ABSTRACT
Centralspindlin, a complex of the kinesin-6-family member MKLP1 and MgcRacGAP (also known as Kif23 and Racgap1, respectively), is required for cytokinesis and cell–cell junctions. During anaphase, Centralspindlin accumulates at overlapping central spindle microtubules and directs contractile ring formation by recruiting the GEF Ect2 to the cell equator to activate RhoA. We found that MgcRacGAP localized to the plus ends of equatorial astral microtubules during cytokinesis in Xenopus laevis embryos. How MgcRacGAP is stabilized at microtubule plus ends is unknown. We identified an SxIP motif in X. laevis MgcRacGAP that is conserved with other proteins that bind to EB1 (also known as Mapre1), a microtubule plus-end tracking protein. Mutation of the SxIP motif in MgcRacGAP resulted in loss of MgcRacGAP tracking with EB3 (also known as Mapre3) on growing microtubule plus ends, abnormal astral microtubule organization, redistribution of MgcRacGAP from the contractile ring to the polar cell cortex, and mislocalization of RhoA and its downstream targets, which together contributed to severe cytokinesis defects. Furthermore, mutation of the MgcRacGAP SxIP motif perturbed adherens junctions. We propose that the MgcRacGAP SxIP motif is functionally important both for its role in regulating adherens junction structure during interphase and for regulating Rho GTPase activity during cytokinesis.
KEY WORDS: Centralspindlin, SxIP motif, Cytokinesis, Cell–cell junctions, Microtubules, Xenopus laevis
Highlighted Article: This article demonstrates that Centralspindlin is tethered to microtubule plus ends via an SxIP motif, and disruption of this motif causes defects in cytokinesis and cell–cell junctions.
INTRODUCTION
Cytokinesis is the last step of cell division that partitions the DNA and cellular components into two daughter cells. In animal cells, the position of the division plane is dictated by signals from the central spindle, which are thought to be delivered to the equatorial cortex by astral microtubules (MTs) (Rappaport, 1961). A key target of these signals is the small GTPase RhoA, which becomes activated in a focused zone at the equatorial cortex and is essential for cytokinesis (Bement et al., 2005; Yüce et al., 2005). At anaphase, Centralspindlin, composed of the kinesin-6-family member MKLP1 (also known as Kif23 in humans) and the GAP protein MgcRacGAP (Mgc; also known as Racgap1 in humans), accumulates on the antiparallel overlapping MTs of the central spindle (Hirose et al., 2001; Jantsch-Plunger et al., 2000; Mishima et al., 2002). Mgc recruits the RhoA GEF Ect2 (Mishima et al., 2004; Somers and Saint, 2003; Su et al., 2011) and, together, Centralspindlin and Ect2 localize to the equatorial cortex to regulate active RhoA (Bement et al., 2006; Miller and Bement, 2009). Mgc and Ect2 have also been shown to regulate RhoA-mediated actomyosin contractility at adherens junctions (Breznau et al., 2015; Ratheesh et al., 2012). Interestingly, the localization of Mgc and Ect2 at both locations is thought to be MT dependent, mediated by the motor activity of MKLP1 (Douglas et al., 2010; Hutterer et al., 2009; Ratheesh et al., 2012).
By live imaging in gastrula-stage Xenopus laevis embryos, we found that Mgc also localizes at the plus ends of non-overlapping MTs at the equatorial cell cortex. These findings are consistent with the reported localization of Centralspindlin at astral MTs in fixed HeLa cells or live Drosophila S2 cells (Nishimura and Yonemura, 2006; Vale et al., 2009). Equatorial astral MTs are thought to be important for delivering the cleavage furrow-stimulating signal and may be uniquely stabilized, allowing them to fulfill this role (Canman et al., 2003; Foe and von Dassow, 2008; Shannon et al., 2005). Despite their importance, the mechanism that stabilizes equatorial astral MTs at the cortex is not understood. Furthermore, how Mgc is localized at MT plus ends at the equatorial cortex and cell–cell junctions is unknown.
End-binding protein 1 (EB1) and its family members (EB2, EB3) (also known as Mapre1, Mapre2 and Mapre 3 in humans) are core components of the network of proteins that bind to and track MT plus ends (+TIPs). EB1 can autonomously recognize and track growing MT ends, and acts as an adaptor to recruit and bind other +TIP proteins (Kumar and Wittmann, 2012; Montenegro Gouveia et al., 2010). One way that EB1 recruits other proteins to MT plus ends is by binding to SxIP motifs (where x indicates any amino acid), which are found in a diverse group of proteins (Honnappa et al., 2009; Jiang et al., 2012). Binding is dependent upon the SxIP motif and electrostatic interactions between positively charged residues that flank the SxIP motif and negatively charged residues on EB1 (Honnappa et al., 2009). +TIP proteins that bind to MTs through EB1 can anchor MTs to specific cellular structures and control MT dynamics, including those during cytokinesis (Kumar and Wittmann, 2012; Mimori-Kiyosue et al., 2005, 2006; Strickland et al., 2005). We identified a putative SxIP motif in Mgc and propose that it is functionally important for the localization of Mgc to the plus ends of equatorial astral MTs during cytokinesis and for its MT-dependent localization to cell–cell junctions. Here, we test whether the Mgc SxIP motif is required to tether Mgc to MT plus ends and whether this is functionally important for proper regulation of Rho GTPase activity, cytokinesis and cell–cell junction structure.
RESULTS
MgcRacGAP localizes to microtubule plus ends at the equatorial cortex as cytokinesis initiates
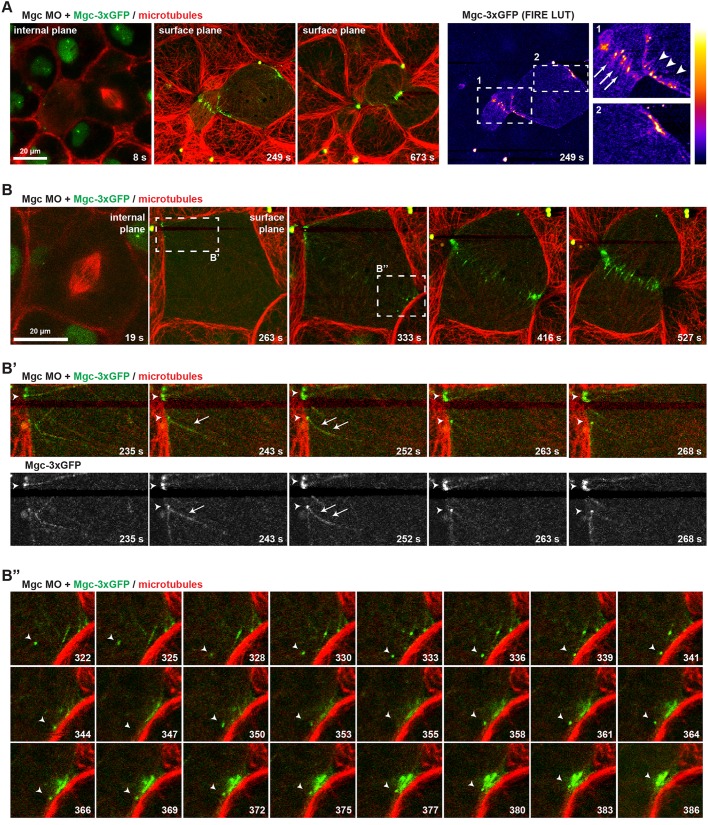
To test how Mgc interacts with MTs that extend toward the equatorial cell cortex during cytokinesis, we examined the localization of Mgc (Mgc–3×GFP) and of an mCherry (mChe)-tagged probe for MTs (2×mChe–EMTB) in dividing gastrula-stage X. laevis embryos [Nieuwkoop and Faber (NF) stage 10-11] by live confocal microscopy. Endogenous Mgc was knocked down with a previously characterized morpholino oligonucleotide (MO), which effectively depletes Mgc protein levels and causes strong cytokinesis defects that can be rescued by expression of WT Mgc (MgcWT) mRNA (Breznau et al., 2015; Miller and Bement, 2009). MO-resistant Mgc–3×GFP was expressed at near-endogenous levels and localized at the overlapping anti-parallel MTs of the central spindle (Fig. 1A; Fig. S1) as expected. Just before cleavage furrow ingression initiated, Mgc–3×GFP localized in discrete puncta at the equatorial cortex, then became increasingly enriched at the ingressing cleavage furrow (Fig. 1A; Movie 1). Mgc–3×GFP decorated individual equatorial astral MTs (Fig. 1B,B″; Movie 2) and accumulated in strong puncta at the plus ends of MTs, which appeared to make stable contacts with the cell cortex (Fig. 1B′). Additionally, some Mgc–3×GFP puncta exhibited directed motility, presumably along astral MTs, as Mgc puncta coalesced at the cleavage furrow during early cytokinesis (Fig. 1B″). These results indicate that in addition to its important role in bundling central spindle MTs (Green et al., 2012), Mgc also localizes to MT plus ends at the equatorial cortex.
Fig. 1.
MgcRacGAP localizes to microtubule plus ends at the equatorial cortex as cytokinesis initiates. (A) Still images from a single z-plane time-lapse movie of a X. laevis embryo co-injected with Mgc MO+Mgc–3×GFP and a probe for MTs (2×mChe–EMTB). Mgc–3×GFP (green) localizes in the nucleus of interphase cells, at overlapping central spindle MTs, as well as at individual MTs at the equatorial cortex prior to furrowing. A FIRE look-up table (LUT) plugin was applied to the Mgc–3×GFP channel to highlight Mgc localization, and enlarged regions are shown on the right (arrows, overlapping central spindle MTs; arrowheads, individual MTs at equatorial cortex). (B) Still images from a single z-plane time-lapse movie of a X. laevis embryo co-injected with Mgc MO+Mgc–3×GFP and 2×mChe–EMTB. The dashed boxes in B indicate regions that are enlarged in B′ and B″. The frames in B′ show Mgc–3×GFP decorating equatorial astral MTs (arrow) and Mgc–3×GFP puncta at MT plus ends (arrowheads). The frames in B″ show directed movement of an Mgc–3×GFP puncta (arrowheads), apparently along astral MTs, as Mgc–3×GFP forms clusters at the equatorial cell cortex.
MgcRacGAP tracks with EB3 at growing MT plus ends
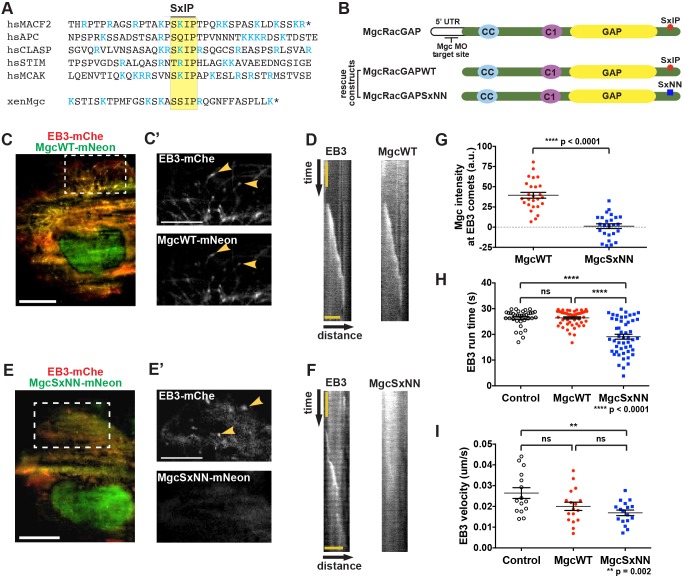
To determine the mechanism by which Mgc associates with MT plus ends, we hypothesized that it might interact with a +TIP. The EB homology domain of EB1 has a hydrophobic cleft that binds other +TIP proteins containing a conserved SxIP motif (Honnappa et al., 2009) (Fig. 2A). We analyzed the amino acid sequence of X. laevis Mgc and identified a putative SxIP motif in the C-terminus of Mgc, as well as other conserved sequence features (Jiang et al., 2012), including flanking basic residues and an arginine residue at the +1 position (Fig. 2A). In order to test whether the putative SxIP motif in Mgc is responsible for its stability on astral MT plus ends (Fig. 1A,B), we mutated the isoleucine and proline residues to asparagine (MgcSxNN) (Fig. 2B). This mutation has been shown to abrogate binding of SxIP proteins to the EB homology domain (Honnappa et al., 2009).
Fig. 2.
MgcRacGAP tracks with EB3 at growing microtubule plus ends. (A) Amino acid sequence alignment surrounding the SxIP motif (yellow box) of five known EB1-binding proteins and the SxIP motif in the C-terminus of X. laevis Mgc. Basic arginine (R) and lysine (K) residues are shown in blue. Stop codons are indicated by *. (B) Mgc domain diagram. Endogenous Mgc is knocked down with MOs that target the 5′UTR; MgcWT and MgcSxNN constructs do not contain the endogenous 5′ UTR sequence, so they are not targeted by the MOs. The location of the putative SxIP motif in Mgc (red circle) and the SxNN mutation (blue square) are indicated. (C) TIRF microscopy image of a Cos-7 cell transfected with EB3–mChe (red) and MgcWT–mNeon (green). Scale bar: 20 µm. (C′) An enlarged view of the boxed region in C showing that EB3–mChe and MgcWT–mNeon accumulate on the plus ends of microtubules (yellow arrowheads). Scale bar: 5 µm. (D) Representative kymographs following an EB3–mChe comet in cells co-expressing EB3–mChe and MgcWT–mNeon. Scale bar: 4.3 µm (distance), 1 sec (time). (E) TIRF microscopy image of a Cos-7 cell expressing EB3–mChe (red) and MgcSxNN–mNeon (green). Scale bar: 20 µm. (E′) An enlarged view of the boxed region in E showing EB3–mChe comets (yellow arrowheads) and the strongly reduced corresponding signal in the MgcSxNN–mNeon channel. Scale bar: 5 µm. (F) Representative kymographs following an EB3–mChe comet in cells co-expressing EB3–mChe and MgcSxNN–mNeon. The total time represented is 6.5 s; scale bar: 4.3 µm (distance). (G–I) Quantification of (G) Mgc–mNeon intensity at EB3 comets. MgcWT=26 kymographs, 9 cells; MgcSxNN=25 kymographs, 8 cells. (H) EB3–mChe run duration on MT plus ends. Control=35 EB3 comets, 7 cells; MgcWT=50 EB3 comets, 10 cells; MgcSxNN=50 EB3 comets, 10 cells. (I) Velocity of EB3–mChe comets. Control=15 EB3 comets, 7 cells; MgcWT=17 EB3 comets, 9 cells; MgcSxNN=17 EB3 comets, 9 cells. Mean±s.e.m. (unpaired Student's t-test). All n values represent cumulative values from three independent experiments. ns, not significant.
To test whether the Mgc SxIP motif plays a role in its ability to track the plus ends of growing MTs, we co-expressed mNeon-Green (mNeon)-tagged X. laevis MgcWT or MgcSxNN with human EB3–mChe in flat Cos-7 cells, which allow for imaging of individual MT dynamics via total internal reflection fluorescence (TIRF) microscopy. MgcWT–mNeon colocalized with EB3–mChe comets at growing MT plus ends in interphase Cos-7 cells (Fig. 2C,C′; Movie 3) and tracked along growing MT plus ends over time (Fig. 2D). In contrast, MgcSxNN–mNeon showed a more diffuse cellular distribution and reduced ability to track with EB3–mChe comets (Fig. 2E,F; Movie 4). To confirm this change in distribution, we quantified the intensity of MgcWT and MgcSxNN at EB3 comets and found that MgcSxNN intensity was significantly reduced compared to that of MgcWT (Fig. 2G), indicating that the Mgc SxIP motif may mediate binding to EB proteins, allowing Mgc to associate with growing MT plus ends. Importantly, MgcWT–mNeon and MgcSxNN–mNeon were expressed at similar levels, and localized to nuclei and midbodies, as expected (Fig. S2A). EB3–mChe expression levels were also comparable among the quantified groups (Fig. S2B,C). We note that although the localization of Mgc at MT plus ends could simply be an artifact of overexpressing X. laevis Mgc in Cos-7 cells, the finding that mutating the SxIP motif strongly reduces MT tracking strengthens the argument that this is a specific interaction. Surprisingly, we found that in cells expressing MgcSxNN–mNeon, the duration of EB3–mChe comets (EB3 run time) was significantly shorter compared to that in cells expressing MgcWT–mNeon (Fig. 2H); however, the EB3–mChe velocity did not differ significantly (Fig. 2I). These data indicate that although the tracking of MgcSxNN with EB3 is strongly reduced, it still has a functional effect on the ability of EB3 to track along MTs (see Discussion).
The MgcRacGAP SxIP motif is necessary for successful cytokinesis and proper astral microtubule structure
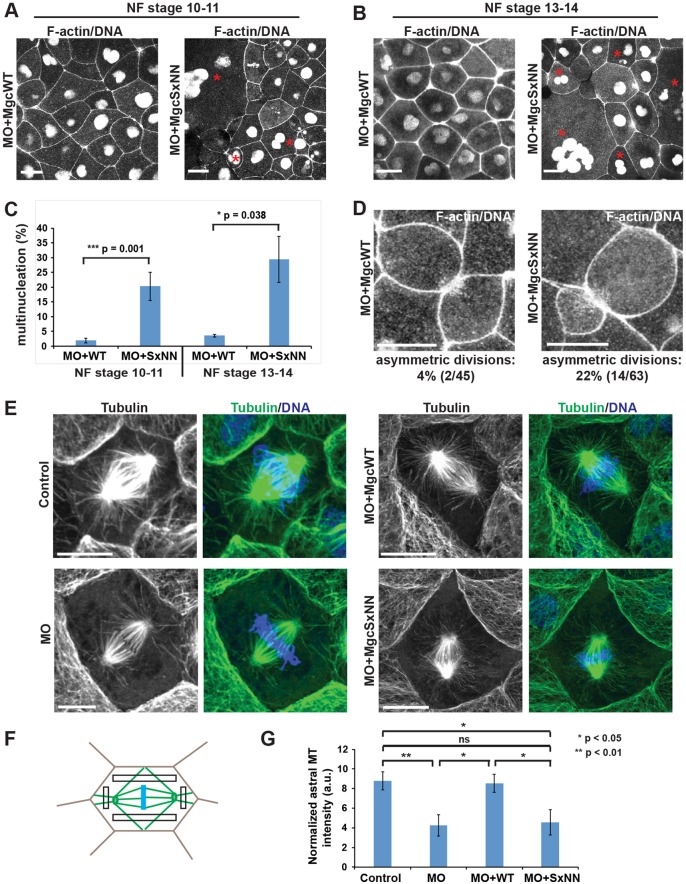
We next sought to determine whether mutating the Mgc SxIP motif has functional consequences on cytokinesis. Previously, we showed that the Mgc-targeting MO caused multinucleation in ∼30% of cells at NF stage 10-11, which can be rescued to near-control multinucleation levels by expressing MO-resistant MgcWT mRNA (Miller and Bement, 2009). Here, we injected MO+MgcWT or MO+MgcSxNN along with probes for F-actin (mChe–UtrCH) and DNA (mChe–H2B) and counted failed cytokinesis events (i.e. multinucleate cells) in images of live gastrula-stage X. laevis embryos. MO+MgcSxNN-expressing embryos exhibited high levels of multinucleation at NF stage 10-11 (20.2±4.7%; mean±s.e.m.; Fig. 3A,C), and further increased multinucleation at NF stage 13-14 (29.4±7.8%; Fig. 3B,C), compared to MO+MgcWT-expressing embryos (1.9±0.8% for NF stage 10-11 and 3.6±0.5% for NF stage 13-14). Similar results were obtained when embryos were fixed and stained with DAPI to quantify multinucleation (Fig. S3A). Moreover, furrow regression could be observed in some MO+MgcSxNN cells by live imaging (Fig. S3B, Movie 5).
Fig. 3.
The MgcRacGAP SxIP motif is necessary for successful cytokinesis and proper astral microtubule structure. (A) Still images from time-lapse movies of X. laevis embryos at NF stage 10-11. Embryos were co-injected with Mgc MO and either MgcWT or MgcSxNN along with a probe for DNA (mChe–H2B) and a probe for F-actin (mChe–UtrCH). Red asterisks indicate multinucleate cells. (B) Still images from time-lapse movies of NF stage 13-14 X. laevis embryos injected as described in A. Red asterisks indicate multinucleate cells. (C) Quantification of the percent of multinucleate cells in MO+MgcWT or MO+MgcSxNN embryos at NF stage 10-11 or NF stage 13-14. Stage 10-11 – MO+MgcWT=230 cells, 13 embryos, 5 experiments; MO+MgcSxNN=203 cells, 12 embryos, 5 experiments; stage 13-14 – MO+MgcWT=109 cells, 3 embryos, 1 experiment; MO+MgcSxNN=60 cells, 4 embryos, 1 experiment. (D) Still images from time-lapse movies of NF stage 10-11 X. laevis embryos injected as described in A showing symmetric (MO+MgcWT) and asymmetric (MO+MgcSxNN) divisions. Quantification of the percent of asymmetric cell divisions in MO+MgcWT or MO+MgcSxNN embryos is indicated below each panel. (E) Images of fixed NF stage 10-11 X. laevis embryos that had been co-injected with a lineage tracer (mChe–membrane, not shown) along with MO alone, MO+MgcWT, or MO+MgcSxNN, then fixed and stained with anti-α-tubulin antibodies to reveal MTs, and DAPI to label the DNA. (F) Schematic of the astral MT intensity quantification strategy. Intensity measurements were taken from the boxed regions and averaged for each cell. (G) Quantification of astral MT intensity, normalized to the cytosol background. Note that in order to observe astral MTs, the spindle MTs sometimes had to be saturated, but the MTs used for astral MT quantification were not saturated. Control=7 cells, 6 embryos; MO=7 cells, 7 embryos; MO+MgcWT=7 cells, 6 embryos; MO+MgcSxNN=8 cells, 6 embryos. Mean±s.e.m. (unpaired Student's t-test). Scale bars: 25 µm (A,B); 10 µm (D,E). All n values represent cumulative values from three independent experiments, unless otherwise indicated. Maximum projection images are shown and were used for quantification.
One reason that the MO+MgcSxNN cells might fail cytokinesis is if their mitotic spindle MT structure is defective. Indeed, we observed that cells in MO+MgcSxNN embryos often underwent asymmetric cell divisions, where the cleavage furrow formed to one side of the central axis of the cell [22% (14/63) in MO+MgcSxNN vs 4% (2/45) in MO+MgcWT], suggesting that spindle structure or positioning might be defective (Fig. 3D; Movie 6).
We reasoned that localization of Mgc to growing MT plus ends might facilitate mitotic spindle stability by connecting the spindle to the cortex via astral MTs, and that defective connections in MO+MgcSxNN-expressing cells would result in asymmetric divisions. To examine the morphology of astral MTs during metaphase, we stained for MTs in MO+MgcWT or MO+MgcSxNN embryos (Fig. 3E). We compared astral MT distribution in early anaphase cells by quantifying α-tubulin intensity (Fig. 3F). Control cells exhibited strong astral MT density connecting the spindle to the cortex (Fig. 3E-G). In contrast, cells that had been injected with the Mgc-targeting MO alone showed a significant decrease in astral MT density (Fig. 3E-G). This phenotype was rescued in MO+MgcWT cells but not in MO+MgcSxNN cells (Fig. 3E-G). Notably, MgcSxNN showed no ability to rescue the astral MT density defects caused by the Mgc-targeting MO. However, MgcSxNN appears to partially rescue the cytokinesis defect, as the ∼20% of MO+MgcSxNN-expressing cells exhibiting multinucleation observed here is not as severe as the ∼30% of MO-expressing cells exhibiting multinucleation observed previously (Miller and Bement, 2009). This suggests that astral MT structure and dynamics may be more sensitive to disruption of the Mgc SxIP motif than cytokinesis as a whole is. Taken together, these results demonstrate that the Mgc SxIP motif is necessary for proper astral MT structure and for successful cytokinesis, suggesting that, similar to other +TIP proteins (Mimori-Kiyosue et al., 2006), Mgc may facilitate local stabilization of astral MTs.
Mutation of the MgcRacGAP SxIP motif disrupts equatorial accumulation of the Centralspindlin complex
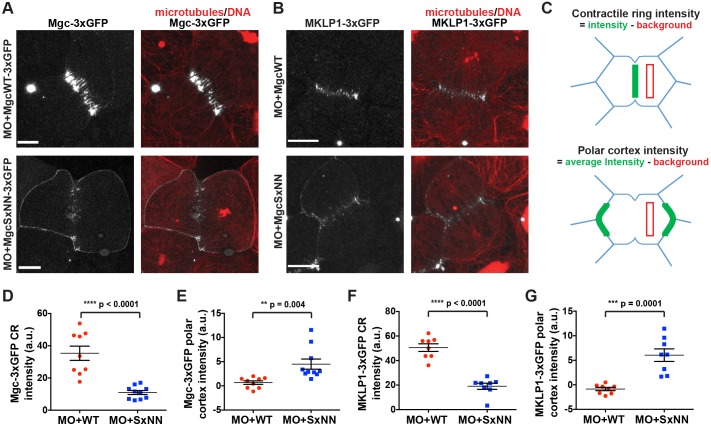
Because equatorial astral MTs are important for regulating the formation of the cleavage furrow by providing tracks for cortical Centralspindlin delivery and local RhoA activation, we next investigated whether accumulation of Centralspindlin at the equatorial cortex was affected when the Mgc SxIP motif was mutated. We injected MO+MgcWT–3×GFP or MO+MgcSxNN–3×GFP along with probes for MTs (2×mChe–EMTB) and DNA (mChe–H2B) and performed live imaging. MgcWT–3×GFP localized to the equatorial cortex of dividing cells and became increasingly enriched at the contractile ring during furrow ingression (Fig. 4A; Movie 7). In contrast, MgcSxNN–3×GFP was not strongly enriched at the contractile ring during cytokinesis, but was largely redistributed around the cell perimeter (Fig. 4A; Movie 7). Note that we still observe a population of MgcSxNN at the contractile ring; this is not surprising because the MgcSxNN mutant should still be able to interact with MKLP1 and bundle overlapping central spindle MTs. Quantification of Mgc signal intensity at 25% ingression (Fig. 4C) showed a significant decrease in MgcSxNN–3×GFP contractile ring intensity (Fig. 4D) and a corresponding increase in MgcSxNN–3×GFP intensity at the polar cortex (Fig. 4E). These results demonstrate that the Mgc SxIP motif is important for its proper localization during cytokinesis.
Fig. 4.
Mutation of the MgcRacGAP SxIP motif disrupts equatorial accumulation of the Centralspindlin complex. (A) Still images from live movies of NF stage 10-11 X. laevis embryos that had been co-injected with MO+MgcWT–3×GFP or MO+MgcSxNN–3×GFP (gray), a probe for MTs (2×mChe–EMTB, red) and a probe for DNA (mChe–H2B, red). (B) Still images from live movies of NF stage 10-11 X. laevis embryos that had been co-injected with MO+MgcWT or MO+MgcSxNN, along with MKLP1–3×GFP (gray), 2×mChe–EMTB (red) and mChe–H2B (red). (C) Schematic of contractile ring (CR) and polar cortex intensity quantification strategy. Quantification was performed at 25% cleavage furrow ingression. (D) Quantification of the Mgc–3×GFP intensity at the contractile ring. MO+MgcWT–3×GFP, 9 cells; MO+MgcSxNN–3×GFP, 10 cells. (E) Quantification of the Mgc–3×GFP intensity along the polar cortex of dividing cells. MO+MgcWT, 9 cells; MO+MgcSxNN, 10 cells. (F) Quantification of the MKLP1–3×GFP intensity at the contractile ring. MO+MgcWT, 8 cells; MO+MgcSxNN, 8 cells. (G) Quantification of the MKLP1–3×GFP intensity along the polar cortex of dividing cells. MO+MgcWT, 8 cells; MO+MgcSxNN, 8 cells. Scale bars: 10 µm. Mean±s.e.m. (unpaired Student's t-test). All n values represent cumulative values from three or more independent experiments. Maximum projection images are shown and were used for quantification.
We next examined the localization of MKLP1 when the Mgc SxIP motif was mutated. We co-injected MO+MgcWT or MO+MgcSxNN along with MKLP1–3×GFP, and probes for MTs and DNA. MO+MgcWT cells showed characteristic MKLP1 accumulation at the equatorial cortex and contractile ring (Fig. 4B,F,G). In contrast, MO+MgcSxNN cells showed reduced MKLP1–3×GFP accumulation at the contractile ring and redistribution to the polar cortex (Fig. 4B,F,G). Taken together, these results indicate that the Mgc SxIP motif is important for proper accumulation of both components of the Centralspindlin complex at the equatorial cortex and contractile ring.
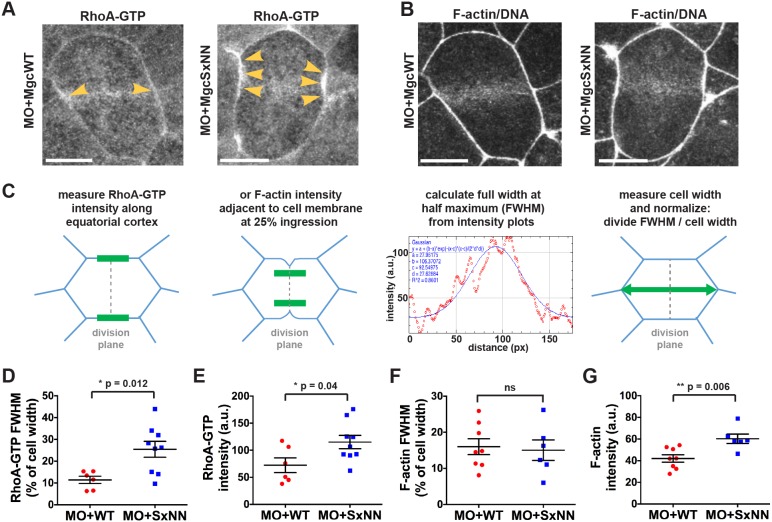
The MgcRacGAP SxIP motif contributes to limiting and focusing RhoA-GTP and F-actin at the cell equator
If Centralspindlin is not properly localized at the equatorial cortex, activation and focusing of the Rho activity zone may be perturbed (Breznau et al., 2015; Miller and Bement, 2009), which could contribute to the increased cell division failure observed in MO+MgcSxNN cells. In order to determine whether the MgcSxNN mutant affects RhoA-GTP accumulation during cytokinesis, we performed live imaging of embryos that had been injected with MO+MgcWT or MO+MgcSxNN along with a probe for active RhoA (GFP–rGBD). In MO+MgcWT-expressing cells, active RhoA accumulated in a focused band at the equatorial cortex following anaphase onset and remained enriched at the furrow throughout cytokinesis (Fig. 5A; Movie 8). In contrast, cells in MO+MgcSxNN embryos exhibited a broader and more intense accumulation of active RhoA at anaphase onset (Fig. 5A; Movie 8). Quantifying the accumulation of active RhoA at anaphase onset using full width at half maximum (FWHM) analysis (Fig. 5C) revealed a significant increase in the breadth and intensity of active RhoA at the equatorial cortex in MO+MgcSxNN- compared to MO+MgcWT-expressing cells (Fig. 5D,E). These results suggest that the Mgc SxIP motif regulates the proper localization of Centralspindlin and thus the ability of Mgc to focus RhoA-GTP at the equatorial cortex during cytokinesis. We propose that when the Mgc SxIP motif is disrupted, causing the redistribution of Mgc away from the equatorial cortex, the ability of Mgc to promote RhoA GTPase flux – locally downregulating and focusing active RhoA at the cytokinetic plane – is diminished, leading to the observed increase in active RhoA.
Fig. 5.
The MgcRacGAP SxIP motif contributes to limiting and focusing RhoA-GTP and F-actin at the cell equator. (A) Still images from time-lapse movies of NF stage 10-11 X. laevis embryos that had been co-injected with MO+MgcWT or MO+MgcSxNN along with a probe for active RhoA (GFP–rGBD), a probe for F-actin (mChe–UtrCH) and a probe for DNA (mChe–H2B). Yellow arrowheads highlight RhoA-GTP accumulation along the equatorial cell cortex during early anaphase. (B) Same cells as in A showing the F-actin accumulation during early anaphase. (C) Schematic of the analysis approach for fitting a Gaussian curve to the intensity plot data to determine FWHM (breadth). The intensity of RhoA-GTP at the equatorial cortex just after anaphase onset and the intensity of F-actin accumulation during early anaphase (at 25% ingression), were measured. The FWHM is graphed as a percentage of cell width. (D) FWHM quantification of the breadth of RhoA-GTP accumulation at the equatorial cortex. MO+MgcWT=6 cells; MO+MgcSxNN=9 cells. (E) RhoA-GTP intensity quantification at the equatorial cortex. MO+MgcWT=6 cells; MO+MgcSxNN=9 cells. (F) FWHM quantification of F-actin contractile ring breadth. MO+MgcWT=8 cells; MO+MgcSxNN=6 cells. (G) F-actin contractile ring intensity quantification. MO+MgcWT=8 cells; MO+MgcSxNN=6 cells. Scale bars: 10 µm. Mean±s.e.m. (unpaired Student's t-test). ns, not significant. All n values represent cumulative values from three or more independent experiments. Maximum projection images are shown and were used for quantification.
To better understand why disrupting the Mgc SxIP motif causes cytokinesis failure, we examined proteins that are activated downstream of RhoA-GTP, starting with F-actin, which can be polymerized via the RhoA-GTP effector formin. Embryos were injected with MO+MgcWT or MO+MgcSxNN along with probes for F-actin (mChe–UtrCH) and DNA (mChe–H2B), and movies of dividing cells were analyzed for defects in F-actin accumulation at the equatorial cortex of dividing cells (Fig. 5B) (Bement et al., 2005; Breznau et al., 2015; Miller and Bement, 2009). Analysis of F-actin intensity plots at an equatorial region adjacent to the cleavage furrow at 25% ingression did not reveal a change in the breadth of F-actin accumulation (Fig. 5F) but did show a significant increase in the intensity of F-actin (Fig. 5G) in MO+MgcSxNN- compared to MO+MgcWT-expressing cells.
Additionally, we examined the effects on Myosin II, which can be activated via phosphorylation by the RhoA-GTP effector ROCK. We overexpressed MgcWT or MgcSxNN and then performed immunofluorescence staining for phosphorylated Myosin II (p-Myo). In cells expressing MgcWT, p-Myo localized as a focused band at the cleavage furrow with a weak signal detected at cell–cell junctions, whereas in cells expressing MgcSxNN, p-Myo accumulated broadly at the contractile ring as well as at cell–cell junctions around the perimeter of the dividing cell (Fig. S3C). When quantified, we detected a significant decrease in the ratio of contractile ring:junction intensity in MgcSxNN cells versus that in MgcWT-expressing or control cells (Fig. S3C). This abnormal accumulation of the key contractile ring components, F-actin and Myosin II, is likely to drive the cytokinesis defects observed when the Mgc SxIP motif is mutated. This is consistent with previous reports demonstrating cytokinesis failure due to an over-robust RhoA activity zone and/or actomyosin contractile ring (DeWard and Alberts, 2009; Manukyan et al., 2015; Miller and Bement, 2009; Reyes et al., 2014). Collectively, these data provide in vivo evidence that the Mgc SxIP motif plays a functional role in regulating cytokinesis by mediating the precise localization of Centralspindlin in order for proper localized RhoA activation and accumulation of downstream targets.
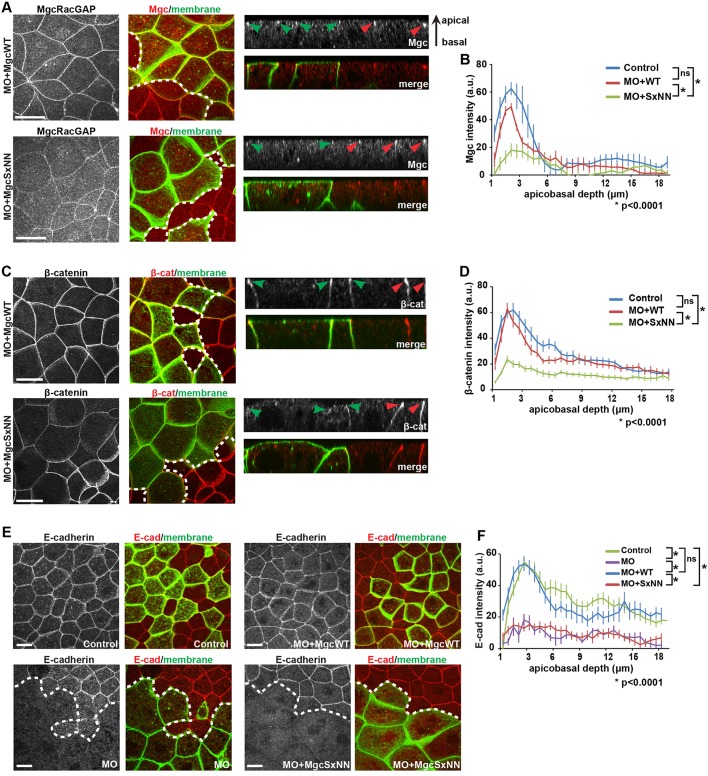
The MgcRacGAP SxIP motif is necessary for proper adherens junction structure
Next, we tested whether mutating the Mgc SxIP motif affects the population of Mgc that localizes to and regulates cell–cell junctions in interphase epithelial cells (Breznau et al., 2015; Guillemot et al., 2014; Ratheesh et al., 2012). Constructs were injected so as to generate mosaic embryos, such that cells expressing MO+MgcWT or MO+MgcSxNN could be compared with neighboring control cells. Then, we performed immunofluorescence staining for Mgc at NF stage 10-11. In MO+MgcWT-injected embryos, Mgc was enriched at apical cell–cell junctions in both MO+MgcWT-injected cells and internal control cells (Fig. 6A,B). In contrast, MO+MgcSxNN-injected cells showed a significant reduction in apical junction localization of Mgc compared with that in internal control cells or MO+MgcWT cells (Fig. 6A,B). MTs are known to regulate the junctional accumulation of Centralspindlin and its binding partner Ect2 (Ratheesh et al., 2012). Therefore, the SxIP motif may be necessary for Mgc tethering and/or stability on MT plus ends for proper delivery of Mgc to cell–cell junctions.
Fig. 6.
The MgcRacGAP SxIP motif is necessary for proper adherens junction structure. (A,C,E) Fixed maximum projection en face and side-view images of mosaic NF stage 10-11 X. laevis embryos. The mosaic injection strategy allows for approximately half of the embryo to express the indicated knockdown and rescue constructs along with GFP-membrane or mChe-membrane as a lineage tracer, while the other half of the embryo serves as an internal control and is not injected. The dotted white line in the merged en face image represents the boundary between the internal control and injected region of mosaic embryos. Antibodies against GFP and one of Mgc (A), β-catenin (C; β-cat) or E-cadherin (E; E-cad) were applied to the embryos after fixation. Side views show the localization of the indicated protein along the apicobasal axis of cell–cell junctions (red arrowheads, internal control regions; green arrowheads, injected regions). (A) Mgc (red) and GFP-membrane (green). (B) Quantification of the normalized Mgc intensity at bicellular junctions from the apical to basal surfaces. Control, 13 cells; MO+MgcWT, 14 cells; MO+MgcSxNN, 19 cells. (C) β-catenin (red) and GFP-membrane (green). (D) Quantification of the normalized β-catenin intensity along the bicellular junctions from the apical to basal surfaces. Control, 17 cells; MO+MgcWT, 16 cells; MO+MgcSxNN, 19 cells. (E) E-cadherin (pseudo-colored red) and mChe-membrane (pseudo-colored green). (F) Quantification of the normalized E-cadherin intensity along the bicellular junctions from the apical to basal surfaces. Control, 19 cells; MO, 7 cells; MO+MgcWT, 15 cells; MO+MgcSxNN, 19 cells. Error bars represent s.e.m. (unpaired Student's t-test). ns, not significant. Scale bars: 20 µm. All n values represent cumulative values from three independent experiments.
Because Mgc contributes to cell–cell junction structure and maintenance, and the GAP activity of Mgc is required for the proper localization of the adherens junction protein β-catenin (Breznau et al., 2015; Guillemot et al., 2014; Ratheesh et al., 2012), we next tested whether β-catenin was properly localized at cell–cell junctions when the Mgc SxIP motif was disrupted. In MO+MgcWT-injected cells, β-catenin was focused at apical junctions similar to controls (Fig. 6C,D). In contrast, β-catenin junctional localization was significantly reduced in MO+MgcSxNN-injected cells compared with control cells or MO+MgcWT cells (Fig. 6C,D), and the cells often appeared apically domed, a phenotype associated with reduced cell–cell junction integrity (Fanning et al., 2012; Yonemura et al., 2010). To further demonstrate that disrupting the Mgc SxIP motif results in defective adherens junctions, we examined the localization of E-cadherin. E-cadherin localization was significantly reduced in MO-injected cells compared with control cells (Fig. 6E,F), consistent with previous results (Breznau et al., 2015). This defect was rescued in MO+MgcWT embryos but was not rescued in MO+MgcSxNN embryos (Fig. 6E,F). Quantification demonstrated that both MO and MO+MgcSxNN-expressing cells exhibit a significant decrease in apical E-cadherin intensity compared with either control or MO+MgcWT embryos (Fig. 6F). Taken together, our data provide strong evidence that the Mgc SxIP motif plays an important role in regulating the localization of Mgc to adherens junctions, as well as the localization of key adherens junction structural components.
DISCUSSION
Centralspindlin localizes to the central spindle at anaphase and is critical for recruitment of the GEF Ect2, formation of the RhoA activity zone and proper completion of cytokinesis. However, mechanisms that regulate Centralspindlin localization at the equatorial cortex – the very place where Centralspindlin and Ect2 locally regulate RhoA activity – are largely unknown. Our study builds on current models by showing that once delivered to MT plus ends via its kinesin-6 MKLP1 partner, X. laevis Mgc can use its SxIP motif to bind to EB proteins, thereby tethering Centralspindlin to growing MT plus ends where it functionally regulates both cytokinesis and cell–cell junction integrity (Fig. 7). We propose that the Mgc–EB protein interaction may influence MT stability and serve to link MT plus ends with the cell membrane either directly (Lekomtsev et al., 2012) or via Ect2 (Su et al., 2011).
Fig. 7.
Model of MgcRacGAP tethering to MT plus ends. We propose that MKLP1 transports Centralspindlin toward MT plus ends, and the Mgc SxIP motif mediates tethering of Centralspindlin to MT plus ends via EB proteins in order to facilitate proper accumulation of Mgc at equatorial astral MTs during cytokinesis and at epithelial cell–cell junctions.
During cell division, astral MTs targeting the equatorial cortex are thought to be important for delivering the cleavage-furrow-stimulating signal and may be uniquely stabilized by an unknown mechanism, allowing them to fulfill this role (Canman et al., 2003; Foe and von Dassow, 2008; Rappaport, 1961; Shannon et al., 2005). Centralspindlin is critical for delivering the furrow-inducing signal because it recruits, focuses and may activate the GEF Ect2 (Loria et al., 2012; Somers and Saint, 2003; Su et al., 2011; Yüce et al., 2005). Ect2 then stimulates RhoA activity, which in turn activates formation of the contractile ring. Our data suggest that in addition to MKLP1-mediated transport to the cortex, the SxIP-based interaction of Mgc with EB proteins serves to tether Centralspindlin to the plus ends of individual equatorial astral MTs (Fig. 7). The Mgc–EB protein interaction is critical for focusing the RhoA activity zone through Mgc-promoted RhoA GTPase flux and for proper formation of the contractile ring. The possibility that redundant mechanisms may exist for localization of Mgc and Ect2 to the equatorial cell cortex is supported by recent evidence from a cell-free reconstituted system showing that RhoA-GTP can still be recruited to overlapping MTs in the absence of MKLP1 (Nguyen et al., 2014).
Proteins recruited to MT plus ends can anchor MTs to specific cellular structures and control MT dynamics (Kumar and Wittmann, 2012; Mimori-Kiyosue et al., 2005, 2006; Strickland et al., 2005). In this respect, tethering of Centralspindlin to the plus ends of equatorial astral microtubules enables it to play a role in regulating MT stability and MT anchoring at the cortex during cytokinesis. Indeed, we found that mutation of the Mgc SxIP motif results in decreased astral microtubule density and promotion of cytokinesis defects. Centralspindlin could anchor astral MTs to the membrane in two ways: through the Mgc C1 membrane-binding domain (Lekomtsev et al., 2012) or through the Mgc interaction with Ect2, which in turn interacts with the plasma membrane through its PH domain and a polybasic cluster (Su et al., 2011).
The effects of mutating the Mgc SxIP motif were not restricted to dividing cells. Recent work has shown that Mgc also regulates cell–cell junctions in interphase epithelial cells (Breznau et al., 2015; Guillemot et al., 2014; Ratheesh et al., 2012). We observed a significant decrease in the ability of the MgcSxNN mutant to track along growing MT plus ends with EB3 when compared to MgcWT in interphase Cos-7 cells. In interphase X. laevis epithelial cells, MgcWT localized to apical cell–cell junctions, whereas MgcSxNN showed reduced junctional localization. This suggests that the Mgc interaction with EB proteins at the plus ends of MTs is required for the delivery of Mgc to cell–cell junctions. Indeed, this is consistent with previous work in cultured epithelial cells showing that junctional localization of both Centralspindlin and its binding partner Ect2 are MT dependent (Ratheesh et al., 2012). Furthermore, we observed a significant decrease in β-catenin and E-cadherin at apical junctions in MgcSxNN-expressing cells, demonstrating that Mgc tethering to MT plus ends is required for proper adherens junction structure in interphase cells.
Although the Mgc SxIP motif is conserved in both alloalleles for X. laevis as well as in X. tropicalis, it is only partially conserved in higher vertebrates (Fig. S4). The SxIP consensus motif is defined as (x1-x2-[ST]-x3-[IL]-P-x4-x5-x6), where x indicates any amino acid, at least one of x1–x4 is R or K, and none of x1–x6 is D or E (Jiang et al., 2012). The X. laevis and X. tropicalis Mgc sequences fit this consensus perfectly. In higher vertebrates, the corresponding TNLG sequence fits the [ST]-X3-[IL] consensus and contains appropriate sequence conservation in the flanking regions; however, the absence of a proline residue makes it unlikely that this is a functional EB protein-binding SxIP motif. It has been previously noted that SxIP motifs can be easily lost or acquired during evolution (Akhmanova and Steinmetz, 2010). It seems likely that in higher vertebrates, Centralspindlin has acquired alternative mechanisms to tether to MT plus ends and/or there may be other proteins or protein complexes that serve to bridge the plus ends of astral MTs to the equatorial cortex in order to promote localized furrowing.
Our data strongly support the notion that the Mgc SxIP motif binds to EB proteins and that the SxNN mutation disrupts this binding. However, careful biochemical studies will be necessary to determine the binding requirements for Centralspindlin and EB proteins throughout the cell cycle. Because Centralspindlin is a heterotetrameric complex (Mishima et al., 2002) and forms higher-order clusters that are important for its function (Hutterer, Glotzer, and Mishima, 2009), it is possible that multiple SxIP motifs within Centralspindlin clusters may facilitate its association with EBs in cells. Indeed, the presence of multiple SxIP motifs that mediate binding to EBs has been noted in other +TIP proteins (Applewhite et al., 2010; Buey et al., 2012; Honnappa et al., 2009; van der Vaart et al., 2011). Interestingly, we found that although mutation of the Mgc SxIP motif strongly reduces the ability of Mgc to track with EB3, the mutant Mgc still affected the EB3 run time along MTs. One possible explanation for this is that the MgcSxNN mutant could be acting as a dominant negative, perhaps by binding to another critical EB-network protein and preventing it from enhancing the EB3 run time. EB interaction networks are complex, and more work needs to be done in the future to resolve this issue.
The ability of a cell to precisely regulate the subcellular localization of proteins that modulate Rho GTPase activity for specific cellular functions is crucial. The GAP protein Mgc is an important regulator of both cytokinesis and cell–cell junctions. Our work shows that the Mgc SxIP motif provides a means of efficient Centralspindlin delivery to localized sites of action at the equatorial cell cortex during cytokinesis and at apical cell–cell junctions. The ability of Mgc to be tethered at specific subcellular locations may be a mechanism that regulates its GTPase specificity, which could help explain the previously reported discrepancies in Mgc GAP specificity toward Rho-family GTPases (Bastos et al., 2012; Breznau et al., 2015; Canman et al., 2008; Jantsch-Plunger et al., 2000; Loria et al., 2012; Miller and Bement, 2009). Key questions that remain to be answered include identifying the binding requirements of Mgc and EB proteins, determining whether Mgc binding to EB proteins is regulated by phosphorylation, testing how Mgc affects MT dynamics and whether Mgc binding to EB proteins might stabilize the dynamics of specific populations of MTs.
MATERIALS AND METHODS
DNA constructs, mRNA preparation and Mgc-targeting MO
pCS2+ vectors encoding X. laevis Mgc (MgcRacGAP and MgcRacGAP–3×GFP) have been described previously (Miller and Bement, 2009). The MgcSxNN mutant was generated by QuikChange (Stratagene, San Diego, CA) site-directed mutagenesis. GFP–rGBD (Benink and Bement, 2005), MKLP1–3×GFP (Miller and Bement, 2009), EB3–mChe (Cai et al., 2009), mChe–UtrCH (Burkel et al., 2007), mChe-membrane and GFP-membrane (Reyes et al., 2014), mChe–H2B (Reyes et al., 2014) and 2×mChe–EMTB (von Dassow et al., 2009) have been described previously. To generate the Mgc–mNeon constructs, X. laevis MgcWT or MgcSxNN were cloned into the mNeonGreen vector (Allele Biotechnology, San Diego, CA) using BglII and EcoRI restriction sites. mRNAs were transcribed in vitro from pCS2+-based vectors using the mMessage mMachine SP6 kit (Ambion, Waltham, MA). Mgc antisense MO.2 (Gene Tools, Philomath, OR), which targets the 5′UTR, has been described previously (Miller and Bement, 2009).
Antibodies
The Xenopus-specific polyclonal Mgc antibody has been described previously (Miller and Bement, 2009). Other antibodies used were anti-β-catenin (Abcam, Ab2365), anti-E-cadherin (Developmental Studies Hybridoma Bank, Iowa City, IA; 5D3-c), anti-GFP [clone JL-8, Clontech, Mountain View, CA, no. 632381 (mouse) and Invitrogen, A-6455 (rabbit)], anti-mChe [Abcam, no. Ab167453 (mouse) and Ab167453 (rabbit)], anti-α-tubulin (DM1A, Sigma, no. T9026) and anti-phosphorylated-Myosin II light chain (p-Myo) (Cell Signaling Technologies, no. 3671). Secondary antibodies for immunofluorescence were anti-mouse or anti-rabbit IgG conjugated to Alexa-Fluor-488- (Invitrogen, no. A11001 or A11008) or Alexa-Fluor-568- (Invitrogen, no. A11004 or 11011).
Xenopus laevis embryos and microinjections
All studies with X. laevis embryos were conducted in compliance with the US Department of Health and Human Services Guide for the Care and Use of Laboratory Animals and were approved by the University of Michigan IACUC. X. laevis embryos were collected, fertilized, dejellied and microinjected as described previously (Breznau et al., 2015; Reyes et al., 2014) using frogs from Nasco (Fort Atkinson, WI). Knockdown and replacement experiments, and mosaic injections have been described previously (Breznau et al., 2015). Needle concentrations for mRNAs were as follows: Mgc–3×GFP (1 µg/ml), GFP–rGBD (25 µg/ml), mChe-membrane (10 µg/ml), GFP-membrane (10 µg/ml), mChe–H2B (20 µg/ml), mChe–UtrCH (10 µg/ml), MKLP1–3×GFP (1 µg/ml).
Immunofluorescence
For immunofluorescence staining of Mgc, β-catenin or E-cadherin, embryos were fixed at NF stages 10-11 with 2% trichloroacetic acid and stained as described previously (Breznau et al., 2015). Antibody concentrations were as follows: anti-Mgc (1:600), anti-β-catenin (1:200), anti-E-cadherin (1:200), anti-GFP (1:200) and anti-mChe (1:200). Alexa-Fluor-conjugated secondary antibodies were used at 1:200.
For immunofluorescence staining of tubulin or p-Myo, albino embryos at NF stage 10-11 were washed three times with PBS and fixed overnight at room temperature in 3.7% formaldehyde, 0.25% glutaraldehyde, 0.2% Triton X-100 in buffer containing 80 mM K-PIPES, 5 mM EGTA and 1 mM MgCl2, pH 6.8. Embryos were quenched for 6 h (tubulin) or 1 h (p-Myo) with shaking at room temperature in PBS containing 100 mM sodium borohydride, rinsed twice in PBS, bisected keeping the animal hemisphere and transferred to 0.6 ml Eppendorf tubes. Embryos were blocked with TBS+10% FBS, 5% DMSO and 0.1% NP40 for 4 h at 4°C, with solution changes 2-3 times. Embryos were incubated with primary antibodies in blocking solution overnight at 4°C on a Nutator instrument, washed with blocking solution (5 min, 15 min, 2 h, overnight at 4°C), incubated with secondary antibodies in blocking solution overnight at 4°C and washed as above. Embryos were stained with 10 μg/ml DAPI diluted in TBSN (TBS plus 0.1% NP40) for 30 min at room temperature. Embryos were then dehydrated using a dilution series of TBSN:MeOH (one 10 min wash of each: TBSN:MeOH 4:1, 1:1, 1:4, then 100% MeOH), cleared with Murray's Clear (1:2 mixture of benzyl alcohol:benzyl benzoate) and mounted for imaging. Antibody concentrations: anti-phosphorylated-Myosin II light chain (1:200), anti-α-tubulin (1:50), Alexa-Fluor-conjugated secondary antibodies (1:200).
Live and fixed confocal microscopy
Images were collected on an Olympus Fluoview 1000 confocal microscope with FV10-ASW software (Olympus America Inc, Lombard, IL) equipped with a 60× supercorrected PLAPON 60×OSC objective (NA=1.4, working distance=0.12 mm). Live and fixed imaging were performed as described previously (Breznau et al., 2015; Reyes et al., 2014).
Cell culture and transfection
Cos-7 cells (monkey kidney fibroblasts) were purchased from American Type Culture Collection (ATCC) (CRL-1651) and tested for mycoplasma contamination using PCR. They were grown in Dulbecco's modified Eagle's medium (DMEM)+10% (vol/vol) FBS and 2 mM L-glutamine at 37°C with 5% (vol/vol) CO2. Cells were co-transfected with 1 µg of plasmid DNA (0.5 µg of human EB3–mChe and 0.5 µg of either Xenopus MgcWT–mNeon or Xenopus MgcSxNN–mNeon) using Expressfect (Danville Scientific, South Plainfield, NJ). Cells were allowed to express for 18-22 h, then processed for microscopy.
TIRF microscopy
Movies were acquired using a Nikon TiE/B microscope with a 100×1.49 NA oil immersion TIRF objective (Nikon, Melville, NY) equipped with three 20 mW diode lasers (488 nm, 561 nm and 640 nm) combined into a single fiber and controlled via an acousto-optical tunable filter (Agilent, Santa Clara, CA). Images were collected via an EMCCD detector (iXon X3 DU897, 512×512, 16 µm array; Andor, Belfast, UK). For imaging in green and/or red, the microscope used a dual-band laser polychroic mirror (ZT488/561rpc; Chroma Technology, Rockingham, VT), a dual-band sputtered emission filter (ZET488/561m; Chroma) and a dual-band sputtered cleanup filter (ZET488/561×; Chroma), and either a 488 nm (10-15% mW power) or 561 nm (10-15% mW power) laser was used for total internal reflection-based illumination. Images were acquired with 50 ms exposures, and image acquisition was controlled by Nikon Elements software.
Quantification and analysis
Kymograph generation and analysis
Kymographs were generated in ImageJ (open-source software) from raw data using the Multiple Kymograph plug-in (width=3). The segmented line tool (line width=1) was used to trace an EB3–mChe comet in the EB3–mChe channel. The region of interest (ROI) was saved to ROI manager so the same line could be generated in the Mgc–mNeon channel. The channels were split and synchronized, then the saved ROI was generated in the mNeon channel (10 pixels wide) that followed the EB3–mChe comet exactly. A background line (100 pixels long) was drawn on the MT lattice. Normalization was achieved by subtracting the background mean intensity from the mNeon mean intensity. EB3–mChe run duration was determined by observing EB3–mChe comets for the duration of a 30 s live movie (0.103 s interval between frames). The starting frame number where an EB3 comet was first observed was subtracted from the ending frame number. Only comets that started after frame #2 were counted. EB3 velocity was determined using the ImageJ velocity measurement tool and raw data kymographs.
Percent multinucleation measurements
Using Volocity software (PerkinElmer, Waltham, MA), the percent multinucleation was determined as described previously (Breznau et al., 2015).
Percent asymmetric cell division
Live movies of cell division were analyzed using Volocity. The pole-to-pole length of each dividing cell was measured at the widest part of the cell perpendicular to the cleavage furrow at 50% ingression. A cell was determined to have symmetric division if the cleavage furrow was positioned within 4 µm (on either side) of the measured middle of the cell.
Quantification of astral MT intensity
Astral MT intensity of cells in metaphase was measured using ImageJ. Two 40×160 pixel lines were manually placed in the cytosol parallel to the spindle, and two 40×80 pixel lines were manually placed in the cytosol adjacent to the spindle poles. Another 40×80 pixel line was placed in the cytosol away from the spindle to measure background intensity. The background intensity was subtracted from each line, and the average α-tubulin intensity was calculated.
Quantification of Mgc–3×GFP and MKLP1–3×GFP
The intensity of Mgc–3×GFP or MKLP1–3×GFP at the contractile ring at 25% cleavage furrow ingression was determined in ImageJ by drawing a line (width 20 pixels) that encompassed the contractile ring, but did not contain membrane signal. The same-sized line was drawn in both polar regions of the cell and the average polar cortex value was determined. A background intensity line was subtracted from both lines described above.
RhoA-GTP and F-actin intensity plots
For RhoA-GTP, frames from live movies of dividing cells just after anaphase onset were analyzed using Volocity. Only frames where the initial appearance of F-actin at the contractile ring could be detected were used. Intensity plot measurements (175 pixel long×1 pixel wide) along the cell membrane of both sides of the developing furrow were made for each cell and averaged. For F-actin, two intensity plot measurements were taken perpendicular to the division plane at positions adjacent to the cell membrane (175 pixels long×1 pixel wide, 5-10 pixels away from the membrane) at 25% cleavage furrow ingression and averaged. Another 175 pixel line was placed in the cytosol in the polar region of the cell to measure background intensity and was subtracted from the average RhoA-GTP or F-actin intensity for each cell. The longest cell width perpendicular to the division plane was also measured for normalization to cell size.
RhoA-GTP and F-actin full width at half max (FWHM) and intensity measurements
The RhoA-GTP and F-actin background-subtracted intensity plots described above were used for the FWHM and intensity analysis. The x and y values for each intensity plot were imported into ImageJ. Using the curve fitter function, a Gaussian curve was fit to each line. In most cases, Gaussian with offset was used, but in cases where the offset was >50, Gaussian with no offset was used; Gaussian curves with an R2 value <0.55 were excluded from the analysis. The parameter d (or c in the case of no offset) from the Gaussian curves was used to calculate the FWHM using the equation: 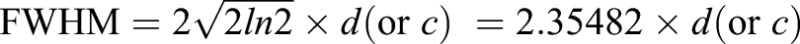 . The FWHM was then divided by the cell width for normalization such that the FWHM is presented as a percentage of cell width. Intensity was determined from the peak of the Gaussian curve: parameter c (or b in the case of no offset).
. The FWHM was then divided by the cell width for normalization such that the FWHM is presented as a percentage of cell width. Intensity was determined from the peak of the Gaussian curve: parameter c (or b in the case of no offset).
Apicobasal intensity intensity plots of junctional proteins
Using ImageJ, the apicobasal intensity values were determined as described previously (Breznau et al., 2015).
Statistical analysis
Unpaired Student's t-tests were used to determine statistical significance of each group pairwise compared to either control or to MO+WT as indicated. Statistics were calculated in GraphPad Prism (La Jolla, CA) software.
Acknowledgements
Thank you to Y. Yue for assistance with TIRF microscopy and data analysis, T. Higashi for help with immunofluorescence experiments, T. R. Arnold and K. M. Dinshaw for mRNAs, and members of the Miller Lab for helpful discussions. Special thanks to W. M. Bement (University of Wisconsin, Madison) in whose lab A.L.M. conducted the initial experiments on Mgc targeting shown in Fig. 1. Thank you to The Company of Biologists - Journal of Cell Science for a Travelling Fellowship awarded to E.B.B. allowing her to travel for training and related experiments to R. Heald's Lab (University of California, Berkeley).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.B.B., K.J.V., A.L.M.; Methodology: E.B.B., M.M., T.L.B., K.J.V., A.L.M.; Formal analysis: E.B.B., M.M., A.L.M.; Investigation: E.B.B., M.M., T.L.B., A.L.M.; Resources: K.J.V., A.L.M.; Writing - original draft: E.B.B.; Writing - review & editing: E.B.B., M.M., T.L.B., K.J.V., A.L.M.; Visualization: E.B.B., A.L.M.; Supervision: A.L.M.; Funding acquisition: A.L.M.
Funding
This work was supported by the National Institutes of Health (R00 GM089765 and R01 GM112794 to A.L.M., R01 GM070862 to K.J.V.). E.B.B. was supported by the National Science Foundation Predoctoral Fellowship, the University of Michigan Rackham Predoctoral Fellowship, and the National Institutes of Health Cellular and Molecular Biology Training Grant [T32-GM007315]. The initial experiments on Mgc targeting shown in Fig. 1 were done with the support of National Institutes of Health grant R01 GM52932 to W. M. Bement. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.195891.supplemental
References
- Akhmanova A. and Steinmetz M. O. (2010). Microtubule +TIPs at a glance. J. Cell Sci. 123, 3415-3419. 10.1242/jcs.062414 [DOI] [PubMed] [Google Scholar]
- Applewhite D. A., Grode K. D., Keller D., Zadeh A. D., Slep K. C. and Rogers S. L. (2010). The spectraplakin Short stop is an actin-microtubule cross-linker that contributes to organization of the microtubule network. Mol. Biol. Cell 21, 1714-1724. 10.1091/mbc.E10-01-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R. N., Penate X., Bates M., Hammond D. and Barr F. A. (2012). CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis. J. Cell Biol. 198, 865-880. 10.1083/jcb.201204107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement W. M., Benink H. A. and von Dassow G. (2005). A microtubule-dependent zone of active RhoA during cleavage plane specification. J. Cell Biol. 170, 91-101. 10.1083/jcb.200501131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement W. M., Miller A. L. and von Dassow G. (2006). Rho GTPase activity zones and transient contractile arrays. BioEssays 28, 983-993. 10.1002/bies.20477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benink H. A. and Bement W. M. (2005). Concentric zones of active RhoA and Cdc42 around single cell wounds. J. Cell Biol. 168, 429-439. 10.1083/jcb.200411109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breznau E. B., Semack A. C., Higashi T. and Miller A. L. (2015). MgcRacGAP restricts active RhoA at the cytokinetic furrow and both RhoA and Rac1 at cell-cell junctions in epithelial cells. Mol. Biol. Cell 26, 2439-2455. 10.1091/mbc.E14-11-1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey R. M., Sen I., Kortt O., Mohan R., Gfeller D., Veprintsev D., Kretzschmar I., Scheuermann J., Neri D., Zoete V. et al. (2012). Sequence determinants of a microtubule tip localization signal (MtLS). J. Biol. Chem. 287, 28227-28242. 10.1074/jbc.M112.373928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel B. M., von Dassow G. and Bement W. M. (2007). Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil. Cytoskeleton 64, 822-832. 10.1002/cm.20226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., McEwen D. P., Martens J. R., Meyhofer E. and Verhey K. J. (2009). Single molecule imaging reveals differences in microtubule track selection between Kinesin motors. PLoS Biol. 7, e1000216 10.1371/journal.pbio.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman J. C., Cameron L. A., Maddox P. S., Straight A., Tirnauer J. S., Mitchison T. J., Fang G., Kapoor T. M. and Salmon E. D. (2003). Determining the position of the cell division plane. Nature 424, 1074-1078. 10.1038/nature01860 [DOI] [PubMed] [Google Scholar]
- Canman J. C., Lewellyn L., Laband K., Smerdon S. J., Desai A., Bowerman B. and Oegema K. (2008). Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science 322, 1543-1546. 10.1126/science.1163086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWard A. D. and Alberts A. S. (2009). Ubiquitin-mediated degradation of the formin mDia2 upon completion of cell division. J. Biol. Chem. 284, 20061-20069. 10.1074/jbc.M109.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas M. E., Davies T., Joseph N. and Mishima M. (2010). Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr. Biol. 20, 927-933. 10.1016/j.cub.2010.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning A. S., Van Itallie C. M. and Anderson J. M. (2012). Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell 23, 577-590. 10.1091/mbc.E11-09-0791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E. and von Dassow G. (2008). Stable and dynamic microtubules coordinately shape the myosin activation zone during cytokinetic furrow formation. J. Cell Biol. 183, 457-470. 10.1083/jcb.200807128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. A., Paluch E. and Oegema K. (2012). Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28, 29-58. 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Guillemot L., Guerrera D., Spadaro D., Tapia R., Jond L. and Citi S. (2014). MgcRacGAP interacts with cingulin and paracingulin to regulate rac1 activation and development of the tight junction barrier during epithelial junction assembly. Mol. Biol. Cell 25, 1995-2005. 10.1091/mbc.e13-11-0680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K., Kawashima T., Iwamoto I., Nosaka T. and Kitamura T. (2001). MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J. Biol. Chem. 276, 5821-5828. 10.1074/jbc.M007252200 [DOI] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J. A., Buey R. M., Lawera A. et al. (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366-376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Hutterer A., Glotzer M. and Mishima M. (2009). Clustering of centralspindlin is essential for its accumulation to the central spindle and the midbody. Curr. Biol. 19, 2043-2049. 10.1016/j.cub.2009.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V., Gönczy P., Romano A., Schnabel H., Hamill D., Schnabel R., Hyman A. A. and Glotzer M. (2000). CYK-4, A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149, 1391-1404. 10.1083/jcb.149.7.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Toedt G., Montenegro Gouveia S., Davey N. E., Hua S., van der Vaart B., Grigoriev I., Larsen J., Pedersen L. B., Bezstarosti K. et al. (2012). A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22, 1800-1807. 10.1016/j.cub.2012.07.047 [DOI] [PubMed] [Google Scholar]
- Kumar P. and Wittmann T. (2012). +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 22, 418-428. 10.1016/j.tcb.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekomtsev S., Su K.-C., Pye V. E., Blight K., Sundaramoorthy S., Takaki T., Collinson L. M., Cherepanov P., Divecha N. and Petronczki M. (2012). Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature 492, 276-279. 10.1038/nature11773 [DOI] [PubMed] [Google Scholar]
- Loria A., Longhini K. M. and Glotzer M. (2012). The RhoGAP domain of CYK-4 has an essential role in RhoA activation. Curr. Biol. 22, 213-219. 10.1016/j.cub.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manukyan A., Ludwig K., Sanchez-Manchinelly S., Parsons S. J. and Stukenberg P. T. (2015). A complex of p190RhoGAP-A and anillin modulates RhoA-GTP and the cytokinetic furrow in human cells. J. Cell Sci. 128, 50-60. 10.1242/jcs.151647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. L. and Bement W. M. (2009). Regulation of cytokinesis by Rho GTPase flux. Nat. Cell Biol. 11, 71-77. 10.1038/ncb1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S. et al. (2005). CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141-153. 10.1083/jcb.200405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Sasaki H., Matsui C., Akhmanova A., Tsukita S. and Vorobjev I. (2006). Mammalian CLASPs are required for mitotic spindle organization and kinetochore alignment. Genes Cells 11, 845-857. 10.1111/j.1365-2443.2006.00990.x [DOI] [PubMed] [Google Scholar]
- Mishima M., Kaitna S. and Glotzer M. (2002). Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2, 41-54. 10.1016/S1534-5807(01)00110-1 [DOI] [PubMed] [Google Scholar]
- Mishima M., Pavicic V., Grüneberg U., Nigg E. A. and Glotzer M. (2004). Cell cycle regulation of central spindle assembly. Nature 430, 908-913. 10.1038/nature02767 [DOI] [PubMed] [Google Scholar]
- Montenegro Gouveia S., Leslie K., Kapitein L. C., Buey R. M., Grigoriev I., Wagenbach M., Smal I., Meijering E., Hoogenraad C. C., Wordeman L. et al. (2010). In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr. Biol. 20, 1717-1722. 10.1016/j.cub.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Nguyen P. A., Groen A. C., Loose M., Ishihara K., Wuhr M., Field C. M. and Mitchison T. J. (2014). Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science 346, 244-247. 10.1126/science.1256773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y. and Yonemura S. (2006). Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J. Cell Sci. 119, 104-114. 10.1242/jcs.02737 [DOI] [PubMed] [Google Scholar]
- Rappaport R. (1961). Experiments concerning the cleavage stimulus in sand dollar eggs. J. Exp. Zool. 148, 81-89. 10.1002/jez.1401480107 [DOI] [PubMed] [Google Scholar]
- Ratheesh A., Gomez G. A., Priya R., Verma S., Kovacs E. M., Jiang K., Brown N. H., Akhmanova A., Stehbens S. J. and Yap A. S. (2012). Centralspindlin and alpha-catenin regulate Rho signalling at the epithelial zonula adherens. Nat. Cell Biol. 14, 818-828. 10.1038/ncb2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes C. C., Jin M., Breznau E. B., Espino R., Delgado-Gonzalo R., Goryachev A. B. and Miller A. L. (2014). Anillin regulates cell-cell junction integrity by organizing junctional accumulation of Rho-GTP and actomyosin. Curr. Biol. 24, 1263-1270. 10.1016/j.cub.2014.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K. B., Canman J. C., Ben Moree C., Tirnauer J. S. and Salmon E. D. (2005). Taxol-stabilized microtubules can position the cytokinetic furrow in mammalian cells. Mol. Biol. Cell 16, 4423-4436. 10.1091/mbc.E04-11-0974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers W. G. and Saint R. (2003). A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell 4, 29-39. 10.1016/S1534-5807(02)00402-1 [DOI] [PubMed] [Google Scholar]
- Strickland L. I., Wen Y., Gundersen G. G. and Burgess D. R. (2005). Interaction between EB1 and p150glued is required for anaphase astral microtubule elongation and stimulation of cytokinesis. Curr. Biol. 15, 2249-2255. 10.1016/j.cub.2005.10.073 [DOI] [PubMed] [Google Scholar]
- Su K.-C., Takaki T. and Petronczki M. (2011). Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev. Cell 21, 1104-1115. 10.1016/j.devcel.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Vale R. D., Spudich J. A. and Griffis E. R. (2009). Dynamics of myosin, microtubules, and Kinesin-6 at the cortex during cytokinesis in Drosophila S2 cells. J. Cell Biol. 186, 727-738. 10.1083/jcb.200902083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B., Manatschal C., Grigoriev I., Olieric V., Gouveia S. M., Bjelić S., Demmers J., Vorobjev I., Hoogenraad C. C., Steinmetz M. O. et al. (2011). SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J. Cell Biol. 193, 1083-1099. 10.1083/jcb.201012179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G., Verbrugghe K. J. C., Miller A. L., Sider J. R. and Bement W. M. (2009). Action at a distance during cytokinesis. J. Cell Biol. 187, 831-845. 10.1083/jcb.200907090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A. and Shibata M. (2010). alpha-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533-542. 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- Yüce O., Piekny A. and Glotzer M. (2005). An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 170, 571-582. 10.1083/jcb.200501097 [DOI] [PMC free article] [PubMed] [Google Scholar]