ABSTRACT
PP2A comprising B56 regulatory subunit isoforms (PP2AB56) is a serine/threonine phosphatase essential for mitosis. At the kinetochore, PP2AB56 both stabilizes microtubule binding and promotes silencing of the spindle assembly checkpoint (SAC) through its association with the SAC protein BubR1. Cells depleted of the B56 regulatory subunits of PP2A are delayed in activation of Cdc20-containing APC/C (APC/CCdc20), which is an essential step for mitotic exit. It has been hypothesized that this delay arises from increased production of the mitotic checkpoint complex (MCC), an APC/CCdc20 inhibitor formed at unattached kinetochores through SAC signaling. In contrast to this prediction, we show that depletion of B56 subunits does not increase the amount or stability of the MCC. Rather, delays in APC/CCdc20 activation in B56-depleted cells correlate with impaired Cdc20 binding to APC/C. Stimulation of APC/CCdc20 assembly does not require binding between PP2AB56 and BubR1, and thus this contribution of PP2AB56 towards mitotic exit is distinct from its functions at kinetochores. PP2AB56 associates with APC/C constitutively in a BubR1-independent manner. A mitotic phosphorylation site on Cdc20, known to be a substrate of PP2AB56, modulates APC/CCdc20 assembly. These results elucidate the contributions of PP2AB56 towards completion of mitosis.
KEY WORDS: Mitosis, PP2A, B56, APC/C, Cdc20
Highlighted Article: Cdc20-dependent activation of the APC/C is essential for chromosome segregation and exit from mitosis. We show that the PP2AB56 phosphatase is required for the assembly of APC/CCdc20.
INTRODUCTION
Error-free chromosome segregation during mitosis requires that replicated sister chromatids attach to microtubules at opposite ends of the mitotic spindle before sister chromatid separation and exit from mitosis. The fidelity of chromosome segregation depends on dynamic phosphorylation, with contributions from multiple serine/threonine kinases, including Cdk1–cyclin-B, Plk1, Aurora B and Mps1 (also known as TTK), leading to the generation of thousands of mitosis-specific phosphorylation events (Olsen et al., 2010). One critical way in which phosphorylation influences mitosis is by controlling the activity of the anaphase-promoting complex/cyclosome (APC/C), an essential E3 ubiquitin ligase. When APC/C is bound to its co-activator Cdc20 (the complex is denoted APC/CCdc20 hereafter), it ubiquitylates securin and cyclin B, leading to their proteasomal degradation. The destruction of securin and cyclin B triggers sister chromatid separation and mitotic exit, respectively, leading to chromosome segregation, and completion of mitosis and cytokinesis (reviewed in Pines, 2011; Sivakumar and Gorbsky, 2015).
Mitosis-specific phosphorylation controls the assembly of APC/CCdc20 and modulates the timing of its activation. APC/CCdc20 is modified at >100 sites, with both stimulatory and inhibitory phosphorylation sites identified (Chung and Chen, 2003; Craney et al., 2016; D'Angiolella et al., 2003; Fujimitsu et al., 2016; Hein and Nilsson, 2016; Jia et al., 2016; Kimata et al., 2008; Labit et al., 2012; Qiao et al., 2016; Schwab et al., 2001; Tang et al., 2004; Yudkovsky et al., 2000; Zhang et al., 2016). Perhaps the best-understood impact of phosphoregulation on APC/C activation is via the spindle assembly checkpoint (SAC), a signal transduction cascade initiated at unattached kinetochores. This checkpoint depends on multiple kinases, including Mps1, and culminates in the inhibition of APC/CCdc20 activity (reviewed in Foley and Kapoor, 2013; London and Biggins, 2014). The SAC inhibits APC/CCdc20 by catalyzing the association of Cdc20 with the SAC proteins Mad2 (also known as MAD2L1), BubR1 (also known as BUB1B) and Bub3, to form the mitotic checkpoint complex (MCC) (Sudakin et al., 2001). Within the MCC, Cdc20 is inhibited because it cannot recognize degron sequences (Chao et al., 2012). Additionally, the MCC can bind to and inhibit a second molecule of Cdc20 already bound to the APC/C (Alfieri et al., 2016; Izawa and Pines, 2015; Yamaguchi et al., 2016). Serine/threonine phosphatases facilitate SAC inactivation at the kinetochore to permit APC/CCdc20 activation (Espert et al., 2014; Meadows et al., 2011; Nijenhuis et al., 2014; Rosenberg et al., 2011), but less is known about whether and how phosphatases impact APC/CCdc20 assembly in mitosis.
Recent data indicate that the PP2A serine/threonine phosphatases containing a B56 isoform regulatory subunit (hereafter denoted PP2AB56) play essential roles in mitotic progression. The PP2AB56 holoenzyme is a heterotrimer consisting of a catalytic subunit (encoded by PPP2CA and PPP2CB) bound to two non-enzymatic subunits, the scaffold subunit (encoded by PPP2R1A and PPP2R1B) and the B56 regulatory subunit. In human cells, there are five B56 isoforms (α, β, γ, δ and ε; encoded by PPP2R5A, PPP2R5B, PPP2R5C, PPP2R5D and PPP2R5E, respectively), which share a pseudo-HEAT repeat domain (Cho and Xu, 2007; Xu et al., 2006). The five isoforms can function redundantly in mitosis (Foley et al., 2011). PP2AB56 substrate selectivity is achieved, at least in part, through protein–protein interactions conferred by B56 subunit binding to a short linear motif sequence of LxxIxE (where L, I and E denote leucine, isoleucine and glutamic acid, respectively, and x indicates any amino acid) (Hertz et al., 2016). PP2AB56 dephosphorylates substrates of multiple mitotic kinases, including Mps1, Plk1 and Aurora B, and this activity presumably underlies its myriad of mitotic functions, including preservation of sister chromatid cohesion at the centromere (Chen et al., 2007; Kitajima et al., 2006; Riedel et al., 2006; Tang et al., 2006), stabilization of microtubule binding at the kinetochore (Foley et al., 2011), silencing of the SAC at the kinetochore (Espert et al., 2014; Nijenhuis et al., 2014), recruitment of E2 ubiquitin-conjugating enzymes to APC/CCdc20 (Craney et al., 2016), and assembly of the central spindle in anaphase (Bastos et al., 2014).
During mitosis, a critical pool of PP2AB56 is the fraction associated with the SAC protein BubR1. The direct binding of B56 subunits to BubR1 is mediated by an LxxIxE motif present in BubR1's kinetochore attachment regulatory domain (KARD) (Suijkerbuijk et al., 2012). Deletion or mutation of BubR1's KARD prevents PP2AB56 kinetochore targeting and is known to impair mitosis in two ways. First, kinetochore–microtubule attachments are destabilized (Kruse et al., 2013; Suijkerbuijk et al., 2012; Xu et al., 2013). Second, SAC silencing at the kinetochore is delayed (Espert et al., 2014; Nijenhuis et al., 2014). Collectively, these data establish a critical role for the direct binding between BubR1 and PP2AB56 in mitotic progression, and hence, the timing of APC/CCdc20 activity.
Here, we examine human tissue culture cells that had been depleted of B56 subunits during mitosis. Consistent with previous reports (Espert et al., 2014; Nijenhuis et al., 2014), we find that B56-depleted cells exhibit delayed APC/CCdc20 activation and have increased recruitment of SAC proteins to the kinetochore. Given that SAC signaling at the kinetochore is rate-limiting for the inhibition of APC/CCdc20 by MCC (Collin et al., 2013; Dick and Gerlich, 2013), we expected a possible increase in or stabilization of the MCC in B56-depleted cells. However, we find no evidence that cells depleted of B56 subunits have increased levels or stability of the MCC. Rather, we show that PP2AB56 is required to stimulate Cdc20 binding to APC/C in mitosis. PP2AB56 is associated with APC/C throughout the cell cycle, in a BubR1-independent manner. We show that residue Ser92 on Cdc20, previously shown to be a substrate of PP2AB56 (Craney et al., 2016; Jia et al., 2016), has a role in modulating APC/CCdc20 assembly. Taken together, these data reveal a role for PP2AB56 in stimulating the assembly of APC/CCdc20 complexes in mitosis.
RESULTS
PP2AB56 promotes mitotic exit
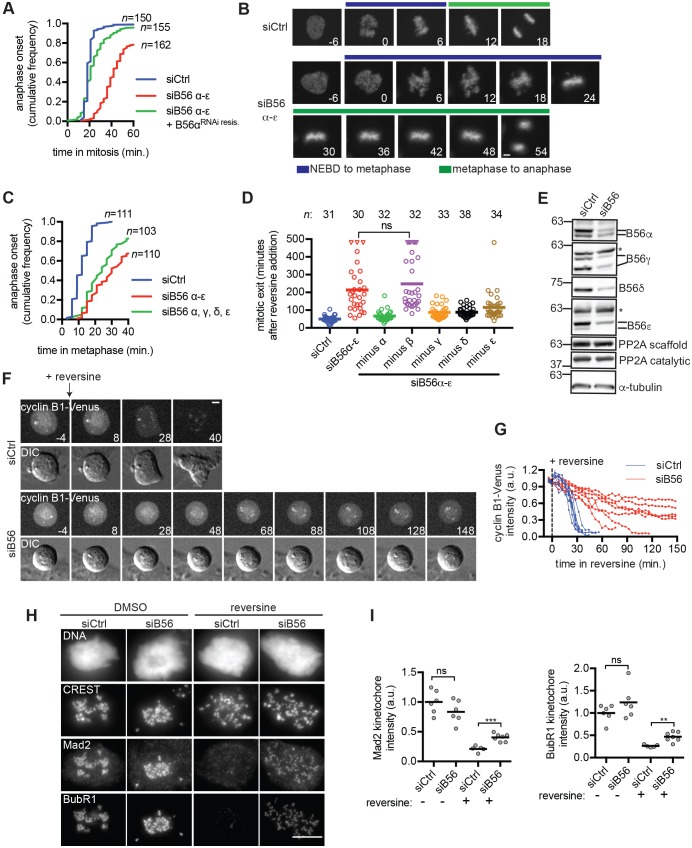
To determine the impact of B56 depletion on the kinetics of chromosome segregation, we used a validated set of five siRNAs targeting B56α through B56ε (siB56α-ε) (Foley et al., 2011). siB56α-ε-transfected cells (hereafter referred to as siB56α-ε cells) were delayed in mitosis compared to cells treated with non-targeting control siRNA (siCtrl), and this delay was rescued by expression of RNA interference (RNAi)-resistant B56α (Fig. 1A). As expected (Foley et al., 2011), siB56α-ε cells required more time to achieve metaphase chromosome alignment (Fig. 1B, blue bars). Additionally, siB56α-ε cells were delayed between metaphase and anaphase (Fig. 1B, green bars, quantified in Fig. 1C). Consistent with recent reports (Espert et al., 2014; Nijenhuis et al., 2014), nocodazole-arrested siB56α-ε cells were delayed in mitotic exit following addition of reversine, an inhibitor of the kinase Mps1 (Santaguida et al., 2010) (Fig. 1D). This delay was rescued by expression of RNAi-resistant B56ε (Fig. S1A,B), and it required co-transfection of siRNAs targeting B56α, B56γ, B56δ and B56ε, but not B56β (Fig. 1D). Because siRNA against B56β was not essential for inducing a delay in anaphase onset (Fig. 1C), further experiments to deplete PP2AB56 were performed with siRNA targeting B56α, B56γ, B56δ and B56ε (hereafter referred to as siB56), resulting in a partial depletion of each subunit (Fig. 1E).
Fig. 1.
PP2AB56 promotes the timing of anaphase and mitotic exit. (A–C) PP2AB56 depletion delays anaphase onset. (A,B) RPE1 cells expressing RNAi-resistant B56α and/or H2B–mCherry were transfected with siRNA targeting B56α, B56β, B56γ, B56δ and B56ε (siB56α-ε) or control siRNA (siCtrl) and imaged live. (A) The duration of mitosis was measured for individual cells. Plotted is the fraction of cells that entered anaphase as a function of time in mitosis. (B) Representative micrographs. Numbers indicate time (min) relative to nuclear envelope breakdown (NEBD). The time to metaphase (blue bar) and time from metaphase to anaphase (green bar) is indicated. (C) RPE1 cells expressing H2B–mCherry were treated with siCtrl, siB56α-ε, or a mixture of siRNAs against B56α, B56γ, B56δ and B56ε (siB56) and imaged live. The cumulative anaphase onset is plotted. (D) Depletion of PP2AB56 delays mitotic exit. RPE1 cells were treated with siCtrl, siB56α-ε or mixtures in which an siRNA against a single B56 isoform had been omitted. Cells were treated with nocodazole for 2 h and then imaged live before and after reversine addition. The median time of mitotic exit (line) and exit time of individual cells (open circles) is plotted. Triangles indicate cells that remained in mitosis at the end of the imaging period. (E) siCtrl and siB56 mitotic HeLa lysates were probed by western blotting. Asterisk indicates non-specific band. (F,G) Cyclin B1 proteolysis is delayed in siB56 cells. (F) RPE1 CCNB1Venus/+ siCtrl or siB56 cells were treated as described in D and imaged live. Still images from differential interference contrast (DIC) and fluorescent imaging are shown. Numbers indicate time (min) relative to addition of reversine. (G) Plotted is the fluorescence intensity relative to reversine addition. Each line indicates a single cell and the last time point plotted is either mitotic exit or the experimental end-point (150 min). (H,I) siB56 cells have increased recruitment of Mad2 and BubR1 when Mps1 is inhibited. RPE1 siCtrl or siB56 cells were incubated with nocodazole and MG132, and treated with or without reversine before processing for immunofluorescence microscopy. (H) Maximum intensity projection images of representative cells used for quantification shown in I. (I) Quantification of kinetochore recruitment. Each circle represents the average kinetochore signal of one cell. Line indicates mean; a.u., arbitrary units; n.s., not significant (P>0.05), **P<0.005, ***P<0.0005, Student's two-tailed t-test. Results are representative of three independent experiments. n indicates number of cells analyzed from three experiments (A,C) or single experiment (D). Scale bars: 5 μm.
In siB56-transfected cells (hereafter referred to as siB56 cells), the delay in mitotic exit after Mps1 inhibition (Fig. 1D) could arise in multiple ways, including defects in SAC inactivation, APC/CCdc20-dependent proteolysis of cyclin B and/or dephosphorylation of Cdk1–cyclin-B substrates. We determined that siB56 cells were not delayed when mitotic exit was triggered by addition of the Cdk1 inhibitor RO-3306 (Vassilev et al., 2006) (Fig. S1C), suggesting that siB56 cells are proficient in dephosphorylating Cdk1 substrates. This finding is consistent with the B55 family of PP2A regulatory subunits mediating Cdk1–cyclin-B substrate dephosphorylation in human cells (Schmitz et al., 2010). Next, we analyzed the rate of cyclin B1 proteolysis induced by Mps1 inhibition in CCNB1Venus/+ RPE1 cells, in which one allele of cyclin B1 is expressed as a fusion with the fluorescent Venus protein (Collin et al., 2013). Cyclin B1 proteolysis was inefficient in siB56 cells (Fig. 1F,G), suggesting a potential defect in APC/CCdc20 activation. Finally, we used quantitative immunofluorescence microscopy to compare kinetochore localization of the SAC proteins Mad2 and BubR1. In the presence of nocodazole and the proteasome inhibitor MG132 (to prevent mitotic exit), the kinetochore targeting of Mad2 and BubR1 were similar in siCtrl and siB56 cells (Fig. 1H,I). Reversine addition reduced the levels of both Mad2 and BubR1 at the kinetochore, although kinetochores in siB56 cells did retain more Mad2 and BubR1 compared to siCtrl cells (Fig. 1H,I). The latter result is consistent with previous work indicating that PP2AB56 promotes BubR1 eviction at the kinetochore after Mps1 inhibition (Espert et al., 2014; Nijenhuis et al., 2014). However, it was unclear whether changes in the localization of SAC proteins at the kinetochore are the only reason that siB56 cells are delayed in mitotic exit following Mps1 inhibition.
PP2AB56 depletion does not alter the amount or stability of the mitotic checkpoint complex
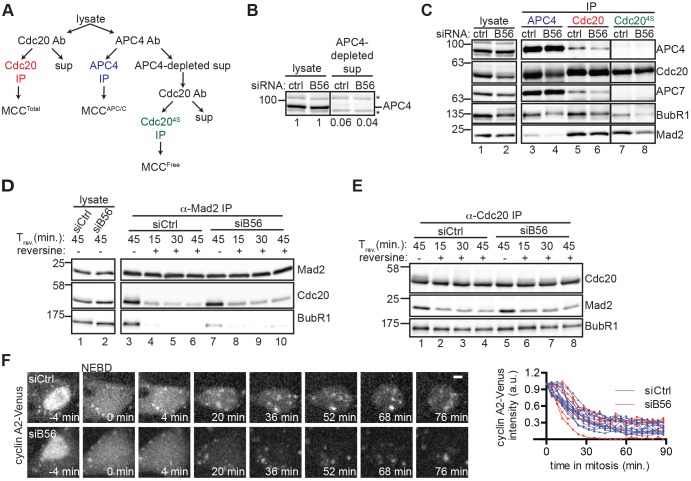
If persistent SAC activation at the kinetochore is the only defect in mitotic exit in siB56 cells, then we would predict an increase in the amount and/or stability of the MCC. We examined this possibility in three ways. First, we used immunoprecipitation (IP) to compare the levels of MCC in siCtrl and siB56 cells. We used an established approach (Collin et al., 2013), outlined in Fig. 2A, performing a Cdc20 IP of whole-cell lysate to compare total MCC amounts (MCCTotal), an APC4 (also known as ANAPC4) IP of whole-cell lysate to compare the pool of MCC bound to APC/C (MCCAPC/C) and, finally, a Cdc20 IP from APC4-depleted supernatant (Cdc204S IP, Fig. 2B) to compare the pool of MCC in excess of the APC/C (MCCFree). In siB56 cells, there was no increase in BubR1 or Mad2 in Cdc20 IPs, indicating that MCCTotal is not increased by B56 depletion (Fig. 2C, compare lanes 5 and 6). APC4 IPs from siB56 cells contained less BubR1, Mad2 and Cdc20 compared to siCtrl cells, suggesting that there is also no increase in MCCAPC/C (Fig. 2C, compare lanes 3 and 4). Finally, Cdc204S IPs from siB56 cells had reduced levels of BubR1 and Mad2 compared to siCtrl cells (Fig. 2C, compare lanes 7 and 8). Thus, by three assessments of the relative amounts of MCC, we observe no increase in MCC in siB56 cells. However, we did note decreased association between APC/C and Cdc20 (Fig. 2C, compare lanes 5 and 6), discussed below.
Fig. 2.
B56 depletion does not increase the amount or stability of the mitotic checkpoint complex. (A) Schematic of experiment to isolate different pools of MCC from whole-cell lysates, including total MCC (Cdc20 IP), MCC bound to APC/C (co-precipitation in APC4 IP), and MCC free of APC4 [isolated by subjecting APC4-depleted supernatant (4S) to Cdc20 IP (Cdc204S IP)]. (B) Comparison of APC4 levels in siCtrl and siB56 whole-cell lysates versus APC4-depleted supernatant by quantitative western blotting. Numbers indicate the relative amount of APC4 in lysate versus APC4-depleted supernatant. *, non-specific band. (C) Whole-cell lysate (lanes 1 and 2) and IPs performed as outlined in A (lanes 3-8) were probed by western blotting. In B,C, solid vertical lines indicate that intervening lanes have been removed. (D,E) MCC is not stabilized in siB56 cells. Mitotic HeLa siCtrl and siB56 cells were incubated with MG132 and reversine or DMSO before lysis and then analyzed directly (D, lanes 1,2) or split for immunoprecipitation of Mad2 (D, lanes 3-10) and Cdc20 (E) and probed by western blotting. (F) Cyclin A proteolysis is not delayed in siB56 cells. RPE1 CCNA2Venus/+ siCtrl or siB56 cells in nocodazole were imaged live. (Left) Micrographs of Venus (cyclin A2) are shown with time (min) relative to nuclear envelope breakdown (NEBD). (Right) The fraction of cyclin-A2–Venus fluorescence remaining as a function of time in mitosis is plotted. Each line indicates a single cell. Scale bar: 5 μm. Results in B–F are representative of three independent experiments. Ab, antibody; sup, supernatant; Trev, time before or after reversine addition.
Next, we compared the stability of MCC after reversine addition. siCtrl and siB56 cells were collected by mitotic shake-off and treated with reversine and MG132. HeLa cells were used in this and further large-scale IP experiments requiring RNAi. In the absence of reversine, Mad2 IPs from siB56 cells contained similar amounts of Cdc20 and less BubR1 compared to siCtrl cells, consistent with there being no increase in the amount of MCC (Fig. 2D, compare lanes 3 and 7). Reversine disassociated Cdc20 and BubR1 from Mad2 with similar kinetics in siCtrl and siB56 cells (Fig. 2D, compare lanes 3-6 vs 7-10). Similarly, in Cdc20 IPs, reversine-induced disassociation of Mad2 from Cdc20 was unchanged by B56 depletion (Fig. 2E, compare lanes 1-4 vs 5-8). These data suggest that the MCC is not stabilized in siB56 cells. However, we note that BubR1 remained associated with Cdc20 upon reversine addition in both siCtrl and siB56 cells (Fig. 2E), possibly reflecting a requirement for proteasome activity in the complete disassembly of MCC (Reddy et al., 2007; Stegmeier et al., 2007; Varetti et al., 2011). Thus, we cannot exclude the possibility that the BubR1–Cdc20 association is altered by B56 depletion.
In our third approach, we compared MCC production at the single cell level (Collin et al., 2013). The basis of this assay is that APC/CCdc20-dependent ubiquitylation targets cyclin A2 for proteolysis at the onset of mitosis. Cyclin A2 and MCC compete for Cdc20 access and therefore the rate of cyclin A2 proteolysis is inversely proportional to the level of MCC (Collin et al., 2013; Di Fiore and Pines, 2010). Thus, if aberrant SAC activity at the kinetochore leads to production of more MCC in siB56 cells, then cyclin A2 proteolysis will be delayed. To test this possibility, RPE1 CCNA2Venus/+ siCtrl or siB56 cells were imaged before and after entry into mitosis. Nocodazole was included to eliminate indirect effects on SAC activity due to defects in kinetochore–microtubule attachment caused by B56 depletion (Foley et al., 2011). We found that cyclin-A2–Venus was degraded with similar kinetics in siCtrl and siB56 cells (Fig. 2F), suggesting that at the single cell level, there is no increase in the amount of MCC. In summary, we find no evidence that the delay in mitotic exit in siB56 cells results from increased production or stabilization of the MCC.
PP2AB56 promotes APC/C–Cdc20 association
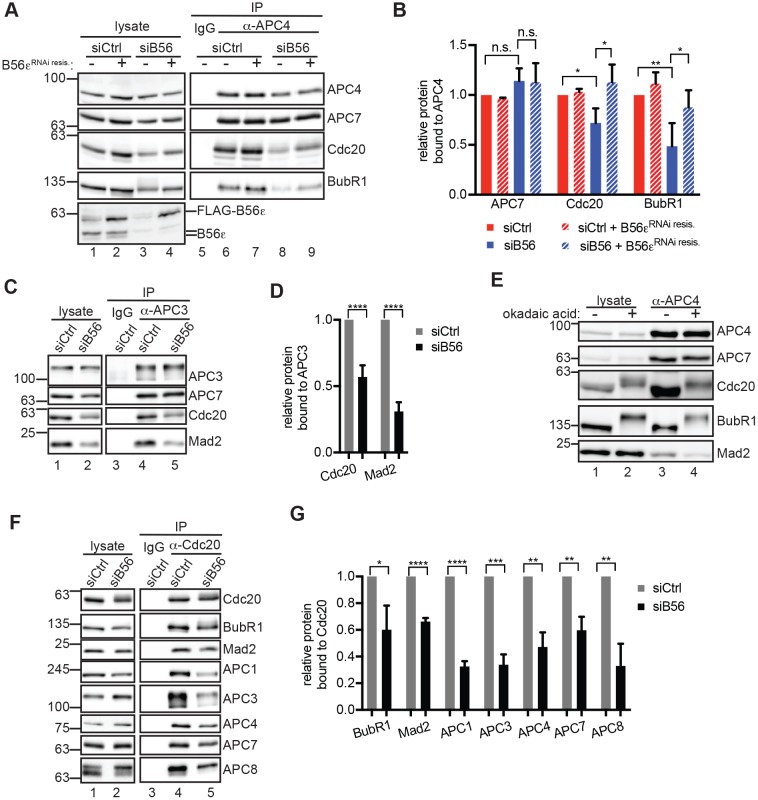
Interestingly, APC4 IPs from siB56 cells had reduced levels of Cdc20 and BubR1 (Fig. 3A, compare lanes 6 and 8; quantified in Fig. 3B). As a control for APC/C assembly, we verified that association between APC4 and another APC/C subunit, APC7 (also known as ANAPC7), was unchanged. This decrease in Cdc20 and BubR1 binding to APC4 was rescued by RNAi-resistant B56ε, demonstrating that the defect is specifically due to the reduction in PP2AB56 (Fig. 3A, compare lanes 8 and 9, quantified in Fig. 3B). Similarly, APC3 (also known as CDC27) IPs from siB56 cells contained less Cdc20 and Mad2 (Fig. 3C, compare lanes 4 and 5, quantified in Fig. 3D). We noted that the decrease in APC/C–Cdc20 association differs from that in recent observations (Craney et al., 2016), possibly due to differences in RNAi (in this study we eliminated siRNA against B56β) or the addition of MG132 by Craney and colleagues, which stabilizes APC/CCdc20 complexes (Kelly et al., 2014). Okadaic acid, which inhibits all PP2A holoenzymes as well as PP1, PP4 and PP6 serine/threonine phosphatases (Swingle et al., 2007), also reduced the association of Cdc20, BubR1 and Mad2 with APC4 (Fig. 3E, compare lanes 3 and 4). Thus, we conclude that phosphatase activity stimulates Cdc20 association with APC/C in mitosis.
Fig. 3.
PP2AB56 stimulates Cdc20 association with APC/C. (A–D) B56 depletion reduces the association of Cdc20 with APC/C. (A,B) HeLa Flp-In cells expressing doxycycline-inducible RNAi-resistant HA–FLAG–B56ε (B56εRNAi resis) were treated with siRNA, induced with doxycycline and collected by mitotic shake-off. (A) Lysates (lanes 1-4) and control IgG (lane 5) or APC4 IPs (lane 6-9) were analyzed by western blotting. (B) The experiment in A was performed three times and the results quantified. The value in siCtrl samples without B56ε induction was set to 1. (C,D) siCtrl and siB56 HeLa cells were collected by mitotic shake-off. (C) Lysates (lanes 1,2) and control IgG (lane 3) or APC3 (lane 4-5) IPs were probed by western blotting for the indicated proteins. (D) Quantification of three replicate experiments as described in C. (E) RPE1 cells were collected by mitotic shake-off, treated with DMSO or okadaic acid for 30 min and then used for IP of APC4 (lanes 3,4). (F,G) siB56 cells have less APC/C associated with Cdc20. (F) siCtrl and siB56 lysates (lanes 1,2) were subjected to Cdc20 (lanes 4,5) or control IgG (lane 3) IP. (G) The experiment in F was performed three times and quantified. Bars indicate mean±s.d. n.s., not significant (P>0.05); *P<0.05, **P<0.005, ***P<0.0005, ****P<0.00005, Student's two-tailed t-test.
We observed that B56 depletion also results in a decrease in Cdc20 levels (Fig. 3A, compare lanes 1 and 3). In lysates that had been treated with MG132 before lysis, this decrease was not observed (Fig. 2D, compare lanes 1 and 2), suggesting that B56 depletion may enhance proteasome-dependent degradation of Cdc20 (Reddy et al., 2007). Therefore, to determine if reduced co-precipitation of Cdc20 with APC/C is simply due to decreased levels of Cdc20, we performed Cdc20 IPs from siCtrl and siB56 cells. B56 depletion reduced Cdc20 association with APC1 (also known as ANAPC1), APC3, APC4, APC7 and APC8 (also known as CDC23) (Fig. 3F, compare lanes 4 and 5, quantified in Fig. 3G). Cdc20 association with BubR1 and Mad2 was also reduced in siB56 cells. Taken together, these results indicate that PP2AB56 is required to promote the association between APC/C and Cdc20. Decreased association between APC/C and Cdc20 would be expected to contribute to the delay in mitotic exit in siB56 cells, synergizing with increased phosphorylation of SAC proteins at the kinetochore (Espert et al., 2014; Nijenhuis et al., 2014) and defects in E2 ubiquitin-conjugating enzyme recruitment to APC/CCdc20 (Craney et al., 2016).
PP2AB56-dependent stimulation of APC/CCdc20 assembly does not require the KARD in BubR1
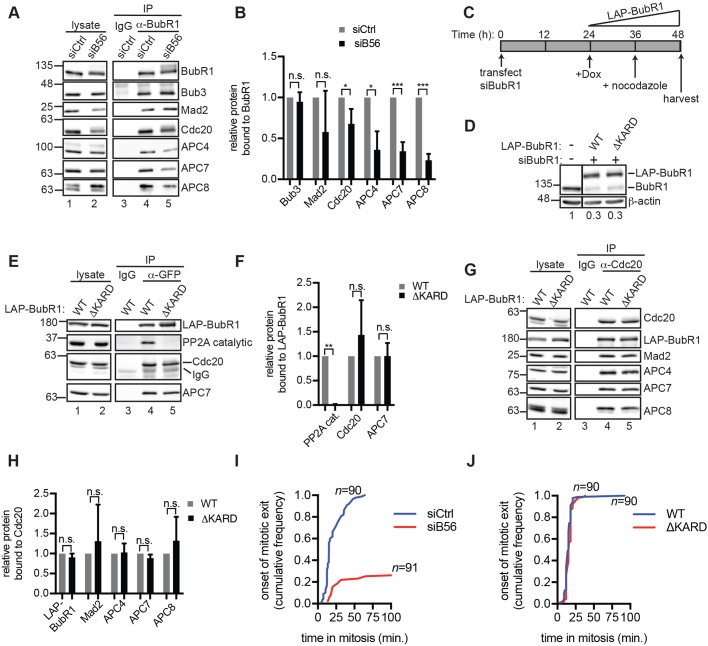
We reasoned that PP2AB56 binding to the BubR1 KARD could potentially mediate its role in promoting APC/CCdc20 assembly. To test this possibility, we first examined BubR1-associated proteins in siCtrl and siB56 cells. BubR1 IPs from siB56 cells had similar levels of the BubR1 constitutive binding partner Bub3, but revealed reductions in Cdc20, APC4, APC7 and APC8 association (Fig. 4A compare lanes 4 and 5, quantified in Fig. 4B). The reduction in APC/C co-precipitation with BubR1 is consistent with reduced Cdc20 binding, as BubR1 association with APC/CCdc20 occurs mainly through Cdc20 (Alfieri et al., 2016). Next, we asked whether eliminating the binding of BubR1 to PP2AB56 is sufficient to reduce APC/C–Cdc20 association. As outlined in Fig. 4C, we used siRNA to deplete endogenous BubR1 by 70% (Fig. 4D) and then engineered cells to express localization and affinity purification (LAP)-tagged BubR1 wild-type (WT) or a mutant in which the KARD had been deleted (ΔKARD) (Suijkerbuijk et al., 2012). We confirmed that BubR1-depleted cells were deficient in SAC activation and that expression of LAP–BubR1 restored SAC activity (Fig. S2). LAP–BubR1ΔKARD did not interact with the catalytic subunit of PP2A as expected (Kruse et al., 2013; Suijkerbuijk et al., 2012; Xu et al., 2013). However, LAP-BubR1ΔKARD bound to similar levels of Cdc20 and APC7 as LAP–BubR1WT (Fig. 4E, compare lanes 4 and 5, quantified in Fig. 4F). Thus, abrogating the association of BubR1 with PP2AB56 is insufficient to reduce the interaction of BubR1 with APC/CCdc20. Moreover, Cdc20 IPs from BubR1-depleted cells rescued with WT and ΔKARD LAP–BubR1 constructs contained similar levels of APC4, APC7 and APC8 (Fig. 4G, compare lanes 4 and 5, quantified in Fig. 4H). Thus, we conclude that, unlike B56 depletion, eliminating the association of PP2AB56 with BubR1 is insufficient to impair APC/CCdc20 assembly. However, we cannot exclude the possibility that the residual BubR1 after RNAi (Fig. 4D), which is too low to sustain mitotic arrest (Fig. S2), is nevertheless sufficient for APC/CCdc20 assembly.
Fig. 4.
PP2AB56-dependent stimulation of APC/CCdc20 assembly does not require direct binding between B56 and BubR1. (A) BubR1 association with APC/C is reduced in siB56 cells. Mitotic siB56 or siCtrl HeLa cells were used for BubR1 (lanes 4,5) or control (lane 3) IgG IPs and analyzed by western blotting. (B) Quantification of three replicate experiments as in (A). (C–H) Deletion of the KARD in BubR1 does not alter APC/C–Cdc20 association. (C) Schematic for BubR1 depletion and rescue in HeLa cells. (D) BubR1-rescue lysates were analyzed alongside lysates from untreated cells by quantitative western blotting. Endogenous BubR1 signals were normalized to the actin signal, and the value in the untreated sample was set to 1. Solid line indicates intervening lanes have been cropped. (E–H) Lysates (lanes 1,2) and IPs (lanes 3-5) of GFP (E) or Cdc20 (G) were analyzed by western blotting. The experiments in E,G were performed three times and the quantifications are shown in F and H, respectively. (I,J) Comparison of mitotic exit delay in siB56 cells versus BubR1-rescue cells. siCtrl and siB56 cells (I) or BubR1-rescue cells (J) were incubated in nocodazole and reversine and imaged live. Plotted is the fraction of cells that exited mitosis as a function of time after mitotic entry. n, total number of cells analyzed from three independent experiments. Bars are mean±s.d. n.s., not significant (P>0.05); *P<0.05, **P<0.005, ***P<0.0005, Student's two-tailed t-test.
Our observation that the role of PP2AB56 in promoting APC/C–Cdc20 association does not require its interaction with BubR1 indicates that B56 depletion will yield a more severe effect on mitotic exit than deletion of the BubR1 KARD. To test this possibility, we scored the duration of mitotic arrest in cells that entered mitosis in the presence of nocodazole and reversine. As expected, inhibition of Mps1 induced rapid exit from mitosis in siCtrl cells with a median exit time of 15 min. By contrast, 75% of siB56 cells remained arrested in mitosis after 100 min (Fig. 4I). Under identical inhibitor conditions, BubR1-depleted cells that had been rescued with LAP–BubR1ΔKARD rapidly exited mitosis, with a median exit time of 16 min, which was indistinguishable from that of BubR1-depleted cells that had been rescued with LAP–BubR1WT (Fig. 4J). Based on these data, we conclude that the association of PP2AB56 with BubR1 is not critical for promoting APC/CCdc20 assembly or exit from mitosis. Moreover, because the BubR1 KARD is required for PP2AB56 targeting to the kinetochore (Kruse et al., 2013; Suijkerbuijk et al., 2012; Xu et al., 2013), our results imply that the cytoplasmic pool of PP2AB56 is sufficient for promoting mitotic exit.
PP2AB56 interacts with APC/C independently of BubR1
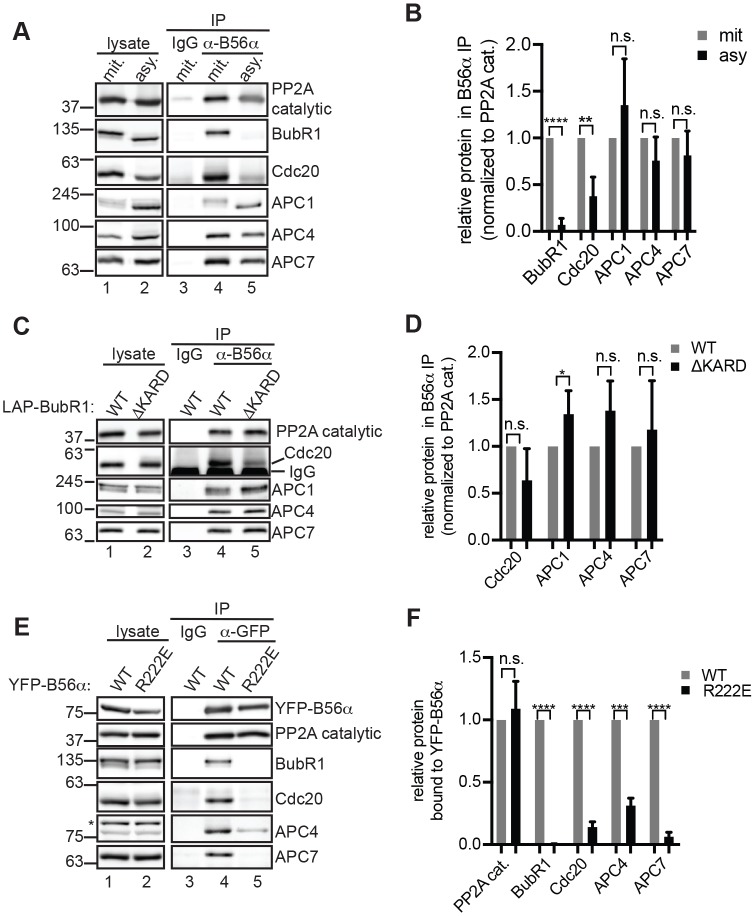
In IPs of endogenous B56α, we observed co-precipitation of APC/C subunits, including APC1, APC4 and APC7, as well as of Cdc20 and BubR1 (Fig. 5A, compare lanes 3 and 4). Interestingly, while the association of B56α with Cdc20 and BubR1 was mitosis specific, as expected (Kruse et al., 2013), the interaction with APC/C subunits was observed in IPs from mitotic and asynchronous cells (Fig. 5A, compare lanes 4 and 5, quantified in Fig. 5B). Considering that BubR1 association with B56α was reduced by 93±7% (mean±s.d.) in asynchronous cells compared to mitotic cells while association with APC1, APC4 and APC7 was unchanged, we reasoned that B56α association with the APC/C might occur independently of its association with BubR1. To test this possibility, we performed B56α IPs in BubR1-depleted cells that had been rescued with LAP–BubR1WT or LAP–BubR1ΔKARD. Indeed, deletion of the KARD had no significant effect on the association of B56α with Cdc20, APC4 and APC7, while APC1 association was slightly increased (Fig. 5C, compare lanes 4 and 5, quantified in Fig. 5D). From these results, we conclude that the APC/C and PP2AB56 interact constitutively and that this association does not depend on PP2AB56 targeting to BubR1.
Fig. 5.
B56α interacts with the APC/C constitutively and independently of BubR1. (A,B) B56α co-precipitates with APC/C. (A) Mitotic (mit.) or asynchronous (asy.) HeLa cells were subjected to IP for B56α (lanes 4,5) or with control IgG (lane 3) and probed by western blotting for the indicated proteins. (B) Quantification of three replicate experiments as described in A. Values were normalized to those for the PP2A catalytic subunit (PP2A cat.). (C,D) Mitotic interaction of B56α and APC/C does not require the KARD of BubR1. BubR1 depletion and rescue was performed as described in Fig. 4C, and cells were subjected to IP for B56α (lanes 4,5) or with control IgG (lane 3) and probed by western blotting. (D) Quantification of three replicate experiments as described in C. (E,F) An LxxIxE motif mediates association of APC/C with B56α. Expression of YFP–B56α WT or an R222E mutant was induced by doxycycline in HeLa FLP-In lines, and mitotic cells were collected. (E) IPs for GFP (for YFP-tagged constructs; lanes 4,5) and control (lane 3) IPs were analyzed by western blotting. Asterisk indicates non-specific band. (F) Quantification of three replicate experiments described in E. Data are mean±s.d. n.s., not significant (P>0.05); *P<0.05, **P< 0.005, ***P<0.0005, ****P<0.00005, Student's two-tailed t-test.
Next, we asked if an LxxIxE docking site mediates PP2AB56 and APC/C association. To this end, we compared the ability of B56α WT and a point mutant (R222E) that abrogates binding to the LxxIxE motif (Hertz et al., 2016) to bind to APC/C. YFP-tagged B56α WT or R222E expression was induced in HeLa cells, and mitotic lysates were used for IP. We confirmed that the R222E mutation abolished BubR1 association but preserved PP2A catalytic subunit binding (Fig. 5E, compare lanes 4 and 5). Importantly, the association with Cdc20, APC4 and APC7 was reduced by the R222E mutation (Fig. 5E, compare lanes 4 and 5, quantified in Fig. 5F), indicating that the region of B56α required for LxxIxE docking is essential for the association between APC/C and PP2AB56. An LxxIxE motif has not been identified in human APC/C subunits, and thus further work will be required to identify the docking site for PP2AB56 on APC/C.
Phosphorylation at Ser92 on Cdc20 controls its association with the APC/C
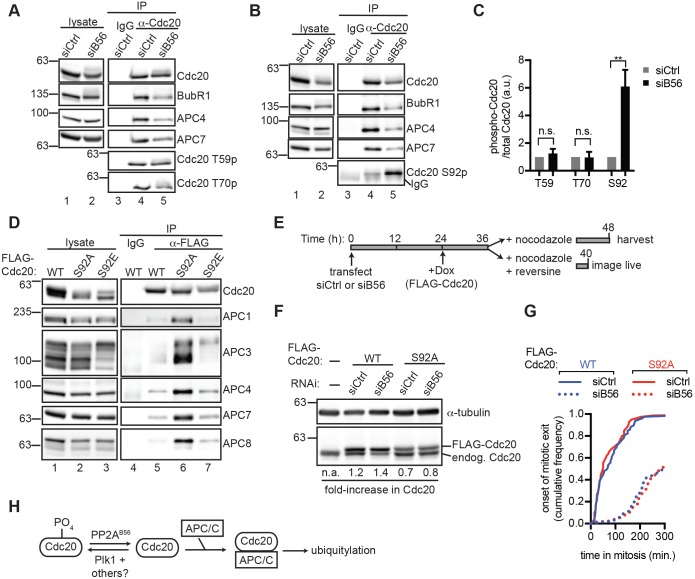
We considered the possibility that increased phosphorylation of Cdc20 in siB56 cells might contribute to the defect in APC/CCdc20 assembly. Indeed, phosphorylation of Cdc20 at sites adjacent to the C-box, a motif essential for APC/C activation (Chang et al., 2014; Kimata et al., 2008), reduces Cdc20 binding to the APC/C (Hein and Nilsson, 2016; Labit et al., 2012). To test this possibility, we utilized antibodies specific to Cdc20 that had been phosphorylated on Thr59 or Thr70 (Hein and Nilsson, 2016). We observed no increase in phosphorylation at either site in Cdc20 IPs from siB56 cells compared to those from siCtrl cells, although in the same experiment, siB56 cells exhibited decreased BubR1, APC4 and APC7 association (Fig. 6A, compare lanes 4 and 5, quantified in Fig. 6C). Thus, reduced APC/C–Cdc20 association in siB56 cells is not due to hyperphosphorylation of Thr59 and Thr70, although we cannot exclude potential contributions from additional phosphorylation sites near the C-box of Cdc20.
Fig. 6.
Residue Ser92 on Cdc20 is a PP2AB56 substrate that influences Cdc20 binding to APC/C. (A,B) siCtrl and siB56 cells were collected by mitotic shake-off. Lysates (lanes 1,2), and control IgG (lane 3) or Cdc20 IPs (lanes 4,5) were analyzed by western blotting for the indicated proteins. (C) Quantification of three replicate experiments described in A,B. Data are mean±s.d. n.s., not significant (P>0.05), **P<0.005, Student's two-tailed t-test. (D) Ser92 is a determinant of Cdc20 binding to APC/C. HeLa cells expressing doxycycline-inducible FLAG–Cdc20 wild type (WT), or the mutants S92A or S92E were collected by mitotic shake-off, lysed and analyzed directly (lanes 1-3) or used in a FLAG (lanes 5-7) or control IgG (lane 4) IP and probed for the indicated proteins by western blotting. (E–G) Cdc20S92A expression does not rescue mitotic exit delays in siB56 cells. (E) Schematic of B56 depletion and FLAG-Cdc20 induction in HeLa cells. (F) Mitotic siCtrl and siB56 cells expressing FLAG–Cdc20 constructs were harvested as outlined in E and analyzed alongside control mitotic HeLa lysates by western blotting. endog, endogenous. (G) Cells were imaged live and duration of mitosis was scored. The cumulative fraction of cells that exited mitosis as a function of time after mitotic entry is plotted. 50 cells were scored per condition. Experiments in D,F,G were performed in triplicate. (H) Schematic of PP2AB56-dependent stimulation of APC/C association with Cdc20.
We confirmed that phosphorylation of Ser92 in Cdc20, a substrate of the kinase Plk1, is dephosphorylated by PP2AB56 (Craney et al., 2016; Jia et al., 2016) (Fig. 6B, compare lanes 4 and 5, quantified in Fig. 6C). Mutations that abolish Ser92 phosphorylation prevent delivery of Ube2S to APC/C and impair APC/C activation in vitro (Craney et al., 2016; Jia et al., 2016). We therefore wanted to know whether Ser92 phosphorylation modulates Cdc20 binding to the APC/C. A non-phosphorylatable Cdc20S92A mutant exhibited increased binding to APC/C subunits compared to Cdc20WT or Cdc20S92E, a mutant that mimics phosphorylation (Fig. 6D, compare lanes 5-7). Finally, we combined B56 depletion with Cdc20 overexpression to determine whether Cdc20S92A rescues the delay in mitotic exit in siB56 cells (Fig. 6E). We determined that FLAG–Cdc20WT was expressed at higher levels than FLAG–Cdc20S92A (Fig. 6F), precluding direct comparisons between the two cell lines. However, in both cell lines, B56 depletion delayed mitotic exit (Fig. 6G), indicating that the presence of Cdc20S92A is insufficient to rescue the mitotic exit delays in siB56 cells. The lack of a rescue could arise from insufficient expression of FLAG–Cdc20, a requirement for the hydroxyl group on Ser92 of Cdc20, and/or contributions from other PP2AB56 substrates towards mitotic exit.
DISCUSSION
PP2AB56 is essential for mitosis, but because its specificity arises through protein–protein interactions rather than through the recognition of a consensus phosphorylation site, predicting its substrates and functions during mitosis remains challenging. Here, we have analyzed the defects associated with mitotic exit delays in siB56 cells. Previous work has established that siB56 cells are delayed in mitotic exit when Mps1 is inactivated in nocodazole-treated cells (Espert et al., 2014; Nijenhuis et al., 2014). While we confirm that B56 depletion causes increased kinetochore recruitment of SAC proteins in Mps1-inhibited cells, our data suggest that such changes may be insufficient to alter the level and/or stability of the MCC. Rather, we find that PP2AB56 is required to stabilize the association of Cdc20 with the APC/C, and in this way, promotes exit from mitosis (Fig. 6H). APC/C subunits associate with PP2AB56 constitutively, and this association is independent of BubR1 binding to PP2AB56. We identify residue Ser92 on Cdc20, a mitosis-specific Plk1 and PP2AB56 substrate (Craney et al., 2016; Jia et al., 2016), as a determinant of APC/CCdc20 assembly.
Substrate selectivity of the PP2AB56 holoenzyme arises, at least in part, from the affinity of B56 subunits for the LxxIxE motif (Hertz et al., 2016). We show that PP2AB56 associates constitutively with APC/C subunits and that this association depends on the ability of B56 subunits to recognize an LxxIxE motif. In mitosis, docking of PP2AB56 to the LxxIxE motif in the KARD of BubR1 localizes PP2AB56 to the kinetochore, stabilizes kinetochore–microtubule attachments (Kruse et al., 2013; Suijkerbuijk et al., 2012; Xu et al., 2013) and stimulates release of BubR1 from the kinetochore after Mps1 inactivation (Espert et al., 2014; Nijenhuis et al., 2014). By contrast, we find that deletion of the BubR1 KARD has no effect on the association of Cdc20 with APC/C or on the timing of mitotic exit, suggesting that the role of PP2AB56 in promoting APC/C–Cdc20 association and exit from mitosis is likely to operate in parallel to its kinetochore-centered functions. Moreover, our findings imply that impaired SAC silencing at the kinetochore cannot account for the delay in mitotic exit in siB56 cells. Rather, we propose that the critical dephosphorylation event(s) mediated by PP2AB56 can proceed in the cytoplasm and converge on APC/CCdc20 assembly and function.
Our results do not preclude a role for the SAC or kinetochore targeting of other factors in the production of the preceding phosphorylation events that limit APC/CCdc20 assembly. Indeed, Plk1, which phosphorylates Ser92 on Cdc20 (Craney et al., 2016; Jia et al., 2016), cooperates with Mps1 to activate the SAC at unattached kinetochores (Espeut et al., 2015; von Schubert et al., 2015). Moreover, both Cdc20 and the APC/C localize to unattached kinetochores (Acquaviva et al., 2004; Kallio et al., 2002). Therefore, it is possible that the relevant phosphorylation events that reduce Cdc20 binding to the APC/C, which likely include Plk1-dependent phosphorylation of Cdc20 at Ser92, could be generated at unattached kinetochores through SAC signaling. Moreover, we have previously shown that Plk1 targeting to the kinetochore is increased in siB56 cells (Foley et al., 2011). Thus, it is possible that the increase in phosphorylation at Ser92 that we observe in siB56 cells arises from both increased Plk1 levels at the kinetochore and decreased PP2AB56 levels in the cytoplasm.
B56 subunits target PP2A phosphatase activity to substrates to stimulate exit from mitosis in multiple ways, including through APC/CCdc20 assembly, as we show here, as well as by E2 ubiquitin-conjugating enzyme recruitment to the APC/C (Craney et al., 2016) and through SAC signaling at the kinetochore (Espert et al., 2014; Nijenhuis et al., 2014). A key challenge will be to discern how these roles intersect synergistically to exert temporal control over chromosome segregation alongside other mitotic functions of PP2AB56, which include stabilization of kinetochore–microtubule attachment (Foley et al., 2011) and preservation of sister chromatid cohesion (Chen et al., 2007; Kitajima et al., 2006; Riedel et al., 2006; Tang et al., 2006). Importantly, the assembly of PP2AB56 holoenzymes is predicted to be disrupted by recurrent cancer-associated heterozygous missense mutations in the scaffold subunit of PP2A (Cho and Xu, 2007; Xu et al., 2006). These mutations are associated with tumors that undergo whole genome duplication (Zack et al., 2013), and it will be important to identify if any of the mitotic functions of PP2AB56 are sensitive to these changes.
MATERIALS AND METHODS
Cell culture, transfection and inhibitor treatments
Cells were grown at 37°C in a humidified atmosphere with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (HeLa, AmphoPack-293) or 1:1 DMEM:F12 (RPE1) supplemented with 10% fetal bovine serum, penicillin-streptomycin, non-essential amino acids and L-glutamine. RPE1 and HeLa cell lines were authenticated annually by short tandem repeat analysis and tested for mycoplasma contamination every 6 months. For immunofluorescence microscopy, cells were grown on no. 1.5 glass coverslips (Thermo Fisher Scientific) coated with poly-D-lysine (Sigma). For live imaging, cells were grown in glass-bottom dishes with no. 1.5 cover glass (Cellvis, Mountain View, CA). RPE1 cells lines stably expressing H2B–mCherry were generated as described previously (Foley et al., 2011). RNAi-resistant LAP–BubR1 WT and ΔKARD, and RNAi-resistant B56ε were introduced into HeLa FLP-In cells by site-specific recombination. The following chemicals were dissolved in DMSO and used at the indicated concentrations: nocodazole (3.3 μM, except in Fig. 4I,J, used at 0.8 μM, Sigma), MG132 (10 μM, Selleck Biochemicals, Houston, TX), reversine (1 μM, except Fig. 4I,J, used at 0.5 μM, Cayman Chemical, Ann Arbor, MI), RO-3306 (10 μM, Sigma), okadaic acid (1 μM, Santa Cruz Biotechnology, Dallas, TX) and doxycycline (1 μg/ml, Thermo Fisher Scientific).
For siRNA transfection, Lipofectamine RNAi Max (Thermo Fisher Scientific) was used following the manufacturer's reverse transfection protocol. A non-targeting siRNA (GE Life Sciences, Marlborough, MA, cat. # D-001810-01, sequence 5ʹ-UGGUUUACAUGUCGACUAAUU-3ʹ) was used for control transfections. The siB56 mix was prepared as described for ‘pool two’ previously (Foley et al., 2011) omitting B56β siRNA, except as indicated in Fig. 1A-D. For microscopy analyses, 1.8×105 RPE1 cells were transfected with 100 pmol siRNA and 5.0 µl Lipofectamine RNAi Max. For HeLa cells, the transfection reagents were halved. For biochemical experiments, 5×106 RPE1 cells were transfected with 1600 pmol siRNA and 80 µl Lipofectamine RNAi Max. For siRNA treatment for biochemical analyses in HeLa cells, 5×106 cells were transfected with 800 pmol siRNA and 40 µl Lipofectamine RNAi Max. Cells were imaged, fixed or harvested at 42-48 h post transfection. Mitotic cells were collected by manual detachment following 14-16 h of incubation in nocodazole. The mitotic index was enumerated by scoring individual cells for the presence or absence of mitotic chromatin (condensed chromosomes) and was typically >90% in siCtrl and siB56 samples. BubR1 depletion and rescue experiments were performed as described previously (Suijkerbuijk et al., 2012). RNAi-resistant-B56α-expressing RPE1 cells (Foley et al., 2011), CCNA2Venus/+ and CCNB1Venus/+ RPE1 cells (Collin et al., 2013), and HeLa cells expressing FLAG–Cdc20 wild type or the S92A and S92E mutants (Jia et al., 2016) have been described previously.
Plasmids
An RNAi-resistant B56ε expression construct was generated using site-directed mutagenesis of a PPP2R5E Gateway entry clone (Foley et al., 2011).
Antibodies
All antibodies were used at 1 µg/ml for immunofluorescence, 0.1 µg/ml for western blotting, and 2-4 µg of IgG was used per immunoprecipitation unless otherwise specified. Antibodies were generated using peptide sequences from APC4, BubR1 and B56α. Peptides were synthesized by EZBiolab, Carmel, IN (APC4: CEIVIKVEKLDPELDS; BubR1: CVKKEGGALSEAMSLE) or by Tufts University, Medford, MA (B56α: CKKALEKQNSAYNMHSILSNTSAE) to 90% purity. 4 mg of peptide was coupled to maleimide-activated keyhole limpet hemocyanin (Thermo Fisher Scientific). Custom antibodies were raised against recombinant GST–GFP. Antigens were injected into New Zealand White rabbits using an institutionally approved protocol and animal-care facility (Pocono Rabbit Farm and Laboratory, Canadensis, PA). For peptide-reactive antibodies, sera in 1× PBS were loaded onto sulfo-link columns (Thermo Fisher Scientific) conjugated to the immunizing peptide. For antibodies against GFP, serum was loaded onto a HiTrap NHS-activated high-pressure column (GE Life Sciences) coupled to GST–GFP. After washing with PBS, antibodies were eluted with 0.2 M glycine, pH 2.5 and subsequently neutralized with Tris-HCl pH 8.0, followed by dialysis into PBS. A custom antibody against B56ε (peptide sequence CRGLRRDGIIPT) was generated by Bethyl Laboratories, Montgomery, TX. To validate custom antibodies for use in IP, we confirmed that pre-incubation of the antibody with the immunizing peptide blocked IP of the target protein. To validate the anti-GFP antibody for western blotting, we confirmed that the antibody recognized recombinant GFP protein and further confirmed its ability to detect induction of GFP- or YFP-tagged proteins in human cells. For antibodies against B56α and B56ε, which were also used for western blotting, we confirmed that the signal attributed to the target protein was reduced by gene-specific RNAi.
Commercial antibodies used in this study include those against: α-tubulin (mouse DM1α–FITC conjugate; Sigma, F2168); β-actin (Sigma, A5316); APC1 (Bethyl Laboratories, A301-653A); APC3 (BD Biosciences, 610455, western blot only); APC7 (Bethyl Laboratories, A302-551A); APC8 (Bethyl Laboratories, A301-182A); BubR1 (BD Transduction Laboratories, 612503; immunofluorescence and western blot only); PP2A catalytic subunit (BD Transduction Laboratories, 610555); PP2A scaffold (Santa Cruz Biotechnology, sc-6112); Mad2 (Bethyl Laboratories, A300-301A); B56γ (Bethyl Laboratories, A303-814A); B56δ (Bethyl Laboratories, A301-098A); and human CREST antiserum (Immunovision, Springsdale, AR, HCT-0100; 1:5000). Antibodies against APC3 (used for IP only). APC4 (used for western blot only) and Cdc20 phosphorylated at Thr59 and Thr70 were gifts from Jakob Nilsson (University of Copenhagen, Denmark) (Hein and Nilsson, 2016). The antibody against Cdc20 phosphorylated at Ser92 was a gift from Hongtao Yu (University of Texas Southwestern Medical Center, Dallas, TX) (Jia et al., 2016).
Immunofluorescence microscopy
Cells on coverslips were fixed and permeabilized by a 5 min incubation at 37°C in 20 mM PIPES, pH 6.8, 10 mM EGTA, 1 mM MgCl2, 4% paraformaldehyde and 0.2% Triton X-100. Coverslips were then washed with TBS+0.1% Triton X-100 and blocked in 2% donkey serum (Jackson Immunoresearch Laboratories, West Grove, PA) for 30 min and stained. After washing in TBS+0.1% Triton X-100, samples were stained with fluorescence-conjugated secondary antibodies (Jackson Immunoresearch Laboratories). Samples were mounted onto slides in Prolong Gold with DAPI (Thermo Fisher Scientific) and sealed with nail polish. Samples were imaged on DeltaVision Image Restoration System (GE Healthcare, Chicago, IL) based on an Olympus IX-70 microscope with a 100×1.4 NA oil objective and a CoolSnap QE cooled CCD camera (Photometrics, Tucson, Arizona). z-stacks were acquired with 0.2 μm spacing. Image projections, crops and measurements were performed in Fiji (Schindelin et al., 2012). Kinetochore regions were defined by a CREST antibody signal, and corresponding regions in other channels were used to calculate the average kinetochore intensity signal per plane. Local background was defined by the DNA region minus the kinetochore region and was subtracted from the kinetochore signal. For each cell, the kinetochore signal was averaged over all z-slices.
Live-cell imaging
Cells in glass-bottom dishes were mounted on a Nikon Eclipse TiE/B microscope operated through NIS Elements software (Nikon, Melville, NY), equipped with 20×0.75 NA and 40×1.3 NA air objectives, a piezo stage, Perfect Focus, a spinning disk confocal head (Yokogawa, Sugar Land, TX) with 488 nm laser line and EMCCD and sCMOS cameras. Cells were maintained in an environmental enclosure with temperature-controlled stage with 5% CO2 delivery (Tokai Hit, Fujinomiya, Japan). For mitotic timing measurements, cells were imaged at 20× by differential interference contrast (DIC) imaging, and/or by fluorescence imaging. A minimum of 30 cells per condition was analyzed in each experiment. For cyclin-A2–Venus imaging, cells in nocodazole were imaged at 20× magnification by confocal microscopy. Two z-planes were taken with 5 μm spacing every 4 min. For cyclin-B1–Venus imaging, cells in nocodazole were visualized at 40× magnification by confocal microscopy and DIC before and after reversine addition. Eight to ten cells per condition were analyzed per experiment. The non-cellular background fluorescence signal was subtracted and the mean fluorescence intensity of a 4×4 μm region was measured. Values were normalized to the fluorescence intensity of the first image after nuclear envelope breakdown or the image taken before reversine addition.
Cell lysis and immunoprecipitation
Lysates were prepared by suspension in buffer B (30 mM HEPES, pH 7.8, 140 mM NaCl, 6 mM MgCl2, 5% glycerol) at 4°C supplemented with 2 mM DTT, 1% ProBlock Gold Mammalian Protease Inhibitor Cocktail (GoldBio Technology, St. Louis, MO) and 1× PhosSTOP (Roche, Indianapolis, IN) followed by nitrogen cavitation (Parr Instruments, Moline, IL) for 5 min at 2000 psi on ice, and centrifugation at 20,000 g for 15 min. Proteins were immunoprecipitated with antibody bound to Protein A or Protein G Dynabeads (Thermo Fisher Scientific) for 45-60 min at 4°C, and washed three times with buffer B at 4°C. In Fig. 6F, cells were suspended in 50 mM HEPES, 100 mM NaCl, 2 mM DTT, 0.01% Brij 35, 1 mM MnCl2 and lysates were treated with λ protein phosphatase (New England BioLabs, Ipswich, MA) for 30 min at 30°C. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose for western blotting via chemiluminescence using an ImageQuant LAS500 (GE Life Sciences). Intensity measurements from western blots were performed in Fiji. For presentation of input data, the input lanes were cropped and scaled to the minimum and maximum pixel intensity in the input samples using Fiji. The IP lanes were processed in the same manner. Thus, input and IP samples are scaled differently. To indicate this, the input and IP portions of the blot are separated in the figure panels. Cropped and scaled images were imported into Illustrator (Adobe, San Jose, CA) for figure assembly.
Acknowledgements
We thank Prasad Jallepalli for scientific discussion and critical reading of the manuscript and Geert Kops, Jakob Nilsson, Jonathon Pines and Hongtao Yu for reagents.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.A.F.; Formal analysis: E.A.F., V.R.-B., H.K., S.D.; Investigation: E.A.F., S.J.L., V.R.-B., H.K., S.D.; Writing - original draft: E.A.F.; Writing - review & editing: E.A.F.; Visualization: E.A.F.; Supervision: E.A.F.; Funding acquisition: E.A.F.
Funding
This work was supported by a Gerstner Young Investigator Award to E.A.F. the Geoffrey Beene Cancer Research Center of Memorial Sloan Kettering Cancer Center, and the Functional Genomics Initiative at Memorial Sloan Kettering Cancer Center. We acknowledge the Memorial Sloan Kettering Cancer Center Support Grant (P30 CA008748) from the National Cancer Institute. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.201608.supplemental
References
- Acquaviva C., Herzog F., Kraft C. and Pines J. (2004). The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat. Cell Biol. 6, 892-898. 10.1038/ncb1167 [DOI] [PubMed] [Google Scholar]
- Alfieri C., Chang L., Zhang Z., Yang J., Maslen S., Skehel M. and Barford D. (2016). Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature 536, 431-436. 10.1038/nature19083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R. N., Cundell M. J. and Barr F. A. (2014). KIF4A and PP2A-B56 form a spatially restricted feedback loop opposing Aurora B at the anaphase central spindle. J. Cell Biol. 207, 683-693. 10.1083/jcb.201409129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Zhang Z., Yang J., McLaughlin S. H. and Barford D. (2014). Molecular architecture and mechanism of the anaphase-promoting complex. Nature 513, 388-393. 10.1038/nature13543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W. C. H., Kulkarni K., Zhang Z., Kong E. H. and Barford D. (2012). Structure of the mitotic checkpoint complex. Nature 484, 208-213. 10.1038/nature10896 [DOI] [PubMed] [Google Scholar]
- Chen F., Archambault V., Kar A., Lio P., D'Avino P. P., Sinka R., Lilley K., Laue E. D., Deak P., Capalbo L. et al. (2007). Multiple protein phosphatases are required for mitosis in Drosophila. Curr. Biol. 17, 293-303. 10.1016/j.cub.2007.01.068 [DOI] [PubMed] [Google Scholar]
- Cho U. S. and Xu W. (2007). Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445, 53-57. 10.1038/nature05351 [DOI] [PubMed] [Google Scholar]
- Chung E. and Chen R.-H. (2003). Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat. Cell Biol. 5, 748-753. 10.1038/ncb1022 [DOI] [PubMed] [Google Scholar]
- Collin P., Nashchekina O., Walker R. and Pines J. (2013). The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat. Cell Biol. 15, 1378-1385. 10.1038/ncb2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craney A., Kelly A., Jia L., Fedrigo I., Yu H. and Rape M. (2016). Control of APC/C-dependent ubiquitin chain elongation by reversible phosphorylation. Proc. Natl. Acad. Sci. USA 113, 1540-1545. 10.1073/pnas.1522423113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V., Mari C., Nocera D., Rametti L. and Grieco D. (2003). The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev. 17, 2520-2525. 10.1101/gad.267603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B. and Pines J. (2010). How cyclin A destruction escapes the spindle assembly checkpoint. J. Cell Biol. 190, 501-509. 10.1083/jcb.201001083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick A. E. and Gerlich D. W. (2013). Kinetic framework of spindle assembly checkpoint signalling. Nat. Cell Biol. 15, 1370-1377. 10.1038/ncb2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert A., Uluocak P., Bastos R. N., Mangat D., Graab P. and Gruneberg U. (2014). PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J. Cell Biol. 206, 833-842. 10.1083/jcb.201406109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J., Lara-Gonzalez P., Sassine M., Shiau A. K., Desai A. and Abrieu A. (2015). Natural loss of Mps1 kinase in nematodes uncovers a role for polo-like kinase 1 in spindle checkpoint initiation. Cell Rep. 12, 58-65. 10.1016/j.celrep.2015.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E. A. and Kapoor T. M. (2013). Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14, 25-37. 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E. A., Maldonado M. and Kapoor T. M. (2011). Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 13, 1265-1271. 10.1038/ncb2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K., Grimaldi M. and Yamano H. (2016). Cyclin-dependent kinase 1-dependent activation of APC/C ubiquitin ligase. Science 352, 1121-1124. 10.1126/science.aad3925 [DOI] [PubMed] [Google Scholar]
- Hein J. B. and Nilsson J. (2016). Interphase APC/C-Cdc20 inhibition by cyclin A2-Cdk2 ensures efficient mitotic entry. Nat. Commun. 7, 10975 10.1038/ncomms10975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz E. P. T., Kruse T., Davey N. E., López-Méndez B., Sigurðsson J. O., Montoya G., Olsen J. V. and Nilsson J. (2016). A conserved motif provides binding specificity to the PP2A-B56 phosphatase. Mol. Cell 63, 686-695. 10.1016/j.molcel.2016.06.024 [DOI] [PubMed] [Google Scholar]
- Izawa D. and Pines J. (2015). The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature 517, 631-634. 10.1038/nature13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Li B. and Yu H. (2016). The Bub1–Plk1 kinase complex promotes spindle checkpoint signalling through Cdc20 phosphorylation. Nat. Commun. 7, 10818 10.1038/ncomms10818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M. J., Beardmore V. A., Weinstein J. and Gorbsky G. J. (2002). Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 158, 841-847. 10.1083/jcb.200201135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A., Wickliffe K. E., Song L., Fedrigo I. and Rape M. (2014). Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Mol. Cell 56, 232-245. 10.1016/j.molcel.2014.09.010 [DOI] [PubMed] [Google Scholar]
- Kimata Y., Baxter J. E., Fry A. M. and Yamano H. (2008). A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol. Cell 32, 576-583. 10.1016/j.molcel.2008.09.023 [DOI] [PubMed] [Google Scholar]
- Kitajima T. S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S. A. and Watanabe Y. (2006). Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441, 46-52. 10.1038/nature04663 [DOI] [PubMed] [Google Scholar]
- Kruse T., Zhang G., Larsen M. S. Y., Lischetti T., Streicher W., Kragh Nielsen T., Bjørn S. P. and Nilsson J. (2013). Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 126, 1086-1092. 10.1242/jcs.122481 [DOI] [PubMed] [Google Scholar]
- Labit H., Fujimitsu K., Bayin N. S., Takaki T., Gannon J. and Yamano H. (2012). Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J. 31, 3351-3362. 10.1038/emboj.2012.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N. and Biggins S. (2014). Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 28, 140-152. 10.1101/gad.233700.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows J. C., Shepperd L. A., Vanoosthuyse V., Lancaster T. C., Sochaj A. M., Buttrick G. J., Hardwick K. G. and Millar J. B. A. (2011). Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell 20, 739-750. 10.1016/j.devcel.2011.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijenhuis W., Vallardi G., Teixeira A., Kops G. J. P. L. and Saurin A. T. (2014). Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat. Cell Biol. 16, 1257-1264. 10.1038/ncb3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A. et al. (2010). Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Pines J. (2011). Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 12, 427-438. 10.1038/nrm3132 [DOI] [PubMed] [Google Scholar]
- Qiao R., Weissmann F., Yamaguchi M., Brown N. G., VanderLinden R., Imre R., Jarvis M. A., Brunner M. R., Davidson I. F., Litos G. et al. (2016). Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 113, E2570-E2578. 10.1073/pnas.1604929113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S. K., Rape M., Margansky W. A. and Kirschner M. W. (2007). Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446, 921-925. 10.1038/nature05734 [DOI] [PubMed] [Google Scholar]
- Riedel C. G., Katis V. L., Katou Y., Mori S., Itoh T., Helmhart W., Gálová M., Petronczki M., Gregan J., Cetin B. et al. (2006). Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441, 53-61. 10.1038/nature04664 [DOI] [PubMed] [Google Scholar]
- Rosenberg J. S., Cross F. R. and Funabiki H. (2011). KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21, 942-947. 10.1016/j.cub.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D'Alise A. M., Taylor S. S. and Musacchio A. (2010). Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190, 73-87. 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M. H. A., Held M., Janssens V., Hutchins J. R. A., Hudecz O., Ivanova E., Goris J., Trinkle-Mulcahy L., Lamond A. I., Poser I. et al. (2010). Live-cell imaging RNAi screen identifies PP2A–B55α and importin-β1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12, 886-893. 10.1038/ncb2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Neutzner M., Möcker D. and Seufert W. (2001). Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 20, 5165-5175. 10.1093/emboj/20.18.5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S. and Gorbsky G. J. (2015). Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 16, 82-94. 10.1038/nrm3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Rape M., Draviam V. M., Nalepa G., Sowa M. E., Ang X. L., McDonald E. R. III, Li M. Z., Hannon G. J., Sorger P. K. et al. (2007). Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 446, 876-881. 10.1038/nature05694 [DOI] [PubMed] [Google Scholar]
- Sudakin V., Chan G. K. T. and Yen T. J. (2001). Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154, 925-936. 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk S. J. E., Vleugel M., Teixeira A. and Kops G. J. (2012). Integration of Kinase and Phosphatase Activities by BUBR1 Ensures Formation of Stable Kinetochore-Microtubule Attachments. Dev. Cell 23, 745-755. 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Swingle M., Ni L. and Honkanen R. E. (2007). Small-molecule inhibitors of ser/thr protein phosphatases: specificity, use and common forms of abuse. Methods Mol. Biol. 365, 23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Shu H., Oncel D., Chen S. and Yu H. (2004). Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell 16, 387-397. 10.1016/j.molcel.2004.09.031 [DOI] [PubMed] [Google Scholar]
- Tang Z., Shu H., Qi W., Mahmood N. A., Mumby M. C. and Yu H. (2006). PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev. Cell 10, 575-585. 10.1016/j.devcel.2006.03.010 [DOI] [PubMed] [Google Scholar]
- Varetti G., Guida C., Santaguida S., Chiroli E. and Musacchio A. (2011). Homeostatic control of mitotic arrest. Mol. Cell 44, 710-720. 10.1016/j.molcel.2011.11.014 [DOI] [PubMed] [Google Scholar]
- Vassilev L. T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D. C. and Chen L. (2006). Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA 103, 10660-10665. 10.1073/pnas.0600447103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schubert C., Cubizolles F., Bracher J. M., Sliedrecht T., Kops G. J. P. L. and Nigg E. A. (2015). Plk1 and Mps1 cooperatively regulate the spindle assembly checkpoint in human cells. Cell Rep. 12, 66-78. 10.1016/j.celrep.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Xu Y., Xing Y., Chen Y., Chao Y., Lin Z., Fan E., Yu J. W., Strack S., Jeffrey P. D. and Shi Y. (2006). Structure of the protein phosphatase 2A holoenzyme. Cell 127, 1239-1251. 10.1016/j.cell.2006.11.033 [DOI] [PubMed] [Google Scholar]
- Xu P., Raetz E. A., Kitagawa M., Virshup D. M. and Lee S. H. (2013). BUBR1 recruits PP2A via the B56 family of targeting subunits to promote chromosome congression. Biol. Open 2, 479-486. 10.1242/bio.20134051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., VanderLinden R., Weissmann F., Qiao R., Dube P., Brown N. G., Haselbach D., Zhang W., Sidhu S. S., Peters J.-M. et al. (2016). Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol. Cell 63, 593-607. 10.1016/j.molcel.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky Y., Shteinberg M., Listovsky T., Brandeis M. and Hershko A. (2000). Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem. Biophys. Res. Commun. 271, 299-304. 10.1006/bbrc.2000.2622 [DOI] [PubMed] [Google Scholar]
- Zack T. I., Schumacher S. E., Carter S. L., Cherniack A. D., Saksena G., Tabak B., Lawrence M. S., Zhang C.-Z., Wala J., Mermel C. H. et al. (2013). Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134-1140. 10.1038/ng.2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Chang L., Alfieri C., Zhang Z., Yang J., Maslen S., Skehel M. and Barford D. (2016). Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature 533, 260-264. 10.1038/nature17973 [DOI] [PMC free article] [PubMed] [Google Scholar]