that ssDNA formed during DNA repair, replication and transcription is susceptible to mutagenesis by these enzymes [9–12]. In concordance with these studies, high mutation loads attributable to the activity of APOBEC3A (A3A) and APOBEC3B (A3B) have been reported in a variety of cancers [8, 13–18].
ssDNA is primarily generated endogenously via two mechanisms (reviewed in [5, 6]). In the first mechanism, damaged persistent ssDNA cannot be fixed by excision repair because due to the lack of the undamaged complementary DNA strand available for templated resynthesis of excised damaged nucleotide(s). For example, several kilobases of ssDNA can be generated during resection of a double-strand break in the genome. Such ssDNA is highly susceptible to DNA damage and as such, high mutation density has been associated with break sites in yeast and in cancer genomes. The second category comprises transiently formed ssDNA during DNA metabolic processes such as replication and transcription. The presence of an undamaged strand allows templated excision repair of lesions in the ssDNA caused by ssDNA-specific damaging agents. However, such transiently formed ssDNA would be highly mutable in cells with excision repair defects and even in cells with normal repair if such a repair is delayed resulting in error-prone replication over the damage.
During DNA replication, the discontinuous mode of synthesis of the lagging strand renders it transiently single stranded and susceptible to damage accumulation. Consistently, in yeast strains with a defective S-phase checkpoint complex, treatment with the genotoxic drug methylmethane sulfonate (MMS) resulted in higher DNA damage and clustered mutagenesis on the lagging strand [18]. Similarly, in bacteria, in yeast and in cancers, it has been demonstrated that APOBEC3A (A3A) and/or APOBEC3B (A3B) predominantly deaminate cytosines on the transiently single-stranded lagging strand during normal DNA replication [9, 19–21]. This effect is further exacerbated upon replication stress, supporting the hypothesis that ssDNA formed during DNA replication is a major source of APOBEC-induced mutations [9].
Highly transcribed genes are known to be at-risk for accumulation of mutations and elevated mitotic recombination frequencies (reviewed in [22]). In yeast and in bacteria, mutation rates in reporter genes under a strong regulatable promoter were found to increase with higher expression level of the gene [23, 24]. Moreover, during transcription, hybridization of the newly formed RNA with the template DNA strand can result in the formation of R-loops – RNA:DNA hybrids with ssDNA generated via extrusion of the non-template strand – which are susceptible to both endogenous and exogenous DNA damaging agents. Consequently, bacterial strains challenged with DNA damaging agents demonstrate a bias in mutation loads towards the non-transcribed strand of reporter genes and expressed genomic regions [25–28]. Transcription-associated mutagenesis due to ssDNA-specific cytidine deaminases was also found to be further augmented in human cells and yeast strains with increased R-loop formation [29, 30].
Previously it was shown in yeast that tRNA genes are targeted with higher efficiency by activation-induced cytidine deaminase (AID) and the hyper-active mutant form of APOBEC3G (A3G) [12] compared to the highly transcribed mRNA genes. The formation and stabilization of these RNA:DNA hybrid structures was shown to be further augmented in tRNA genes in strains lacking either RNase H activity or containing defective Sen1 helicase [31, 32]. Therefore, it is highly likely that ssDNA in non-transcribed strand of R-loops formed in tRNA genes could be the origin for increased mutagenesis by APOBEC enzymes as well as by other ssDNA damaging agents. However, this hypothesis was not directly evaluated and the factors governing ssDNA mutagenesis and repair at tRNA genes were largely unknown.
In this study, we confirmed that in comparison with the remainder of the genome, tRNA genes are mutated at much higher frequencies by A3B and established that there are pronounced differences in repair efficiencies in these regions. We found that hypermutation of tRNA genes is transcription-dependent, and that it is confined to the non-transcribed strand. We also developed a selective system in yeast to assess the amounts of A3B-induced mutagenic lesions and their repair in each of these genomic targets, enabling us to determine the relative contributions and target-specific efficiencies of the repair pathways preventing hypermutation in transiently single stranded DNA of tRNA and mRNA genes.
2. Results
2.1. tRNA genes are preferentially targeted for damage and mutagenesis by APOBEC3B
We expressed human A3B under an inducible TET-promoter on a centromeric vector in haploid yeast strains [9]. In strains deficient in Ung1 activity, where all uracils formed by APOBEC-driven cytidine deamination would result in C→T (or G→A) transitions, mutations would indicate the sites of A3B enzymatic activity. The yeast strains utilized in our assays additionally carried the trp1-289 allele with the UAG (amber) nonsense mutation in the gene for phosphoribosylanthranilate isomerase (TRP1) resulting in tryptophan auxotrophy [33]. A3B expression led to a 191-fold increase in the reversion frequency of this allele, and a 79-fold increase in the rate of canavanine resistance (CanR) caused by mutations leading to inactivation of the Can1 arginine permease compared to strains without A3B (Table 1). Whole genome sequencing of 23 Trp+ revertants from independent cultures expressing A3B indicated that C→T mutations in the CCA anticodon of the Trp-tRNA genes, leading to the formation of Trp-tRNAs with CTA anticodon, were responsible for the Trp+ phenotype as opposed to direct reversion of the stop codon within the TRP1 gene (Supplementary Table 1a and1b). The A3B mutagenesis motif is tCw (mutated base capitalized, w – corresponds to either A or T), however cCa motifs can also be targeted by A3B [9, 17]. The CCA→CTA anticodon change would allow the mutated Trp-tRNA to pair with the UAG stop codon in the trp1-289 mRNA, thereby enabling the protein synthesis machinery to read through the stop codon and produce a full length polypeptide, thus restoring prototrophy for tryptophan.
Table 1.
A3B-induced CAN1 mutation and trp1-289 reversion rates.
| Genotype | A3B | CAN1 mutation rate X 10−7 (95% confidence intervals) | trp1-289 reversion rate X 10−7 (95% confidence intervals) |
|---|---|---|---|
| UNG1 | Yes | 100 (60–150) | 7 (3–20) |
| UNG1 | No | 2 (1–3) | 1 (0.4–1) |
| Δung1 | Yes | 500 (430–620) * | 2100 (1700–2700) * |
| Δung1 | No | 6 (5–10) | 11 (9–13) |
| Δung1, Δrnh1 | Yes | 500 (160–670) * | 1500 (1100–6300) * |
| Δung1, Δrnh1 | No | 5 (4–9) | 11 (6–17) |
| Δung1, Δrnh35 | Yes | 540 (380–690) * | 1800 (900–2100) * |
| Δung1, Δrnh35 | No | 10 (6–22) | 10 (8–20) |
| Δung1, Δrnh1, Δrnh35 | Yes | 910 (550–2400) # | 3900 (3200–12000) # |
| Δung1, Δrnh1, Δrnh35 | No | 10 (7–11) | 11 (9–13) |
| Δapn1 | Yes | 120 (100–190) | 21 (16–32) * |
| Δapn1 | No | 7 (6–12) | 3 (2–9) |
| Δapn2 | Yes | 150 (150–190) * | 19 (15–22) * |
| Δapn2 | No | 4 (3–7) | 1 (0.4 – 2) |
| Δapn1, Δapn2 | Yes | 120 (60–190) | 21 (15–40) * |
| Δapn1, Δapn2 | No | 9 (5–13) | 2 (0.5–2) |
| Δmms2 | Yes | 190 (170–210) * | 29 (24–34) * |
| Δmms2 | No | 16 (12–23) | 4 (3–6) |
| Δrad51 | Yes | 410 (270–980) * | 103 (61–176) * |
| Δrad51 | No | 37 (29–50) | 10 (8–19) |
| Δapn1, Δrad51 | Yes | 680 (450–1000) * | 370 (210–890) * |
| Δapn1, Δrad51 | No | 100 (100–290) | 39 (22–51) |
| Δmms2, Δapn1 | Yes | 320 (190–420) * | 125 (80–160) * |
| Δmms2, Δapn1 | No | 63 (40–140) | 19 (10–75) |
Denotes rates significantly higher than UNG1 strain with A3B (P<0.05)
Denotes rates significantly higher than Δung1 RNH1 RNH35 strain with A3B (P<0.05)
To confirm that mutations within Trp-tRNA CCA anticodon cause reversion to tryptophan in Δung1 strains, we tested six Trp+ strains with C→T mutations in the anticodons in each of the following Trp-tRNA genes - tW(CCA)G1, tW(CCA)K, tW(CCA)P and tW(CCA)M. First, the A3B-containing plasmid was lost, and subsequently each strain was crossed with an isogenic Trp− strain. As expected for mutations in tRNA genes, all the suppressor mutations were dominant, resulting in Trp+ diploids. Furthermore, we observed a 2:2 meiotic segregation ratio of the Trp+ and Trp- phenotypes in all 6 tetrads obtained from each of the 6 diploid strain, indicating that mutation in a single gene defines the reversion. 24 segregants from each diploid strain, 12 Trp+ and 12 Trp−, were tested for mutations in the Trp-tRNA anticodon in the Trp+ isolates. In all Trp+ isolates tested, C→T mutations were present in the anticodon of the Trp-tRNA genes and were absent in the Trp− isolates (Supplementary table 2). We did not observe any mutations in either the tW(CCA)G2 or the tW(CCA)J gene by whole-genome sequencing, therefore these genes were excluded from testing co-segregation of mutations with Trp+ phenotype. However, anticodon mutations in these genes could also presumably lead to suppression. Therefore, the estimated target size for A3B-induced reversion to Trp+ phenotype is estimated to be 6 nucleotides (1 in each Trp-tRNA gene) (Supplementary Table 1a and 1b).
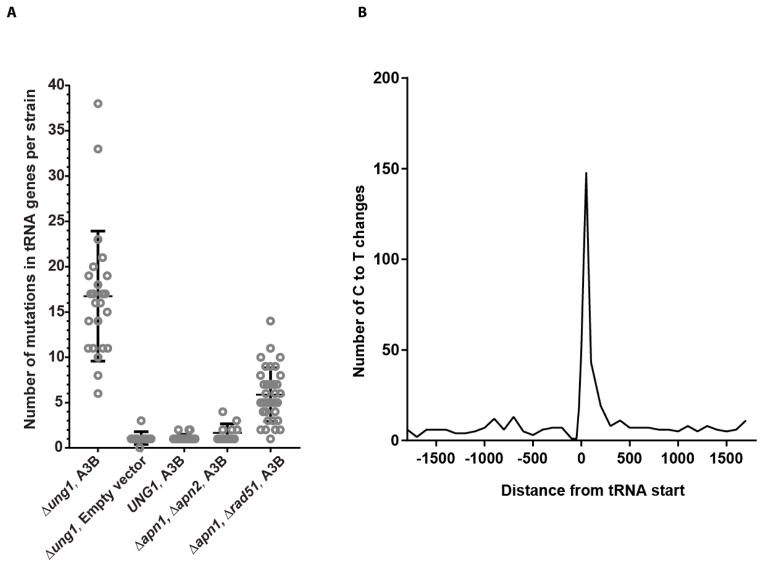
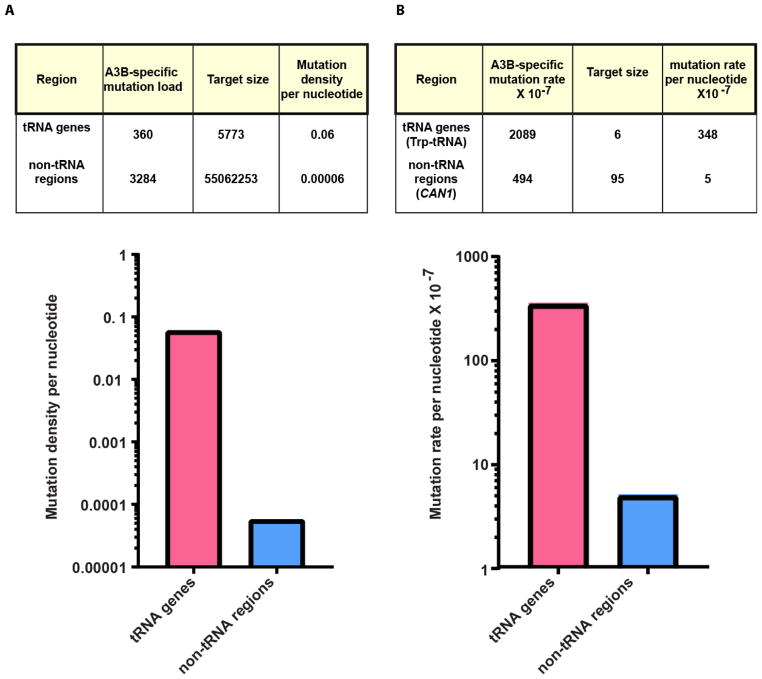
In addition to Trp-tRNA genes, multiple other tRNA genes (6 to 37 per strain) were also mutated in the genomes of trp-revertants expressing A3B in the Δung1 background as opposed to only one or no tRNA genes mutated in strains with the empty vector control (Fig. 1A). Moreover, alignment of all tRNA start positions and measurement of the number of C→T or G→A changes in 100 bp bins, demonstrates a clear peak in mutations up to 200 bases into tRNA genes (Fig. 1B). Most A3B-induced mutations occurred in tC and cC motifs of tRNA genes (383 /388) and non-tRNA areas of genomes (3284/3352). Previous observations indicated that APOBEC3B cytidine deamination has a preference towards tRNA genes in the yeast genome [12]. Accounting for the estimated number of cC and tC A3B mutation motifs in both areas, we observed that this preference is extremely high. The mutation frequency per nucleotide in tRNA genes is approximately 1,000-fold greater than in the remaining yeast genome (Fig. 2A).
Fig. 1. A3B-induced mutation load and spread in tRNA genes. (A).
The number of mutations in tRNA genes in each isolate sequenced and the median are depicted. (B) A3B-induced mutations in the Δung1 background are enriched within tRNA genes and spread up to 200 bases within the genes.
Fig. 2. A3B-induced mutation density is higher in tRNA genes than the remaining genome. (A).
Density of A3B-mutations in tC or cC motifs per tc or cc dinucleotide present in tRNA genes and non-tRNA genomic regions in the Δung1 background is calculated (mutated residue is capitalized). (B) The mutation rates per targeted nucleotide in CAN1 leading to CanR (non-tRNA) and Trp-tRNA(tRNA) genes leading to Trp+ phenotype in the Δung1 background are depicted. The target size was calculated as the number of tC and cC motifs multiplied by the total number of strains sequenced.
2.2. trp1-289 – reporter of A3B-induced ssDNA damage and mutagenesis in tRNA genes
High preference of A3B cytidine deamination for tRNA genes is also clearly seen with the trp1-289 reversion system, with rates of A3B-induced trp1-289 reversions exceeding that of CanR mutation by 4-fold. Increased reversion to Trp+ phenotype with this reporter has also been seen for various other ssDNA mutating cytidine deaminases [34]. Based on the assumption that mutations in the anticodon CCA to CTA in any of the six Trp-tRNA genes in the genome can result in Trp+ phenotype, we suggest that the selectable target size for reversion to Trp+ by A3B is only 6 nucleotides. On the other hand, the CanR phenotype can arise by any mutation that can inactivate the Can1 protein and permit selection on media containing canavanine. To determine the selectable target size of A3B-induced CanR mutant alleles, we calculated the total number of unique mutations made by A3B in proliferating cells in CAN1 in the study by Hoopes et.al [9]. In this dataset built from 383 CanR mutations, mutations were found in 99 different nucleotide positions (95 were C→T or G→A base substitutions within the context of cC or tC) (Supplementary Table 3). Therefore, the target size for A3B-induced CanR mutations is 95 nucleotides. Accounting for calculated target sizes, the rate per selectable nucleotide for reversion to Trp+ is 70-fold higher than that for CAN1 inactivation (Fig. 2B), suggesting that tRNA genes are the better preferred target for A3B cytidine deamination. The discrepancy in the ratio of mutation frequencies per targeted nucleotide in the reporter systems as compared to the ratio obtained from genome-wide analysis likely reflects the selection of trp-revertants in a non-canonical A3B motif. Regardless of this discrepancy, comparison of A3B-induced mutagenesis resulting in trp1-289 reversion with mutagenesis resulting in CanR is a useful tool for identifying genetic controls and defining mechanisms of damage repair and mutation prevention in tRNA genes.
2.3. A3B-induced cytidine deamination in tRNA genes does not show replication bias
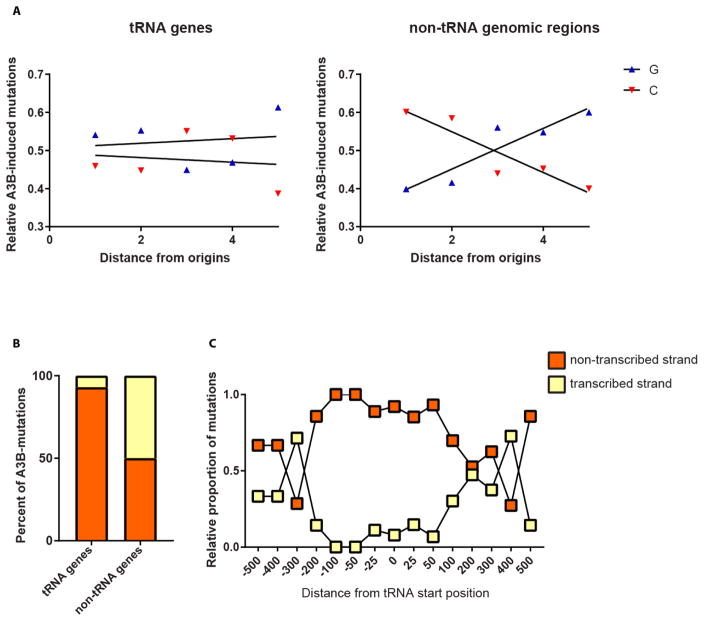
Recently, it was shown that A3B in yeast and in E. coli deficient in uracil DNA glycosylase preferentially mutagenizes the lagging strand during DNA replication, presumably targeting transient ssDNA formed at initiation of every Okazaki fragment ([9, 19]. To assess the role of DNA replication in A3B-induced tRNA mutations, we determined total numbers of C→T and G→A mutations binned by the relative distances between the origins of replication in the yeast genome, similar to [9]. Analysis of the total mutations across the genome in Δung1 strains with constitutive APOBEC3B expression showed a preponderance of C→T changes to the left of the origins and mostly G→A changes to the right of the origins, indicating that the direction of the fork progression is a key determinant of the strand in which cytosine deamination occurs (Fig. 3A). This bias in mutagenesis consistent with the lagging strand preference has also been shown for CAN1 in these strains in a previous study [9]. On the other hand, mutations in tRNA genes did not exhibit such a bias, pointing towards a replication-independent mode of A3B-driven cytidine deamination and mutagenesis in these genes (Fig. 3A).
Fig. 3. Replication and transcription-dependent strand-bias in A3B-induced mutations in tRNA genes and other genomic regions.
(A) Relative number of A3B-induced C→T and G→A changes in tRNA genes and non-tRNA genomic regions are plotted against the fractional distance between neighboring origins of replication in the yeast genome. The X-axis denotes the distance between neighboring origins of replication divided into 5 bins. (B) The percentages of A3B-induced mutations in the transcribed and non-transcribed strands of tRNA genes and other genes in the Δung1 strain are depicted. (C) The relative proportion of C→T changes in the transcribed and non-transcribed strands along the distance of tRNA genes in the Δung1 strain is shown. On the X-axis, the relative distances from tRNA gene start position is plotted in increments of 100 bases. The negative values denote regions upstream of tRNA genes, while the positive values represent genomic positions downstream of tRNA start positions.
2.4. A3B-mediated deamination is predominantly targeted to the non-transcribed strand of tRNA genes
Examination of the mutations in tRNA genes demonstrated that the majority (90%) of C→T changes in these genes were present on the non-transcribed strand. On the other hand, mutations in mRNA genes were not biased towards the non-transcribed strand with 40% of the mutations present on the non-transcribed strand, and 60% on the transcribed strand (Fig. 3B). We can see this preference of C→T mutations for the non-transcribed strand up to 100 bp upstream of tRNA genes (Fig. 3C). To determine if other cytidine deaminases demonstrate a similar bias towards the non-transcribed strand of tRNA genes, we analyzed previously published datasets of mutations in yeast genomes caused by A3A, A3B, A3G* and AID* (* = hyperactive mutants), where such a bias was not reported [12]. Similar to A3B, we noticed that A3A (93% of C→T changes in non-transcribed strand) and AID*, to a lesser extent, but still statistically significant (60% of C→T changes in the non-transcribed strand), demonstrate a bias towards mutagenizing the non-transcribed strand of tRNA genes. No bias is detectable for mutagenesis in mRNA genes. However, A3G* mutagenizes both strands of tRNA genes and mRNA genes to an equal extent (Supplementary Table 3). These data suggest that transcription of tRNA genes leads to prolonged exposure of the non-transcribed strand which is targeted by the APOBEC enzymes. The differences between the enzymes in their strand bias could indicate differences in their ability to bind RNA. A3G* was previously shown to be inefficient at binding RNA [12], as such, this enzyme, unlike A3A, A3B and AID, may not be actively targeted to transcriptionally active tRNA genes. On the other hand, the ability to bind RNA would place A3A, A3B and AID in close proximity to the transiently single-stranded non-transcribed strand during transcription, allowing increased deamination and mutagenesis.
2.5. Formation of transcription-associated R-loops increases access of A3B to transient ssDNA in the non-transcribed strand
tRNA genes have been shown to be hotspots for RNA:DNA hybrid formation [31, 32]. Formation and stabilization of such DNA secondary structures may lead to prolonged exposure of single stranded DNA which is a substrate for the APOBEC enzymes. To test if R-loop formation influences the ability of A3B to target tRNA genes, we deleted RNH1 and RNH35, the redundant ribonucleases that remove RNA from RNA:DNA hybrids in yeast. We found a 1.9-fold increase in trp1-289 reversion rates and a 1.8-fold increase in CAN1 mutation rate in the rnh1, rnh35 double mutants with A3B expression (Table 1). The increase in A3B-induced mutations points towards co-transcriptionally formed RNA:DNA hybrids as a source of ssDNA at tRNA genes. Previously, it has been demonstrated that strains with increased predisposition to RNA:DNA hybrid formation are susceptible to AID-induced cytidine deamination in the non-transcribed strand of RNA polymerase II transcribed genes [30]. The increase in mutagenesis at CAN1 locus in the rnh1, rnh35 double mutants with A3B expression is concordant with this observation and implies that in the double mutant there is an overall increase in R-loop formation, leading to increase in exposed single stranded DNA across the genome which can then be targeted by A3B.
2.6. Transcription-associated mutagenesis by A3B is repressed by functional repair pathways
A3B deaminates dC to create dU in DNA. If unrepaired, uracils in DNA result in C→T transitions marking positions of cytidine deamination by A3B. However, uracils can be recognized and removed by uracil DNA-glycosylase (Ung1 in yeast) resulting in the formation of abasic sites. When present in double stranded DNA, abasic sites can be removed and repaired by BER or by other repair pathways which use the complementary DNA strand as a template for re-synthesis [35]. If abasic sites generated by Ung1 are not repaired, they can be bypassed during replication by translesion polymerases leading mostly to C→T or to C→G substitutions in DNA [36]. Thus A3B induced mutations in UNG1 yeast would mark the positions where accurate repair of abasic sites did not occur. To determine the proficiency of repair of abasic sites in tRNA and in mRNA genes, we compared the rates of reversion to Trp+ and CAN1 inactivation when A3B was constitutively expressed in UNG1 wild-type and Δung1 strains. We found a 300-fold reduction in rates of reversion to Trp+ in the UNG1 wild-type strain compared to the Δung1 strain. However, mutation rate at the CAN1 locus showed only a 5-fold decrease in strains with proficient Ung1 protein, indicating that Trp-tRNA genes were repaired more efficiently than CAN1 (Table 1). To assess genome-wide bias in repair efficacy , we analyzed the number of C→T or C→G mutations in tC or cC motifs (mutated residue is capitalized) in tRNA genes and non-tRNA genomic regions, in whole genome sequenced Trp+ isolates derived from the wild-type or the Δung1 strain with A3B expression. To avoid biasing our analysis due to selection of the Trp+ phenotype, we only analyzed mutations in tRNA genes that were not within the CCA anticodon of the Trp-tRNA genes. We found that although in non-tRNA regions 18% of the A3B-induced mutagenic lesions were not accurately repaired, less than 1% of the lesions in tRNA genes were left unrepaired and resulted in mutations (Table 2). These data suggest that abasic sites in tRNA genes are repaired more efficiently than the remainder of the genome.
Table 2.
Genome-wide repair efficiency of A3B-induced lesions in tRNA and non-tRNA genomic regions in Ung1 defective and repair proficient strains.
| Genotype | Genomic region | Mutations | Number of strains sequenced | Unrepaired lesions (%)b | Repaired lesions (%)c |
|---|---|---|---|---|---|
| ung1::natMX3 | non- tRNA | 3284 | 23 | 100 | 0 |
| ung1::natMX3 | tRNA | 359 | 23 | 100 | 0 |
| UNG1 | non- tRNA | 580 | 24 | 17 | 83 |
| UNG1 | tRNA | 3 a | 24 | 1 | 99 |
Numbers denote only those mutations in tRNA genes that did not lead to selection of Trp+ phenotype.
Calculated as (100-Repaired lesions (%))
Calculated as described in methods.
2.7. Base excision repair and error-free DNA damage tolerance machinery work synergistically to repair co-transcription damage in tRNA genes
To determine the repair pathways responsible for preventing mutagenesis by abasic sites in tRNA genes, we deleted the AP-endonucleases APN1 and APN2 in strains with proficient Ung1 protein. Deletion of either or both genes in strains expressing A3B did not alter CAN1 mutation rates, however, rates of reversion to Trp+ were augmented 3-fold in both Δapn1, Δapn2 single mutant strains and in the Δapn1Δapn2 double mutants (Table 1). The lack of a dramatic increase in mutation rates suggests the existence of a second pathway for repair of abasic sites in yeast.
However, for the trp1-289 selective system there could be an alternative explanation involving abasic sites bypass in tRNA genes leading mostly to C→G changes which would not create nonsense-suppressor tRNA and would cause underestimation of tRNA mutation rate in UNG1 wild-type and Δapn1Δapn2 strains. To rule out this possibility, we examined genome-wide mutation spectra in 17 Trp+ isolates induced by A3B expression in Δapn1Δapn2 double mutants and in 24 Trp+ isolates derived from UNG1 wild-type strains (described in the previous section). Due to selection of Trp+ phenotype, 30 of 41 Trp+ isolates carried a C→T change in 1 of the 6 Trp-tRNA CCA anticodons. Altogether, the isolates carried 13 unselected mutations (not in Trp-tRNA anticodon) in tRNA genes 4 of which (all in Δapn1Δapn2) were C→G changes (Supplementary Table 1a). Thus, C→G substitutions are not the predominant change and would therefore not abrogate reversion to Trp+ phenotype. Moreover, the total number of tRNA genes mutated in these samples ranges from 1 to 4 in each strain, unlike the Δung1 strain (Fig. 1A), further substantiating the conclusion that another repair pathway likely corrects damage in tRNA genes.
We further analyzed defects in the error-free DNA damage tolerance machinery. Upon encountering a replication blocking lesion, Rad5, Mms2 and Ubc13 polyubiquitinate PCNA and allow for homologous recombination dependent template switching using Rad51 and synthesis over the undamaged newly synthesized DNA strand, culminating in error-free lesion bypass (reviewed in [37]). Deletion of MMS2 or RAD51 in strains expressing A3B led to a 2 and 4-fold increase in CAN1 mutation rates, and a 4- and 15-fold increase in rates of reversion to Trp+, respectively. The rates of reversion to Trp+ were further amplified in Δapn1Δmms2 (18-fold increase over the wild-type strain) and Δapn1Δrad51 (53-fold increase over the wild-type strain). However, CAN1 mutation rates were not altered in the Δapn1Δmms2 and Δapn1Δrad51 as compared to the single mutant Δmms2 or Δrad51 strains (Table 1).
We note that mutation rates for reversion to Trp+ in the Δrad51Δapn1 strains with A3B are still 6-fold lower than the Δung1 strains with A3B (Table 1). To determine if Apn2 was responsible for correction of the remaining abasic sites in tRNA genes, we created the Δrad51Δapn1Δapn2 strain. However, when transformed with A3B-containing plasmids, this strain did not yield any transformants, indicating inviability of this triple mutant with A3B.
In accordance with increased rates of reversion to Trp+ in the Δapn1Δrad51 strains with A3B, genome-wide sequencing of 35 Trp+ isolates demonstrated that mutations in tRNA genes are also augmented in these strains. There were 1 to 13 mutations in tRNA genes per isolate (Fig. 1A), with 186 out of 203 mutations in tC or cC motifs (Supplementary Table 1a).
These data suggest that transcription-associated abasic sites formed in tRNA genes, by the subsequent actions of A3B and Ung1, are primarily repaired by the BER machinery, however in BER-deficient strains, these sites can block the replication machinery and are bypassed by the error-free DNA damage tolerance pathway, which thereby provides a second level of mutation prevention. In contrast, BER does not play an active role in repair of abasic sites formed by the consecutive actions of A3B and Ung1 in CAN1.
3. Discussion
Transcriptionally active tRNA genes have been shown to be prone to accumulate RNA:DNA hybrids and consequently DNA damage and rearrangements [31, 32, 38]. We demonstrate here that these regions are also hypermutable by A3B, the ssDNA-specific cytidine deaminase. Analysis of the mutation density per targeted nucleotide indicated that tRNA genes were mutated 1000-fold more frequently than the non-tRNA genomic regions. We found that mutagenic lesions in tRNA genes can be efficiently repaired by the redundant BER and the error-free DNA damage tolerance pathways, while BER was found to be inept at preventing mutations in protein coding genes of proliferating yeast cells. Surprisingly, overall repair efficacy in non-tRNA portions of the genome was much lower. Our data suggest that the mutagenic potential of A3B-induced lesions in the genome is dependent on the susceptibility of the target region and the repair pathways’ functionality within the given context.
Since mutagenesis caused by cytidine deamination in tRNA genes was heavily biased to the non-transcribed strand, unlike mRNA genes, increased formation of R-loops in tRNA genes can explain their higher propensity to lesions. In support of this, inactivation of redundant RNase H1 and H2, known to elevate R-loop formation, further increases the already high rate of A3B-induced mutations in tRNA genes (Table 1). Moreover, the bias for mutagenesis of the non-transcribed strand by cytidine deaminases appears to depend upon their ability to bind RNA (Supplementary Table 4). This phenomenon would target these enzymes to actively transcribed tRNAs and increased stability of RNA:DNA hybrids in these regions would augment the susceptibility of the single-stranded non-transcribed strand to mutagenesis.
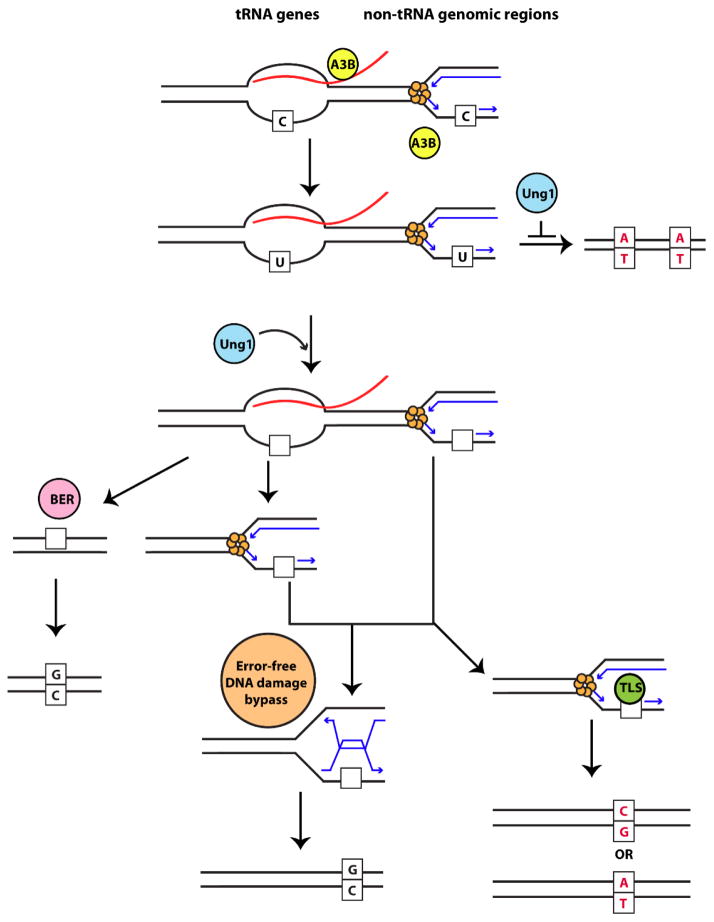
The susceptibility of the tRNA genomic regions to lesions appears to be counteracted by very high efficiency of mutation avoidance provided by redundant repair pathways schematically presented in Fig. 4. Cytosine deamination by the APOBEC enzymes results in the formation of uracils (dU) in DNA. These residues are detected and removed by the uracil N-glycosylase Ung1 in yeast, leading to the formation of mutagenic abasic sites (Reviewed in [35]).
Fig. 4. Model for A3B-induced mutagenesis and repair in tRNA genes and non-tRNA genomic regions.
ssDNA mutated by A3B (yellow circle) in tRNA genes is stabilized by RNA:DNA hybrids formed during transcription (red = RNA) while in other genomic regions ssDNA is formed on the lagging strand during DNA replication (blue arrows). Moreover, the RNA transcript at tRNA genes could directly recruit A3B to the ssDNA formed during transcription. In the absence of Ung1 (blue circle), replication across dU residues leads to a C→T transition (red). Ung1 (blue circle) can act on dU residues and form abasic sites in DNA (empty squares). In tRNA genes, removal of RNA leads to reannealing of the strands, and base excision repair machinery (pink circle) can repair the abasic site in an error-free manner. On the other hand, persistent RNA:DNA hybrids, would prevent repair by BER, and abasic site would later be presented as a block to the replication fork. Abasic sites at the fork, can either be undergo error-free repair by the error-free DNA damage bypass pathway (orange circle) or are bypassed by translesion synthesis polymerases (green circle) which can lead to C→T or C→G substitutions (red).
Recognition of abasic sites in double stranded DNA by the AP-endonucleases Apn1 and Apn2, and their cleavage initiates the base excision repair (BER) pathway to repair the damage using the undamaged second DNA strand as a template. Unrepaired and persistent abasic sites can block the progression of the DNA replication machinery and can be bypassed error-free by template switching and synthesis using the undamaged DNA strand, or error-prone synthesis over the DNA damage by translesion polymerases [37]. On the other hand, re-initiation of synthesis downstream of unrepaired abasic sites could lead to a single stranded gap, and upon a second round of replication double-strand breakage (DSB) occurs stimulating mitotic recombination. Consequently, in strains lacking Apn1 and Apn2 or the downstream BER enzymes, treatment with DNA damaging agents leads to accumulation of abasic sites in DNA, and results in DSB formation and cytotoxicity [39, 40]. Also, expression of AID in yeast strains with proficient Ung1 is recombinogenic, most likely reflecting the unstable nature of abasic sites formed upon excision of dU residues in DNA [41]. Abasic sites on the transcribed strand are also capable of blocking the transcription machinery and are highly mutagenic in nature [42, 43]. In the absence of BER, abasic sites encountered by the RNA-polymerase II machinery on the transcribed strand are repaired by the transcription-coupled nucleotide excision repair machinery with restoration of wild-type sequence, while those on the non-transcribed strand are repaired by the Ntg1 and Ntg2 lyases [44]. Based on the studies presented above and others, it is becoming evident that the context in which abasic sites are formed influences the downstream repair pathway choice.
Through our current study, we described a convenient selective reporter of mutagenesis in tRNA genes in combination with whole genome sequencing, which allowed us to compare damage propensity and repair efficiency between tRNA and mRNA genes. We showed that mutations in tRNA genes are repaired more efficiently than the remaining genomic regions. Our analysis of the repair pathways operating in the two genomic regions demonstrated that in Ung1 proficient strains, abasic sites formed in tRNA genes due to cytidine deamination during transcription, are repaired by the activity of both BER or bypassed by the error-free DNA damage tolerance pathway comprising of Mms2, Ubc13, Rad5 and the homologous recombination machinery.
The difference in repair of abasic sites in tRNA genes and mRNA genes likely reflects the modes of ssDNA formation in these regions. In non-tRNA genomic regions, we saw a preponderance of C→T changes in the lagging strand, implying that A3B mutates ssDNA in Okazaki fragments during replication. However, the lack of a complementary undamaged DNA strand precludes repair by BER and the abasic sites could lead to blockage of the replication machinery. Conversely, A3B mutagenizes tRNA genes at RNA:DNA hybrids formed during transcription. Resolution of the RNA:DNA hybrids, and reannealing of the abasic site-containing strand with the undamaged strand provides an ideal substrate for BER. However, in strains with defective BER the abasic site would persist until the next round of replication and would also present as a block to the replisome. The paused replication machinery at tRNA genes or non-tRNA regions, would use the template switching mechanism, employing the newly synthesized leading strand for error-free repair of the abasic site and replication restart (Fig. 4).
Our work demonstrates that tRNA genes can be highly susceptible to transcription- associated mutagenesis when repair pathways are compromised. The key to preventing mutagenesis in tRNA genes is the robust activity of uracil DNA glycosylase, which opens the gateway to redundant mutation avoidance pathways. Mutated tRNAs could lead to the production of defective and misfolded proteins during translation which in turn could aggregate in cells leading to proteotoxic stress [45]. In concordance with the toxic nature of such defective proteins, mutations in the protein synthesis machinery leading to widespread mistranslation have been associated with a variety of neurological diseases and cancers [46]. On the other hand, mutations in tRNA genes could provide phenotypic diversity and provide selective benefits to cells during evolutionary processes [47, 48]. Elucidation of the mechanisms underlying mutagenesis and repair of DNA damage in tRNA genes is therefore crucial in understanding their role in disease and evolution.
4. Materials and Methods
4.1. Yeast strains
The yeast strains used in this study were derivatives of the YSR128 strain (MATα ura3Δ can1Δ ade2Δ leu2-3,112 trp1-289 lys2::ADE2-URA3-CAN1) [18]. Briefly, the CAN1, URA3 and ADE2 genes were deleted from their native positions, and inserted into the LYS2 locus. Previously we determined that relocating CAN1 from its native position to LYS2 did not alter mutation rates within CAN1 [18]. UNG1 was replaced with natMX3 cassette, conferring resistance to nourseothricin [49]. RNH1, APN2, RAD51 and MMS2 were disrupted by the kanMX cassette, conferring resistance to geneticin [49], APN1 and RNH35 were replaced with the BSD gene, leading to resistance of yeast strains to blasticidin [50]. The complete strain list is provided in Supplementary Table 5. PCR using the disruption cassettes was performed with primers carrying overhangs for the 3’ and 5’ regions of the genes mentioned above, and one-step integration was used to replace the ORFs of the genes with the selectable markers.
4.2. Measurement of tRNA-specific and CAN1 mutation rate
Yeast strains were transformed with an A3B-containing expression vector, or an empty vector carrying the hphMX selective cassette conferring resistance to hygromycin [9]. The strains carrying the plasmids were streaked out for single colonies on complete media supplemented with 300 μg/ml hygromycin. After 2 days of growth, 8 to 12 patches of independent colonies were made on hygromycin-containing media and the strains were allowed to grow overnight at 30°C. The complete patches were re-suspended in water, and appropriate dilutions were plated out on complete media to determine viability of the strains, synthetic media lacking tryptophan for assaying trp1-289 reversion frequency and synthetic media without arginine supplemented with 60 mg/L L-canavanine for CANR mutation rate estimation.
Mutation rate (μ) for each culture was calculated based on the mutant frequency (f) and total number of cells in a culture (N) resolving by iterations the transcendent equation (1)
derived in [51] for accumulation of mutants in a culture grown from a single cell or from small number of cells in non-selective growth conditions. Since this method is sensitive to mutation jackpots, the median rate was used as a measure of mutability. The 95% confidence intervals for a median rate were determined as described in [52]. Since we cannot rule out that mutation accumulation during cell division is not uniform, we also provide mutation frequencies (f) for the genotypes tested (Supplementary Table 6). Comparison of mutation frequencies in different genotypes leads to the same conclusions as comparison of rates.
With small N, equation (1) transforms into equation (2)
which reflects the notion that in cultures with 1–4 mutants they are coming from independent events rather than from propagation of a smaller number of mutants. Thus for the genotypes and/or conditions with very small mutation frequencies, especially with large number of cultures lacking mutants the rate was estimated approximately equal to the total frequency of mutants in in all cultures. In these cases, confidence intervals were calculated in assumption that the total number of mutants in all cultures has Poisson distribution.
p-values for differences between mutation rates were calculated by comparing the sets of mutation rates calculated for all replicates of the genotypes being tested, using the two sided Mann-Whitney U test.
4.3. Whole genome sequencing of the yeast strains
The A3B and empty plasmid containing strains were streaked out on complete media with hygromycin. 12 to 24 individual colonies were plated on media lacking tryptophan. The colonies were also inoculated in YPD overnight for DNA extraction (Trp− isolates). Genomic DNA was also extracted for the Trp+ revertants obtained from each isolate. DNA libraries were prepared using the KAPA hyper kit (KAPA Biosystems, Wilmington, MA), and paired end 100 bp or125 bp reads from the HiSeq 2500 sequencer or 150 bp reads from the HiSeq 4000 sequencer (Illumina, San Diego, CA). The mutation analysis was done using the CLC-genomics workbench (Qiagen, Redwood City, CA) as follows. The sequences were aligned to the reference genome from ySR128 [18] using default parameters, and were realigned using variants detected from the “low frequency variant detection” tool. Duplicate mapped reads were removed and the “fixed ploidy variant detection” tool was used with ploidy annotated as 1, and required variant probability of 90%. Only homozygous mutations were considered. To remove mutation calls accrued in the parental strains, the variants in each Trp+ isolate were compared against the parental Trp− isolates. Only base substitutions that were present in the Trp+ strains but not in the parent strains, were analyzed further.
4.4. Analysis of replication and transcription strand bias
Replication strand bias in all A3B-specific mutations in the Δung1 strain was analyzed as described previously [9]. To assess transcription associated strand bias, the variants were annotated for their presence in mRNA or tRNA genes using GRanges in Bioconductor [53]. C→T changes were assessed for their presence on either the same or complementary strands in mRNA or tRNA genes. A one-sided binomial test was performed using the expectation that mutations were enriched on the non-transcribed strand. The Benjamini-Hochberg method was used to correct P-values for multiple testing.
4.5. Analysis of mutation density relative to tRNA start position
Bedtools closestBed [54] was used to compare mutation coordinates and report the distance of mutations from the closest tRNA gene start position. The mutations were binned in 100 base pair bins with 0 referring to mutations in the start position of tRNAs, negative distances implying mutations present upstream and positive distances implying mutations present downstream to the start position.
4.6. Analysis of mutation density per targeted nucleotide in selected genomic regions
Bedtools getFasta [54] was used to get the fasta sequence for tRNA genes. To analyze the number of tc or cc dinucleotide motifs in either the tRNA genes or the whole yeast genome, SeqinR [55] was used. The number of tc or cc dinucleotides in the non-tRNA genomic regions were calculated by subtracting the number of motifs in the yeast genome with those in the tRNA genes. To avoid a bias due to selection of trp-revertants, we only calculated the mutation density for unselected mutations and eliminated the C→T changes in the cCa anticodon of Trp-tRNA genes. Mutation density per targeted nucleotide in the genomic regions was calculated as:
The efficiency of correction of A3B-induced lesions in Ung1 proficient and BER deficient strains was calculated as:
Wherein, mutations in Δung1 isolates are representative of the uncorrected lesions induced by A3B in the given genomic targets.
Supplementary Material
Highlights.
APOBEC3B mutagenic activity is 1000-fold more in tRNA than in mRNA genes
APOBEC3B mutagenic lesions in tRNA genes are targeted to non-transcribed strand
Reversion of trp1-289 can be used as a proxy for hypermutation in tRNA genes
Efficacy of AP-site repair in tRNA genes is 99%, unlike only 83% in mRNA genes
Base excision repair and error-free lesion bypass prevent tRNA genes’ hypermutation
Acknowledgments
We are thankful to Dr. C. J. Sakofsky, Dr. S. N. Andres and Dr. S. Vijayraghavan for critically reading this manuscript and providing their feedback. We are also immensely grateful to Dr. K. Chan for providing help with the data analyses.
Funding This work was supported by the US National Institute of Health Intramural Research Program Project Z1AES103266 to D.A.G.; and NIH grant R00ES022633-03 from NIEHS and Breast Cancer Research Program Breakthrough Award BC141727 from the Department of Defense awarded S.A.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burch LH, Yang Y, Sterling JF, Roberts SA, Chao FG, Xu H, Zhang L, Walsh J, Resnick MA, Mieczkowski PA, Gordenin DA. Damage-induced localized hypermutability. Cell Cycle. 2011;10:1073–1085. doi: 10.4161/cc.10.7.15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan K, Gordenin DA. Clusters of Multiple Mutations: Incidence and Molecular Mechanisms. Annu Rev Genet. 2015;49:243–267. doi: 10.1146/annurev-genet-112414-054714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR, PedBrain I, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoopes JI, Cortez LM, Mertz TM, Malc EP, Mieczkowski PA, Roberts SA. APOBEC3A and APOBEC3B Preferentially Deaminate the Lagging Strand Template during DNA Replication. Cell Rep. 2016;14:1273–1282. doi: 10.1016/j.celrep.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lada AG, Kliver SF, Dhar A, Polev DE, Masharsky AE, Rogozin IB, Pavlov YI. Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes. PLoS Genet. 2015;11:e1005217. doi: 10.1371/journal.pgen.1005217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor BJ, Wu YL, Rada C. Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. Elife. 2014;3:e03553. doi: 10.7554/eLife.03553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns MB, Leonard B, Harris RS. APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed J. 2015;38:102–110. doi: 10.4103/2319-4170.148904. [DOI] [PubMed] [Google Scholar]
- 15.Harris RS. Molecular mechanism and clinical impact of APOBEC3B-catalyzed mutagenesis in breast cancer. Breast Cancer Res. 2015;17:8. doi: 10.1186/s13058-014-0498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, Ramakrishna M, Shlien A, Simonic I, Xue Y, Tyler-Smith C, Campbell PJ, Stratton MR. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, Kim J, Kwiatkowski DJ, Fargo DC, Mieczkowski PA, Getz G, Gordenin DA. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhagwat AS, Hao W, Townes JP, Lee H, Tang H, Foster PL. Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli. Proc Natl Acad Sci U S A. 2016;113:2176–2181. doi: 10.1073/pnas.1522325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seplyarskiy VB, Soldatov RA, Popadin KY, Antonarakis SE, Bazykin GA, Nikolaev SI. APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome Res. 2016;26:174–182. doi: 10.1101/gr.197046.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haradhvala NJ, Polak P, Stojanov P, Covington KR, Shinbrot E, Hess JM, Rheinbay E, Kim J, Maruvka YE, Braunstein LZ, Kamburov A, Hanawalt PC, Wheeler DA, Koren A, Lawrence MS, Getz G. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell. 2016;164:538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim N, Abdulovic AL, Gealy R, Lippert MJ, Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst) 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pybus C, Pedraza-Reyes M, Ross CA, Martin H, Ona K, Yasbin RE, Robleto E. Transcription-associated mutation in Bacillus subtilis cells under stress. J Bacteriol. 2010;192:3321–3328. doi: 10.1128/JB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francino MP, Chao L, Riley MA, Ochman H. Asymmetries generated by transcription-coupled repair in enterobacterial genes. Science. 1996;272:107–109. doi: 10.1126/science.272.5258.107. [DOI] [PubMed] [Google Scholar]
- 26.Francino MP, Ochman H. Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol Biol Evol. 2001;18:1147–1150. doi: 10.1093/oxfordjournals.molbev.a003888. [DOI] [PubMed] [Google Scholar]
- 27.Klapacz J, Bhagwat AS. Transcription promotes guanine to thymine mutations in the non-transcribed strand of an Escherichia coli gene. DNA Repair (Amst) 2005;4:806–813. doi: 10.1016/j.dnarep.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Fix D, Canugovi C, Bhagwat AS. Transcription increases methylmethane sulfonate-induced mutations in alkB strains of Escherichia coli. DNA Repair (Amst) 2008;7:1289–1297. doi: 10.1016/j.dnarep.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Fan M, Kalis S, Wei L, Scharff MD. A source of the single-stranded DNA substrate for activation-induced deaminase during somatic hypermutation. Nat Commun. 2014;5:4137. doi: 10.1038/ncomms5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Gonzalez B, Aguilera A. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci U S A. 2007;104:8409–8414. doi: 10.1073/pnas.0702836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10:e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10:e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chabelskaya S, Gryzina V, Moskalenko S, Le Goff C, Zhouravleva G. Inactivation of NMD increases viability of sup45 nonsense mutants in Saccharomyces cerevisiae. BMC Mol Biol. 2007;8:71. doi: 10.1186/1471-2199-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lada AG, Krick CF, Kozmin SG, Mayorov VI, Karpova TS, Rogozin IB, Pavlov YI. Mutator effects and mutation signatures of editing deaminases produced in bacteria and yeast. Biochemistry (Mosc) 2011;76:131–146. doi: 10.1134/s0006297911010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Chan K, Resnick MA, Gordenin DA. The choice of nucleotide inserted opposite abasic sites formed within chromosomal DNA reveals the polymerase activities participating in translesion DNA synthesis. DNA Repair (Amst) 2013;12:878–889. doi: 10.1016/j.dnarep.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosal G, Chen J. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res. 2013;2:107–129. doi: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratt-Hyatt MJ, Kapadia KM, Wilson TE, Engelke DR. Increased recombination between active tRNA genes. DNA Cell Biol. 2006;25:359–364. doi: 10.1089/dna.2006.25.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma W, Resnick MA, Gordenin DA. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res. 2008;36:1836–1846. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heacock M, Poltoratsky V, Prasad R, Wilson SH. Evidence for abasic site sugar phosphate-mediated cytotoxicity in alkylating agent treated Saccharomyces cerevisiae. PLoS One. 2012;7:e47945. doi: 10.1371/journal.pone.0047945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poltoratsky VP, Wilson SH, Kunkel TA, Pavlov YI. Recombinogenic phenotype of human activation-induced cytosine deaminase. J Immunol. 2004;172:4308–4313. doi: 10.4049/jimmunol.172.7.4308. [DOI] [PubMed] [Google Scholar]
- 42.Tornaletti S, Maeda LS, Hanawalt PC. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem Res Toxicol. 2006;19:1215–1220. doi: 10.1021/tx060103g. [DOI] [PubMed] [Google Scholar]
- 43.Yu SL, Lee SK, Johnson RE, Prakash L, Prakash S. The stalling of transcription at abasic sites is highly mutagenic. Mol Cell Biol. 2003;23:382–388. doi: 10.1128/MCB.23.1.382-388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim N, Jinks-Robertson S. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2010;30:3206–3215. doi: 10.1128/MCB.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reverendo M, Soares AR, Pereira PM, Carreto L, Ferreira V, Gatti E, Pierre P, Moura GR, Santos MA. TRNA mutations that affect decoding fidelity deregulate development and the proteostasis network in zebrafish. RNA Biol. 2014;11:1199–1213. doi: 10.4161/rna.32199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- 47.Yona AH, Bloom-Ackermann Z, Frumkin I, Hanson-Smith V, Charpak-Amikam Y, Feng Q, Boeke JD, Dahan O, Pilpel Y. tRNA genes rapidly change in evolution to meet novel translational demands. Elife. 2013;2:e01339. doi: 10.7554/eLife.01339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santos MA, Perreau VM, Tuite MF. Transfer RNA structural change is a key element in the reassignment of the CUG codon in Candida albicans. EMBO J. 1996;15:5060–5068. [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 50.Kimura M, Kamakura T, Tao QZ, Kaneko I, Yamaguchi I. Cloning of the blasticidin S deaminase gene (BSD) from Aspergillus terreus and its use as a selectable marker for Schizosaccharomyces pombe and Pyricularia oryzae. Mol Gen Genet. 1994;242:121–129. doi: 10.1007/BF00391004. [DOI] [PubMed] [Google Scholar]
- 51.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dixon WJ, Massey FJ, Jr Australian Pancreatic Cancer Genome, I.B.C. Consortium, I.M.-S. Consortium, Consortium IM-S. Introduction of Statistical Analysis. McGraw-Hill; New York: 1969. p. 349. [Google Scholar]
- 53.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lobry DCaJR. U.B.a.M.P.a.H.E.R.a.M. Vendruscolo, editor. Structural approaches to sequence evolution: Molecules, networks, populations. Springer Verlag; New York: 2007. Seqin{R} 1.0-2: a contributed package to the {R} project for statistical computing devoted to biological sequences retrieval and analysis; pp. 207–232. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.