Abstract
Infection with hepatitis A virus (HAV) is a major cause of acute hepatitis globally and it is important to identify the mechanisms of HAV replication. Glucose-regulated protein 78 (GRP78) is an endoplasmic reticulum (ER) chaperone and serves a role in unfolded protein response pathways. Previous studies have demonstrated that GRP78 functions as an endogenous antiviral factor. In the present study, two loss-of-function studies using GRP78 were completed to elucidate the role of GRP78 in HAV infection. HAV replication was observed to be enhanced by deficient GRP78 although GRP78-deficiency also led to lower expression of ER stress molecules downstream of GRP78. Therefore, GRP78 appears to be a potential novel defensive molecule against HAV in hepatocytes.
Keywords: CRISPR/Cas9, endoplasmic reticulum stress, glucose-regulated protein 78, hepatitis A virus
Introduction
The hepatitis A virus (HAV) belongs to the Hepatovirus genus of the Picornaviridae family. HAV is a positive single-stranded RNA virus ~7.6 kb in length. Although effective prophylactic vaccines have been available for a number of years, HAV infection remains a major cause of acute hepatitis globally. HAV infection may lead to acute liver failure, resulting in certain patients requiring a liver transplant (1,2). Therefore, it is important to improve the understanding of the pathogenesis of hepatitis A.
Glucose-regulated protein 78 (GRP78) is an endoplasmic reticulum (ER) chaperone and serves a role in signaling unfolded protein response (UPR). Viral infection induces ER stress and interferon responses, and certain viruses interact with GRP78 (3). GRP78 also acts as a master control interacting with the following three mediators: PKR-like ER kinase (PERK), activating transcription factor (ATF)-6 and the ER transmembrane protein kinase/endoribonuclease (IRE1) (3). Downstream of IRE1 is X-box-binding protein 1 (XBP1) and C/EBP homologous proteins, while downstream of PERK, growth arrest and DNA damage gene 34 exist. They function as effector molecules activated by ER stress (4).
A previous study (5) demonstrated that GRP78 functions as an endogenous anti-hepatitis B virus (HBV) factor, which works through the interferon-β-mediated signaling pathway in hepatocytes. Ma et al (5) reported that suppression of GRP78 increases HBV replication and HBV antigen expression. Treatment with thapsigargin, an unfolded response inducer, may decrease hepatitis C virus replication (6). However, the role of GRP78 in HAV infection is not well known. The present study investigated the association between HAV replication and ER stress marker GRP78 expression.
Materials and methods
Cell lines and HAV strain
Huh7 human hepatoma cells, kindly provided by Professor R. Bartenschlager (University of Mainz, Mainz, Germany), were grown in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, Merck KGaG, Darmstadt, Germany) supplemented with 10% fetal calf serum (FCS; Sigma-Aldrich; Merck KGaG) at 5% CO2 and 37°C (7). The HAV HA-11-1299 genotype IIIA strain was used for HAV infection in all experiments (8). This HAV cell culture-adapted strain was established in Department of Infection and Immunity, Jichi Medical University School of Medicine, Shimotsuke, Tochigi, Japan (8).
Infection with HAV in Huh7 and its derived cells
HAV infection was performed as previously described (8). Briefly, cells were plated for 24 h prior to infection at a density of 1×106 cells/well in 6-well plates (AGC Techno Glass, Shizuoka, Japan). The cells were washed twice with phosphate-buffered saline (PBS) and infected with HAV HA-11-1299 genotype IIIA at a multiplicity of infection (MOI) of 0.1 in DMEM supplemented with 2% FCS (8). At 24 h after infection, the cells were washed three times with PBS, followed by the exchange of DMEM supplemented with 2% FCS. At 96 h after infection, total cellular RNA was extracted for the quantification of HAV RNA (8).
RNA extraction and quantification of HAV RNA
Cellular RNA extraction and the quantification of HAV RNA were performed as previously described (8). In brief, total cellular RNA was extracted using an RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. cDNA was synthesized using the Prime Script RT reagent (Perfect Real Time; Takara Bio, Inc., Otsu, Japan), and reverse transcription was performed at 37°C for 15 min, followed by 85°C for 5 sec. The primer sets for HAV and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) measurements have been described previously (8). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min, with Power SYBR Green Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) on a StepOne Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Specificity was confirmed by melting curve analysis. Each experiment was performed in triplicate. Data were analyzed based on the ∆∆Cq method (8,9).
Knockdown of GRP78
Small interfering RNA (siRNA) against GRP78 (si-GRP78) and control siRNA (si-C) were obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). These siRNA were validated in a previous study (4). These siRNAs (50 nM) were electroporated into Huh7 cells using the GenePulser Xcell system (Bio-Rad Laboratories, Hercules, CA, USA) at 850 µF and 220 V, according to the manufacturer's protocol (10).
Knockout of GRP78 by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated genome editing
Human GRP78 CRISPR/Cas9 knockout plasmids were purchased from Santa Cruz Biotechnology, Inc. Huh7 cells were electroporated with GRP78 CRISPR/Cas9 knockout plasmids using the GenePulser Xcell system. Surviving cells were reseeded at 0.5 cells per well into a 96-well plate, 48 h after transfection. Expression of GRP78 in the expanded colonies was detected by western blot analysis using anti-GRP78 antibodies to select GRP78-depleted colonies. Clone #5 was subjected to HAV infection because GRP78 was well knocked out in this clone.
Western blot analysis
Western blot analysis was performed as previously described (4). In brief, cells were lysed with a sodium dodecyl sulfate (SDS) lysis buffer [0.5 M Tris-HCl (pH 6.8), 10% SDS, 2-mercaptoethanol, glycerol and 1% bromophenol blue]. All reagents were purchased from Sigma-Aldrich; Merck KGaG. Protein concentration was determined by the Bradford method (4). Cellular proteins (5 µg per each well) were separated by a gradient gel of 5–20% SDS-PAGE and transferred onto nitrocellulose membranes (ATTO Corporation, Tokyo, Japan). The membranes were probed with primary antibodies against GRP78 (#3177), ATF4 (#11815; all 1:1,000; all from Cell Signaling Technology, Danvers, MA, USA), ATF6 (sc-166659), XBP1 (sc-80154) or GAPDH (sc-25778; all 1:1,000; all from Santa Cruz Biotechnology, Inc.) at 4°C for 16 h. Anti-mouse IgG HRP-linked antibody (NA931; dilution, 1:7,000; GE Healthcare Life Sciences, Chalfont, UK) or anti-rabbit IgG HRP-linked antibody (#7074; dilution, 1:3,500; Cell Signaling Technology) were used as secondary antibodies, incubated at room temperature for 1 h. Proteins were visualized using an enhanced chemiluminescent ECL Western blot substrate (Amersham ECL Prime Western Blotting Detection Reagent; GE Healthcare Life Sciences) and scanned with an image analyzer LAS-4000 (Fujifilm Corporation, Tokyo, Japan) and Image Gauge (version 3.1; Fujifilm) (4).
Statistical analysis
Data are expressed as the mean ± standard deviation, unless otherwise stated. Statistical analyses were performed using Student's t-test. P<0.05 was considered to represent a statistically significant difference.
Results and Discussion
Effects of the knockdown of GRP78 on HAV replication
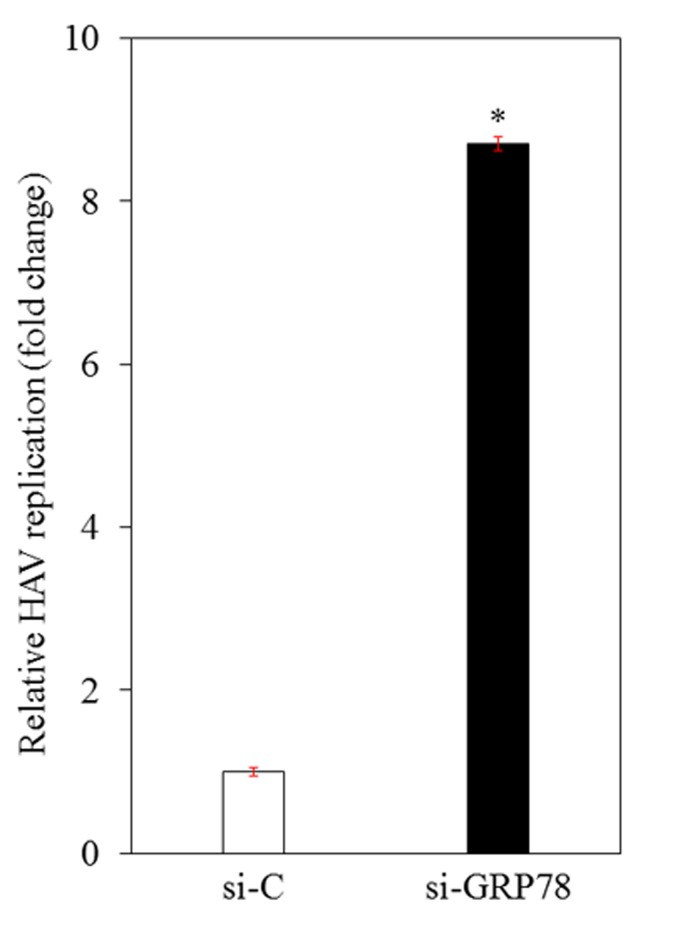
The effects of the knockdown of GRP78 on HAV replication were investigated. si-GRP78 and si-C were respectively transfected into Huh7 cells. These siRNAs were validated as in previous studies (4,11). At 24 h after transfection, cells were infected with HAV HA11-1299 genotype IIIA at a MOI of 0.1. HAV RNA levels were measured 96 h after infection using RT-qPCR. This confirmed that HAV replication was significantly increased 8.7-fold in Huh7 cells transfected with si-GRP78, compared with those with si-C (Fig. 1).
Figure 1.
Effects of GRP78 knockdown on HAV replication in Huh7 cells. Cells were transfected with si-GRP78 or si-C 1 day prior to HAV infection. Cellular RNA was extracted and subjected to reverse transcription-quantitative polymerase chain reaction 96 h after HAV HA-11-1299 genotype IIIA strain infection at a multiplicity of infection of 0.1. Data are expressed as the mean ± standard deviation. *P<0.05 vs. si-C-transfected controls using Student's t-test. All experiments were performed in triplicate. GRP78, glucose regulated protein 78; HAV, hepatitis A virus; si-GRP78, siRNA against GRP78; si-C, control siRNA.
Effects of knockout of GRP78 on HAV replication
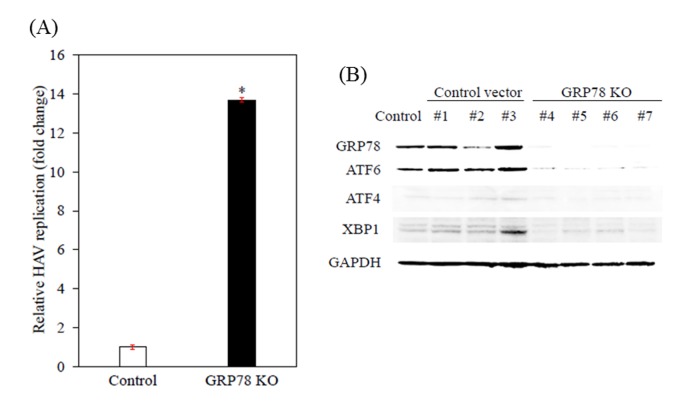
To confirm the effects of GRP78 on HAV replication, GRP78 was knocked out using a CRISPR/Cas9 system in Huh7 cells (12–14), and whether HAV replication was increased by the knockout of GRP78 was examined. As observed in Fig. 2A, Huh7-knockout clones exhibited significant enhancement of HAV replication compared with the parent Huh7 cells. As presented in Fig. 2B, GRP78-knockout Huh7 cells also had lower expression levels of ATF6, ATF4 and XBP1, which are located in ER stress pathways downstream of GRP78 and are of particular interest. Thus, the knockout of GRP78 appears to lead to reduced expression of other ER stress molecules such as ATF4, ATF6 and XBP1.
Figure 2.
Effects of GRP78 KO on HAV replication in Huh7 cells. (A) Effect of KO of GRP78 on HAV replication in control Huh7 cells and GRP78 KO cells. Cellular RNA was extracted and subjected to reverse transcription-quantitative polymerase chain reaction 96 h after HAV HA-11-1299 genotype IIIA strain infection at a multiplicity of infection of 0.1. Data are expressed as mean ± standard deviation. *P<0.05 vs. si-C-transfected controls using Student's t-test. All experiments were performed in triplicate. (B) Effects of knockout of GRP78 on endoplasmic reticulum stress molecules. Western blot analysis was performed using specific primary antibodies. Clone #5 was subjected to HAV infection. GRP78, glucose regulated protein 78; HAV, hepatitis A virus; KO, knockout; si-C, control siRNA; ATF, activating transcription factor; XBP, X-box-binding protein.
In the present study, HAV replication was increased by the knockdown or complete knockout of GRP78. A previous study (5) demonstrated that GRP78, an ER stress marker, is one of the intracellular antiviral factors against HBV. The silencing of GRP78 has also been shown to inhibit Dengue viral entry and multiplication in HepG2 cells (15).
A study involving GRP78-knockout mice (16) indicated that the level of XBP1s was reduced in GRP78-deficient mouse embryonic fibroblasts with impaired adipogenesis. In PERK-knockout mice (17) and XBP1-knockout mice (18), dysregulation of GRP78 occurred during the early ablation of these genes. In the present study in addition to a previous study (11), GRP78-deficiency in hepatic cells leads to the downregulation of ATF4, ATF6 and XBP1.
Recent genome-editing technology makes it possible for Cas9 nucleases to be directed by short RNAs to induce precise cleavage at endogenous genomic loci in human cells and to be converted into a nicking enzyme to facilitate homology-directed repair with minimal mutagenic activity (13,14). Targeting HBV covalently closed circular DNA with CRISPR/Cas9 may efficiently inhibit viral replication (19,20). In general, there are two forms of antiviral agents, namely direct-acting antivirals and host-targeting agents (21). GRP78 is potentially a candidate host-targeting agent.
HAV infection may lead to the swelling of the perinuclear space and the ER (22). HAV interacts with the host cell and modifies the ER-Golgi intermediate compartment structure (23). HAV appears to induce ER stress and, confronted with the UPR, may lead to the type of apoptotic cell death observed with other picornaviruses (24). Coxackievirus A9 infection of permissive cells requires GRP78 and major histocompatibility complex class I molecules, which are essential for virus internalization (25). It has been reported that the expression of GRP78 is activated by an enterovirus 71-dependent mechanism and that enterovirus 71 infection induces ER stress, but modifies the outcome to assist viral replication (26).
The results of the present study support the recent study (27) and indicate that GRP78 is an attractive target for controlling HAV infection. In conclusion, GRP78 may be an intracellular anti-HAV factor, and is a novel line of defense against HAV in hepatocytes.
Acknowledgements
The authors thank Professor Ralf Bartenschlager for providing the Huh7 cells. The present study was supported by grants from the Ministry of Health, Labour and Welfare of Japan. This research was partially supported by the Research Program on Hepatitis from the Japan Agency for Medical Research and Development (AMED).
References
- 1.Collier MG, Khudyakov YE, Selvage D, Adams-Cameron M, Epson E, Cronquist A, Jervis RH, Lamba K, Kimura AC, Sowadsky R, et al. Outbreak of hepatitis A in the USA associated with frozen pomegranate arils imported from Turkey: An epidemiological case study. Lancet Infect Dis. 2014;14:976–981. doi: 10.1016/S1473-3099(14)70883-7. [DOI] [PubMed] [Google Scholar]
- 2.Miyamura T, Ishii K, Kanda T, Tawada A, Sekimoto T, Wu S, Nakamoto S, Arai M, Fujiwara K, Imazeki F, et al. Possible widespread presence of hepatitis A virus subgenotype IIIA in Japan: Recent trend of hepatitis A causing acute liver failure. Hepatol Res. 2012;42:248–253. doi: 10.1111/j.1872-034X.2011.00919.x. [DOI] [PubMed] [Google Scholar]
- 3.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 4.Kanda T, Jiang X, Nakamura M, Haga Y, Sasaki R, Wu S, Nakamoto S, Imazeki F, Yokosuka O. Overexpression of the androgen receptor in human hepatoma cells and its effect on fatty acid metabolism. Oncol Lett. doi: 10.3892/ol.2017.5973. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, Yu J, Chan HL, Chen YC, Wang H, Chen Y, Chan CY, Go MY, Tsai SN, Ngai SM, et al. Glucose-regulated protein 78 is an intracellular antiviral factor against hepatitis B virus. Mol Cell Proteomics. 2009;8:2582–2594. doi: 10.1074/mcp.M900180-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagawa M, Sakamoto N, Tanabe Y, Koyama T, Itsui Y, Takeda Y, Chen CH, Kakinuma S, Oooka S, Maekawa S, et al. Suppression of hepatitis C virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology. 2005;129:1031–1041. doi: 10.1053/j.gastro.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 7.Kanda T, Yokosuka O, Imazeki F, Tanaka M, Shino Y, Shimada H, Tomonaga T, Nomura F, Nagao K, Ochiai T, et al. Inhibition of subgenomic hepatitis C virus RNA in Huh-7 cells: Ribavirin induces mutagenesis in HCV RNA. J Viral Hepat. 2004;11:479–487. doi: 10.1111/j.1365-2893.2004.00531.x. [DOI] [PubMed] [Google Scholar]
- 8.Kanda T, Sasaki R, Nakamoto S, Haga Y, Nakamura M, Shirasawa H, Okamoto H, Yokosuka O. The sirtuin inhibitor sirtinol inhibits hepatitis A virus (HAV) replication by inhibiting HAV internal ribosomal entry site activity. Biochem Biophys Res Commun. 2015;466:567–571. doi: 10.1016/j.bbrc.2015.09.083. [DOI] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Wu S, Kanda T, Imazeki F, Nakamoto S, Tanaka T, Arai M, Roger T, Shirasawa H, Nomura F, Yokosuka O. Hepatitis B virus e antigen physically associates with receptor-interacting serine/threonine protein kinase 2 and regulates IL-6 gene expression. J Infect Dis. 2012;206:415–420. doi: 10.1093/infdis/jis363. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X, Kanda T, Nakamoto S, Miyamura T, Wu S, Yokosuka O. Involvement of androgen receptor and glucose-regulated protein 78 kDa in human hepatocarcinogenesis. Exp Cell Res. 2014;323:326–336. doi: 10.1016/j.yexcr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alhoot MA, Wang SM, Sekaran SD. RNA interference mediated inhibition of dengue virus multiplication and entry in HepG2 cells. PLoS One. 2012;7:e34060. doi: 10.1371/journal.pone.0034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu G, Ye R, Jung DY, Barron E, Friedline RH, Benoit VM, Hinton DR, Kim JK, Lee AS. GRP78 plays an essential role in adipogenesis and postnatal growth in mice. FASEB J. 2013;27:955–964. doi: 10.1096/fj.12-213330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng D, Wei J, Gupta S, McGrath BC, Cavener DR. Acute ablation of PERK results in ER dysfunctions followed by reduced insulin secretion and cell proliferation. BMC Cell Biol. 2009;10:61. doi: 10.1186/1471-2121-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurczak MJ, Lee AH, Jornayvaz FR, Lee HY, Birkenfeld AL, Guigni BA, Kahn M, Samuel VT, Glimcher LH, Shulman GI. Dissociation of inositol-requiring enzyme (IRE1α)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. J Biol Chem. 2012;287:2558–2567. doi: 10.1074/jbc.M111.316760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong C, Qu L, Wang H, Wei L, Dong Y, Xiong S. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral Res. 2015;118:110–117. doi: 10.1016/j.antiviral.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Xu ZW, Liu S, Zhang RY, Ding SL, Xie XM, Long L, Chen XM, Zhuang H, Lu FM. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J Gastroenterol. 2015;21:9554–9565. doi: 10.3748/wjg.v21.i32.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanda T, Nakamoto S, Wu S, Nakamura M, Jiang X, Haga Y, Sasaki R, Yokosuka O. Direct-acting antivirals and host-targeting agents against the hepatitis A virus. J Clin Transl Hepatol. 2015;3:205–210. doi: 10.14218/JCTH.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klinger MH, Kämmerer R, Hornei B, Gauss-Müller V. Perinuclear accumulation of hepatitis A virus proteins, RNA and particles and ultrastructural alterations in infected cells. Arch Virol. 2001;146:2291–2307. doi: 10.1007/s007050170003. [DOI] [PubMed] [Google Scholar]
- 23.Seggewiß N, Kruse HV, Weilandt R, Domsgen E, Dotzauer A, Paulmann D. Cellular localization and effects of ectopically expressed hepatitis A virus proteins 2B, 2C, 3A and their intermediates 2BC, 3AB and 3ABC. Arch Virol. 2016;161:851–865. doi: 10.1007/s00705-015-2723-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhu G, Zheng Y, Zhang L, Shi Y, Li W, Liu Z, Peng B, Yin J, Liu W, He X. Coxsackievirus A16 infection triggers apoptosis in RD cells by inducing ER stress. Biochem Biophys Res Commun. 2013;441:856–861. doi: 10.1016/j.bbrc.2013.10.142. [DOI] [PubMed] [Google Scholar]
- 25.Triantafilou K, Fradelizi D, Wilson K, Triantafilou M. GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J Virol. 2002;76:633–643. doi: 10.1128/JVI.76.2.633-643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jheng JR, Lau KS, Tang WF, Wu MS, Horng JT. Endoplasmic reticulum stress is induced and modulated by enterovirus 71. Cell Microbiol. 2010;12:796–813. doi: 10.1111/j.1462-5822.2010.01434.x. [DOI] [PubMed] [Google Scholar]
- 27.Nwe Win N, Kanda T, Nakamura M, Nakamoto S, Okamoto H, Yokosuka O, Shirasawa H. Free fatty acids or high-concentration glucose enhances hepatitis A virus replication in association with a reduction in glucose-regulated protein 78 expression. Biochem Biophys Res Commun. 2017;483:694–699. doi: 10.1016/j.bbrc.2016.12.080. [DOI] [PubMed] [Google Scholar]