Abstract
Previous studies identified that phosphodiesterase 4D (PDE4D) gene polymorphism might be associated with cerebral infarction or ischemic stroke, and hemorrhagic stroke in human populations. However, as yet, no meta-analysis has revealed any detailed association. We retrospectively reviewed studies regarding the relationship of PDE4D gene polymorphism with ischemic stroke (IS) published during the period January 2003 to September 2012. According to the inclusion criteria, 9 of 105 initial studies were included in the subsequent analysis. The PubMed, Embase and CNKI of China were searched to identify the relevant studies. A total of 186 young patients with IS were included for the meta-analysis and 232 matched control subjects were enrolled and results were presented. The association of PDE4D gene polymorphism with IS in various populations was examined. The results suggested that single nucleotide polymorphism (SNP), SNP 83 in PDE4D gene was significantly related with susceptibility to IS. The meta-analysis also showed that PDE4D gene was associated with an enhanced risk of IS. The meta-analysis suggested that PDE4D SNP 87 constitutes an independent risk factor for IS development. To the best of our knowledge, the present meta-analysis reveals a number of possible associations between PDE4D gene polymorphism and IS.
Keywords: PDE-gene polymorphism, cerebral infarction, ischemic stroke, meta-analysis
Introduction
Phosphodiesterase 4D (PDE4D) gene polymorphism is associated with cerebral infarction or ischemic stroke (IS), and hemorrhagic stroke in human populations worldwide (1). Variants of the PDE4D gene are associated with stroke, especially with the combination of cardioembolic and carotid stroke identified in the population of Iceland (2). In the year 2002, the researchers in Iceland studied the gene in the DeCode study and identified a locus that was associated with stroke on 5q12 using a genome-wide association study (3). The authors of that study found that PDE4D gene within their region of interest was associated with stroke, especially with cardiogenic and carotid stroke. The meta analysis of molecular and epidemiological studies of PDE, PDE-gene polymorphisms and cerebral infarction risks or susceptibilities in China along with other populations worldwide have been studied and described (4). Nevertheless, whether patients with the PDE4D gene are susceptible to stroke remains to be determined.
The PDE4D gene variation and cerebral infarction or IS risks were also found in a Chinese population. A case-control study of a Chinese population comprising 639 stroke patients, including 253 with cerebral thrombosis, 171 with lacunar infarction, 215 with intra cerebral hemorrhage, and 887 healthy controls, was conducted (5). Single nucleotide polymorphisms (SNPs), rs966221, rs456009 and rs2910829 in PDE4D were selected and based on the significant association with stroke as previously reported in a Western population (6). These were genotyped using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), and were confirmed by DNA sequencing. The results showed only SNP 83 (rs966221) was associated with stroke. The allele C of rs966221 was a risk allele, which increased the risk for atherothrombotic strokes [odds ratio (OR)=1.51, 95% confidence interval (CI): 1.09–2.10] and was independent of conventional risk factors. The haplotype analysis revealed that the haplotype G-C-C was associated with increased risk for atherothrombotic stroke (OR=1.80, 95% CI: 1.300–2.491). The results verified that SNP 83 of PDE4D was a genetic risk factor for the atherothrombotic strokes in a Chinese population.
The human PDE4D gene spans 1.5 Mb regions on chromosome 5q12 and contains 24 exons and 9 splice variants. The PDE4D is a cyclic adenosine monophosphate, cyclic adenosine monophosphate (cAMP)-specific key signal transduction molecule present in many tissues and cell types. Previous findings showed that the PDE4D gene was a susceptibility gene for IS primarily in elderly populations. However, to the best of our knowledge, few studies have shown the role of PDE4D gene polymorphisms in a young cohort in a Chinese population. Genetic variants of PDE4D gene have been found showing the association with an enhanced risk for IS in a young Chinese population.
In the present study, we retrospectively reviewed studies focusing on the relationship of the PDE4D gene polymorphism with IS published during the period January 2003 to September 2012.
Materials and methods
A meta-analysis of molecular and epidemiological studies of PDE-gene polymorphisms and cerebral infarction risks or susceptibility in coastal residents of Wenling, China was completed. A case-control study of a Chinese population comprising 639 stroke patients including 253 with cerebral thrombosis, 171 with lacunar infarction, 215 with intra cerebral hemorrhage, and 887 healthy controls was conducted (5).
A case-control study of 205 consecutive Korean patients with non-cardiogenic IS, and 103 healthy controls that were used as stroke-free without neurological and radiological problems were included. A meta-analysis on 6 SNPs of the PDE4D gene was used to investigate the association between this gene and IS by integrating the results of previous studies, comprising 11,834 cases and 15,233 controls (7–14). Results of a multi-centre study involving a large number of samples from 2,599 IS patients and 2,093 control subjects from the south and west regions of Sweden were identified, to replicate previous studies regarding IS risk and rs12188950, the allele associated with stroke (7–14).
In total, 105 studies from January 2003 to September 2012 were identified by the literature search, of which 9 studies met the inclusion criteria. A total of 186 young patients with IS were included in the meta-analysis as well as 232 matched control subjects. The results of the meta-analysis on PDE4D gene polymorphism and IS in various populations were evaluated. From the 96 excluded studies, 7 studies were duplicate publications; 36 were reviews, editorials or comments; 32 were from other ethnic populations; 4 studies did not focus on SNP 83; and the remaining 17 were studies, which were not related to the subject of this meta-analysis. A total of 186 young patients, aged 18–45 years were studied for the present meta-analysis with IS and 232 matched control subjects were enrolled and results presented. The results of the meta-analysis studies are shown from various countries of the world on PDE4D gene polymorphism and IS in various populations. The meta-analysis results suggested that SNP, specifically SNP 83, in the PDE4D gene was significantly associated with susceptibility to IS in Chinese populations. The results of the meta-analysis also showed that PDE4D SNP 87 may not be an independent risk factor for IS development. However, the meta-analysis revealed that PDE4D gene was associated with an enhanced risk for IS in a Chinese population. Results of the meta-analysis suggest that PDE4D SNP 87 may or may not be an independent risk factor for IS development. More current meta-analysis studies have identified possible associations between PDE4D gene polymorphism and IS as compared to other studies.
Results
A meta-analysis of candidate gene polymorphisms and IS in six study populations was examined in association with lymphotoxin-α (LTA) in non-hypertensive patients (15). As IS is a multifactorial disease with a strong genetic component including pathways of lipid metabolism, systemic chronic inflammation, blood coagulation, blood pressure regulation, and cell adhesion and is involved in stroke pathophysiology, candidate gene polymorphisms in these pathways are considered genetic risk factors. Wang et al (15) genotyped 105 simple deletions and SNPs from 64 candidate genes in 3,550 patients and 6,560 controls from six case-control association studies performed in United States, Europe and China (15). Genotyping was carried out using the same immobilized probe typing system, and meta-analyses for each study were based on the summary logistic regressions. The primary analyses were fixed-effects meta-analyses adjusting for age and sex with additive, dominant and recessive models of inheritance. The results showed 7 polymorphisms with a nominal additive association, and none remained statistically significant after adjustment for multiple comparisons. After stratification for hypertension, LTA polymorphisms that were in strong linkage disequilibrium were significantly associated among non-hypertensive individuals: for LTA 252A>G (additive model), OR=1.41, 95% CI: 1.20–1.65; P=0.00002; for LTA 26Thr>Asn, OR=1.19, 95% CI: 1.06–1.34; P=0.003. LTA 252A>G remained significant after adjustment for multiple testing using either the false discover rate or by permutation testing. The two SNPs showed no association in hypertensive subjects (e.g., LTA 252A>G, OR=0.93, 95% CI: 0.84–1.03; P=0.17). These observations indicated an important role of LTA-mediated inflammatory processes in the pathogenesis of IS.
The meta-analysis study was performed on PDE4D gene and risk of non-cardiogenic IS in a Korean population (16). Various evidence on different populations suggested the association between PDE4D gene and IS. The PDE4D genotype and IS relationship was investigated in a case-control study of 205 consecutive Korean patients with non-cardiogenic IS (7–14). The 103 healthy controls were considered stroke-free without neurological and radiological problems. The genotyped PDE4D SNP (SNP 41, rs152312) served as a candidate marker and was selected for susceptibility to IS, because SNP 41 showed the most significant association with stroke in a previous meta-analysis and a study on Iceland with regard to PDE4D gene (7–14). The results showed no significant differences between the cases and controls in the distribution of the PDE4D SNP 41 genotypes. The results from the adjusted conditional logistic regression analysis, adjusted for age, hypertension, diabetes and smoking status showed no significant association between PDE4D SNP 41 genotypes and an increased risk of non-cardiogenic IS. It was found that PDE4D gene was not a major risk factor for non-cardiogenic IS in a Korean population, which supported the suggestion that the causative genetic variants of IS may differ in eastern and western populations.
The study of the meta-analysis was performed for the association between variation in the PDE4D gene and ischemic cerebral infarction risk in Asian populations (17). In the year 2003, it was found in the populations of Iceland, PDE4D was associated with increased risk of IS (3). These results were different in Asian populations. A recent meta-analysis showed that no genetic variant of PDE4D demonstrated a reproducible association with IS, when analyzed with respect to Caucasian populations. In the present study, the Asian populations were included to determine the association between PDE4D and ischemic infarction random risk. The case-control studies were collected from English and Chinese populations related to the association between PDE4D and ischemic infarction. For the Asian populations selection criteria and exclusion criteria were determined, and a fixed or effects model was used on the basis of heterogeneity. For each gene variant, the strength of the genetic association, a pooled OR and 95% CI were calculated and publication bias was evaluated. It was found that none of the Asian subjects were heterozygous for SNP (SNP 45 or SNP 41). Only 7 studies showed that SNP 83 and SNP 87 had sufficient data to allow inclusion into the meta-analyses. The statistically significant associations with ischemic infarction in the Asian populations were determined for SNP 83 C allele (OR=1.22, 95% CI: 1.03–1.43) and SNP 83 CC genotype (OR=1.42, 95% CI: 1.14–1.77), but not for SNP 83 CC + CT genotypes or SNP 87. The results showed that PDE4D was associated with ischemic infarction risk in Asian individuals, and SNP 83 was an important biomarker.
The meta-analysis studies of homogeneous subgroups have shown the association between PDE4D gene variants and IS (18). Results of the Icelandic study showed a significant positive association between PDE4D gene variants and stroke. However, subsequent studies have shown conflicting results, possibly due to small sample sizes and the heterogeneity of these studies (7–14). A meta-analysis on six SNPs of the PDE4D gene was performed to investigate the association between this gene and IS by integrating the results of previous studies, comprising 11,834 cases and 15,233 controls (7–14). The pooled genotypic OR, and OR for each SNP was determined under three genetic models i.e., dominant, recessive, and co-dominant using fixed- and random-effects models with consideration for heterogeneity and publication bias across the studies. The results of the SNPs included in this study, SNP56 (rs702553) showed the most significant association with IS in a meta-analysis comprising seven homogenous studies. The overall OR of the TT genotype compared to the AA genotype was (OR=1.29, 95% CI: 1.03–1.61; P=0.022). For SNP, SNP 83 (rs966221), a protective effect was observed of the ancestral allele T only in Asian populations (ORTT=0.79, 95% CI: 0.69–0.90; P=0.0005). The meta-analysis showed a significant association of PDE4D gene variants with the risk of IS. Further research is important for the evaluation of possible ethnic-specific effects in various populations. In the Chinese population, the results of the meta-analysis of PDE4D gene polymorphism which was associated with ischemic and hemorrhagic stroke (1).
A large-sample assessment for the possible association between IS and rs12188950 in the PDE4D gene in a European population from Sweden was described (19). In previous studies, there was some ambiguity in findings regarding the possible associations between IS, IS and SNPs, and SNPs in the PDE4D gene region. The SNP rs12188950 (or SNP 45) was studied in this regard. A multi-centre study was performed involving a large number of sample of 2,599 IS patients and 2,093 control subjects from the south and west regions of Sweden to replicate previous studies regarding IS risk and rs12188950 (3). The human participants were enrolled from Lund Stroke Register (LSR), Malmö Diet and Cancer Study (MDC) and Sahlgrenska Academy Study on Ischemic Stroke (SAHLSIS). The subgroups of subjects were also assessed with hypertension and participants <55 years of age, and the TOAST subgroups large vessel disease, small vessel disease and CE (3). The univariate ORs and ORs controlling for hypertension, diabetes, and current smoking were calculated. A meta-analysis was performed including 10,500 patients and 10,102 control subjects from 17 publications, including the present study (7–14). The pooled data from LSR, MDC and SAHLSIS, were assessed, and there were no associations between IS and rs12188950 for all the participants (OR=0.93, 95% CI: 0.83–1.05). Significant associations were not evident for hypertensive participants or participants aged <55 years, or when separately evaluating patients the three different TOAST subgroups. The meta-analysis showed no significant overall estimate (OR=0.96, 95% CI: 0.89–1.04) with a significant heterogeneity for random effect (P=0.042) (3). No effect from rs12188950 on IS was found from either the pooled multi-centre data or from the performed meta-analysis. In the present study, there was no association between the examined subgroups and rs12188950 (19). A genetic study is needed in this respect. As stroke is the second most common cause of mortality and major cause of disability in the world, the SNP 83 in the PDE4D gene has been suggested as a risk factor in IS, but direct evidence from genetic association studies are inconclusive in the Chinese population (20).
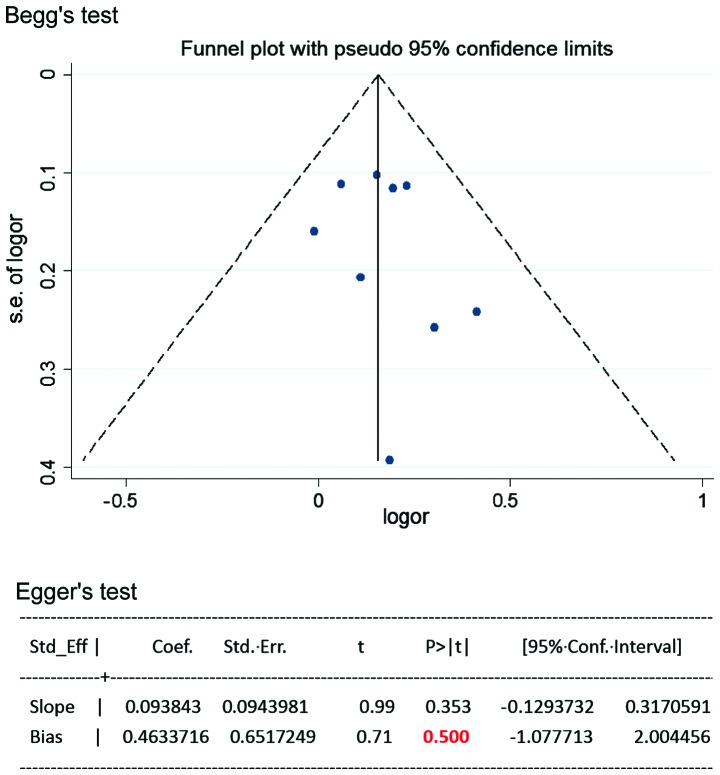
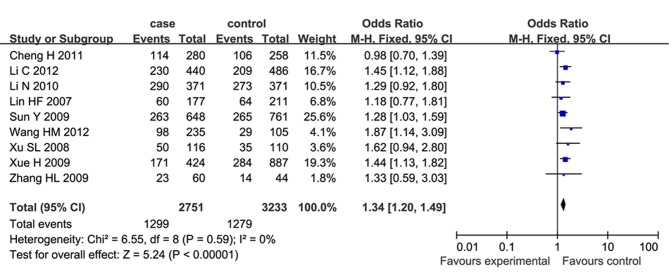
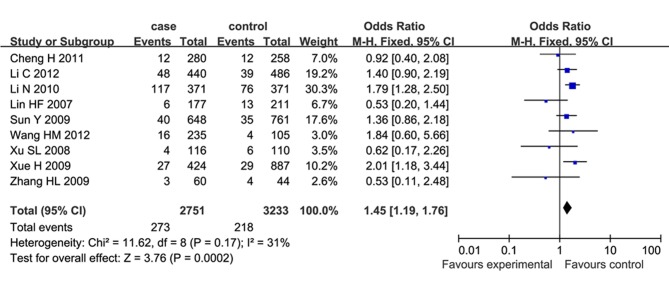
Meta-analysis of case-control studies on the relationship between SNP 83 in PDE4D gene and susceptibility to IS in a Chinese population published domestically and abroad from January 2003 to September 2012 were performed (7–14). For the current meta-analysis, 9 case-control studies were selected, and the results showed that a significant association between SNP 83 and IS was identified under the dominant model (OR=1.34, 95% CI: 1.20–1.49) and recessive model (OR=1.45, 95% CI: 1.19–1.76) in the Chinese population. In the subgroup meta-analysis, SNP 83 and atherothrombotic stroke, rather than lacunar stroke, showed a significant association under the dominant model (OR=1.69, 95% CI: 1.41–2.01) and recessive model (OR=1.47, 95% CI: 1.04–2.06). These results suggested that SNP 83 in PDE4D gene was significantly associated with susceptibility to IS in Chinese populations. The PDE4D gene was found at the locus on the chromosome 5q12 that encodes cAMP-specific 30, 50-cyclic PDE4D, belonging to a super family of PDEs (PDE-4 family). The data for statistical analysis were analyzed and processed using Review Manager 5.0 and Stata 10.0. The heterogeneity among studies was examined with the I2 statistic and I2>50%, indicating significant heterogeneity between the studies. Based on the test of heterogeneity, a pooled OR was calculated by using the fixed or random effect model, along with the 95% CI to measure the strength of the genetic association. Publication bias was examined by visual inspection of the funnel plot, Begg's test and Egger's regression test (P<0.05 was considered representative of statistically significant publication bias) (Fig. 1). In total, 105 studies were identified in the literature search, of which nine studies met the inclusion criteria. From the 96 excluded studies, 7 studies were duplicate publications; 36 were reviews, editorials or comments; 32 were from other ethnic populations; 4 studies were not involved with SNP 83; and 17 were irrelevant articles. Meta-analysis results suggested that SNP, specifically SNP 83 in the PDE4D gene was significantly associated with susceptibility to IS in the Chinese populations. For SNP 83, the heterogeneity among the studies, measured by I2 statistic, was not significant under the dominant model (I2=0) and recessive model (I2=31%). Therefore, the fixed effect model was used for calculating the pooled OR for the the two models. A significant association between SNP 83 and IS was identified under the dominant model (OR=1.34, 95% CI: 1.20–1.49; P<0.00001) and recessive model (OR=1.45, 95% CI: 1.19–1.76; P=0.0002), as shown in Figs. 2 and 3 [adapted from Liu et al (20)].
Figure 1.
Publication bias were evaluated by Begg's test and Egger's regression test. P-value of bias equals 0.500, indicating no significant publication bias among the included studies.
Figure 2.
Forrest plot on the association between SNP 83 and ischemic stroke under the dominant model (CC+CT/TT) (8): Many meta-analysis studies were included as described in (1,9–16). CI, confidence interval; SNP, single nucleotide polymorphism.
Figure 3.
Forrest plot on the association between SNP 83 and ischemic stroke under the recessive model (CC/CT+TT) (8). CI, confidence interval; SNP, single nucleotide polymorphism.
The sensitivity analysis was also performed with seven case-control studies whose genotype frequencies in the control groups were consistent with the Hardy-Weinberg equilibrium. The heterogeneity between studies was not significant (I2<0.5) and the fixed effect model was used for calculating the pooled OR (20). The sensitivity analysis revealed that a significant positive association was identified between SNP 83 and IS under the dominant model (OR=1.35, 95% CI: 1.20–1.51; P<0.00001) and the recessive model (OR=1.54, 95% CI: 1.26–1.88; P<0.0001). Our meta-analysis revealed that the risk of developing IS in carriers with C allele on SNP 83 was 1.34-fold higher than that in the individuals without C allele in the Chinese populations. Additionally, the risk in individuals with CC genotype was 1.45-fold higher than that in individuals without CC genotype.
Since there are different genetic studies in the populations of various continents, including North America, Europe, Asia, and Africa that show variation in genetic results, the manner in which we explain the associations between SNP, SNP 83 in PDE, PDE4D gene and IS in the Chinese population remained to be determined. Genetic experiments of DNA from blood showed variations from different populations globally. PDE4D gene selectively degraded the second messenger, cAMP in vascular smooth muscle cells and activated macrophages, and a key signaling molecule, that mediates cell proliferation and migration associated with atherosclerosis and plaque stability (7,8). cAMP is responsible for neuron survival and is a selective inhibitor of cAMP-specific PDE-4, which can promote the survival of newborn hippocampal neurons after ischemia (9,10). SNP, SNP 83, which is located at the 5′ end of PDE4D gene may affect the transcription, splicing, message stability, or message transport of one or more isoforms studied (11). Based on the TOAST classification, IS can be divided into five subtypes: i) Large-artery atherosclerosis (LAA), ii) small-vessel occlusion (SVO), iii) cardioembolism (CE), iv) stroke of other determined etiology, and v) stroke of undetermined etiology (12). For the Chinese population, five studies explored the association between SNP 83 and LAA, and six studies explored the association between SNP 83 and SVO. Thus, the meta-analysis was conducted in these two subgroups. The significant association of SNP 83 was found with IS only in the LAA subgroup. This can be explained by the role of the PDE4D gene in the process of atherosclerosis. For the remaining three subtypes of IS, the data were not sufficient to conduct the meta-analysis of the LAA subgroup. Thus the meta-analysis should be interpreted within the context of its limitations. A retrospective case-control design should be followed for meta-analysis studies that may be biased and artifact than prospective studies.
Any variant in or near the SNP 83 in the PDE4D gene can miss the true association. The different linkage disequilibrium patterns with the causal variant can lead to variable results in different populations. It is possible that the potential roles of SNP 83 in the PDE4D gene are diluted or masked by other gene-gene or gene-environment interactions. Publication bias is an important source of bias in any meta-analysis study, which can cause false-positive results. In meta-analysis, under the dominant model, there was no evidence of publication bias found by visual inspection of the funnel plot, Begg's test or Egger's regression test. However, under the recessive model, the publication bias was found by the visual inspection of funnel plot and Egger's regression test. Thus, the meta-analysis results were interpreted with caution for the association of SNP 83 with IS under the recessive model. It has been suggested that SNP 83 in PDE4D gene is associated with developing IS in the Chinese population. Recently, genetic variants of PDE4D gene have been found for the association with an enhanced risk for IS in a young Chinese population (13).
Previous studies showed that the PDE4D gene was a susceptibility gene for the IS primarily in elderly populations. However, to the best of our knowledge, few studies have shown the role of PDE4D gene polymorphisms in a young cohort in the Chinese population. A total of 186 young patients, aged 18–45 years were studied for meta-analysis with IS and 232 matched control subjects were included. The SNPs rs918592 and rs2910829 in the PDE4D gene were genotyped by PCR-RFLP method. The OR and 95% CIs were calculated to test the association between the genetic factors and IS. The results showed that rs918592A/A genotype frequency and A allele frequency, rs2910829 CT/TT genotype frequency and T allele frequency of young IS group were significantly higher as compared to the control group (P<0.05). The frequency of Hap (A-T) was markedly higher in the young patients than in the controls (OR=4.047, 95% CI: 3.521–4.652). The Hap (A-C) and Hap (G-C) were associated with a decreased risk of IS (OR=0.640, 95% CI: 0.452–0.906; OR=0.675, 95% CI: 0.466–0.978, respectively). These findings suggested that the rs918592 and rs2910829 polymorphisms and haplotypes of PDE4D gene were significantly associated with the IS in young Chinese population. The recent detailed study of a meta-analysis of gene polymorphisms associated with the IS in South Asians have been described in a study from London, UK (14). The burden of stroke is higher in the South Asian subcontinent with ethnicity of South Asians that confer a greater risk of IS than European ancestry. The genes associated with IS in European populations have been studied but it is not known for Asian populations. A meta-analysis study was performed of known genetic polymorphisms associated with South Asian IS, and compared effect size of the methylenetetrahydrofolate reductase, methylene tetrahydrofolate reductase (MTHFR) C677T-stroke association with effect sizes predicted from homocysteine-stroke association. Databases of PubMed, Embase and CNKI were searched up to August 2012 for published case control studies which investigated genetic polymorphisms associated with IS in South Asians. The pooled ORs were calculated using random-effects model for each gene-disease association. Twenty-six studies were identified with 2,529 cases of strokes and 2,881 controls which investigated 33 independent genetic polymorphisms in 22 genes. The 10 studies described MTHFR C677T (108 with TT genotype and 2,018 with CC genotype) -homocysteine relationship and 6 studies with 735 stroke cases and 713 controls, described the homocysteine-IS relationship. The risk association of ORs was calculated for ACE I/D (OR=5.00, 95% CI: 1.17–21.37; P=0.03), PDE4D SNP 83 (OR=2.20, 95% CI: 1.21–3.99; P=0.01), PDE4D SNP 32 (OR=1.57, 95% CI: 1.01–2.45; P=0.045) and IL-10 G1082A (OR=1.44, 95% CI: 1.09–1.91; P=0.01). The results showed a significant association between elevated plasma homocysteine levels and MTHFR/677 TT genotypes in healthy South Asians (mean difference (ΔX)=5.18 µmol/l, 95% CI: 2.03–8.34; P=0.001). The genetic etiology of IS in South Asians showed a broad similarity to the risk conferred in Europeans. However, the dataset was considerably smaller to suggest the same clinical considerations for the risk profiling of the study.
Recently, the identification of genetic contribution to IS by screening of SNPs in stroke patients using a case control study design has been described (21). Stroke is the second most common cause of mortality and disability worldwide and is a multifactorial disease that is influenced by environmental as well as genetic factors. Several meta-analysis studies have been reported from different populations and different ethnic regions worldwide that have shown variable results with regard to the association of apolioprotein E, MTHFR, endothelial nitric oxide synthase, factor V (F5) Leiden, cytochrome P450 4F2, β-fibrinogen and PDE4D gene in stroke (21). There is substantial evidence from European descent genetic studies that have shown that genetic risk of stroke varies as per specific subtypes of IS in the population. The hypothesis was studied that encoding gene polymorphisms was associated with stroke and to determine whether risk varied as per specific subtypes of stroke. A case-control study was performed and 600 cases with diagnosis of stroke and 600 age- and gender-matched controls were recruited (5). The controls were matched at a 1:1 ratio. The baseline and demographic data were obtained from a standardized data collection form. Blood (4 ml) was collected in EDTA-coated vials and were used for DNA isolation. The genotyping was carried out using PCR-RFLP, and the RFLP results were reconfirmed by PCR product of each genotype in triplet for all the selected polymorphism which was used for DNA sequencing. The data were analyzed using conditional logistic regression to determine OR associated with the above genes. The meta-analysis study of association of the abovementioned gene polymorphisms with IS in a North Indian population was performed (22). The present study was useful in determining the genetic component of stroke and to observe whether variations in genetic risks as per different subtypes of stroke are significant. It is known that PDE4D gene is located on the short arm of chromosome number 5q12 and consists of 24 exons. The gene expresses nine different functional protein isoforms through alternative splicing or the use of differential promoters. It is also known that different PDE4D variants are expressed in various tissues which include brain, lungs, kidneys, monocytes, B and T lymphocytes and vascular smooth muscles (23). PDE4D is the family of enzyme that breaks the phosphodiester bond of cAMP, degrades these and maintains the appropriate level and duration of action of cAMP within the cell. cAMP is a secondary cell signaling molecule that involves genes in the preoduction of inflammatory mediators by several types of inflammatory cells and arthrosclerosis. PDE4D degrades cAMP, and is responsible for inhibiting the inflammatory process by blocking the messaging through cAMP to genes that produce inflammatory proteins. The polymorphism in this gene affects the catalytic efficiency of PDE4D. The decoding group in 2002 from Iceland found the results of a genome widescreen for stroke susceptibility genes (11). The 260 PDE4D SNPs were examined, and 6 were significantly associated with stroke and observed after multiple comparisons.
In the year 2006, scientists in Iceland identified the relationship between PDE4D gene and IS (24). To estimate this association, the results of many researchers are contradictory. A meta-analysis study was performed to confirm these results with a large sample size (25). Databases of PubMed, Embase and CNKI were searched to identify the relevant studies to January 2013. The OR with 95% CI were used to calculate the association between the SNP 83 polymorphism and IS risk. The 25 publications comprising 8,878 cases and 12,306 controls were included in this meta-analysis study. The results showed a significant association between SNP 83 and IS risk, especially in Asian and Chinese populations, but not in Caucasians (dominant model: OR=0.87, 95% CI: 0.69–1.11; recessive model: OR=0.95, 95% CI: 0.84–1.07; allele model: OR=0.95, 95% CI: 0.84–1.08; co-dominant model 1: OR=0.96, 95% CI: 0.85–1.08; co-dominant model 2: OR=0.95, 95% CI: 0.83–1.09). Among the overall and Chinese populations, the cumulative meta-analysis indicated a stable trend of association between SNP 83 and IS from the year 2009 to 2012.
Discussion
An association between SNP 83 and IS has been found in the overall population and in the Asian and Chinese populations, but not among Caucasian populations. The recent study of Liang et al focused on the association between PDE4D SNP 87 and IS by evaluating the meta-analysis of various populations (26). The results on the association between PDE4D SNP 87 and the risk of IS are contentious and debatable. The new meta-analysis studies indicated some possible associations while other studies did not. Based on a comprehensive search of PubMed, Embase, and CNKI databases, 18 eligible articles were identified and examined for the relationship between PDE4D SNP 87 and IS risk in various populations. The strength of the association was established using ORs with 95% CIs. The overall analysis showed that PDE4D SNP 87 did not affect the risk of IS. The null association persisted in the subgroup analyses by ethnicity and sample size. The results of the meta-analysis suggested that PDE4D SNP 87 may not be an independent risk factor for IS development. However, additional genetic research is needed to confirm the association of PDE4D gene polymorphism and risk of IS in Chinese population. The meta-analysis also showed that the D allele is a modest but independent risk factor for IS onset (27). Thus, the meta-analysis is useful in the study of allelic association for detecting the risks of candidate genes in polygenic disorders.
Further studies are needed to confirm the association of PDE4D gene polymorphism and risk of IS in the Chinese population. The results on the association between PDE4D SNP 87 and the risk of IS are contentious and debatable.
Acknowledgements
This study was supported by the Project of Wenling Municipal Science and Technology Bureau (no. 2012WLCA0060).
References
- 1.Xue H, Wang H, Song X, Li W, Sun K, Zhang W, Wang X, Wang Y, Hui R. Phosphodiesterase 4D gene polymorphism is associated with ischaemic and haemorrhagic stroke. Clin Sci (Lond) 2009;116:335–340. doi: 10.1042/CS20080162. [DOI] [PubMed] [Google Scholar]
- 2.Song Q, Cole JW, O'Connell JR, Stine OC, Gallagher M, Giles WH, Mitchell BD, Wozniak MA, Stern BJ, Sorkin JD, et al. Phosphodiesterase 4D polymorphisms and the risk of cerebral infarction in a biracial population: the Stroke Prevention in Young Women Study. Hum Mol Genet. 2006;15:2468–2478. doi: 10.1093/hmg/ddl169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gretarsdottir S, Sveinbjörnsdottir S, Jonsson HH, Jakobsson F, Einarsdottir E, Agnarsson U, Shkolny D, Einarsson G, Gudjonsdottir HM, Valdimarsson EM, et al. Localization of a susceptibility gene for common forms of stroke to 5q12. Am J Hum Genet. 2002;70:593–603. doi: 10.1086/339252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng H, Wang L, Shi T, Shang Y, Jiang L. Association of insulin degrading enzyme gene polymorphisms with Alzheimer's disease: A meta-analysis. Int J Neurosci. 2015;125:328–335. doi: 10.3109/00207454.2014.941440. [DOI] [PubMed] [Google Scholar]
- 5.Li N, He Z, Xu J, Liu F, Deng S, Zhang H. Association of PDE4D and IL-1 genepolymorphism with ischemic stroke in a Han Chinese population. Brain Res Bull. 2010;81:38–42. doi: 10.1016/j.brainresbull.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Jørgensen C, Yasmeen S, Iversen HK, Kruuse C. Phosphodiesterase4D (PDE4D) - A risk factor for atrial fibrillation and stroke? J Neurol Sci. 2015;359:266–274. doi: 10.1016/j.jns.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama T, Asai S, Sato N, Soma M. PDE4D gene in the STRK1 region on 5q12: Susceptibility gene for ischemic stroke. Curr Med Chem. 2007;14:3171–3178. doi: 10.2174/092986707782793871. [DOI] [PubMed] [Google Scholar]
- 8.Naghavi M, Libby P, Falk E, Casscells SW, Litovsky S, Rumberger J, Badimon JJ, Stefanadis C, Moreno P, Pasterkamp G, et al. From vulnerable plaque to vulnerable patient: a call for new definitions and risk assessment strategies: Part I. Circulation. 2003;108:1664–1672. doi: 10.1161/01.CIR.0000087480.94275.97. [DOI] [PubMed] [Google Scholar]
- 9.Rydel RE, Greene LA. cAMP analogs promote survival and neurite outgrowth in cultures of rat sympathetic and sensory neurons independently of nerve growth factor; Proc Natl Acad Sci USA; 1988; pp. 1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki T, Kitagawa K, Omura-Matsuoka E, Todo K, Terasaki Y, Sugiura S, Hatazawa J, Yagita Y, Hori M. The phosphodiesterase inhibitor rolipram promotes survival of newborn hippocampal neurons after ischemia. Stroke. 2007;38:1597–1605. doi: 10.1161/STROKEAHA.106.476754. [DOI] [PubMed] [Google Scholar]
- 11.Gretarsdottir S, Thorleifsson G, Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T, Gudmundsdottir T, Bjarnadottir SM, Einarsson OB, Gudjonsdottir HM, et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 12.Adams HP, Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, Marsh EE., III Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.STR.24.1.35. [DOI] [PubMed] [Google Scholar]
- 13.He Y, Yang DZ, Yu H, Li MY, Feng QC, Zheng H. Genetic variants of phosphodiesterase 4D gene are associated with an enhanced risk for ischemic stroke in young Chinese population. Neurol India. 2013;61:21–25. doi: 10.4103/0028-3886.108131. [DOI] [PubMed] [Google Scholar]
- 14.Yadav S, Hasan N, Marjot T, Khan MS, Prasad K, Bentley P, Sharma P. Detailed analysis of gene polymorphisms associated with ischemic stroke in South Asians. PLoS One. 2013;8:e57305. doi: 10.1371/journal.pone.0057305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Cheng S, Brophy VH, Erlich HA, Mannhalter C, Berger K, Lalouschek W, Browner WS, Shi Y, Ringelstein EB, et al. RMS Stroke SNP Consortium: A meta-analysis of candidate gene polymorphisms and ischemic stroke in 6 study populations: Association of lymphotoxin-alpha in nonhypertensive patients. Stroke. 2009;40:683–695. doi: 10.1161/STROKEAHA.108.524587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MK, Kim JT, Choi SM, Lee SH, Park MS, Cho KH. Phosphodiesterase 4D gene and risk of noncardiogenic ischemic stroke in a Korean population. J Korean Med Sci. 2009;24:307–310. doi: 10.3346/jkms.2009.24.S2.S307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Li X, Li J, Ou R, Sheng W. Meta-analysis of association between variation in the PDE4D gene and ischemic cerebral infarction risk in Asian populations. Neurogenetics. 2010;11:327–333. doi: 10.1007/s10048-010-0235-8. [DOI] [PubMed] [Google Scholar]
- 18.Yoon D, Park SK, Kang D, Park T, Park JW. Meta-analysis of homogeneous subgroups reveals association between PDE4D gene variants and ischemic stroke. Neuroepidemiology. 2011;36:213–222. doi: 10.1159/000327915. [DOI] [PubMed] [Google Scholar]
- 19.Lövkvist H, Olsson S, Höglund P, Melander O, Jern C, Sjögren M, Engström G, Smith JG, Hedblad B, Andsberg G, et al. A large-sample assessment of possible association between ischaemic stroke and rs12188950 in the PDE4D gene. Eur J Hum Genet. 2012;20:783–789. doi: 10.1038/ejhg.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Zhu R, Li L, Deng S, Li Q, He Z. Genetic polymorphism in PDE4D gene and risk of ischemic stroke in Chinese population: A meta-analysis. PLoS One. 2013;8:e66374. doi: 10.1371/journal.pone.0066374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar A, Sagar R, Kumar P, Sahu JK, Grover A, Srivastava AK, Vivekanandhan S, Prasad K. Identification of genetic contribution to ischemic stroke by screening of single nucleotide polymorphisms in stroke patients by using a case control study design. BMC Neurol. 2013;13:136. doi: 10.1186/1471-2377-13-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bevan S, Dichgans M, Gschwendtner A, Kuhlenbäumer G, Ringelstein EB, Markus HS. Variation in the PDE4D gene and ischemic stroke risk: a systematic review and meta-analysis on 5200 cases and 6600 controls. Stroke. 2008;39:1966–1971. doi: 10.1161/STROKEAHA.107.509992. [DOI] [PubMed] [Google Scholar]
- 23.Omori K, Kotera J. Overview of PDEs and their regulation. Circ Res. 2007;100:309–327. doi: 10.1161/01.RES.0000256354.95791.f1. [DOI] [PubMed] [Google Scholar]
- 24.Zee RY, Brophy VH, Cheng S, Hegener HH, Erlich HA, Ridker PM. Polymorphisms of the phosphodiesterase 4D, cAMP-specific (PDE4D) gene and risk of ischemic stroke: a prospective, nested case-control evaluation. Stroke. 2006;37:2012–2017. doi: 10.1161/01.STR.0000230608.56048.38. [DOI] [PubMed] [Google Scholar]
- 25.Yan Y, Luo X, Zhang J, Su L, Liang W, Huang G, Wu G, Huang G, Gu L. Association between phosphodiesterase 4D polymorphism SNP83 and ischemic stroke. J Neurol Sci. 2014;338:3–11. doi: 10.1016/j.jns.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Liang W, Zhang D, Mang J, He J, Liu H, Shao Y, Han F, Xu Z. Association between phosphodiesterase 4D (PDE4D) SNP 87 and ischemic stroke: A meta-analysis. Int J Clin Exp Med. 2015;8:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma P. Meta-analysis of the ACE gene in ischaemic stroke. J Neurol Neurosurg Psychiatry. 1998;64:227–230. doi: 10.1136/jnnp.64.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]