Abstract
Intracranial aneurysm (IA) remains one of the most devastating neurological conditions. However, the pathophysiology of IA formation and rupture still remains unclear. The purpose of the present study was to identify the crucial microRNA (miRNA/miR) and genes involved in IAs and elucidate the mechanisms underlying the development of IAs. In the present study, novel miRNA regulation activities in IAs were investigated through the integration of public gene expression data of miRNA and mRNA using the Gene Expression Omnibus database, combined with bioinformatics prediction. A total of 15 differentially expressed miRNA and 1,447 differentially expressed mRNA between IAs and controls were identified. A number of miRNA-target gene pairs (770), whose expression levels were inversely correlated, were used to construct a regulatory network of miRNA-target genes in IAs. The biological functions and pathways of these target genes were revealed to be associated with IAs. Specific miRNA and genes, such as hsa-let-7f, hsa-let-7d, hsa-miR-7, RPS6KA3, TSC1 and IGF1 may possess key roles in the development of IAs. The integrated analysis in the present study may provide insights into the understanding of underlying molecular mechanisms of IAs and novel therapeutic targets.
Keywords: mRNA expression data, miRNA expression data, intracranial aneurysms, regulatory network, miRNA target genes
Introduction
Intracranial aneurysms (IAs), also referred to as cerebral aneurysms, are balloons or sac-like dilatations of arteries inside the brain. To date, IAs remain to be one of the most devastating neurological conditions with a prevalence of 2–3% in the general population (1). Unruptured IAs are typically asymptomatic; however, in the event that IAs rupture, this process results in hemorrhage to the subarachnoid space, which is a devastating condition that has been indicated to have a mortality rate of 30–40, and 50% of survivors are left disabled (2).
Previous research on the etiology of IAs indicated that the formation of IAs is assumed to be caused by diverse environmental and genetic factors, such as cigarette smoking, excessive alcohol consumption, hypertension, female gender and family history of IAs (3–5). However, the pathophysiology of IA formation and rupture still remains to be fully elucidated.
Microarray-based gene expression analyses have implied several mechanisms underlying the development of IAs (6–10). Extracellular matrix turnover factors and inflammatory factors, such as interleukin (IL)-1β, IL-6, IL-8, IL-18, interferon-γ, tumor necrosis factor-α and major histocompatibility complex class II gene, have essential roles in the development, progression, and rupture of aneurysms (11,12). Several pathways, including those associated with inflammatory responses, the immune system, extracellular matrix, and apoptosis are considered to be crucial in the formation, progression, and rupture of IAs (8,13).
MicroRNA (miRNA/miR) are small, non-coding, single-stranded RNA, which are implicated in the post-transcriptional regulation of gene expression of either mRNA degradation or inhibiting translation, followed by protein synthesis repression (14). Furthermore, miRNA may modulate pathways and mechanisms of IAs via the control in gene expression. Previous studies have demonstrated that miRNA are involved in vascular remodeling and atherosclerosis (15,16). In addition, a previous study revealed that a subset of inflammation-related miRNA were specifically upregulated in stroke patients with intracerebral hemorrhage and indicated that miR-16, and miR-25 were independent factors for IA occurrence by screening the miRNA expression level of 40 IA patients (20 unruptured and 20 ruptured) and 20 healthy volunteers via microarray assays and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis (17).
In the present study, bioinformatic methods were used to merge miRNA and mRNA expression data separately, using data available on the Gene Expression Omnibus database (GEO), to identify differentially expressed mRNA and miRNA between IAs and normal tissues. Subsequently, differentially expressed miRNA target genes were detected by bioinformatics prediction, inversely correlated analysis of miRNA and mRNA expressions were conducted and a miRNA-target gene regulatory network was constructed. The present findings may contribute to future investigations aimed at elucidaing the mechanisms of IAs.
Materials and methods
Eligible gene expression profiles
IA expression profiling studies were searched on the GEO database (ncbi.nlm.gov/geo), which serves as a public repository for gene expression datasets to meet the growing demand for a public repository for high-throughput gene expression data (18). IA expression profiling studies were only retained if they compared miRNA or mRNA expression profiling between IAs and normal tissues.
Differential analysis of miRNA and mRNA
Raw microarray data from each study was downloaded, and preprocessed with log2 transformation and Z-score normalization. The Linear Models for Microarray Data package in R (r-project.org) was used to identify the differently expressed probe sets between IAs and controls using the two-tailed Student's t-test and P-values of individual microarray studies were obtained. MetaMA package in R (r-project.org) was used to combine P-values from multiple microarray studies and false discovery rate (FDR) was calculated for multiple comparisons using the Benjamini & Hochberg method (19). We selected differently expressed mRNA with criterion of FDR <0.01 and a criterion of FDR <0.01 for differently expressed miRNA. Heat map analysis was performed using the ‘heatmap.2’ function of the R/Bioconductor package ‘gplots’ (20).
Identification of differently expressed miRNA target genes
To understand the potential association between differentially expressed mRNA and miRNA obtained in the present study, the transcriptional targets of the identified miRNA were predicted using the online tools of miRWalk (umm.uni-heidelberg.de/apps/zmf/mirwalk/) (21) based on six bioinformatic algorithms (DIANAmT, miRanda, miRDB, miRWalk, PICTAR and TargetScan). Putative targets that were common in the prediction of ≥4 algorithms were selected to match with those identified to be dysregulated in IAs. As miRNA tend to decrease the expression of their target mRNA, differentially expressed target genes whose expression levels were inversely correlated with that of miRNA were to subjected to further investigation (22–24).
Functional annotation
Functional enrichment analysis is essential to uncover biological functions of miRNA target genes. To gain insights into the biological functions of miRNA target genes, Gene Ontology (GO) classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using GENECODIS online software (genecodis.cnb.csic.es) (25). GO, which includes three categories (biological process, molecular function and cellular component), provides a common descriptive framework of gene annotation and classification for analyzing gene set data. KEGG pathway enrichment analysis was also performed to detect the potential pathway of miRNA target genes based on the KEGG pathway database, which is a recognized and comprehensive database including various types of biochemistry pathways (26). FDR <0.05 was set as the cut-off for selecting significantly enriched functional GO terms and KEGG pathway.
Construction of the regulatory network of miRNA-target gene in IA
miRNA-target gene interaction networks in IA with miRNA-target gene interacting pairs with expression levels that were inversely correlated were investigated. miRNA regulation networks were visualized using Cytoscape software (27).
Results
Differentially expressed miRNA and mRNA in IA
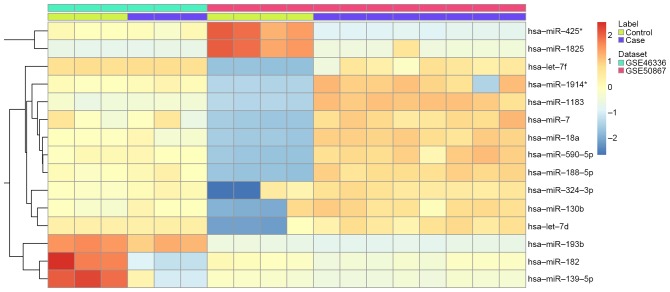
In the present study, two miRNA and five mRNA expression profiling studies (7,10,13,17,28) of IA were collected (Table I). Following normalization of the original miRNA and mRNA expression datasets, 15 miRNA were regarded as significantly differentially expressed under the threshold of FDR<0.01. A total of 10 upregulated and five downregulated miRNA were identified (Table II). The upregulated miRNA with the lowest FDR was hsa-miR-188-5p, and the downregulated gene with the lowest FDR was hsa-miR-425. The significantly differentially expressed miRNA were displayed in a heat map (Fig. 1) and 1,447 genes were identified to be significantly differentially expressed between IAs and controls; 682 genes were upregulated and 765 genes were downregulated.
Table I.
Characteristics of mRNA and miRNA expression profiling of IAs.
| Authors | Year | GEO ID | Platform | Samples (control:case) | Country | Refs. |
|---|---|---|---|---|---|---|
| mRNA expression profile | ||||||
| Nakaoka et al | 2014 | GSE54083 | GPL4133Agilent-014850 Whole Human Genome Microarray 4×44K G4112F | 10:13 | Japan | (28) |
| Jiang et al | 2013 | GSE46337 | GPL6480Agilent-014850 Whole Human Genome Microarray 4×44K G4112F | 2: 2 | China | – |
| Li et al | 2011 | GSE26969 | GPL570Affymetrix Human Genome U133 Plus 2.0 Array | 3:3 | China | (7) |
| Pera et al | 2010 | GSE15629 | GPL6244Affymetrix Human Gene 1.0 ST Array | 5:14 | Poland | (10) |
| Weisheimer et al | 2007 | GSE6551 | GPL570Affymetrix Human Genome U133 Plus 2.0 Array/GPL2507 Sentrix Human-6 Expression BeadChip | 5:5 | USA | (13) |
| miRNA expression profile | ||||||
| Li et al | 2013 | GSE50867 | GPL17725Agilent-031945 human_miRNA_v14 [miRBase release 16.0 miRNA ID version] | 4:8 | China | (17) |
| Jiang et al | 2013 | GSE46336 | GPL16770Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray (miRBase release 16.0 miRNA ID version) | 3:3 | China | – |
miRNA, microRNA; GEO, Gene Expression Omnibus database.
Table II.
List of differentially expressed miRNA.
| miRNA | FDR | Fold-change |
|---|---|---|
| Upregulated miRNA | ||
| hsa-miR-188-5p | 2.79E-09 | 2.9750 |
| hsa-miR-1183 | 9.74E-08 | 3.4515 |
| hsa-miR-18a | 6.43E-06 | 2.6047 |
| hsa-miR-7 | 5.18E-05 | 2.6007 |
| hsa-miR-590-5p | 2.85E-04 | 2.3965 |
| hsa-let-7d | 1.77E-03 | 2.3005 |
| hsa-let-7f | 2.17E-03 | 2.3227 |
| hsa-miR-130b | 7.24E-03 | 2.0646 |
| hsa-miR-324-3p | 8.20E-03 | 1.8100 |
| hsa-miR-1914a | 8.20E-03 | 2.4691 |
| Downregulated miRNA | ||
| hsa-miR-425a | 1.98E-05 | −3.4574 |
| hsa-miR-182 | 5.39E-04 | −1.3745 |
| hsa-miR-1825 | 1.80E-03 | −3.1043 |
| hsa-miR-139-5p | 2.17E-03 | −1.5437 |
| hsa-miR-193b | 9.22E-03 | −1.3729 |
Target predictions were not available via the miRWalk database. miRNA, microRNA; FDR, false discovery rate.
Figure 1.
Heat-map image representing 15 miRNA that were significantly upregulated or downregulated (false discovery rate <0.01) in intracranial aneurysms compared with normal controls.
Regulatory network of miRNAs and target genes in IA
The miRWalk database was used to predict putative targets of significantly upregulated or downregulated miRNA in IAs. Comparing those putative targets with the list of differentially expressed genes in IAs, miRNA-target gene pairs with inversely correlated expression levels were selected. As a result, 531 miRNA-target gene pairs for the upregulated miRNA, with 29 pairs validated by experiments, and 211 miRNA-target gene pairs for the down regulated miRNA, with nine pairs validated by experiments, were identified (Table III). The target predictions of hsa-miR-1914 and hsa-miR-425 were not available in miRWalk databases.
Table III.
miRNA-mRNA pairs with inversely correlated expression levels.
| miRNA | Regulation (miRNA) | Target counts | Target mRNA |
|---|---|---|---|
| hsa-miR-188-5p | Up | 59 | AEBP2, ATP6V1G1, BAG5, BCL9, BET1, BIN2, CAPN2, CD80, CDC25B, CDON, CLU, CPSF2, CYP1A1, CYYR1, DAAM1, DLC1, FBXO11, FBXO9, FNBP1, FNBP4, FUBP3, GLI2, HMGB1, IL28RA, IL2RA, ING5, KPTN, MX2, MYT1, N6AMT1, PAX8, PCDH9, PER2, PLVAP, PROS1, PSMF1, RPS6KA3, SCN3A, SH3BGRL2, SLC22A3, SPG20, SPOP, SYNJ2BP, SYT11, SYTL2, TACC1, TCL1A, TIAM1, TSN, UPF2, UPF3A, USP14, USP47, UVRAG, ZNF185, ZNF451, ITLN1, GEMIN4, PCNA |
| hsa-miR-1183 | Up | 9 | AEBP2, AIM1, ARG2, CRIM1, DARS, F8, POT1, ROCK2, SLC9A6 |
| hsa-miR-18a | Up | 63 | TP53, GEMIN4, CD8A, ADCY1, AEBP2, AIM1, C1RL, C9orf5, CBX7, CLOCK, CNTN4, CRELD1, CRIM1, CTLA4, DAAM2, DUSP3, EFS, EGLN2, EYA4, FBXO9, FNBP1, GAS7, GFOD1, IGF1, IL28RA, INSR, ITIH5, KIF3B, MC2R, MS4A2, NDUFS1, NEDD4, NPY1R, OSTM1, PCNP, PCSK2, PLEKHG1, POT1, POU6F1, PRMT6, PSMF1, RERG, RPS6KA3, RRAS, SH3BGRL2, SH3BP4, SLC22A3, SLC6A7, SON, SOX8, SRI, STARD7, STAT6, TSC1, TXNIP, UQCRB, USP24, VPS4B, WASF2, ZHX2, ZNF169, ZNF430, ZNF451 |
| hsa-miR-7 | Up | 94 | AIM1, ARID2, ATP6V1G1, BAG5, C1GALT1, CAMK1G, CAMK2D, CAPN2, CBX7, CD5, CD8A, CDC14B, CDON, CISH, CLNS1A, COLEC12, CRIM1, CRY2, CXCL12, DCK, DMXL1, EVC, FAM20B, FBLN5, FBXO21, GAS7, GATA4, GATA6, GGA2, GNG4, GRIK3, HN1, IGF1, INHBB, INSR, ITIH5, JAM3, KIF3B, LDB3, LGALS8, LIFR, LUC7L2, MAOA, MC2R, MEIS2, MFAP4, MIPOL1, MRPS36, N6AMT1, NEDD4, NEIL1, PARD6G, PAX8, PIGH, PIK3R3, POLE3, POU6F1, PPP2R2B, PPP2R3A, ProSAPiP1, PSORS1C2, PUM2, RAB5B, RAP1A, RFC5, RIMS3, ROCK2, RPS6KA3, SEMA6A, SLC22A3, SLC9A6, SNAP29, SQSTM1, SS18L1, STEAP2, SYNJ2BP, SYNPR, TCERG1, TCL1A, TGFA, THAP6, TIAM1, TMOD1, TMOD2, TOX, TRDN, TRIM52, TRPV1, TSC1, TSN, UTRN, ZFYVE21, ZNF185, ZNF319 |
| hsa-miR-590-5p | Up | 49 | ANXA1, BBS7, C14orf101, CAMK2D, CAPN2, CNTFR, COL4A4, CRIM1, CTCF, CUBN, DNAJA2, DNM1L, ENPP4, FAT3, FBXO11, FUBP3, FZD6, GFOD1, INSR, IRAK1BP1, ITIH5, KIAA0240, KL, KLF8, LIFR, MAOA, MATN2, MS4A2, OLFM3, PCNA, PELI1, PER2, POT1, PPP1R3D, RABIF, REPS1, RIOK1, RNF38, RPS6KA3, SLMAP, SMARCE1, SRI, TIAM1, TPK1, TRUB1, TSC1, USP24, WSB1, ZNF295 |
| hsa-let-7d | Up | 55 | TAF9, CLU, IGF1, XPO1, TP53, ACTA2, CISH, IFNG, FMR1, ABCB9, ADCY9, AIM1, C14orf28, C1QTNF1, C1RL, CALM1, CD80, CDC14B, CDC25B, CDCA8, COL14A1, COL4A4, CRY2, DPF2, DUSP19, ENPP4, FRAS1, GAS7, GFOD1, GGA2, GNG5, ICOS, IL28RA, INSR, KIAA1609, LDB3, LEPROTL1, LIFR, MC2R, MTHFD2, MUC4, P2RX1, PRDM12, PSORS1C2, ROBO4, RPS6KA3, SCARA3, SPOP, SYT11, TNFSF10, TPK1, TRIM39, TSC1, USP24, UTRN |
| hsa-let-7f | Up | 68 | ABCB9, ADCY1, ADCY9, AIM1, ATP6V1G1, BMPR1A, C14orf28, C1QTNF1, C1RL, CALM1, CD80, CD8A, CDC14B, CDC25B, CDCA8, CISH, CLU, COL14A1, COL4A4, COL4A5, CRY2, DIABLO, DLC1, DPF2, DUSP19, EGLN2, ENPP4, FMR1, FRAS1, GALR1, GAS7, GFOD1, GGA2, GNG5, ICOS, IFNG, IGF1, IL28RA, IL2RA, INSR, KIAA1609, LDB3, LEPROTL1, MC2R, MEIS2, MTHFD2, MUC4, NTRK3, P2RX1, PAX8, PSORS1C2, RIMS3, RNF38, ROBO4, RPS6KA3, SCARA3, SIPA1L2, SPOP, SYNJ2BP, SYT11, TNFSF10, TP53, TPK1, TSC1, USP24, USP47, UTRN, XPO1 |
| hsa-miR-130b | Up | 73 | ADCY1, ADD2, AIG1, ANK2, BAI3, C1GALT1, CALM2, CAMK2D, CCR6, CDON, CLOCK, CNTN4, COL4A4, CRY1, CXCL12, DEDD2, DNM1L, DOK5, DUSP19, EFS, ENPP4, EPHB4, FAM20B, FAT3, FBXO9, FMR1, FNBP1, FZD6, GGA2, HOXA3, IGF1, IL28RA, INHBB, ITPR1, LDB3, LEPROTL1, METAP1, MLLT10, MRRF, MTMR4, MYT1, NEIL1, PCYOX1, PELI1, PLSCR4, POU6F1, PPIA, PPP1R12A, PTPRG, RAB5B, RNF38, RPS6KA3, RYR2, SCARA3, SCN3A, SLC9A6, SLMAP, SOX21, SPG20, SRPX, STOM, TACC1, TBC1D8, TGFA, THAP6, TSC1, TXNIP, USP47, VPS4B, WDR1, WRN, ZAK, ZNF430 |
| hsa-miR-130b | Up | 73 | ADCY1, ADD2, AIG1, ANK2, BAI3, C1GALT1, CALM2, CAMK2D, CCR6, CDON, CLOCK, CNTN4, COL4A4, CRY1, CXCL12, DEDD2, DNM1L, DOK5, DUSP19, EFS, ENPP4, EPHB4, FAM20B, FAT3, FBXO9, FMR1, FNBP1, FZD6, GGA2, HOXA3, IGF1, IL28RA, INHBB, ITPR1, LDB3, LEPROTL1, METAP1, MLLT10, MRRF, MTMR4, MYT1, NEIL1, PCYOX1, PELI1, PLSCR4, POU6F1, PPIA, PPP1R12A, PTPRG, RAB5B, RNF38, RPS6KA3, RYR2, SCARA3, SCN3A, SLC9A6, SLMAP, SOX21, SPG20, SRPX, STOM, TACC1, TBC1D8, TGFA, THAP6, TSC1, TXNIP, USP47, VPS4B, WDR1, WRN, ZAK, ZNF430 |
| hsa-miR-324-3p | Up | 83 | ADAMTS17, ADCY1, ANKRD11, ARHGAP10, ARHGEF17, BARHL1, BRD2, C1QTNF1, CAMK1G, CBX7, CD34, CD5, CD8A, CDC14B, CDC25B, CLU, COL14A1, COL21A1, CRY2, CXCL12, CYGB, DIABLO, DLC1, DNAJB2, EFS, EGLN2, ELTD1, EPHB4, ESAM, FBXO9, FNBP1, GAS7, GATA4, GFOD1, GGA2, GRIK3, HOXD4, KCND1, KCNS1, KIAA1609, KIF3B, LEPROTL1, LGALS8, LUC7L2, MYLK2, MYT1, NGFR, NTRK3, NUDC, PAX8, PIK3R3, PODN, PPAP2B, PPP1R3D, PRKAR1A, ProSAPiP1, PSMF1, PSORS1C2, RAB5B, RARG, RBM3, RGS3, RGS6, RIMS3, RPL13A, RPS6KA3, SLAMF7, SLC6A7, SOX21, SOX8, SS18, SYT11, TRPV3, TSC1, USP22, USP47, UVRAG, VASP, WASF2, YWHAG, ZNF319, ZNF451, ZNF510 |
| hsa-miR-182 | Down | 84 | CDKN1A, MYCN, BAX, EGR1, DOK4, MBNL2, ARF4, PCDH8, XPR1, DDAH1, GALNT2, KITLG, PAPPA, KCNK10, SLC2A3, THBS1, THBS2, ZIC3, GPR68, HOOK3, ADAM9, NAV1, LHFPL2, SIRPB1, VAT1, YKT6, KHDRBS3, MAL2, SLC36A4, COL5A1, SLC31A1, ASB6, CYBB, RDH10, DDX3X, DLAT, FBLN1, C6orf89, MRAS, NLGN4Y, LPHN1, MESDC2, SYNE2, FN1, KIAA0368, NUDT13, D4S234E, KCNH5, HTR2C, KIAA2022, IL16, KIF5C, LRP1, MCL1, MKLN1, OLR1, PC, CECR1, WHSC1L1, PCDHB4, SH3GLB2, THAP10, USP31, PTPRE, NTN4, RNASE6, RNASEL, RRBP1, SCNN1G, SLC11A1, SLC22A5, SRPK1, SSTR2, TBL1X, SLC35A2, TNFSF9, CCND2, NAV2, ACVR1B, SYNGR2, GCM2, KCNK6, KIF23, KIAA0247 |
| hsa-miR-1825 | Down | 6 | CDH2, ABCA1, PANX1, SERPINE1, NLK, C2orf3 |
| hsa-miR-139-5p | Down | 80 | MBNL2, DDX3X, ABCA1, FOS, GALNT3, ENAH, TBL1X, PTPRU, DPYSL4, YKT6, ABHD2, BAZ2A, SLC6A14, SLC35A4, MAL2, AP2M1, CCR5, COL11A1, CX3CR1, RDH10, ARX, DSC2, EFNA3, EREG, C6orf89, MRAS, FN1, DDAH1, NR5A1, GALNT2, D4S234E, GLI3, GNAL, USP25, NME7, EHD4, HOXA7, KIAA2022, IGFBP5, HCN1, IL16, JAK3, KCNA3, KIF5C, LHCGR, MARK1, MBNL1, MCL1, KITLG, NF2, PPAT, DIRAS2, BCAS3, PPP2R4, DOK4, PRKCA, CYP4F11, RFXAP, RNASEL, RPL15, SCD, SMOC1, SLC39A8, SGCD, SRPK1, TGFB1, THBS1, TPM3, UFD1L, CUL3, PPFIBP1, KIAA1755, STX11, TNFRSF10D, CCND2, SLC28A2, GCM2, TM9SF4, SPOCK2, TAGLN2 |
| hsa-miR-193b | Down | 47 | SOX9, MCL1, BCL2L10, STMN1, MKLN1, PTPRU, ABHD2, BAZ2A, AP2M1, SLC31A1, CRK, CX3CR1, E2F1, DDAH1, KCNE1L, GALNT2, TNFRSF21, GCLC, APOA2, IGFBP5, MMP14, MYCN, NF2, NUMA1, SERPINE1, RNF141, PPAT, DIRAS2, PPP2R4, PRKCA, KIAA1199, PTPRE, SGCD, SLC20A2, SOX12, TRAF1, ZIC3, AXIN2, HAVCR2, KIAA1755, TNFRSF10B, CCND2, NAV1, SOCS3, ACVRL1, BCAR1, KIAA0195 |
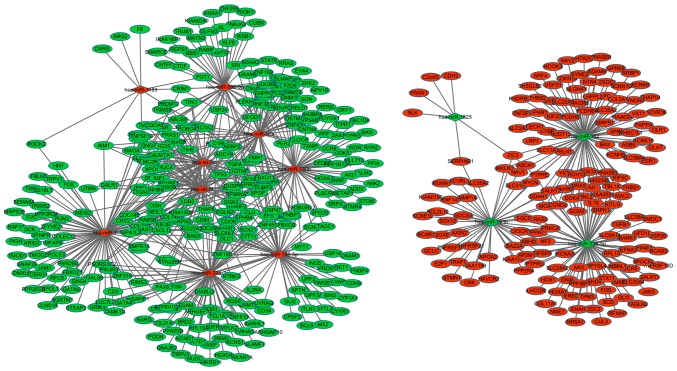
Using the 770 miRNA-target gene pairs, a miRNA-target gene regulatory network was constructed. In the miRNA-target gene regulatory network, the top ten miRNA, which included hsa-miR-7, hsa-miR-182, hsa-miR-324-3p, hsa-miR-139-5p, hsa-miR-130b, hsa-let-7f, hsa-miR-18a, hsa-miR-188-5p, hsa-let-7d and hsa-miR-590-5p, were identified to regulate the greatest number of target genes, and the target genes, such as RPS6KA3, TSC1, AIM1, GAS7, GFOD1, GGA2, IGF1, IL28RA and INSR, were regulated by the greatest number of miRNA (Fig. 2).
Figure 2.
Regulatory network between miRNA and target genes in intracranial aneurysms. Diamonds and ellipses represent miRNA and genes, respectively. Red and green colors represent the relatively high and low expression, respectively. miRNA, microRNA.
GO classification and KEGG pathways of miRNA target genes
GO classification and KEGG pathway enrichment analysis were performed for miRNA target genes that were differently expressed. Peptide transport (GO, 0015833; FDR, 2.63E-01) and amide transport (GO, 0042886; FDR, 1.31E-01) were indicated to be significantly enriched for biological processes. Molecular functions, ATP binding (GO, 0005524; FDR, 8.93E-02) and adenyl ribonucleotide binding (GO, 0032559; FDR, 6.09E-02) were also significantly enriched. Furthermore, cellular component, collagen trimer (GO, 0005581; FDR, 1.29E-02) and endoplasmic reticulum lumen (GO, 0005788; FDR, 1.30E-01) were significantly enriched (Table IV). The most significant pathway in the present KEGG analysis was focal adhesion (FDR, 1.07E-08). Pathways in cancer (FDR, 2.49E-08) and cytokine-cytokine receptor interaction (P=5.88E-08) were also indicated to be highly enriched (Table V).
Table IV.
Top 15 GO functional annotations of differentially expression miRNA target genes.
| GO ID | GO term | Count | P-value | FDR |
|---|---|---|---|---|
| Biological process | ||||
| GO:0015833 | Peptide transport | 2 | 5.21E-05 | 2.63E-01 |
| GO:0042886 | Amide transport | 2 | 5.21E-05 | 1.31E-01 |
| GO:0042327 | Positive regulation of phosphorylation | 19 | 1.13E-04 | 1.91E-01 |
| GO:0044712 | Single-organism catabolic process | 15 | 2.38E-04 | 3.00E-01 |
| GO:0010562 | Positive regulation of phosphorus metabolic process | 20 | 2.38E-04 | 2.40E-01 |
| GO:0045937 | Positive regulation of phosphate metabolic process | 20 | 2.38E-04 | 2.00E-01 |
| GO:0044763 | Single-organism cellular process | 320 | 2.73E-04 | 1.97E-01 |
| GO:0060191 | Regulation of lipase activity | 5 | 3.88E-04 | 2.45E-01 |
| GO:0030574 | Collagen catabolic process | 6 | 5.20E-04 | 2.92E-01 |
| GO:0032963 | Collagen metabolic process | 6 | 5.20E-04 | 2.62E-01 |
| GO:0044259 | Multicellular organismal macromolecule metabolic process | 6 | 5.20E-04 | 2.39E-01 |
| GO:0044243 | Multicellular organismal catabolic process | 6 | 5.20E-04 | 2.19E-01 |
| GO:0044236 | Multicellular organismal metabolic process | 6 | 5.20E-04 | 2.02E-01 |
| GO:0042325 | Regulation of phosphorylation | 20 | 6.27E-04 | 2.26E-01 |
| GO:0032924 | Activin receptor signaling pathway | 2 | 7.48E-04 | 2.52E-01 |
| Cellular component | ||||
| GO:0005581 | Collagen trimer | 8 | 2.31E-05 | 1.29E-02 |
| GO:0005788 | Endoplasmic reticulum lumen | 7 | 4.65E-04 | 1.30E-01 |
| Molecular function | ||||
| GO:0005524 | ATP binding | 7 | 9.26E-05 | 8.93E-02 |
| GO:0032559 | Adenyl ribonucleotide binding | 7 | 1.26E-04 | 6.09E-02 |
| GO:0030554 | Adenyl nucleotide binding | 7 | 1.26E-04 | 4.06E-02 |
| GO:0043492 | ATPase activity, coupled to movement of substances | 2 | 1.47E-04 | 3.53E-02 |
| GO:0015399 | Primary active transmembrane transporter activity | 2 | 1.47E-04 | 2.83E-02 |
| GO:0015405 | P-P-bond-hydrolysis-driven transmembrane transporter activity | 2 | 1.47E-04 | 2.36E-02 |
| GO:0016820 | Hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | 2 | 1.47E-04 | 2.02E-02 |
| GO:0042626 | ATPase activity, coupled to transmembrane movement of substances | 2 | 1.47E-04 | 1.77E-02 |
| GO:0048185 | Activin binding | 2 | 2.69E-04 | 2.88E-02 |
| GO:0016361 | Activin receptor activity, type I | 2 | 2.69E-04 | 2.59E-02 |
| GO:0017002 | Activin-activated receptor activity | 2 | 2.69E-04 | 2.36E-02 |
| GO:0051117 | ATPase binding | 3 | 3.01E-04 | 2.42E-02 |
| GO:0019901 | Protein kinase binding | 12 | 3.39E-04 | 2.51E-02 |
| GO:0032549 | Ribonucleoside binding | 7 | 4.08E-04 | 2.81E-02 |
| GO:0032550 | Purine ribonucleoside binding | 7 | 4.08E-04 | 2.62E-02 |
GO, gene otology; FDR, false discovery rate; ATP, adenosine triphosphate.
Table V.
KEGG pathway enrichment analysis of differentially expressed microRNA target genes (Top 15).
| KEGG ID | KEGG term | Count | FDR | Genes |
|---|---|---|---|---|
| hsa04510 | Focal adhesion | 19 | 1.07E-08 | COL4A4, IGF1, PPP1R12A, ROCK2, CCND2, BCAR1, COL4A5, RAP1A, THBS1, MYLK2, PRKCA, CRK, PIK3R3, CAPN2, THBS2, VASP, COL5A1, COL11A1, FN1 |
| hsa05200 | Pathways in cancer | 23 | 2.49E-08 | COL4A4, AXIN2, KITLG, FOS, FZD6, TGFA, IGF1, GLI3, COL4A5, E2F1, TPM3, PRKCA, CRK, PIK3R3, CDKN1A, TGFB1, BAX, GLI2, TP53, PAX8, TRAF1, FN1, EGLN2 |
| hsa04060 | Cytokine-cytokine receptor interaction | 20 | 5.88E-08 | INHBB, TNFRSF10D, KITLG, BMPR1A, TNFRSF10B, CNTFR, IL28RA, TNFRSF21, CX3CR1, IFNG, LIFR, TGFB1, IL2RA, NGFR, CCR5, ACVR1B, CXCL12, TNFSF9, TNFSF10, CCR6 |
| hsa05146 | Amoebiasis | 13 | 1.03E-07 | ARG2, COL4A4, GNAL, ADCY1, COL4A5, RAB5B, IFNG, PRKCA, PIK3R3, TGFB1, COL5A1, COL11A1, FN1 |
| hsa04062 | Chemokine signaling pathway | 15 | 2.66E-06 | TIAM1, ADCY1, ROCK2, BCAR1, ADCY9, CX3CR1, RAP1A, CRK, PIK3R3, JAK3, GNG5, CCR5, CXCL12, GNG4, CCR6 |
| hsa05214 | Glioma | 8 | 8.41E-05 | CAMK2D, TGFA, IGF1, E2F1, PRKCA, PIK3R3, CDKN1A, TP53 |
| hsa05144 | Malaria | 4 | 1.01E-04 | IFNG, THBS1, TGFB1, THBS2 |
| hsa04350 | TGF-beta signaling pathway | 4 | 1.01E-04 | IFNG, THBS1, TGFB1, THBS2 |
| hsa04670 | Leukocyte transendothelial migration | 10 | 1.06E-04 | JAM3, ROCK2, BCAR1, ESAM, RAP1A, PRKCA, PIK3R3, CYBB, VASP, CXCL12 |
| hsa04115 | p53 signaling pathway | 8 | 1.18E-04 | TNFRSF10B, IGF1, CCND2, THBS1, SERPINE1, CDKN1A, BAX, TP53 |
| hsa04971 | Gastric acid secretion | 8 | 1.36E-04 | KCNK10, CAMK2D, ADCY1, ADCY9, MYLK2, PRKCA, ITPR1, SSTR2 |
| hsa04310 | Wnt signaling pathway | 11 | 1.42E-04 | NLK, CAMK2D, AXIN2, FZD6, ROCK2, CCND2, PRKCA, DAAM2, TBL1X, TP53, DAAM1 |
| hsa04722 | Neurotrophin signaling pathway | 10 | 1.56E-04 | CAMK2D, RAP1A, CRK, PIK3R3, YWHAG, NGFR, BAX, RPS6KA3, TP53, NTRK3 |
| hsa04630 | Jak-STAT signaling pathway | 11 | 1.58E-04 | CNTFR, CCND2, I L28RA, IFNG, LIFR, STAT6, SOCS3, PIK3R3, JAK3, IL2RA, CISH |
| hsa04020 | Calcium signaling pathway | 6 | 1.61E-04 | CAMK2D, ADCY1, ADCY9, MYLK2, PRKCA, ITPR1 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate; TGF, transforming growth factor; STAT, signal transducer and activator of transcription.
Discussion
IAs are considered to be the most fatal cerebrovascular system disease and are characterized by the apoptosis of smooth muscle cells, degeneration of vessel walls, and activation of the immune system in the aortic wall (29). miRNA have been revealed to be critical modulators in vascular biology and disease, such as atherosclerosis, arterial remodeling, angiogenesis, and smooth muscle cell regeneration (30). Additionally, miRNA may have vital roles in IA development by regulating downstream genes.
Studies have examined the miRNA and mRNA expression profiles in IA to identify differentially-expressed miRNA and genes. However, inconsistent results were obtained due to platform differences, tissue sampling and control selection in gene expression profiles (9,31–33). In the present study, miRNA and mRNA expression data were integrated to identify differentially expressed miRNA and mRNA between IAs and normal tissues. Subsequently, 770 miRNA-target gene pairs with inversely correlated expression levels were identified via bioinformatics prediction were selected to construct a miRNA-target gene regulatory network in IA.
In the miRNA-target gene regulatory network, the top ten miRNA (hsa-miR-7, hsa-miR-182, hsa-miR-324-3p, hsa-miR-139-5p, hsa-miR-130b, hsa-let-7f, hsa-miR-18a, hsa-miR-188-5p, hsa-let-7d and hsa-miR-590-5p) were identified to regulate the greatest number of target genes. Target genes, such as RPS6KA3, TSC1, AIM1, GAS7, GFOD1, GGA2, IGF1, IL28RA and INSR, were indicated to be regulated by the greatest number of miRNA. hsa-let-7d and hsa-let-7f, two members of the let family, which are enriched in endothelium, were revealed to be differently expressed between six intracranial aneurysmal samples and normal superficial temporal arteries by genome-wide microRNA screening (32). hsa-miR-7, which is brain-enriched, may be implicated in the pathogenesis of glioblastoma, characterized by microvascular proliferation (34–36). Furthermore, hsa-miR-7 may function as a tumor suppressor gene to regulate glioblastoma microvascular endothelial cell proliferation by targeting RAF1. To the best of our knowledge, no reports of hsa-miR-7 in IAs have been published. In the present study, 94 targets of hsa-miR-7 were indicated to be significantly enriched in the mTOR signaling pathway and may modulate the apoptosis of muscle cell differentiation in IAs.
RPS6KA3, the target gene regulated by the greatest number of miRNA, has been indicated to be expressed in high levels in regions with high synaptic activity (37). Moreover, RPS6KA3 has been suggested to be associated with Coffin-Lowry syndrome, which causes severe mental problems sometimes associated with abnormalities of growth, cardiac abnormalities, kyphoscoliosis, as well as auditory and visual abnormalities (38). Molecular evidence from previous studies has revealed that RPS6KA3 may regulate neurotransmitter release by activating phospholipase D production of lipids required for exocytosis and that RPS6KA3 may also function as a proto-oncogene in multiple types of cancer targeted by corresponding miRNA (39,40).
Mutations in either tuberous sclerosis (TSC)1 or TSC2 suppressor genes are able to provoke tuberous sclerosis complex, which is an autosomal dominant disorder promoting the development of benign tumors in multiple organ systems, including the skin, brain, and kidneys, via increasing mammalian target of rapamycin (mTOR) activity (41,42). TSC1 also has a role in arterial remodeling events by affecting the inflammatory and the growth-promoting response of angiotensin II (43). Insulin-like growth factor 1 (IGF1) expression has been indicated in the vasculature and lower IGF1 expression levels increased the risk of cardiovascular and abdominal aortic aneurysm in a previous study (44). Histological analysis in a swine aneurysm model has demonstrated that IGF1 is upregulated (4-fold) in thrombus organization (45).
With regards to the pathways that the identified target genes were involved in, focal adhesion was the most significant pathway revealed in KEGG analysis. This finding was consistent with a previous study by Shi et al (8), in which Illumina microarray analysis was performed on human the aneurysm wall of IAs. Based on the fact that IAs arise from progressive wall degeneration and remodeling in brain artery walls, focal adhesion may be involved in the pathogenesis of IA.
In conclusion, the present study identified 15 differentially expressed miRNA and 1,447 differentially expressed mRNA between IAs and normal tissues and constructed a regulatory network including 770 miRNA-target gene pairs with inversely correlated expression levels. In this network, several miRNA and genes that may possess key roles in IAs were discovered, such as miRNA hsa-let-7f, hsa-let-7d and hsa-miR-7, and genes, including RPS6KA3, TSC1 and IGF1. The biological pathway of focal adhesion may be involved in the pathogenesis of IA. The findings in the present study may contribute to future investigations aimed at elucidating the mechanisms of IAs.
References
- 1.Brown RD. Unruptured intracranial aneurysms. Semin Neurol. 2010;30:537–544. doi: 10.1055/s-0030-1268858. [DOI] [PubMed] [Google Scholar]
- 2.Nieuwkamp DJ, Setz LE, Algra A, Linn FH, de Rooij NK, Rinkel GJ. Changes in case fatality of aneurysmal subarachnoid haemorrhage over time, according to age, sex and region: A meta-analysis. Lancet Neurol. 2009;8:635–642. doi: 10.1016/S1474-4422(09)70126-7. [DOI] [PubMed] [Google Scholar]
- 3.van Gijn J, Kerr RS, Rinkel GJ. Subarachnoid haemorrhage. Lancet. 2007;369:306–318. doi: 10.1016/S0140-6736(07)60153-6. [DOI] [PubMed] [Google Scholar]
- 4.Tromp G, Weinsheimer S, Ronkainen A, Kuivaniemi H. Molecular basis and genetic predisposition to intracranial aneurysm. Ann Med. 2014;46:597–606. doi: 10.3109/07853890.2014.949299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalouhi N, Ali MS, Starke RM, Jabbour PM, Tjoumakaris SI, Gonzalez LF, Rosenwasser RH, Koch WJ, Dumont AS. Cigarette smoke and inflammation: Role in cerebral aneurysm formation and rupture. Mediators Inflamm. 2012;2012:271582. doi: 10.1155/2012/271582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krischek B, Kasuya H, Tajima A, Akagawa H, Sasaki T, Yoneyama T, Ujiie H, Kubo O, Bonin M, Takakura K, et al. Network-based gene expression analysis of intracranial aneurysm tissue reveals role of antigen presenting cells. Neuroscience. 2008;154:1398–1407. doi: 10.1016/j.neuroscience.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Yang X, Jiang F, Dusting GJ, Wu Z. Transcriptome-wide characterization of gene expression associated with unruptured intracranial aneurysms. Eur Neurol. 2009;62:330–337. doi: 10.1159/000236911. [DOI] [PubMed] [Google Scholar]
- 8.Shi C, Awad IA, Jafari N, Lin S, Du P, Hage ZA, Shenkar R, Getch CC, Bredel M, Batjer HH, Bendok BR. Genomics of human intracranial aneurysm wall. Stroke. 2009;40:1252–1261. doi: 10.1161/STROKEAHA.108.532036. [DOI] [PubMed] [Google Scholar]
- 9.Marchese E, Vignati A, Albanese A, Nucci CG, Sabatino G, Tirpakova B, Lofrese G, Zelano G, Maira G. Comparative evaluation of genome-wide gene expression profiles in ruptured and unruptured human intracranial aneurysms. J Biol Regul Homeost Agents. 2010;24:185–195. [PubMed] [Google Scholar]
- 10.Pera J, Korostynski M, Krzyszkowski T, Czopek J, Slowik A, Dziedzic T, Piechota M, Stachura K, Moskala M, Przewlocki R, Szczudlik A. Gene expression profiles in human ruptured and unruptured intracranial aneurysms: What is the role of inflammation? Stroke. 2010;41:224–231. doi: 10.1161/STROKEAHA.109.562009. [DOI] [PubMed] [Google Scholar]
- 11.Kao HW, Lee KW, Kuo CL, Huang CS, Tseng WM, Liu CS, Lin CP. Interleukin-6 as a prognostic biomarker in ruptured intracranial aneurysms. PLoS One. 2015;10:e0132115. doi: 10.1371/journal.pone.0132115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sathyan S, Koshy LV, Srinivas L, Easwer HV, Premkumar S, Nair S, Bhattacharya RN, Alapatt JP, Banerjee M. Patho genesis of intracranial aneurysm is mediated by proinflammatory cytokine TNFA and IFNG and through stochastic regulation of IL10 and TGFB1 by comorbid factors. J Neuroinflammation. 2015;12:135. doi: 10.1186/s12974-015-0354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinsheimer S, Lenk GM, van der Voet M, Land S, Ronkainen A, Alafuzoff I, Kuivaniemi H, Tromp G. Integration of expression profiles and genetic mapping data to identify candidate genes in intracranial aneurysm. Physiol Genomics. 2007;32:45–57. doi: 10.1152/physiolgenomics.00015.2007. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rooij E. The art of microRNA research. Circ Res. 2011;108:219–234. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 17.Li P, Zhang Q, Wu X, Yang X, Zhang Y, Li Y, Jiang F. Circulating microRNAs serve as novel biological markers for intracranial aneurysms. J Am Heart Assoc. 2014;3:e000972. doi: 10.1161/JAHA.114.000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 20.Reimers M, Carey VJ. Bioconductor: An open source framework for bioinformatics and computational biology. Methods Enzymol. 2006;411:119–134. doi: 10.1016/S0076-6879(06)11008-3. [DOI] [PubMed] [Google Scholar]
- 21.Dweep H, Sticht C, Pandey P, Gretz N. MiRWalk-database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Luo Z, Zhang L, Li Z, Li X, Li G, Yu H, Jiang C, Dai Y, Guo X, Xiang J, Li G. An in silico analysis of dynamic changes in microRNA expression profiles in stepwise development of nasopharyngeal carcinoma. BMC Med Genomics. 2012;5:3. doi: 10.1186/1755-8794-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, Sales G, Deliliers GL, Bicciato S, Lombardi L, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009;114:e20–e26. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- 24.Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Rønneberg JA, Johnsen H, Navon R, Rødland E, et al. MiRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3:A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40(Web Server issue):W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altermann E, Klaenhammer TR. Pathwayvoyager: Pathway mapping using the kyoto encyclopedia of genes and genomes (KEGG) database. BMC Genomics. 2005;6:60. doi: 10.1186/1471-2164-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakaoka H, Tajima A, Yoneyama T, Hosomichi K, Kasuya H, Mizutani T, Inoue I. Gene expression profiling reveals distinct molecular signatures associated with the rupture of intracranial aneurysm. Stroke. 2014;45:2239–2245. doi: 10.1161/STROKEAHA.114.005851. [DOI] [PubMed] [Google Scholar]
- 29.Brown RD, Jr, Broderick JP. Unruptured intracranial aneurysms: Epidemiology, natural history, management options and familial screening. Lancet Neurol. 2014;13:393–404. doi: 10.1016/S1474-4422(14)70015-8. [DOI] [PubMed] [Google Scholar]
- 30.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu L, Fan J, Wang S, Zhang D, Wang R, Zhao Y, Zhao J. Gene expression profiles in intracranial aneurysms. Neurosci Bull. 2014;30:99–106. doi: 10.1007/s12264-013-1398-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D, Han L, Wu X, Yang X, Zhang Q, Jiang F. Genome-wide microRNA changes in human intracranial aneurysms. BMC Neurol. 2014;14:188. doi: 10.1186/s12883-014-0188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei L, Gao YJ, Wei SP, Zhang YF, Zhang WF, Jiang JX, Sun ZY, Xu W. Transcriptome network-based method to identify genes associated with unruptured intracranial aneurysms. Genet Mol Res. 2013;12:3263–3273. doi: 10.4238/2013.September.3.2. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z, Liu Y, Li L, Xu Z, Bi B, Wang Y, Li JY. MiR-7-5p is frequently downregulated in glioblastoma microvasculature and inhibits vascular endothelial cell proliferation by targeting RAF1. Tumour Biol. 2014;35:10177–10184. doi: 10.1007/s13277-014-2318-x. [DOI] [PubMed] [Google Scholar]
- 35.Dong L, Li Y, Han C, Wang X, She L, Zhang H. MiRNA microarray reveals specific expression in the peripheral blood of glioblastoma patients. Int J Oncol. 2014;45:746–756. doi: 10.3892/ijo.2014.2459. [DOI] [PubMed] [Google Scholar]
- 36.Visani M, de Biase D, Marucci G, Cerasoli S, Nigrisoli E, Reggiani ML Bacchi, Albani F, Baruzzi A, Pession A, PERNO study group Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I–III. Mol Oncol. 2014;8:417–430. doi: 10.1016/j.molonc.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeniou M, Ding T, Trivier E, Hanauer A. Expression analysis of RSK gene family members: The RSK2 gene, mutated in coffin-lowry syndrome, is prominently expressed in brain structures essential for cognitive function and learning. Hum Mol Genet. 2002;11:2929–2940. doi: 10.1093/hmg/11.23.2929. [DOI] [PubMed] [Google Scholar]
- 38.Nishimoto HK, Ha K, Jones JR, Dwivedi A, Cho HM, Layman LC, Kim HG. The historical coffin-lowry syndrome family revisited: Identification of two novel mutations of RPS6KA3 in three male patients. Am J Med Genet A. 2014;164A:2172–2179. doi: 10.1002/ajmg.a.36488. [DOI] [PubMed] [Google Scholar]
- 39.Polioudakis D, Abell NS, Iyer VR. MiR-191 regulates primary human fibroblast proliferation and directly targets multiple oncogenes. PLoS One. 2015;10:e0126535. doi: 10.1371/journal.pone.0126535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song R, Liu Q, Hutvagner G, Nguyen H, Ramamohanarao K, Wong L, Li J. Rule discovery and distance separation to detect reliable miRNA biomarkers for the diagnosis of lung squamous cell carcinoma. BMC Genomics. 2014;15(Suppl 9):S16. doi: 10.1186/1471-2164-15-S9-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan JA, Zhang H, Roberts PS, Jozwiak S, Wieslawa G, Lewin-Kowalik J, Kotulska K, Kwiatkowski DJ. Pathogenesis of tuberous sclerosis subependymal giant cell astrocytomas: Biallelic inactivation of TSC1 or TSC2 leads to mTOR activation. J Neuropathol Exp Neurol. 2004;63:1236–1242. doi: 10.1093/jnen/63.12.1236. [DOI] [PubMed] [Google Scholar]
- 42.Habib SL, Yadav A, Mahimainathan L, Valente AJ. Regulation of PI 3-K, PTEN, p53 and mTOR in malignant and benign tumors deficient in tuberin. Genes Cancer. 2011;2:1051–1060. doi: 10.1177/1947601912445376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyon P, van Zuylen WJ, Servant MJ. Role of IκB kinase-β in the growth-promoting effects of angiotensin II in vitro and in vivo. Arterioscler Thromb Vasc Biol. 2013;33:2850–2857. doi: 10.1161/ATVBAHA.113.302487. [DOI] [PubMed] [Google Scholar]
- 44.Yeap BB, Chubb SA, McCaul KA, Flicker L, Ho KK, Golledge J, Hankey GJ, Norman PE. Associations of IGF1 and its binding proteins with abdominal aortic aneurysm and aortic diameter in older men. Eur J Endocrinol. 2012;166:191–197. doi: 10.1530/EJE-11-0725. [DOI] [PubMed] [Google Scholar]
- 45.Lee D, Yuki I, Murayama Y, Chiang A, Nishimura I, Vinters HV, Wang CJ, Nien YL, Ishil A, Wu BM, Viñuela F. Thrombus organization and healing in the swine experimental aneurysm model. Part I. A histological and molecular analysis. J Neurosurg. 2007;107:94–108. doi: 10.3171/JNS-07/07/0094. [DOI] [PubMed] [Google Scholar]