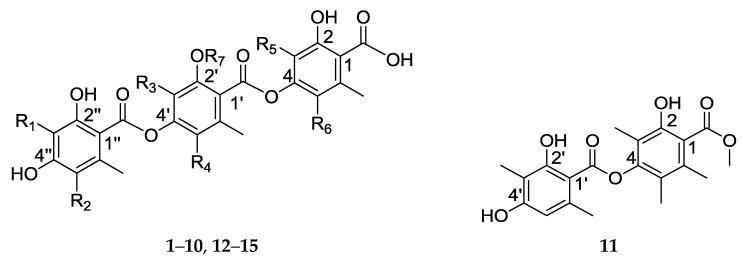
Abstract
Eleven new depsides—thielavins W–Z (1–4) and thielavins Z1–Z7 (5–11)—and also four known thielavins—A, H, J, and K (12–15)—were isolated from the ethyl acetate extract of a marine-derived fungal strain Thielavia sp UST030930-004. All of these compounds were evaluated for antifouling activity against cyprids of the barnacle Balanus (=Amphibalanus) amphitrite. The results showed that compounds 1–3 and 6–13 were active, with EC50 values ranging from 2.95 ± 0.59 to 69.19 ± 9.51 μM, respectively. The inhibitive effect of compounds 1–3 and 7 was reversible. This is the first description of the antifouling activity of thielavins against barnacle cyprids.
Keywords: marine-derived fungus, Thielavia sp., antifouling, Balanus (=Amphibalanus) amphitrite, thielavins
1. Introduction
Marine microorganisms are being explored as potentially important sources of environmentally friendly antifouling (AF) compounds because they can be cultivated to produce diverse chemical compounds under optimal culture conditions [1]. Several potent antifouling compounds have been isolated from various microorganisms, including polyethers from the marine bacterium Winogradskyella Poriferorum [2], 12-methyltetradecanoid acid and butenolides from Streptomyces sp. UST040711-290 and Streptomyces Albidoflavus [3,4], 3-chloro-2,5-dihydroxybenzyl alcohol from the marine fungus Ampelomyces sp. [5], and Diindol-3-ylmethanes from Pseudovibrio Denitrificans [6]. Of these compounds, butenolides are regarded as the most potent natural antifoulants [7]. The binding targets of the butenolide 5-octylfuran-2(5H)-one have been identified in the barnacle Balanus (=Amphibalanus) amphitrite, the bryozoan Bugula neritina and the marine bacterium Vibrio sp. UST020129-010 [8]. These studies clearly demonstrate that marine microbes are a promising source of effective antifouling compounds.
In the present study, we report 11 new depsides—thielavins W–Z, Z1–Z7 (1–11)—and four known thielavins (Figure 1)—thielavins A, H, J, and K (12–15) [9]—which we isolated from the marine-derived fungal strain Thielavia sp. UST030930-004, and we evaluated the antifouling activity of each against the barnacle B. amphitrite cyprids.
Figure 1.
Structures of compounds 1–15.
| R1 | R2 | R3 | R4 | R5 | R6 | R7 | |
|---|---|---|---|---|---|---|---|
| 1 | H | H | Me | Me | Me | H | H |
| 2 | H | H | Me | Me | Me | H | Me |
| 3 | Me | H | Me | Me | H | H | Me |
| 4 | Me | H | H | Me | Me | H | Me |
| 5 | Me | H | Me | H | Me | H | Me |
| 6 | Me | H | Me | H | Me | Me | Me |
| 7 | Me | H | Me | Me | Me | H | H |
| 8 | H | H | Me | H | Me | H | Me |
| 9 | H | H | Me | H | Me | Me | Me |
| 10 | Me | Cl | Me | Me | Me | Me | Me |
| 12 | Me | H | Me | Me | Me | Me | H |
| 13 | H | H | Me | Me | Me | Me | H |
| 14 | Me | H | Me | Me | Me | H | Me |
| 15 | Me | H | Me | Me | Me | Me | Me |
2. Results and Discussion
2.1. Identification of the Fungus
The internal transcribed spacer (ITS) sequence of the fungal isolate UST030930-004 (GenBank accession number KJ716558) had a similarity of 97% with Thielavia terrestris NRRL 8126 (CP003011), indicating that isolate UST030930-004 is a Thielavia sp. closely related to this taxon.
2.2. Structure Elucidation
Fifteen compounds were isolated from the ethyl acetate extract of Thielavia sp. UST030930-004, including the 11 new compounds—thielavins W–Z (1–4) and thielavins Z1–Z7 (5–11)—together with 4 known compounds—thielavins A, H, J, and K (12–15). Their chemical structures were determined using MS, 1D- and 2D-NMR spectroscopy.
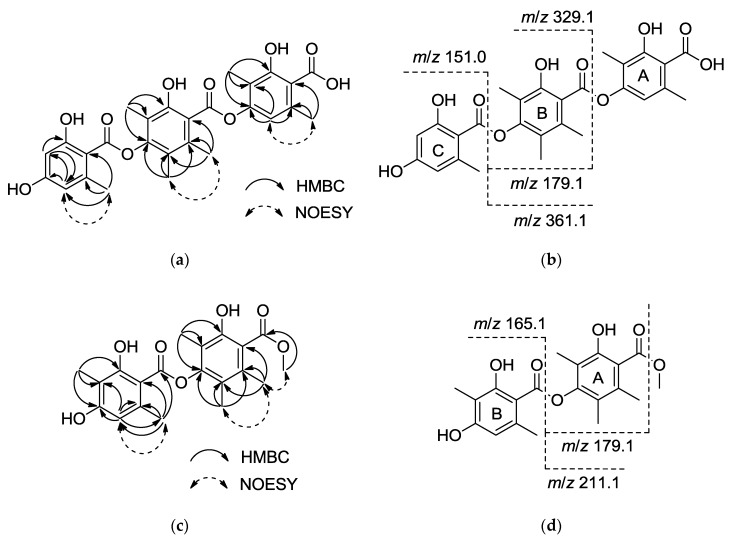
Compound 1 was isolated as a white amorphous powder. The positive HRESIMS provided [M + H]+ at m/z 511.1604, corresponding to a molecular formula of C27H26O10 (calcd. for C27H27O10 [M + H]+ 511.1599), and the UV spectrum revealed absorption at λmax 218.1, 267.8, and 304.2 nm. The 1H, 13C-NMR (Table 1) and HSQC spectra of 1 indicated the presence of 6 methyl groups at δH 2.52 (3H, s), 2.40 (3H, s), 2.29 (3H, s), 2.06 (3H, s), 2.06 (3H, s), 1.99 (3H, s), 18 olefinic carbons, including fifteen quaternary carbons at δC 138.7, 165.4, 114.2, 149.5, 114.7, 131.8, 150.7, 116.5, 149.1, 120.8, 121.6, 141.0, 161.4, 160.8, and 107.6, and 3 tertiary carbons at δC 110.3, 100.6, 110.3. Three carbonyl signals were also observed at δC 172.0, 167.1, and 166.7. These data suggested that the compound is similar to thielavin H (13) [9]. The only difference between 1 and thielavin H was that one methyl group in thielavin H was absent in 1. The structure of 1 was established based on HMBC correlations and in-source collision-induced dissociation (ISCID) fragment ions. The HMBC correlations observed from the methyl group δH 1.99 (3-Me) to δC 165.4 (C-2), 114.2 (C-3) and 149.5 (C-4), from δH 2.52 (6-Me) to δC 138.7 (C-1), 110.3 (C-5) and 114.7 (C-6), and from an olefinic proton δH 6.18 (H-5) to C-4 and C-6 indicated the presence of a 2,4-dioxygenated-3,6-dimethylbenzene substructure (A). The second substructure (B) was confirmed by long-range couplings from δH 2.06 (3′-Me) to δC 150.7 (C-2′), 116.5 (C-3′) and 149.1(C-4′), and from δH 2.06 (5′-Me) to δC 120.8 (C-5′), 121.6 (C-6′) and C-4′, and couplings from another methyl group δH 2.29 (6′-Me) to δC 131.8 (C-1′), C-5′ and C-6′. The HMBC correlations from δH 2.40 (6″-Me) to δC 141.0 (C-1″), 110.3 (C-5″) and 107.6 (C-6″), correlations from an olefinic proton δH 6.26 (H-3″) to δC 161.4 (C-2″), C-5″, and correlations from olefinic proton δH 6.26 (H-5″) to δC 100.6 (C-3″), 160.8 (C-4″) and C-6″, indicated the presence of a 2,4-dioxygenated-6-methylbenzene substructure (C). From the deduction above, the remaining three carbonyl groups can only be assigned to C-1, C-1′ and C-1″, and these carbonyl groups may be interchangeable as no HMBC correlations were observed (Figure 2a). The positive ISCID MS/MS gave ion peaks at m/z 329.1, 151.0, 361.1 and 179.1, suggesting the absence of [M − A]+, [M − A − B]+, [M − C + 2H]+ and [M − A − C + 2H]+ fragments (Figure 2b), indicating that the order of the substructures is A − B − C (Figure 2b). Thus the structure of 1 was determined (shown in Figure 1) and named thielavin W.
Table 1.
The 1H (500 MHz) and 13C-NMR (125 MHz) NMR data for compounds 1–4 (δ in ppm, DMSO-d6).
| Position | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δH | δC | |
| 1 | 138.7 | 139.0 | 142.1 | 138.7 | ||||
| 2 | 165.4 | 164.8 | 161.2 | 165.1 | ||||
| 3 | 114.2 | 114.3 | 6.36 d 2.0 | 107.0 | 114.4 | |||
| 4 | 149.5 | 150.8 | 151.5 | 149.8 | ||||
| 5 | 6.18 s | 110.3 | 6.21 s | 111.2 | 6.29 d 2.0 | 111.3 | 6.41 s | 111.3 |
| 6 | 114.7 | 115.5 | 117.5 | 115.3 | ||||
| 1′ | 131.8 | 132.2 | 132.4 | 135.5 | ||||
| 2′ | 150.7 | 153.2 | 153.2 | 154.5 | ||||
| 3′ | 116.5 | 121.8 | 121.6 | 7.01 s | 104.2 | |||
| 4′ | 149.1 | 149.5 | 148.9 | 149.9 | ||||
| 5′ | 120.8 | 125.8 | 125.6 | 121.2 | ||||
| 6′ | 121.6 | 126.2 | 126.4 | 121.7 | ||||
| 1″ | 141.0 | 141.0 | 139.5 | 139.3 | ||||
| 2″ | 161.4 | 161.4 | 162.7 | 162.2 | ||||
| 3″ | 6.26 s | 100.6 | 6.27 s | 100.6 | 108.7 | 114.4 | ||
| 4″ | 160.8 | 160.6 | 160.6 | 160.9 | ||||
| 5″ | 6.26 s | 110.3 | 6.27 s | 110.2 | 6.44 s | 111.3 | 6.14 s | 111.3 |
| 6″ | 107.6 | 107.2 | 102.8 | 103.5 | ||||
| 1-C=O | 172.0 * | 172.5 * | 170.7 * | 171.9 * | ||||
| 2-OMe | ||||||||
| 3-Me | 1.99 s | 9.4 | 1.99 s | 9.0 | 1.98 s | 8.1 | ||
| 5-Me | ||||||||
| 6-Me | 2.52 s | 23.1 | 2.40 s | 23.0 | 2.56 s | 24.1 | 2.55 s | 23.8 |
| 1′-C=O | 167.1 * | 166.9 * | 169.3 * | 169.7 * | ||||
| 2′-OMe | 3.78 s | 62.0 | 3.79 s | 62.0 | 3.82 s | 56.3 | ||
| 3′-Me | 2.06 s | 10.2 | 2.13 s | 10.0 | 2.08 s | 9.9 | ||
| 5′-Me | 2.06 s | 12.5 | 2.12 s | 12.7 | 2.06 s | 12.6 | 2.03 s | 12.0 |
| 6′-Me | 2.29 s | 16.7 | 2.31 s | 16.6 | 2.29 s | 16.5 | 2.32 s | 16.8 |
| 1″-C=O | 166.7 * | 165.9 * | 165.9 * | 165.7 * | ||||
| 3″-Me | 1.99 s | 8.1 | 1.96 s | 9.1 | ||||
| 5″-Me | ||||||||
| 6″-Me | 2.40 s | 21.8 | 2.54 s | 21.8 | 2.55 s | 21.8 | 2.52 s | 23.1 |
NMR assignments marked by an asterisk (*) are interchangeable.
Figure 2.
(a) Key HMBC and NOESY correlations; (b) ESIMS in-source fragmentation of compound 1; (c) Key HMBC and NOESY correlations; (d) ESIMS in-source fragmentation of compound 11.
Comparison of the UV, NMR, HRESIMS and ISCID MS/MS data shows that compounds 2–10 have the same core structure as compound 1. Their structures were determined using the procedure described for compound 1, and named thielavins X–Z and Z1–Z5, respectively. All of the 1H and 13C-NMR assignments of the new compounds 2–10 are summarized in Table 1, Table 2, Table 3 and Table 4.
Table 2.
The 1H (500 MHz) and 13C-NMR (125 MHz) NMR data for compounds 5–8 (δ in ppm, DMSO-d6).
| Position | 5 | 6 | 7 | 8 | ||||
|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δH | δC | |
| 1 | 139.1 | 139.1 | 138.9 | 138.9 | ||||
| 2 | 156.2 | 156.7 | 162.0 | 165.2 | ||||
| 3 | 115.4 | 115.4 | 116.5 | 114.2 | ||||
| 4 | 151.0 | 150.7 | 151.7 | 149.5 | ||||
| 5 | 6.40 s | 113.2 | 118.6 | 6.64 s | 115.8 | 6.16 s | 109.8 | |
| 6 | 114.1 | 120.4 | 112.1 | 115.8 | ||||
| 1′ | 134.3 | 134.8 | 132.0 | 134.0 | ||||
| 2′ | 156.2 | 156.2 | 150.7 | 156.1 | ||||
| 3′ | 122.0 | 122.2 | 116.5 | 122.0 | ||||
| 4′ | 150.6 | 149.3 | 148.8 | 150.8 | ||||
| 5′ | 7.10 s | 120.1 | 7.12 s | 120.4 | 120.7 | 7.00 s | 119.9 | |
| 6′ | 125.7 | 125.4 | 121.7 | 126.0 | ||||
| 1″ | 139.1 | 139.1 | 139.5 | 140.4 | ||||
| 2″ | 162.0 | 162.0 | 162.7 | 160.0 | ||||
| 3″ | 108.6 | 108.6 | 108.6 | 6.25 d 2.0 | 100.7 | |||
| 4″ | 160.9 | 160.9 | 161.0 | 161.1 | ||||
| 5″ | 6.40 s | 111.1 | 6.41 s | 111.0 | 6.42 s | 111.2 | 6.24 d 2.0 | 109.9 |
| 6″ | 103.6 | 103.7 | 102.8 | 108.3 | ||||
| 1-C=O | 172.7 * | 171.5 * | 173.0 * | 171.7 * | ||||
| 2-OMe | ||||||||
| 3-Me | 2.03 s | 9.1 | 2.08 s | 9.9 | ||||
| 5-Me | 2.10 s | 12.6 | 2.10 s | 9.2 | 1.97 s | 9.3 | ||
| 6-Me | 2.52 s | 23.6 | 2.40 s | 17.4 | ||||
| 1′-C=O | 169.4 * | 169.4 * | 2.50 s | 22.9 | 2.53 s | 23.1 | ||
| 2′-OMe | 3.82 s | 61.9 | 3.79 s | 61.9 | 169.5 * | 167 * | ||
| 3′-Me | 2.11 s | 9.6 | 2.13 s | 9.6 | 3.81 s | 62.0 | ||
| 5′-Me | 2.02 s | 10.1 | 2.14 s | 9.6 | ||||
| 6′-Me | 2.4 s | 18.7 | 2.40 s | 19.2 | 2.01 s | 12.4 | ||
| 1″-C=O | 165.1 * | 165.0 * | 2.30 s | 16.7 | 2.39 s | 18.6 | ||
| 3″-Me | 1.98 s | 8.1 | 1.98 s | 8.1 | 166.4 * | 165.4 * | ||
| 5″-Me | 1.98 s | 8.0 | ||||||
| 6″-Me | 2.52 s | 23.0 | 2.53 s | 23.6 | ||||
NMR assignments marked by an asterisk (*) are interchangeable.
Table 3.
The 1H (500 MHz) and 13C-NMR (125 MHz) NMR data for compounds 9–11 (δ in ppm, DMSO-d6).
| Position | 9 | 10 | 11 | |||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | 136.8 | 138.3 | 132.1 | |||
| 2 | 161.3 | 160.0 | 150.7 | |||
| 3 | 113.3 | 116.5 | 116.0 | |||
| 4 | 148.6 | 151.8 | 148.5 | |||
| 5 | 115.5 | 120.6 | 120.1 | |||
| 6 | 117.5 | 115.7 | 121.4 | |||
| 1′ | 134.5 | 134.1 | 139.4 | |||
| 2′ | 156.6 | 155.0 | 162.6 | |||
| 3′ | 122.2 | 122.7 | 108.6 | |||
| 4′ | 150.9 | 150.3 | 161.0 | |||
| 5′ | 7.01 s | 120.2 | 126.4 | 6.40 s | 111.0 | |
| 6′ | 125.7 | 127.5 | 102.7 | |||
| 1″ | 140.3 | 136.2 | ||||
| 2″ | 161.1 | * 160.8 | ||||
| 3″ | 6.23 s | 100.5 | 112.8 | |||
| 4″ | 161.2 | * 157.9 | ||||
| 5″ | 6.23 s | 109.9 | 107.2 | |||
| 6″ | 108.3 | 116.2 | ||||
| 1-C=O | 171.8 * | UD | 168.7 * | |||
| 1-COOMe | 3.83 s | 51.9 | ||||
| 2-OMe | ||||||
| 3-Me | 2.01 s | 9.8 | 2.62 s | 9.6 | 1.96 s | 9.7 |
| 5-Me | 2.04 s | 12.5 | 2.42 s | 12.6 | 1.96 s | 12.3 |
| 6-Me | 2.58 s | 17.4 | 2.84 s | 17.8 | 2.13 s | 16.6 |
| 1′-C=O | 167.1 * | UD | 168.9 | |||
| 2′-OMe | 3.78 s | 62.0 | 3.90 s | 61.9 | ||
| 3′-Me | 2.16 s | 9.7 | 2.36 s | 9.7 | 1.97 s | 7.8 |
| 5′-Me | 2.21 s | 12.6 | ||||
| 6′-Me | 2.40 s | 19.1 | 2.44 s | 16.7 | 2.54 s | 24.0 |
| 1″-C=O | 165.8 * | UD | ||||
| 3″-Me | 2.57 s | 9.2 | ||||
| 5″-Me | ||||||
| 6″-Me | 2.39 s | 21.4 | 2.94 s | 19.4 | ||
NMR assignments marked by an asterisk (*) are interchangeable. UD: undetected. 13C-NMR signals were not observed due to a limited amount of compound 10.
Table 4.
Physicochemical properties of new thielavins.
| 1 | 2 | 3 | 4 | |
|---|---|---|---|---|
| Molecular Formula | C27H27O10 | C28H29O10 | C28H29O10 | C28H29O10 |
| HRESIMS (pos) Obsd. (m/z) | 511.1604 | 525.1727 | 525.1765 | 525.1757 |
| Cald. (m/z) | 511.1599 | 525.1755 | 525.1755 | 525.1755 |
| ISCID (m/z) | 493.2 | 507.2 | 507.1 | |
| 361.1 | 375.2 | 361.1 | 361.1 | |
| 329.1 | 343.1 | 357.1 | 343.1 | |
| 179.1 | 193.1 | 193.1 | 179.1 | |
| 151.0 | 151.0 | 165.1 | 165.1 | |
| UV | 218.1 | 218.1 | 218.1 | 218.1 |
| 267.8 | 267.8 | 276.1 | 276.1 | |
| 304.2 | 306.5 | 304.7 | 306.5 | |
| 5 | 6 | 7 | 8 | |
| Molecular Formula | C28H29O10 | C29H31O10 | C28H29O10 | C27H27O10 |
| HRESIMS (pos) Obsd. (m/z) | 525.1747 | 539.1916 | 525.1758 | 511.1564 |
| Cald. (m/z) | 525.1755 | 539.1912 | 525.1755 | 511.1599 |
| ISCID (m/z) | 507.2 | 479.1 | ||
| 361.1 | 375.2 | 361.1 | 361.1 | |
| 343.1 | 343.1 | 343.1 | 329.1 | |
| 179.1 | 179.1 | 179.1 | 179.1 | |
| 165.1 | 165.1 | 165.1 | 151.0 | |
| UV | 218.1 | 218.1 | 214.6 | 218.1 |
| 276.1 | 276.8 | 276.1 | 267.8 | |
| 306.9 | 305.2 | 309.1 | 305.2 | |
| 9 | 10 | 11 | ||
| Molecular Formula | C28H29O10 | C30H32ClO10 | C20H23O7 | |
| HRESIMS (pos) Obsd. (m/z) | 525.1778 | 587.1719/589.1646 | 375.1464 | |
| Cald. (m/z) | 525.1755 | 587.1679 | 375.1438 | |
| ISCID (m/z) | 389.1 | |||
| 375.2 | 391.1/393.1 | |||
| 329.1 | 199.0/201.0 | 211.1 | ||
| 179.1 | 193.1 | 179.1 | ||
| 151.0 | 165.1 | |||
| UV | 213.4 | 215.8 | 215.8 | |
| 267.8 | 276.1 | 276.1 | ||
| 307.0 | 307.0 | 308.9 |
Compound 11 was obtained as a white amorphous powder. Positive HRESIMS gave [M + H]+ as m/z 375.1464, and the molecular formula was established as C20H22O7 (calcd. for C20H23O7 [M + H]+ 375.1438). The maximum UV absorption occurred at λmax 215.8, 276.1, and 308.9 nm, and the 1H NMR (Table 3) revealed the presence of 5 quaternary methyl signals [δH 1.96 (3H, s), 1.96 (3H, s), 2.13 (3H, s), 1.97 (3H, s), and 2.54 (3H, s)], 1 methoxy signal [δH 3.83 (3H, s)], and 1 olefinic proton signal [δH 6.40 (1H, s)]. The 13C-NMR and HSQC results (Table 3) indicated the presence of 12 olefinic carbons, including 11 quaternary carbons at δC 162.6, 161.0, 150.7, 148.5, 139.4, 132.1, 121.4, 120.1, 116.0, 108.6, and 102.7, 1 tertiary carbon at δC 111.0 and 2 carbonyl groups at δC 168.7 and 168.9. Based on these data, compound 11 was proposed as a thielavin derivative consisting of two hydroxybenzoic acid groups with a methyl ester terminus. HMBC revealed correlations (Figure 2c) between δH 1.96 (3-Me) and δC 150.7 (C-2), 116.0 (C-3) and 148.5 (C-4), between δH 1.96 (s, 5-Me) and δC 120.1 (C-5), 121.4 (C-6), C-4, and between δH 2.13 (6-Me) to δC 132.1 (C-1), C-5 and C-6, and also revealed long range correlations between δH 3.83 (1-COOMe) and δC 168.7 (1-C=O). NOESY results show the correlation from protons 6-Me to methoxy proton (1-COOMe), indicating the presence of a 2-hydroxyl-4-oxygenated-3,5,6-trimethylbenzoic methyl ester subunit, corresponding to the ion fragment of m/z 179.1. The HMBC correlations (Figure 2c) from δH 1.97 (3′-Me) to δC 162.6 (C-2′), 108.6 (C-3′) and 161.0 (C-4′), from δH 6.40 (H-5′) to δC 102.7 (C-6′), 24.0 (6′-Me), C-3′ and C-4′, and from δH 2.54 (6′-Me) to δC 139.4 (C-1′), 111.0 (C-5′) and C-6′, suggested the presence of a 2-hydroxyl-4-oxygenated-3,6-dimethylbenzoyl subunit, corresponding to an ion fragment with m/z 211.1 (Figure 2d). These two units should be connected by an ester bond, but there was no correlation between the two units and the connection was only supported by ion fragments [M − B + 2H]+, [M – Ome − B + H]+ and [M – Ome − A]+ at m/z 211.1, 179.1 and 165.1, respectively. Thus, the structure of 11 was determined (shown in Figure 1) and named thielavin Z7.
Compounds 12–15 were identified as thielavin A (12), thielavin H (13), thielavin J (14), and thielavin K (15), by comparison of their spectral data with those reported in the literature [9,10].
2.3. Anti-Larval Settlement and Recovery Activities
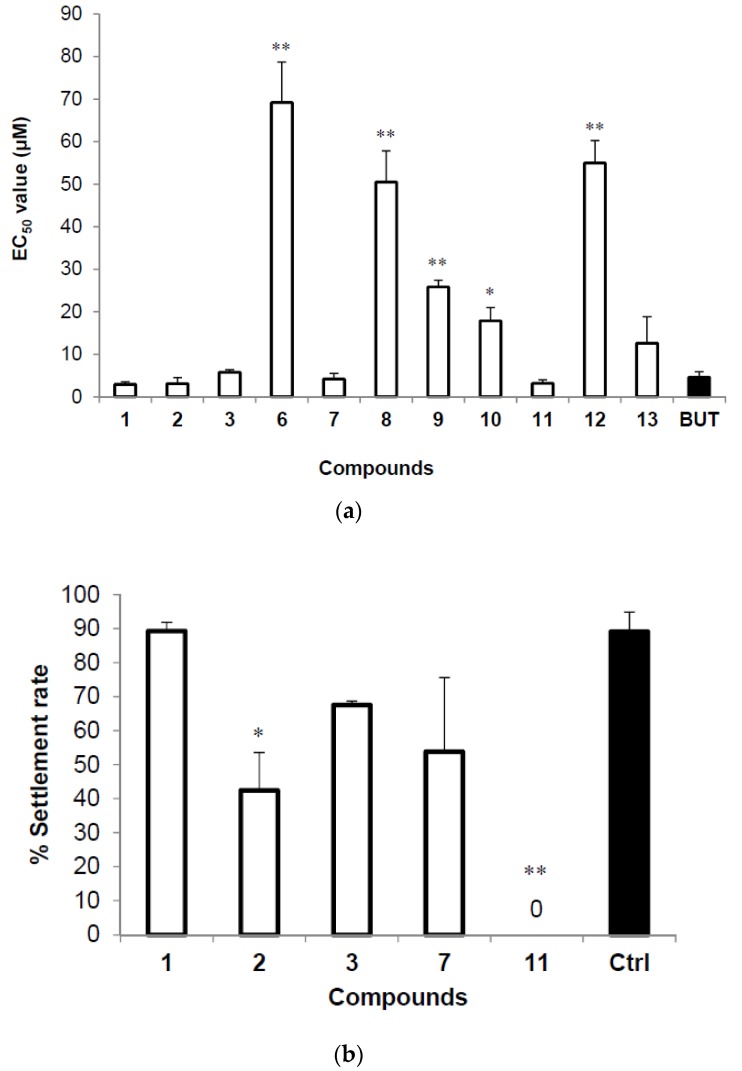
The anti-larval settlement activities of compounds 1–15 against cyprid larvae of B. amphitrite are summarized in Table 5. Compounds 1–3 and compounds 6–13 deterred larval settlement (Figure 3a). Due to the poor solubility of this group of compounds, we could not determine the LC50 values. Compounds 1–3, 7, and 11 also showed narcotic effects against cyprids of B. amphitrite at a concentration of 10 μM. Many organic molecules cause narcosis in barnacle larvae [11,12]. With increasing concentrations of active thielavins, cyprids lost their phototactic response, showed reduced appendage activity and were completely immobilized at a concentration of 10 μM. However, when the cyprids were transferred into 0.22 μM filtered seawater FSW following exposure to thielavins for 24 h, some recovered quickly from the chemical shock and completed their attachment and metamorphosis. The recovery rates of cyprids treated with 10 μM of compounds 1–3, 7 and 11 demonstrated that larvae had the highest recovery rate from treatment with compound 1, while no larvae recovered from treatment with compound 11 for 24 h (Figure 3b). Of all of the compounds, compound 1 showed excellent antifouling activity, and cyprids treated with this compound had the highest recovery rate. Thus, compound 1 is a promising natural antifoulant.
Table 5.
Anti-larval settlement activity of compounds from the fungus Thielavia sp. UST030930-004 against Balanus amphitrite. EC50 means the settlement of 50% of the larval population was inhibited, compared with the negative control. Data presented are the mean ± SE of three independent experiments performed in triplicate. Butenolide was served as the positive control.
| Compounds | EC50 (μM) | Compounds | EC50 (μM) |
|---|---|---|---|
| 1 | 2.95 ± 0.59 | 9 | 25.86 ± 1.56 |
| 2 | 3.13 ± 1.37 | 10 | 17.86 ± 3.14 |
| 3 | 5.78 ± 0.60 | 11 | 3.20 ± 0.83 |
| 6 | 69.19 ± 9.51 | 12 | 54.99 ± 5.23 |
| 7 | 4.23 ± 1.30 | 13 | 12.64 ± 6.20 |
| 8 | 50.50 ± 7.35 | Butenolide | 4.62 ± 1.30 |
Figure 3.
(a) EC50 of thielavin compounds against cyprids of Balanus amphitrite; Butenolide served as a positive control. (b) Recovery of Balanus amphitrite cyprid settlement in filtered seawater after a 24 h treatment with 10 μM of the thielavins we identified. The results were observed 48 h after being removed to filtered seawater. Values are presented as means ± SE of triplicate experiments. Asterisks indicate significant differences from control using Tukey’s test (* p < 0.05, ** p < 0.01, oneway ANOVA).
3. Materials and Methods
3.1. General Experimental Procedure
1H/13C and 2D-NMR spectral data were obtained using Varian Inova 500 MHz NMR spectrometers (Varian, Palo Alto, CA, USA). High-resolution mass spectra were acquired from UPLC-TOF-MS. The UPLC system was a Waters ACQUITY UPLC system (Waters, Manchester, UK) equipped with 150 mm × 2.1 mm Waters Acquity BEH C18 1.7-μm UPLC column and coupled to a Bruker micrOTOF-Q II mass spectrometer (Brucker Daltonics GmbH, Bremen, Germany). UV spectra were measured with a Shimadzu UV-2600 UV–vis spectrophotometer (Shimadzu, Kyoto, Japan) in an ACN solution. Semi-preparative reversed-phase HPLC was performed on a Waters 2695 liquid chromatography (Waters, Milford, CT, USA) with Luna C18(2) column (250 mm × 10 mm, 5 μm 100 Å, Phenomenex, Torrance, CA, USA). Column chromatography was performed on Rp-18 silica gel (Merck, Darmstadt, Germany).
3.2. Isolation and Identification of the Fungus
The fungal strain UST030930-004 was isolated from 12-d biofilms developed at the pier of the Hong Kong University of Science and Technology (Hong Kong, China) in Port Shelter. The biofilms were developed on polystyrene dishes submerged in seawater for 12 days in September 2003 and scraped from the dishes using a sterile glass coverslip. The scraped samples were suspended in 1 mL and 10 mL of autoclaved Ringer solution separately. For each sample, 200 μL of the mixture was spread on a Corn Meal Agar plate (Oxoid Ltd. Hampshire, UK) containing the antibiotics streptomycin and penicillin (final concentrations of 100 and 50 mg·L−1) to inhibit bacterial contamination. These plates were incubated at 27 °C for 7 to 14 days. The hyphal tip was transferred to new agar plates, incubated at 27 °C, and replated until a pure culture was obtained.
The total genomic DNA of the fungal isolate UST030930-004 was extracted as described by Lai et al. [13], and the internal transcribed spacer (ITS) gene sequences were amplified by the polymerase chain reaction (PCR) using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGAT ATGC-3′). The ITS region was sequenced and compared with reference sequences in GenBank by BLAST search, showing a similarity of 97% with Thielavia terrestris NRRL 8126 (CP003011). The ITS sequence of the fungal isolate UST030930-004 has been submitted to GenBank (Accession no. KJ716558). The fungal strain UST030930-004 was deposited in the China Center for Type Culture Collection (CCTCC) as CCTCC AF 2014015.
3.3. Fermentation, Bioassay-Guided Isolation, and Purification
The fungal strain UST030930-004 was cultured in a liquid medium containing 24 g·L−1 of potato-dextrose broth (Difco Laboratories, Detroit, MI, USA) and 20 g·L−1 of sea salts. Seed cultures were prepared in 50-mL Falcon tubes (BD Labware, Bedford, MA, USA), each containing 25 mL of medium, and cultivated at 23 °C for 3 days with shaking at 160 rpms. Afterwards, 2.8 L flasks, each containing 1.0 L of the same medium, were used for large-scale fermentation (16 L) with the following conditions: inoculation volume 5% (v/v), temperature 23 °C, rotation speed 160 rpms and a duration of 10 days on a shaker. The culture was filtered through 8 layers of cheesecloth to separate the filtrate and mycelia, which were treated separately. The filtrate was extracted with an equal volume of ethyl acetate (EA) three times, while the mycelial pellet was suspended in 80% acetone and sonicated using an ultrasonicator (Branson B-12, Danbury, CT, USA). The wet residue obtained after rotary evaporation to remove acetone was partitioned with EA. The filtrate and mycelial EA extracts were combined and evaporated in vacuo at 35 °C to dryness. The EA extract (22.0 g) was subjected to reversed-phase C18 flash chromatography and then eluted with solvents using a step gradient of H2O-MeOH to obtain fractions 1–5 (Fr. 1–5). Fr. 1–5 were tested for anti-larval settlement activity against Balanus amphitrite, and those showing potential activity were further purified. Fr. 3 was purified using semi-preparative HPLC (Phenomenex Luna C18 (2) 250 × 10 mm column) with MeOH/H2O (63:37, v/v) containing 5 mM ammonium acetate at the flow rate of 3 mL/min. In total, we obtained pure compounds 1 (2.4 mg), 2 (2.4 mg), 3 (3.9 mg), 4 (1.6 mg), 5 (4.2 mg), 6 (3.6 mg), 7 (2.0 mg), 8 (1.6 mg), 9 (0.8 mg), 10 (0.7 mg), 11 (0.7 mg), 12 (3.6 mg), 13 (4.4 mg), 14 (6.9 mg) and 15 (2.3 mg).
3.4. Spectral Data
Thielavin W (1): White powder, UV ACN λmax 218.1, 267.8, 304.2 nm; 1H and 13C-NMR data, see Table 1; HRESIMS m/z 511.1604 [M + H]+ (calcd. for C27H27O10 511.1599); ISCID data, see Table 4.
Thielavin X (2): White powder, UV ACN λmax 218.1, 267.8, 306.5 nm; 1H and 13C-NMR data, see Table 1; HRESIMS m/z 525.1727 [M + H]+ (calcd. for C28H29O10 525.1755); ISCID data, see Table 4.
Thielavin Y (3): White powder, UV ACN λmax 218.1, 276.1, 304.7 nm; 1H and 13C-NMR data, see Table 1; HRESIMS m/z 525.1765 [M + H]+ (calcd. for C28H29O10 525.1755); ISCID data, see Table 4.
Thielavin Z (4): White powder, UV ACN λmax 218.1, 276.1, 306.5 nm; 1H and 13C-NMR data, see Table 1; HRESIMS m/z 525.1757 [M + H]+ (calcd. For C28H29O10 525.1755); ISCID data, see Table 4.
Thielavin Z1 (5): White powder, UV ACN λmax 218.1, 276.1, 306.9 nm; 1H and 13C-NMR data, see Table 2; HRESIMS m/z 525.1747 [M + H]+ (calcd. for C28H29O10 525.1755); ISCID data, see Table 4.
Thielavin Z2 (6): White powder, UV ACN λmax 218.1, 267.8, 305.2 nm; 1H and 13C-NMR data, see Table 2; HRESIMS m/z 539.1916 [M + H]+ (calcd. for C29H31O10 539.1912); ISCID data, see Table 4.
Thielavin Z3 (7): White powder, UV ACN λmax 214.6, 276.1, 309.1 nm; 1H and 13C-NMR data, see Table 2; HRESIMS m/z 525.1758 [M + H]+ (calcd. for C28H29O10 525.1755); ISCID data, see Table 4.
Thielavin Z4 (8): White powder, UV ACN λmax 218.1, 267.8, 305.2 nm; 1H and 13C-NMR data, see Table 2; HRESIMS m/z 511.1564 [M + H]+ (calcd. for C27H27O10 511.1599); ISCID data, see Table 4.
Thielavin Z5 (9): White powder, UV ACN λmax 213.4, 267.8, 307.0 nm; 1H and 13C-NMR data, see Table 3; HRESIMS m/z 525.1778 [M + H]+ (calcd. for C28H29O10 525.1755); ISCID data, see Table 4.
Thielavin Z6 (10): White powder, UV ACN λmax 215.8, 276.1, 307.0 nm; 1H and 13C-NMR data, see Table 3; HRESIMS m/z 587.1719/589.1646 [M + H]+ 3:1 (calcd. for C30H32ClO10 587.1679); ISCID data, see Table 4.
Thielavin Z7 (11): White powder, UV ACN λmax 215.8, 276.1, 308.9 nm; 1H and 13C-NMR data, see Table 3; HRESIMS m/z 375.1464 [M + H]+ (calcd. for C20H23O7 375.1438); ISCID data, see Table 4.
3.5. Anti-Larval Settlement and Larval Recovery Assay
To prepare the larval culture of the barnacle B. amphitrite, adult brood stocks of B. amphitrite were collected from piling at the Pak Sha Wan Pier, Hong Kong (22°36′ N, 114°25′ E). Barnacle larvae were obtained and reared to cyprid stage as previously described by Harder et al. [14]. The cyprids were filtered and maintained in (FSW) at 4 °C overnight, before being used in the settlement assay. A stock solution of each extract or purified compound was prepared at 50 mg·mL−1 in dimethyl sulfoxide (DMSO) and further diluted to varying concentrations (25, 12.5, 6.25, 3.125, 1.562, 0.781 mg·mL−1) immediately before each bioassay. One milliliter of the test solutions was added to each well of a 24-well plate (Nunc, Thermo Scientific, Rochester, NY, USA), and approximately 15 larvae were gently pipetted into each well. For the negative controls, 1.0 mL of FSW together with 1.0 μL of DMSO, instead of the testing solution, was added to each well prior to the addition of larvae. The plates were incubated at 28 °C for 48 h in the dark. At the end of the incubation, the numbers of settled and swimming larvae were counted directly under a microscope, and where appropriate, potential toxic effects were noted. The number of settled larvae was expressed as a percentage of the total number of larvae per well. Three replicates of each extract concentration or purified compound were evaluated. EC50 represents the concentration of a compound that inhibits settlement of 50% of the larval population compared with the negative control, while the LC50 represents the concentration of a compound that is required to kill 50% of the larvae in a tested population. The experiments were performed in triplicate.
In the recovery assay, cyprids used in the negative controls were placed in new polypropylene containers containing FSW for 24 h, as described in the literature [15]. One milliliter of 10 μM of the test solutions was added to each well of a 24-well plate, and approximately 15 larvae were added to each well. After a 24-h incubation at 28 °C in the dark, the tested solutions were removed, and the treated larvae were washed 3 times with FSW prior to being plated in a new 24-well plate. For the negative controls, swimming larvae in a polypropylene container were transferred to 1.0 mL of FSW containing 1.0 μL of DMSO. The plates were maintained at 28 °C for 48 h before determining the percentage of attached cyprids of B. amphitrite. Three replicates of each experimental concentration were assessed.
4. Conclusions
Thielavins are para-depside-type compounds originally isolated from Thielavia terricola [10]. To date, 23 thielavins (thielavin A–V) have been discovered, including two thielavin Qs [16,17,18]. The thielavins act as indoleamine 2,3-dioxygenase, prostaglandin synthetase, glucose-6-phosphatase, cell wall transglycosylation and telomerase inhibitors [9,10,16,19,20]. We have not found any description of the antifouling activity of thielavin compounds. In the present study, we isolated 15 thielavins from a marine-derived fungus Thielavia sp. UST030930-004, including 11 new ones, and 11 of them had antifouling activities. Of all of the compounds, Thielavin W (1) showed excellent antifouling activity, and cyprids treated with this compound had the highest recovery rate. Thus, Thielavin W is a promising natural antifoulant.
Acknowledgments
We are grateful for the financial support from the China Ocean Mineral Resources Research and Development Association (DY135), Youth Innovation Promotion Association CAS (QC2015388), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB06010204), Hainan Special Project for Ocean Exploration (XH201410), the Natural Science Foundation of Hainan Province (414186), and Institute of Deep-Sea Science and Engineering (SIDSSE-QN-201303).
Supplementary Materials
The following are available at www.mdpi.com/1660-3397/15/5/128/s1. All 1D, 2D-NMR, HRESIMS and ISCID-MS/MS spectra of compounds 1–11 are included.
Author Contributions
Zhuang Han performed the isolation, purification and characterization of all the compounds. Yong-Xin Li, Ling-Li Liu gave suggestions on experiments and structure elucidation. Liang Lu, Xian-Rong Guo and Xi-Xiang Zhang helped take NMR spectra. Xiao-Yong Zhang identified the fungus strain. Shu-Hua Qi, Ying Xu and Pei-Yuan Qian designed and guided the experiment.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Dobretsov S., Xiong H., Xu Y., Levin L.A., Qian P.Y. Novel antifoulants: Inhibition of larval attachment by proteases. Mar. Biotechnol. 2007;9:388–397. doi: 10.1007/s10126-007-7091-z. [DOI] [PubMed] [Google Scholar]
- 2.Dash S., Nogata Y., Zhou X., Zhang Y., Xu Y., Guo X., Qian P.Y. Poly-ethers from Winogradskyella poriferorum: Antifouling potential, time-course study of production and natural abundance. Bioresour. Technol. 2011;102:7532–7537. doi: 10.1016/j.biortech.2011.05.034. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y., Li H., Li X., Xiao X., Qian P.Y. Inhibitory effects of a branched-chain fatty acid on larval settlement of the polychaete Hydroides elegans. Mar. Biotechnol. 2009;11:495–504. doi: 10.1007/s10126-008-9161-2. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y., He H., Schulz S., Liu X., Fusetani N., Xiong H., Qian P.Y. Potent antifouling compounds produced by marine Streptomyces. Bioresour. Technol. 2010;101:1331–1336. doi: 10.1016/j.biortech.2009.09.046. [DOI] [PubMed] [Google Scholar]
- 5.Kwong T.F.N., Miao L., Li X., Qian P.Y. Novel antifouling and antimicrobial compound from a marine-derived fungus Ampelomyces sp. Mar. Biotechnol. 2006;8:634–640. doi: 10.1007/s10126-005-6146-2. [DOI] [PubMed] [Google Scholar]
- 6.Wang K.L., Xu Y., Lu L., Li Y., Han Z., Zhang J., Shao C.L., Wang C.Y., Qian P.Y. Low-Toxicity diindol-3-ylmethanes as potent antifouling compounds. Mar. Biotechnol. 2015;17:624–632. doi: 10.1007/s10126-015-9656-6. [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Zhang F., Xu Y., Matsumura K., Han Z., Liu L., Lin W., Jia Y., Qian P.Y. Structural optimization and evaluation of butenolides as potent antifouling agents: Modification of the side chain affects the biological activities of compounds. Biofouling. 2012;28:857–864. doi: 10.1080/08927014.2012.717071. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y.F., Zhang H., He L., Liu C., Xu Y., Qian P.Y. Butenolide inhibits marine fouling by altering the primary metabolism of three target organisms. ACS Chem. Biol. 2012;7:1049–1058. doi: 10.1021/cb200545s. [DOI] [PubMed] [Google Scholar]
- 9.Sakemi S., Hirai H., Ichiba T., Inagaki T., Kato Y., Kojima N., Nishida H., Parker J.C., Saito T., Tonai-Kachi H., et al. Thielavins as glucose-6-phosphatase (G6Pase) inhibitors: Producing strain, fermentation, isolation, structural elucidation and biological activities. J. Antibiot. 2002;55:941–951. doi: 10.7164/antibiotics.55.941. [DOI] [PubMed] [Google Scholar]
- 10.Kitahara N., Endo A., Furuya K., Takahashi S. Thielavin A and B, New inhibitors of prostaglandin biosynthesis produced by Thielavia terricola. J. Antibiot. 1981;34:1562–1568. doi: 10.7164/antibiotics.34.1562. [DOI] [PubMed] [Google Scholar]
- 11.Crisp D.J., Christie A.O., Ghobashy A.F.A. Narcotic and toxic action of organic compounds on barnacle larvae. Comp. Biochem. Physiol. 1967;22:629–649. doi: 10.1016/0010-406X(67)90758-X. [DOI] [Google Scholar]
- 12.Stupak M.E., García M.T., Pérez M.C. Non-toxic alternative compounds for marine antifouling paints. Int. Biodeterior. Biodegrad. 2003;52:49–52. doi: 10.1016/S0964-8305(03)00035-0. [DOI] [Google Scholar]
- 13.Lai X., Cao L., Tan H., Fang S., Huang Y., Zhou S. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 2007;1:756–762. doi: 10.1038/ismej.2007.51. [DOI] [PubMed] [Google Scholar]
- 14.Harder T.N., Thiyagarajan V., Qian P.Y. Effect of cyprid age on the settlement of Balanus amphitrite Darwin in response to natural biofilms. Biofouling. 2001;17:211–219. doi: 10.1080/08927010109378480. [DOI] [Google Scholar]
- 15.Han Z., Sun J., Zhang Y., He F., Xu Y., Matsumura K., He L.S., Qiu J.W., Qi S.H., Qian P.Y. iTRAQ-based proteomic profiling of the barnacle Balanus amphitrite in response to the antifouling compound meleagrin. J. Proteome Res. 2013;12:2090–2100. doi: 10.1021/pr301083e. [DOI] [PubMed] [Google Scholar]
- 16.Jang J.P., Jang J.H., Oh M., Son S., Kim S.M., Kim H.M., Shin K.S., Oh H., Soung N.K., Hong Y.S., et al. Inhibition of indoleamine 2, 3-dioxygenase by thielavin derivatives from a soil fungus, Coniochaeta sp. 10F058. J. Antibiot. 2013;67:331–333. doi: 10.1038/ja.2013.134. [DOI] [PubMed] [Google Scholar]
- 17.de Medeiros L.S., Murgu M., de Souza A.Q. Antimicrobial depsides produced by Cladosporium uredinicola, an endophytic fungus isolated from Psidium guajava fruits. Helv. Chim. Acta. 2011;94:1077–1084. doi: 10.1002/hlca.201000387. [DOI] [Google Scholar]
- 18.de Medeiros L.S., Abreu L.M., Nielsen A., Ingmer H., Larsen T.O., Nielsen K.F., Rodrigues-Filho E. Dereplication-guided isolation of depsides thielavins S–T and lecanorins D–F from the endophytic fungus Setophoma sp. Phytochemistry. 2015;111:154–162. doi: 10.1016/j.phytochem.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 19.Mani N., Sanchet P., Jiang Z.D., McNaney C., DeCenzo M., Knighti B., Stankis M., Kuranda M., Rothstein D.M. Screening systems for detecting inhibitors of cell wall transglycosylation in Enterococcus. Cell wall transglycosylation inhibitors in Enterococcus. J. Antibiot. 1998;51:471–479. doi: 10.7164/antibiotics.51.471. [DOI] [PubMed] [Google Scholar]
- 20.Togashi K.I., Ko H.R., Ahn J.S., Osada H. Inhibition of telomerase activity by fungus metabolites, CRM646-A and thielavin B. Biosci. Biotechnol. Biochem. 2001;65:651–653. doi: 10.1271/bbb.65.651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.