 Design strategies, response modes and bioapplications of spectroscopic probes are briefly reviewed, which may have important guiding significance for readers.
Design strategies, response modes and bioapplications of spectroscopic probes are briefly reviewed, which may have important guiding significance for readers.
Abstract
Spectroscopic (chromogenic, fluorescent, or chemiluminescent) probes have been widely used in many fields due to their high sensitivity and unrivaled spatiotemporal resolution. This area is an old one but always full of activity, because the rapid development of science and technology requires not only new probes for specific purposes (e.g., subcellular imaging) but also the update of current probes with more satisfactory properties. Based on our experiences and including existing knowledge, in this mini-review we briefly discuss the design strategies, response modes, and bioapplications of small molecular spectroscopic probes, in particular their advantages and disadvantages as well as possible research trends, which may be helpful to those who are interested in this continually growing research area.
1. Introduction
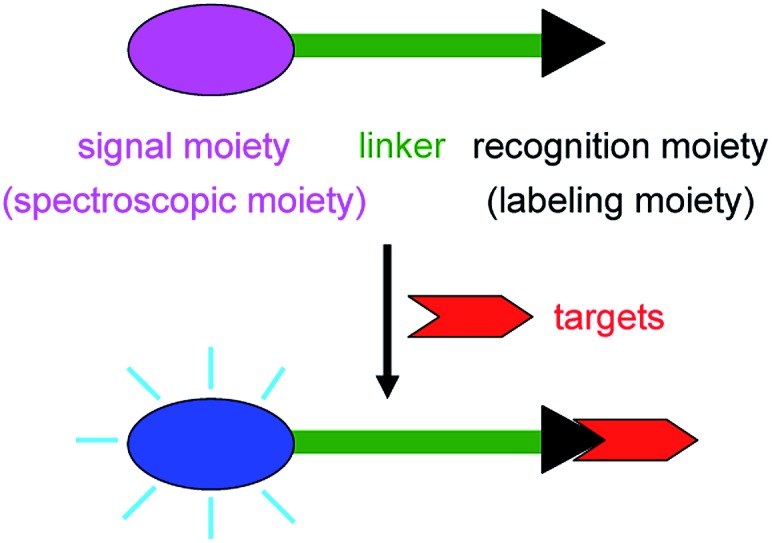
Spectroscopic probes can be defined as the reagents that interact with analytes or environmental factors, accompanying the changes of their spectroscopic (absorption, fluorescent, or chemiluminescent) properties; based on such changes the targets can thus be determined.1 Usually, as shown in Fig. 1, spectroscopic probes are composed of three moieties: (1) a signal or spectroscopic moiety, whose change in spectroscopic property should be as large as possible upon reaction with the target of interest; (2) a recognition or labeling moiety responsible for the selective reaction with the target; and (3) an appropriate linker to connect the two aforementioned moieties (in some cases the two moieties are directly integrated without any linker).1 They have been extensively investigated and widely used in many fields because of their high sensitivity and unrivaled spatiotemporal resolution.2–20 Sometimes a molecular event may be visualized directly by the naked eye (for example, the color change of the KI-starch test paper from white to blue reflects the occurrence of the molecular event of I– oxidized to I2 by an oxidant). Spectroscopic probes are an old area but always full of activity, because the rapid development of science and technology requires not only new probes for specific purposes (e.g., subcellular imaging) but also the update of current probes with more satisfactory properties. In this mini-review we will briefly discuss the design strategies, response modes, and bioapplications of spectroscopic probes, including their current research status, problems, and possible trends, which may be helpful to those who are interested in this continually growing research area.
Fig. 1. Structure and response mechanism of a spectroscopic probe.
It should be pointed out that a comprehensive review on each of the above respects would make the length of the mini-review too long, which is not our aim; moreover, such a kind of comprehensive review has been reported quite a lot already.2–20 Differently from the previous publications, in this mini-review we will only summarize and discuss the advantages and disadvantages (including the existing problems) of spectroscopic probes along the following key links: design strategies, response modes, and bioapplications. To the best of our knowledge, a short but comprehensive commentary and summary from this point of view has not been reported in the literature. Herein, we will make such an attempt according to our experiences, including existing knowledge, which may have important guiding significance for readers.
2. Design strategies
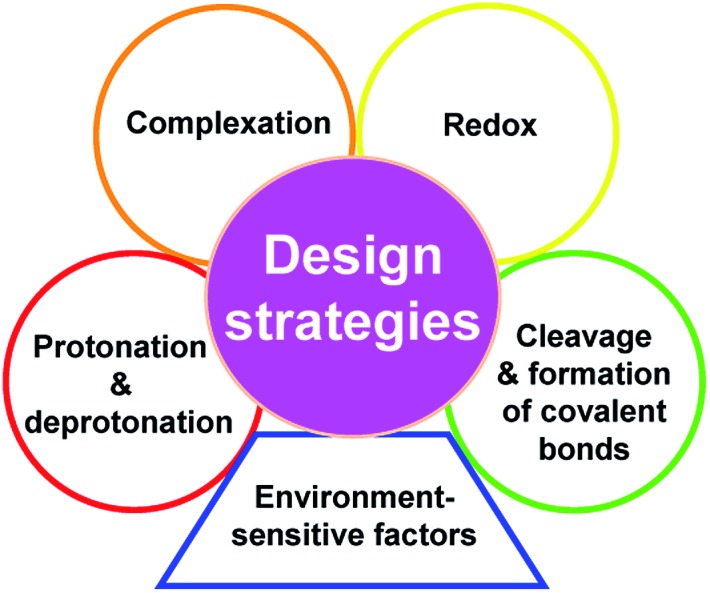
According to the different purposes of detection, there are usually two kinds of strategies available for designing a spectroscopic probe. The former is mainly based on four different chemical reactions, which include: (1) protonation–deprotonation, (2) complexation, (3) redox reaction, and (4) cleavage and formation of covalent bonds; the latter is based on a suitable physical environmental factor, which can be polarity, viscosity, temperature, etc. The design strategies are broad, but they are helpful for the reader to understand the essence of spectroscopic probes on the whole (Fig. 2).
Fig. 2. Design strategies available for a spectroscopic probe.
2.1. Protonation/deprotonation-based spectroscopic probes
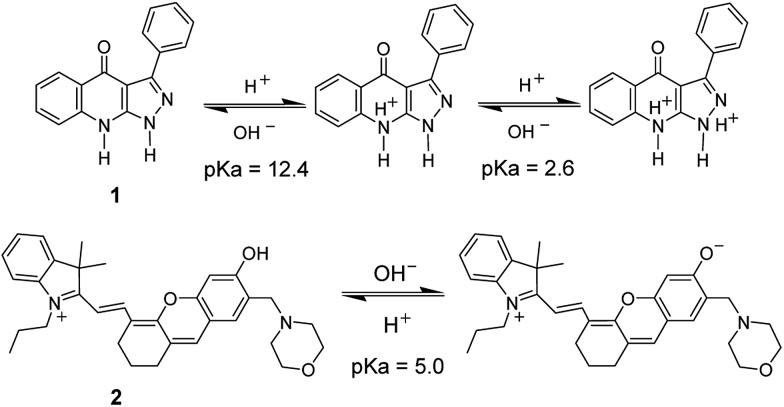
Protonation/deprotonation reactions are mainly used to design spectroscopic pH probes based on H+-sensitive groups such as –COOH, –OH, and amino groups, whose protonation/deprotonation can change the charge distribution in the whole molecule, thereby causing spectroscopic alteration (Fig. 3).21,22 Generally, pH probes can achieve accurate pH determination over a range of only about two pH units, that is, pK a ± 1, which, however, can meet the requirement for normal biosystems because of their rather small pH variation.2 In some cases, pH probes with a wide pH response range are needed, and an effective strategy to design such a pH probe is to assemble several proton acceptors (e.g., N and/or O) in the different positions of a conjugated molecule to form multiple H+ binding sites.
Fig. 3. Structures of two pH probes (1 and 2) and their responses to H+ via protonation/deprotonation.
For pH probes, the elimination of interferences from commonly coexisting metal ions is a challenging subject, because the typically used pH-sensitive groups, such as –COOH, –OH, and an amino group, also have a strong affinity for them, which may result in complexation and nonspecific response of the probe to H+. To overcome this issue and raise the selectivity, the arrangement of proton acceptors in the designed pH probe should not provide a suitable cavity or a convenient formation of five- and six-membered ring complexes for metal ions (e.g., probe 1 in Fig. 3).21a
2.2. Complexation-based spectroscopic probes
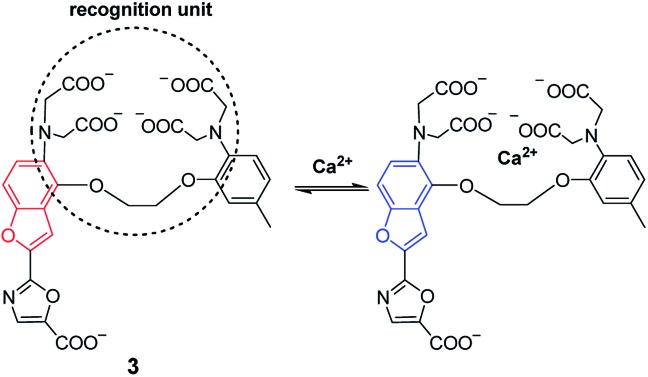
Complexation reactions are mainly utilized to design spectroscopic probes for metal ions, whose response mechanisms may be explained in terms of a suitable photophysical process like internal charge transfer (e.g., probe 3 (ref. 2 and 23) in Fig. 4), photoinduced electron transfer, or fluorescence resonance energy transfer. For detailed information about the photophysical processes, the reader is directed to some comprehensive reviews on the subject.2,20 However, it should be mentioned that these photophysical processes depend on the energy levels of molecular orbitals, which are often difficult to determine accurately. This may lead to the poor prediction of analytical performance of a designed probe, and sometimes previous experience still plays an important role.
Fig. 4. Complexation-based Ca2+ probe (3).
The complexation-based probes usually show rapid spectroscopic response and good reversibility, and are well suited for monitoring dynamic changes in the analyte concentration.
Because many metal ions have similar coordinating ability and may interfere with each other (e.g., Mg2+ vs. Ca2+, and Na+ vs. K+), how to construct a specific recognition unit for a given analyte is the key issue for designing complexation-based spectroscopic probes. Common strategies available for this purpose include: (a) a matchable ring/cavity size for a given ion (e.g., crown ethers); (b) suitable ligands for the convenient formation of five- and six-membered ring complexes with metal ions, like the probes with an EGTA (ethylene glycol tetraacetic acid) moiety for Ca2+; and (c) the hard–soft acid–base principle, such as the soft base of a sulfur atom with a high affinity for the soft acid of Hg2+ ions.2,24
Besides, it should be noted that complexation-based probes often suffer from the influence of pH because the coordinating ability of the Lewis base donors in the recognition unit depends largely on their protonation states. This is why the pH influence must be examined and a pH buffer is usually needed in detection systems. Otherwise, in the selectivity study it is difficult to make sure that the change in spectroscopic signal is induced by a potential interfering species, or by the pH change that results from the added species.
2.3. Redox-based spectroscopic probes
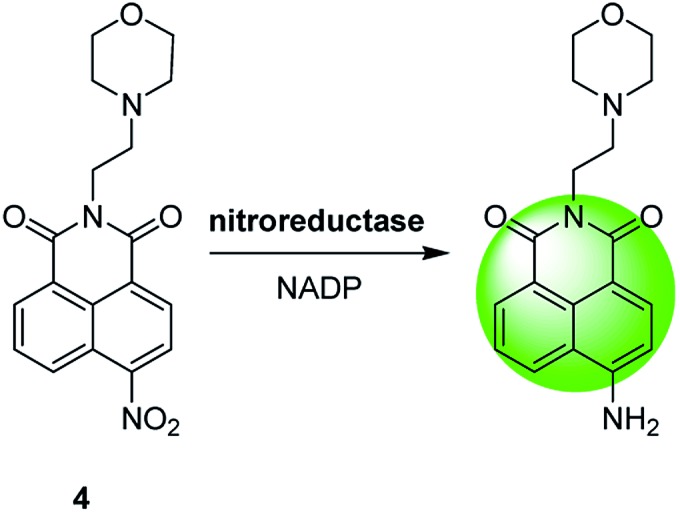
Redox reactions are mainly used to develop spectroscopic probes for analytes with redox properties, such as reactive oxygen species (ROS) and oxidoreductases (these kind of enzymes usually utilize nicotinamide adenine dinucleotide phosphate, NADP, as a cofactor).2,25,26 The response mechanism of these probes is based on the following fact: redox reactions may lead to the inhibition of a photophysical process (e.g., photoinduced electron transfer) or the conversion between a non-fluorescent molecule and a fluorescent one. Fig. 5 shows a typical oxidoreductase probe (4), in which the redox reaction with nitroreductase causes a pull–push conversion from the electron-deficient nitro group (a common quencher) to the electron-rich amino one, thereby generating a selective fluorescence off–on response.25
Fig. 5. Redox-based spectroscopic probe (4) for lysosomal nitroreductase.
Most ROS have a rather short lifetime and extremely low basal level in biosystems. Moreover, some ROS possess similar strong oxidability, and can produce significant cross-interference. Therefore, developing the probes with fast trapping ability, high sensitivity and selectivity for ROS is a great challenge. Specific cleavage of chemical bonds by ROS has led to the successful design of some spectroscopic probes, but probes with excellent analytical performance are still rare for ROS assays. Similarly, there is a serious lack of ultrasensitive detection of many oxidoreductases, because they usually exist at trace levels in biosystems.
2.4. Spectroscopic probes based on the cleavage and formation of covalent bonds
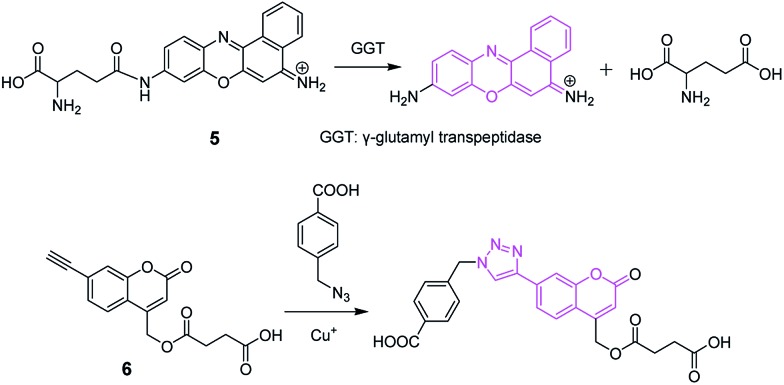
The cleavage or formation of covalent bonds is used to design spectroscopic probes of various substances (including inorganic, organic or biological substances; e.g., Fig. 6).27,28 Between them, the cleavage of covalent bonds has aroused much interest during the past decade. This type of probe may be designed through the following strategy: a fluorochrome is attached to a quencher via a cleavable active bond by virtue of various chemical reactions such as protection–deprotection. The resulting probes often have no or weak fluorescence; upon reaction with an analyte, the active bond in the probe is cleaved and the fluorochrome is released, causing the recovery of fluorescence. Probe 5 (Fig. 6) is such an example, which has been used to image the change of endogenous γ-glutamyl transpeptidase in cells regulated by a potential anticancer drug of sodium butyrate.27 The formation of covalent bonds has long been employed in the pre- and post-column derivatizations in chromatography as well as the labeling of biomolecules, but now is mainly utilized to construct the probes for the selective detection of analytes without chromatographic separation, as exemplified by a Cu+ probe (6).28
Fig. 6. Spectroscopic probes (5 and 6) based on the cleavage and formation of covalent bonds.
Probes that are based on the cleavage and formation of covalent bonds usually have higher selectivity and signal/background ratio, when compared to complexation-based probes, which provides an alternate route to the specific determination of analytes. The disadvantages of this kind of probes are their relatively slow reaction rate and irreversibility in most cases,29 which make them unsuitable for dynamic detection of analyte change.
2.5. Environment-sensitive spectroscopic probes
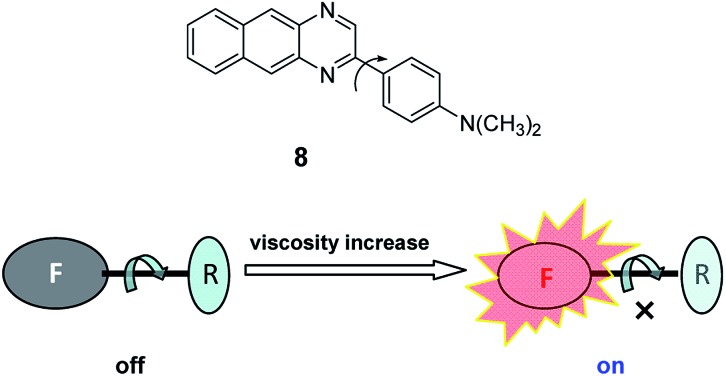
Physical environmental factors, such as polarity, viscosity, and temperature, are also often employed to develop spectroscopic probes with environment-sensitive features.30 If a fluorochrome has a larger dipole moment in the excited state than in the ground state, its energy in the excited state may be lowered via the reorganization of nearby solvent molecules. The higher the polarity of the solvent, the lower the energy of the excited state and the larger the red-shift of the emission spectrum. Neutral red belongs to such a fluorochrome, and thus can be used to construct a polarity-sensitive spectroscopic probe for detecting the local polarity in a protein (7; Fig. 7).31 Fig. 8 shows a viscosity-sensitive fluorescence probe (8),32 whose response mechanism is based on the fact that the viscosity increase inhibits the internal rotation of the probe, which thereby reduces the related non-radiative de-excitation and causes fluorescence enhancement.
Fig. 7. Polarity-sensitive spectroscopic probe (7).
Fig. 8. Viscosity-sensitive fluorescence probe (8) and its response mechanism.
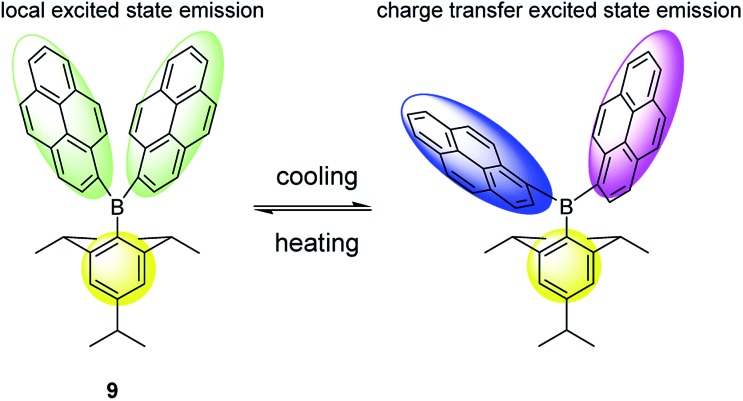
Increase of temperature usually results in a decrease in the fluorescence quantum yield, because the non-radiative processes (collisions with solvent molecules, intramolecular vibration, etc.) become more efficient at higher temperature. Differently from this phenomenon, an arylboron compound (9; Fig. 9) which was recently proposed as a temperature-sensitive probe,33 exhibits strong fluorescence at high temperature (e.g., >50 °C), and more importantly its color changes with temperature. This property of arylboron compounds may be rather useful to develop probes with high fluorescence quantum yields even at high temperature. However, high selectivity for a unique environmental factor (e.g., detecting polarity without interference from viscosity) proves also very challenging.
Fig. 9. Temperature-sensitive probe (9) based on emission mechanism change.
As can be seen from the above discussion, each kind of probe has its own merits and shortcomings. On the basis of specific practical requirements, how to choose a suitable design strategy for a given analyte/environmental factor is the key issue for a researcher.
3. Response modes
There are three basic kinds of response modes in fluorescence spectroscopy: on–off, off–on, and ratiometric; other modes (e.g., fluorescence anisotropy and lifetime) are also used occasionally.
3.1. On–off
The fluorescence on–off response, also called fluorescence quenching, refers to the decrease of fluorescence intensity of a probe, which can be caused by a variety of processes such as excited state reactions, energy transfer, complex-formation and intermolecular collision. At present, the fluorescence detection and imaging of trace levels of analyte is still difficult and thus attracts much more attention. Because of this, a fluorescence on–off probe shows no obvious advantage in the detection of an analyte due to its high background signal. As shown in Fig. 10, it is relatively difficult for an analyte at a concentration around its detection limit to cause a detectable fluorescence signal change in contrast to the high background signal.
Fig. 10. Fluorescence on–off response mode. An analyte at a concentration around its detection limit only causes a dull contrast (compare the left two images).
3.2. Off–on
The fluorescence off–on response, also called fluorescence enhancement, refers to the increase of the fluorescence intensity of a probe upon reaction. Because of its low background signal, the fluorescence off–on probe has the advantage of high detection sensitivity. As shown in Fig. 11, an analyte at a concentration around its detection limit can produce a detectable signal change relative to the near-dark background. However, this kind of probe, when compared to the ratiometric probes (infra vide), is found to suffer from the influence of several variants, such as probe concentration, instrumental efficiency, and environmental conditions. Therefore, when applied to intracellular analysis, the off–on probes are rather more suited to qualitative analysis due to their higher sensitivity.
Fig. 11. Fluorescence off–on response mode. An analyte at a concentration around its detection limit can produce a sharp contrast (compare the left two images).
3.3. Ratiometric
The fluorescence ratiometric response usually refers to a shift in the emission or excitation spectra of a probe upon reaction with an analyte (Fig. 12). Based on this shift, a ratiometric measurement can be achieved by calculating the ratio of fluorescence intensities at the two wavelengths. The advantage of ratiometric measurements lies in the ability to eliminating efficiently the interfering influences from many factors (e.g., probe bleaching, changes in focus, variations in laser intensity, etc.) by division manipulation, which is very suitable for an accurate quantitative analysis.
Fig. 12. Fluorescence ratiometric response mode.
Compared to the off–on probes, nevertheless, ratiometric probes have two drawbacks. The first one is the lower sensitivity, which usually results from the high fluorescence background (from the wavelength shift) and the division manipulation; the second one is the complicated and time-consuming ratio measurement (including data processing).
In addition, the following operations are not recommended: (1) ratiometric measurements are achieved using a mixture of a fluorescence intensity-sensitive probe and a fluorescence intensity-insensitive probe [because some factors such as the different stability (i.e., concentration change) of the two probes can not be cancelled out as in the case of a single ratiometric probe]; (2) the ratiometric mode is used for absorbance measurements (because the division manipulation often decreases sensitivity). In fact, absorbance is measured against a corresponding reagent blank, which already eliminates the influences from some common invariants that are encountered in fluorescence.34
3.4. Others
In addition, fluorescence anisotropy and lifetime are also used occasionally as a detection mode/signal. For instance, the mass increase of a spectroscopic probe usually raises anisotropy, and this behavior is often employed to investigate the interaction between the probe and an analyte; rare earth metal complexes have a long fluorescence lifetime, and can be used for time-resolved fluorescence measurements to increase sensitivity and accuracy.
4. Bioapplications
Herein we mainly discuss the requirements for the following bioapplications: in vitro, cells, and in vivo.
4.1. In vitro
The in vitro application of spectroscopic probes mostly focuses on the analysis of body fluids, including plasma/serum, urine, cerebrospinal fluid, saliva, etc. These body fluids (in particular plasma/serum) contain a large amount of albumin, which usually has to be removed prior to analysis with current spectroscopic methods. Therefore, spectroscopic probes with proteins anti-interference are in urgent demand to achieve direct analysis without pre-separation. Moreover, the availability of some body fluids is rather limited, which requires highly sensitive probes.
4.2. Cells
Imaging and analysis of cells including subcellular organelles has become an important research topic in the last decade. The contents of various substances (ions, reactive oxygen species, amino acids, enzymes, etc.) in cells are quite different. For instance, alkali and alkaline earth metal ions such as Na+, K+, Mg2+, and Ca2+ normally exist at rather high concentrations (at or over mM levels) except for intracellular Ca2+ (about μM levels). Therefore, the major requirement for a reliable analysis of these ions is high selectivity rather than sensitivity. In contrast, for trace substances such as some proteinases and ROS, sensitivity becomes the key demand. Unfortunately, many newly developed probes are still unable to detect related endogenous species at the basal level due to their low sensitivity, and the imaging and detection experiments were only performed by addition of excessive exogenous analyte, which actually seems less significant (certainly such probes are still useful for the study of an analyte's function). In a word, the number of probes that can meet the above basic requirements is still limited.
On the other hand, many spectroscopic probes, such as pH probes, can bind with proteins in cells, causing a change in their pK a values and thus a large error in pH measurements. To decrease the microenvironmental influence, the interference from coexisting substances should be studied carefully, and the pH calibration curve in cells (cytosol) instead of in plain buffer solution should be established by using a H+/K+ ionophore of nigericin35 for the accurate determination of intracellular pH values. Moreover, in interference studies of a probe, the tested substances and their concentrations should be chosen according to the characteristics of a sample to be analyzed. For cells, common substances (some inorganic salts, glucose, glutathione, some amino acids, etc.) often exist at or over mM levels, and these substances should be tested at relatively high concentrations; whereas the other substances, like reactive oxygen species and proteinases, should be examined at their relatively low physiological concentrations. In addition, the photostability of a spectroscopic probe is sometimes overemphasized, because, in fact, cells themselves can not tolerate light irradiation for a long period of time.
4.3. In vivo
The general requirement for in vivo analysis is similar to that for cell studies, except for the indispensability of long excitation and emission wavelengths. To realize in vivo imaging, probes with near infrared (NIR) fluorescence features are in pressing need, because they have the advantages of deep tissue penetration, and low autofluorescence and biological damage. Current NIR probes are mainly derived from cyanine dyes, which have poor stabilities and high background fluorescence as a result of ready autoxidation and photooxidation. Interestingly, hemicyanines, formed from their precursors, unstable cyanines, have recently been found to still possess near-infrared spectroscopic features and higher stability,22 which may provide an important molecular platform for developing stable NIR probes.
5. Future perspectives and conclusions
According to the above discussions and the current status in this field, future researches may focus on the following aspects:
(1) Design of chiral spectroscopic probes for the analysis of biological chiral compounds. This area has not yet drawn much attention but is full of challenges.
(2) Synthesize spectroscopic probes with long analytical wavelength features (e.g., NIR wavelengths) to increase sample penetration ability and reduce background signal and damage to biological species. Current NIR probes are still far from more practical requirements.
(3) Develop in situ spectroscopic traps/probes to achieve real-time analysis of unstable species.
(4) Prepare spectroscopic probes for the assay of gaseous molecules.
(5) Develop sensitive ratiometric probes for the accurate detection of more intracellular substances.
(6) Study spectroscopic probe arrays to achieve high-throughput detection.
(7) Design excellent two-photon and super-resolution probes to promote the development of corresponding new fluorescent techniques. At present, super-resolution fluorescence analysis is severely restricted by the lack of small molecular probes with high fluorescence quantum yield, anti-bleaching and fast switching properties.
It should be born in mind that the purpose of developing a new spectroscopic probe is for the analysis and detection of a target of interest, therefore the most important evaluation standard is whether the developed probe shows an excellent analytical performance (e.g., high sensitivity, selectivity, and applicability such as good water-solubility for biosystems) or not. On the above basis, the simpler both the design and synthesis of the probe the better. This rule means that a probe even with a long and complicated synthetic route but poor performance would not be a good choice. In contrast, the idea of obtaining a superior probe by either the use of new synthetic chemistry or the smart modification of existing fluorochromes should be encouraged. Regretfully an irrational phenomenon in this field seems to be that excessive attention is paid more to the time-consuming and complicated synthetic methods of a new probe instead of its analytical performance and validated bioapplication.
In summary, we have briefly analyzed the existing problems in several key issues of spectroscopic probes. As can be seen, solving these challenges still needs a long-term and hard effort, but the above discussions may have an important significance in guiding readers, who are ready to promote the steady development of better spectroscopic probes.
Acknowledgments
This work is supported by grants from the 973 Program (No. 2015CB932001 and 2015CB856301), the NSF of China (No. 21535009, 21435007 and 21321003), and the Chinese Academy of Sciences (XDB14030102).
Biographies

Jin Zhou
Jin Zhou received his BSc in Materials Chemistry in 2010 from Shandong Agricultural University and MSc in Analytical Chemistry in 2013 from Zhejiang Normal University. Currently, he is doing his PhD studies under the guidance of Professor Huimin Ma at the Key Laboratory of Analytical Chemistry for Living Biosystems, Institute of Chemistry, Chinese Academy of Sciences (ICCAS). His research interests focus on the design and synthesis of small molecular and nanoscaled fluorescent probes for bioimaging, biosensors and pharmaceutical analysis.

Huimin Ma
Huimin Ma is a Professor of Chemistry at ICCAS. He received his BSc in 1983 from Shandong University and PhD in Analytical Chemistry in 1990 from ICCAS, followed by postdoctoral research at Wuhan University. He was granted a research fellowship from the Alexander von Humboldt Foundation, working at Bremen University with Prof. Wolfram Thiemann. He has contributed over 100 journal articles as well as 20 invited book chapters and reviews, and is a member of the Editorial Board of several journals. His research interests focus on the design and synthesis of new spectroscopic probes and their analytical uses.
References
- Shi W., Ma H. M., Chem. Commun., 2012, 48 , 8732 , , and references therein . [DOI] [PubMed] [Google Scholar]
- Li X. H., Gao X. H., Shi W., Ma H. M. Chem. Rev. 2014;114:590. doi: 10.1021/cr300508p. [DOI] [PubMed] [Google Scholar]
- (a) You Y., Nam W. Chem. Sci. 2014;5:4123. [Google Scholar]; (b) Liu J., Sun Y. Q., Zhang H. X., Huo Y. Y., Shi Y. W., Guo W. Chem. Sci. 2014;5:3183. [Google Scholar]; (c) Sun X. L., Xu Q. L., Kim G., Flower S. E., Lowe J. P., Yoon J., Fossey J. S., Qian X. H., Bull S. D., James T. D. Chem. Sci. 2014;5:3368. [Google Scholar]
- Niu L. Y., Chen Y. Z., Zheng H. R., Wu L. Z., Tung C. H., Yang Q. Z. Chem. Soc. Rev. 2015;44:6143. doi: 10.1039/c5cs00152h. [DOI] [PubMed] [Google Scholar]
- (a) Kowada T., Maeda H., Kikuchi K. Chem. Soc. Rev. 2015;44:4953. doi: 10.1039/c5cs00030k. [DOI] [PubMed] [Google Scholar]; (b) Yuan L., Jin F. P., Zeng Z. B., Liu C. B., Luo S. L., Wu J. S. Chem. Sci. 2015;6:2360. doi: 10.1039/c4sc03883e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wang F. Y., Zhou L., Zhao C. C., Wang R., Fei Q., Luo S. H., Guo Z. Q., Tian H., Zhu W. H. Chem. Sci. 2015;6:2584. doi: 10.1039/c5sc00216h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Ashton T. D., Jolliffe K. A., Pfeffer F. M. Chem. Soc. Rev. 2015;44:4547. doi: 10.1039/c4cs00372a. [DOI] [PubMed] [Google Scholar]; (b) Tran M. N., Rarig R. F., Chenoweth D. M. Chem. Sci. 2015;6:4508. doi: 10.1039/c5sc01601k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Hamilton G. R. C., Sahoo S. K., Kamila S., Singh N., Kaur N., Hyland B. W., Callan J. F. Chem. Soc. Rev. 2015;44:4415. doi: 10.1039/c4cs00365a. [DOI] [PubMed] [Google Scholar]; (b) Hu J. J., Wong N. K., Lu M. Y., Chen X. M., Ye S., Zhao A. Q., Gao P., Kao R. Y. T., Shen J. G., Yang D. Chem. Sci. 2016;7:2094. doi: 10.1039/c5sc03855c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Zhou X., Lee S., Xu Z. C., Yoon J. Chem. Rev. 2015;115:7944. doi: 10.1021/cr500567r. [DOI] [PubMed] [Google Scholar]; (b) Li Y. H., Wu W., Yang J. F., Yuan L., Liu C. H., Zheng J., Yang R. H. Chem. Sci. 2016;7:1920. doi: 10.1039/c5sc04415d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Morgan M. T., McCallum A. M., Fahrni C. J. Chem. Sci. 2016;7:1468. doi: 10.1039/c5sc03643g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. L., James T. D. Chem. Rev. 2015;115:8001. doi: 10.1021/cr500562m. [DOI] [PubMed] [Google Scholar]
- Carter K. P., Young A. M., Palmer A. E. Chem. Rev. 2014;114:4564. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Fuentes P., Lippard S. J. Acc. Chem. Res. 2015;48:2927. doi: 10.1021/acs.accounts.5b00388. [DOI] [PubMed] [Google Scholar]
- Aron A. T., Ramos-Torres K. M., Cotruvo Jr J. A., Chang C. J. Acc. Chem. Res. 2015;48:2434. doi: 10.1021/acs.accounts.5b00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin V. S., Chen W., Xian M., Chang C. J. Chem. Soc. Rev. 2015;44:4596. doi: 10.1039/c4cs00298a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Tang Y. H., Lee D. Y., Wang J. L., Li G. H., Yu J. H., Lin W. Y., Yoon J. Chem. Soc. Rev. 2015;44:5003. doi: 10.1039/c5cs00103j. [DOI] [PubMed] [Google Scholar]; (b) Zheng K. B., Lin W. Y., Tan L., Chen H., Cui H. J. Chem. Sci. 2014;5:3439. [Google Scholar]; (c) Divya K. P., Sreejith S., Ashokkumar P., Yuzhan K., Peng Q. W., Maji S. K., Tong Y., Yu H., Zhao Y. L., Ramamurthyc P., Ajayaghosh A. Chem. Sci. 2014;5:3469. [Google Scholar]
- Yin C. X., Huo F. J., Zhang J. J., Martinez-Manez R., Yang Y. T., Lv H. G., Li S. D. Chem. Soc. Rev. 2013;42:6032. doi: 10.1039/c3cs60055f. [DOI] [PubMed] [Google Scholar]
- (a) Yin J., Hua Y., Yoon J. Chem. Soc. Rev. 2015;44:4619. doi: 10.1039/c4cs00275j. [DOI] [PubMed] [Google Scholar]; (b) Hirayama T., Okuda K., Nagasawa H. Chem. Sci. 2013;4:1250. [Google Scholar]
- Yun S. W., Kang N. Y., Park S. J., Ha H. H., Kim Y. K., Lee J. S., Chang Y. T. Acc. Chem. Res. 2014;47:1277. doi: 10.1021/ar400285f. [DOI] [PubMed] [Google Scholar]
- Yuan L., Lin W. Y., Zheng K. B., Zhu S. S. Acc. Chem. Res. 2013;46:1462. doi: 10.1021/ar300273v. [DOI] [PubMed] [Google Scholar]
- Lou Z. R., Li P., Han K. L. Acc. Chem. Res. 2015;48:1358. doi: 10.1021/acs.accounts.5b00009. [DOI] [PubMed] [Google Scholar]
- (a) de Silva A. P., Gunaratne H. Q. N., Gunnlaugsson T., Huxley A. J. M., McCoy C. P., Rademacher J. T., Rice T. E. Chem. Rev. 1997;97:1515. doi: 10.1021/cr960386p. [DOI] [PubMed] [Google Scholar]; (b) Valeur B., Leray I. Coord. Chem. Rev. 2000;205:3. [Google Scholar]; (c) Carter K. P., Young A. M., Palmer A. E. Chem. Rev. 2014;114:4564. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Su M. H., liu Y., Ma H. M., Ma Q. L., Wang Z. H., Yang J. L., Wang M. X. Chem. Commun. 2001:960. [Google Scholar]; (b) He L. W., Lin W. Y., Xu Q. Y., Ren M. G., Wei H. P., Wang J. Y. Chem. Sci. 2015;6:4530. doi: 10.1039/c5sc00348b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Q., Chen S. M., Shi W., Li L. H., Ma H. M. Angew. Chem., Int. Ed. 2014;53:10916. doi: 10.1002/anie.201405742. [DOI] [PubMed] [Google Scholar]
- Grynkiewicz G., Poenie M., Tsien R. Y. J. Biol. Chem. 1985;260:3440. [PubMed] [Google Scholar]
- Hancock R. D., Martell A. E. Chem. Rev. 1989;89:1875. [Google Scholar]
- Zhou J., Shi W., Li L. H., Gong Q. Y., Wu X. F., Li X. H., Ma H. M. Chem.–Asian J. 2016 doi: 10.1002/asia.201600012. [DOI] [Google Scholar]
- Toone E. J., Advances in Enzymology and Related Areas of Molecular Biology, Protein Evolution, Wiley-Interscience, Durham, 1st edn, 2006, vol. 75. [Google Scholar]
- Li L. H., Shi W., Wang Z., Gong Q. Y., Ma H. M. Anal. Chem. 2015;87:8353. doi: 10.1021/acs.analchem.5b01535. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Fahrni C. J. J. Am. Chem. Soc. 2004;126:8862. doi: 10.1021/ja049684r. [DOI] [PubMed] [Google Scholar]
- (a) Jiang X., Yu Y., Chen J., Zhao M., Chen H., Song X., Matzuk A. J., Carroll S. L., Tan X., Sizovs A., Cheng N., Wang M. C., Wang J. ACS Chem. Biol. 2015;10:864. doi: 10.1021/cb500986w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lippert A. R. J. Inorg. Biochem. 2014;133:136. doi: 10.1016/j.jinorgbio.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Wang K., Ma H. M. Progr. Chem. 2010;22:1633. [Google Scholar]
- Wang X. C., Wang S. J., Ma H. M. Analyst. 2008;133:478. doi: 10.1039/b717230c. [DOI] [PubMed] [Google Scholar]
- Wang K., Shi W., Jia J., Chen S. M., Ma H. M. Talanta. 2009;77:1795. doi: 10.1016/j.talanta.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Feng J., Tian K. J., Hu D. H., Wang S. Q., Li S. Y., Zeng Y., Li Y., Yang G. Q. Angew. Chem., Int. Ed. 2011;50:8072. doi: 10.1002/anie.201102390. [DOI] [PubMed] [Google Scholar]
- (a) Ma H. M., Huang Y. X., Liang S. C. Microchim. Acta. 1998;128:181. [Google Scholar]; (b) Ma H. M., Huang Y. X., Liang S. C. Talanta. 1996;43:21. doi: 10.1016/0039-9140(95)01700-3. [DOI] [PubMed] [Google Scholar]
- (a) Tafani M., Cohn J. A., Karpinich N. O., Rothman R. J., Russo M. A., Farber J. L. J. Biol. Chem. 2002;277:49569. doi: 10.1074/jbc.M208915200. [DOI] [PubMed] [Google Scholar]; (b) Shi W., Li X. H., Ma H. M. Angew. Chem., Int. Ed. 2012;51:6432. doi: 10.1002/anie.201202533. [DOI] [PubMed] [Google Scholar]