Abstract
Background
Children with congenital heart defects (CHDs) are at high risk for myocardial failure after operative procedures with cardiopulmonary bypass (CPB). Recent studies suggest that microRNAs (miRNA) are involved in the development of CHDs and myocardial failure. Therefore, the aim of this study was to determine alterations in the miRNA profile in heart tissue after cardiac surgery using CPB.
Methods
In total, 14 tissue samples from right atrium were collected from patients before and after connection of the CPB. SurePrint™ 8 × 60K Human v21 miRNA array and quantitative reverse transcription-polymerase chain reaction (RT-qPCR) were employed to determine the miRNA expression profile from three patients before and after connection of the CPB. Enrichment analyses of altered miRNA expression were predicted using bioinformatic tools.
Results
According to miRNA array, a total of 90 miRNAs were significantly altered including 29 miRNAs with increased and 61 miRNAs with decreased expression after de-connection of CPB (n = 3) compared to before CPB (n = 3). Seven miRNAs had been validated using RT-qPCR in an independent cohort of 11 patients. Enrichment analyses applying the KEGG database displayed the highest correlation for signaling pathways, cellular community, cardiovascular disease and circulatory system.
Conclusion
Our result identified the overall changes of the miRNome in right atrium tissue of patients with CHDs after CPB. The differentially altered miRNAs lay a good foundation for further understanding of the molecular function of changed miRNAs in regulating CHDs and after CPB in particular.
Electronic supplementary material
The online version of this article (doi:10.1186/s12967-017-1213-9) contains supplementary material, which is available to authorized users.
Keywords: MicroRNA, Congenital heart disease, Atrial myocardium, Cardiopulmonary bypass
Background
Congenital heart diseases (CHDs) are the most common congenital organ malformations in newborns [1]. Nearly 10 of 1000 newborns have congenital heart and vessel defects with a wide variation with regards to the severity of defects and the need for immediate therapeutic intervention [1–3]. Advances in diagnostic and therapeutic methods have improved the long-term survival and morbidities of these patients [3–5]. However, considerable mortality rates in neonates rather than older children in association with corrective cardiac surgery remain challenging [1–5]. Neonates needing surgical intervention in early life still have significantly higher mortality than infants and older children [6, 7]. Corrective cardiac surgery is one of the most major cause of mortality in neonates and infants [8]. The morbidities and risk factors associated with cardiopulmonary bypass (CPB) are well documented in adults and children [9–12]. Data on the mechanisms of adaptation and remodeling in the myocardium of neonates and small children undergoing cardiac surgery are still scarce. Therefore, a better understanding of the molecular and genetic mechanisms underlying congenital heart diseases and the specific adapting mechanisms during stress and ischemia is fundamental. Recent studies in adults with different cardiac morbidities have suggested an important role for microRNAs (miRNAs) in the pathogenesis of numerous cardiovascular diseases [13, 14]. MiRNAs are a class of small non-coding RNAs that regulate gene expression post-transcriptionally via sequence-specific interaction with the 3′ UTR of target mRNAs, resulting in mRNA degradation and/or inhibition of translation [15]. In signaling and transcriptional processes of cardiac biology, miRNAs can act as ‘molecular switches’ of post-transcriptional regulation of gene expression by regulating the expression of multiple proteins that function at different steps in cellular processes such as growth, differentiation, metabolism and apoptosis [16] as well as cardiac remodeling including increased angiogenesis and decreased fibrosis [17]. These miRNAs-regulated pathways are contributing to the development of the cardiac remodeling process and their activation following a myocardial injury suggest a functional role for these signaling pathways in cardiac repair. MiRNAs known to be involved in myocardial development include miR-208a, miR-21, miR-145, miR-1, miR-133a and miR-29 [18–20]. Moreover, their aberrant expression has been linked to cardiac remodeling and hypertrophy [18–20]. However, the role of miRNAs in the myocardium under non-physiological conditions of CPB is not well understood. In this study, we aimed to characterize the miRNA profile in the atrial myocardium before and after cardiac surgery by means of CPB in patients (infants and children) with CHDs.
Methods
Study population and sample collection
Institutional Review Board approval (No. 156/14) was obtained before initiation of this study. Children aged from 5 days to 10.4 years who had cardiac surgery for CHDs were included in this pilot study after written informed consent was obtained. Selected demographics and preoperative characteristics of the children including diagnoses, surgical procedure and duration of hospitalization are shown in Table 1. Patients were excluded from the study when parents declined participation, complications occurred during surgery or scarring prevented secure extraction of a tissue sample. All patients received midazolam (0.1 mg/kg) for premedication. After preoxygenation with 100% oxygen, anesthesia was induced with thiopental and fentanyl in titrated doses. Atracurium (0.5 mg/kg) was given to facilitate nasotracheal intubation with a cuffed tube. Thereafter anesthesia was maintained with continuous infusions of fentanyl (5–10 µg/kg/h) and midazolam (0.1–0.2 mg/kg/h). The lungs were ventilated with oxygen/air to maintain normocapnia at pH of 7.35–7.45. Volume replacement was done to maintain physiological CVP and arterial pressure. Aortic and atrial cannulation were performed after systemic heparin administration with intravenous bolus. Before cannulation, the CPB was filled with priming solution which consisted of Sterofundin ISO, Gelafundin, Ringer’s lactate and tranexamic acid and then mixed with packed red blood cells (PRBCs) from donors. In every case the Heart–Lung Machine: JOSTRA HL 20 (Maquet, Rastatt, Germany) was used. Blood cardioplegia of 8–10 °C was given according to the Calafiore scheme [21]. Atrial myocardial tissue samples from all patients were collected from the right auricular appendage or the right atrium before cannulation and after de-cannulation of the right atrial cannula. Tissue samples of interest were transferred to siliconized microcentrifuge tubes and immediately frozen in liquid nitrogen. These samples were stored in liquid nitrogen at −180 °C until processing.
Table 1.
Characteristics of patients included in the study
| Patient Nr. | Age at surgery | Gender | Weight (kg) | Diagnosis | Surgical procedure | Duration of CPB (min) | Lowest body temperature (°C) | Duration of hospitalization (days) |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.5 year | Male | 5.5 | ASD II | Closure of the ASD | 45 | 35.1 | 12 |
| 2 | 0.9 year | Female | 8.5 | VSD | Closure of the VSD | 41 | 34.4 | 10 |
| 3 | 10.4 year | Female | 33.0 | Doubly committed VSD, double chambered right ventricle | Closure of the VSD | 82 | 34.0 | 10 |
| 4 | 0.5 year | Male | 5.2 | VSD, ASD II | Closure of the ASD | 79 | 29.4 | 11 |
| 5 | 2.0 year | Female | 11 | ASD II | Closure of the ASD | 32 | 34.8 | 9 |
| 6 | 1.1 year | Male | 9.4 | ASD I | Closure of the ASD | 63 | 33.9 | 12 |
| 7 | 1.0 year | Female | 6.2 | Malalignment-VSD | Closure of the VSD | 87 | 31.8 | 13 |
| 8 | 5 days | Male | 3.3 | HLHS | Norwood I, Blalock-Taussig shunt | 136 | 24.2 | 25 |
| 9 | 7 days | Male | 4.1 | d-TGA | Switch | 112 | 24.0 | 24 |
| 10 | 11 days | Male | 2.1 | TAPVC | Correction of TAPVC | 124 | 22.7 | 54 |
| 11 | 1.2 year | Male | 7.6 | ASD II | Closure of the ASD | 34 | 34.5 | 8 |
| 12 | 1.7 year | Male | 10.1 | ASD I, mitral insufficiency | Closure of the ASD | 41 | 32.8 | 8 |
| 13 | 3.5 year | Female | 13.0 | ASD II | Closure of the ASD | 24 | 34.8 | 7 |
| 14 | 1.4 year | Female | 10.6 | ASD II | Closure of the ASD | 23 | 35.0 | 8 |
ASD II, Atrial septal defect type II; VSD, membranous ventricular septal defects; HLHS, hypoplastic left heart syndrome; d-TGA, dextro-transposition of the great arteries; TAPVC, total anomalous pulmonary venous connection
Isolation of total RNA, including miRNAs, from myocardial tissue samples
Myocardial (right atrium) tissue samples were cut into small pieces, crushed and homogenized using the ball mill TissueLyser LT (Qiagen, Hilden, Germany) with an oscillation frequency of 50 Hz. Thereafter, total RNA including miRNA was extracted using the miRNeasy Mini kit on a QIAcube robot (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Concentration and purity of the samples were measured using the NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Massachusetts, USA). RNA integrity was assessed with an Agilent 2100 Bioanalyzer using RNA 6000 Nano kit (Agilent Technologies, California, USA). DNase I (Thermo Fisher Scientific, Massachusetts, USA) treatment was carried out according to the manufacturer’s instructions to remove any DNA contamination. Conventional PCR with exon spanning primers for Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was performed to exclude residual DNA in the samples [22].
MiRNA microarray
MiRNA expression profiles of three samples from patients with CHD, before CPB (n = 3) and after CPB (n = 3) were hybridized on a SurePrint G3 miRNA Array v21.0 (Agilent Technologies, California, USA). The patients were chosen based on their age at the time of surgery to largely cover the range of ages included in our study. Briefly, 100 ng input of RNA from each sample was dephosphorylated by incubation with calf intestinal phosphatase and denatured using 100% DMSO. Samples were labeled with pCp-Cy3 by using T4 ligase. Each labeled RNA sample was hybridized onto an individual 8 × 60K format Agilent miRNA array slide v21. Arrays were washed and dried according to manufacturer’s recommendations and scanned at a resolution of 3 μm double-pass mode using an Agilent scanner. Data were acquired using Agilent AGW Feature Extraction software version 10.10.11 (Agilent Technologies, California, USA).
Reverse transcription qPCR of miRNA (array validation)
To validate the microarray results in the study, relative quantitative real-time PCR was performed on an ABI StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, USA) using SYBR Green I on nine differentially expressed miRNAs. Complementary DNA (cDNA) was generated by reverse transcription of 200 ng of total RNA using miScript RT II Kit (Qiagen, Hilden, Germany). Briefly, 200 ng of total RNA containing miRNAs was mixed with miScript 4 µL HiSpec Buffer, 2 µL nucleic mix, 2 µL miScript Reverse Transcriptase mix and RNase-free water to a final volume of 20 µL. Following the reverse transcription reaction, the cDNA was diluted 1:10 and then mixed with 10 µL QuantiTect SYBR Green PCR Master Mix, 2 µL miScript Universal Primer, 2 µL miScript Primer Assay for the selected nine miRNAs namely miR-328-5p, miR-4750-5p, miR-210-5p, miR-423-3p, miR-143-3p, miR-564, miR-770-5p, miR-874-5p, miR-93-5p and RNU6B as endogenous control and RNase-free water to a final volume of 20 µL. The primer sequences used in the study are shown in Additional file 1: Table S1. Reactions were assembled with the QIAgility automated pipetting system (Qiagen, Hilden, Germany). All PCR data were analyzed with SDS Relative Quantification Software version 2.3 (Applied Biosystems, Foster City, USA).
Target prediction and functional analysis
Enriched KEGG pathway analyses were performed using DIANA-miRPath v.3.0 software based on predicted targets by DIANA-microT-CDS [23]. Targets of miRNAs with a score of more than 0.8 were selected. Only KEGG pathways with P value <0.05 and a false discovery rate (FDR) <0.05 were retained. The effect of miRNAs on target genes and networks has been evaluated using miRTargetLink software [24].
Reverse transcription qPCR of miRNA and mRNA (functional network validation)
We used miScript Primer Assays for 9 miRNAs (miR-744-5p, miR-648, miR-193b-3p, miR-212-3p, miR-143-3p, miR-93-5p, miR-222-3p, miR-423-3p and miR-766-3p) and QuantiTect Primer assays for 9 target genes (CDKN1A, MYC, PTEN, ESR1, ETS1, SOD2, MGMT, KRAS and HNF4A) (Qiagen, Hilden, Germany) to validate the different expression levels of the miRNA and their target genes, which are determined by miRTargetLink prediction software. In brief, 350 ng of total RNA including miRNA were converted into cDNA using the miScript II RT Kit. During the reverse transcription step, 5× miScript HiFlex Buffer was used to promote conversion of RNA into cDNA. The resulting cDNA was diluted to obtain a miRNA concentration of 1.5 ng/µL and a mRNA concentration of 5 ng/µL for. All reverse transcription PCR (RT-PCR) experiments were performed using the QIAgility™ automated PCR setup (Qiagen, Hilden, Germany). RT-qPCR analysis was done on a StepOnePlus™ Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). GAPDH and RNU6B were chosen as reference genes for mRNA and miRNA normalization, respectively. In addition, we included a no-template control (NTC) and no-reverse transcriptase control (NRT) in each run.
Statistical analysis
The freely available R software (R Development Core Team, 2010) was used to analyze the differences in miRNA expression in the atrial myocardial tissue samples from patients with CHD samples before and after CPB. After applying the Agilent Feature Extraction image analysis software on our hybridized microarray slides, we collected the computed total gene signals (TGS) for each miRNA, and performed quantile normalization and a log (base 2) transformation of the TGS values. Differential levels of miRNAs were analyzed by employing a paired two-tailed t-test for miRNAs that showed a significant change in the two groups. P-values below a threshold of 0.05 were considered statistically significant. We used the relative quantitative method of 2−ΔΔCq to measure the expression differences of specifically selected miRNAs in RT-qPCR [25]. Two-tailed paired T-test was used to evaluate the fold change of miRNA levels.
Results
Patient characteristics
A total of 14 patients who had cardiac surgery for CHDs were included in the present study. Preoperative characteristics of the children including diagnoses, surgical procedure and duration of hospitalization are shown in Table 1.
Differentially expressed miRNAs
In order to characterize miRNA expression profiles in the atrial myocardial tissue of patient after CPB, we performed microarray assays using SurePrint G3 miRNA arrays that contain 2549 human miRNAs annotated in miRBase version 21.0. Atrial myocardial tissue from patients with CHD before cardiac surgery by means of CPB and after CPB were collected (three patients in total). By applying a paired two-tailed t-test, microarray assays showed that miRNAs were expressed differentially in atrial myocardial tissue after surgical treatment with CPB. A total of 90 miRNAs were significantly altered in the two considered groups with a P value of <0.05. Out of the 90 altered miRNAs, 29 were up-regulated in the samples after CPB compared to samples before CPB, while 61 miRNA were down-regulated (Table 2). As shown in Table 2, the majority of altered miRNA (49 out of 90) fell into the range of 1.50–1.99 fold up- or down-regulation. In addition, 17 miRNAs including six up-regulated and another eleven down-regulated miRNAs displayed expression level with changes ≥2.0-fold and 24 miRNA including eight up-regulated and another sixteen down-regulated miRNAs displayed expression level with changes <1.5 fold between two groups. Using hierarchic clustering with the euclidian distance measure, we analyzed how the expression profiles of patients before and after CPB relate to each other. For this task, we used the 50 miRNAs with the highest variance of miRNA levels out of the 2.549 miRNAs. Figure 1 shows the heatmap of the hierarchic clustering from the differentially expressed miRNA by microarray. In general, the hierarchical clustering exhibited a clear separation of the examined groups based on miRNA expression profiles. The first cluster contains mostly patients before CPB and the second most of the patients after CPB.
Table 2.
Significantly expressed miRNAs in the atrial myocardial tissue of patients with CHD after CPB (n = 3) compared to before CPB (n = 3) as determined by microarray
| miRNA | Median before CPB | Median after CPB | STDV before CPB | STDV after CPB | Fold change | Regulation | P-value |
|---|---|---|---|---|---|---|---|
| miR-4750-5p | 1.67 | 8.60 | 2.59 | 4.24 | 5.26 | Up | 0.0399 |
| miR-6134 | 1.00 | 3.08 | 0.46 | 0.78 | 3.03 | Up | 0.0215 |
| miR-6873-3p | 2.49 | 6.29 | 0.29 | 1.31 | 2.50 | Up | 0.0441 |
| miR-4747-5p | 1.07 | 2.38 | 0.78 | 1.05 | 2.22 | Up | 0.0151 |
| miR-5195-5p | 1.48 | 3.04 | 0.49 | 0.91 | 2.04 | Up | 0.0213 |
| miR-6074 | 1.50 | 3.00 | 1.14 | 1.52 | 2.00 | Up | 0.0252 |
| miR-6751-3p | 2.89 | 5.68 | 0.33 | 1.12 | 1.96 | Up | 0.0323 |
| miR-328-5p | 15.50 | 28.90 | 4.82 | 3.56 | 1.85 | Up | 0.0263 |
| miR-6792-5p | 2.42 | 4.40 | 0.56 | 1.34 | 1.82 | Up | 0.0485 |
| miR-4514 | 1.65 | 2.93 | 0.80 | 0.61 | 1.79 | Up | 0.0333 |
| miR-4538 | 3.31 | 5.83 | 1.60 | 1.37 | 1.75 | Up | 0.0033 |
| miR-6870-5p | 2.31 | 3.91 | 1.63 | 1.88 | 1.69 | Up | 0.0102 |
| miR-7156-5p | 1.25 | 2.12 | 0.61 | 0.61 | 1.69 | Up | 0.0300 |
| miR-4447 | 1.64 | 2.73 | 0.44 | 0.19 | 1.67 | Up | 0.0369 |
| miR-6769b-5p | 20.56 | 33.29 | 3.95 | 4.51 | 1.61 | Up | 0.0442 |
| miR-7846-3p | 5.11 | 8.12 | 1.69 | 2.65 | 1.59 | Up | 0.0398 |
| miR-1261 | 1.90 | 2.98 | 1.58 | 2.01 | 1.56 | Up | 0.0353 |
| miR-6740-5p | 39.53 | 61.77 | 6.38 | 14.38 | 1.56 | Up | 0.0489 |
| miR-6746-5p | 3.75 | 5.79 | 1.03 | 1.20 | 1.54 | Up | 0.0265 |
| miR-645 | 1.56 | 2.42 | 0.93 | 0.70 | 1.54 | Up | 0.0455 |
| miR-2113 | 1.46 | 2.20 | 0.47 | 0.74 | 1.52 | Up | 0.0437 |
| miR-497-3p | 1.46 | 2.14 | 0.29 | 0.51 | 1.47 | Up | 0.0280 |
| miR-648 | 1.64 | 2.39 | 0.55 | 0.44 | 1.47 | Up | 0.0497 |
| miR-5088-5p | 9.24 | 13.26 | 1.59 | 2.19 | 1.43 | Up | 0.0379 |
| miR-3945 | 3.43 | 4.81 | 0.97 | 0.98 | 1.41 | Up | 0.0342 |
| miR-4468 | 2.07 | 2.84 | 0.55 | 0.62 | 1.37 | Up | 0.0289 |
| miR-1471 | 6.69 | 9.07 | 2.16 | 2.11 | 1.35 | Up | 0.0464 |
| miR-5006-5p | 21.99 | 28.90 | 2.45 | 3.83 | 1.32 | Up | 0.0108 |
| miR-198 | 3.59 | 4.49 | 1.04 | 1.32 | 1.25 | Up | 0.0404 |
| miR-770-5p | 9.59 | 3.17 | 1.11 | 1.81 | 3.03 | Down | 0.0095 |
| miR-4261 | 35.83 | 14.45 | 1.98 | 7.10 | 2.48 | Down | 0.0183 |
| miR-874-5p | 15.30 | 6.57 | 1.76 | 3.12 | 2.33 | Down | 0.0102 |
| miR-550a-5p | 5.57 | 2.44 | 1.67 | 1.10 | 2.29 | Down | 0.0114 |
| miR-3651 | 53.15 | 23.24 | 5.23 | 9.24 | 2.29 | Down | 0.0455 |
| miR-6865-3p | 8.00 | 3.55 | 1.51 | 1.76 | 2.26 | Down | 0.0108 |
| miR-222-3p | 14.15 | 6.51 | 1.03 | 2.80 | 2.17 | Down | 0.0308 |
| miR-3607-3p | 8.69 | 4.10 | 0.70 | 1.25 | 2.12 | Down | 0.0424 |
| miR-1304-3p | 9.97 | 4.71 | 1.98 | 0.36 | 2.12 | Down | 0.0469 |
| miR-6508-5p | 9.27 | 4.41 | 2.31 | 1.58 | 2.10 | Down | 0.0358 |
| miR-6800-3p | 10.18 | 5.06 | 2.12 | 2.07 | 2.01 | Down | 0.0182 |
| miR-6861-3p | 6.13 | 3.09 | 0.74 | 0.86 | 1.99 | Down | 0.0139 |
| miR-212-3p | 25.83 | 13.01 | 2.76 | 6.93 | 1.99 | Down | 0.0294 |
| miR-6737-3p | 11.31 | 5.76 | 2.96 | 1.67 | 1.96 | Down | 0.0293 |
| miR-6820-5p | 37.19 | 19.53 | 0.20 | 4.42 | 1.90 | Down | 0.0238 |
| miR-6792-3p | 6.60 | 3.47 | 1.06 | 1.47 | 1.90 | Down | 0.0366 |
| miR-3616-3p | 6.92 | 3.68 | 1.31 | 1.75 | 1.88 | Down | 0.0082 |
| miR-3162-3p | 10.75 | 5.79 | 2.89 | 2.53 | 1.86 | Down | 0.0029 |
| miR-423-3p | 9.21 | 4.94 | 1.09 | 1.21 | 1.86 | Down | 0.0056 |
| miR-5190 | 10.43 | 5.68 | 0.59 | 0.86 | 1.84 | Down | 0.0172 |
| miR-4725-5p | 8.95 | 4.90 | 1.35 | 2.34 | 1.83 | Down | 0.0421 |
| miR-744-5p | 9.67 | 5.32 | 1.56 | 1.98 | 1.82 | Down | 0.0308 |
| miR-6829-5p | 48.68 | 26.75 | 3.55 | 8.52 | 1.82 | Down | 0.0360 |
| miR-6889-3p | 8.18 | 4.51 | 1.73 | 1.02 | 1.82 | Down | 0.0432 |
| miR-4433a-5p | 8.38 | 4.62 | 2.04 | 1.02 | 1.81 | Down | 0.0276 |
| miR-6813-3p | 7.19 | 4.06 | 2.29 | 1.01 | 1.77 | Down | 0.0343 |
| miR-4649-3p | 9.50 | 5.47 | 3.12 | 2.42 | 1.74 | Down | 0.0093 |
| miR-6763-3p | 7.94 | 4.62 | 1.52 | 1.52 | 1.72 | Down | 0.0434 |
| miR-532-3p | 18.58 | 10.78 | 0.43 | 3.99 | 1.72 | Down | 0.0487 |
| miR-6769a-5p | 11.01 | 6.45 | 1.97 | 1.02 | 1.71 | Down | 0.0219 |
| miR-193b-3p | 74.91 | 43.92 | 26.81 | 34.56 | 1.71 | Down | 0.0341 |
| miR-331-5p | 2.64 | 1.58 | 0.28 | 0.62 | 1.67 | Down | 0.0446 |
| miR-6858-3p | 8.53 | 5.15 | 2.59 | 2.47 | 1.66 | Down | 0.0017 |
| miR-564 | 17.68 | 10.75 | 2.27 | 3.32 | 1.65 | Down | 0.0099 |
| miR-6723-5p | 26.08 | 16.14 | 2.83 | 6.35 | 1.62 | Down | 0.0352 |
| miR-28-3p | 4.85 | 3.02 | 0.21 | 0.58 | 1.61 | Down | 0.0187 |
| miR-339-3p | 13.22 | 8.22 | 1.98 | 2.36 | 1.61 | Down | 0.0275 |
| miR-23c | 5.83 | 3.62 | 1.59 | 0.81 | 1.61 | Down | 0.0378 |
| miR-93-5p | 114.26 | 70.98 | 9.72 | 24.81 | 1.61 | Down | 0.0488 |
| miR-6752-3p | 5.76 | 3.67 | 1.11 | 0.79 | 1.57 | Down | 0.0329 |
| miR-766-3p | 10.39 | 6.64 | 1.38 | 2.07 | 1.56 | Down | 0.0125 |
| miR-1306-5p | 4.68 | 3.05 | 0.44 | 0.67 | 1.54 | Down | 0.0074 |
| miR-6798-3p | 4.49 | 2.92 | 0.87 | 1.04 | 1.54 | Down | 0.0141 |
| miR-664a-3p | 17.21 | 11.20 | 1.39 | 3.72 | 1.54 | Down | 0.0362 |
| miR-208a-5p | 45.35 | 29.37 | 7.47 | 13.45 | 1.54 | Down | 0.0450 |
| miR-6880-3p | 6.55 | 4.43 | 2.43 | 1.74 | 1.48 | Down | 0.0272 |
| miR-143-3p | 327.88 | 223.74 | 43.81 | 69.68 | 1.47 | Down | 0.0425 |
| miR-624-5p | 3.66 | 2.49 | 0.37 | 0.53 | 1.47 | Down | 0.0447 |
| miR-210-5p | 4.05 | 2.86 | 0.41 | 0.39 | 1.42 | Down | 0.0101 |
| miR-6073 | 5.18 | 3.67 | 1.17 | 1.43 | 1.41 | Down | 0.0140 |
| miR-374c-5p | 11.99 | 8.54 | 1.55 | 2.75 | 1.40 | Down | 0.0378 |
| miR-4484 | 27.73 | 20.62 | 1.81 | 1.80 | 1.34 | Down | 0.0000 |
| miR-4664-3p | 5.01 | 3.75 | 0.50 | 0.61 | 1.34 | Down | 0.0096 |
| miR-6731-3p | 4.38 | 3.26 | 1.21 | 1.48 | 1.34 | Down | 0.0399 |
| miR-4769-3p | 6.46 | 5.11 | 1.50 | 1.35 | 1.26 | Down | 0.0213 |
| miR-15a-3p | 3.49 | 2.79 | 0.34 | 0.28 | 1.25 | Down | 0.0114 |
| hsa-let-7a-3p | 3.24 | 2.59 | 0.35 | 0.29 | 1.25 | Down | 0.0172 |
| miR-26b-3p | 3.15 | 2.54 | 0.72 | 0.55 | 1.24 | Down | 0.0244 |
| miR-940 | 32.01 | 26.08 | 7.78 | 6.20 | 1.23 | Down | 0.0218 |
| miR-3189-3p | 3.61 | 3.01 | 0.44 | 0.33 | 1.20 | Down | 0.0153 |
| miR-29a-5p | 3.05 | 2.53 | 0.16 | 0.28 | 1.20 | Down | 0.0166 |
Each value represents the median of three patients before and after CBP and ±standard deviation (STDV). Statistical analysis was performed with paired-two-tailed t-test (P < 0.05)
Fig. 1.
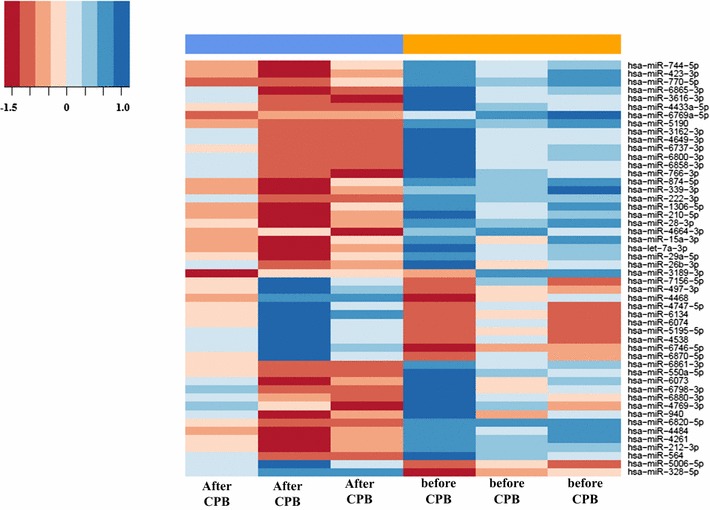
Unsupervised hierarchical clustering (Euclidian distance, complete linkage) of the three patients (six samples, three before and three after CPB) based on expression of the 50 with highest variance. The heatmap shows miRNAs with high expression in blue, miRNAs with low expression in red. The blue and orange lines indicate the two main clusters of samples
Validation of differentially expressed miRNAs using RT-qPCR
To validate the data obtained from the miRNA microarray, RT-qPCR was performed to re-examine the expression level of nine miRNAs, namely miR-328-5p, miR-4750-5p, miR-210-5p, miR-423-3p, miR-143-3p, miR-564, miR-770-5p, miR-874-5p and miR-93-5p. These miRNAs were chosen based on their differential expression level in each patient group before and after CPB and/or on their known associations with heart diseases. In detail, we selected two miRNAs with highest (miR-4750-5p) and moderate (miR-328-5p) fold change among the up-regulated miRNAs and two miRNAs with highest (miR-770-5p) and moderate (miR-564) fold change among the down-regulated miRNAs. In addition, we selected six miRNAs (miR-328-5p, miR-210-5p, miR-423-3p, miR-143-3p, miR-874-5p and miR-93-5p) with low or moderate expression levels based on the array analysis and with known association with cardiac pathologies and its related process [26–33]. For this validation step, we analyzed a new cohort of 11 samples taken from patients with CHD undergoing corrective surgery by CPB. The RT-qPCR showed the same direction of expression changes as the microarray analysis for seven miRNAs (Fig. 2). The significance of the differences in the expression was confirmed for seven miRNAs namely miR-210-5p, miR-423-3p, miR-143-3p, miR-564, miR-770-5p, miR-874-5p and miR-93-5p) (P < 0.05) in the atrial myocardial samples from patients with CHDs after CPB compared to before CPB by RT-qPCR (Fig. 3). By including two out of the seven validated miRNAs namely miR-143-3p and miR-93-5p, a clear distinction between the groups based on the clustering dendrogram was, however, not possible (Additional file 2: Figure S1). We next analyzed the expression levels of the nine miRNAs before CPB and after CPB separately for neonate and infant patients. As for the atrial myocardial samples from neonates with CHDs, we found a significantly different expression for miR-874-5p with a P-value of 0.040 between prior CPB and after CPB. In addition, we found two miRNAs with border-line P-values (miR-423-3p, P-value = 0.060 and miR-93-5p, P-value = 0.070) (Fig. 4). As for atrial myocardial samples from infants with CHDs, we found significantly different expression of six miRNAs (miR-210-5p with a P-value of 0.014, miR-423-3p with a P-value of 0.034, miR-143-3p with a P-value of 0.018, miR-564 with a P-value of 0.024, miR-770-5p with a P-value of 0.025, and miR-874-5p with a P-value of 0.040) between prior and after CPB (Fig. 4). In addition, we analyzed the relative expression level of different miRNAs depending on the age of each patient before and after CPB. As shown in Additional file 3: Figure S2, the relative expression level (2−ΔΔCt) for certain miRNAs decreased with age. In addition, we found a remarkable difference between the miRNAs analyzed.
Fig. 2.
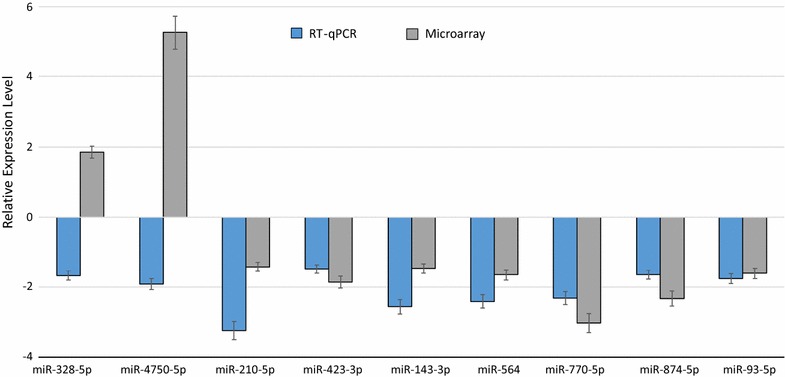
Relative fold change of the miRNAs in the atrial myocardial tissue of patients with CHD after CBP (n = 11) compared to before CPB (n = 11) as determined by RT-qPCR and after CBP (n = 3) compared to before CPB (n = 3) as determined by microarray (P < 0.05). Paired-two-tailed t-tests and ±standard deviation (STDV) were used to evaluate differences in expression in both assays. Relative expression level of 2−ΔΔCt and quantile normalization were used for RT-qPCR and microarray data, respectively
Fig. 3.
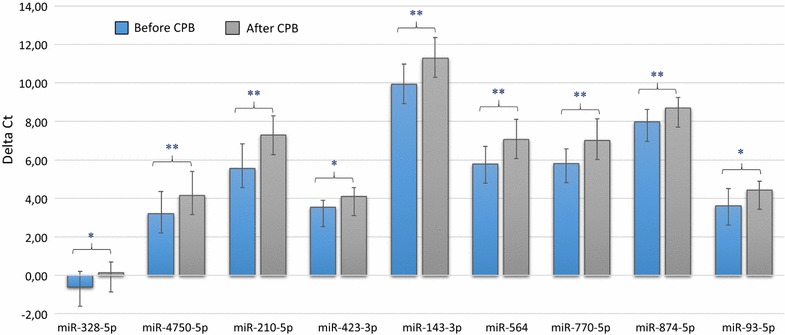
Validation of nine differentially expressed miRNAs in the atrial myocardial tissue of patients with CHD after CBP (n = 11) compared to before CPB (n = 11) as determined by RT-qPCR (P < 0.05). Mean ΔCt before CPB and after CPB (Lower ΔCt, higher expression level). RNAU6B as an endogenous control for normalization, paired-two-tailed t-tests and ±standard deviation (STDV) were used to evaluate differences in expression
Fig. 4.
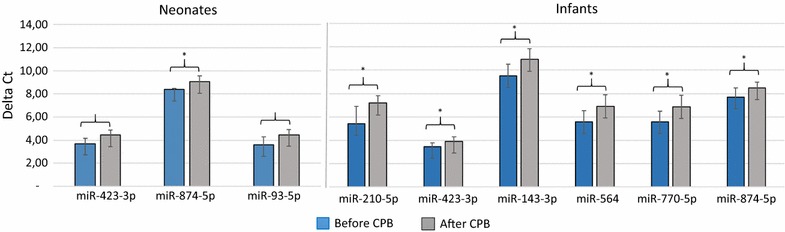
Significantly expressed miRNAs in the atrial myocardial tissue of neonates and infants with CHD after CBP compared to before CPB as determined by RT-qPCR (P < 0.05). Mean ΔCt before CPB and after CPB (Lower ΔCt, higher expression level). RNAU6B as an endogenous control for normalization, paired-two-tailed t-tests and ±standard deviation (STDV) were used to evaluate differences in expression
Comparative pathway analysis
We used DIANA-mirPath algorithm to gain insights into the biological pathways of the miRNAs that were altered in the atrial myocardial tissue from patients with CHD after CPB compared to before CPB. Based on the deregulated miRNAs by microarray analysis, we identified seventy KEGG pathways that were significantly enriched (P < 0.05, FDR corrected) for targets of the deregulated miRNAs, respectively (Additional file 1: Table S2). As shown in Table 3, the target genes of the altered miRNAs are mostly involved in “Signaling transduction”, “Cellular community”, “Cardiovascular disease”, “Circulatory system” and many others pathways. Using miRTargetLink, the resulting network is drawn schematically in Fig. 5. Besides the nine miRNAs, nine genes were targeted by one, two or more of the selected miRNAs in the “Strong” category. A strong interaction was observed between miRNAs and genes are highlighted in ‘‘Green’’ in the resulting network (Fig. 5).
Table 3.
The KEGG pathways significantly enriched for target genes of deregulated miRNAs in the atrial myocardial tissue from patients with CHD after CPB (n = 3) compared to before CPB (n = 3) ( P value < 0.05)
| Mode of interaction | KEGG pathway | P-value | No. genes | No. miRNAs |
|---|---|---|---|---|
| Signaling transduction | Hippo signaling pathway | 1.11E + 05 | 114 | 70 |
| Phosphatidylinositol signaling system | 1.23E + 06 | 66 | 63 | |
| TGF-beta signaling pathway | 3.29E + 06 | 64 | 53 | |
| cGMP-PKG signaling pathway | 3.29E + 06 | 128 | 74 | |
| FoxO signaling pathway | 0.001 | 98 | 72 | |
| ErbB signaling pathway | 0.0027 | 67 | 62 | |
| MAPK signaling pathway | 0.003 | 179 | 77 | |
| Ras signaling pathway | 0.0035 | 155 | 76 | |
| Sphingolipid signaling pathway | 0.0145 | 85 | 64 | |
| Calcium signaling pathway | 0.0174 | 124 | 71 | |
| AMPK signaling pathway | 0.0189 | 86 | 67 | |
| cAMP signaling pathway | 0.019 | 137 | 71 | |
| TNF signaling pathway | 0.0229 | 76 | 63 | |
| Rap1 signaling pathway | 0.0243 | 143 | 70 | |
| Cardiovascular diseases and Circulatory system | Adrenergic signaling in cardiomyocytes | 1.20E + 06 | 112 | 71 |
| Dilated cardiomyopathy | 0.0154 | 65 | 57 | |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 0.0174 | 53 | 54 | |
| Vascular smooth muscle contraction | 0.0196 | 83 | 65 | |
| Cellular community | Adherens junction | 3.20E − 03 | 56 | 58 |
| Focal adhesion | 0.0044 | 146 | 74 | |
| Gap junction | 0.0106 | 65 | 61 | |
| Cell adhesion molecules (CAMs) | 0.0248 | 98 | 68 |
Fig. 5.
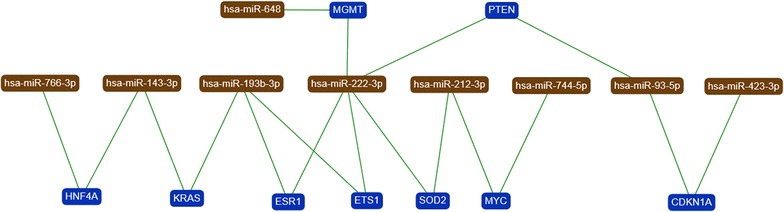
Target network with deregulated miRNAs indicated in brown and target genes in blue. Only genes targeted by at least two or ore miRNAs are included. Strong interaction was observed between miRNAs and genes are highlighted by “green” edges in the resulting network
Validation of miRNAs and mRNAs that were differentially expressed in atrial myocardial tissues
To confirm the differential expression of miRNAs and their targeted mRNAs before and after CPB, we analyzed eight of the down-regulated miRNAs, one of the up-regulated miRNAs and nine target mRNAs by RT-qPCR. With the exception of miR-222-3p, the remaining eight miRNAs showed the same direction of expression changes in the RT-PCR and in the microarray analysis. These findings are summarized in Additional file 4: Figure S3. Out of the eight miRNAs, miR-648 was up-regulated and the seven remaining miRNAs were down regulated (miR-744-5p, miR-193b-3p, miR-212-3p, miR-143-3p, miR-93-5p, miR-423-3p and miR-766-3p). As for the mRNA analysis, 6 out of 9 analyzed target mRNAs showed the expected expression changes. In detail, for five down-regulated miRNAs the according mRNA targets (ESR1, ETS1, MYC, PTEN and CDKN1A) were up-regulated. For the up-regulated miRNA miR-222-3p the according mRNA target (MGMT) was down-regulated. Due to low number of samples, we could not determine the significance of the expression differences. For the targets SOD2 and KRAS we did not find expression changes between prior CPB and after CPB. For the target HNF4A, we could not determine the fold changes.
Discussion
In our study, we found evidence for an overall altered miRNA expression pattern in the atrial myocardial tissue of patients with CHD according to corrective cardiac surgery by using the cardiopulmonary bypass (CPB). MiRNA expression analysis using microarrays indicated 90 miRNAs with significantly differential expression, including 29 miRNAs with significantly up- and 61 miRNAs with significantly down-regulated expression after CPB. In the validation phase, by using a new cohort of samples, seven miRNAs were validated. These data show that miRNA expression levels in atrial myocardial tissue changes in the course of cardiac surgery using the CPB. MiRNAs specifically expressed or enriched in smooth, skeletal, and/or cardiac muscle that plays a physiological role in normal heart development and function are termed Myo-miRs [34]. In particular, miR-208, a member of the miR-208 family, is differentially expressed during heart developmental and plays an essential role in normal cardiac conduction [34, 35]. Both over-expression and under-expression of miR-208a has been associated with cardiac arrhythmia [36], with apoptosis in ischemic cardiomyocytes [37] and generally with heart diseases [38]. Down-regulation of miR-208a in atrial myocardial tissues of patients with CHD after CPB underscore the integral function of this miRNA in regulating cell morphology and contractility. The cardiomyogenesis regulator miR-143 is highly expressed during vascular smooth muscle cell differentiation and is involved in normal function and formation of the cardiac chamber via regulation of myocardial cell morphology [39]. The down-regulation of miR-143 causes ventricular collapse by affecting cell morphology and contractility [39]. Furthermore, miR-143 targets ETS Transcription Factor (Elk1), an activator of vascular smooth muscle cells proliferation [40].
Deregulation of a number of miRNAs in our study have been previously reported in right and left atrial appendages of patients with rheumatic mitral valve disease (RMVD), including miR-222-3p, miR-4484 and miR-940 in left atrial appendage [41], miR-5190 and miR-23c in right atrial appendages [42] and miR-143-3p in both, in the LAA and RAA of patients with RMVD [43]. MiR-222 has a documented function in regulating cell proliferation and is involved in the vascular smooth muscle cells differentiation [38, 44]. In neonatal cardiomyocytes, miR-222 induces cellular hypertrophy and proliferation and inhibits apoptosis after ischemic injury [45]. In addition, miR-222 increases cell proliferation and inhibits cardiomyogenic differentiation in right ventricular outflow tract (RVOT) myocardial tissues from infants with nonsyndromic tetralogy of fallot (TOF) [46]. Our finding for miR-222 also suggests that this miRNA protects cardiac structure and functions after corrective surgery by CPB. Previous studies have identified that many miRNAs are expressed at low levels under normal condition and expressed strongly during pathological stress like miR-212, a cardiomyocyte-specific miRNA, which is strongly activated during heart failure. Its up-regulation expression leads to pathological cardiac hypertrophy [47]. In agreement with our expression direction, Zhou et al. [48] reported down-regulation of miR-423-3p and miR-532-3p in ischemia–reperfusion injury heart grafts. MiR-423-5p was suggested as a diagnostic biomarker to distinguish heart failure patients from healthy controls. The plasma concentration patients with heart failure may reflect the severity of dilated cardiomyopathy [49]. Similarly, reduced expression of miR-93 was observed in the left atrium compared to right atrial tissue of patients with sinus rhythm [50] indicating that miR-93 promotes angiogenesis in the ischemic tissue by coordinating the functional pathways of cell proliferation and apoptosis [51]. The up-regulation of miR-328 expression suppresses its target gene SERCA2a (ATPase Sarcoplasmic/Endoplasmic Reticulum Ca2+ Transporting 2) in cardiac myocytes to indirectly activate the calcineurin/NFATc3 signaling pathway, leading to cardiac hypertrophy [33]. In addition, miR-328 was up-regulated in the atria of patients with atrial fibrillation (AF) [52]. Two miRNAs, miR-624 and miR-339, were deregulated in ischemic heart disease and coronary artery disease [53, 54]. MiR-744 was deregulated in patients with chronic congestive heart failure [55]. Several miRNAs were detected in cardiac tissue at different stages of development and are highly expressed in the fetal heart, including miR-212, miR-210 and miR-423 [41]. Moreover, Huang et al. found that miR-210 is involved in cellular hypoxia, regulation of angiogenesis and apoptosis [36]. Similarly, miR-423-5p, miR-193b-3p, and miR-550a-5p showed an altered expression level in patients with heart failure [56, 57].
Bioinformatics analysis by DIANA-mirPath predicted several KEGG biological pathways that were significantly enriched for the differentially deregulated miRNAs in the atrial myocardial samples (Table 3). Enrichment analysis displayed the highest correlation for signaling pathways, providing further evidence that myocardial target proteins are involved in the signaling pathways. Pathways related to the cardiovascular system and vascular smooth muscle contraction and its related processes, including cardiomyocytes proliferation, differentiation and apoptosis have been identified [58–63]. Recent studies implicating the Hippo signaling pathway promotes cardiomyocyte proliferation by activating the insulin-like growth factor [64] and Wnt signaling pathways [65]. Similarly, ErbB signaling pathway plays an important role in proper heart morphogenesis and also in all developmental stages of the heart [66]. These results demonstrate that the deregulated miRNAs in atrial myocardial tissue play an important role in participating in many signaling pathways that control heart function. We employed bioinformatics tools to gain further insights into the impact of the deregulated miRNAs on target genes. As indicated in Fig. 5 and Additional file 4: Figure S3 specific genes are likely affected by the several deregulated miRNAs including the genes for O-6-methylguanine-DNA methyltransferase (MGMT) and superoxide dismutase 2 (SOD2). The expression of these genes is enhanced in myocardial remodeling, cardiac hypertrophy and/or failure indicating that severe stress effects plays a critical role in in the pathogenesis of myocardial remodeling and failure [67, 68]. Phosphatase and tensin homolog (PTEN) plays a role in promoting cardiomyocyte proliferation and regeneration and protect the heart from hypertrophy and heart failure under biomechanical stress [69–71]. The transcription factors v-Myc Avin myelocytomatosis viral oncogene homologue (c-Myc) also play a regulatory role in stimulating cardiac myocytes proliferation and differentiation during fetal development [72].
We would like to point out that this study has also a number of limitations. One important point is that the study population was small and heterogeneous; age at surgery ranged from 5 days of life to 10.4 years and we did not differ between male and female gender. Moreover, the examined CHDs reached from simple ASDs to complex CHDs like HLHS or TAPVC and complicated the interpretation of the data. In fact, we were able to find several miRNAs which were elevated in all patients despite the heterogeneous study population. Another limitation is that all samples were taken from the right atrium. Acquisition of the samples happened before cannulation and after de-cannulation. However, for example, it is not clear which impact the suture before excision of the sample on the miRNA expression has. A sampling of a myocardial specimen from the right or left ventricle before the connection to CPB is not possible. This can be done during the corrective surgery after the cardiac arrest and may not represent the physiological conditions before the connection to CPB and cardiac arrest. Thus, the tissue sampling from the atrial appendage may facilitate the examining of the myocardial conditions before and after surgery using the CPB without increasing risks. In addition, this side of the heart may represent nearly a uniform myocardial region in all patients with CHD. Other settings of myocardial sampling will not be approved by the ethic committee of our institution. The precise mechanisms underlying the expressions of such miRNA should be evaluated in a larger cohort of patients including subgroups of uniform diagnosis of congenital heart defects. Nevertheless, we conclude that the connection to CPB and hypothermic cardiac arrest may induce several physiological responses in the myocardium, which in part is reflected by changed miRNA expression levels. Furthermore, it is unclear which effect the cardiac surgery itself as well as the usage of the CPB has on the miRNA expression. Several non-physiological circumstances occur during this process; for example, cardioplegia induces cardiac arrest, hypothermia is used, the heart is mechanical unloaded and reperfusion occurs after CPB. In addition, the CPB means contact of the blood with a foreign body surface. Whether this circumstance alone already causes changes in the miRNA profile has to be cleared.
Conclusion
In summary, we report miRNAs in the atrial myocardial tissue with significantly altered expression levels in patients with CHD after cardiac surgery with CPB. These altered miRNAs include cardiac-specific miRNAs which have been described in various cardiac pathologies including congenital heart disease. The overlap between the miRNAs identified in our study and the miRNAs that are involved in various levels of cardiac development by either up- or down-regulation strongly supports the idea that these miRNAs play an essential role in the pathophysiology of congenital heart diseases. Although it is important to bear in mind, that the miRNA expression profiles were obtained from a small number of atrial myocardial tissue samples, the altered expression levels have been further confirmed RT-qPCR for nine deregulated in a cohort of 11 independent atrial myocardial tissue samples. The alteration of the expression of miRNAs may provide new insights into the underlying mechanisms of cardiac surgery with CPB and provide potential novel mechanism-based therapeutic strategies for CHD.
Additional files
Additional file 1: Table S1. RT-qPCR primer sequences used in the study. Table S2. MiRNA associated pathways in the atrial myocardial tissue of patients with CHD after CPB (n=3) compared to before CPB (n=3): KEGG Pathways with predicted interaction enrichments for deregulated miRNAs by microarray (P value < 0.05).
Additional file 2: Figure S1. Unsupervised hierarchical clustering (Euclidian distance, complete linkage) of 3 patients with 6 samples including 3 samples before and 3 samples after CPB. The clustering was done based on the expression of the highest variance miRNAs as determined by RT-qPCR. MiRNAs with high expression are shown in blue and miRNAs with low expression in red. The blue and orange lines indicate the two main clusters of samples.
Additional file 3: Figure S2. Influence of age on the expression of 9 differentially expressed miRNA as determined by RT-qPCR. The relative expression level of 2-ΔΔCt is indicated on the y-axis and the patient age at the time of surgery on the x-axis. The different miRNAs are color coded as indicated. RNAU6B was used as endogenous control for normalization and paired-two-tailed t-tests was used to evaluate differences in expression.
Additional file 4: Figure S3. Functional target network validation of 9 differentially expressed miRNAs and 9 predicted target mRNAs. The comparison was done between atrial myocardial tissue of CHD patients before CBP (n=10) and after CPB (n=10). RNAU6B and GAPDH were used as an endogenous controls for normalization of miRNA and mRNA, respectively. Paired-two-tailed t-tests and ± standard deviation (STDV) were used to evaluate differences in expression.
Additional file 5. 1) Information letter to patients and parents, 2) consent form for anonymous publication, participation & storage of medical data and 3) tissue sample form.
Authors’ contributions
MA performed experimental work, particularly the miRNA array experiment, RT-qPCR validation and wrote the manuscript, MP, helped in the study design, diagnosed patients and helped in writing, NL, helped in RT-qPCR validation experiments and edited the manuscript. JM, helped in experimental work and samples collection. IM, isolated RNA, assessed the quality and quantity of RNA, GG, patients’ diagnosis and samples collection, JP, patients’ diagnosis and samples collection, HJS, patients’ diagnosis and samples collection, UG, patients’ diagnosis and samples collection, TP, patients’ diagnosis and samples collection, AK, performed bioinformatics analysis, EM, designed the study, coordinated the molecular biology experiment and article editing, HAK, designed the study, diagnosed patients and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Consent for publication
Consent has been provided, the information letter and consent form are available in additional file 5.
Ethics approval and consent to participate
Institutional Review Board approval/Ethikvotum Ärztekammer des Saarlandes: Nr. 156/14.
Funding
This study was funded by the Fördergemeinschaft Kinderherzen in Bonn, the Competence Network for Congenital Heart Defects, which received funding from the Federal Ministry of Education and Research, Grant No. 01GI0601 (2014), and the German Centre for Cardiovascular Research (DZHK), Grant No. 81X2800112 (2015).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AF
atrial fibrillation
- ASD
atrial septal defect
- CDKN1A
cyclin dependent kinase inhibitor 1A
- CHD
congenital heart defect
- c-Myc
v-Myc avin myelocytomastosis viral oncogene homologue
- CPB
cardiopulmonary bypass
- DMSO
dimethyl sulfoxide
- d-TGA
dextro-position of the great arteries
- Elk1
eTS transcription factor
- ESR1
estrogen receptor 1
- ETS1
eTS proto-oncogene 1
- FDA
false discovery rate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HLHS
hypoplastic left heart syndrome
- HNF4A
hepatocyte nuclear factor 4 alpha
- KEGG
Kyoto encyclopedia of genes and genomes
- KRAS
KRAS proto-oncogene, GTPase
- LAA
left atrial appendages
- MGMT
O-6-methylguanine-DNA methyltransferase
- miEAA
miRNA enrichment analysis and annotation
- miRNA
microRNA
- PRBCs
packed red blood cells
- PTEN
phosphatase and tensin homolog
- RAA
right atrial appendages
- RMVD
rheumatic mitral valve disease
- RT-qPCR
quantitative reverse transcription-polymerase chain reaction
- RVOT
right ventricular outflow tract
- SERCA2a
ATPase sarcoplasmic/endoplasmic reticulum Ca2 + transporting 2
- SOD2
superoxide dismutase 2
- TAPVC
total anomalous pulmonary venous connection
- TGS
total gene signals
- TOF
tetralogy of fallot
- VSD
ventricular septal defect
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12967-017-1213-9) contains supplementary material, which is available to authorized users.
Masood Abu-Halima, Martin Poryo, Eckart Meese and Hashim Abdul-Khaliq contributed equally to this work
Contributor Information
Masood Abu-Halima, Phone: +49 6841 16- 26270, Email: masood@daad-alumni.de.
Martin Poryo, Email: Martin.Poryo@uniklinikum-saarland.de.
Nicole Ludwig, Email: n.ludwig@mx.uni-saarland.de.
Janine Mark, Email: janine.mark@uks.eu.
Ina Marsollek, Email: i.marsollek@mx.uni-saarland.de.
Christian Giebels, Email: christian.giebels@uks.eu.
Johannes Petersen, Email: joh.petersen@uke.de.
Hans-Joachim Schäfers, Email: h-j.schaefers@uks.eu.
Ulrich Grundmann, Email: ulrich.grundmann@uks.eu.
Thomas Pickardt, Email: pickardt@kompetenznetz-ahf.de.
Andreas Keller, Email: andreas.keller@ccb.uni-saarland.de.
Eckart Meese, Email: hgemee@uks.eu.
Hashim Abdul-Khaliq, Email: Hashim.Abdul-Khaliq@uniklinikum-saarland.de.
References
- 1.van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, Roos-Hesselink JW. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Schwedler G, Lindinger A, Lange PE, Sax U, Olchvary J, Peters B, Bauer U, Hense HW. Frequency and spectrum of congenital heart defects among live births in Germany: a study of the competence network for congenital heart defects. Clin Res Cardiol. 2011;100(12):1111–1117. doi: 10.1007/s00392-011-0355-7. [DOI] [PubMed] [Google Scholar]
- 3.Erikssen G, Liestol K, Seem E, Birkeland S, Saatvedt KJ, Hoel TN, Dohlen G, Skulstad H, Svennevig JL, Thaulow E, et al. Achievements in congenital heart defect surgery: a prospective, 40-year study of 7038 patients. Circulation. 2015;131(4):337–346. doi: 10.1161/CIRCULATIONAHA.114.012033. [DOI] [PubMed] [Google Scholar]
- 4.Raissadati A, Nieminen H, Jokinen E, Sairanen H. Progress in late results among pediatric cardiac surgery patients: a population-based 6-decade study with 98% follow-up. Circulation. 2015;131(4):347–353. doi: 10.1161/CIRCULATIONAHA.114.011190. [DOI] [PubMed] [Google Scholar]
- 5.Jones B, Muscara F, Lloyd O, McKinlay L, Justo R. Neurodevelopmental outcome following open heart surgery in infancy: 6-year follow-up. Cardiol Young. 2015;25(5):903–910. doi: 10.1017/S1047951114001140. [DOI] [PubMed] [Google Scholar]
- 6.Dollat C, Vergnat M, Laux D, Stos B, Baruteau A, Capderou A, Demontoux S, Hamann M, Mokhfi E, Van Aerschot I, et al. Critical congenital heart diseases in preterm neonates: is early cardiac surgery quite reasonable? Pediatr Cardiol. 2015;36(6):1279–1286. doi: 10.1007/s00246-015-1158-9. [DOI] [PubMed] [Google Scholar]
- 7.Kumar TK, Charpie JR, Ohye RG, Hirsch-Romano JC, Donohue JE, Yu S, Sood V, Wilkinson DA, Nelson K, Mitchell E, et al. Timing of neonatal cardiac surgery is not associated with perioperative outcomes. J Thorac Cardiovasc Surg. 2014;147(5):1573–1579. doi: 10.1016/j.jtcvs.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Ades AM, Dominguez TE, Nicolson SC, Gaynor JW, Spray TL, Wernovsky G, Tabbutt S. Morbidity and mortality after surgery for congenital cardiac disease in the infant born with low weight. Cardiol Young. 2010;20(1):8–17. doi: 10.1017/S1047951109991909. [DOI] [PubMed] [Google Scholar]
- 9.Cheng HH, Wypij D, Laussen PC, Bellinger DC, Stopp CD, Soul JS, Newburger JW, Kussman BD. Cerebral blood flow velocity and neurodevelopmental outcome in infants undergoing surgery for congenital heart disease. Ann Thorac Surg. 2014;98(1):125–132. doi: 10.1016/j.athoracsur.2014.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang N, Cole T, Tsang V, Elliott M, de Leval M. Risk stratification in paediatric open-heart surgery. Eur J Cardiothorac Surg. 2004;26(1):3–11. doi: 10.1016/j.ejcts.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 11.Kussman BD, Wypij D, Laussen PC, Soul JS, Bellinger DC, DiNardo JA, Robertson R, Pigula FA, Jonas RA, Newburger JW. Relationship of intraoperative cerebral oxygen saturation to neurodevelopmental outcome and brain magnetic resonance imaging at 1 year of age in infants undergoing biventricular repair. Circulation. 2010;122(3):245–254. doi: 10.1161/CIRCULATIONAHA.109.902338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wypij D, Newburger JW, Rappaport LA, duPlessis AJ, Jonas RA, Wernovsky G, Lin M, Bellinger DC. The effect of duration of deep hypothermic circulatory arrest in infant heart surgery on late neurodevelopment: the Boston Circulatory Arrest Trial. J Thorac Cardiovasc Surg. 2003;126(5):1397–1403. doi: 10.1016/S0022-5223(03)00940-1. [DOI] [PubMed] [Google Scholar]
- 13.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 14.Mishra PK, Tyagi N, Kumar M, Tyagi SC. MicroRNAs as a therapeutic target for cardiovascular diseases. J Cell Mol Med. 2009;13(4):778–789. doi: 10.1111/j.1582-4934.2009.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 16.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18(4):510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal U, George A, Bhutani S, Ghosh-Choudhary S, Maxwell JT, Brown ME, Mehta Y, Platt MO, Liang Y, Sahoo S, et al. Experimental, systems, and computational approaches to understanding the microRNA-mediated reparative potential of cardiac progenitor cell-derived exosomes From pediatric patients. Circ Res. 2017;120(4):701–712. doi: 10.1161/CIRCRESAHA.116.309935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105(35):13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 20.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31(6):659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 21.Calafiore AM, Teodori G, Mezzetti A, Bosco G, Verna AM, Di Giammarco G, Lapenna D. Intermittent antegrade warm blood cardioplegia. Ann Thorac Surg. 1995;59(2):398–402. doi: 10.1016/0003-4975(94)00843-V. [DOI] [PubMed] [Google Scholar]
- 22.Abu-Halima M, Ludwig N, Hart M, Leidinger P, Backes C, Keller A, Hammadeh M, Meese E. Altered micro-ribonucleic acid expression profiles of extracellular microvesicles in the seminal plasma of patients with oligoasthenozoospermia. Fertil Steril. 2016;106:1061–1069. doi: 10.1016/j.fertnstert.2016.06.030. [DOI] [PubMed] [Google Scholar]
- 23.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamberg M, Backes C, Fehlmann T, Hart M, Meder B, Meese E, Keller A. MiRTargetLink–miRNAs, genes and interaction networks. Int J Mol Sci. 2016;17(4):564. doi: 10.3390/ijms17040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Contu R, Latronico MV, Condorelli G. Circulating microRNAs as potential biomarkers of coronary artery disease: a promise to be fulfilled? Circ Res. 2010;107(5):573–574. doi: 10.1161/CIRCRESAHA.110.227983. [DOI] [PubMed] [Google Scholar]
- 27.Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM, Lynch JM, Hullinger TG, Seto AG, van Rooij E. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail. 2013;15(6):650–659. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 28.Endo K, Naito Y, Ji X, Nakanishi M, Noguchi T, Goto Y, Nonogi H, Ma X, Weng H, Hirokawa G, et al. MicroRNA 210 as a biomarker for congestive heart failure. Biol Pharm Bull. 2013;36(1):48–54. doi: 10.1248/bpb.b12-00578. [DOI] [PubMed] [Google Scholar]
- 29.Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. 2012;14(2):147–154. doi: 10.1093/eurjhf/hfr155. [DOI] [PubMed] [Google Scholar]
- 30.Latronico MV, Catalucci D, Condorelli G. MicroRNA and cardiac pathologies. Physiol Genom. 2008;34(3):239–242. doi: 10.1152/physiolgenomics.90254.2008. [DOI] [PubMed] [Google Scholar]
- 31.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, Pinto YM. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106(6):1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 32.Wang K, Liu F, Zhou LY, Ding SL, Long B, Liu CY, Sun T, Fan YY, Sun L, Li PF. miR-874 regulates myocardial necrosis by targeting caspase-8. Cell Death Dis. 2013;4:e709. doi: 10.1038/cddis.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Li X, Gao X, Zhang R, Zhang Y, Liang H, Xu C, Du W, Zhang Y, Liu X, et al. MicroRNA-328 as a regulator of cardiac hypertrophy. Int J Cardiol. 2014;173(2):268–276. doi: 10.1016/j.ijcard.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 34.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17(5):662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91(3):827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 36.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Investig. 2009;119(9):2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tony H, Meng K, Wu B, Yu A, Zeng Q, Yu K, Zhong Y. MicroRNA-208a dysregulates apoptosis genes expression and promotes cardiomyocyte apoptosis during ischemia and its silencing improves cardiac function after myocardial infarction. Med Inflamm. 2015;2015:479123. doi: 10.1155/2015/479123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winbanks CE, Ooi JY, Nguyen SS, McMullen JR, Bernardo BC. MicroRNAs differentially regulated in cardiac and skeletal muscle in health and disease: potential drug targets? Clin Exp Pharmacol Physiol. 2014;41(9):727–737. doi: 10.1111/1440-1681.12281. [DOI] [PubMed] [Google Scholar]
- 39.Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, Washko D, Guner-Ataman B, Burns CG, Burns CE. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development. 2010;137(11):1887–1896. doi: 10.1242/dev.050526. [DOI] [PubMed] [Google Scholar]
- 40.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116(3):258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 42.Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. 2005;73(3):427–433. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 43.Liu H, Qin H, Chen GX, Liang MY, Rong J, Yao JP, Wu ZK. Comparative expression profiles of microRNA in left and right atrial appendages from patients with rheumatic mitral valve disease exhibiting sinus rhythm or atrial fibrillation. J Transl Med. 2014;12:90. doi: 10.1186/1479-5876-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104(4):476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, Xiao C, Bezzerides V, Bostrom P, Che L, et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21(4):584–595. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Chang JJ, Xu F, Ma XJ, Wu Y, Li WC, Wang HJ, Huang GY, Ma D. MicroRNA deregulation in right ventricular outflow tract myocardium in nonsyndromic tetralogy of fallot. Can J Cardiol. 2013;29(12):1695–1703. doi: 10.1016/j.cjca.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 1078;2012:3. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L, Zang G, Zhang G, Wang H, Zhang X, Johnston N, Min W, Luke P, Jevnikar A, Haig A, et al. MicroRNA and mRNA signatures in ischemia reperfusion injury in heart transplantation. PLoS ONE. 2013;8(11):e79805. doi: 10.1371/journal.pone.0079805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan KL, Zhang HF, Shen J, Zhang Q, Li XL. Circulating microRNAs levels in Chinese heart failure patients caused by dilated cardiomyopathy. Indian Heart J. 2013;65(1):12–16. doi: 10.1016/j.ihj.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slagsvold KH, Johnsen AB, Rognmo O, Hoydal M, Wisloff U, Wahba A. Comparison of left versus right atrial myocardium in patients with sinus rhythm or atrial fibrillation-an assessment of mitochondrial function and microRNA expression. Physiol Rep. 2014;2(8):e12124. doi: 10.14814/phy2.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hazarika S, Farber CR, Dokun AO, Pitsillides AN, Wang T, Lye RJ, Annex BH. MicroRNA-93 controls perfusion recovery after hindlimb ischemia by modulating expression of multiple genes in the cell cycle pathway. Circulation. 2013;127(17):1818–1828. doi: 10.1161/CIRCULATIONAHA.112.000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McManus DD, Lin H, Tanriverdi K, Quercio M, Yin X, Larson MG, Ellinor PT, Levy D, Freedman JE, Benjamin EJ. Relations between circulating microRNAs and atrial fibrillation: data from the Framingham Offspring Study. Heart Rhythm. 2014;11(4):663–669. doi: 10.1016/j.hrthm.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen F, Zhao X, Peng J, Bo L, Fan B, Ma D. Integrated microRNA-mRNA analysis of coronary artery disease. Mol Biol Rep. 2014;41(8):5505–5511. doi: 10.1007/s11033-014-3426-9. [DOI] [PubMed] [Google Scholar]
- 54.Saddic LA, Chang TW, Sigurdsson MI, Heydarpour M, Raby BA, Shernan SK, Aranki SF, Body SC, Muehlschlegel JD. Integrated microRNA and mRNA responses to acute human left ventricular ischemia. Physiol Genom. 2015;47(10):455–462. doi: 10.1152/physiolgenomics.00049.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cakmak HA, Coskunpinar E, Ikitimur B, Barman HA, Karadag B, Tiryakioglu NO, Kahraman K, Vural VA. The prognostic value of circulating microRNAs in heart failure: preliminary results from a genome-wide expression study. J Cardiovasc Med. 2015;16(6):431–437. doi: 10.2459/JCM.0000000000000233. [DOI] [PubMed] [Google Scholar]
- 56.Huang X, Zuo J. Emerging roles of miR-210 and other non-coding RNAs in the hypoxic response. Acta Biochim Biophys Sin. 2014;46(3):220–232. doi: 10.1093/abbs/gmt141. [DOI] [PubMed] [Google Scholar]
- 57.Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, Lim JY, Chong JP, Ng JY, Chen YT, Chan MM, et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail. 2015;17(4):393–404. doi: 10.1002/ejhf.223. [DOI] [PubMed] [Google Scholar]
- 58.Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harbor Perspect Biol. 2013;5(3):a008292. doi: 10.1101/cshperspect.a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galasso G, De Rosa R, Piscione F, Iaccarino G, Vosa C, Sorriento D, Piccolo R, Rapacciuolo A, Walsh K, Chiariello M. Myocardial expression of FOXO3a-Atrogin-1 pathway in human heart failure. Eur J Heart Fail. 2010;12(12):1290–1296. doi: 10.1093/eurjhf/hfq102. [DOI] [PubMed] [Google Scholar]
- 60.Holweg CT, Baan CC, Niesters HG, Vantrimpont PJ, Mulder PG, Maat AP, Weimar W, Balk AH. TGF-beta1 gene polymorphisms in patients with end-stage heart failure. J Heart Lung Transpl. 2001;20(9):979–984. doi: 10.1016/S1053-2498(01)00296-0. [DOI] [PubMed] [Google Scholar]
- 61.Tsai EJ, Kass DA. Cyclic GMP signaling in cardiovascular pathophysiology and therapeutics. Pharmacol Ther. 2009;122(3):216–238. doi: 10.1016/j.pharmthera.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci USA. 2013;110(34):13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Communal C, Colucci WS. The control of cardiomyocyte apoptosis via the beta-adrenergic signaling pathways. Arch Mal Coeur Vaiss. 2005;98(3):236–241. [PubMed] [Google Scholar]
- 64.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4(196):ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanchez-Soria P, Camenisch TD. ErbB signaling in cardiac development and disease. Semin Cell Dev Biol. 2010;21(9):929–935. doi: 10.1016/j.semcdb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fritz G, Kaina B. Stress factors affecting expression of O6-methylguanine-DNA methyltransferase mRNA in rat hepatoma cells. Biochem Biophys Acta. 1992;1171(1):35–40. doi: 10.1016/0167-4781(92)90137-o. [DOI] [PubMed] [Google Scholar]
- 68.Siwik DA, Tzortzis JD, Pimental DR, Chang DL, Pagano PJ, Singh K, Sawyer DB, Colucci WS. Inhibition of copper-zinc superoxide dismutase induces cell growth, hypertrophic phenotype, and apoptosis in neonatal rat cardiac myocytes in vitro. Circ Res. 1999;85(2):147–153. doi: 10.1161/01.RES.85.2.147. [DOI] [PubMed] [Google Scholar]
- 69.Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, Hu X, Wang G, Lin Z, Wang S, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112(12):1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hassink RJ, Pasumarthi KB, Nakajima H, Rubart M, Soonpaa MH, de la Riviere AB, Doevendans PA, Field LJ. Cardiomyocyte cell cycle activation improves cardiac function after myocardial infarction. Cardiovasc Res. 2008;78(1):18–25. doi: 10.1093/cvr/cvm101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480(7377):372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jackson T, Allard MF, Sreenan CM, Doss LK, Bishop SP, Swain JL. The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol Cell Biol. 1990;10(7):3709–3716. doi: 10.1128/MCB.10.7.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. RT-qPCR primer sequences used in the study. Table S2. MiRNA associated pathways in the atrial myocardial tissue of patients with CHD after CPB (n=3) compared to before CPB (n=3): KEGG Pathways with predicted interaction enrichments for deregulated miRNAs by microarray (P value < 0.05).
Additional file 2: Figure S1. Unsupervised hierarchical clustering (Euclidian distance, complete linkage) of 3 patients with 6 samples including 3 samples before and 3 samples after CPB. The clustering was done based on the expression of the highest variance miRNAs as determined by RT-qPCR. MiRNAs with high expression are shown in blue and miRNAs with low expression in red. The blue and orange lines indicate the two main clusters of samples.
Additional file 3: Figure S2. Influence of age on the expression of 9 differentially expressed miRNA as determined by RT-qPCR. The relative expression level of 2-ΔΔCt is indicated on the y-axis and the patient age at the time of surgery on the x-axis. The different miRNAs are color coded as indicated. RNAU6B was used as endogenous control for normalization and paired-two-tailed t-tests was used to evaluate differences in expression.
Additional file 4: Figure S3. Functional target network validation of 9 differentially expressed miRNAs and 9 predicted target mRNAs. The comparison was done between atrial myocardial tissue of CHD patients before CBP (n=10) and after CPB (n=10). RNAU6B and GAPDH were used as an endogenous controls for normalization of miRNA and mRNA, respectively. Paired-two-tailed t-tests and ± standard deviation (STDV) were used to evaluate differences in expression.
Additional file 5. 1) Information letter to patients and parents, 2) consent form for anonymous publication, participation & storage of medical data and 3) tissue sample form.
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.


