Abstract
Both common and rare variants contribute to the genetic architecture of pulmonary fibrosis. Genome-wide association studies have identified common variants, or those with a minor allele frequency of >5%, that are linked to pulmonary fibrosis. The most widely replicated variant (rs35705950) is located in the promoter region of the MUC5B gene and has been strongly associated with idiopathic pulmonary fibrosis (IPF) and familial interstitial pneumonia (FIP) across multiple different cohorts. However, many more common variants have been identified with disease risk and in aggregate account for approximately one-third of the risk of IPF. Moreover, several of these common variants appear to have prognostic potential. Next generation sequencing technologies have facilitated the identification of rare variants. Recent whole exome sequencing studies have linked pathogenic rare variants in multiple new genes to FIP. Compared with common variants, rare variants have lower population allele frequencies and higher effect sizes. Pulmonary fibrosis rare variants genes can be subdivided into two pathways: telomere maintenance and surfactant metabolism. Heterozygous rare variants in telomere-related genes co-segregate with adult-onset pulmonary fibrosis with incomplete penetrance, lead to reduced protein function, and are associated with short telomere lengths. Despite poor genotype-phenotype correlations, lung fibrosis associated with pathogenic rare variants in different telomere genes is progressive and displays similar survival characteristics. In contrast, many of the heterozygous rare variants in the surfactant genes predict a gain of toxic function from protein misfolding and increased endoplasmic reticulum (ER) stress. Evidence of both telomere shortening and increased ER stress have been found in sporadic IPF patients, suggesting that the mechanisms identified from rare variant genetic studies in unique individuals and families are applicable to a wider spectrum of patients. The ability to sequence large cohorts of individuals rapidly has the potential to further our understanding of the relative contributions of common and rare variants in the pathogenesis of pulmonary fibrosis. The UK 100,000 Genomes Project will provide opportunities to interrogate both common and rare variants and to investigate how these biological signals provide diagnostic and prognostic information in the era of stratified medicine.
INTRODUCTION
Interstitial lung diseases (ILDs) are a diverse set of diseases affecting the tissue around the alveoli and can be caused by environmental exposures, drug reactions and connective tissue diseases, and are subcategorised based on their specific radiographic, histopathological and clinical characteristics. A subset of ILDs is characterised by both interstitial abnormalities and irreversible pulmonary fibrosis. Fibrosing ILDs not linked to underlying systemic disorders or identifiable environmental insults are known as fibrosing idiopathic interstitial pneumonias (IIPs), the most common of which is idiopathic pulmonary fibrosis (IPF). IPF is characterised by relentless scarring of the lungs and poor prognosis, with a median survival time from diagnosis of 3 years,1,2 few medical therapies and no cure outside of lung transplantation.3,4 It has long been observed that pulmonary fibrosis at times appears to cluster in families, in which case the disease is called familial interstitial pneumonia (FIP). Recent studies have deepened our appreciation of the role of inherited risk in the development of IPF, FIP and IIP, leading to new hypotheses regarding disease pathogenesis.
Genetic variants, common and rare, contribute to the genetic architecture of pulmonary fibrosis. The advancement of next generation sequencing technologies and genome-wide studies have facilitated the discovery and study of genetic variants and disease risk. The ability to sequence large numbers of individuals has the potential to advance our understanding of the relative contributions of common and rare variants in the pathogenesis of pulmonary fibrosis. Here we review the current understanding of genetic risk and pulmonary fibrosis and its clinical implications in the era of stratified medicine.
COMMON VARIANTS IN FIBROSING IIP
Although numerous rare variants have been associated with risk of FIP, there is evidence that common variants (defined as minor allele frequency (MAF) >5%) also play a role in FIP risk (figure 1). Notably, the common variants associated with FIP risk are the same as those associated with sporadic IPF risk and, in aggregate, account for one-third of the risk of developing familial or sporadic IIP. The concordance of common variants strongly suggests a shared genetic aetiology for both sporadic and familial pulmonary fibrosis. However, common variants are themselves much more common than pulmonary fibrosis in the population, and therefore, while these common variants provide important insights and clues into the role of genetic risk and familial and sporadic IPF, there remains much to understand about gene by gene and gene by environment interactions that lead to incomplete penetrance.
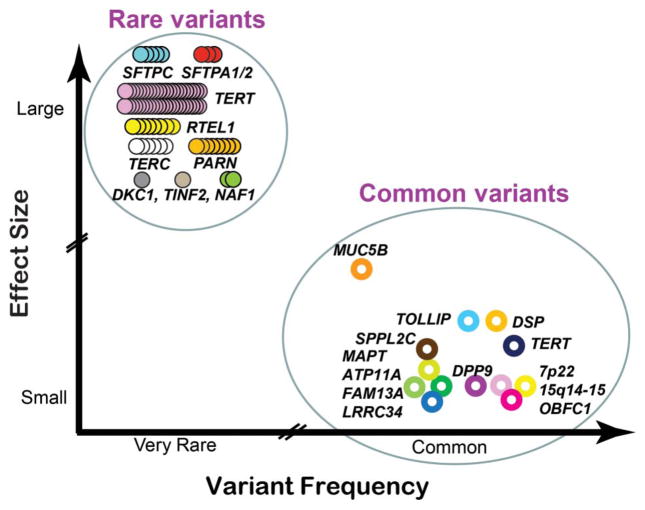
Figure 1.
Common and rare variants associated with fibrotic idiopathic interstitial pneumonia, idiopathic pulmonary fibrosis and familial interstitial pneumonia. Variants are distinguished by allele frequency and effect size. Common and rare variants have allele frequencies of >0.05 and <0.001, respectively. Effect sizes are rough estimates (see text). The common variants are indicated by open circles, each representing the most highly associated single nucleotide polymorphism at a particular locus. The rare variants are indicated by multiple closed circles, each representing a different pathogenic allele described for unrelated affected individuals or families. Distinct genetic loci are represented by the various colours. Adapted with permission from Macmillan Publishers Ltd: Nature,97 copyright (2009).
MUC5B PROMOTER POLYMORPHISM
In 2011, Seibold et al used genome-wide linkage analysis followed by targeted genetic sequencing in FIP and sporadic IPF cases to conclude that a single nucleotide polymorphism (SNP) rs35705950 (G>T transversion) on the p-terminus of chromosome 11 within the promoter region of the MUC5B gene, a highly conserved, regulatory genetic region, is associated with both FIP and IPF. The rs35705950 variant is also associated with significantly higher MUC5B expression in healthy lung tissue (37.4-fold increased for those with the T allele), and MUC5B gene expression was more than 14-fold higher in fibrotic lung tissue than in unaffected controls irrespective of genotype.5 The ORs for disease for those who were heterozygous and those who were homozygous for the minor allele for this SNP were 6.8 (95% CI 3.9 to 12.0) and 20.8 (95% CI 3.8 to 113.7), respectively, for FIP (genotypic association test, p=3.7×10−12).5 Similarly, the ORs for heterozygous and homozygous individuals were 9.0 (95% CI 6.2 to 13.1) and 21.8 (95% CI 5.1 to 93.5), respectively, for sporadic IPF (genotypic association test, p=4.6×10−31).5 MUC5B encodes Mucin 5B, a glycosylated macromolecular mucus component secreted by airway epithelial cells. It is the major gel-forming mucin in mucus and lubricates saliva, lung mucus and cervical mucus, and based on recent studies, is important to innate immune function.6 MUC5B can be localised to the honeycomb cyst, a characteristic structure seen in IPF,7 strengthening the case for a pathological role for MUC5B in lung fibrosis.
The association between the rs35705950 minor allele and IPF has been validated in seven independent cohorts;8–14 it remains the strongest and most replicated single genetic risk factor for IPF. Adding to its credibility as a genetic risk factor specific to IIPs, rs35705950 has been examined in cohorts of subjects with ILD secondary to systemic sclerosis,11,12 asbestosis, sarcoidosis,11 acute respiratory distress syndrome (ARDS), COPD and asthma,15 and these investigations have shown no association between genotype and these other pulmonary disorders.
Furthermore, rs35705950 is likely to be a significant risk factor for pulmonary fibrosis in some populations outside of the non-Hispanic white (NHW) population in which it was originally described: in a Mexican cohort, the variant was a risk factor for IPF (OR=7.36, p=0.0001), yet was rare in a Korean IPF cohort.16 Among Asians, the MAF of rs35705950 is low, but in Japanese and Chinese studies, subjects with IPF had a significantly higher frequency of the risk allele than controls.13,17 The rs35705950 MAF across different ethnic groups appears to reflect disease prevalence in these groups, since NHWs have been observed to be at higher risk of pulmonary fibrosis than Hispanics, Asians and Africans.18 Indeed, in sub-Saharan African populations, in which pulmonary fibrosis is thought to be rare, the MUC5B promoter polymorphism is not present,19 so although the rs35705950 variant may have been discovered to be important in familial and sporadic forms of pulmonary fibrosis in the NHW population, it appears to be important in other groups as well, and broader study of this common variant in other ethnic groups will be important in terms of understanding its role in disease pathogenesis. Given the broad prevalence differences by ancestry, it is possible that the gain-of-function MUC5B promoter variant confers a beneficial biological effect, perhaps improved host defence, during childhood or through childbearing years, leading to positive selection in the NHW population.
Also, the 2011 study by Seibold et al5 was the first to implicate secreted airway mucins in fibrogenesis, thereby suggesting that airway epithelia could play an important role in disease development. Furthermore, it identified a specific common genetic variant, rs35705950, that could be utilised to identify individuals at high risk for the development of disease.5 However, the variant’s MAF of 9.1% in the control cohort (vs 33.8% in FIP and 37.5% in sporadic IPF) suggests that the variant may not be sufficient to cause a disease as rare as FIP (or IPF) despite the significant risk it confers. Indeed, other large cohort studies, such as the those examining the Framingham Heart Study population, have shown that in NHWs, 19% of those without any evidence of interstitial lung abnormalities (ILAs) or fibrosis carry at least one T allele (MAF of 9.7%).20 Therefore, there may be significant gene by gene or gene by environment interactions that are critical in the translation of increased risk to the development of fibrosis;21 these potential interactions are an area of active investigation.
Follow-up studies have shown that the same variant is associated with radiographic evidence of ILAs, hypothesised to be an early form of disease.22,23 For each copy of the rs35705950 minor allele, the odds of ILAs (which included ground-glass or reticular abnormalities, centrilobular nodularity, non-emphysematous cysts, honeycombing and traction bronchiectasis) were 2.8 times higher.20 This is notable since these ILAs progress over time and have been linked to increased mortality, specifically from respiratory causes.22,23 Intriguingly, the Framingham Heart Study investigations have also shown that 2% of NHWs over 50 years of age have definite radiographic evidence of pulmonary fibrosis, higher than what had been reported in the past as the prevalence of disease.20,24 These associations between rs35705950, FIP, IPF and ILAs are provocative as they suggest that common variants could be used alone or in concert with other genetic or demographic parameters to guide screening, earlier diagnosis and early intervention.20–23,25,26
OTHER COMMON GENETIC VARIANTS AND FIBROSING IIP
Although rs35705950 is the most robustly replicated common genetic variant associated with pulmonary fibrosis (familial or sporadic), other common variants have been found to be associated with disease utilising genome-wide approaches. A 2008 genome-wide association study (GWAS) identified a common variant in TERT, rs2736100, associated with IPF in a Japanese cohort.27 In 2013, Fingerlin et al8 published a case-control GWAS in 1616 individuals with fibrotic IIP and 4683 control subjects followed by a replication study of 876 cases and 1890 controls. The study included individuals from FIP families (one-per-family, n=390 in the discovery cohort) in addition to sporadic cases of IIP. This large GWAS confirmed disease associations with TERT (chromosome 5p15), MUC5B (11p15) and the 3q26 region near TERC; however, it also identified seven new loci associated with fibrotic IIP, including FAM13A (4q22), DSP (6p24), OBFC1 (10q24), ATP11A (13q4), DPP9 (19p13), and regions on chromosomes 7q22 and 15q14–15.8 Excluding MUC5B, these loci account for approximately one-third of disease risk.8 Although these genes have disparate reported biological functions, in aggregate they implicate host defence (MUC5B, ATP11A), cell-cell adhesion (DSP, DPP9) and DNA repair (TERT, TERC, OBFC1) pathways in disease pathogenesis.8,21,25,28
As was observed in the case of the MUC5B promoter polymorphism rs35705950, the ORs for loci identified in the Fingerlin et al study did not differ in post hoc analyses comparing the familial to sporadic cases, suggesting that when it comes to risk conferred by common variants, FIP is similar to sporadic IIP.8,21 However, many of the individual variants identified in this GWAS had MAFs approaching 50% even in the control cohorts (eg, rs2076295 in the DSP locus on chromosome 6, MAF of 44% in the control cohort); given the rarity of fibrotic IIP as a phenotype, the relationship of individual common variants to the development of disease phenotype may involve gene by gene or gene by environment interactions that have yet to be elucidated.
Another GWAS performed in subjects with sporadic IPF also confirmed the association of the MUC5B promoter polymorphism with IPF and also identified new disease-associated variants, including one in Toll-interacting protein (TOLLIP) and signal peptidase-like 2C (SPPL2C).10 This study identified these risk variants and also showed that rs5743890 in TOLLIP was associated with increased IPF mortality.10,29
COMMON VARIANTS AND DISEASE OUTCOMES
The ability of genotype to predict phenotype is an important clinical application of the finding that common variants are associated with disease. With respect to the common MUC5B promoter polymorphism, retrospective analysis of large clinical trial data illustrates that sporadic IPF subjects with the risk variant (T) had improved survival at 2 years when compared to those without the variant in single-variable proportional hazards models as well as in models adjusting for age, sex, lung function and treatment status; outcome modelling accuracy improved significantly when the rs35705950 genotype was included.30 This observation regarding improved survival for those carrying the rs35705950 variant was noted in two independent study cohorts (INSPIRE and a cohort from the University of Chicago) and supports the hypothesis that there are at least two phenotypes of IPF defined by the T allele at rs35705950.30 Given the importance of rs35705950 in FIP as well as sporadic IPF, it is likely that this variant could have similar prognostic power in FIP cases as well, although this has not formally been studied.
Similarly, the TOLLIP variant described by Noth and colleagues to be associated with IPF was also associated with differential survival. However, the rs5743890 minor allele (G), although associated with decreased risk of disease overall, was, within analysis of diseased subjects alone, associated with increased mortality.10 An analysis of the PANTHER-IPF clinical trial data also showed that when TOLLIP and MUC5B risk variants were examined retrospectively in the trial cohorts, of those who received N-acetylcysteine (NAC), those with the TT genotype for rs3750920 (TOLLIP) had decreased risk of the trial’s composite endpoint of death, transplantation, hospitalisation or greater than 10% decrement in forced vital capacity (FVC).29 Of the subjects that received NAC, those with the CC genotype for rs3750920 had increased risk of the trial’s composite endpoint.29 While NAC has not been shown to be effective in all patients with IPF, it is possible that a subset of patients defined by TOLLIP genotyping could have some therapeutic response to the drug while others may have clinical decline in response to the same drug, highlighting the importance of pharmacogenetics in this disease and the need for genotype-stratified prospective clinical trials to confirm this finding.29,31
The relationship between common variants, disease risk and survival may be more complicated than initially hypothesised, since validated risk variants (eg, rs35705950) are strongly associated with disease risk but simultaneously associated with better clinical outcomes within IPF patients.30 Similarly, as Noth and colleagues described, the rs5743890 variant appears protective in term of disease risk, yet associated with increased mortality within the diseased subject group.10 It may be that such common variants predispose individuals to clinically milder forms of disease. These findings suggest that ‘IPF’ may itself be a heterogeneous phenotype that could be more granularly defined by genetic markers and risk variants, implying an important future role for genetics-based personalisation of care.
Therefore, further prospective study of the relationship between genotype and therapeutic response will be critical to tailoring individual patient treatment in the future. Currently, although the studies described point to intriguing genotype-phenotype relationships, clinical use of these findings remains uncertain, especially as gene-gene interactions are poorly understood in terms of how they relate to disease phenotype and therapeutic response. At this time, therapeutic decision-making should be done on the basis of published, prospective, randomised, controlled trials and well-described risks and benefits, which have been studied independently of genotype.3,4
Future clinical trials should take into account genotype-phenotype relationships, especially given the findings described by Oldham et al29 and Peljto et al,30 since both these retrospective studies suggest that primary outcomes (ie, mortality and therapeutic response such as lung function decrement) can be influenced by common variant genotypes.
RARE VARIANTS IN FIBROSING INTERSTITIAL PNEUMONIA
For decades, physicians have observed that pulmonary fibrosis can segregate in families with an autosomal dominant pattern of inheritance affecting individuals over multiple generations. In some cases, a rare variant can be identified that co-segregates with disease and is predictive of overt or subclinical disease. These families represent a monogenetic form of FIP, attributable to a rare variant in a single gene. In contrast with the common variants discussed above, these variants are generally absent or infrequently present in control populations (MAF of <0.1%) and are termed ‘rare’ or alternatively as ‘novel’, ‘singletons’ or ‘ultra-rare’ variants, or those that are ‘private’ to the family or individual in which they were discovered. The rarity of these alleles is consistent with the population frequency of FIP (roughly estimated at ~1 in 100 000) and with evolutionary theory predicting that deleterious disease alleles should be rare.32 Individual rare variants are more highly penetrant than common variants and generally have larger ORs.33 For many ultra-rare variants, ORs are several orders of magnitude larger than for common variants or cannot be calculated because allele frequencies are so small to non-existent in control populations.
While association studies demonstrate genome-wide significance for common variants, this measure of significance cannot be achieved for individual rare variants given their low allele frequencies. Genome-wide measures of the significance of the rare variant FIP genes are represented either by linkage of an identical-by-descent genomic region with pulmonary fibrosis within large extended families34–36 or by gene burden tests demonstrating excess numbers of variants in cases versus controls.35 While common variants are usually characterised by a single tagging SNP, the rare variants linked to FIP include multiple different genetic changes within a gene that each predict a deleterious change in the genomic, RNA or protein sequence. Various in silico tools exist for predicting variant effects on protein function based upon alignments of evolutionarily conserved sequences or predicting effects on splicing or posttranslational modifications. Additional proof in support of the pathogenicity of individual rare variants is made through segregation analysis in families or examination of protein function by molecular, biochemical or pathophysiological assays.
RARE VARIANTS IN GENES INVOLVED WITH TELOMERE MAINTENANCE
Telomeres are composed of repetitive nucleotides sequences (TTAGGG) at the ends of chromosomes that protect the chromosome from progressive shortening during cell replication. Dysfunction of telomere maintenance machinery components results in short telomeres, activation of the DNA damage response and cellular senescence. Multiple genes in the telomere maintenance pathway have been implicated in adult-onset familial pulmonary fibrosis, including those that affect telomerase catalytic enzyme activity (TERT, TERC),34,37 its biogenesis (DKC1, PARN, NAF1)35,38,39 or the telomere end (RTEL1, TINF2).35,40–42 All pathogenic rare variants lead to a reduction in protein function, which is manifested as decreased telomerase catalytic activity,34,37,43 decreased enzyme processivity,44 aberrant T-circle formations40 or shortened telomere lengths.35,38,39,41,42 These heterozygous germline mutations lead to telomere shortening in somatic cells, including blood leukocytes, oral mucosal epithelial cells and lung epithelial cells.45,46
Collectively, rare variants of deleterious function are found more often in the TERT gene than in any other gene associated with pulmonary fibrosis. Distinct novel variants have been discovered in different patients and families that span the full length of the gene. Pathogenic rare variants in TERT are found in ~15% of familial cohorts34,37 and in ~3% of cases of sporadic IPF.34,45 The development of pulmonary fibrosis is age-dependent, being uncommon in TERT variant carriers less than 40 years old. Incomplete penetrance is observed, with ~60% of male and ~50% of female TERT rare variant carriers over 60 years of age having overt disease.43 Most mutation carriers who develop pulmonary fibrosis report a prior history of smoking or another fibrogenic exposure, suggesting a role of the environment in modulating the risk of genetically susceptible individuals.
FIP and IPF are the most common pulmonary manifestations associated with rare variants in telomere genes,47 although a variety of ILD subtypes, including those of known and unknown causes,48 as well as an increased risk for emphysema, 49 have been described. Heterogeneous diagnoses of family members with identical mutations are common and found in up to 80% of kindreds.48 Despite poor genotype-pulmonary phenotype correlations, lung fibrosis associated with rare variants in four different telomere genes (TERT, TERC, RTEL1 and PARN) is progressive and displays similar survival characteristics.48 Asymptomatic rare variant TERT carriers exhibit subclinical signs of pulmonary fibrosis including increased quantitative tissue volumes measured from high resolution CT scans of the chest, reduced diffusion capacity at rest, and reduced recruitment of diffusion capacity with exercise.50 Consistent with the findings of incomplete penetrance in kindreds, 34,35 the lag time between subclinical to overt disease is highly variable even for individuals with the same telomerase mutation.51
Additive effects of pathogenic variants in the telomere-related genes can be seen in human patients. Whereas monoallelic mutations are associated with adult-onset pulmonary fibrosis, biallelic mutations in TERT, RTEL1 and PARN are found in rare children and young adults with more severe disease presenting as dyskeratosis congenita (DC) or Hoyeraal-Hreidarsson syndrome with mucocutaneous manifestations, bone marrow failure, developmental delay, enterocolitis and/or immunodeficiency.52–59 Frequently, DC-associated phenotypes are often found in adults with these ‘short telomere syndromes’.60 Patients with rare variants in TERC appear to have a higher prevalence of severe manifestations of bone marrow dysfunction (leukopenia, thrombocytopenia, aplastic anaemia, myelodysplastic syndrome) than those with rare variants in the other telomere genes.48 And the presence of a personal or family history of a spectrum of short-telomere phenotypes (cytopenias, macrocytosis, liver dysfunction, early greying of hair, predisposition to leukaemia, and myelodysplastic syndrome) in conjunction with pulmonary fibrosis is often predictive of short telomere lengths or a rare variant in a telomere-related gene.50,61,62
Genetic anticipation can be seen in families with telomere-related gene mutations, with earlier and more severe onset of disease in successive generations, and is related to progressive telomere shortening.48,63–65 Consistent with these observations, the mean age at the time of ILD diagnosis is correlated with telomere length, with earlier ages at the time of pulmonary fibrosis diagnosis for TERC rare variant carriers with shorter telomere lengths than PARN mutation carriers with longer telomeres.48
The lung transplantation outcomes of patients with rare variants in telomerase have been described.66–68 The incidence of haematological complications, renal disease and infections in this patient group suggests an increased risk for certain adverse effects which are unmasked by transplantation or post-transplant medication regimens. Pre-transplant bone marrow biopsies may identify those at increased risk for severe haematological complications.61
SHORT TELOMERE LENGTHS IN SPORADIC IPF
Patients with sporadic, or non-familial, forms of various ILDs also have shorter telomere lengths than would be predicted by their age. Nearly 25% of sporadic IPF patients have age-adjusted telomere lengths of <10th percentile.45,46 Telomere length is a prognostic biomarker in sporadic IPF; patients with the shortest telomere lengths have the worst transplant-free survival even after adjusting for age, gender and pulmonary function.69 This finding has been validated in multiple independent cohorts.69,70 Thus, the molecular mechanism of telomere shortening which was discovered through genetic studies of rare families is relevant in patients with sporadic disease.
RARE VARIANTS IN GENES INVOLVED WITH SURFACTANT METABOLISM
Surfactant produced by the type II alveolar epithelial cell is rich in lipids and proteins. The surfactant proteins are translated within the endoplasmic reticulum (ER) and transported to lamellar bodies where they are stored prior to secretion into the alveolar space. Pulmonary surfactant serves two main functions: (1) to reduce surface tension and prevent atelectasis, and (2) to opsonise pathogens and modulate the innate immune response.71 Rare variants in the genes encoding surfactant protein A and C (SFTPA1, SFTPA2 and SFTPC) have been associated with adult-onset pulmonary fibrosis. Biallelic rare variants in the genes encoding surfactant protein B (SFTPB) and the lamellar body transporter of cellular proteins and phospholipids (ABCA3) are more commonly associated with neonatal respiratory distress syndrome and paediatric diffuse parenchymal lung disease. Rare case reports of adults with homozygous ABCA3 mutations have been described.72,73
SFTPC heterozygous rare variants have been described in patients presenting from infancy to late adulthood with a range of pulmonary fibrosis subtypes. These FIP kindreds demonstrate an autosomal dominant pattern of inheritance. Over 40 different missense, splice site or frameshift mutations within the SFTPC gene have been associated with familial or sporadic disease.74 The mechanism of most SFTPC mutations is thought to be due to a gain of toxic function from protein misfolding. The SP-C pre-protein contains a BRICHOS domain that is critical for proper folding and trafficking within the secretory pathway.75 Some rare variants within the BRICHOS domain cause misfolding of the pre-SP-C, which accumulates within the ER resulting in activation of the unfolded protein response, increased ER stress and type II alveolar cell toxicity.75–77 One of the rare variants that has been described across different cohorts predicts a missense change, Ile72Thr (c.218T>C), within the linker domain which leads to impaired trafficking and disruption of the autophagy pathway.78,79 Although rare variants SFTPC mutations are rarely found (1–2%) in most FIP cohorts,80–83 they have been found at a higher incidence (25%) in a Dutch cohort.84
Surfactant protein A is encoded by two closely related genes, SFTPA1 and SFTPA2, located on chromosome 10q22.3.85 Mutations in both genes have been associated with FIP and lung adenocarcinoma. All pathogenic heterozygous rare variants in the SFTPA1 and SFTPA2 genes encode missense mutations located within the carbohydrate recognition domain of the protein.36,86,87 These mutations result in reduced secretion of mature protein and increased ER stress,36,86,88 markers of which have been found in alveolar epithelial cells of sporadic IPF patients adjacent to regions of lung fibrosis,89,90 suggesting that increased ER stress may lead to a cellular substrate with increased vulnerability to fibrosis after injury. Thus, the pathway identified by studying rare patients and families with surfactant mutations is relevant to a broader IPF patient population.
IMPLICATIONS FOR PERSONALISED MEDICINE
There is ample evidence in support of both common and rare variants’ contribution to the genetic architecture of pulmonary fibrosis. Recent advances in genetic sequencing and bioinformatics have made it much easier to detect genetic variants rapidly. The UK’s 100,000 Genomes Project is an effort to sequence genomes from 70 000 individuals in the National Health Service with a focus on cancer and rare diseases, including IIPs. As this study is completed, hypotheses regarding the relevance of common and rare variants can be tested in this real-life human experiment.
Major challenges lie ahead to determine if individual sequence data can be translated into clinically relevant information. For common variants, a major challenge will determine how these variants can inform an individual patient’s risk, since a tagging SNP could be a proxy for a linked causal variant. Or perhaps interplay between one or more common SNP(s) and environmental triggers is critical in disease risk or response to therapy. For rare variants, the major challenge will be to determine which are clinically meaningful given the plethora of rare variants per human genome.91–93 The American College of Medical Genetics and the European Society of Human Genetics have outlined standards for interpreting genetic variants,94,95 using suggested terminology such as pathogenic, likely pathogenic, uncertain significance, likely benign and benign. These designations highlight the uncertainty regarding the clinical implications of individual variants and the need for new scoring systems for predicting pathogenicity. A newly identified rare variant in an individual IPF patient is likely to be called a variant of unclear significance (VUS) if it has never before been reported. Will the identification of a VUS lead to a ‘genetic purgatory’ 96 for the patient and the medical team if there is little certainty regarding predictions of risk for the patient or family members? The burden of proof for pathogenicity of individual rare variants often requires segregation analyses in families and/or demonstration of impaired assays of protein function, data not obtained by sequencing alone.
The ILDs are a diverse collection of disorders, and the genetic studies substantiate this heterogeneity at a molecular level. Studies to date have demonstrated that the inclusion of genetic information leads to stratification of pulmonary fibrosis patients with regard to rate of progression and responsiveness to therapeutics. Given the potential impact of rare and common variants on disease outcome, one must consider the prognostic potential of these sequence changes and their inclusion in interventional trials.
Large population sequencing projects will provide opportunities to describe the penetrance and effect sizes of a spectrum of common and rare variants with regard to subclinical and overt pulmonary phenotypes. With datasets collected from large numbers of well-phenotyped individuals, interactions between common and rare variants, genes and pathways can be interrogated across both healthy and diseased subjects. Thus, the UK’s 100,000 Genomes Project will lead to an era of studying individual genotypes in conjunction with classic phenotypes, creating new ways to molecularly characterise pulmonary fibrosis.
Acknowledgments
Funding
The U.S. Department of Veterans Affairs (grant no. Merit Award 1I01BX001534 (DAS)) and the National Institutes of Health (grant nos. P01-HL092870 (DAS), R01-HL097163 (DAS), R01HL093096 (CKG), R25-ES025476 (DAS), R33-HL120770 (DAS), T32HL098040 (CAN), UL1TR001105 (CAN) and UH2-HL123442 (DAS)).
Footnotes
Competing interests
The authors have in the past received and currently receive funding from the National Institutes of Health.
Provenance and peer review
The authors have in the past received and currently receive funding from the National Institutes of Health and the U.S. Department of Veterans Affairs.
References
- 1.Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson AL, Swigris JJ, Lezotte DC, et al. Mortality from pulmonary fibrosis increased in the United States from 1992 to 2003. Am J Respir Crit Care Med. 2007;176:277–84. doi: 10.1164/rccm.200701-044OC. [DOI] [PubMed] [Google Scholar]
- 3.Richeldi L, du Bois RM, Raghu G, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2071–82. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 4.King TE, Bradford WZ, Castro-Bernardini S, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2083–92. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 5.Seibold MA, Wise AL, Speer MC, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–12. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy MG, Livraghi-Butrico A, Fletcher AA, et al. Muc5b is required for airway defence. Nature. 2014;505:412–16. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seibold MA, Smith RW, Urbanek C, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS ONE. 2013;8:e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fingerlin TE, Murphy E, Zhang W, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–20. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Noth I, Garcia JGN, et al. A variant in the promoter of MUC5B and idiopathic pulmonary fibrosis. N Engl J Med. 2011;364:1576–7. doi: 10.1056/NEJMc1013504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noth I, Zhang Y, Ma SF, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1:309–17. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stock CJ, Sato H, Fonseca C, et al. Mucin 5B promoter polymorphism is associated with idiopathic pulmonary fibrosis but not with development of lung fibrosis in systemic sclerosis or sarcoidosis. Thorax. 2013;68:436–41. doi: 10.1136/thoraxjnl-2012-201786. [DOI] [PubMed] [Google Scholar]
- 12.Borie R, Crestani B, Dieude P, et al. The MUC5B variant is associated with idiopathic pulmonary fibrosis but not with systemic sclerosis interstitial lung disease in the European Caucasian population. PLoS ONE. 2013;8:e70621. doi: 10.1371/journal.pone.0070621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horimasu Y, Ohshimo S, Bonella F, et al. MUC5B promoter polymorphism in Japanese patients with idiopathic pulmonary fibrosis. Respirology. 2015;20:439–44. doi: 10.1111/resp.12466. [DOI] [PubMed] [Google Scholar]
- 14.Wei R, Li C, Zhang M, et al. Association between MUC5B and TERT polymorphisms and different interstitial lung disease phenotypes. Transl Res. 2014;163:494–502. doi: 10.1016/j.trsl.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang IV, Schwartz DA. Epigenetics of idiopathic pulmonary fibrosis. Transl Res. 2015;165:48–60. doi: 10.1016/j.trsl.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peljto AL, Selman M, Kim DS, et al. The MUC5B promoter polymorphism is associated with idiopathic pulmonary fibrosis in a Mexican cohort but is rare among Asian ancestries. Chest. 2015;147:460–4. doi: 10.1378/chest.14-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Zhuang Y, Guo W, et al. Mucin 5B promoter polymorphism is associated with susceptibility to interstitial lung diseases in Chinese males. PLoS ONE. 2014;9:e104919. doi: 10.1371/journal.pone.0104919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ley B, Collard HR. Epidemiology of idiopathic pulmonary fibrosis. Clin Epidemiol. 2013;5:483–92. doi: 10.2147/CLEP.S54815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.NCBI. dbSNP entry for rs35705950. http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?rs=357.
- 20.Hunninghake GM, Hatabu H, Okajima Y, et al. MUC5B promoter polymorphism and interstitial lung abnormalities. N Engl J Med. 2013;368:2192–200. doi: 10.1056/NEJMoa1216076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathai SK, Schwartz DA, Warg LA. Genetic susceptibility and pulmonary fibrosis. Curr Opin Pulm Med. 2014;20:429–35. doi: 10.1097/MCP.0000000000000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Putman RK, Hatabu H, Araki T, et al. Association between interstitial lung abnormalities and all-cause mortality. JAMA. 2016;315:672. doi: 10.1001/jama.2016.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Araki T, Putman RK, Hatabu H, et al. Development and progression of interstitial lung abnormalities in the Framingham Heart Study. Am J Respir Crit Care Med. doi: 10.1164/rccm.201512-2523OC. Published Online First: 17 Jun 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghu G, Weycker D, Edelsberg J, et al. Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2006;174:810–16. doi: 10.1164/rccm.200602-163OC. [DOI] [PubMed] [Google Scholar]
- 25.Mathai SK, Yang IV, Schwarz MI, et al. Incorporating genetics into the identification and treatment of Idiopathic Pulmonary Fibrosis. BMC Med. 2015;13:191. doi: 10.1186/s12916-015-0434-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putman RK, Rosas IO, Hunninghake GM. Genetics and early detection in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;189:770–8. doi: 10.1164/rccm.201312-2219PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mushiroda T, Wattanapokayakit S, Takahashi A, et al. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. J Med Genet. 2008;45:654–6. doi: 10.1136/jmg.2008.057356. [DOI] [PubMed] [Google Scholar]
- 28.Yang IV, Fingerlin TE, Evans CM, et al. MUC5B and idiopathic pulmonary fibrosis. Ann Am Thorac Soc. 2015;12(Suppl 2):S193–9. doi: 10.1513/AnnalsATS.201503-110AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oldham JM, Ma SF, Martinez FJ, et al. TOLLIP, MUC5B, and the response to N-acetylcysteine among individuals with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2015;192:1475–82. doi: 10.1164/rccm.201505-1010OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peljto AL, Zhang Y, Fingerlin TE, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. JAMA. 2013;309:2232–9. doi: 10.1001/jama.2013.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raghu G, Anstrom KJ, King TE, et al. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012;366:1968–77. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–10. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 33.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsakiri KD, Cronkhite JT, Kuan PJ, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci USA. 2007;104:7552–7. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart BD, Choi J, Zaidi S, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet. 2015;47:512–17. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Kuan PJ, Xing C, et al. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet. 2009;84:52–9. doi: 10.1016/j.ajhg.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armanios MY, Chen JJ, Cogan JD, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–26. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 38.Kropski JA, Mitchell DB, Markin C, et al. A novel dyskerin (DKC1) mutation is associated with familial interstitial pneumonia. Chest. 2014;146:e1–7. doi: 10.1378/chest.13-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanley SE, Gable DL, Wagner CL, et al. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis-emphysema. 2016;8:351ra107. doi: 10.1126/scitranslmed.aaf7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cogan JD, Kropski JA, Zhao M, et al. Rare variants in RTEL1 are associated with familial interstitial pneumonia. Am J Respir Crit Care Med. 2015;191:646–55. doi: 10.1164/rccm.201408-1510OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kannengiesser C, Borie R, Ménard C, et al. Heterozygous RTEL1 mutations are associated with familial pulmonary fibrosis. Eur Respir J. 2015;46:474–85. doi: 10.1183/09031936.00040115. [DOI] [PubMed] [Google Scholar]
- 42.Alder JK, Stanley SE, Wagner CL, et al. Exome sequencing identifies mutant TINF2 in a family with pulmonary fibrosis. Chest. 2015;147:1361–8. doi: 10.1378/chest.14-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diaz de Leon A, Cronkhite JT, Katzenstein AL, et al. Telomere lengths, pulmonary fibrosis and telomerase (TERT) mutations. PLoS ONE. 2010;5:e10680. doi: 10.1371/journal.pone.0010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alder JK, Cogan JD, Brown AF, et al. Ancestral mutation in telomerase causes defects in repeat addition processivity and manifests as familial pulmonary fibrosis. PLoS Genet. 2011;7:e1001352. doi: 10.1371/journal.pgen.1001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cronkhite JT, Xing C, Raghu G, et al. Telomere shortening in familial and sporadic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178:729–37. doi: 10.1164/rccm.200804-550OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alder JK, Chen JJ, Lancaster L, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci USA. 2008;105:13051–6. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armanios M. Telomerase and idiopathic pulmonary fibrosis. Mutat Res. 2012;730:52–8. doi: 10.1016/j.mrfmmm.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newton CA, Batra K, Torrealba J, et al. Telomere-related lung fibrosis is diagnostically heterogeneous but uniformly progressive. Eur Respir J. doi: 10.1183/13993003.00308-2016. Published Online First: 18 Aug 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanley SE, Chen JJL, Podlevsky JD, et al. Telomerase mutations in smokers with severe emphysema. J Clin Invest. 2015;125:563–70. doi: 10.1172/JCI78554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diaz De Leon A, Cronkhite JT, Yilmaz C, et al. Subclinical lung disease, macrocytosis, and premature graying in kindreds with telomerase (TERT) mutations. Chest. 2011;140:753–63. doi: 10.1378/chest.10-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Chemaly S, Ziegler SG, Calado RT, et al. Natural history of pulmonary fibrosis in two subjects with the same telomerase mutation. Chest. 2011;139:1203–9. doi: 10.1378/chest.10-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aspesi A, Vallero S, Rocci A, et al. Compound heterozygosity for two new TERT mutations in a patient with aplastic anemia. Pediatr Blood Cancer. 2010;55:550–3. doi: 10.1002/pbc.22589. [DOI] [PubMed] [Google Scholar]
- 53.Walne AJ, Vulliamy T, Kirwan M, et al. Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am J Hum Genet. 2013;92:448–53. doi: 10.1016/j.ajhg.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du HY, Pumbo E, Manley P, et al. Complex inheritance pattern of dyskeratosis congenita in two families with 2 different mutations in the telomerase reverse transcriptase gene. Blood. 2008;111:1128–30. doi: 10.1182/blood-2007-10-120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ballew BJ, Yeager M, Jacobs K, et al. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum Genet. 2013;132:473–80. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tummala H, Walne A, Collopy L, et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J Clin Invest. 2015;125:2151–60. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dhanraj S, Gunja SMR, Deveau AP, et al. Bone marrow failure and developmental delay caused by mutations in poly(A)-specific ribonuclease (PARN) J Med Genet. 2015;52:738–48. doi: 10.1136/jmedgenet-2015-103292. [DOI] [PubMed] [Google Scholar]
- 58.Moon DH, Segal M, Boyraz B, et al. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat Genet. 2015;47:1482–8. doi: 10.1038/ng.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Guen T, Jullien L, Touzot F, et al. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum Mol Genet. 2013;22:3239–49. doi: 10.1093/hmg/ddt178. [DOI] [PubMed] [Google Scholar]
- 60.Stanley SE, Armanios M. The short and long telomere syndromes: paired paradigms for molecular medicine. Curr Opin Genet Dev. 2015;33:1–9. doi: 10.1016/j.gde.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.George G, Rosas IO, Cui Y, et al. Short telomeres, telomeropathy, and subclinical extrapulmonary organ damage in patients with interstitial lung disease. Chest. 2015;147:1549–57. doi: 10.1378/chest.14-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parry EM, Alder JK, Qi X, et al. Syndrome complex of bone marrow failure and pulmonary fibrosis predicts germline defects in telomerase. Blood. 2011;117:5607–11. doi: 10.1182/blood-2010-11-322149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Armanios M, Chen JL, Chang YPC, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci USA. 2005;102:15960–4. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vulliamy T, Marrone A, Szydlo R, et al. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet. 2004;36:447–9. doi: 10.1038/ng1346. [DOI] [PubMed] [Google Scholar]
- 65.Fernandez BA, Fox G, Bhatia R, et al. A Newfoundland cohort of familial and sporadic idiopathic pulmonary fibrosis patients: clinical and genetic features. Respir Res. 2012;13:64. doi: 10.1186/1465-9921-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silhan LL, Shah PD, Chambers DC, et al. Lung transplantation in telomerase mutation carriers with pulmonary fibrosis. Eur Respir J. 2014;44:178–87. doi: 10.1183/09031936.00060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tokman S, Singer JP, Devine MS, et al. Clinical outcomes of lung transplant recipients with telomerase mutations. J Heart Lung Transplant. 2015;34:1318–24. doi: 10.1016/j.healun.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Borie R, Kannengiesser C, Hirschi S, et al. Severe hematologic complications after lung transplantation in patients with telomerase complex mutations. J Heart Lung Transplant. 2015;34:538–46. doi: 10.1016/j.healun.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Stuart BD, Lee JS, Kozlitina J, et al. Effect of telomere length on survival in patients with idiopathic pulmonary fibrosis: an observational cohort study with independent validation. Lancet Respir Med. 2014;2:557–65. doi: 10.1016/S2213-2600(14)70124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dai J, Cai H, Li H, et al. Association between telomere length and survival in patients with idiopathic pulmonary fibrosis. Respirology. 2015;20:947–52. doi: 10.1111/resp.12566. [DOI] [PubMed] [Google Scholar]
- 71.Han S, Mallampalli RK. The role of surfactant in lung disease and host defense against pulmonary infections. Ann Am Thorac Soc. 2015;12:765–74. doi: 10.1513/AnnalsATS.201411-507FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campo I, Zorzetto M, Mariani F, et al. A large kindred of pulmonary fibrosis associated with a novel ABCA3 gene variant. Respir Res. 2014;15:43. doi: 10.1186/1465-9921-15-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doan ML, Guillerman RP, Dishop MK, et al. Clinical, radiological and pathological features of ABCA3 mutations in children. Thorax. 2008;63:366–73. doi: 10.1136/thx.2007.083766. [DOI] [PubMed] [Google Scholar]
- 74.Whitsett JA, Wert SE, Weaver TE. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu Rev Med. 2010;61:105–19. doi: 10.1146/annurev.med.60.041807.123500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mulugeta S, Nguyen V, Russo SJ, et al. A surfactant protein C precursor protein BRICHOS domain mutation causes endoplasmic reticulum stress, proteasome dysfunction, and caspase 3 activation. Am J Respir Cell Mol Biol. 2005;32:521–30. doi: 10.1165/rcmb.2005-0009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tanjore H, Blackwell TS, Lawson WE. Emerging evidence for endoplasmic reticulum stress in the pathogenesis of idiopathic pulmonary fibrosis. AJP Lung Cell Mol Physiol. 2012;302:L721–9. doi: 10.1152/ajplung.00410.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kropski JA, Lawson WE, Young LR, et al. Genetic studies provide clues on the pathogenesis of idiopathic pulmonary fibrosis. Dis Model Mech. 2013;6:9–17. doi: 10.1242/dmm.010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hawkins A, Guttentag SH, Deterding R, et al. A non-BRICHOS SFTPC mutant (SP-CI73T) linked to interstitial lung disease promotes a late block in macroautophagy disrupting cellular proteostasis and mitophagy. Am J Physiol Lung Cell Mol Physiol. 2015;308:L33–47. doi: 10.1152/ajplung.00217.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abou Taam R, Jaubert F, Emond S, et al. Familial interstitial disease with I73T mutation: a mid- and long-term study. Pediatr Pulmonol. 2009;44:167–75. doi: 10.1002/ppul.20970. [DOI] [PubMed] [Google Scholar]
- 80.Thomas AQ, Lane K, Phillips J, et al. Heterozygosity for a surfactant protein C gene mutation associated with usual interstitial pneumonitis and cellular nonspecific interstitial pneumonitis in one kindred. Am J Respir Crit Care Med. 2002;165:1322–8. doi: 10.1164/rccm.200112-123OC. [DOI] [PubMed] [Google Scholar]
- 81.Coghlan MA, Shifren A, Huang HJ, et al. Sequencing of idiopathic pulmonary fibrosis-related genes reveals independent single gene associations. BMJ Open Respir Res. 2014;1:e000057. doi: 10.1136/bmjresp-2014-000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ono S, Tanaka T, Ishida M, et al. Surfactant protein C G100S mutation causes familial pulmonary fibrosis in Japanese kindred. Eur Respir J. 2011;38:861–9. doi: 10.1183/09031936.00143610. [DOI] [PubMed] [Google Scholar]
- 83.Lawson WE, Grant SW, Ambrosini V, et al. Genetic mutations in surfactant protein C are a rare cause of sporadic cases of IPF. Thorax. 2004;59:977–80. doi: 10.1136/thx.2004.026336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Moorsel CHM, Van Oosterhout MFM, Barlo NP, et al. Surfactant protein C mutations are the basis of a significant portion of adult familial pulmonary fibrosis in a Dutch cohort. Am J Respir Crit Care Med. 2010;182:1419–25. doi: 10.1164/rccm.200906-0953OC. [DOI] [PubMed] [Google Scholar]
- 85.Whitsett JA, Wert SE, Weaver TE. Diseases of pulmonary surfactant homeostasis. Annu Rev Pathol. 2015;10:371–93. doi: 10.1146/annurev-pathol-012513-104644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nathan N, Giraud V, Picard C, et al. Germline SFTPA1 mutation in familial idiopathic interstitial pneumonia and lung cancer. Hum Mol Genet. 2016;25:1457–67. doi: 10.1093/hmg/ddw014. [DOI] [PubMed] [Google Scholar]
- 87.van Moorsel CHM, Ten Klooster L, van Oosterhout MFM, et al. SFTPA2 mutations in familial and sporadic idiopathic interstitial pneumonia. Am J Respir Crit Care Med. 2015;192:1249–52. doi: 10.1164/rccm.201504-0675LE. [DOI] [PubMed] [Google Scholar]
- 88.Maitra M, Wang Y, Gerard RD, et al. Surfactant protein A2 mutations associated with pulmonary fibrosis lead to protein instability and endoplasmic reticulum stress. J Biol Chem. 2010;285:22103–13. doi: 10.1074/jbc.M110.121467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lawson WE, Crossno PF, Polosukhin VV, et al. Endoplasmic reticulum stress in alveolar epithelial cells is prominent in IPF: association with altered surfactant protein processing and herpesvirus infection. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1119–26. doi: 10.1152/ajplung.00382.2007. [DOI] [PubMed] [Google Scholar]
- 90.Korfei M, Ruppert C, Mahavadi P, et al. Epithelial endoplasmic reticulum stress and apoptosis in sporadic idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178:838–46. doi: 10.1164/rccm.200802-313OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–76. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.MacArthur DG, Balasubramanian S, Frankish A, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–8. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Claustres M, Kožich V, Dequeker E, et al. Recommendations for reporting results of diagnostic genetic testing (biochemical, cytogenetic and molecular genetic) Eur J Hum Genet. 2014;22:160–70. doi: 10.1038/ejhg.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ackerman MJ. Genetic purgatory and the cardiac channelopathies: exposing the variants of uncertain/unknown significance issue. Heart Rhythm. 2015;12:2325–31. doi: 10.1016/j.hrthm.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 97.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]